ABSTRACT
Colistin is a last-resort antibiotic for multidrug-resistant Gram-negative infections. Recently, the ninth allele of the mobile colistin resistance (mcr) gene family, designated mcr-9, was reported. However, its clinical and public health significance remains unclear. We queried genomes of carbapenem-resistant Enterobacterales (CRE) for mcr-9 from a convenience sample of clinical isolates collected between 2012 and 2017 through the Georgia Emerging Infections Program, a population- and laboratory-based surveillance program. Isolates underwent phenotypic characterization and whole-genome sequencing. Phenotypic characteristics, genomic features, and clinical outcomes of mcr-9-positive and -negative CRE cases were then compared. Among 235 sequenced CRE genomes, 13 (6%) were found to harbor mcr-9, all of which were Enterobacter cloacae complex. The median MIC and rates of heteroresistance and inducible resistance to colistin were similar between mcr-9-positive and -negative isolates. However, rates of resistance were higher among mcr-9-positive isolates across most antibiotic classes. All cases had significant health care exposures. The 90-day mortality was similarly high in both mcr-9-positive (31%) and -negative (7%) CRE cases. Nucleotide identity and phylogenetic analysis did not reveal geotemporal clustering. mcr-9-positive isolates had a significantly higher number of median [range] antimicrobial resistance (AMR) genes (16 [4 to 22] versus 6 [2 to 15]; P < 0.001) than did mcr-9-negative isolates. Pangenome tests confirmed a significant association of mcr-9 detection with mobile genetic element and heavy metal resistance genes. Overall, the presence of mcr-9 was not associated with significant changes in colistin resistance or clinical outcomes, but continued genomic surveillance to monitor for emergence of AMR genes is warranted.
IMPORTANCE Colistin is a last-resort antibiotic for multidrug-resistant Gram-negative infections. A recently described allele of the mobile colistin resistance (mcr) gene family, designated mcr-9, has been widely reported among Enterobacterales species. However, its clinical and public health significance remains unclear. We compared characteristics and outcomes of mcr-9-positive and -negative CRE cases. All cases were acquired in the health care setting and associated with a high rate of mortality. The presence of mcr-9 was not associated with significant changes in colistin resistance, heteroresistance, or inducible resistance but was associated with resistance to other antimicrobials and antimicrobial resistance (AMR), virulence, and heavy metal resistance (HMR) genes. Overall, the presence of mcr-9 was not associated with significant phenotypic changes or clinical outcomes. However, given the increase in AMR and HMR gene content and potential clinical impact, continued genomic surveillance of multidrug-resistant organisms to monitor for emergence of AMR genes is warranted.
KEYWORDS: healthcare epidemiology, next-generation sequencing, CRE, MDR, AMR, antimicrobial resistance, carbapenem-resistant Enterobacterales, multidrug resistance
INTRODUCTION
With the rise of carbapenem-resistant organisms over the past few decades (1, 2), polymyxins (colistin or polymyxin B) remain last-resort antibiotics for multidrug-resistant (MDR) Gram-negative infections (3). While concerns regarding their efficacy and nephrotoxicity (4) have relegated polymyxins to the second or third line of the antibiotic armamentarium (5), these agents remain listed as critically important antibiotics by the WHO and are widely used globally.
In 2015, a colistin resistance gene localized on a plasmid was designated mobilized colistin resistance 1 (mcr-1) (6). The mcr-1 gene encodes a transferase that adds a phosphoethanolamine residue to cell membrane lipid A, altering the binding site of colistin and consequently leading to colistin resistance (6). Since its initial description in Escherichia coli (6), multiple mcr alleles (mcr-2 to mcr-10.1) have been described (7). The mcr-9 allele was reported in 2019 and is most similar to mcr-3 among previously described mcr alleles (8). A recent search of publicly available sequence databases revealed a wide global distribution of mcr-9-harboring isolates, across six continents and in at least 9 Enterobacterales species (7, 9). However, it is most commonly detected among Enterobacter species (9).
In the United States, the initial wave of carbapenem-resistant Enterobacterales (CRE) was predominantly driven by proliferation of KPC-harboring Klebsiella pneumoniae (1, 10); however, as demonstrated in recent reports, a second wave of CRE in the United States seems to be driven by the rise of Enterobacter species (1, 11). Genomic analysis indicates this second wave is associated with a high degree of clonal diversity among isolates (12). With the increasing spread of carbapenem-resistant Enterobacter, the dissemination of mcr-9 is highly probable. Despite its global distribution, the impact of mcr-9 on colistin phenotypic susceptibility remain unclear. Moreover, its association with patient clinical outcomes or potential for outbreaks of public health concern is yet to be examined.
The Centers for Disease Control and Prevention (CDC)-funded Georgia Emerging Infections Program (GA EIP) performs active, population- and laboratory-based surveillance for CRE isolated from sterile sites or urine in metropolitan Atlanta, GA (population ~4 million). We aimed to estimate the frequency of mcr-9 among CRE cases within the GA EIP catchment area and to compare clinical outcomes and microbiological, genomic, and clinical characteristics of mcr-9-positive and mcr-9-negative cases.
RESULTS
The mcr-9 allele was infrequently detected among GA EIP isolates between 2012 and 2017.
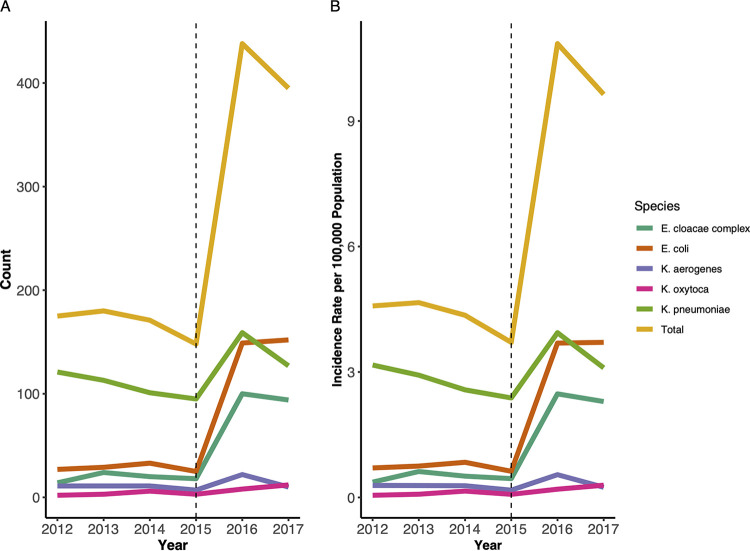
Between 2012 and 2017, the GA EIP identified 1,507 incident CRE cases; 716 (47.5%) were K. pneumoniae, 415 (27.5%) were Escherichia coli, 270 (17.9%) were Enterobacter cloacae complex, 72 (4.8%) were Klebsiella aerogenes (formerly Enterobacter), and 34 (2.3%) were Klebsiella oxytoca. The overall crude annual CRE incidence across GA EIP increased from 4.6 to 9.6 per 100,000 population from 2012 to 2017. Carbapenem-resistant E. cloacae complex incidence, in particular, increased from 0.37 to 2.3 per 100,000 population during the study period. This increase coincided with revision of the CDC case surveillance definition for CRE (13) (Fig. 1A and B and see also Table S1 and Fig. S1 in the supplemental material).
FIG 1.
Carbapenem-resistant Enterobacterales (CRE) count (A) and crude annual incidence per 100,000 population (B) by species across the Georgia Emerging Infections Program from 2012 to 2017. Beginning in 2016, the phenotypic CRE case definition was changed to resistance to ≥1 carbapenem (now including ertapenem) with no cephalosporin parameter.
A convenience sample of 384 isolates which met the GA EIP CRE case definition was sent to the CDC for further characterization. Of the 384, 235 (61%) underwent whole-genome sequencing (WGS). Among 235 sequenced CRE isolates, 13 (6%) were found to harbor mcr-9, all of which were E. cloacae complex. All remaining sequenced mcr-9-negative E. cloacae complex isolates (n = 14) were included as a comparative group, yielding a total number of 27 E. cloacae complex isolates.
Microbiology characteristics.
Following collection, isolates underwent reference antimicrobial susceptibility testing by broth microdilution (BMD) at the CDC. Of the E. cloacae complex isolates that underwent WGS from 2012 to 2017, 22 isolates (81.7%, 22/27) were confirmed to be carbapenem resistant. Carbapenem resistance rates were similar between mcr-9-positive and -negative cases (84.6% [11/13] versus 78.6% [11/14]; P = 1.00). Fluoroquinolone resistance was significantly higher among mcr-9-positive isolates than mcr-9-negative isolates (100% [13/13] versus 57.1% [8/14]; P = 0.03). This contributed to a higher proportion of isolates being classified as difficult-to-treat resistance (DTR) (14). Overall, 48.1% (13/27) were classified as harboring DTR (14, 15). with mcr-9 isolates having higher-rates of DTR (61.5% [8/13] versus 35.7% [5/14]; P = 0.34). Similarly, rates of aminoglycoside, tetracycline, and trimethoprim-sulfamethoxazole resistance were higher among mcr-9-positive isolates than mcr-9-negative isolates (Table S2).
The median (range of concentrations tested) colistin MIC for all E. cloacae complex isolates was 0.5 μg/mL (≤0.25 to >8.0). Proportions of resistance, heteroresistance, and inducible resistance were 11.1% (3/27), 48.1% (13/27), and 14.8% (4/27), respectively. There was no significant difference in colistin MIC, heteroresistance, or inducible resistance between mcr-9-positive and -negative isolates (Table 1). Of the three E. cloacae complex isolates which were colistin resistant by BMD, none were mcr-9 positive, although one was positive for mcr-10.1.
TABLE 1.
Carbapenem-resistant E. cloacae complex clinical and microbiological characteristics
| Characteristic | No. (%) for category: |
P value | ||
|---|---|---|---|---|
| All (n = 27) | mcr-9 positive (n = 13) | mcr-9 negativea (n = 14) | ||
| Culture source | 0.33 | |||
| Urine | 24 (88.9) | 12 (92.3) | 12 (85.7) | |
| Blood | 2 (7.4) | 1 (7.7) | 1 (7.1) | |
| Peritoneal fluid | 1 (3.7) | 0 (0.0) | 1 (7.1) | |
| Yr | 0.62 | |||
| 2012 | 1 (3.7) | 1 (7.7) | 0 (0.0) | |
| 2013 | 8 (29.6) | 2 (15.4) | 6 (42.9) | |
| 2014 | 2 (7.4) | 1 (7.7) | 1 (7.1) | |
| 2015 | 3 (11.1) | 3 (23.1) | 0 (0.0) | |
| 2016 | 9 (33.3) | 4 (30.8) | 5 (35.7) | |
| 2017 | 4 (14.9) | 2 (15.4) | 2 (14.3) | |
| Infection onset | 0.71 | |||
| Hospital onset | 3 (11.1) | 2 (15.4) | 1 (7.1) | |
| Healthcare-associated community onset | 10 (37.0) | 4 (30.8) | 6 (42.9) | |
| Long-term care facility onset | 14 (51.9) | 7 (53.8) | 7 (50.0) | |
| Microbiology characteristic | ||||
| Colistin MIC (median [range])b | 0.5 [<0.25–>0.8] | 0.5 [<0.25–1.00] | 0.5 [<0.25–>8.0] | 0.11 |
| Resistant | 3 (11.1) | 0 | 3 (21.4) | 0.25 |
| Heteroresistant | 13 (48.1) | 5 (38.5) | 8 (57.1) | 0.56 |
| Inducible resistance | 4 (14.8) | 2 (15.4) | 2 (14.3) | 1.00 |
| Difficult-to-treat resistance | 13 (48.1) | 8 (61.5) | 5 (35.7) | 0.34 |
| Outcomes | ||||
| Hospitalization within 29 days after culture | 10 (37.0) | 5 (38.5) | 5 (35.7) | 0.86 |
| ICU admissionc,d | 4 (40.0) | 2 (40.0) | 2 (40.0) | 1.00 |
| In-hospital mortalityd | 1 (10.0) | 1 (20.0) | 0 (0.0) | 0.59 |
| 90-day mortality | 5 (18.5) | 4 (30.8) | 1 (7.1) | 0.28 |
One mcr-10-positive isolate.
MIC units are micrograms per milliliter.
Any ICU admission 7 days before or 6 days after specimen collection.
Among 10 hospitalized patients, 5 in each group (mcr-9 positive and negative).
Clinical characteristics of mcr-9-positive and -negative CRE cases.
E. cloacae complex isolates were commonly isolated from urine (88.9%, 24/27), followed by blood (7.4%, 2/27) and peritoneal fluid (3.7%, 1/27). All cases had significant health care exposures, with 14 cases (51.9%) of long-term care facility onset, 10 (37.0%) of health care-associated community onset, and 3 (11.1%) of hospital onset. Ten patients (37.0%) were hospitalized at time of culture or within 29 days of CRE culture. Among the 10 hospitalized patients, 3 (30.0%) patients were admitted to the intensive care unit (ICU) within 7 days of culture and one patient died during the period of hospitalization (10.0%). Among hospitalized patients with available follow-up data (n = 7), 42.8% (3/7) were readmitted within 30 days. Overall unadjusted all-cause 90-day mortality was 18.5% (5/27). No clinical characteristics and outcomes were significantly different, and most were numerically similar among mcr-9-positive and -negative cases. Ninety-day mortality was higher among mcr-9-positive cases (30.7% [4/13] versus 7.1% [1/14]) than mcr-9-negative cases, but this was not statistically significant (P = 0.28).
Genomic analysis.
To expand the analytic genome set, a comparator cohort of nine publicly available clinical and environmental carbapenem-resistant E. cloacae complex (three mcr-9 positive, six mcr-9 negative) genomes were downloaded from the National Center for Biotechnology Information (NCBI) and included in further comparative genomic analyses. Review of associated metadata revealed the isolates to have been collected between January 2012 and December 2016 at the National Institutes of Health (NIH) Clinical Center (Bethesda, MD) (PRJNA430442) (16). In addition, 582 publicly available global E. cloacae complex genomes were included in the phylogenetic analysis.
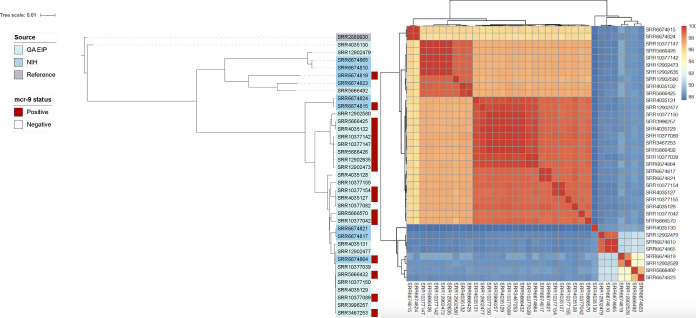
The 13 mcr-9-positive E. cloacae complex isolates were obtained from 10 distinct GA EIP facilities across the study period (2012 to 2017). Average nucleotide identity pairwise comparisons revealed three distinct clusters (Fig. 2). The clustering did not appear to be related to geographical location or year of isolation. NIH isolates clustered with GA EIP isolates, and GA EIP isolates did not cluster by facility or year (Fig. S2). Their distribution throughout the genome phylogeny suggested that the mcr-9-positive isolates were largely genetically distinct from one another. Among the total set of 609 genomes, 16% (98/609) were mcr-9 positive. Both the local GA EIP and global mcr-9-positive genomes were distributed throughout the genome phylogeny. A notable exception were genomes DRX055644 to DRX055660, which were sequenced as part of an ongoing outbreak at a burn center (17); many of the genomes had a pairwise single nucleotide polymorphism (SNP) distance of <50 SNPs (Fig. S3 and Table S4).
FIG 2.
Phylogeny (left) and average nucleotide identity heatmap (right) of mcr-9-positive (n = 13) and mcr-9-negative (n = 14) E. cloacae complex genomes from the Georgia Emerging Infections Program in addition to 9 available E. cloacae complex genomes (three mcr-9 positive, six mcr-9 negative) from the National Institutes of Health. A phylogenetic tree based on a core gene alignment containing 1,904 genes defined using Roary v3.13.0 was generated using IQtree v2.0.3. A maximum likelihood tree was generated by running 1,000 bootstrap replicates under the generalized time-reversible model of evolution. The tree was visualized and annotated using Interactive Tree of Life (iTOL) v4. Pairwise comparisons of average nucleotide identity on the assembled genomes were performed with the Mashmap method using fastANI v1.32. GA EIP, Georgia Emerging Infections Program; NIH, National Institutes of Health.
Median [range] antimicrobial resistance (AMR) gene content (excluding mcr-9) was significantly higher among mcr-9-positive isolates than mcr-9-negative isolates (16 [4 to 22] versus 6 [2 to 15]; P < 0.001) (Fig. 3). Among the three isolates with elevated colistin MICs, no point mutations in pmrA/pmrB, a two-component system regulator of lipopolysaccharide (the target site of colistin) modifications, were detected. Pangenome-wide association tests revealed a significant association of mcr-9 detection with the detection of mobile genetic element (MGE)-associated genes such as repB, parM, and hns2; heavy metal resistance (HMR) genes such as arsC2, arsB2, fieF2, pcoE2, and merA; and virulence genes such as hipA (Table 2, Table S2, and Fig. S3). Taken together, these comparative genomic analyses across two sites with mcr-9-positive E. cloacae complex isolate draft genomes confirmed the colocalization of mcr-9 with plasmid-mobilized heavy metal resistance genes but did not provide evidence of a high-identity outbreak cluster in space or time.
FIG 3.
Antimicrobial resistance gene heatmap of mcr-9-positive (n = 13) and mcr-9-negative (n = 14) E. cloacae complex genomes from the Georgia Emerging Infections Program in addition to 9 available E. cloacae complex genomes (three mcr-9 positive, six mcr-9 negative) from the National Institutes of Health. Genomes were annotated using Prodigal v2.6.3, and antimicrobial resistance gene content was assessed using AMRFinder. Antimicrobial resistance gene presence/absence heatmaps were created using the package pheatmap on R version 4.0.2 (Vienna, Austria) and the RStudio interface version 1.3.1073 (Boston, MA, USA).
TABLE 2.
Highest-ranking genes for association with mcr-9 presence
| Genea | Comment | Odds ratio | Bonferroni-adjusted P value |
|---|---|---|---|
| smc | Chromosome partition protein Smc | ∞ | 1.60E−06 |
| dcm2 | DNA-cytosine methyltransferase | ∞ | 1.60E−06 |
| pcoE2 | Putative copper-binding protein PcoE | ∞ | 2.88E−05 |
| hns2 | DNA-binding protein H-NS, plasmid | ∞ | 3.36E−05 |
| group_10390 | Tn3 family transposase ISEc63 | ∞ | 3.36E−05 |
| hipA | Serine/threonine-protein kinase toxin HipA | ∞ | 3.36E−05 |
| rcnR2 | Transcriptional repressor RcnR | ∞ | 0.00030219 |
| hha2 | Hemolysin expression-modulating protein Hha | ∞ | 0.00036934 |
| uvrD2 | DNA helicase II | ∞ | 0.00036934 |
| parM | Plasmid segregation protein | ∞ | 0.00036934 |
| higB-1 | Toxin HigB-1 | ∞ | 0.00036934 |
| group_1846 | Tn3 family transposase ISEc63 | ∞ | 0.00036934 |
| dam2 | DNA adenine methylase | ∞ | 0.00036934 |
| repB | RepFIB replication protein A | ∞ | 0.00036934 |
| traC | Protein TraC | ∞ | 0.00036934 |
| group_7173 | Stable plasmid inheritance protein | ∞ | 0.00036934 |
| yjcD | Putative ATP-dependent DNA helicase YjcD | 304 | 0.00054522 |
| umuD2 | Protein UmuD | ∞ | 0.00283164 |
| dsbC_2 | Thiol:disulfide interchange protein DsbC | ∞ | 0.00283164 |
| virB | Virulence regulon transcriptional activator VirB | ∞ | 0.00283164 |
| umuC_3 | Protein UmuC | ∞ | 0.00283164 |
| group_7174 | IS110 family transposase ISEsa2 | 142.5 | 0.00540586 |
| merA | Mercuric reductase | ∞ | 0.01698985 |
| fieF_2 | Ferrous-iron efflux pump FieF | 90.7 | 0.03283809 |
| arsB_2 | Arsenical pump membrane protein | 67.5 | 0.04368339 |
| group_7063 | ISNCY family transposase ISEsa1 | 67.5 | 0.04368339 |
| group_8953 | ISNCY family transposase ISBcen27 | 67.5 | 0.04368339 |
| arsH | NADPH-dependent FMNb reductase ArsH | 67.5 | 0.04368339 |
| arsC2 | Arsenate reductase | 67.5 | 0.04368339 |
Hypothetical proteins not included.
FMN, flavin mononucleotide.
DISCUSSION
Among 235 CRE isolates collected through a comprehensive, population-based surveillance program targeting the most common CRE species, we found a low prevalence of mcr-9, all of which was detected in Enterobacter cloacae isolates. Our phylogenetic analyses revealed a genetically diverse mcr-9-positive CRE population, suggesting sporadic carriage rather than clonal spread. Using multimodal phenotypic testing, we were unable to detect impacts of mcr-9 on colistin susceptibility; however, genomic analysis revealed an association with increased AMR, HMR, and virulence genes. In addition, our mcr-9-containing CRE isolates were exclusively acquired in health care settings, with a trend toward increased mortality. Since their initial description, recognition of mcr genes associated with colistin resistance has spread rapidly across the globe (7). Our study of mcr-9-harboring CRE cases provides unique insights into the phenotypic and genomic implications of mcr-9 and is one of the first to examine clinical outcomes.
Whether mcr-9 confers colistin resistance has been debated (18). The first isolate identified to harbor mcr-9 was also susceptible to colistin, but the allele was found to confer resistance to colistin when cloned into a colistin-susceptible E. coli strain and expressed under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-induced promoter. However, this was only at 1, 2, and 2.5 mg/L, not at 5 mg/L, of colistin (8). Kieffer et al. later reported that mcr-9 expression was inducible in the presence of colistin when located upstream of the two-component sensor kinase system qseBC (18). This two-component signaling network allows bacteria to sense and respond to their changing environments. In particular, the qseC and qseB genes encode a histidine kinase sensor (qseC) and its cognate partner (qseB). The qseBC system has been shown to interact with pmrA/pmrB, to induce resistance to colistin (19). However, a study of mcr-9-containing isolates from retail meat conducted by the National Antimicrobial Resistance Monitoring System (NARMS) found all 105 isolates (99 Salmonella enterica and 6 E. coli) tested to be susceptible to colistin, including 10 isolates with qseBC (20), indicating that the previously demonstrated impact of qseBC on mcr-9 expression and colistin resistance may be dependent on strain backgrounds, as originally demonstrated in E. coli (18). Among clinical CRE isolates, we found the presence of mcr-9 was not associated with frank or inducible colistin resistance. Furthermore, our study is the first to examine the association of mcr-9 with heteroresistance. Heteroresistance is a largely unrecognized form of antibiotic resistance where only a subset of cells within a bacterial population are resistant to a given drug (21). These resistant cells can be selected for in the presence of the antibiotic and cause colistin treatment failures in vivo (22). In a multisite surveillance study of colistin heteroresistance among CRE, Enterobacter spp. and, in particular, E. cloacae displayed the highest proportion of colistin heteroresistance (23). However, here we found no association between mcr-9 and colistin heteroresistance.
Carbapenem-resistant Gram-negative bacteria are a public health threat broadly prioritized by public health organizations (24). Given the limited therapeutic options, morbidity and mortality rates are increased disproportionately compared to infections caused by susceptible bacteria. We observed high 90-day mortality rates, but these were similar to reported CRE mortality rates at other U.S. academic centers (1). However, there was a nonsignificant numerically higher rate of mortality associated with mcr-9-positive isolates. This association should be further evaluated in larger studies, with adjustment for potentially confounding variables associated with mortality such as severity of illness, age, and comorbidities as our study’s small size may underestimate differences in mortality (1). This finding may be related to the increase in phenotypic resistance and AMR gene content associated with mcr-9. A similar finding was previously reported describing 1,035 mcr-9-containing isolates in which 97% (1003/1035) were classified as multidrug resistant (MDR) (7). This increased AMR gene content renders isolates not only carbapenem resistant but also with DTR, further limiting therapeutic options (14). DTR is a clinically relevant and functional classification of resistance which signifies in vitro resistance to all high-efficacy, low-toxicity (or first-line) agents and is associated with worse clinical outcomes than those of carbapenem-resistant phenotypes (14). Moreover, genome-wide association studies revealed a difference in key virulence genes such as hipA, a eukaryote-like serine threonine kinase that inhibits cell growth and induces bacterial persistence (25).
We found the presence of mcr-9 to be associated with HMR genes such as arsA and merA, conferring arsenic and mercury resistance, respectively. There has been increasing evidence for the coselection of AMR and HMR genes through either coresistance or cross-resistance (26). Coresistance occurs when AMR and HMR genes are carried on the same mobile genetic element. IncH12 plasmids, which frequently harbor HMR genes (8, 27), have been found to be the predominant replicon type carrying mcr-9 and frequently demonstrated in our isolates. Hospital wastewater is an increasingly recognized reservoir for resistant Gram-negative organisms that cause health care-associated infections (28), and HMR genes may allow for continued persistence in the environment (29). While the community setting is starting to represent an increased source of multidrug-resistant infections (30), the health care setting still represents a major risk for MDR acquisition, as was seen among our cohort.
Our study combines detailed epidemiological, clinical, phenotypic, and genomic data to examine the significance of mcr-9 but has some limitations. First, we could not do a full interrogation of mcr-9-containing plasmids, due to limitations of short-read sequencing. However, prior studies have significantly characterized the genomic background of mcr-9-containing plasmids (9). Second, our study did not include Salmonella species, which are a major reservoir for mcr-9, or other Enterobacterales species such as Citrobacter (7), and our findings may not be generalizable to these species. However, our data set of 235 includes the most common and significant clinical CRE species (1, 24) and is one of the few studies carried out on clinical human isolates (20). Third, while we assessed for the presence of the pmrA/pmrB regulatory system (31), we did not include the assessment of the two-component system qsceBC which has been shown to influence mcr-9 expression and colistin MIC results. Fourth, while our overall cohort is from a population-based surveillance program, the collected and sequenced isolates represent a convenience sample, which may limit generalizability, and our sample size was not powered to control for important variables such as source of infection, severity of illness, and treatment received and to detect clinical outcomes.
In conclusion, mcr-9 may not have actionable public health implications as do other mcr alleles, most of which consistently display colistin resistance. However, given the increased AMR and HMR gene content, continued genomic surveillance of multidrug-resistant organisms to monitor for the emergence of AMR genes such as mcr-9 is prudent, especially as changes in the up- or downstream genetic context or the accumulation of mutations may impact its ability to confer colistin resistance.
MATERIALS AND METHODS
CRE cases were identified by routine queries on automated testing instruments in the clinical labs that serve residents of the GA EIP catchment area. Clinical characteristics were obtained through medical record review, all-cause mortality data were obtained through the Georgia Vital Statistics records, and hospital readmission data were obtained through the Department of Public Health’s hospital discharge data sets. Georgia EIP surveillance activities are reviewed and approved by the Emory University Institutional Review Board (IRB00089004).
From 2012 to 2015, a CRE case was defined as an isolate of E. coli, E. cloacae complex, K. (formerly Enterobacter) aerogenes, K. pneumoniae, or K. oxytoca collected from a normally sterile body site (e.g., bloodstream) or urine that tested nonsusceptible to ≥1 carbapenem (imipenem, meropenem, or doripenem) and resistant to all third-generation cephalosporins tested (ceftriaxone, ceftazidime, and cefotaxime) by testing performed at the local collection microbiology laboratory. Beginning in 2016, the phenotypic case definition was changed to resistance to ≥1 carbapenem (now including ertapenem) with no cephalosporin parameter. Antibiotic susceptibility interpretations were determined using the current Clinical and Laboratory Standards Institute breakpoints (32). Fluoroquinolone resistance was defined as nonsusceptibility (intermediate or resistant) to ≥1 fluoroquinolone. DTR was defined as intermediate or resistant to all reported agents in carbapenem, β-lactam, and fluoroquinolone categories (14, 15).
An incident CRE case was defined as the first CRE isolate from a patient during a 30-day period that met the surveillance definition. All incident CRE cases underwent medical record review using a standardized abstraction form. Both inpatient and outpatient medical records were reviewed for patient demographics, underlying clinical comorbidities, location of culture collection, specimen source, associated infectious syndromes, relevant health care exposures, and patient outcomes. Ninety-day mortality was determined based on matching to vital records.
A convenience sample of CRE isolates is collected annually and submitted to the CDC for further characterization. Isolates that are collected and matched to an incident case with a completed case report form are eligible for shipment. All isolates undergo repeat reference BMD at the CDC followed by whole-genome sequencing using an Illumina MiSeq benchtop sequencer.
All mcr-9-positive and a comparative control group of mcr-9-negative E. cloacae complex isolates underwent additional population analysis profiling and inducible resistance testing at the Emory Investigational Clinical Microbiology Core as previously described (21). Briefly, all isolates were tested via the population analysis profile (PAP) method. This consists of plating overnight cultures of each isolate onto solid cation-adjusted Mueller-Hinton (MH) agar with or without colistin concentrations of 0.5, 1, 2, 4, 16, 32, and 100 μg/mL. Surviving colonies were enumerated and used to detect colistin-resistant subpopulations characteristic of heteroresistance (21). Inducible resistance testing was performed as previously described (33). Briefly, a single colony of each clinical isolate was grown in cation-adjusted MH broth overnight at 37°C, and cultures were diluted 1:100 in MH broth containing serially increasing concentrations of colistin, starting at the one-half MIC value of the respective isolate and doubling every 24 h until bacterial growth was completely inhibited (with no bacterial growth after spreading 100 μL of the culture on MH agar plates supplemented with the corresponding concentration of colistin). The concentration of colistin at which bacterial growth was completely inhibited was recorded as the final colistin concentration.
Bioinformatic methods.
Fastq files of Enterobacter cloacae complex isolates of interest were downloaded from the Sequence Read Archive (SRA) repository maintained by the National Center for Biotechnology Information (NCBI) using the fasterq-dump tool from the SRA Toolkit v2.5.7 (https://hpc.nih.gov/apps/sratoolkit.html). Illumina reads were quality filtered using Trimmomatic (34) and assembled de novo using SPAdes v3.13 (35). Pairwise comparisons of average nucleotide identity on the assembled genomes were performed with the Mashmap method using fastANI v1.32 (36). Gene sequences were predicted with Prodigal v2.6.3 (37) and annotated with Prokka v1.14.6 (38). Antimicrobial resistance and virulence gene content was assessed using AMRFinder Plus (39). The presence of plasmids and point mutations in housekeeping genes associated with colistin resistance was assessed using the ResFinder and PlasmidFinder web interface with default settings and the E. coli database (40, 41). AMR gene presence/absence heatmaps were created using the package pheatmap on R version 4.0.2 (Vienna, Austria) and the RStudio interface version 1.3.1073 (Boston, MA, USA). Pangenome-wide comparison of core genomes of mcr-9-positive to mcr-9-negative genomes was completed using Scoary (42). Additional publicly available global E. cloacae complex genomes were downloaded, assembled, and analyzed using the Bactopia pipeline (43). Core genes were defined using PIRATE (44). A phylogenetic tree based on a core gene alignment was generated using IQtree v2.0.3 (45). A maximum likelihood tree was generated by running 1,000 bootstrap replicates under the generalized time-reversible model of evolution. The tree was visualized and annotated using Interactive Tree of Life (iTOL) v4 (46). The core genome pairwise SNP distance for each sample is also calculated with snp-dists (47).
Statistical analysis.
Annual incidence rates for CRE cases were calculated using the annual U.S. census estimates of the surveillance area population as the denominator. Descriptive analyses were performed to summarize specimen information, health care exposures, outcomes, and microbiological results of incident cases; χ2 and Wilcoxon rank sum tests were used to compare groups when applicable. Gene differences were assessed by a P value adjusted with Bonferroni’s method for multiple-comparison correction. Statistical analysis was performed using R version 4.0.2 (Vienna, Austria) and the RStudio interface version 1.3.1073 (Boston, MA, USA). A two-sided P value of <0.05 was considered statistically significant.
Data availability.
All local GA sequence data are available on NCBI under BioProject PRJNA288601 (GA isolates). Accession numbers for global accessions are found in Table S5 in the supplemental material.
ACKNOWLEDGMENTS
We thank the EIP and MuGSI staff and the CDC laboratory staff for their dedication and work to collect, validate, clean, and maintain the data used in this analysis. We thank Alison Laufer Halpin for supervising the sequencing efforts for these isolates.
EIP Surveillance of the Multi-site Gram-Negative Surveillance Initiative (MuGSI) was funded through the Centers for Disease Control and Prevention’s Emerging Infections Program (U50CK000485). Research reported in this publication was supported in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (K23AI144036). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplemental material is available online only.
Contributor Information
Ahmed Babiker, Email: ahmed.babiker@emory.edu.
Michael H. Woodworth, Email: michael.holmes.woodworth@emory.edu.
Cheryl P. Andam, University at Albany, State University of New York
REFERENCES
- 1.Babiker A, Clarke LG, Saul M, Gealey JA, Clancy CJ, Nguyen MH, Shields RK. 2021. Changing epidemiology and decreased mortality associated with carbapenem-resistant Gram-negative bacteria, 2000-2017. Clin Infect Dis 73:e4521–e4530. doi: 10.1093/cid/ciaa1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babiker A, Evans DR, Griffith MP, McElheny CL, Hassan M, Clarke LG, Mettus RT, Harrison LH, Doi Y, Shields RK, Van Tyne D. 2020. Clinical and genomic epidemiology of carbapenem-nonsusceptible Citrobacter spp. at a tertiary health care center over 2 decades. J Clin Microbiol 58:e00275-20. doi: 10.1128/JCM.00275-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. 2019. Critically important antimicrobials for human medicine, 6th revision. World Health Organization, Geneva, Switzerland. Accessed May 7, 2021.
- 4.Eljaaly K, Bidell MR, Gandhi RG, Alshehri S, Enani MA, Al-Jedai A, Lee TC. 2021. Colistin nephrotoxicity: meta-analysis of randomized controlled trials. Open Forum Infect Dis 8:ofab026. doi: 10.1093/ofid/ofab026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strich JR, Ricotta E, Warner S, Lai YL, Demirkale CY, Hohmann SF, Rhee C, Klompas M, Palmore T, Powers JH, Dekker JP, Adjemian J, Matsouaka R, Woods CW, Danner RL, Kadri SS. 2021. Pharmacoepidemiology of ceftazidime-avibactam use: a retrospective cohort analysis of 210 US Hospitals. Clin Infect Dis 72:611–621. doi: 10.1093/cid/ciaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism mcr-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 7.Ling Z, Yin W, Shen Z, Wang Y, Shen J, Walsh TR. 2020. Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J Antimicrob Chemother 75:3087–3095. doi: 10.1093/jac/dkaa205. [DOI] [PubMed] [Google Scholar]
- 8.Carroll LM, Gaballa A, Guldimann C, Sullivan G, Henderson LO, Wiedmann M. 2019. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. mBio 10:e00853-19. doi: 10.1128/mBio.00853-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Dai X, Zeng J, Gao Y, Zhang Z, Zhang L. 2020. Characterization of the global distribution and diversified plasmid reservoirs of the colistin resistance gene mcr-9. Sci Rep 10:8113. doi: 10.1038/s41598-020-65106-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson BM, El Chakhtoura NG, Patel S, Saade E, Donskey CJ, Bonomo RA, Perez F. 2017. Carbapenem-eesistant Enterobacter cloacae in patients from the US Veterans Health Administration, 2006-2015. Emerg Infect Dis 23:878–880. doi: 10.3201/eid2305.162034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Annavajhala MK, Gomez-Simmonds A, Uhlemann AC. 2019. Multidrug-resistant Enterobacter cloacae complex emerging as a global, diversifying threat. Front Microbiol 10:44. doi: 10.3389/fmicb.2019.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chea N, Bulens SN, Kongphet-Tran T, Lynfield R, Shaw KM, Vagnone PS, Kainer MA, Muleta DB, Wilson L, Vaeth E, Dumyati G, Concannon C, Phipps EC, Culbreath K, Janelle SJ, Bamberg WM, Guh AY, Limbago B, Kallen AJ. 2015. Improved phenotype-based definition for identifying carbapenemase producers among carbapenem-resistant Enterobacteriaceae. Emerg Infect Dis 21:1611–1616. doi: 10.3201/eid2109.150198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kadri SS, Lai YL, Ricotta EE, Strich JR, Babiker A, Rhee C, Klompas M, Dekker JP, Powers JH, III, Danner RL, Adjemian J, NIH Antimicrobial Resistance Outcomes Research Initiative (NIH-ARORI) . 2019. External validation of difficult-to-treat resistance prevalence and mortality risk in Gram-negative bloodstream infection using electronic health record data from 140 US hospitals. Open Forum Infect Dis 6:ofz110. doi: 10.1093/ofid/ofz110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kadri SS, Adjemian J, Lai YL, Spaulding AB, Ricotta E, Prevots DR, Palmore TN, Rhee C, Klompas M, Dekker JP, Powers JH, III, Suffredini AF, Hooper DC, Fridkin S, Danner RL, National Institutes of Health Antimicrobial Resistance Outcomes Research Initiative (NIH–ARORI) . 2018. Difficult-to-treat resistance in Gram-negative bacteremia at 173 US hospitals: retrospective cohort analysis of prevalence, predictors, and outcome of resistance to all first-line agents. Clin Infect Dis 67:1803–1814. doi: 10.1093/cid/ciy378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weingarten RA, Johnson RC, Conlan S, Ramsburg AM, Dekker JP, Lau AF, Khil P, Odom RT, Deming C, Park M, Thomas PJ, Henderson DK, Palmore TN, Segre JA, Frank KM, NISC Comparative Sequencing Program . 2018. Genomic analysis of hospital plumbing reveals diverse reservoir of bacterial plasmids conferring carbapenem resistance. mBio 9:e02011-17. doi: 10.1128/mBio.02011-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanamori H, Parobek CM, Juliano JJ, van Duin D, Cairns BA, Weber DJ, Rutala WA. 2017. A prolonged outbreak of KPC-3-producing Enterobacter cloacae and Klebsiella pneumoniae driven by multiple mechanisms of resistance transmission at a large academic burn center. Antimicrob Agents Chemother 61:e01516-16. doi: 10.1128/AAC.01516-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kieffer N, Royer G, Decousser J-W, Bourrel A-S, Palmieri M, Ortiz De La Rosa J-M, Jacquier H, Denamur E, Nordmann P, Poirel L. 2019. mcr-9, an inducible gene encoding an acquired phosphoethanolamine transferase in Escherichia coli, and its origin. Antimicrob Agents Chemother 63:e00965-19. doi: 10.1128/AAC.00965-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breland EJ, Zhang EW, Bermudez T, Martinez CR, III, Hadjifrangiskou M. 2017. The histidine residue of QseC is required for canonical signaling between QseB and PmrB in uropathogenic Escherichia coli. J Bacteriol 199:e00060-17. doi: 10.1128/JB.00060-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyson GH, Li C, Hsu C-H, Ayers S, Borenstein S, Mukherjee S, Tran T-T, McDermott PF, Zhao S. 2020. The mcr-9 gene of Salmonella and Escherichia coli is not associated with colistin resistance in the United States. Antimicrob Agents Chemother 64:e00573-20. doi: 10.1128/AAC.00573-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicoloff H, Hjort K, Levin BR, Andersson DI. 2019. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat Microbiol 4:504–514. doi: 10.1038/s41564-018-0342-0. [DOI] [PubMed] [Google Scholar]
- 22.Band VI, Satola SW, Burd EM, Farley MM, Jacob JT, Weiss DS, Hunstad DA, Hultgren SJ. 2018. Carbapenem-resistant Klebsiella pneumoniae exhibiting clinically undetected colistin heteroresistance leads to treatment failure in a murine model of infection. mBio 9:e02448-17. doi: 10.1128/mBio.02448-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Band VI, Satola SW, Smith RD, Hufnagel DA, Bower C, Conley AB, Rishishwar L, Dale SE, Hardy DJ, Vargas RL, Dumyati G, Kainer MA, Phipps EC, Pierce R, Wilson LE, Sorensen M, Nilsson E, Jordan IK, Burd EM, Farley MM, Jacob JT, Ernst RK, Weiss DS. 2021. Colistin heteroresistance is largely undetected among carbapenem-resistant Enterobacterales in the United States. mBio 12:e02881-20. doi: 10.1128/mBio.02881-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Centers for Disease Control and Prevention. 2019. Antibiotic resistance threats in the United States, 2019. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 25.Germain E, Castro-Roa D, Zenkin N, Gerdes K. 2013. Molecular mechanism of bacterial persistence by HipA. Mol Cell 52:248–254. doi: 10.1016/j.molcel.2013.08.045. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen CC, Hugie CN, Kile ML, Navab-Daneshmand T. 2019. Association between heavy metals and antibiotic-resistant human pathogens in environmental reservoirs: a review. Front Environ Sci Eng 13:46. doi: 10.1007/s11783-019-1129-0. [DOI] [Google Scholar]
- 27.Campos-Madueno EI, Moser AI, Risch M, Bodmer T, Endimiani A. 2021. Exploring the global spread of Klebsiella grimontii isolates possessing blaVIM-1 and mcr-9. Antimicrob Agents Chemother 65:e0072421. doi: 10.1128/AAC.00724-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kizny Gordon AE, Mathers AJ, Cheong EYL, Gottlieb T, Kotay S, Walker AS, Peto TEA, Crook DW, Stoesser N. 2017. The hospital water environment as a reservoir for carbapenem-resistant organisms causing hospital-acquired infections—a systematic review of the literature. Clin Infect Dis 64:1435–1444. doi: 10.1093/cid/cix132. [DOI] [PubMed] [Google Scholar]
- 29.Kamathewatta K, Bushell R, Rafa F, Browning G, Billman-Jacobe H, Marenda M. 2020. Colonization of a hand washing sink in a veterinary hospital by an Enterobacter hormaechei strain carrying multiple resistances to high importance antimicrobials. Antimicrob Resist Infect Control 9:163. doi: 10.1186/s13756-020-00828-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jernigan JA, Hatfield KM, Wolford H, Nelson RE, Olubajo B, Reddy SC, McCarthy N, Paul P, McDonald LC, Kallen A, Fiore A, Craig M, Baggs J. 2020. Multidrug-resistant bacterial infections in U.S. hospitalized patients, 2012–2017. N Engl J Med 382:1309–1319. doi: 10.1056/NEJMoa1914433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen HD, Groisman EA. 2013. The biology of the PmrA/PmrB two-component system: the major regulator of lipopolysaccharide modifications. Annu Rev Microbiol 67:83–112. doi: 10.1146/annurev-micro-092412-155751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clinical and Laboratory Standards Institute. 2021. Performance standards for antimicrobial susceptibility testing, 31st informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 33.Luo Q, Niu T, Wang Y, Yin J, Wan F, Yao M, Lu H, Xiao Y, Li L. 2019. In vitro reduction of colistin susceptibility and comparative genomics reveals multiple differences between MCR-positive and MCR-negative colistin-resistant Escherichia coli. Infect Drug Resist 12:1665–1674. doi: 10.2147/IDR.S210245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. 2020. Using SPAdes de novo assembler. Curr Protoc Bioinformatics 70:e102. doi: 10.1002/cpbi.102. [DOI] [PubMed] [Google Scholar]
- 36.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. 2018. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 39.Feldgarden M, Brover V, Gonzalez-Escalona N, Frye JG, Haendiges J, Haft DH, Hoffmann M, Pettengill JB, Prasad AB, Tillman GE, Tyson GH, Klimke W. 2021. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci Rep 11:12728. doi: 10.1038/s41598-021-91456-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zankari E, Allesøe R, Joensen KG, Cavaco LM, Lund O, Aarestrup FM. 2017. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother 72:2764–2768. doi: 10.1093/jac/dkx217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brynildsrud O, Bohlin J, Scheffer L, Eldholm V. 2016. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol 17:238. doi: 10.1186/s13059-016-1108-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petit RA, III, Read TD. 2020. Bactopia: a flexible pipeline for complete analysis of bacterial genomes. mSystems 5:e00190-20. doi: 10.1128/mSystems.00190-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bayliss SC, Thorpe HA, Coyle NM, Sheppard SK, Feil EJ. 2019. PIRATE: a fast and scalable pangenomics toolbox for clustering diverged orthologues in bacteria. Gigascience 8:giz119. doi: 10.1093/gigascience/giz119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Letunic I, Bork P. 2021. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seemann T. 2018. snp-dists - pairwise SNP distance matrix from a FASTA sequence alignment. https://github.com/tseemann/snp-dists.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.02522-21-s0001.pdf, PDF file, 1.4 MB (1.4MB, pdf)
Data Availability Statement
All local GA sequence data are available on NCBI under BioProject PRJNA288601 (GA isolates). Accession numbers for global accessions are found in Table S5 in the supplemental material.