ABSTRACT
Gene duplications significantly impact the gene repertoires of both eukaryotic and prokaryotic microorganisms. The genomes of pathogenic Escherichia coli strains share a group of duplicated genes whose function is mostly unknown. The irmA gene is one of the duplicates encoded in several pathogenic E. coli strains. The function of its gene product was investigated in the uropathogenic E. coli strain CFT073, which contains a single functional copy. The IrmA protein structure mimics that of human interleukin receptors and likely plays a role during infection. The enteroaggregative E. coli strain 042 contains two functional copies of the irmA gene. In the present work, we investigated their biological roles. The irmA_4509 allele is expressed under several growth conditions. Its expression is modulated by the global regulators OxyR and Hha, with optimal expression at 37°C and under nutritional stress conditions. Expression of the irmA_2244 allele can only be detected when the irmA_4509 allele is knocked out. Differences in the promoter regions of both alleles account for their differential expression. Our results show that under several environmental conditions, the expression of the IrmA protein in strain 042 is dictated by the irmA_4509 allele. The irmA_2244 allele appears to play a backup role to ensure IrmA expression when the irmA_4509 allele loses its function.
IMPORTANCE Gene duplications occur in prokaryotic genomes at a detectable frequency. In many instances, the biological function of the duplicates is unknown, and hence, the significance of the presence of multiple copies of these genes remains unclear. In pathogenic E. coli isolates, the irmA gene can be present either as a single copy or in two or more copies. We focused our work on studying why a different pathogenic E. coli strain encodes two functional copies of the irmA gene. We show that under several environmental conditions, one of the alleles dictates IrmA expression, and the second remains silent. The latter allele is only expressed when the former is silenced. The presence of more than one functional copy of the irmA gene in some pathogenic E. coli strains can result in sufficient expression of this virulence factor during the infection process.
KEYWORDS: enteroaggregative E. coli, 042, gene duplications, aec69, irmA
INTRODUCTION
Gene duplications occur in both eukaryotes and prokaryotes and play a key role in the generation of the functional diversity of genes (1–6). In bacteria, gene duplications have been associated with the adaptation of cells to a changing environment (7, 8) and have been found to occur more frequently among transferred genes than among indigenous genes (9). The incidence of gene duplications can be different across different bacterial genera and species. These duplications can also be strain-specific, species-specific, and genus-specific (10, 11). In E. coli, a group of duplicated ORFs is common to almost all the currently sequenced pathogenic strains (10). Several of these ORFs code for proteins of unknown function. One of the duplicated genes in several E. coli strains is the aec69 gene. In a recent work, the aec69 gene and its gene product were studied in the uropathogenic E. coli strain CFT073 (12). The aec69 gene was renamed irmA in the latter work. The IrmA protein forms a dimer whose structure is similar to those of human cytokine receptors such as the interleukin-2 or the interleukin-4 receptors. In addition, purified IrmA can bind to their cognate cytokines. IrmA likely plays a role during the infection process of the E. coli strains that express it. Most of these strains belong to the uropathogenic (UPEC), enterohemorrhagic (EHEC) and enteroaggregative (EAEC) pathotypes (12). The irmA gene maps close to the flu gene, which codes for the Ag43 outer membrane protein (13). Previous reports have shown that flu can also be present in multiple copies (14, 15). In some E. coli strains, such as EC958 (which belongs to the ST131 clonal type), the flu yeeR irmA gene cluster is present only in a single copy. In other strains, such as the UPEC strain CFT073, it is present in two copies.
Based on the amplification of the flu yeeR irmA transcripts of strains EC958 and CFT073 by RT–PCR, as well as the determination of the 5′ transcriptional start points by RACE, it was concluded that irmA forms part of a single flu yeeR irmA transcriptional unit. In the CFT073 strain, transcription of one of the flu yeeR irmA operons is disrupted because of the presence of insertion sequences between flu and yeeR genes (12). Previous studies have shown that the expression of Ag43 varies across phases and is controlled by an epigenetic mechanism involving the global regulatory protein OxyR and the Dam methyltransferase (16); additionally, it was shown that irmA expression is subjected to OxyR repression (12).
The E. coli strain 042 is the EAEC prototype. It caused diarrhea in a volunteer trial (17). The genome sequence of this strain is available (18), and its virulence factors have been characterized. The 042 strain harbors the IncFIIA virulence plasmid pAA2 (18, 19), which encodes the virulence master regulator AggR and other virulence determinants, such as the aggregative adherence fimbriae AAF/II (20, 21). When analyzing the 042 genomic sequence, we noticed that it contains several genes that are present in two or more copies, including irmA (10). Unlike strain CFT073, no insertion sequences disrupt the flu yeeR irmA region in strain 042, and only minor nucleotide changes can be detected. In this work, we aimed to gain insight into the biological role of both irmA alleles. We also show that these alleles are differentially expressed. One allele remains silent when the other is expressed. We show that, unlike strain CFT073, transcription of the irmA alleles in the 042 strain is dependent upon a promoter that maps to the intergenic yeeR irmA region. The biological reason for the existence of duplicates of the irmA genes in some pathogenic E. coli strains is discussed.
RESULTS
Analysis of IrmA protein expression dictated by the irmA_2244 and irmA_4509 alleles in the E. coli strain 042.
To detect IrmA expression, we first purified the IrmA protein (Fig. S1) and obtained polyclonal antibodies. Expression of the IrmA protein was tested in the culture supernatants of the 042 wt strain and its single oxyR and double irmA_2244 irmA_4509 mutant derivatives (Fig. 1). In accordance with the data previously obtained for E. coli strains CFT703 and EC958 (12), IrmA expression was barely detectable in the 042 wt strain, but its expression was significantly increased in the oxyR mutant derivative (Fig. 1A).
FIG 1.
Immunodetection of the IrmA protein and expression analysis of the irmA_2244 and irmA_4509 alleles. (A) Western blot analysis of protein extracts from the 042 wt strain, its double irmA derivative and its oxyR derivative. Purified IrmA protein was used as a control. (B) Immunodetection of the IrmA protein by using anti-Flag antibodies in samples from cultures grown to an OD of 0.4 and 2.0. Strains 042 wt (used as a negative control), 042 irmA_2244xFLAG and 042 irmA_4509xFLAG were used for the assay. (C) Transcription of the irmA_2244 and irmA_4509 alleles, measured as the β-galactosidase activity of the irmA_2244::lacZ and irmA_4509::lacZ transcriptional fusions in samples collected at the exponential and early stationary phases of growth (OD600 0.4 and 2.0, respectively). The values obtained are based on triplicate experiments. ns, not significant; ***, P < 0.001.
To improve the detection of the IrmA protein and to assess the differential expression of the gene product controlled by the irmA_2244 or irmA_4509 allele, we inserted a Flag-tag downstream of the irmA_2244 and irmA_4509 genes. We also constructed irmA::lacZ transcriptional fusions with both irmA alleles. IrmA expression was then assessed in the 042 ΔLC (042 derivative lacking the lacZ and cat genes) strain grown in LB medium at 37°C, both by immunodetecting the IrmA protein with anti-Flag monoclonal antibodies and by analyzing the transcription of both irmA alleles of strain 042 ΔLC (Fig. 1B and C). The results showed that although one of the irmA alleles (irmA_4509) is expressed in the exponential growth phase and at the onset of the stationary phase in cultures grown in LB medium at 37°C, the other (irmA_2244) is not expressed.
Expression of the irmA_2244 allele is silent under several in vitro growth conditions.
We next investigated whether expression of the irmA_2244 allele could be detected when strain 042 was grown outside normal conditions (LB medium at 37°C). First, we analyzed the effect of the growth temperature and the culture medium (rich and minimal). With respect to the irmA_4509 allele, its expression is increased at high temperature (37°C) and when cells were grown in minimal medium (Fig. 2A to D). Again, no expression was detected for the irmA_2244 allele.
FIG 2.
Effect of different environmental conditions on the expression of the irmA_2244 and irmA_4509 alleles. Expression was measured either by immunodetecting the IrmA protein with anti-FLAG monoclonal antibodies (A, C, E) or by measuring the β-galactosidase activity of the corresponding irmA::lacZ transcriptional fusion (B, D, F). The values obtained are based on triplicate experiments. The effect of the growth temperature (A, B), culture medium (C, D) and presence of oxygen (E, F) on the expression of both irmA alleles is shown. Aer., aerobiosis; Anaer., anaerobiosis. **, P < 0.01; ***, P < 0.001.
The next environmental factor studied was the presence of oxygen in the medium. Samples from both aerated cultures and anaerobic cultures were used for IrmA immunodetection and for the analysis of the transcription of the corresponding irmA::lacZ fusion. For the irmA_4509 allele, anaerobic conditions resulted in reduced expression of the gene. Again, expression of the irmA_2244 allele could not be detected (Fig. 2E and F).
We also analyzed whether nutritional stress could influence the expression of the irmA alleles in strain 042. First, we obtained the growth curves of strain 042 ΔLC in minimal M9 medium with different glucose concentrations (Fig. 3). With a glucose concentration of 0.5 g/l, growth ceased at the mid-exponential growth phase. We used cultures grown with either 4 or 0.5 g/l glucose to compare the expression of both irmA alleles in strain 042 ΔLC grown with excess glucose or under glucose-limited conditions. With respect to the irmA_4509 allele, exhaustion of the carbon source resulted in a significant increase in IrmA expression. Again, no expression of the irmA_2244 allele could be detected (Fig. 3B and C).
FIG 3.
Effect of nutritional stress on the expression of the irmA_2244 and irmA_4509 alleles. (A) Growth curves of strain 042 ΔLC in M9 minimal medium with different glucose concentrations. (B) Immunodetection of the IrmA_2244xFLAG and IrmA_4509xFLAG proteins with anti-FLAG monoclonal antibodies. (C) β-galactosidase activity of the strains harboring the irmA_2244::lacZ and irmA_4509::lacZ transcriptional fusions. The values obtained are based on triplicate experiments. ns, not significant; *, P < 0.05; **, P < 0.01.
Regulation of the expression of the irmA alleles by the H-NS/Hha and OxyR systems.
One of the genes that is also duplicated in strain 042 is the hha gene. Its gene product (the Hha protein) interacts with the well-characterized nucleoid-associated protein H-NS to modulate gene expression (22). We found a strong correlation between the presence of additional copies of the hha gene (i.e., the hha2 gene, locus tag ec042_4516, located upstream of the irmA_4509 gene) and the yeeR irmA cluster in several pathogenic E. coli strains, including strain 042 (10). This result suggests that the irmA genes of strain 042 can be regulated by Hha (and/or its HhaII allele) and by H-NS. To confirm this result, we constructed the corresponding hns, hha and hha hha2 mutant derivatives of strains 042 irmA_2244xFLAG and 042 irmA_4509xFLAG and analyzed IrmA expression in the different strains. With respect to the expression of the irmA_4509 allele, its expression was increased in the double hha hha2 mutant but not in the hns mutant (Fig. 4A and B). For the irmA_2244 allele, no effect was found (Fig. 4A).
FIG 4.
Effect of the Hha and H-NS proteins and of the oxidative stress on the expression of the irmA_2244 and irmA_4509 alleles. Immunodetection of the IrmA_2244xFLAG and IrmA_4509xFLAG proteins with anti-FLAG monoclonal antibodies in cell extracts from the 042 strain harboring either the irmA_2244xFLAG or irmA_4509xFLAG derivatives, alone (wt) or combined with hha, hhahha2 (Hha null mutant) and hns derivatives (A and B). (C) Immunodetection of the IrmA_2244xFLAG and IrmA_4509xFLAG proteins with anti-FLAG monoclonal antibodies in cell extracts from the 042 wt strain and its irmA_2244xFLAG, irmA_2244xFLAG oxyR, irmA_4509xFLAG and irmA_4509xFLAG oxyR derivatives.
We also decided to analyze whether the previously reported repression of the irmA genes by OxyR (12) responds to oxidative stress. We established optimal conditions to generate oxidative stress in the 042 strain (see Materials and Methods section for details) and analyzed the effect of oxidative stress on the expression of both irmA alleles (Fig. 4C). No expression of the irmA_2244 allele could be detected under oxidative stress conditions or in the oxyR mutant derivative of strain 042. In contrast, the expression of the irmA_4509 allele was significantly increased in the oxyR mutant derivative, but the effect was independent of oxidative stress.
Expression of the irmA_2244 allele can be detected in an irmA_4509 mutant derivative of strain 042.
Under all of the above reported conditions, we were unable to detect the expression of the irmA_2244 allele. We then decided to test the individual expression of each of the alleles in the absence of the other. Expression was monitored by immunodetecting the IrmA protein produced by the corresponding irmA allele with polyclonal anti-IrmA antibodies and by measuring the transcriptional activity of the already obtained irmA::lacZ fusions.
The expression of the irmA_4509 allele in the 042 ΔirmA_2244 strain was similar to that measured in the 042 wt strain (Fig. 5A). Expression of the irmA_2244 allele in the 042 ΔirmA_4509 strain could barely be detected by Western blotting but not at the transcriptional level. When the oxyR allele was introduced into the 042 ΔirmA_2244 and 042 ΔirmA_4509 strains, this resulted, as expected, in increased expression of IrmA. In this genetic background, the expression of the 042 irmA_2244 allele could be detected both by Western blotting and by measuring the transcription of the irmA_2244::lacZ fusion (Fig. 5A to D). These results were corroborated by detecting the expression of the Flag-tagged alleles in the corresponding mutants by using anti-Flag monoclonal antibodies (Fig. 5B–C).
FIG 5.
Expression of each irmA allele alone in the 042 strain. (A) Immunodetection of the IrmA_2244 and IrmA_4509 proteins in cell extracts from the wt, irmA_2244, and irmA_4509 strains, as well as their oxyR derivatives, using polyclonal anti-IrmA antibodies. (B) Immunodetection of the IrmA_2244xFLAG protein in the wt strain and its irmA_2244xFLAG and irmA_2244xFLAG irmA_4509 derivatives by using anti-Flag monoclonal antibodies. (C) Immunodetection of the IrmA_4509xFLAG protein in the wt strain and its irmA_4509xFLAG and irmA_4509xFLAG irmA_2244 derivatives by using anti-Flag monoclonal antibodies. (D, E) β-galactosidase activity of the strains harboring the irmA_2244::lacZ and irmA_4509::lacZ transcriptional fusions in the 042 ΔLC (indicated as wt), irmA_2244, and irmA_4509 strains, as well as their oxyR derivatives. The values obtained are based on triplicate experiments. ns, not significant; ***, P < 0.001; ****, P < 0.0001.
irmA transcripts are initiated downstream of the yeeR gene in strain 042.
The results reported above show that under a wide range of conditions, the expression of the irmA_2244::lacZ allele is significantly lower than that of the irmA_4509::lacZ allele. We then compared the corresponding regulatory regions of both genes to identify modifications to the nucleotide sequences that might account for the observed differences in gene expression. Nucleotide differences can be found either in the flu regulatory region or in the intergenic yeeR irmA region (Fig. S2).
As the irmA gene has been reported in strain CFT073 to be cotranscribed with flu, we obtained flu::lacZ gene fusions by cloning the flu promoter corresponding to nucleotides −105 to +22 into the promoterless plasmid pUJ8 and analyzed β-galactosidase expression in four clones harboring each of the constructs. The same expression pattern was obtained for all clones of both constructs (Fig. 6). For each gene fusion construct, two of four clones showed very low β-galactosidase activity, and the other two clones showed high β-galactosidase activity. Considering that flu is subjected to phase variation, these results can be interpreted as the different clones expressing the flu operon in the on/off phases. As these results did not support the observed differences in the expression of the irmA_2244 and irmA_4509 alleles, we searched for other differences in the DNA sequence downstream of the flu gene. Some alterations in the nucleotide sequence appeared in the intergenic region between yeeR and irmA, where a putative promoter sequence was detected bioinformatically (Fig. S3). Hence, we decided to determine the transcriptional start point for both irmA alleles in strain 042.
FIG 6.
β-galactosidase activity of plasmidic transcriptional fusions with the flu promoter. (A) flup2242 fusion. (B) flup4511 fusion. The values obtained are based on triplicate experiments with four independent transformants for each of the plasmids.
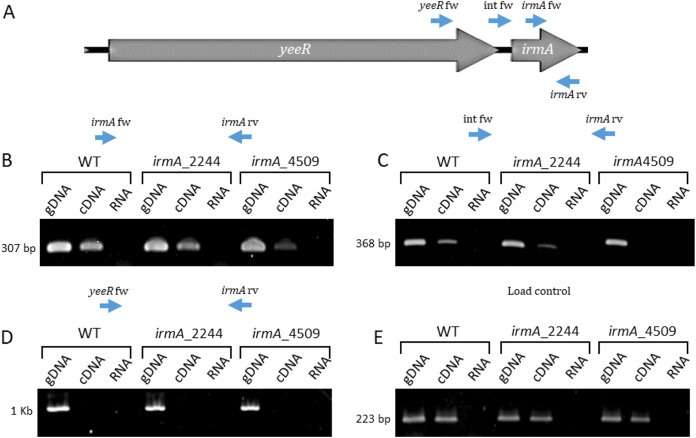
We first analyzed the transcription of the yeeR irmA region of strains 042 wt, 042 irmA_2244 and 042 irmA_4509 by walking RT–PCR. We used a fixed reverse primer within irmA and three forward primers located within irmA, in the yeeR irmA intergenic region or in the yeeR coding sequence. Although amplification products could be obtained for primer pairs located in the irmA-irmA and irmA-intergenic regions, no amplification product could be obtained when the primer pairs mapping in the irmA and yeeR regions were used (Fig. 7). These results strongly suggest that both irmA alleles are transcribed from their own promoters located downstream of the yeeR gene. To confirm this result, we determined the transcriptional start point for both the irmA_2244 and irmA_4509 alleles by 5′ RACE (Fig. 8). The transcriptional start point was found to occur downstream of the −10 sequence of the bioinformatically identified promoter (Fig. 11). The distance that exists between the bioinformatically identified −10 sequence and the identified transcriptional start point is too long. Hence, it cannot be ruled out that the true −10 sequence of the irmA promoter corresponds to the ACCTTTTA sequence of the irmA_4509 promoter region.
FIG 7.
Analysis of the transcription of the irmA operon by walking RT–PCR. (A) Diagram of the oligonucleotides used in the experiment. (B) Amplification products obtained with the oligonucleotide pair irmA fw-irmA rv. (C) Amplification products obtained with the oligonucleotide pair Int fw-irmA rv. (D) Amplification products obtained with the oligonucleotide pair yeeR fw-irmA rv. (E) Load control amplifying the gapA gene (gapA fw-gapA rv). The expected size of the amplicons is indicated. gDNA: genomic DNA, cDNA: complementary DNA. The 042 wt, irmA_2244 and irmA_4509 strains were used for the analysis.
FIG 8.
Identification of the irmA promoter by BPROM software and the transcriptional start point of the irmA alleles by 5′ RACE. The transcriptional start point identified by 5′ RACE is marked with +1, and the putative -10 and −35 boxes found by using BPROM software are highlighted in yellow. The stop codon of the yeeR gene is colored purple, and the start codon of the irmA gene is labeled green.
FIG 11.
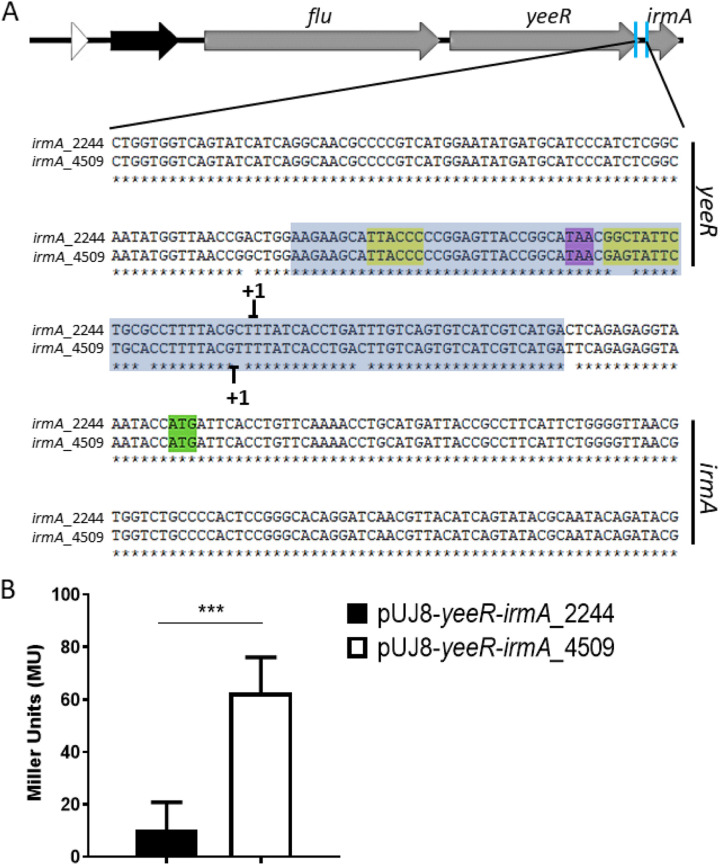
Differences in the intergenic region between the yeeR and irmA genes account for the differential expression of both irmA alleles. (A) Intergenic yeeR/irmA region. Selected point mutations that have been introduced into the sequence are colored blue and orange. The transcriptional start points of the irmA alleles identified by 5′ RACE are marked with +1, and the putative −10 and −35 boxes found by using BPROM software and are highlighted in yellow. The stop codon of yeeR is colored purple, and the start codon of irmA is colored green. (B) β-galactosidase activity of plasmid transcriptional fusions performed with the wt irmA_2244, wt irmA_4509 and mutant irmA_4509 promoters. (C) Immunodetection of the IrmA_4509xFLAG protein in the wt strain and its irmA_4509xFLAG and irmA_4509xFLAG p4509 derivatives. A protein extract from strain 042 wt was used as a negative control. The values obtained are based on triplicate experiments. ***, P < 0.001.
To further support that irmA transcription is initiated at the intergenic yeeR irmA region, we mutated the flu promoter sequence corresponding to the flu4511 allele (p4511) in strain 042 irmA_4509xFLAG and compared the IrmA expression in strains 042 wt, 042 irmA_4509xFLAG and 042 irmA_4509xFLAG p4511. Deletion of the flu p4511 promoter did not influence IrmA expression (Fig. 9), but significantly reduced flu transcription levels (Fig. S4).
FIG 9.

Influence of deletion of the flu_4511 promoter (p4511) on the expression of the irmA_4509 gene. Immunodetection of IrmA_4509xFLAG in the 042 wt strain and its irmA_4509xFLAG Δp4511 derivatives. Protein extract from the 042 wt strain was used as a negative control.
Alterations in the irmA promoter regions account for the differential expression of the irmA_2244::lacZ and irmA_4509::lacZ alleles.
As detailed above, there are different nucleotide changes in the intergenic region between yeeR and irmA (Fig. 8), with some mapping in the bioinformatically identified irmA promoter. We hypothesized that these changes are responsible for the reduced expression of the irmA_2244 allele. To support this assumption, we cloned the intergenic region of both the irmA_2244 and irmA_4509 alleles (Fig. 10A) in the promoterless vector pUJ8. The constructs were transformed into strain 042 ΔLC, and β-galactosidase activity was determined for both constructs (Fig. 10B). In accordance with the previous data obtained, the construct harboring the intergenic region of the yeeR-irmA_4509 allele showed much higher β-galactosidase activity than the construct that incorporated the intergenic region corresponding to the irmA_2244 allele (Fig. 10B). To demonstrate that nucleotide changes in the promoter region account for the observed reduced expression of the irmA_2244 allele, we used a site-directed mutagenesis approach to modify the yeeR-irmA_4509 intergenic region, specifically nucleotides +1, −10, −19 and −20 (Fig. 11A), thus partially transforming the yeeR-irmA_4509 intergenic region into the yeeR-irmA_2244 intergenic region. Strain 042 ΔLC harboring plasmid pUJ8-mut*prom4509 was used to detect irmA transcription, and strain 042 irmA_4509xFLAG p4509 (lacking the intergenic region between yeeR and irmA) was used to detect IrmA expression by Western blotting (Fig. 11B and C). Individual point mutations did not have a significant effect on irmA expression (data not shown), but the combined alterations in the 042 irmA_4509 promoter region resulted in reduced expression of IrmA.
FIG 10.
Transcriptional activity of the intergenic region between the yeeR and irmA genes. (A) Intergenic yeeR/irmA region. The cloned sequence is marked with a blue box. The transcriptional start points of the irmA genes identified by 5′ RACE are marked with +1. The putative -10 and −35 boxes found by using BPROM software are highlighted in yellow. The stop codon of the yeeR gene is colored purple, and the start codon of the irmA gene is colored green. (B) β−galactosidase activity of plasmid transcriptional fusions with the intergenic yeeR-irmA region of both irmA alleles. The values obtained are based on triplicate experiments. ***, P < 0.001.
The flu yeeR irmA cluster in pathogenic E. coli strains.
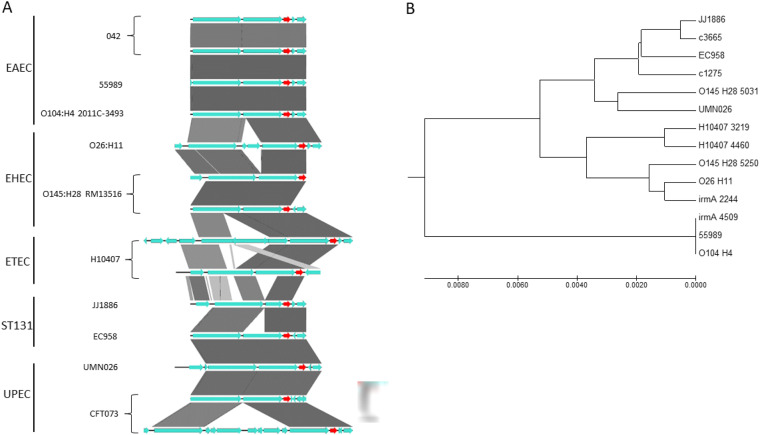
We also bioinformatically analyzed the flu yeeR irmA gene cluster in E. coli strains belonging to different pathotypes. Duplications, deletions, and insertions between flu and yeeR occur in several strains (Fig. 12A). Of the 10 strains analyzed, 4 harbored duplications of the flu yeeR irmA gene cluster. We also performed a phylogenetic analysis of the intergenic yeeR-irmA and the nucleotide sequence of the irmA gene (Fig. 12B). The irmA_4509 allele shows close proximity to the corresponding irmA alleles of the EAEC strains 55989 and 0104:H4 and shows a large distance from the irmA_2244 allele.
FIG 12.
Bioinformatic analysis of the flu-yeeR-irmA region. (A) Alignment of the flu-yeeR-irmA region of 10 pathogenic strains of E. coli with different pathotypes. The pathotype to which they belong is shown on the left. The irmA gene is shown in red. The figure was made with Easyfig. (B) Phylogenetic analysis of the intergenic yeer-irmA region and irmA gene nucleotide sequence. The evolutionary history was inferred using the UPGMA method. The optimal tree with a total of branch length equal to 0.03874617 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed by using the maximum composite likelihood method in units of the number of base substitutions per site.
DISCUSSION
When gene duplications occur, their fate (fixation or elimination) may depend on the occurrence of positive selection (23). Different hypotheses have been proposed to justify the selective advantage of duplicates. First, duplication results in a higher expression level, which can be favorable (24). In this case, expression of the ancestral system should be far from optimal, and increasing the expression level should be beneficial. This can occur in extreme environmental circumstances but not in stable environments to which the organism should be adapted (25). Therefore, it is not surprising that several of the reported examples of gene duplications in bacteria correspond to those living in stressful environments (24, 26–28).
Second, the presence of the second copy of a gene may allow the accumulation of beneficial mutations (29, 30). In addition to these two situations, duplications can also be positively selected when errors in gene expression occur (31). Errors can be either of genotypic or phenotypic origin. Errors in phenotype may be due to the existence of stochastic fluctuations in gene expression (32). The accuracy in the response of noisy genes can be improved by their corresponding duplicates (33–35). Duplicates can also behave as molecular backups for their paralogs (36).
The flu yeeR irmA cluster shows high genetic variability in Escherichia coli strains. It is widely distributed among the different pathotypes. Even for a single pathotype such as UPEC, the cluster can be present either as a single copy or as a duplicate (12), and insertion elements may disrupt the structure of one of the copies, as is the case for strain CFT073. We show in this work that IS-mediated disruptions of flu yeeR irmA are frequent among strains belonging to different pathotypes. Insertions and deletions occur mainly within the flu yeeR region. Nevertheless, other strains, such as EAEC strain 042, contain two copies of the cluster. These copies map in a large duplicated region in the EAEC strain 042. In virulent E. coli isolates, the presence or absence of specific genetic determinants, which can also be present in one or more copies, argues for their corresponding gene products being expressed only under very specific infection conditions, which in turn supports the sophisticated evolution of pathogenic E. coli.
In contrast to the results obtained for the flu yeeR irmA determinant of the UPEC strains EC958 and CFT073, transcription of the irmA_4509 allele occurs independently of flu and is tightly regulated by several factors. As described for the irmA gene of strains EC958 and CFT073 (12), transcription of the irmA_4509 allele of the 042 strain is repressed by OxyR, but OxyR-mediated repression does not appear to respond to the occurrence of oxidative stress conditions. Environmental factors reported to influence the expression of virulence factors also influence the expression of the irmA_4509 gene (i.e., growth temperature, culture medium). The stress generated by the exhaustion of the carbon source also induces irmA_4509 expression. High temperature, reduced nutrient supply, and nutritional stress are conditions that strain 042 must cope with in the intestine.
With respect to the regulation of the expression of the irmA_4509 gene by the H-NS/Hha system, we show here that this gene is subjected to Hha-mediated regulation but not to H-NS-mediated regulation. In several examples of different virulence factors, the Hha protein was found to fine tune the H-NS regulatory activity (22, 37–39). Nevertheless, Hha-mediated regulation of gene expression independent of H-NS has been reported previously, but the mechanism by which this happens remains to be elucidated (40).
The irmA_2244 allele appears to be silent under all the environmental conditions tested that lead to irmA_4509 expression in strain 042. This phenomenon may be due to the existence of nucleotide substitutions in the irmA promoter region. We support this assumption by demonstrating that alterations in the sequence of the irmA_4509 promoter region that transform it into the irmA_2244 promoter region abolish irmA_4509 expression.
In the 042 strain, expression of the irmA_2244 allele was detected in irmA_4509 mutants. The mechanism underlying irmA_2244 induction when irmA_4509 expression ceases remains to be elucidated. The observed dependence of the expression of one of the duplicated alleles on the expression of the other suggests that the irmA_2244 allele is a backup for the irmA_4509 allele in the 042 strain. Several virulent E. coli strains encode two copies of the irmA gene. In some of these strains, one of the copies has lost its function because of the insertion of foreign DNA sequences. Whether both copies of the irmA gene remain functional may rely on the relevance of the IrmA protein itself in the infective process of the corresponding strain. In the EAEC 042 strain, the existence of the irmA_2244 allele ensures IrmA expression even when the function of the irmA_4509 allele is lost.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this work are listed in Tables S1 and S2. The bacterial strains were routinely grown in Luria-Bertani (LB) medium (10 g l−1 NaCl, 10 g l−1 tryptone and 5 g l−1 yeast extract) or in M9 minimal medium supplemented with glucose at a final concentration of 0.4% with vigorous shaking at 200 rpm (Innova 3100, New Brunswick Scientific). The antibiotics used were chloramphenicol (Cm) (25 μg mL−1), tetracycline (Tc) (15 μg mL−1), carbenicillin (Cb) (100 μg mL−1) and kanamycin (Km) (50 μg mL−1) (Sigma–Aldrich).
Genetic manipulations.
All enzymes used to perform standard molecular and genetic procedures were used according to the manufacturer’s recommendations.
Deletions of the irmA, hns, hha, oxyR, and flu genes were performed in strain 042 by using the λ Red recombination method, as previously described (41). The antibiotic resistance determinants of the plasmids pKD3/pKD4 was amplified using the corresponding oligonucleotides (P1/P2 series, see Table S3). The mutants were confirmed by PCR using the corresponding oligonucleotides (P1up/P2down series, see Table S3). When necessary, the antibiotic resistance cassette was eliminated using the FLP/FRT-mediated site-specific recombination method, as previously described (42).
A transcriptional lacZ fusion was made with the irmA_2244 and irmA_4509 genes of strain 042 ΔLC. The antibiotic resistance determinant from kanamycin was eliminated using an FLP/FRT-mediated site-specific recombination method, as previously described, thus generating strains irmA_2244 and irmA_4509. An FRT-generated site was used to integrate the lacZ reporter gene encoded in plasmid pKG136 (43), generating transcriptional irmA_2244::lacZ and irmA_4509::lacZ fusions.
Recombinational transfer of the Flag sequence into the irmA_2244 and irmA_4509 genes was achieved by following a previously described methodology (44). The template vector encoding the Flag sequence and Kmr cassette used was the pSUB11 plasmid. The primers used for construction of the Flag-tagged derivative were the 3× P1/P2 series (Table S3). The correct insertion of the Flag-tag was confirmed by PCR using oligonucleotides from the P1up/P2down series (see Table S3).
Transcriptional fusions stored in plasmidic vectors were generated to study the regulatory regions of genes of interest using the promoterless plasmid pUJ8. The promoter regions to be studied were amplified by PCR using Phusion Hot Start II DNA polymerase (Thermo Scientific) and the corresponding oligonucleotides (Fw/Rv series, see Table S3). The EcoRI and BamHI restriction sites were added at the 5′ and 3′ ends, respectively. Upon amplification, both the insert and the vector were digested and ligated into the EcoRI/BamHI site. The plasmids generated were Sanger sequenced and termed pUJ8-yeeR-irmA_2244, pUJ8-yeeR-irmA_4509 and pUJ8-mut*prom4509.
Site-directed mutagenesis.
Point mutations were generated in the transcriptional fusions obtained in the plasmid pUJ8, and oligonucleotides with sizes between 25 and 45 nucleotides were designed. These oligonucleotides contained the desired nucleotide change in the center of their sequence (Fw/Rv series, see Table S3). The amplification reaction was performed with Phusion Hot Start II DNA polymerase (Thermo Scientific) to obtain the desired nucleotide mutation with the following program: 3 min at 98°C for initial denaturation, followed by denaturation at 98°C for 30 sec, hybridization at 55°C for 30 sec and extension at 68°C for 30 sec/kb. The PCR cycle was repeated 18 times, and a final extension was carried out for 10 min at 68°C. Once the amplification reaction had finished, the PCR product was digested with DpnI, thus eliminating the methylated template plasmid. Next, strain DH5α was transformed by electroporation and seeded on LB plates supplemented with carbenicillin. Transformants were selected and checked for the predicted point mutations upon plasmid extraction and Sanger sequencing with the pUJ8 P1up and lacZR primers (Table S3).
Oligonucleotides.
The oligonucleotides (from 5′ to 3′) used in this work are listed in Table S3.
β-Galactosidase assay.
β-Galactosidase activity measurements were performed as previously described (45). The values are given as Miller units. Student's t test was used to determine statistical significance, and the values were obtained by using GraphPad Prism 8 software. A P value of less than 0.05 was considered significant.
Oxidative stress assay.
The optimal conditions to induce oxidative stress in strain 042 were established by modifying the protocol described in (46). Briefly, 10 mL of an overnight culture was grown in LB medium under shaking at 37°C. The next day, a 1:100 dilution was made in 50 mL of M9 medium supplemented with 4 g/L glucose, and cultures were grown at 37°C under shaking. Upon reaching an OD600nm of 1, oxidative stress was induced by adding 0.05% H2O2 and incubating for 2 h at 37°C under shaking. Then, 1 mL of the culture was centrifuged, the supernatant was removed, and the pellet was resuspended in Laemmli buffer (glycerol 5% vol/vol, β-mercaptoethanol 2.5%, SDS 1.15% p/v, Tris–HCl 31 mM pH 6.6 and bromophenol blue 0.05%).
Polyclonal antibody production.
For polyclonal antibody production, we purified the His-tagged IrmA protein. Then, the DNA region containing the complete irmA coding sequence was amplified by PCR using the genomic DNA from strain 042 as a template and the primers irmA pLATE31CT fw and irmA pLATE31CT rv together with Thermo Scientific Phusion Hot Start II High-fidelity DNA polymerase following the manufacturer’s recommendations. The DNA was then purified using a Thermo Scientific Gen eJet PCR purification kit and ligated into the pLATE31 vector to add a His-tag to the C-terminal end following the manufacturer´s instructions (Thermo Scientific LICator LIC cloning and expression system). The resulting plasmid, termed pLATE31-irmA, was Sanger sequenced. BL21DE3 cells were used for heterologous recombinant expression of the IrmA protein. Cells transformed with the pLATE31-irmA plasmid were grown in LB medium supplemented with carbenicillin at a final concentration of 100 μg/mL at 37°C until an OD600nm of 0.4 was reached. Then, protein expression was induced by adding 1 mM IPTG to a final concentration for 3 h. Cells were then centrifuged at 7,500 × g for 30 min at 4°C. The pellet was subsequently resuspended in buffer A20 (20 mM HEPES pH 7.9, 100 mM KCl, 5 mM MgCl2, 10% glycerol, and 20 mM imidazole) plus protease inhibitor (Complete Ultra Tablets, Mini, EDTA-free, EASYpack, Roche). The cells were then disrupted by sonication, and the soluble fraction was collected after centrifugation at 12,000 × g for 30 min at 4°C. The supernatant was used for protein purification by immobilized-metal affinity chromatography (IMAC) using HisPur Ni-NTA Superflow Agarose (Thermo Scientific). The recombinant IrmA protein was eluted from Ni-NTA resin by increasing the concentration of imidazole using buffer A200 (20 mM HEPES pH 7.9, 100 mM KCl, 5 mM MgCl2, 10% glycerol, and 200 mM imidazole). The purified His-tagged IrmA protein was adjusted to 1 mg/mL and inoculated into rabbits according to standard protocols (Unitatd’Experimentació Animal de Farmàcia–CCiTUB. Universitat de Barcelona, Barcelona, Spain). After immunization, preimmune serum and serum collected after the immunization period were tested against the IrmA protein by Western blotting.
Electrophoresis and Western blotting of proteins.
Whole-cell protein extracts were prepared in Laemmli buffer (glycerol 5% vol/vol, β-mercaptoethanol 2.5%, SDS 1.15% p/v, Tris–HCl 31 mM pH 6.6 and bromophenol blue 0.05%). Protein samples were analyzed by Tris-Glycine-SDS triphasic gels with 16.5% polyacrylamide. Proteins were transferred from the gels to PVDF membranes by using a semidry electrophoretic transfer cell (Bio–Rad) at 15 V for 40 min. For Western blot analysis, a monoclonal antibody directed against the Flag epitope (Sigma–Aldrich) or a polyclonal antibody directed against the IrmA protein were diluted 1:10.000 or 1:1000 in a solution of PBS, 0.2% Triton and 3% skimmed milk. The membranes containing the proteins were incubated with the diluted antibody for 16 h at 4°C. The membranes were then washed for 10 min with PBS and 0.2% Triton X-100 (Sigma–Aldrich) solution. The washing step was repeated three times. Thereafter, the membranes were incubated with horseradish peroxidase-conjugated goat anti-mouse IgG (Promega) or anti-rabbit IgG (Promega) diluted 1:2.500 in a solution of PBS and 0.2% Triton X-100 for 45 min at room temperature. Again, three washing steps with PBS and 0.2% Triton solution were performed for 30 min each, and immunodetection of the specific protein was performed by enhanced chemiluminescence by using the Molecular Imager ChemiDoc XRS system and Quantity One software (Bio–Rad).
Isolation of RNA.
For RNA isolation, bacterial cells were grown to an OD600nm of 2.0 at 37°C. Then, 5 mL of cells was mixed with a 0.2 × volume of a stop solution (95% ethanol, 5% phenol), shaken and centrifuged for 10 min at 6,000 × g. Bacterial pellets were subsequently frozen at −80°C until use. Total RNA was extracted from the bacterial pellets by using Tripure Isolation Reagent (Roche) according to the manufacturer’s instructions. Potential traces of DNA were removed by digestion with Turbo DNA-free (Thermo Scientific) according to the manufacturer’s instructions. RNA concentration and quality were measured using a NanoDrop 1000 (Thermo Scientific).
Walking RT–PCR.
The walking RT–PCR assay was performed following the methodology previously described in (47).
5′ RACE.
5′ RACE experiments were performed as previously described in (48).
Bioinformatics analysis.
The flu-yeeR-irmA alignment was generated using Easyfig (49). The evolutionary history was inferred using the UPGMA method (50). The optimal tree with a total branch length equal to 0.03874617 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method (51) and are in units of the number of base substitutions per site. The analysis involved 14 nucleotide sequences. The codon positions included were 1st + 2nd + 3rd+Noncoding. All positions containing gaps and missing data were eliminated. There was a total of 956 positions in the final data set. Evolutionary analyses were conducted in MEGA7 (52).
The prediction analysis of the irmA promoter was performed using the BPROM software (https://www.softberry.com/berry.phtml?topic=bprom&group=programs&subgroup=gfindb).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from Fundació “La Marató TV3,” Spain (project 201818 10), and BIO2016-76412-C2-1-R and PID2019-107479RB-I00 (AEI/FEDER, UE) from the Ministerio de Economía, Industria y Competitividad, and CERCA Program/Generalitat de Catalunya to A.J. M.B. was the recipient of an FI fellowship from the Generalitat de Catalunya. The funders had no role in data collection, interpretation, study design, or the decision to publish these data.
Footnotes
Supplemental material is available online only.
Contributor Information
M. Hüttener, Email: mhuttener@me.com.
A. Juárez, Email: ajuarez@ub.edu.
Sandeep Tamber, Health Canada.
REFERENCES
- 1.He X, Zhang J. 2005. Gene complexity and gene duplicability. Curr Biol 15:1016–1021. doi: 10.1016/j.cub.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 2.Conant GC, Wolfe KH. 2008. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- 3.Serres MH, Kerr AR, McCormack TJ, Riley M. 2009. Evolution by leaps: gene duplication in bacteria. Biol Direct 4:46. doi: 10.1186/1745-6150-4-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Innan H, Kondrashov F. 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet 11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- 5.Gao Y, Zhao H, Jin Y, Xu X, Han G-Z. 2017. Extent and evolution of gene duplication in DNA viruses. Virus Res 240:161–165. doi: 10.1016/j.virusres.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 6.Arun PVPS, Miryala SK, Chattopadhyay S, Thiyyagura K, Bawa P, Bhattacharjee M, Yellaboina S. 2016. Identification and functional analysis of essential, conserved, housekeeping and duplicated genes. FEBS Lett 590:1428–1437. doi: 10.1002/1873-3468.12192. [DOI] [PubMed] [Google Scholar]
- 7.Kondrashov FA. 2012. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc Biol Sci 279:5048–5057. doi: 10.1098/rspb.2012.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliott KT, Cuff LE, Neidle EL. 2013. Copy number change: evolving views on gene amplification. Future Microbiol 8:887–899. doi: 10.2217/fmb.13.53. [DOI] [PubMed] [Google Scholar]
- 9.Hooper SD, Berg OG. 2003. Duplication is more common among laterally transferred genes than among indigenous genes. Genome Biol 4:R48. doi: 10.1186/gb-2003-4-8-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernabeu M, Sánchez-Herrero JF, Huedo P, Prieto A, Hüttener M, Rozas J, Juárez A. 2019. Gene duplications in the E. coli genome: common themes among pathotypes. BMC Genomics 20:313. doi: 10.1186/s12864-019-5683-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez-Herrero JF, Bernabeu M, Prieto A, Hüttener M, Juárez A. 2020. gene duplications in the genomes of Staphylococci and Enterococci. Front Mol Biosci 7. doi: 10.3389/fmolb.2020.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moriel DG, Heras B, Paxman JJ, Lo AW, Tan L, Sullivan MJ, Dando SJ, Beatson SA, Ulett GC, Schembri MA. 2016. Molecular and structural characterization of a novel Escherichia coli interleukin receptor mimic protein. mBio 7. doi: 10.1128/mBio.02046-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Woude MW, Henderson IR. 2008. Regulation and function of Ag43 (flu). Annu Rev Microbiol 62:153–169. doi: 10.1146/annurev.micro.62.081307.162938. [DOI] [PubMed] [Google Scholar]
- 14.Roche AJ, McFadden JP, Owen P. 2001. Antigen 43, the major phase-variable protein of the Escherichia coli outer membrane, can exist as a family of proteins encoded by multiple alleles. Microbiology 147:161–169. doi: 10.1099/00221287-147-1-161. [DOI] [PubMed] [Google Scholar]
- 15.Restieri C, Garriss G, Locas M-C, Dozois CM. 2007. Autotransporter-encoding sequences are phylogenetically distributed among Escherichia coli clinical isolates and reference strains. Appl Environ Microbiol 73:1553–1562. doi: 10.1128/AEM.01542-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henderson IR, Meehan M, Owen P. 1997. A novel regulatory mechanism for a novel phase-variable outer membrane protein of Escherichia coli. Adv Exp Med Biol 412:349–355. doi: 10.1007/978-1-4899-1828-4_56. [DOI] [PubMed] [Google Scholar]
- 17.Nataro JP, Deng Y, Cookson S, Cravioto A, Savarino SJ, Guers LD, Levine MM, Tacket CO. 1995. Heterogeneity of enteroaggregative Escherichia Coli virulence demonstrated. J Infect Dis 171:465–468. doi: 10.1093/infdis/171.2.465. [DOI] [PubMed] [Google Scholar]
- 18.Chaudhuri RR, Sebaihia M, Hobman JL, Webber MA, Leyton DL, Goldberg MD, Cunningham AF, Scott-Tucker A, Ferguson PR, Thomas CM, Frankel G, Tang CM, Dudley EG, Roberts IS, Rasko DA, Pallen MJ, Parkhill J, Nataro JP, Thomson NR, Henderson IR. 2010. Complete genome sequence and comparative metabolic profiling of the prototypical enteroaggregative Escherichia coli strain 042. PLoS One 5:e8801. doi: 10.1371/journal.pone.0008801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nataro JP, Scaletsky IC, Kaper JB, Levine MM, Trabulsi LR. 1985. Plasmid-mediated factors conferring diffuse and localized adherence of enteropathogenic Escherichia coli. Infect Immun 48:378–383. doi: 10.1128/iai.48.2.378-383.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nataro JP, Yikang D, Yingkang D, Walker K. 1994. AggR, a transcriptional activator of aggregative adherence fimbria I expression in enteroaggregative Escherichia coli. J Bacteriol 176:4691–4699. doi: 10.1128/jb.176.15.4691-4699.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Czeczulin JR, Balepur S, Hicks S, Phillips A, Hall R, Kothary MH, Navarro-Garcia F, Nataro JP. 1997. Aggregative adherence fimbria II, a second fimbrial antigen mediating aggregative adherence in enteroaggregative Escherichia coli. Infect Immun 65:4135–4145. doi: 10.1128/iai.65.10.4135-4145.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madrid C, Balsalobre C, García J, Juárez A. 2007. The novel Hha/YmoA family of nucleoid-associated proteins: use of structural mimicry to modulate the activity of the H-NS family of proteins. Mol Microbiol 63:7–14. doi: 10.1111/j.1365-2958.2006.05497.x. [DOI] [PubMed] [Google Scholar]
- 23.Rodrigo G, Fares MA. 2018. Intrinsic adaptive value and early fate of gene duplication revealed by a bottom-up approach. Elife 7. doi: 10.7554/eLife.29739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riehle MM, Bennett AF, Long AD. 2001. Genetic architecture of thermal adaptation in Escherichia coli. Proc Natl Acad Sci USA 98:525–530. doi: 10.1073/pnas.98.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.King OD, Masel J. 2007. The evolution of bet-hedging adaptations to rare scenarios. Theor Popul Biol 72:560–575. doi: 10.1016/j.tpb.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slayden RA, Dawson CC, Cummings JE. 2018. Toxin–antitoxin systems and regulatory mechanisms in Mycobacterium tuberculosis. Pathog Dis 76. doi: 10.1093/femspd/fty039. [DOI] [PubMed] [Google Scholar]
- 27.Harding T, Roger AJ, Simpson AGB. 2017. Adaptations to high salt in a halophilic protist: differential expression and gene acquisitions through duplications and gene transfers. Front Microbiol 8. doi: 10.3389/fmicb.2017.00944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slack A, Thornton PC, Magner DB, Rosenberg SM, Hastings PJ. 2006. On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS Genet 2:e48. doi: 10.1371/journal.pgen.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bergthorsson U, Andersson DI, Roth JR. 2007. Ohno’s dilemma: evolution of new genes under continuous selection. Proc Natl Acad Sci USA 104:17004–17009. doi: 10.1073/pnas.0707158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hittinger CT, Carroll SB. 2007. Gene duplication and the adaptive evolution of a classic genetic switch. Nature 449:677–681. doi: 10.1038/nature06151. [DOI] [PubMed] [Google Scholar]
- 31.Nowak MA, Boerlijst MC, Cooke J, Smith JM. 1997. Evolution of genetic redundancy. Nature 388:167–171. doi: 10.1038/40618. [DOI] [PubMed] [Google Scholar]
- 32.Balázsi G, van Oudenaarden A, Collins JJ. 2011. Cellular decision making and biological noise: from microbes to mammals. Cell 144:910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elowitz MB, Levine AJ, Siggia ED, Swain PS. 2002. Stochastic gene expression in a single cell. Science 297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 34.Raser JM, O'Shea EK. 2004. Control of stochasticity in eukaryotic gene expression. Science 304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodrigo G, Poyatos JF. 2016. Genetic redundancies enhance information transfer in noisy regulatory circuits. PLoS Comput Biol 12:e1005156. doi: 10.1371/journal.pcbi.1005156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li W-H. 2003. Role of duplicate genes in genetic robustness against null mutations. Nature 421:63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- 37.Nieto JM, Madrid C, Miquelay E, Parra JL, Rodríguez S, Juárez A. 2002. Evidence for direct protein-protein interaction between members of the enterobacterial Hha/YmoA and H-NS families of Proteins. J Bacteriol 184:629–635. doi: 10.1128/JB.184.3.629-635.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madrid C, García J, Pons M, Juárez A. 2007. Molecular evolution of the H-ns protein: interaction with Hha-like proteins is restricted to Enterobacteriaceae. J Bacteriol 189:265–268. doi: 10.1128/JB.01124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prieto A, Bernabeu M, Aznar S, Ruiz-Cruz S, Bravo A, Queiroz MH, Juárez A. 2018. Evolution of bacterial global modulators: role of a novel H-NS paralogue in the enteroaggregative Escherichia coli strain 042. mSystems 3. doi: 10.1128/mSystems.00220-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Solórzano C, Srikumar S, Canals R, Juárez A, Paytubi S, Madrid C. 2015. Hha has a defined regulatory role that is not dependent upon H-NS or StpA. Front Microbiol 6. doi: 10.3389/fmicb.2015.00773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 43.Ellermeier CD, Janakiraman A, Slauch JM. 2002. Construction of targeted single copy lac fusions using λ Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153–161. doi: 10.1016/s0378-1119(02)00551-6. [DOI] [PubMed] [Google Scholar]
- 44.Uzzau S, Figueroa-Bossi N, Rubino S, Bossi L. 2001. Epitope tagging of chromosomal genes in Salmonella. Proc Natl Acad Sci USA 98:15264–15269. doi: 10.1073/pnas.261348198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller JH. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, NYC. [Google Scholar]
- 46.Wallecha A, Correnti J, Munster V, van der Woude M. 2003. Phase variation of Ag43 is independent of the oxidation state of OxyR. J Bacteriol 185:2203–2209. doi: 10.1128/JB.185.7.2203-2209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hüttener M, Prieto A, Aznar S, Dietrich M, Paytubi S, Juárez A. 2018. Tetracycline alters gene expression in Salmonella strains that harbor the Tn10 transposon. Environ Microbiol Rep 10:202–209. doi: 10.1111/1758-2229.12621. [DOI] [PubMed] [Google Scholar]
- 48.Prieto A, Bernabeu M, Falgenhauer L, Chakraborty T, Hüttener M, Juárez A. 2020. Overexpression of the third H-NS paralogue H-NS2 compensates fitness loss in hns mutants of the enteroaggregative Escherichia coli strain 042. Sci Rep 10:18131. doi: 10.1038/s41598-020-75204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sneath PHA, Sokal RR. 1973. Numerical Taxonomy: the Principles and Practice of Numerical Classification. W. H. Freeman & Co., Ltd., San Francisco, USA. [Google Scholar]
- 51.Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA 101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.00454-22-s0001.pdf, PDF file, 1.6 MB (1.7MB, pdf)