ABSTRACT
Streptococcus pneumoniae (the pneumococcus) is a human pathogen of global importance, classified into serotypes based on the type of capsular polysaccharide produced. Serotyping of pneumococci is essential for disease surveillance and vaccine impact measurement. However, the accuracy of serotyping methods can be affected by previously undiscovered variants. Previous studies have identified variants of serotype 14, a highly invasive serotype included in all licensed vaccine formulations. However, the potential of these variants to influence serotyping accuracy and evade vaccine-induced protection has not been investigated. In this study, we screened 1,386 nasopharyngeal swabs from children hospitalized with acute respiratory infection in Papua New Guinea for pneumococci. Swabs containing pneumococci (n = 1,226) were serotyped by microarray to identify pneumococci with a divergent serotype 14 capsule locus. Three serotype 14 variants (‘14-like’) were isolated and characterized further. The serotyping results of these isolates using molecular methods varied depending on the method, with 3/3 typing as nontypeable (PneumoCaT), 3/3 typing as serotype 14 (seroBA), and 2/3 typing as serotype 14 (SeroCall and quantitative PCR). All three isolates were nontypeable by phenotypic methods (Quellung and latex agglutination), indicating the absence of capsule. Illumina and nanopore sequencing were employed to examine their capsule loci and revealed unique mutations. Lastly, when incubated with sera from vaccinated individuals, the 14-like isolates evaded serotype-specific opsonophagocytic killing. Our study highlights the need for phenotypic testing to validate serotyping data derived from molecular methods. The convergent evolution of capsule loss underscores the importance of studying pneumococcal population biology to monitor the emergence of pneumococci capable of vaccine escape, globally.
IMPORTANCE Pneumococcus is a pathogen of major public health importance. Current vaccines have limited valency, targeting a subset (up to 20) of the more than 100 capsule types (serotypes). Precise serotyping methods are therefore essential to avoid mistyping, which can reduce the accuracy of data used to inform decisions around vaccine introduction and/or maintenance of national vaccination programs. In this study, we examine a variant of serotype 14 (14-like), a virulent serotype present in all currently licensed vaccine formulations. Although these 14-like pneumococci no longer produce a serotype 14 capsule, widely used molecular methods can mistype them as serotype 14. Importantly, we show that 14-like pneumococci can evade opsonophagocytic killing mediated by vaccination. Despite the high accuracy of molecular methods for serotyping, our study reemphasizes their limitations. This is particularly relevant in situations where nonvaccine type pneumococci (e.g., the 14-likes in this study) could potentially be misidentified as a vaccine type (e.g., serotype 14).
KEYWORDS: serotype, capsule, pneumococcus, Streptococcus pneumoniae, variant, pneumococcal conjugate vaccine
INTRODUCTION
Streptococcus pneumoniae (the pneumococcus) is a bacterial pathogen of global importance, as it causes a range of diseases, including pneumonia, sepsis, and meningitis (1). Pneumococci are also carried in the nasopharynx, which is typically an asymptomatic event but is important for transmission and progression to disease (2, 3). The capsular polysaccharide is a major virulence determinant and is the basis for pneumococcal classification. Over 100 pneumococcal serotypes have been described, but currently licensed pediatric vaccines only confer protection against a subset (up to 20) of these serotypes. Pneumococcal serotyping is essential for disease surveillance and vaccine impact. Molecular approaches used to infer the serotype have increased in popularity and rely on a preexisting database of reference capsule locus (cps) sequences (4). Unfortunately, there are limited genomic data from pneumococci from low and middle-income countries, particularly in the Asia-Pacific region. We have previously discovered new genetic variants from this region that can be ‘mistyped’ using common molecular methods (5). This reduces the accuracy of the serotyping data used to make decisions around vaccine introduction and around the monitoring of vaccine impact.
The burden of pneumococcal disease in Papua New Guinea (PNG) is among the highest, worldwide (6, 7). High-density pneumococcal carriage with a diverse range of serotypes occurs early in life (6, 7). In contrast with most other settings, randomized controlled trials in PNG showed pneumococcal vaccines have thus far had a limited impact on vaccine-type nasopharyngeal carriage (6, 7). Therefore, it is important to identify pneumococcal variants with the potential to evade vaccine-induced protection in PNG.
Serotype 14 is a highly invasive serotype (8) that is targeted by all currently licensed pneumococcal vaccines (PCV10, Pneumosil, PCV13, PCV20, and PPSV23). Variants of serotype 14 have been identified in Australia and South Africa. These variants are unable to be serotyped by phenotypic methods (‘nontypeable’) because mutations in the serotype 14 capsule locus mean that they no longer produce a capsule (9–11). However, an in-depth investigation of these variants has not been conducted. Molecular typing methods are now common, but the potential for mistyping these variants as serotype 14 via such methods is unclear. Additionally, the capacity of these variants to evade the antibody response elicited by vaccination has not been examined.
Here, we investigate variants of pneumococcal serotype 14 that we isolated from nasopharyngeal swabs in PNG. We report the source of the divergence and the phenotypic consequences on capsule production. Importantly, we investigate these variants for their potential to be mistyped by popular serotyping methods as well as whether they can evade vaccine-induced protection.
RESULTS
We examined nasopharyngeal swabs from children hospitalized with acute respiratory infection as part of the PneuCAPTIVE study (12). We tested 1,386 swabs for pneumococci using lytA quantitative PCR (qPCR). Of these, 1,287 (92.86%) were positive (or equivocal) for the lytA gene. Following culture, α-hemolytic growth was obtained from 1,226 of the swabs, which were then serotyped via DNA microarray. Among these, three (0.24%) swabs contained a putative variant of serotype 14. These ‘14-likes’ comprised 3.29% of swabs where serotype 14 was detected (n = 91). A pneumococcal isolate from each of these samples was purified (PMP1437, PMP1438, and PMP1514). Each isolate was a distinct, novel genotype as defined by microarray arrayCGH, Multi-Locus Sequence Typing (MLST), and Global Pneumococcal Sequence Cluster (GPSC) (Table 1).
TABLE 1.
Strain information of serotype 14 variant (14-like) pneumococci identified in this study
| MLST |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Isolate | Source | aroE | gdh | gki | recP | spi | xpt | ddl | Sequence type | GPSC |
| PMP1437 | Nasopharynx of child with acute respiratory infection | 7 | 5 | 792a | 4 | 6 | 1 | 1 | 17329a | 1000b |
| PMP1438 | Nasopharynx of child with acute respiratory infection | 1 | 5 | 4 | 8 | 9 | 277 | 1123a | 17330a | 999b |
| PMP1514 | Nasopharynx of child with acute respiratory infection | 1 | 5 | 793a | 5 | 9 | 1 | 1 | 17331a | 998b |
Novel allele and/or sequence type identified in this study.
Novel GPSC type identified in this study.
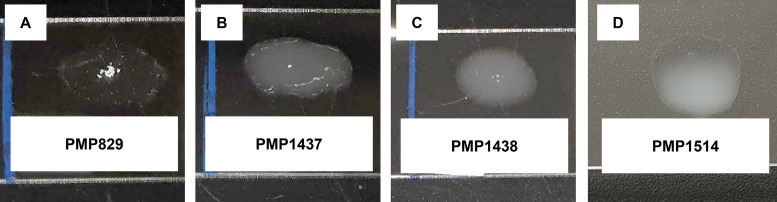
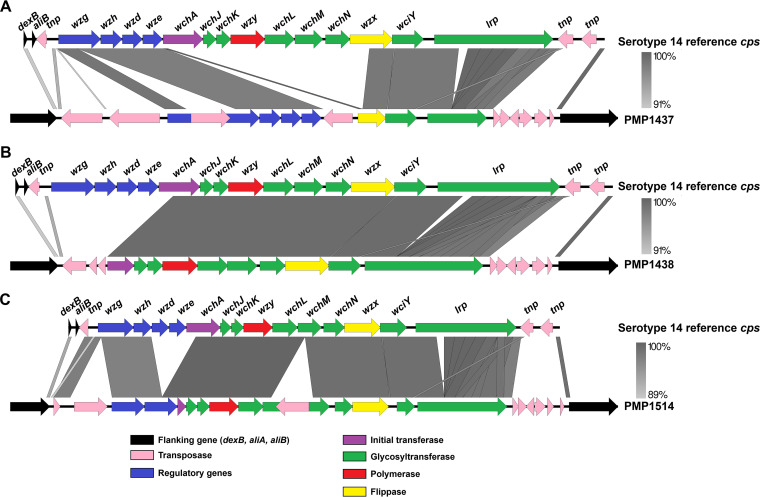
To determine if these 14-like pneumococci are ‘nontypeable’, similarly to those previously identified in Australia and South Africa (10, 11), we applied phenotypic serotyping methods. All three isolates were nontypeable using both Quellung and latex agglutination (Fig. 1 and Table 2). To verify the presence of a defective serotype 14 cps locus in these isolates, we performed whole-genome sequencing. The reads (Illumina and Nanopore) were combined, and the resultant assemblies for each isolate (GenBank accession: OM328061-OM328063) were used to interrogate the cps locus (4). Each 14-like isolate has mutations in their cps locus. PMP1437 has a 6.9 kb deletion, including wchA, wchJ, wchK, wzy, wchL, wchM, wchN, and part of wzx as well as a 1.3 kb insertion of an IS1380 element in the wzg gene. The cps locus in PMP1438 has a 4.2 kb deletion consisting of wzg, wzh, wzd, wze, and part of wchA. Lastly, PMP1514 has a 1.8 kb deletion, including wzd, wze, and part of wchA as well as disruption of the wchM gene with an IS1380 element (Fig. 2). These findings were consistent with the microarray results.
FIG 1.
Representative latex agglutination reactions of a serotype 14 positive-control strain PMP829 (A) and 14-like isolates PMP1437 (B), PMP1438 (C), PMP1514 (D). Bacterial suspensions were mixed with a latex reagent that contained serotype 14 antibodies from type 14 antisera from Statens Serum Institut adsorbed to polystyrene latex beads. A positive reaction is indicated by visible agglutination and clearing of the suspension, whereas a negative reaction lacks agglutination and remains uniform and white/opaque.
TABLE 2.
Summary of serotyping results of serotype 14 variant (14-like) pneumococci from PNG
| Serotype resulta |
|||||||
|---|---|---|---|---|---|---|---|
| Molecular serotyping methods |
Phenotypic serotyping methodse |
||||||
| Isolate | Microarrayb | PneumoCaT (% coverage)c | seroBA | SeroCall | Serotype 14 qPCR (ct value)d | Quellung | Latex agglutination |
| PMP1437 | 14-like | Nontypeable (44.50%) |
14 | Nontypeable | Negative (no ct) |
Nontypeable | Nontypeable |
| PMP1438 | 14-like | Nontypeable (68.52%) |
14 | 14 | Positive (22.75) |
Nontypeable | Nontypeable |
| PMP1514 | 14-like | Nontypeable (84.21%) |
14 | 14 | Positive (19.78) |
Nontypeable | Nontypeable |
The number outside parentheses refers to the serotype call made by that method, e.g., ‘14’ indicates ‘serotype 14’.
Serotype by DNA microarray was initially determined from a swab following a culture amplification step and was subsequently repeated on the isolates derived from these samples to validate that the 14-like strains were isolated.
The number inside parentheses represents the percent coverage against the serotype 14 cps locus, which was the top cps locus match for all three isolates.
The number in parentheses represents the mean cycle threshold (ct) value obtained by quantitative PCR (qPCR) (wzy gene target) from duplicate wells.
Phenotypic serotyping was performed with all SSI pools and type 14 sera.
FIG 2.
Comparison of the capsular polysaccharide loci of 14-like pneumococci PMP1437 (A), PMP1438 (B), and PMP1514 (C) to the reference serotype 14 sequence strain 34359 (4). Schematics of the sequence comparison were generated using Easyfig version 2.2.5 (30).
To determine the potential for the 14-like pneumococci to be mistyped, we applied four molecular methods: three whole-genome sequencing methods (PneumoCaT [13], seroBA [14], and SeroCall [15]) and singleplex qPCR using the CDC serotype 14-specific qPCR primers and probe (16). PneumoCaT made no serotype call (nontypeable), while seroBA designated these isolates as serotype 14 (Table 2). Interestingly, SeroCall and qPCR designated PMP1438 and PMP1514 as serotype 14, whereas PMP1437 was designated as nontypeable by these methods (Table 2).
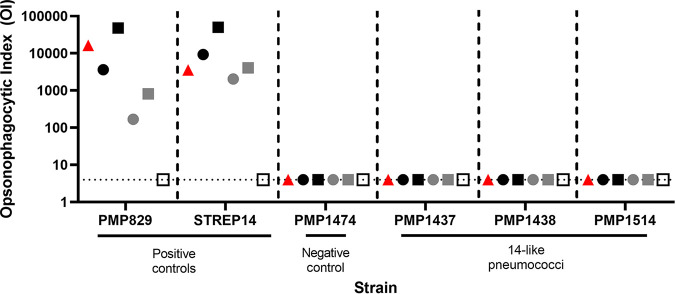
As the 14-like pneumococci appear to be unable to synthesize capsule, we hypothesized they would be able to escape opsonophagocytic killing mediated by the serotype 14 antibodies elicited by vaccination. To test this, we performed opsonophagocytic assays using sera (with a range of serotype 14-specific IgG values) from individuals vaccinated with PCV7 and/or PPSV23, both of which target serotype 14. All three 14-like isolates evaded serotype-specific opsonophagocytic killing, in contrast with the capsule producing serotype 14 control strains (PMP829 and STREP14) (Fig. 3).
FIG 3.
Opsonophagocytic indices of serotype 14 (PMP829, STREP14), nonencapsulated (PMP1474) and 14-like (PMP1437, PMP1438, and PMP1514) pneumococci. Isolates were incubated with either 007SP reference sera (red triangles) or sera from individuals vaccinated with PCV7 and/or PPSV23. Each symbol represents an individual serum sample with black, gray, and white colors representing serum samples with high (>10 μg/mL), medium (1 to 5 μg/mL), and low (<0.5 μg/mL) serotype 14 IgG titers, respectively. Times specified in brackets below are the ages of the children at the time of vaccination. Red triangle, 007SP reference sera; black circle, 1 dose PCV7 (2 years); black square, 1 dose PPSV23 (12 months) + 1 dose PCV7 (2 years); gray circle, 1 dose PCV7 (2 years); gray square, 2 doses PCV7 (14 weeks and 2 years); white square, 2 doses PCV7 (6 and 14 weeks) + 1 dose PPSV23 (12 months). OI 4 (dotted line) or less is defined as negative for opsonophagocytic killing. Nonspecific killing in this assay was −25%, −12%, −2%, 21%, 2%, and 20% for PMP829, STREP14, PMP1474, PMP1437, PMP1438, and PMP1514, respectively.
DISCUSSION
S. pneumoniae serotype 14 is one of the major capsule types associated with pneumonia and invasive pneumococcal disease (IPD), and it is included in all licensed pediatric and adult pneumococcal vaccines (PCV10, Pneumosil, PCV13, PCV20, and PPSV23) as well as in the new vaccines under development (17). Thus, accurate identification of this serotype is paramount for disease surveillance, including the monitoring of vaccine impact. In this study, we investigate variants of serotype 14 isolated from three children in PNG who were hospitalized with an acute respiratory infection. These variants have mutations in the cps locus, resulting in the loss of capsule production. These belong to the Group I lineage of nonencapsulated pneumococci, consisting of pneumococci that possess either a partial or full cps locus but also possess mutations that lead to the inability to produce capsule (18). This group includes pneumococci with defective serotype 14 cps loci as well as various cps loci of other serotypes (10, 11, 18).
Although similar variants have been identified previously (10, 11), we have conducted an in-depth investigation which revealed that these 14-like pneumococci exhibit discrepant serotyping results, depending on the method employed. Despite the inability of these pneumococci to produce capsule, most molecular serotyping methods assessed in this study designated these isolates as serotype 14. Not only were discrepant serotyping results obtained by different methods, but the mistyping potential was also further complicated by inconsistent serotype calls of each isolate by the same method (observed for the SeroCall and qPCR methods) (Table 2). The latter was due to mutations of different length and location across the cps locus of the different isolates (e.g., the deletion in PMP1437 included wzy, the target for the qPCR method, whereas this gene is intact in PMP1438 and PMP1514).
Although overlooking mutations in the cps is a known limitation of most molecular methods for serotyping, the occurrence of these variants in the population raises concerns around the potential for reducing the accuracy of serotyping data used to make decisions around vaccination programs. This is particularly relevant for the variants described in the present study, as their mistyping (as serotype 14) would incorrectly classify them as a vaccine serotype, despite their ability to evade the opsonizing serotype 14 antibodies induced by vaccination (Fig. 3). Thus, although molecular methods for serotyping generally perform well, displaying a high concordance with phenotype, it is important to note their limitations, particularly in the serotyping of isolates from countries where limited information on genetic variation in the pneumococcal population is available.
All three 14-like isolates identified in this study belong to different genetic lineages (Table 1), and the mutations in their cps loci differ in size and location (Fig. 2). This suggests convergent evolution, where mutations have occurred on multiple occasions within independent lineages of serotype 14 pneumococci. It is plausible that capsule loss may be advantageous under specific circumstances, such as during initial adherence (19) or in reducing the metabolic burden on the bacterium.
Although nonencapsulated pneumococci rarely cause pneumonia or IPD, they can cause other diseases including conjunctivitis and otitis media (20). Additionally, nonencapsulated pneumococci play important roles in shaping the pneumococcal population. Hiller et al. (21) showed recombination between a nonencapsulated and a serotype 14 strain during a carriage episode. Interestingly, the nonencapsulated recombinant had enhanced biofilm-forming capability compared with its serotype 14 parent. Biofilm formation is important for colonization and persistence in the nasopharynx. Nonencapsulated pneumococci are more likely to undergo genetic recombination compared with capsulated strains (20). Chewapreecha et al. (22) identified higher recombination frequencies in nonencapsulated pneumococci compared with serotype 14 isolates of the same genetic background. In PNG, multiple serotype carriage is high (6), providing optimal conditions for horizontal gene transfer between co-colonizing strains. Therefore, although the 14-like pneumococci are unlikely to be directly involved in disease, they have the potential to drive genetic diversity in the PNG pneumococcal population, making these variants a concern if they are not efficiently targeted by vaccination.
Pneumococcal serotyping is essential in obtaining reliable data for disease surveillance and in monitoring vaccine impact. As this information is used by decision-makers to inform policy, the accuracy of serotyping methods is vital. Our study highlights the potential for serotype diversity and mistyping, particularly where there is limited genomic information, such as in low- and middle-income settings. Therefore, it is essential that when putative variants with divergent capsule loci are detected by molecular serotyping methods, these results are validated by phenotypic approaches. In this situation, isolates should be examined further by Quellung and/or latex agglutination, with the final serotype call being based on the phenotype. Lastly, our study highlights the need to monitor the pneumococcal population for variants that could escape vaccine-induced protection, including those in IPD and pneumonia surveillance programs.
MATERIALS AND METHODS
Pneumococcal identification and molecular serotyping from nasopharyngeal samples.
Cases of moderate to severe pneumonia (2 to 59 months of age) at the Eastern Highlands Provincial Hospital and community health care clinics within the Goroka town, as well as their contacts and caregivers, were recruited as part of the PneuCAPTIVE study (12). Criteria for inclusion in the study have been described previously (12). Ethical approval for this study was obtained from the PNG Institute of Medical Research Institutional Review Board (IRB no. 1510), PNG Medical Research Advisory Committee (MRAC 15.19/16.09), and The Royal Children’s Hospital Human Resources Ethics Committee (HREC reference no. 35249). Following written informed consent, nasopharyngeal swabs were collected from patients in accordance with World Health Organization guidelines (23). Swabs were placed in 1 mL skim milk, tryptone, glucose, and glycerol medium (24), then vortexed and aliquoted prior to storing at −80°C. Samples were then shipped to the Murdoch Children’s Research Institute on dry ice and stored at −80°C until use. Swabs were screened for pneumococci using qPCR, targeting the lytA gene (25) as described previously (26). Pneumococci from swabs were cultured on selective media (horse blood agar supplemented with 5 μg/mL gentamicin) for DNA extraction and DNA microarray to infer the serotype(s) as described previously (27), using Senti-SPv1.5 microarray slides (BUGS Bioscience).
Bacterial isolates.
Pneumococcal isolates were purified from nasopharyngeal swabs by plating sample aliquots on horse blood agar supplemented with 5 μg/mL gentamicin. An α-hemolytic colony from each sample was purified. These isolates (PMP1437, PMP1438, and PMP1514) were confirmed as pneumococci by optochin sensitivity testing and whole-genome sequencing (multilocus sequence typing and pathogenwatch ID).
Whole-genome sequencing and molecular serotyping.
For all molecular methods, genomic DNA was extracted from pneumococcal isolates using the QIAcube HT with the QIAamp 96 DNA QIAcube HT Kit (Qiagen) as described previously (27). Quantitative PCR was performed, targeting the serotype 14 wzy gene. Primer and probe sequences, concentrations, and cycling conditions were kept as described previously (16). DNA from the 14-like isolates (and the serotype 14 positive-control strain PMP829) was extracted as described above. The qPCRs were performed using the GoTaq Probe qPCR Master Mix (Promega), in which each reaction consisted of 1x master mix, 300 nM forward and reverse primer, 100 nM probe (6-FAM dye and BHQ1 quencher), and 0.5 ng/μL genomic DNA. The reactions were run under the following cycling conditions: 95°C for 10 min, then 40 cycles of 95°C for 15 sec and 60°C for 1 min. Isolates were defined as serotype 14 if a cycle threshold (ct) value less than 40 was obtained.
For whole-genome sequencing, library preparation was performed using the Illumina DNA Prep kit (Illumina) and sequenced in 2 × 150 bp paired end reads on the NovaSeq platform. For molecular serotyping, reads were run through PneumoCaT 1.2 (13), seroBA 1.0.2 (14), (recommended k-mer size of 71) or SeroCall 1.0 (15), using the default parameters.
To fully characterize the cps locus in the isolates from this study, long read nanopore sequencing was conducted. Genomic DNA (1 to 2 μg) was prepared using the Ligation Sequencing Kit with Native Barcoding Expansion (Oxford Nanopore Technologies), followed by sequencing on a MinION SpotON flow cell (R9.4.1) (Oxford Nanopore Technologies). Short- and long-read data were then filtered with Trim Galore v.0.6.4 (28) and qcat v.1.1.0 (Oxford Nanopore Technologies), respectively. The combining and assembly of short- and long-reads was performed with Unicycler v.0.4.8, using the default settings (29). Schematic comparisons of the 14-like capsule locus sequences to the serotype 14 reference (4) were conducted using Easyfig version 2.2.5 (30).
Phenotypic serotyping.
Serotyping by Quellung and latex agglutination was performed against all SSI pneumococcal pools and type 14 antisera. For Quellung serotyping (31), antisera from the Statens Serum Institut (SSI) (https://ssidiagnostica.com) was used. Pneumococci were cultured on horse blood agar plates and incubated at 37°C with 5% CO2 overnight. A slightly turbid bacterial suspension (2 McFarland standard) was prepared from these cultures in saline. On a glass microscope slide, 1 μL of the suspension was mixed with 1 μL of the antisera of interest. The mixture was then examined under a microscope (400× magnification). A positive reaction with the antisera of interest was defined as cells with a ‘swollen’ appearance that were more visible under the microscope.
For serotyping by latex agglutination (32), latex reagents were prepared by adsorbing SSI antisera to polystyrene latex beads as described previously (33). A saline suspension of the pneumococcal culture was prepared (4 or 5 McFarland standard). On a glass microscope slide, 10 μL of the bacterial suspension was mixed with 10 μL of the latex reagent and rotated on an orbital shaker at ~140 rpm for 2 min. A positive reaction was indicated by visible agglutination and clearing of the suspension.
Opsonophagocytosis assay.
Sera from a cohort of healthy individuals vaccinated with either one or two doses of PCV7 and/or PPSV23 from the Fiji Pneumococcal Project (FiPP) (34) were inactivated at 56°C for 30 min and serially diluted 1:2, with 10 μL placed into each well. Aliquots of each pneumococcal strain were thawed and diluted to ~5 × 104 CFU/mL, and 20 μL of this suspension was added to each well and then incubated for 15 min at 37°C, 5% CO2 to allow opsonization to occur. Into each well, 10 μL of complement and ~4 × 105 differentiated HL60 cells (a neutrophil cell line, previously washed twice with Hanks’ balanced salt solution + 0.2% BSA) were added and incubated, shaking at 37°C, at 220 rpm for 45 min to promote killing of opsonized pneumococci. After chilling on ice, 5 μL of each reaction were spotted, allowed to form drips onto Todd-Hewitt agar supplemented with 0.5% (wt/vol) yeast extract, and allowed to dry prior to the addition of an overlay with 2,3,5-triphenyltetrazolium chloride (TTC) and incubation at 37°C, 5% CO2 overnight. The next day, the number of pneumococcal colonies was counted and used to determine the viable count and to calculate the opsonophagocytic index (OI), defined as the interpolated dilution of serum that shows 50% of serotype-specific killing of pneumococci. The lower limit of detection in the assay is 4. The OIs of samples that do not kill 50% of bacteria were reported as 4 for analysis purposes. A positive response was defined as an OI of >4. Nonspecific killing was assessed by comparing the viable count of pneumococci incubated with complement versus heat-inactivated complement.
Data availability.
The 14-like capsule locus sequences have been deposited in GenBank (accession nos. OM328061-OM328063).
ACKNOWLEDGMENTS
We thank the staff involved in the recruitment and sample collection for the PneuCAPTIVE study in PNG as well as all of the participants and their families enrolled in the study.
This study was supported by a Robert Austrian Research Award in Pneumococcal Vaccinology awarded to S.M. by ISPPD (funded by Pfizer) as well as by the National Health and Medical Research Council (NHMRC) Centre of Research Excellence for Pneumococcal Disease Control in the Asia-Pacific (GNT1196415). This work was also supported by the Victorian Government’s Operational Infrastructure Support Program. P.V.L. is a recipient of an NHMRC Career Development Fellowship (GNT1146198). During the study period, C.C.B. was a recipient of a NHMRC Career Development Fellowship (GNT1111596) and an Investigator Award (GNT1173163). F.M.R. holds a NHMRC Investigator Award (GNT1177245). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conceptualization: S.M. and C.S. Methodology and study design: S.M., R.L.F., F.M.R., C.C.B., W.S.P., J.H., P.V.L., and C.S. Data analysis: S.M., L.S., and P.V.L. Experimental investigation: S.M., L.S., A.W.H., B.D.O., L.K.B., S.B., N.M., S.P., J.H., S.W.L., and S.D.B. Funding acquisition: S.M., F.M.R., and C.S. Writing of the original manuscript draft: S.M. and C.S. Review and editing of the manuscript for submission: S.M., L.S., A.W.H., B.D.O., L.K.B., S.B., N.M., R.L.F., S.W.L., S.D.B., F.M.R., C.C.B., W.S.P., S.P., J.H., P.V.L., and C.S.
C.S. is lead investigator on a Merck Investigator Studies Program grant funded by MSD on pneumococcal serotype epidemiology in children and is an investigator on a clinical research collaboration on PCV vaccination in Mongolia, funded by Pfizer.
Footnotes
[This article was published on 12 July 2022 with Fiona M. Russell’s surname misspelled as “Russelll” in the byline. The byline has been updated in the current version, posted on 4 August 2022.]
Contributor Information
Sam Manna, Email: sam.manna@mcri.edu.au.
Christopher N. LaRock, Emory University School of Medicine
REFERENCES
- 1.Wahl B, O'Brien KL, Greenbaum A, Majumder A, Liu L, Chu Y, Lukšić I, Nair H, McAllister DA, Campbell H, Rudan I, Black R, Knoll MD. 2018. Burden of Streptococcus pneumoniae and Haemophilus influenzae type b disease in children in the era of conjugate vaccines: global, regional, and national estimates for 2000–15. Lancet Glob Heal 6:e744–e757. doi: 10.1016/S2214-109X(18)30247-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simell B, Auranen K, Käyhty H, Goldblatt D, Dagan R, O'Brien KL, Pneumococcal Carriage Group . 2012. The fundamental link between pneumococcal carriage and disease. Expert Rev Vaccines 11:841–855. doi: 10.1586/erv.12.53. [DOI] [PubMed] [Google Scholar]
- 3.Weiser JN, Ferreira DM, Paton JC. 2018. Streptococcus pneumoniae: transmission, colonization and invasion. Nat Rev Microbiol 16:355–367. doi: 10.1038/s41579-018-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, Donohoe K, Harris D, Murphy L, Quail MA, Samuel G, Skovsted IC, Kaltoft MS, Barrell B, Reeves PR, Parkhill J, Spratt BG. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet 2:e31. doi: 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manna S, Ortika BD, Dunne EM, Holt KE, Kama M, Russell FM, Hinds J, Satzke C. 2018. A novel genetic variant of Streptococcus pneumoniae serotype 11A discovered in Fiji. Clin Microbiol Infect 24:428.e1–428.e7. doi: 10.1016/j.cmi.2017.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Britton KJ, Pickering JL, Pomat WS, de Gier C, Nation ML, Pell CL, Granland CM, Solomon V, Ford RL, Greenhill A, Hinds J, Moore HC, Richmond PC, Blyth CC, Lehmann D, Satzke C, Kirkham LAS, 10v13vPCV trial team . 2021. Lack of effectiveness of 13-valent pneumococcal conjugate vaccination against pneumococcal carriage density in Papua New Guinean infants. Vaccine 39:5401–5409. doi: 10.1016/j.vaccine.2021.07.085. [DOI] [PubMed] [Google Scholar]
- 7.Pomat WS, Van Den Biggelaar AHJ, Wana S, Francis JP, Solomon V, Greenhill AR, Ford R, Orami T, Passey M, Jacoby P, Kirkham LA, Lehmann D, Richmond PC, 10v13v PCV Trial Team . 2019. Safety and immunogenicity of pneumococcal conjugate vaccines in a high-risk population: a randomized controlled trial of 10-valent and 13-valent pneumococcal conjugate vaccine in Papua New Guinean infants. Clin Infect Dis 68:1472–1481. doi: 10.1093/cid/ciy743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Vidal C, Ardanuy C, Gudiol C, Cuervo G, Calatayud L, Bodro M, Duarte R, Fernández-Sevilla A, Antonio M, Liñares J, Carratalà J. 2012. Clinical and microbiological epidemiology of Streptococcus pneumoniae bacteremia in cancer patients. J Infect 65:521–527. doi: 10.1016/j.jinf.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 9.Salter SJ, Hinds J, Gould KA, Lambertsen L, Hanage WP, Antonio M, Turner P, Hermans PWM, Bootsma HJ, O'Brien KL, Bentley SD. 2012. Variation at the capsule locus, cps, of mistyped and non-typable Streptococcus pneumoniae isolates. Microbiology (Reading) 158:1560–1569. doi: 10.1099/mic.0.056580-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marsh R, Smith-Vaughan H, Hare KM, Binks M, Kong F, Warning J, Gilbert GL, Morris P, Leach AJ. 2010. The nonserotypeable pneumococcus: phenotypic dynamics in the era of anticapsular vaccines. J Clin Microbiol 48:831–835. doi: 10.1128/JCM.01701-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohale T, Wolter N, Allam M, Ndlangisa K, Crowther-Gibson P, Du Plessis M, von Gottberg A. 2016. Genomic analysis of nontypeable pneumococci causing invasive pneumococcal disease in South Africa, 2003–2013. BMC Genomics 17:1–11. doi: 10.1186/s12864-016-2808-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan J, Nguyen CD, Lai JYR, Dunne EM, Andrews R, Blyth CC, Datta S, Fox K, Ford R, Hinds J, La Vincente S, Lehmann D, Lim R, Mungun T, Newton PN, Phetsouvanh R, Pomat WS, Xeuatvongsa A, von Mollendorf C, Dance DAB, Satzke C, Muholland K, Russell FM, PneuCAPTIVE Protocol Group . 2018. Determining the pneumococcal conjugate vaccine coverage required for indirect protection against vaccine-type pneumococcal carriage in low and middle-income countries: a protocol for a prospective observational study. BMJ Open 8:e021512. doi: 10.1136/bmjopen-2018-021512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapatai G, Sheppard CL, Al-Shahib A, Litt DJ, Underwood AP, Harrison TG, Fry NK. 2016. Whole genome sequencing of Streptococcus pneumoniae: development, evaluation and verification of targets for serogroup and serotype prediction using an automated pipeline. PeerJ 4:e2477. doi: 10.7717/peerj.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Epping L, van Tonder AJ, Gladstone RA, Bentley SD, Page AJ, Keane JA. 2018. SeroBA: rapid high-throughput serotyping of Streptococcus pneumoniae from whole genome sequence data. Microb Genom 4:e000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knight JR, Dunne EM, Mulholland EK, Saha S, Satzke C, Tothpal A, Weinberger DM. 2021. Determining the serotype composition of mixed samples of pneumococcus using whole-genome sequencing. Microb Genom 7:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pimenta FC, Roundtree A, Soysal A, Bakir M, Du Plessis M, Wolter N, Von Gottberg A, McGee L, Da Gloria Carvalho M, Beall B. 2013. Sequential triplex real-time PCR assay for detecting 21 pneumococcal capsular serotypes that account for a high global disease burden. J Clin Microbiol 51:647–652. doi: 10.1128/JCM.02927-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bentley SD, Lo SW. 2021. Global genomic pathogen surveillance to inform vaccine strategies: a decade-long expedition in pneumococcal genomics. Genome Med 13:84. doi: 10.1186/s13073-021-00901-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hathaway LJ, Meier PS, Bättig P, Aebi S, Mühlemann K. 2004. A homologue of aliB is found in the capsule region of nonencapsulated Streptococcus pneumoniae. J Bacteriol 186:3721–3729. doi: 10.1128/JB.186.12.3721-3729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammerschmidt S, Wolff S, Hocke A, Rosseau S, Müller E, Rohde M. 2005. Illustration of pneumococcal polysaccharide capsule during adherence and invasion of epithelial cells. Infect Immun 73:4653–4667. doi: 10.1128/IAI.73.8.4653-4667.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keller LE, Robinson DA, McDaniel LS. 2016. Nonencapsulated Streptococcus pneumoniae: emergence and pathogenesis. mBio 7. doi: 10.1128/mBio.01792-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiller NL, Ahmed A, Powell E, Martin DP, Eutsey R, Earl J, Janto B, Boissy RJ, Hogg J, Barbadora K, Sampath R, Lonergan S, Post JC, Hu FZ, Ehrlich GD. 2010. Generation of genic diversity among Streptococcus pneumoniae strains via horizontal gene transfer during a chronic polyclonal pediatric infection. PLoS Pathog 6:e1001108. doi: 10.1371/journal.ppat.1001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chewapreecha C, Harris SR, Croucher NJ, Turner C, Marttinen P, Cheng L, Pessia A, Aanensen DM, Mather AE, Page AJ, Salter SJ, Harris D, Nosten F, Goldblatt D, Corander J, Parkhill J, Turner P, Bentley SD. 2014. Dense genomic sampling identifies highways of pneumococcal recombination. Nat Genet 46:305–309. doi: 10.1038/ng.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Satzke C, Turner P, Virolainen-Julkunen A, Adrian PV, Antonio M, Hare KM, Henao-Restrepo AM, Leach AJ, Klugman KP, Porter BD, Sá-Leão R, Scott JA, Nohynek H, O'Brien KL, WHO Pneumococcal Carriage Working Group . 2013. Standard method for detecting upper respiratory carriage of Streptococcus pneumoniae: updated recommendations from the World Health Organization Pneumococcal Carriage Working Group. Vaccine 32:165–179. doi: 10.1016/j.vaccine.2013.08.062. [DOI] [PubMed] [Google Scholar]
- 24.O'Brien KL, Bronsdon MA, Dagan R, Yagupsky P, Janco J, Elliott J, Whitney CG, Yang YH, Robinson LG, Schwartz B, Carlone GM. 2001. Evaluation of a medium (STGG) for transport and optimal recovery of Streptococcus pneumoniae from nasopharyngeal secretions collected during field studies. J Clin Microbiol 39:1021–1024. doi: 10.1128/JCM.39.3.1021-1024.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carvalho M da GS, Tondella ML, McCaustland K, Weidlich L, McGee L, Mayer LW, Steigerwalt A, Whaley M, Facklam RR, Fields B, Carlone G, Ades EW, Dagan R, Sampson JS. 2007. Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J Clin Microbiol 45:2460–2466. doi: 10.1128/JCM.02498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satzke C, Dunne EM, Choummanivong M, Ortika BD, Neal EFG, Pell CL, Nation ML, Fox KK, Nguyen CD, Gould KA, Hinds J, Chanthongthip A, Xeuatvongsa A, Mulholland EK, Sychareun V, Russell FM. 2019. Pneumococcal carriage in vaccine-eligible children and unvaccinated infants in Lao PDR two years following the introduction of the 13-valent pneumococcal conjugate vaccine. Vaccine 37:296–305. doi: 10.1016/j.vaccine.2018.10.077. [DOI] [PubMed] [Google Scholar]
- 27.Satzke C, Dunne EM, Porter BD, Klugman KP, Mulholland EK, Vidal JE, Sakai F, Strachan JE, Hay BD, Holtzman D, Boelsen K, Habib M, Manning J, Ortika BD, Pell CL, Smyth JA, Antonio M, Klugman KP, O’Brien KL, Robins BR, Anthony SJ, Saha SK, Russell FM, Greenhill AR, Lehmann D, Adrian PV, Madhi SA, Rubin LG, Rizvi A, Hinds J, Gould KA, Kong F, Oftadeh S, Gilbert GL, Feng L, Cao B, Paranhos BG, Telles JN, Messaoudi M, Borrow R, Stanford E, George R, Slotved HC, Brugger SD, Mühlemann K, Hilty M, Rivera OI, de WJ, Charalambous BM, Leung MH, PneuCarriage project group , et al. 2015. The PneuCarriage Project: a multi-centre comparative study to identify the best serotyping methods for examining pneumococcal carriage in vaccine evaluation studies. PLoS Med 12:e1001903. doi: 10.1371/journal.pmed.1001903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J 17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 29.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Habib M, Porter BD, Satzke C. 2014. Capsular serotyping of Streptococcus pneumoniae using the Quellung reaction. J Vis Exp:e51208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Porter BD, Ortika BD, Satzke C. 2014. Capsular serotyping of Streptococcus pneumoniae by latex agglutination. J Vis Exp 51747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ortika BD, Habib M, Dunne EM, Porter BD, Satzke C. 2013. Production of latex agglutination reagents for pneumococcal serotyping. BMC Res Notes 6:49. doi: 10.1186/1756-0500-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Licciardi PV, Toh ZQ, Clutterbuck EA, Balloch A, Marimla RA, Tikkanen L, Lamb KE, Bright KJ, Rabuatoka U, Tikoduadua L, Boelsen LK, Dunne EM, Satzke C, Cheung YB, Pollard AJ, Russell FM, Mulholland EK. 2016. No long-term evidence of hyporesponsiveness following the use of pneumococcal conjugate vaccine in children previously immunised with pneumococcal polysaccharide vaccine. J Allergy Clin Immunol 137:1772–1779.e11. doi: 10.1016/j.jaci.2015.12.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The 14-like capsule locus sequences have been deposited in GenBank (accession nos. OM328061-OM328063).