ABSTRACT
The present study was conducted under the hypothesis that, in field peas, type of plant material, stage of maturity, ensiling, silage additive, and aerobic stress affect the composition and diversity of epiphytic microbial communities. Epiphytic microbial composition and diversity of pea seeds, partial crop peas, and whole crop peas was analyzed at different stages of late maturity, before and after ensiling, and with or without the use of lactic acid bacteria (LAB) as inoculant. Suitable combinations among pea crop variants, maturity stages, and inoculant use for the production of stable silages with sufficient aerobic stability after opening and during feed-out were identified. Genomic DNA was extracted, and 16S and 18S rRNA gene amplicons were sequenced. To assess the quality of the various silages, nutrient concentration, pH value, concentration of lactic acid, short chain fatty acids, and alcohols, and aerobic stability were determined. Pea seeds were barely colonized by epiphytic microorganisms. In partial and whole crop peas, composition and α-diversity (Shannon index) of bacterial communities did not differ between crop variants but differed among maturity stages. Epiphytic eukaryotes were rarely found on partial and whole crop peas. Bacterial composition and α-diversity were affected by ensiling and subsequent aerobic storage. In partial and whole crop peas, plant maturation caused an increase of the relative abundance of naturally occurring LAB (Weissella, Pediococcus, and Lactobacillus spp.). As a possible result, natural LAB support stable ensiling conditions even without the use of inoculants beginning with a maturity of 78 on the BBCH scale. This corresponded with a dry matter (DM) concentration of 341 and 363 g/kg in partial and whole crop peas, respectively. Addition of LAB inoculants, however, reduced ammonia, acetic acid, and butanol concentrations, and supported aerobic stability. Earlier stages of plant maturity (BBCH 76 and 77, 300 g DM/kg or less) were more prone to microbial spoilage. Stable pea seed silages can be produced at a maturity between BBCH 78 (427 g DM/kg) and 79 (549 g DM/kg), but they undoubtedly require LAB inoculation or application of other ensiling agents.
IMPORTANCE Field peas are important protein suppliers for human and animal nutrition. They can be grown in many areas of the world, which may reduce imports of protein plants and has beneficial economic and ecological effects. Ensiling is a method of preserving feed that can be implemented easily and cost-effectively at the farm. Peas harvested as seeds, partial crop, or whole crop at different maturities enable a wide range of applications. The study characterized epiphytic microbial communities on peas in terms of composition and diversity depending on the maturity of the plants and feed conservation by ensiling as they play an essential role for the production of silages. Even if this study did not consider year, site, or cultivar effects, the results would show which part of the plant is probably well suited for the production of stable and high-quality silages and at which stage of maturity.
KEYWORDS: field pea, maturity, silage quality, epiphytic bacteria, eukaryotes
INTRODUCTION
Dry pulses such as field peas (Pisum sativum) have gained importance as protein suppliers for human and animal nutrition, although production levels have varied over the years (1–3). In 2018, the worldwide production of dry peas reached 13.5 Mt (7.9 Mha), whereas in Europe, 5.3 Mt of peas were produced on 2.8 Mha (3). They can be grown locally in many areas and may reduce imports of other protein plants such as soybeans. This has both economic and ecological benefits (2).
Fermentation by lactic acid bacteria (LAB) is a traditional and commonly used method of preservation of foods (4) and animal feeds (5). A major challenge in silage making is the control of rapid respiration of carbohydrates and proteolysis by plant enzymes after cutting. Then, microorganisms change the chemical composition of the raw plant material (4) and that way determine silage quality with beneficial or detrimental outcomes (6–13). Silage quality can be controlled by wilting, sufficient pH reduction (if necessary, using inoculants and/or sugar), and provision of continuous anoxic conditions (to avoid growth of proteolytic bacteria, yeasts, and molds before opening of the silage) (5, 10).
Advanced analyses of the composition and diversity of epiphytic microbial communities and the community dynamics during ensiling can provide a better understanding of the ensiling process (14), which is prospectively helpful to adapt control strategies. Such information is currently not available for field peas. Therefore, the current study was conducted to describe the composition and diversity of epiphytic bacteria and eukaryotes on native and ensiled pea seeds, partial crop peas, and whole crop peas at different stages of plant maturity. The silages were made with and without a LAB preparation as inoculant, and were analyzed before and after being exposed to aerobic stress. The hypothesis was that type of plant material, stage of maturity, ensiling, silage additive, and aerobic stress affect the composition and diversity of epiphytic microbial communities.
RESULTS AND DISCUSSION
Changes in epiphytic microbial composition throughout pea maturation.
A total of 109 samples with minimal 11 and maximal 193 amplicon sequence variants (ASVs; Table S1 in the supplemental material) and 110 samples with minimal 1 and maximal 75 ASVs (Table S2) were analyzed for the composition of bacterial and eukaryotic communities, respectively. At least 355 and at most 437,064 16S rRNA reads per sample were available (Table S3), whereas a minimum of 38 and a maximum of 257,309 18S rRNA reads per sample were obtained (Table S4). The rarefaction curves are shown in Fig. S1.
Before removal of ASVs assigned to archaea, chloroplasts, or mitochondria, 63,181 to 121,740 16S rRNA reads per sample were available in native pea seeds. The number of 16S rRNA reads per sample that remained was 355 to 59,470, and it was especially low at premature stages (355 to 12,543; BBCH 76 to 78). A number of 63,221 to 164,526 18S rRNA reads per sample was available, but only 38 to 447 18S rRNA reads per sample remained after discarding ASVs assigned to the pea plant (i.e., to the phylum Charophyta). This clearly shows that native pea seeds were hardly colonized by bacteria (at least at premature stages) and eukaryotic microorganisms, which can be explained by their protected location in the pod. In native partial and whole crop peas, colonization with epiphytic eukaryotes was, in many samples, low as well. A significant number of 18S rRNA reads per sample remained in partial and whole crop peas only at maturity stages with BBCH 78 and 79 (3,059 to 98,331 reads per sample) (Table S4).
Commensal epiphytes are defined as nonpathogenic microbes (at least for the plant itself) that strictly colonize the plant surface without penetrating plant tissue throughout their life cycles (15). They strike up symbiotic relationships with the plants and other pro- and eukaryotes (16). They support the host plant nutritionally, promote growth, and defend it against biotic and abiotic stressors (16–18). Epiphytic bacterial communities are also antagonists of several pests and pathogens (18–23).
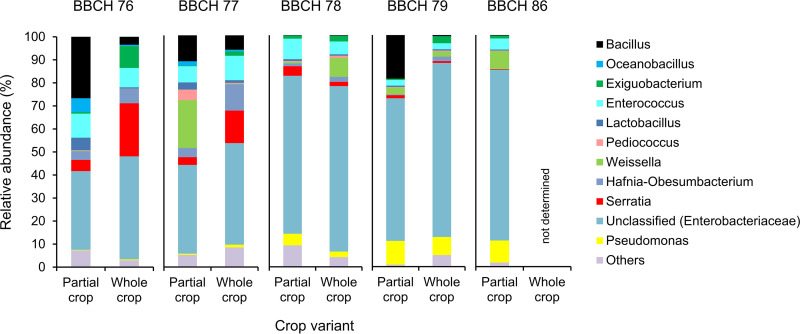
Relative abundance of epiphytic bacteria on native partial and whole crop peas was calculated on the basis of ASVs given in Table S5. The relative abundances are illustrated in Fig. 1 and specified in Table S6. The most abundant epiphytic bacteria belonged to the phyla Firmicutes and Proteobacteria. Next to unclassified representatives of the family Enterobacteriaceae, we found Bacillus, Enterococcus, Pseudomonas, Serratia, and Weissella as dominant genera (Fig. 1).
FIG 1.
Relative abundances of bacterial genera as percent of total bacteria in native partial and whole crop peas at five stages of maturity. BBCH stages were assigned according to Meier (45) and are specified in Table 2. Enterobacteriaceae could not be classified further in every case. Bacteria with a relative abundance lower than 5% are summarized under the term “others.” The number of biological replicates and standard deviations of the means are included in Table S6.
The epiphytic bacterial composition did not differ between partial and whole crop peas (P = 0.208) but differed among the stages of pea maturity (P < 0.001) (Fig. 2). Premature stages (BBCH 76 to 78) and matured stages (BBCH 79 and 86) were clearly differentiated (Fig. 2). Relative abundances of Bacillus, Enterococcus, and Serratia spp. decreased during maturation of the host plant, whereas those of unclassified Enterobacteriaceae and Pseudomonas spp. increased (Fig. 1).
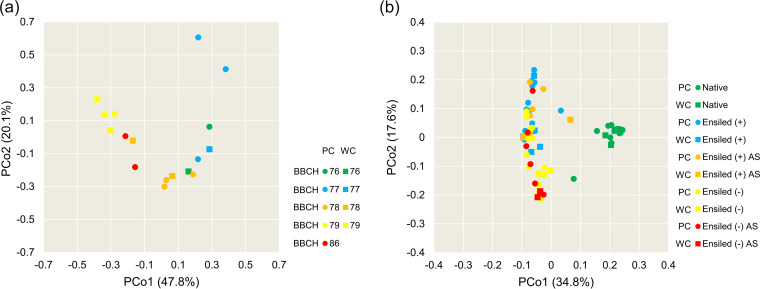
FIG 2.
Principal coordinate analysis (PCoA) based on Bray-Curtis similarity of the epiphytic bacterial composition of native field peas (a) discerned according to crop variant and stage of maturity and of the bacterial composition of native and ensiled field peas (b) discerned according to treatment and crop variant. BBCH maturity stages were assigned according to Meier (45) and are specified in Table 2. + denotes addition of microbial inoculant; – denotes ensiling without addition of inoculant; AS, aerobic storage (i.e., silages were stored 7 days under aerobic conditions), PC, partial crop peas, WC, whole crop peas.
Most of the 18S rRNA reads were assigned to Opisthokonta. Next to some insects, the eukaryotic community on partial and whole crop peas mainly comprised unclassified representatives of the Sporidiobolaceae family, Cladosporium spp., Pichia spp., as well as other unclassified fungi.
Effect of ensiling on microbial composition.
The analysis of native and ensiled peas in one data set revealed that epiphytic bacterial communities differed between crop variants (P < 0.05) and treatments (P < 0.001). The PCoA clearly shows that ensiling changed the composition of the bacterial community, whereas aerobic storage of the silages apparently had little effect (Fig. 2). In the silages made without inoculant, PCoA differentiated bacterial communities between partial and whole crop peas (Fig. 2). The relative abundances reported in the following were calculated on the basis of ASVs documented in Table S5 and Table S7. Native pea seeds were hardly colonized by bacterial or eukaryotic microorganisms.
In pea seed silages, Lactobacillus and Pediococcus spp. were most commonly identified (21 to 91% relative abundance) together with Weissella, Bacillus, and Staphylococcus spp. (maximal 54, 46, and 23% relative abundance, respectively).
Lactic acid bacteria (Weissella, Pediococcus, and Lactobacillus spp.) clearly dominated partial and whole crop pea silages throughout maturation (together 57 to 99% relative abundance). The remainder of epiphytic bacteria mainly comprised Bacillus spp. (maximal 33% relative abundance). Lactobacillus and Pediococcus spp. reached nearly 100% relative abundance after addition of LAB inoculant in most of the silages. On immature partial crop peas (BBCH 77), Bacillus and Acetobacter spp. were found with an abundance of 48 and 22%, respectively. Partial and whole crop peas at BBCH 79 harbored up to 44% Weissella spp. in addition to inoculated LAB.
After opening the silage, oxygen enters and aerobic microorganisms initialize deterioration (24, 25). Consumption of sugars and fermentation products raise silage temperature and the pH (24, 25). When the pH is increasing, Bacillus spp. as well as other aerobic bacteria and eukaryotes grow and further increase the temperature (25). Proliferation of molds completes silage deterioration, which results in dry matter (DM) loss, reduction of DM intake by animals when the silage is fed, and, in dairy applications, reduced milk yields (25). In the present study, aerobic storage did not clearly change the bacterial composition of pea silages (Fig. 2). Lactobacillus and Weissella spp. remained at up to 67 and 73% relative abundance on partial and whole crop peas, respectively. In seeds at BBCH 86, however, we found 81% to 98% relative abundance of Staphylococcus spp. after aerobic storage without artificial addition of LAB.
Application of silage additives, either chemicals or microorganisms, support fermentation before and, preferably, stability after opening of silages (25). Using the inoculant for ensiling led to bacterial communities dominated by Lactobacillus and Weissella spp. (up to 85 and 42% relative abundance, respectively, after silage opening). However, whole crop peas at BBCH 76 hosted 61% Bacillus and up to 39% Solibacillus spp., which matches previous observations (25). Dry matter concentrations of ensiled peas lower than 350 g/kg do not support rapid establishment of LAB in competition with other microorganisms and thus stable fermentation (26). In seeds harvested at BBCH 86 and ensiled with LAB inoculant, Pediococcus spp. remained at 38 to 51% after silage opening, while Staphylococcus spp. were still present with maximal 46% relative abundance.
Referring to eukaryotic communities, we predominantly found unclassified fungi (up to 99% relative abundance) and Pichia spp. (up to 86% relative abundance). Pichia spp. in silages are typically associated with high moisture contents and the presence of LAB as they ferment lactic acid (27). Next, the abundance of Pichia spp. seemed to decline throughout ongoing maturation of the peas. Aerobic storage clearly decreased the abundance of Pichia spp. in silages without inoculation with LAB strains to maximal 31%. Then, Penicillium, Cladosporium, and Hyphopichia/Candida clade spp. as well as unclassified representatives of the Saccharomycetaceae, Sporidiobolaceae, and Aspergillaceae families, were detected in individual samples. They primarily occurred in silages made from immature stages (i.e., BBCH 76 and 77) and after opening the silages. The presence of such eukaryotes is typical for spoiled silages (24, 27). The application of LAB inoculants can improve or impair aerobic stability of the silage, depending on the LAB strains used, the material, and the presence of yeasts that initiate aerobic deterioration (28). Reports on this are not consistent (28), but our results have shown that LAB inoculation widely inhibited the growth of eukaryotes under aerobic storage conditions.
Changes in α-diversity.
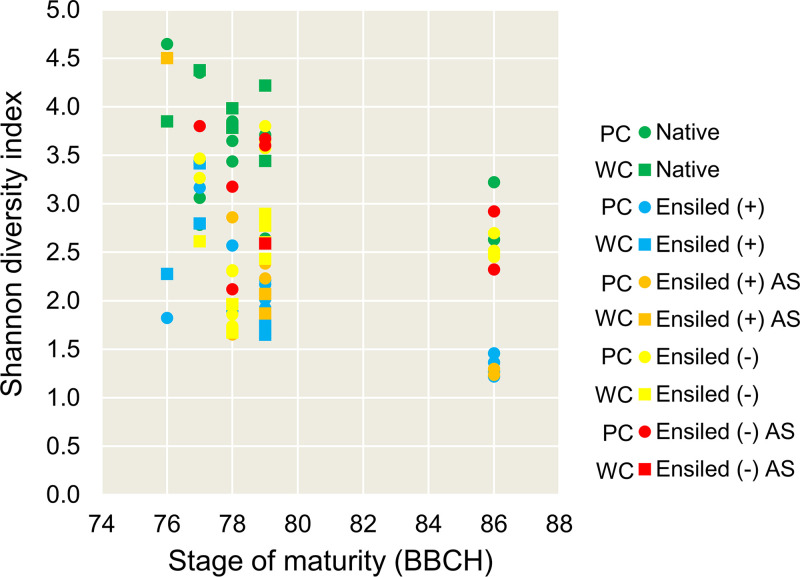
Calculated Shannon diversity index values are shown in Fig. 3. The Kruskal-Wallis test revealed significant differences in α-diversity of bacteria among maturity stages (P < 0.01) and treatments (P < 0.001), whereas no differences were found between the crop variants (P = 0.564). The Shannon index values generally decreased during ongoing maturation of the plants. Shannon diversity was highest in native peas and lowest in pea silages inoculated with LAB (Fig. 3). Aerobic storage of inoculated silages did not change Shannon diversity (Fig. 3). Pea silages made without LAB inoculant showed higher Shannon diversity, which further increased during aerobic storage (Fig. 3). Hence, application of inoculants supported stable bacterial communities dominated by LAB. This was most evident from the beginning of BBCH 79 when DM concentrations of the partial and whole crop peas were higher than 400 g/kg. In premature pea silages (BBCH 76 and 77), Shannon diversity was high even with LAB inoculation (Fig. 3). High water activity at these stages may support the growth of concomitant microorganisms in addition to natural and applied LAB (27, 29).
FIG 3.
α-diversity of epiphytic bacterial communities on native and ensiled field peas based on the Shannon index discerned according to treatment and crop variant. BBCH maturity stages were assigned according to Meier (45) and are specified in Table 2. + denotes addition of microbial inoculant; – denotes ensiling without addition of inoculant; AS, aerobic storage (i.e., silages were stored 7 days under aerobic conditions); PC, partial crop peas; WC, whole crop peas.
Nutrient concentrations of pea maturity stages and ensiling quality traits.
The results of additional analyses of DM, crude nutrient, and detergent fiber concentrations of native and ensiled pea seeds, partial crop peas, and whole crop peas are summarized in Table S8.
Pea seeds ripened much faster than partial or whole crop peas (Table S8), which has to be considered during seed harvesting at premature stages with regard to harvesting technology. Harvest of seeds in which nutrient storage is complete but that are not fully ripened (from BBCH 79 at the latest to BBCH 86) may have phytosanitary benefits (30). Pea seeds had 457 g starch and 133 g sugar per kg DM at BBCH 76, and 519 g starch and 51 g sugar per kg DM at BBCH 86. Together with high protein concentrations in the seeds, which were more or less consistent within the tested maturity range (Table S8), this offers the opportunity to partly replace protein-rich feeds and cereals by peas in diets for ruminants and monogastric livestock (31–33).
Partial crop peas ranged in nutrient density and fiber concentration between pea seeds and whole crop peas (Table S8). They offer a balance in protein and fiber supply. In partial and whole crop peas, crude protein concentration was higher at late maturity (from BBCH 78 on; Table S8). This is usually similar to the concentration of starch (34, 35) and linked to pod development (34). Digestible nonstarch polysaccharides (mostly glucose and xylose residues) are a source of energy the plant provides in addition to energy provided by the seeds, but these components decrease during nutrient storage in the seeds and plant maturation (36).
On a DM basis, amino acid concentrations tended to be lower at later stages of maturity (Table S9). Ensiling modified amino acid composition, likely as a result of microbial degradation and synthesis (37).
Ensiling characteristics and the silages’ concentrations of ammonia, lactic acid, acetic acid, and alcohols are given in Table S10. Propionic acid, butyric acid, valeric acid, and caproic acid concentrations were mostly below detection limit.
The pea seed silages had an acidic, fresh scent and a moist doughy texture. Their pH values ranged from 4.2 to 5.9 and from 4.6 to 6.1 with and without LAB inoculant, respectively (Table S10). Silages from premature seeds (BBCH 76) were spoiled. After aerobic storage, the pH was higher than it was at the time of opening. Pea seeds ensiled at a late maturity stage (BBCH 86) had the highest pH values and lowest aerobic stability (Table S10). After aerobic storage, they were widely molded. Produced ammonia, acetic acid, and butanol levels were lower at late maturity (Table S10). In the pea seed silages, lactic acid was formed at BBCH 76 only after LAB inoculation. Also, at BBCH 78 and 79 more lactic acid was formed after using the additive. At BBCH 86, there was no lactic acid formation, either with or without LAB additive (Table S10). The concentrations of ammonia, acetic acid, and butanol were higher in seed silages without inoculant than with the addition of LAB until BBCH 79 (Table S10).
As with pea seeds, most of partial crop and whole crop pea silages were of good quality, with pH distinctly lower than 5.0. They had an acidic, nutty scent and a moist texture until BBCH 78. At BBCH 79 and 86, the silages were dry and strawlike. The concentration of ammonia was maximal 4.1 g/kg DM (i.e., 14% NH3-N of total nitrogen) and lowest at late maturity (Table S10). Ammonia is in silages mainly associated with reduction of palatability and intake depression (29, 38). In silages from partial and whole crop peas, the concentrations of lactic acid showed a decreasing tendency with increasing maturity (Table S10). In most cases, the lactic acid concentration differed only slightly between silages with and without LAB additive. In some cases, even more lactic acid was formed without LAB additive, which shows the significant presence of natural LAB. Significant quantities of butanol were detected in partial and whole crop pea silages (Table S10). Butanol concentration was low when DM concentration was high. Partial and whole crop pea silages at BBCH 77 and higher were stable for 104 to 168 h under aerobic storage conditions (Table S10). Despite LAB inoculation, silages made from premature materials (BBCH 76) were often mildewed after aerobic storage and had a perceptible scent of ammonia. Growth rates of spoiling bacteria increase along with availability of free water (39), and thus pea plants having less than 350 g DM per kg should be wilted before ensiling to maintain stable lactic acid fermentation (26).
Apart from DM concentration, sugar concentration and the sugar-to-buffering capacity ratio influence pH during ensiling and the appearance of LAB (40). In the native pea crop variants, total sugar concentration ranged from 24 to 76 g/kg DM. It was higher in the premature stages (BBCH 76 to 78) than in ripened materials. Ensiling generally reduced sugar concentration (7 to 37 g/kg DM), because oligomeric sugars are a substrate for lactic acid fermentation (10). In mature seed silages (BBCH 86), no sugar fermentation occurred (36 g/kg DM in native seeds, 36 g/kg DM in control silages, and 21 g/kg DM in silages with added inoculant, respectively), which corresponded to the stable pH (Table S10).
The current study did not consider year, site, or cultivar effects, which undoubtedly contribute to variation in canopy structure and development. However, we summarized results of the study relevant for practice to give an idea how harvesting and preserving of field peas can be controlled and improved (Table 1). The German Agricultural Society (DLG) key for evaluating roughages (41) was used to classify the ensiling success. This considers the measured proportion of acetic acid, butyric acid, and the pH value depending on the DM concentration. As a result, all silages achieved at least 85 out of 100 points and are rated as “good” to “very good.” This excludes silages that were obviously spoiled (e.g., seed silages at BBCH 76). However, the critical NH3-N content, which, according to the DLG key (42), should be a maximum of 10% of the total nitrogen, was often exceeded until BBCH 77, especially in silages that were produced without silage additives. With regard to the preservation of true protein and the protein solubility in the rumen, deamination and the associated formation of biogenic amines must be limited (43). Therefore, dry silage with a DM concentration of more than 500 g/kg is a sensible option for conservation, at least in case of partial or whole crop peas. In such a silage, the fermentation activity of the epiphytic LAB is alone not sufficient to achieve a preservative effect, due to the osmotic conditions. It is rather the combination of dryness and the anaerobic environment that is conserving the material, supported by lactic acid production.
TABLE 1.
Prospectively relevant information for practice on harvesting and preserving field peas based on the results of this study
| Stage of maturity at harvest (BBCH)a | 76 | 77 | 78 | 79 | 86 |
|---|---|---|---|---|---|
| Dry matter concn at harvest (g/kg) | |||||
| Seeds | 310 | 390 | 430 | 550 | 740 |
| Partial crops | 250 | 300 | 340 | 420 | 630 |
| Whole crops | 250 | 300 | 360 | 450 | 590 |
| Nutrient storage in seeds | incomplete | incomplete | almost complete | complete | complete |
| Preservation by ensiling reasonable | |||||
| Seeds | limited | limited | yes | yes | yes |
| Partial and whole crops | limited | limited | yes | yes | yes |
| Use of silage additives required | |||||
| Seeds | yes | yes | yes | yes | yes |
| Partial and whole crops | yes | yes | not mandatory | not mandatory | not mandatory |
| Silage quality to expectb | |||||
| Seeds | low | high | high | limitedc | |
| Partial and whole crops | low | limitedc | high | high | high |
| Aerobic stress toleranced | |||||
| Seeds | high | high | lowc | ||
| Partial and whole crops | lowc | high | high | high | high |
Maturity stages are encoded using the BBCH code for phenological maturity of plants according to Meier (45).
Classified according to DLG keys (55, 56) on the basis of acetic acid, butyric acid, and NH3-N concentration; pH value; and aerobic stability.
Depends on use of silage additive.
Assuming a minimum of 72 h of aerobic stability is sufficient for practical application. This, however, depends on farm size and management.
Conclusions.
The present study has shown that pea seeds were generally little colonized by epiphytic microorganisms. To produce stable silages from pea seeds and to protect them from spoilage after opening, the use of silage additives is required. As such, LAB inoculants can replace natural LAB. In the case of partial and whole crop peas, a significant natural stock of LAB was established during plant maturity. Our results suggest that partial and whole crop peas can be successfully ensiled beginning at BBCH 78 even without the use of inoculants. This could contribute to a reduction in farm costs and increased cultivation of field peas as feed plants. At BBCH 78, nutrient storage in the seeds is almost complete. Significant loss of nutrients is therefore not expected with harvest of pea plants from this stage on. Then, partial crop peas are of particular interest for ruminant nutrition, as they add easily soluble nutrients (protein and starch) as well as fiber to the ration.
MATERIALS AND METHODS
Harvesting and ensiling of peas.
The field pea cultivar “Astronaute” (Fig. 4) was grown and harvested in Köllitsch (Saxony, Germany) in 2018. The cultivar “Astronaute” was chosen because it is one of the most widely used cultivars in Saxony and nationwide in both conventional and organic farming. Specific information on phenological, morphological, and yield characteristics can be obtained online from the Federal Plant Variety Office (44). Representative spots were sampled by hand in June and July at five maturity stages on the basis of the seeds’ DM concentration. At each of these stages, material was collected representing the seeds, partial crops, and whole crops. The investigator additionally determined phenological characteristics of the plants at each stage of maturity and classified them in the BBCH scaling (45). The DM concentrations at individual stages and corresponding BBCH codes are given in Table 2. Whole crops were cut 10 cm above the ground, whereas partial crops were cut at 25 cm height (Fig. 4), which means that approximately 58% (based on plant height) of the upper part of the plant was harvested. Strict use of gloves, cleaning, and disinfection of the equipment avoided contamination of the samples. From the native seeds and crops, Rostock Model Silages were prepared according to Hoedtke and Zeyner (46). Initial DM concentrations were those given in Table 2. The materials were chopped, vacuum-sealed in polyethylene bags (three bags per maturity stage for seeds, partial crops, and whole crops, respectively, with 2 kg material each), and stored at approximately 25°C for a minimum of 59 and maximum of 62 days. Two silage variants were prepared: (i) a control without any inoculant, and (ii) with addition of homofermentative LAB (Lactobacillus plantarum LSI NCIMB 30083 1k20736 and L256 NCIMB 30084 1k20737, and Pediococcus acidilactici P11 DSM 23689 1k1011 and P6 DSM 23688 1k1010 strains; together 1.0 × 1011 CFU per gram fresh matter) and carbohydrate degrading enzymes using a commercial preparation (Josilac classic; Josera GmbH & Co. KG, Kleinheubach, Germany). This LAB preparation was chosen because it is widely used in Germany, has a high practical relevance, is also approved for organic farms, and covers a wide DM range (25 to 40%). The type of the enzymes included in this preparation is not known and is confidential to the manufacturer.
FIG 4.
Partial crop of the pea cultivar “Astronaute” was harvested at approximately 25 cm height (beneath the lowest pods; on this photograph with a maturity referring to BBCH 79); specification of the maturity stage is given in Table 2; photo by C. Kuhnitzsch.
TABLE 2.
Dry matter concentration of pea seeds, partial crop peas, and whole crop peas at harvest at five maturity stages
| Maturity (BBCH)a | Crop variant | Dry matter (g/kg) |
|---|---|---|
| 76 | Seeds | 313 |
| Partial crops | 251 | |
| Whole crops | 249 | |
| 77 | Seeds | 389 |
| Partial crops | 305 | |
| Whole crops | 301 | |
| 78 | Seeds | 427 |
| Partial crops | 341 | |
| Whole crops | 363 | |
| 79 | Seeds | 549 |
| Partial crops | 421 | |
| Whole crops | 447 | |
| 86 | Seeds | 737 |
| Partial crops | 632 | |
| Whole crops | 591 |
Maturity stages are encoded using the BBCH code for phenological maturity of plants according to Meier (45).
DNA extraction.
A quantity of 5 g sample material previously chopped and mixed was incubated for 15 min in 100 mL suspension consisting of 0.58 g NaH2PO4 × 2 H2O, 2.5 g Na2HPO4 × 2 H2O, 4 g NaCl, 1 g tryptic peptone, and 0.3 mL Tween 80 in 1 L water, with a pH of 7.0 as proposed by the Association of German Agricultural Analytic and Research Institutes (VDLUFA) (47). The sample material and the suspension were homogenized for 5 min at 230 rpm using a Seward Stomacher 400 Circulator paddle blender (Seward Ltd., Worthing, United Kingdom). The resulting liquid phase was centrifuged at 18,000 × g for 5 min, transferred to 2 mL tubes, and centrifuged at 10,000 × g for 1 min. Genomic DNA was extracted and purified using the Quick-DNA Fungal/Bacterial Miniprep Kit (Zymo Research Corp., Irvine, CA, USA) following the manufacturer’s instructions. The DNA concentration was determined using the Invitrogen Qubit 3.0 fluorometer and the Qubit dsDNA BR assay kit (Thermo Fisher Scientific Inc., Waltham, MA, USA). Extracted DNA was stored at −20°C.
PCR amplification and amplicon sequencing.
The PCR and purification of the PCR products were carried out in accordance with the instructions provided for the Illumina MiSeq System (48). The amplification of the V3–V4 region of 16S rRNA genes was performed on a PCR SensoQuest Labcycler (SensoQuest GmbH, Göttingen, Germany) using the forward primer V3f (5′-CCTACGGGNGGCWGCAG-3′) and the reverse primer V4r (5′-GGACTACHVGGGTATCTAATCC-3′) (49). For amplification of the V8–V9 region of 18S rRNA genes, the forward primer 1422f (5′-ATAACAGGTCTGTGATGCCCT-3′´) and the reverse primer 1797r (5′-GCCTCCYGCAGGTTCACCTAC-3′) (50) were used. Each sample of 2.5 μL microbial DNA (5 ng/μL in 10 mM Tris-HCl, pH 8.5) was mixed with PCR master mix, consisting of 1 μL each of the barcoded forward and reverse primer (10 pmol/μL; Eurofins Genomics Germany GmbH, Ebersberg, Germany), 12.5 μL of 2 × KAPA HiFi DNA polymerase (Hot Start Ready Mix; KAPA Biosystems Inc., F. Hoffmann-La Roche AG, Basel, Switzerland), and 8 μL of PCR grade water. The PCR was performed under the following conditions: hot start at 95°C for 3 min, followed by 25 cycles of 95, 55, and 72°C, each for 30 s, and a final extension at 72°C for 5 min, then hold at 4°C. The PCR products were analyzed by gel electrophoresis in 1.5% agarose. PCR cleanup was carried out using AMPure XP beads (Beckman Coulter Genomics Inc., Chaska, MN, USA; 20 μL per sample) and a 96-well 0.2 mL PCR plate (Bio-Rad Laboratories Inc., Hercules, CA, USA). Index PCR was performed using index primers of the Nextera XT Index kit (Illumina Inc., San Diego, CA, USA). The library was arranged on a TruSeq Index Plate Fixture (Illumina Inc., San Diego, CA, USA) as given in the manual (48). Sequencing of the amplicons was performed on the Illumina MiSeq system (Illumina Inc., San Diego, CA, USA) using the MiSeq reagent kit v3 (600-cycle), followed by demultiplexing and removal of barcode sequences by the Illumina software.
Chemical analyses.
Dry matter (after freeze-drying), crude ash, crude protein, acid ether extract, starch (determination by polarimetry), and sugars (determination by Luff-Schoorl method), crude fiber, and detergent fibers were analyzed according to VDLUFA (47) using the methods 3.1, 4.1.1, 5.1.1 B, 6.1.1, 6.5.1, 6.5.2, 6.5.3, 7.1.1, 7.2.1, and 8.1, respectively. The neutral detergent fiber was determined after pretreatment with heat stable amylase. Neutral detergent fiber and acid detergent fiber were expressed exclusive of residual ash. The proteins were hydrolyzed with hydrochloric acid, and individual amino acids were analyzed according to VDLUFA (47) method no. 4.11.1 using a Biochrom 30 Amino Acid Analyser with PEEK-Sodium Prewash Column (100 mm × 4.6 mm) and PEEK-Oxidized Feedstuff Column (200 mm × 4.6 mm) (Biochrom Ltd., Cambridge, United Kingdom). For detection of tryptophan, the proteins were hydrolyzed with phosphoric acid and hydrochloric acid. Tryptophan was analyzed according to Fontaine et al. (51) by liquid chromatography (Agilent 1100 Series fitted with 150 mm × 4.6 mm × 5 μm ZORBAX Eclipse XDB-C8 column; Agilent Technologies Inc., Santa Clara, CA, USA). Lactic acid concentrations were determined using liquid chromatography (internal method LKS FMUAA 166; the laboratory was accredited according to DIN EN ISO/IEC 17025:2018). Short chain fatty acids (SCFA) and alcohols produced during the fermentation of pea silages were determined after aqueous extraction by gas chromatography using a Shimadzu GC2010 (Shimadzu Corp., Kyoto, Japan) with flame ionization detector. An SGE BP21 separation column (30 m × 0.53 mm × 0.5 μm) (Trajan Scientific and Medical, Ringwood, AU-VIC, Australia) was used. The extracts were centrifuged at 2,000 × g before injection. The following settings were used for detection of SCFA: on-column injection, 0.5 μL injection volume, 180°C injection temperature, constant pressure of 22.7 kPa (i.e., 29.7 cm/s linear velocity and 3.64 mL/min column flow), 85°C initial oven temperature, raised up by 8°C/min to 200°C and held for 6 min, and 200°C detection temperature; and for detection of alcohols: on-column injection, 0.5 μL injection volume, 180°C injection temperature, constant column flow of 7.7 mL/min, 35°C initial oven temperature held for 2.5 min, raised up by 8°C/min to 50°C, then by 100°C/min to 200°C and held for 2 min, and 200°C detection temperature. Helium was the carrier and makeup gas. The concentration of target analytes was determined on the basis of an external standard calibration. The aerobic stability of the silages was tested following the procedure of Honig (52) and was expressed as time until the temperature difference between material (silage) and environment exceeds 3 K. The ammonia concentrations were determined according to the method of Conway and Byrne (53).
Bioinformatic and statistical analysis.
Bioinformatic analysis of MiSeq amplicon sequences was performed with QIIME 2 version 2019.1 (54) including removal of primers by Cutadapt, quality and length filtering, chimera removal, DADA2 clustering, and taxonomic assignment using the SILVA 132 rRNA database (55, 56). The available number of sequences was normalized among samples by rarefaction, where sampling depth was restricted to 20,000 reads in the 16S rRNA data set and to 12,000 reads in the 18S rRNA data set. Amplicon sequence variants (ASVs) were used for all further calculations and statistical tests. The 16S rRNA ASVs that were assigned to archaea, chloroplasts, or mitochondria were removed from the 16S rRNA data set. The archaea were removed because the used primers do not have reliable coverage (49). The 18S rRNA ASVs assigned to the pea plant itself (i.e., to the phylum Charophyta) were removed from the 18S rRNA data set. Two-way PERMANOVA and a principal coordinate analysis (PCoA) were performed in PAST version 4.01 (57) based on Bray-Curtis similarity considering crop variant and stage of maturity (for native peas) or crop variant and treatment (for pea silages). The Shannon diversity index was calculated using QIIME 2. The Kruskal-Wallis test was used in SAS version 9.4 NPAR1WAY (SAS Institute Inc., Cary, NC, USA) to identify differences in α-diversity (Shannon index) among maturity stages, crop variants, or treatments. The significance level for all statistical tests was set to P < 0.05. Statistical tests were not performed with data referring to pea seeds (16S rRNA and 18S rRNA) and partial and whole crop peas (18S rRNA), because the number of reads available after filtering (see above) did not reveal notable microbial colonization of native materials.
Data availability.
Raw sequence data were deposited at the European Nucleotide Archive (ENA) under the study accession number PRJEB45910.
ACKNOWLEDGMENTS
The project was supported by funds of the Federal Ministry of Food and Agriculture (BMEL) based on a decision of the parliament of the Federal Republic of Germany via the Federal Office for Agriculture and Food (BLE) under the federal program Protein Crop Strategy (grant number 2815EPS058). Moreover, we acknowledge the financial support within the funding program Open Access Publishing by the German Research Foundation (DFG).
We declare no conflicts of interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Annette Zeyner, Email: annette.zeyner@landw.uni-halle.de.
Junhyun Jeon, Yeungnam University.
REFERENCES
- 1.Watts P. 2011. Global pulse industry: state of production, consumption and trade; marketing challenges and opportunities, p 437–464. In Tiwari BK, Gowen A, McKenna B (ed), Pulse foods processing, quality and nutraceutical applications. Elsevier, Philadelphia, PA. [Google Scholar]
- 2.Khan TN, Meldrum A, Croser JS. 2016. Pea overview. Elsevier, Philadelphia, PA. [Google Scholar]
- 3.FAOSTAT. 2022. Food and agriculture statistics. https://www.fao.org/food-agriculture-statistics/en/.
- 4.Tamang JP, Watanabe K, Holzapfel WH. 2016. Review: diversity of microorganisms in global fermented foods and beverages. Front Microbiol 7:377. doi: 10.3389/fmicb.2016.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolsen K, Ashbell G, Weinberg Z. 1996. Silage fermentation and silage additives—review. Asian Australas J Anim Sci 9:483–494. doi: 10.5713/ajas.1996.483. [DOI] [Google Scholar]
- 6.Harris CE, Raymond WF. 1963. The effect of ensiling on crop digestibility. Grass and Forage Sci 18:204–212. doi: 10.1111/j.1365-2494.1963.tb00350.x. [DOI] [Google Scholar]
- 7.Aksu T, Baytok E, Bolat D. 2004. Effects of a bacterial silage inoculant on corn silage fermentation and nutrient digestibility. Small Rum Res 55:249–252. doi: 10.1016/j.smallrumres.2003.12.012. [DOI] [Google Scholar]
- 8.Bachmann M, Kuhnitzsch C, Martens SD, Steinhöfel O, Zeyner A. 2019. Einfluss des Silierens und Toastens auf antinutritive Inhaltsstoffe von Erbsen und Ackerbohnen, p 146–148. In Zeyner A, Kluth H (ed ), Tagung Schweine- und Geflügelernährung. Universität Halle-Wittenberg, Halle, Germany. [Google Scholar]
- 9.Rizzello CG, Losito I, Facchini L, Katina K, Palmisano F, Gobbetti M, Coda R. 2016. Degradation of vicine, convicine and their aglycones during fermentation of faba bean flour. Sci Rep 6:32452. doi: 10.1038/srep32452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gefrom A, Ott EM, Hoedtke S, Zeyner A. 2013. Effect of ensiling moist field bean (Vicia faba), pea (Pisum sativum) and lupine (Lupinus spp.) grains on the contents of alkaloids, oligosaccharides and tannins. J Anim Physiol Anim Nutr (Berl) 97:1152–1160. doi: 10.1111/jpn.12024. [DOI] [PubMed] [Google Scholar]
- 11.Hoedtke S, Gabel M, Zeyner A. 2010. Protein degradation in feedstuffs during ensilage and changes in the composition of the crude protein fraction. Übers Tierern 38:157–179. [Google Scholar]
- 12.Szumacher-Strabel M, Stochmal A, Cieslak A, Kozłowska M, Kuznicki D, Kowalczyk M, Oleszek W. 2019. Structural and quantitative changes of saponins in fresh alfalfa compared to alfalfa silage. J Sci Food Agric 99:2243–2250. doi: 10.1002/jsfa.9419. [DOI] [PubMed] [Google Scholar]
- 13.Kuhnitzsch C, Hofmann T, Bachmann M, Martens SD, Henle T, Zeyner A, Steinhöfel O. 2019. Effect of ensiling and toasting of field pea grains on formation of Maillard polymers from lysine and arginine. Adv Anim Biosci 10:540. [Google Scholar]
- 14.Guo XS, Ke WC, Ding WR, Ding LM, Xu DM, Wang WW, Zhang P, Yang FY. 2018. Profiling of metabolome and bacterial community dynamics in ensiled Medicago sativa inoculated without or with Lactobacillus plantarum or Lactobacillus buchneri. Sci Rep 8:357. doi: 10.1038/s41598-017-18348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zambell CB, White JF, Jr.. 2015. In the forest vine Smilax rotundifolia, fungal epiphytes show site-wide spatial correlation, while endophytes show evidence of niche partitioning. Fungal Divers 75:279–297. doi: 10.1007/s13225-014-0316-3. [DOI] [Google Scholar]
- 16.Alvarenga DO, Fiore MF, Varani AM. 2017. A metagenomic approach to cyanobacterial genomics. Front Microbiol 8:809. doi: 10.3389/fmicb.2017.00809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bacon CW, White JF, Jr.. 2016. Functions, mechanisms and regulation of endophytic and epiphytic microbial communities of plants. Symbiosis 68:87–98. doi: 10.1007/s13199-015-0350-2. [DOI] [Google Scholar]
- 18.Yousaf S, Afzal M, Reichenauer TG, Brady CL, Sessitsch A. 2011. Hydrocarbon degradation, plant colonization and gene expression of alkane degradation genes by endophytic Enterobacter ludwigii strains. Environ Pollut 159:2675–2683. doi: 10.1016/j.envpol.2011.05.031. [DOI] [PubMed] [Google Scholar]
- 19.Choudhary DK, Johri BN. 2009. Interactions of Bacillus spp. and plants—with special reference to induced systemic resistance (ISR). Microbiol Res 164:493–513. doi: 10.1016/j.micres.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Silby MW, Cerdeño-Tárraga AM, Vernikos GS, Giddens SR, Jackson RW, Preston GM, Zhang X-X, Moon CD, Gehrig SM, Godfrey SAC, Knight CG, Malone JG, Robinson Z, Spiers AJ, Harris S, Challis GL, Yaxley AM, Harris D, Seeger K, Murphy L, Rutter S, Squares R, Quail MA, Saunders E, Mavromatis K, Brettin TS, Bentley SD, Hothersall J, Stephens E, Thomas CM, Parkhill J, Levy SB, Rainey PB, Thomson NR. 2009. Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens. Genome Biol 10:R51. doi: 10.1186/gb-2009-10-5-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, Cao Y, Cai Y, Terada F. 2010. Natural populations of lactic acid bacteria isolated from vegetable residues and silage fermentation. J Dairy Sci 93:3136–3145. doi: 10.3168/jds.2009-2898. [DOI] [PubMed] [Google Scholar]
- 22.Vescovo M, Torriani S, Orsi C, Macchiarolo F, Scolari G. 1996. Application of antimicrobial‐producing lactic acid bacteria to control pathogens in ready‐to‐use vegetables. J Appl Bacteriol 81:113–119. doi: 10.1111/j.1365-2672.1996.tb04487.x. [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez-Palacios A, Staempfli HR, Duffield T, Weese JS. 2009. Isolation of bovine intestinal Lactobacillus plantarum and Pediococcus acidilactici with inhibitory activity against Escherichia coli O157 and F5. J Appl Microbiol 106:393–401. doi: 10.1111/j.1365-2672.2008.03959.x. [DOI] [PubMed] [Google Scholar]
- 24.Pahlow G, Muck RE, Driehuis F, Oude Elferink SJWH, Spoelstra SF. 2003. Microbiology of ensiling, p 31–93. In Buxton DR, Muck RE, Harrison JH (ed), Silage science and technology, vol 42. American Society of Agronomy, Madison, WI. [Google Scholar]
- 25.Borreani G, Tabacco E, Schmidt RJ, Holmes BJ, Muck RE. 2018. Silage review: factors affecting dry matter and quality losses in silages. J Dairy Sci 101:3952–3979. doi: 10.3168/jds.2017-13837. [DOI] [PubMed] [Google Scholar]
- 26.Cavallarin L, Tabacco E, Borreani G. 2007. Forage and grain legume silages as a valuable source of proteins for dairy cows. Ital J Anim Sci 6:282–284. doi: 10.4081/ijas.2007.1s.282. [DOI] [Google Scholar]
- 27.Santos MC, Golt C, Joerger RD, Mechor GD, Mourão GB, Kung L. 2017. Identification of the major yeasts isolated from high moisture corn and corn silages in the United States using genetic and biochemical methods. J Dairy Sci 100:1151–1160. doi: 10.3168/jds.2016-11450. [DOI] [PubMed] [Google Scholar]
- 28.Muck RE, Nadeau EMG, McAllister TA, Contreras-Govea FE, Santos MC, Kung L. 2018. Silage review: recent advances and future uses of silage additives. J Dairy Sci 101:3980–4000. doi: 10.3168/jds.2017-13839. [DOI] [PubMed] [Google Scholar]
- 29.Patterson DC, Yan T, Gordon FJ. 1996. The effects of wilting of grass prior to ensiling on the response to bacterial inoculation. 2. Intake and performance by dairy cattle over three harvests. Anim Sci 62:419–429. doi: 10.1017/S135772980001496X. [DOI] [Google Scholar]
- 30.Mihiretu E, Wale M. 2013. Effect of harvesting and threshing time and grain fumigation of field peas (Pisum sativum L.) on pea weevil (Bruchus pisorum L.) (Coleoptera: Bruchidae) development and damage. Eth J Sci Technol 6:13–24. [Google Scholar]
- 31.Corbett RR, Okine EK, Goonewardene LA. 1995. Effects of feeding peas to high-producing dairy cows. Can J Anim Sci 75:625–629. doi: 10.4141/cjas95-092. [DOI] [Google Scholar]
- 32.Soto-Navarro SA, Encinias AM, Bauer ML, Lardy GP, Caton JS. 2012. Feeding value of field pea as a protein source in forage-based diets fed to beef cattle. J Anim Sci 90:585–591. doi: 10.2527/jas.2011-4098. [DOI] [PubMed] [Google Scholar]
- 33.Stein HH, Benzoni G, Bohlke RA, Peters DN. 2004. Assessment of the feeding value of South Dakota-grown field peas (Pisum sativum L.) for growing pigs. J Anim Sci 82:2568–2578. doi: 10.2527/2004.8292568x. [DOI] [PubMed] [Google Scholar]
- 34.Holl FB, Vose JR. 1980. Carbohydrate and protein accumulation in the developing field pea seed. Can J Plant Sci 60:1109–1114. doi: 10.4141/cjps80-161. [DOI] [Google Scholar]
- 35.Fraser MD, Fychan R, Jones R. 2001. The effect of harvest date and inoculation on the yield, fermentation characteristics and feeding value of forage pea and field bean silages. Grass Forage Sci 56:218–230. doi: 10.1046/j.1365-2494.2001.00268.x. [DOI] [Google Scholar]
- 36.Åman P, Graham H. 1987. Whole-crop peas. I. Changes in botanical and chemical composition and rumen in vitro degradability during maturation. Anim Feed Sci Technol 17:15–31. doi: 10.1016/0377-8401(87)90049-6. [DOI] [Google Scholar]
- 37.Cavallarin L, Antoniazzi S, Tabacco E, Borreani G. 2006. Effect of the stage of growth, wilting and inoculation in field pea (Pisum sativum L.) silages. II. Nitrogen fractions and amino acid compositions of herbage and silage. J Sci Food Agric 86:1383–1390. doi: 10.1002/jsfa.2526. [DOI] [Google Scholar]
- 38.Cushnahan A, Gordon FJ. 1995. The effects of grass preservation on intake, apparent digestibility and rumen degradation characteristics. Anim Sci 60:429–438. doi: 10.1017/S1357729800013308. [DOI] [Google Scholar]
- 39.Leibensperger RY, Pitt RE. 1987. A model of clostridial dominance in ensilage. Grass and Forage Sci 42:297–317. doi: 10.1111/j.1365-2494.1987.tb02118.x. [DOI] [Google Scholar]
- 40.Muck RE. 1988. Factors influencing silage quality and their implications for management. J Dairy Sci 71:2992–3002. doi: 10.3168/jds.S0022-0302(88)79897-5. [DOI] [Google Scholar]
- 41.DLG. 2006. Praxishandbuch Futterkonservierung, 7th ed. DLG-Verlag, Frankfurt, Germany. [Google Scholar]
- 42.DLG. 1997. Grobfutterbewertung. Teil B – DLG-Schlüssel zur Beurteilung der Gärqualtität von Grünfuttersilagen auf der Basis der chemischen Untersuchung nach Weißbach und Honig. DLG-Verlag, Frankfurt, Germany. [Google Scholar]
- 43.Kung L, Jr, Shaver RD, Grant RJ, Schmidt RJ. 2018. Silage review: interpretation of chemical, microbial, and organoleptic components of silages. J Dairy Sci 101:4020–4033. doi: 10.3168/jds.2017-13909. [DOI] [PubMed] [Google Scholar]
- 44.Federal Plant Variety Office. Descriptive variety lists.https://www.bundessortenamt.de/bsa/en/variety-testing/descriptive-variety-lists.
- 45.Meier U. 2018. Growth stages of mono- and dicotyledonous plants: BBCH Monograph. Open Agrar Repositorium, Quedlinburg, Germany. doi: 10.5073/20180906-074619. [DOI] [Google Scholar]
- 46.Hoedtke S, Zeyner A. 2011. Comparative evaluation of laboratory-scale silages using standard glass jar silages or vacuum-packed model silages. J Sci Food Agric 91:841–849. doi: 10.1002/jsfa.4255. [DOI] [PubMed] [Google Scholar]
- 47.VDLUFA. 2012. Methodenbuch: Die chemische Untersuchung von Futtermitteln, 3rd ed. VDLUFA-Verlag, Speyer, Germany. [Google Scholar]
- 48.Illumina. 2021. 16S metagenomic sequencing library preparation guide. https://support.illumina.com/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf.
- 49.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bradley IM, Pinto AJ, Guest JS. 2016. Design and evaluation of Illumina MiSeq-compatible, 18S rRNA gene-specific primers for improved characterization of mixed phototrophic communities. Appl Environ Microbiol 82:5878–5891. doi: 10.1128/AEM.01630-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fontaine J, Bech-Andersen S, Butikofer U, de Froidmont-Görtz I. 1998. Determination of tryptophan in feed by HPLC—development of an optimal hydrolysis and extraction procedure by the EU Commission DG XII in three international collaborative studies. Rev Etud Slaves 51:97–108. [Google Scholar]
- 52.Honig H. 1990. Evaluation of aerobic stability, p 72–78. In Lindgren S, Pettersson K (ed), Proceedings of the EUROBAC Conference. Swedish University of Agricultural Sciences, Uppsala, Sweden. [Google Scholar]
- 53.Conway EJ, Byrne A. 1933. An absorption apparatus for the micro-determination of certain volatile substances: the micro-determination of ammonia. Biochem J 27:419–429. [PMC free article] [PubMed] [Google Scholar]
- 54.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glöckner FO. 2014. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res 42:D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hammer Ø, Harper DAT, Ryan PD. 2001. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electronica 4:1–9. http://palaeo-electronica.org/2001_1/past/issue1_01.htm. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and Tables S1 to S4, S6, and S8 to S10. Download spectrum.00953-22-s0001.pdf, PDF file, 0.9 MB (893.3KB, pdf)
Table S5. Download spectrum.00953-22-s0002.xlsx, XLSX file, 0.1 MB (117.6KB, xlsx)
Table S7. Download spectrum.00953-22-s0003.xlsx, XLSX file, 0.05 MB (51.1KB, xlsx)
Data Availability Statement
Raw sequence data were deposited at the European Nucleotide Archive (ENA) under the study accession number PRJEB45910.