Abstract
The sulfolipid sulfoquinovosyldiacylglycerol is present in the photosynthetic membranes of plants and many photosynthetic bacteria. A novel gene, sqdX, essential for sulfolipid biosynthesis in the cyanobacterium Synechococcus sp. strain PCC7942 is proposed to encode the cyanobacterial sulfolipid synthase catalyzing the last reaction of the pathway.
One of the most ubiquitous sulfur-containing bioorganic compounds is the sulfolipid sulfoquinovosyldiacylglycerol (11). It is a typical constituent of photosynthetic membranes in plants and many photosynthetic bacteria (2, 13). Four genes, sqdA, sqdB, sqdC, and sqdD, encoding enzymes of the sulfolipid biosynthetic pathway, as determined by genetic analysis, were first isolated from the purple bacterium Rhodobacter sphaeroides (3, 4, 17). Orthologs of the sqdB gene were subsequently cloned from the cyanobacterium Synechococcus sp. strain PCC7942 (hereafter referred to as Synechococcus) and Arabidopsis thaliana (8, 10). A mutant of R. sphaeroides inactivated in sqdD with sequence similarity to glycosyltransferase genes accumulates UDP-sulfoquinovose (17). Based on this observation, it has been proposed that the sqdD gene product transfers sulfoquinovose from UDP-sulfoquinovose onto a suitable acceptor, presumably diacylglycerol, during the final step of sulfolipid biosynthesis. The recent completion of the genomic sequence of the cyanobacterium Synechocystis sp. strain PCC6803 (hereafter referred to as Synechocystis) (14) provided the opportunity to search for cyanobacterial orthologs of the of R. sphaeroides sqd genes. Although a putative sqdB ortholog was present, none of the other sqd sequences of R. sphaeroides matched sequences in the cyanobacterial genome.
Identification of sqdX.
Genes encoding enzymes in the same biosynthetic pathway are often organized in operons, and novel cyanobacterial genes involved in specific biosynthetic pathways, e.g. vitamin E biosynthesis (18), have been successfully isolated based on this assumption. Employing a similar approach, the entire insert of the previously isolated plasmid pSYB (10) carrying the sqdB gene of Synechococcus was sequenced (GenBank accession no. AF155063), leading to the identification of a new open reading frame (ORF) directly downstream of sqdB (Fig. 1A). This ORF encodes a putative protein of 377 amino acids with no sequence similarity to any of the described sqd gene products of R. sphaeroides (3, 4, 17). Unlike the preceeding sqdB ORF which starts with GTG, the second ORF begins with ATG 15 bp downstream of the sqdB gene. An ideal ribosome binding site is not present, but an AAAG sequence 14 bp upstream of the ATG may serve as such. We designate this ORF sqdX. Analysis of the deduced amino acid sequence of sqdX employing Pfam (protein families database of alignments) (1) revealed a glycosyltransferase group 1 domain between the residues 228 and 347. Members of this family transfer activated sugars, for example UDP-, ADP-, GDP-, or CMP-linked sugars, to a variety of substrates, including fructose-6-phosphate and glycogen (6). Furthermore, based on the analysis with Toppred (19) and TMPred (12), the protein contains two hydrophobic domains, indicating that the sqdX gene product might be a transmembrane protein.
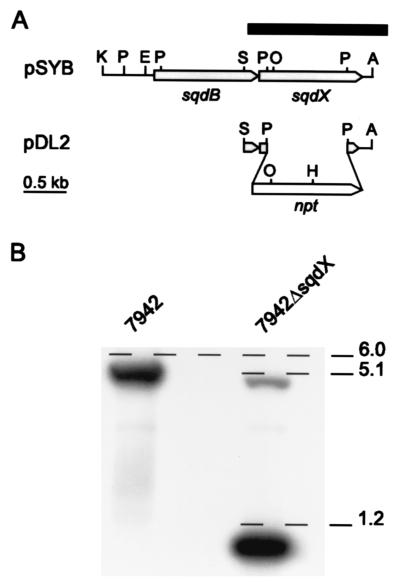
FIG. 1.
Insertional inactivation of sqdX. (A) Restriction and ORF map of the plasmid pSYB and the inactivation plasmid pDL2. The black bar indicates the location of the probe used for hybridization, and the open arrow indicates the neomycin phosphotransferase cassette. Restriction sites: A, BamHI; E, SpeI; H, HindIII; K, KpnI; O, XhoI; P, PstI; S, SalI. (B) Autoradiograph of a Southern blot showing genomic DNA from Synechococcus wild type and 7942ΔsqdX mutant. The DNA was cleaved with HindIII and probed with the fragment indicated in A. The approximate size of the three diagnostic fragments is indicated in kb.
Disruption of sqdX in Synechococcus leads to sulfolipid deficiency.
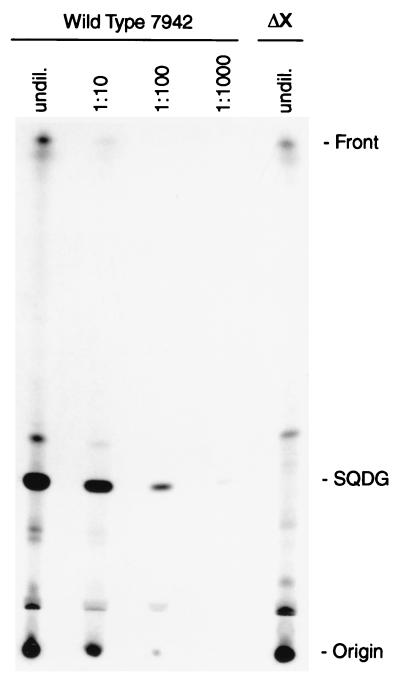
To test whether sqdX is indeed essential for sulfolipid biosynthesis, a gene replacement approach was employed (Fig. 1A). A 1.5-kb SalI/BamHI fragment of the plasmid pSYB (10) containing the entire sqdX gene and the carboxy-terminal coding region of the sqdB gene was subcloned into pBluescript II SK+ (Stratagene). This subclone was digested with PstI to remove a 0.9-kb fragment internal to the sqdX gene. The deleted fragment was replaced with a 1.2-kb PstI fragment carrying the aminoglycoside 3′-phosphotransferase gene of pUC4K (Pharmacia) conferring kanamycin resistance. The resulting plasmid pDL2 (Fig. 1A) was used to transform wild-type Synechococcus as previously described (10). Repeated restreaking of transformants on agar-solidified (1.5% wt/vol) BG-11 medium (7) with 25 μg of kanamycin ml−1 resulted in the complete loss of all wild-type genome copies. This was confirmed by DNA-DNA hybridization of HindIII-digested genomic DNA probed with a fragment covering sqdX (Fig. 1A). Under highly stringent hybridization conditions, the wild type showed a diagnostic fragment approximately 6.0 kb in length which was entirely replaced by fragments of approximately 5.1 and 1.2 kb (Fig. 1B) in the mutant strain designated 7942ΔsqdX. This strain completely lacked sulfolipid based on labeling experiments with [35S]sulfate (Fig. 2) which were performed as previously described (10). Taken together, these results strongly suggest that sqdX encodes an enzyme essential for sulfolipid biosynthesis in Synechococcus.
FIG. 2.
Separation of 35S-labeled lipids from Synechococcus wild type and 7942ΔsqdX mutant (ΔX). Either equal amounts of undiluted lipids (undil.) or serial dilutions of the wild-type extract, as indicated, were spotted. The sulfolipid band is marked SQDG. Compounds were visualized by autoradiography.
Synechococcus and Synechocystis differ in the organization of sqd genes.
A search for the sqdX gene in the recently completed genome sequence of Synechocystis (14) identified the putative ORF slr0384 (Cyanobase [www.kazusa.or.jp/cyanobase/index.html]) encoding a protein with 72% identity over the entire length compared to the sqdX gene product of Synechococcus—starting with the first in-frame ATG (position 2386042 of the genome sequence). However, the sqdB gene of Synechocystis (slr1020) as annotated in the Cyanobase with an amino acid sequence identity of 44% between the two orthologs was located approximately 1.8 Mb downstream from the putative sqdX gene. Unlike in Synechococcus, this arrangement excludes the possibility that the two sqd genes of Synechocystis are cotranscribed. To obtain proof that the putative sqdX gene of Synechocystis indeed encodes an enzyme essential for sulfolipid biosynthesis, we attempted to inactivate ORF slr0384 in Synechocystis. By using primers 5′-CGCGGATCCATGCGTGTTGCCCTGTTT-3′ and 5′-CCCAAGCTTCTAAGCCGCTAACGGAGCGT-3′, the putative sqdX ORF of Synechocystis was PCR cloned into HindIII/BamHI-digested pUC18 plasmid. A SalI fragment derived from pUC4K carrying the aminoglycoside 3′-phosphotransferase gene was inserted into the XhoI site of sqdX in pUC18. Transformation of Synechocystis with this plasmid and selection of sqdX inactivation mutants was done under the same conditions as for Synechococcus (10). However, despite multiple rounds of restreaking, we were unable to obtain a sulfolipid-deficient mutant strain of Synechocystis lacking wild-type genome copies as determined by lipid analysis and DNA-DNA hybridization (data not shown). Either the insertion into the sqdX gene affects the expression of another essential gene near to it, or sulfolipid is essential for the growth of Synechocystis under the employed conditions.
Synechocystis sqdX restores sulfolipid biosynthesis in Synechococcus mutant 7942ΔsqdX.
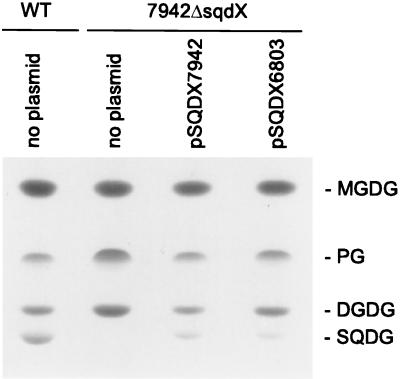
To confirm that the sqdX genes in both cyanobacteria indeed encode functionally homologous proteins, we inserted the sqdX ORFs of Synechococcus and of Synechocystis behind the tac promoter in the mobilizable broad-host-range plasmid pRL59EH (5) and transferred the constructs by conjugation into Synechococcus mutant 7942ΔsqdX as described (20). Because it was not clear where the sqdX ORF of Synechocystis starts, we included sequences upstream of the presumed ATG up to the first in-frame stop codon (positions 2385912 to 2387168 of the genome sequence). The sqdX gene of Synechococcus was PCR cloned from the plasmid pSYB (Fig. 1) by using the primers 5′-AAGGATCCTGCGCTAAAGTCGCACTC-3′ and 5′-ATAAGCTTCGAGCTCAGGCCGCT-3′ into the HindIII/BamHI sites of pRL59EH. In the same way, sqdX of Synechocystis was cloned from genomic DNA using primers 5′-CGGGATCCTATTCTAGGTTGGCCCAC-3′ and 5′-CCCAAGCTTCTAAGCCGCTAACGGAGCGT-3′. Finally, an omega cassette from the plasmid pHP45Ω (16) conferring spectinomycin and streptomycin resistance was inserted into the HindIII sites of these plasmids to provide a suitable selectable marker. The resulting plasmids containing sqdX of Synechococcus or Synechocystis were designated pSQDX7942 or pSQDX6803, respectively. Exconjugants were selected on BG11 medium containing 25 μg of kanamycin ml−1, 10 μg of spectinomycin ml−1 and 1 μg of streptomycin ml−1 and were analyzed by DNA-DNA hybridization to confirm the presence of the proper plasmid constructs (data not shown). Both constructs restored the sulfolipid biosynthetic activity in the Synechococcus mutant 7942ΔsqdX (Fig. 3) as shown by thin-layer chromatography lipid analysis (10). Based on the observed genetic complementation, it can be concluded that both cyanobacterial sqdX genes encode proteins involved in sulfolipid biosynthesis and that sqdX of Synechocystis is an ortholog of the respective gene of Synechococcus, despite different gene organization and different susceptibility for inactivation by insertional mutagenesis.
FIG. 3.
Genetic complementation of the Synechococcus 7942ΔsqdX mutant. Lipid extracts from Synechococcus wild type (WT) and mutant strain 7942ΔsqdX carrying plasmids as indicated were separated by thin-layer chromatography. Lipids were visualized by exposure to iodine vapor. DGDG, diagalactosyldiacylglycerol; MGDG, monogalactosyldiacylglycerol; PG, phosphatidylglycerol; SQDG, sulfoquinovosyldiacylglycerol
Evolution of the sulfolipid biosynthetic pathway.
Unlike sqdB, no sequences with similarity to sqdA, sqdC, or sqdD of R. sphaeroides were detected upon examination of the Synechocystis genomic sequence, suggesting that some aspects of the pathway are evolutionarily highly conserved, while others are not. The bacterial sqdB (3, 10) and the plant SQD1 (8) genes encode highly conserved proteins with similarity to sugar-nucleotide-modifying enzymes which are proposed to be involved in the biosynthesis of the UDP-sulfoquinovose headgroup donor for sulfolipid biosynthesis (9, 15). The underlying reaction seems to be unique to sulfolipid biosynthesis, and thus, the capability to synthesize sulfoquinovose evolved only once. However, the apparent lack of conservation of other genes involved in the final assembly of sulfolipid even between photosynthetic bacteria suggests that different glycosyltransferases may have been recruited to catalyze this reaction in different organisms. Our discovery of the putative glycosyltransferase involved in sulfolipid biosynthesis in cyanobacteria was initially based on the assumption that genes encoding enzymes of the same metabolic pathway are often organized in transcriptional units. This strategy worked in the case of Synechococcus, but would have failed if Synechocystis had been chosen as the model organism. Inactivation of sqdX in Synechococcus led to complete sulfolipid deficiency which could be restored by introducing the sqdX genes of either Synechococcus or Synechocystis in trans. These two results provide sufficient evidence to conclude that the sqdX gene product is essential for cyanobacterial sulfolipid biosynthesis, at least in Synechococcus and presumably also in Synechocystis. Furthermore, the sequence similarity to glycosyltransferases strongly suggests that sqdX encodes the sulfolipid synthase catalyzing the transfer of sulfoquinovose from UDP-sulfoquinovose onto a suitable acceptor, presumably diacylglycerol. Accordingly, the two putative membrane spanning domains predict a tight membrane association of this enzyme. The isolation and genetic characterization of sqdX now provide the means to study the last reaction of sulfolipid biosynthesis in greater detail in cyanobacteria. It remains to be seen whether the sqdX gene product may also serve as a model for plant sulfolipid synthases.
Acknowledgments
This work was supported in part by grant DE-FG02-98ER20305 from the Department of Energy.
We thank Peter Wolk for helpful discussion during the course of this work and his criticism of the manuscript. In addition, we thank Michaele Peters-Kottig for her efforts to inactivate sqdX in Synechocystis and Jamie Hubert for her technical assistance.
REFERENCES
- 1.Bateman A, Birney E, Durbin R, Eddy S R, Finn R D, Sonnhammer E L. Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res. 1999;27:260–262. doi: 10.1093/nar/27.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benning C. Membrane lipids in anoxygenic bacteria. In: Siegenthaler P A, Murata N, editors. Lipids in photosynthesis: structure, function and genetics. Boston, Mass: Kluwer Academic Publishers; 1998. pp. 83–101. [Google Scholar]
- 3.Benning C, Somerville C R. Identification of an operon involved in sulfolipid biosynthesis in Rhodobacter sphaeroides. J Bacteriol. 1992;174:6479–6487. doi: 10.1128/jb.174.20.6479-6487.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benning C, Somerville C R. Isolation and genetic complementation of a sulfolipid-deficient mutant of Rhodobacter sphaeroides. J Bacteriol. 1992;174:2352–2360. doi: 10.1128/jb.174.7.2352-2360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black T D, Wolk C P. Analysis of a Het- mutation in Anabaena sp. PCC7120 implicates a secondary metabolite in the regulation of heterocyst spacing. J Bacteriol. 1994;176:2282–2292. doi: 10.1128/jb.176.8.2282-2292.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell J A, Davies G J, Bulone V, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1997;326:929–939. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castenholz R W. Culturing methods for cyanobacteria. Methods Enzymol. 1988;167:68–93. [Google Scholar]
- 8.Essigmann B, Güler S, Narang R A, Linke D, Benning C. Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1998;95:1950–1955. doi: 10.1073/pnas.95.4.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Essigmann B, Hespenheide B M, Kuhn L A, Benning C. Prediction of the active-site structure and NAD+ binding in SQD1, a protein essential for sulfolipid biosynthesis in Arabidopsis. Arch Biochem Biophys. 1999;369:30–41. doi: 10.1006/abbi.1999.1344. [DOI] [PubMed] [Google Scholar]
- 10.Güler S, Seeliger A, Härtel H, Renger G, Benning C. A null mutant of Synechococcus sp. PCC7942 deficient in the sulfolipid sulfoquinovosyl diacylglycerol. J Biol Chem. 1996;271:7501–7507. doi: 10.1074/jbc.271.13.7501. [DOI] [PubMed] [Google Scholar]
- 11.Heinz E. Recent investigations on the biosynthesis of the plant sulfolipid. In: De Kok L J, editor. Sulfur nutrition and assimilation in higher plants. The Hague, The Netherlands: SPB Academic Publishing; 1993. pp. 163–178. [Google Scholar]
- 12.Hofmann K, Stoffel W. TMbase: a database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- 13.Imhoff J F, Bias-Imhoff U. Lipids, quinones and fatty acids of anoxygenic photosynthetic bacteria. In: Blankeship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 179–205. [Google Scholar]
- 14.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 15.Mulichak A M, Theisen M J, Essigmann B, Benning C, Garavito R M. Crystal structure of SQD1, an enzyme involved in the biosynthesis of the plant sulfolipid headgroup donor UDP-sulfoquinovose. Proc Natl Acad Sci USA. 1999;96:13097–13102. doi: 10.1073/pnas.96.23.13097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 17.Rossak M, Tietje C, Heinz E, Benning C. Accumulation of UDP-sulfoquinovose in a sulfolipid-deficient mutant of Rhodobacter sphaeroides. J Biol Chem. 1995;270:25792–25797. doi: 10.1074/jbc.270.43.25792. [DOI] [PubMed] [Google Scholar]
- 18.Shintani D, DellaPenna D. Elevating the vitamin E content of plants through metabolic engineering. Science. 1998;282:2098–2100. doi: 10.1126/science.282.5396.2098. [DOI] [PubMed] [Google Scholar]
- 19.von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 20.Wolk C P, Vonshak A, Kehoe P, Elhai J. Construction of shuttle vectors capable of conjugative transfer from Escherichia coli to nitrogen-fixing filamentous cyanobacteria. Proc Natl Acad Sci USA. 1984;81:1561–1565. doi: 10.1073/pnas.81.5.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]