Abstract
Previously, we identified a gene (aldA) from Myxococcus xanthus, which we suggested encoded the enzyme alanine dehydrogenase on the basis of similarity to known Ald protein sequences (M. J. Ward, H. Lew, A. Treuner-Lange, and D. R. Zusman, J. Bacteriol. 180:5668–5675, 1998). In this study, we have confirmed that aldA does encode a functional alanine dehydrogenase, since it catalyzes the reversible conversion of alanine to pyruvate and ammonia. Whereas an aldA gene disruption mutation did not significantly influence the rate of growth or spreading on a rich medium, AldA was required for growth on a minimal medium containing l-alanine as the major source of carbon. Under developmental conditions, the aldA mutation caused delayed aggregation in both wild-type (DZ2) and FB (DZF1) strains. Poorly formed aggregates and reduced levels of spores were apparent in the DZ2 aldA mutant, even after prolonged development.
Myxococcus xanthus is a gram-negative bacterium that undergoes a starvation-induced developmental cycle. The developmental program results in the morphogenesis of vegetative rod-shaped cells into spherical myxospores within multicellular aggregates termed fruiting bodies. Starvation for certain amino acids, carbon, energy, or inorganic phosphate is sufficient to initiate the process. The first obvious signs of development occur after approximately 6 h of starvation, at which time the cells start streaming into aggregation centers. Mounds develop as more and more cells enter the aggregates, which then darken into fruiting bodies as the cells differentiate into spores.
Amino acids, rather than sugars, are of particular nutritional importance to M. xanthus, since this species hydrolyzes protein as both an energy and a carbon source. Dworkin (5) showed that M. xanthus can, in fact, grow on a mixture of amino acids, which are present as the only organic constituent of the growth medium. Certain amino acids (methionine, leucine, isoleucine, and valine) are specifically required for growth. The remainder are degraded to acetate, pyruvate, and other tricarboxylic acid cycle intermediates to act as sources of energy, carbon, and nitrogen (1). Conversely, during vegetative growth, glucose is not converted into pyruvate for use as an energy source, and carbon from glucose is not incorporated into the cell (7, 18).
Many of the nutritional conditions and events which initiate development in M. xanthus also initiate development in other bacteria, including Bacillus subtilis. For example, after starvation for amino acids, both B. subtilis and M. xanthus transiently decrease the cellular GTP pool, while increasing (p)ppGpp (guanosine 3′-di[tri]phosphate-5′-diphosphate) levels (10, 11). The enzyme alanine dehydrogenase, which catalyzes the reversible conversion of alanine to pyruvate and ammonia, has also been shown to be required for normal sporulation in B. subtilis (14). During sporulation, this enzyme is thought to be involved in the generation of pyruvate from alanine for the production of energy by metabolism through the tricarboxylic acid cycle. Alanine dehydrogenase activity has previously been reported in M. xanthus (9), and while studying a region of DNA involved in directed motility, we identified an open reading frame which could encode this enzyme (16). In this study, we confirm that this open reading frame, renamed aldA, does indeed encode a functional alanine dehydrogenase and show that cells with a mutation in this gene have developmental defects. These defects seem unlikely to be due to a lack of pyruvate in developing cells but may be the result of an increased cellular pool of l-alanine.
Identification of a potential transcriptional start site for the aldA gene.
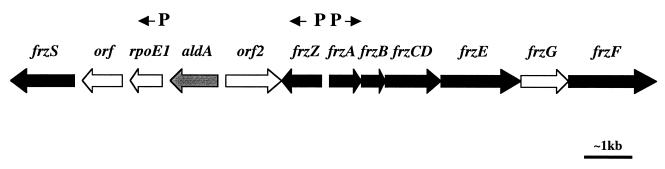
Further analysis of the aldA gene was performed to determine whether the gene, because of its chromosomal location near the frz genes (Fig. 1), might be functionally associated with the Frz signal transduction system and thereby involved with regulating directed motility behavior. The M. xanthus aldA gene was suggested to be 1,125 bp long (GenEMBL accession no. AF049107) based on preferential codon usage, which is a good indicator of translational potential in GC-rich genomes (13). The proposed GTG start codon lies 39 bp (13 codons) downstream of an in-frame TAG stop codon, and no alternative start codons are present in the intervening sequence. A potential ribosome binding site (GGAGG) was located 6 bp upstream of the proposed start codon (16).
FIG. 1.
Map of the region surrounding aldA, showing frz genes (black) both upstream and downstream of the aldA gene (grey). Proposed ςA-dependent promoter sites (P) are shown. Arrows denote the direction of gene transcription.
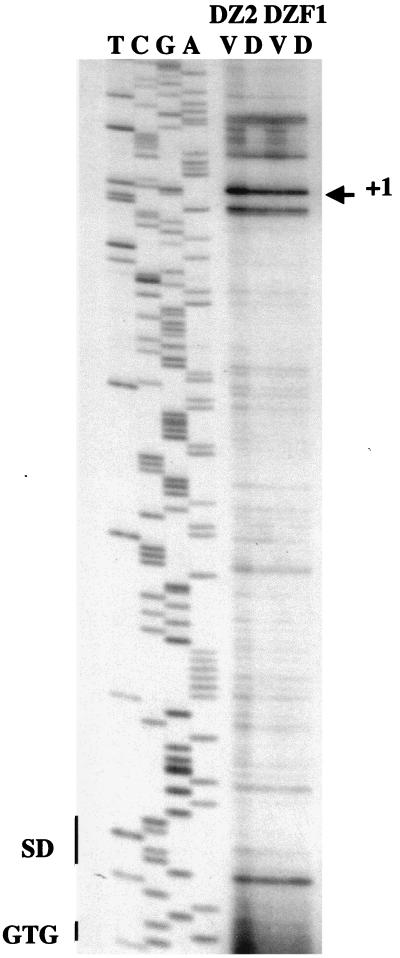
In this study, we have used primer extension analysis to identify potential transcriptional start sites for aldA. Several potential start sites were identified approximately 100 bp upstream of the GTG translational start codon, and the one showing the strongest signal was designated +1 as the most frequently used start site (Fig. 2). Most of the potential start sites were seen to be present in both vegetative (V) and developmental (D; 6 h of starvation) RNA transcripts prepared from both DZ2 (wild type) (4) and DZF1 (FB) (3) strains (Fig. 2). While several additional faint start sites were present in the vegetative extracts (Fig. 2), most of the transcripts appeared to be present at similar levels in both vegetative and developmental samples, suggesting that the aldA gene is transcribed similarly during vegetative growth and early development. However, we were unable to identify a potential promoter upstream of this group of proposed transcriptional start sites. The primer extension analysis also identified four additional transcriptional start sites (not shown) upstream of those indicated in Fig. 2. These results suggest that there may be multiple promoters upstream of aldA. A potential transcriptional terminator structure (CGGATG-4 bp-CATCCG) was identified downstream of the aldA gene, in the aldA-rpoE1 intergenic region (16).
FIG. 2.
Primer extension analysis of the region upstream of the aldA translational start codon, highlighting the strongest of the proposed transcriptional start sites (+1). The translational start codon (GTG) is shown next to the sequencing ladder (TCGA), with the potential Shine-Dalgarno (SD) site (GGAGG) shown upstream. RNA from vegetative (V) and developmental (D) M. xanthus cells was prepared with the RNeasy kit (QIAGEN). RNA was diluted to 1 mg/ml in RNase-free water for primer extension analysis. Primer extensions were performed as described previously (16), using 6 μg of RNA for cDNA synthesis. The oligonucleotide 5′-GGTTTTGATCTCCTTGGG (17 pmol) was used as the primer for this reaction. Sequencing reactions for primer extension analysis were performed, using the same primer, by the dideoxynucleotide chain termination method, using Sequenase (U.S. Biochemical Corp.) and [α-35S]dATP (410 Ci mmol−1; Amersham) on double-stranded DNA. Sequencing and primer extension reactions were run for 2 h on a 6% polyacrylamide gel prepared with National Diagnostics Sequagel reagents.
Construction of aldA mutants.
The aldA gene encodes a protein containing 374 amino acids with strong homology (58% identity) to the enzyme alanine dehydrogenase from B. subtilis (14). Since alanine dehydrogenase activity has previously been identified in crude extracts of M. xanthus (9), insertion mutations were constructed in the aldA gene in order to analyze both wild-type and mutant cell extracts for enzyme activity. M. xanthus DZ2 (wild type) and DZF1 (pilQ1) were used for the construction of aldA mutants DZ24280 (DZ2 aldA) and DZF4281 (DZF1 pilQ1 aldA). Mutants were constructed by cloning a 750-bp internal fragment of the aldA gene into the 3.3-kb kanamycin resistance-encoding vector, pZErO-2 (Invitrogen), to create the plasmid pZALD. The internal fragment of the gene was prepared by PCR, using Taq polymerase (Promega). The following primer pair combination was used for the amplification: forward primer, 5′-CGGACGAGGTCTGGAAGCGC (bp 182 to 201 from the GTG start codon); reverse primer, 5′-AGGTGGACGTCTGCGGCACG (bp 915 to 934 from the GTG start codon). The plasmid pZALD was electroporated into M. xanthus strains. Growth on CYE agar (3) containing 25 μg of kanamycin per ml was used to select for mutants. Since the vector is unable to replicate in M. xanthus, kanamycin resistance could be maintained after integration of the plasmid into the host chromosome only by homologous recombination. Chromosomal DNA was prepared from both mutant and parent strains, and insertions within the aldA gene were confirmed by Southern blotting. The hybridization probe was constructed by PCR amplification of the internal 750-bp fragment of aldA, incorporating the hapten digoxigenin-11-dUTP (Boehringer Mannheim), and detection was performed by enzyme immunoassay and an enzyme-catalyzed color reaction. The aldA gene is present on a 3.3-kb SphI fragment in the wild-type strains. This fragment was missing and replaced by two SphI fragments of approximately 3.7 kb each in the mutant strains (not shown). The positioning of the aldA gene upstream of three genes (rpoE1, orf5, and frzS) in the same transcriptional orientation might suggest that all four genes are part of an operon and therefore that mutations in aldA could have polar effects on downstream gene expression. However, we have previously identified a strong transcriptional start site upstream of the rpoE1 gene (16). This transcriptional start site was present in both vegetative and developmental extracts and was shown to be positioned downstream of a potential ςA-dependent promoter. Therefore, it seems likely that the genes downstream of aldA can be transcribed independently of aldA.
The aldA gene encodes a functional alanine dehydrogenase.
To determine whether the aldA gene encodes a functional alanine dehydrogenase, we prepared crude cell extracts from vegetatively growing cultures and assayed them for enzyme activity by the protocol of Kottel et al. (9). This assay measures the reaction in which pyruvate and ammonia are converted to alanine and requires NADH, which is oxidized to NAD+. Alanine dehydrogenase activity was measured by monitoring the loss in absorbancy at 340 nm in a cuvette containing the following: 5 mM potassium phosphate buffer (pH 7.5), 52 mM NH4Cl, 0.1 mM reduced NAD, 100 μg of cell extract, and 5.2 mM sodium pyruvate. While both the wild-type DZ2 and DZF1 extracts showed rapid oxidation of NADH, as indicated by the loss of absorbance at 340 nm (−0.8 absorbance [Abs]/min/mg of crude protein for DZ2 and −0.73 Abs/min/mg of crude protein for DZF1), the aldA mutant extracts showed only low level residual activity (<−0.08 Abs/min/mg of crude protein). These results show that the aldA gene does, indeed, encode a functional alanine dehydrogenase.
Effects of aldA mutations on vegetative growth and spreading.
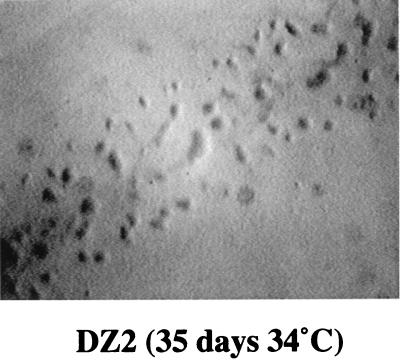
Since alanine dehydrogenase is required for the conversion of alanine to pyruvate, which can then be utilized as an energy and carbon source, we measured the ability of the wild-type DZ2 and the DZ2 aldA mutant to grow on a modified version of A1 minimal medium (1) in which the normal carbon sources, pyruvate and aspartate, were replaced by l-alanine. Figure 3 shows that, after 35 days of incubation, single colonies of the wild-type DZ2 were growing on this medium, whereas no growth was visible on the plates inoculated with the DZ2 aldA mutant. This confirms that the aldA gene is required for growth when l-alanine is the major carbon source.
FIG. 3.
Growth of DZ2 cells streaked on A1 minimal medium containing l-alanine as the major carbon source. Minimal A1 medium was prepared as described by Bretscher and Kaiser (1) with the following modification: the major carbon sources, sodium pyruvate and potassium aspartate, were replaced by 10 mg of l-alanine per ml. Media were solidified, using 0.8% ultrapure agarose (Gibco BRL). Cells were incubated at 34°C for 35 days and photographed with a dissecting scope at a magnification of ×12.
Under nutrient-rich conditions, the aldA mutation was determined to have a minimal effect on growth rate, since both parent and mutant strains showed similar doubling times of just over 4 h in CYE liquid medium. Liquid cultures (50 ml) were grown in 500-ml Erlenmeyer flasks, with a side arm, and doubling times were calculated between Klett units of 50 to 100 (approximately 2 × 108 to 4 × 108 CFU/ml). In a representative experiment, DZ2 had a doubling time of 4 h 3 min, while the DZ2 aldA mutant doubled in 4 h 12 min. Similarly, the DZF1 strain had a doubling time of 4 h 6 min, while the DZF1 aldA mutant had a doubling time of 4 h 9 min.
The aldA mutation was also shown to cause only slight differences in cell spreading behavior on CYE agar. Cells were concentrated to 4 × 109 CFU/ml, prior to spotting (5-mm-diameter spots) on plates. Both DZ2 and DZ2 aldA cells spread identically on plates containing 1.5% agar (spread diameter of 9 mm after 18 h of incubation), whereas a small difference in spreading diameter was apparent on plates containing 0.3% agar (DZ2 spreading diameter, 12 mm; DZ2 aldA spreading diameter, 10 mm). Spreading movement on 1.5% agar was slightly reduced in the DZF1 aldA mutant. After 6 days of growth, the DZF1 colony had a diameter of 12 mm, while that of the DZF1 aldA mutant was only 9 mm (not shown). Spreading in the DZF1 background was not analyzed on low-percentage agars, since the DZF1 strain contains a leaky social motility defect (pilQ1) which results in poor movement on soft agar (12, 15). The slightly reduced spreading phenotypes of the aldA mutants could be due to the marginally reduced growth rates on nutrient-rich CYE medium but seem unlikely to be associated with the Frz signal transduction system and directed motility behavior, since mutations in the frz genes result in significantly reduced and disorganized spreading (17).
Developmental effects of aldA mutations.
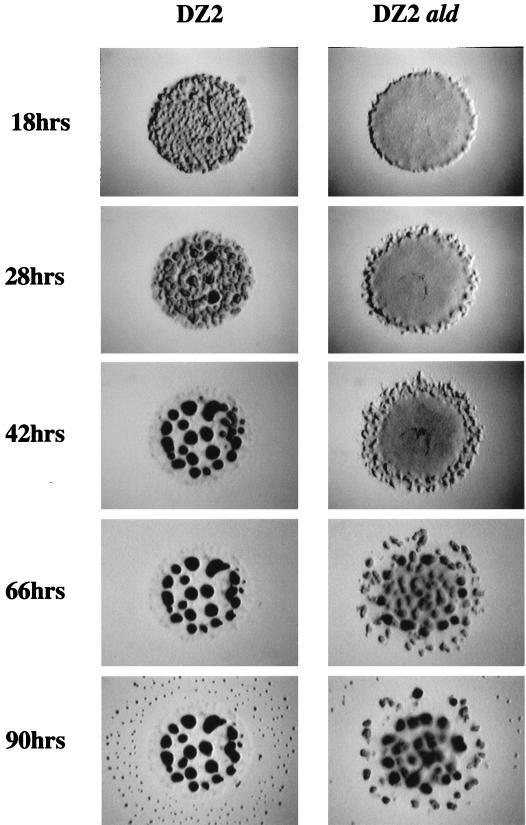
The most striking effect of the aldA mutation was seen under developmental conditions. In the DZ2 background, the aldA mutation delayed aggregation by more than 24 h (Fig. 4). The DZ2 aldA mutant did not form truly wild-type fruiting bodies. The aggregates remained poorly formed and did not darken even after prolonged (7-days) development, and a reduced number of spores was present in the fruiting bodies (approximately 10% of the wild-type number as evaluated microscopically). These spores were also less viable on germination than those of the DZ2 parent (germination was reduced approximately 10-fold when equivalent numbers of spores were plated on CYE agar). This phenotype is distinct from that of cells with mutations in the rpoE1 gene which show developmental defects only when plated at high cell density (16), confirming that the aldA mutant phenotype is unlikely to be due to polar effects on downstream gene expression. In the DZF1 background, aggregation was delayed 8 to 10 h with respect to the parent (not shown). However, after 4 days of development, the DZF1 aldA mutant had formed normal fruiting bodies containing wild-type levels of spores. These spores were shown to germinate at a frequency similar to that of DZF1 spores (data not shown). Currently, it is not known why a mutation in the pilQ gene should partially suppress the developmental defects of the aldA mutant. Mutations in the frz genes also result in both developmental aggregation and strain-dependent sporulation defects. However, since the frz phenotype is highly distinctive and dissimilar to that shown for the aldA mutants, we conclude that, although the aldA and frz genes are positioned close together on the chromosome, these genes are unlikely to be functionally associated. However, it is worth noting that the pilQ1 mutation also partially suppresses frz sporulation deficiencies (8).
FIG. 4.
Developmental aggregation of DZ2 and DZ2 aldA cells on CF fruiting medium. Cells for developmental analyses were concentrated to 4 × 109 CFU/ml, and then 5-μl volumes were spotted onto CF agar. Aggregation patterns were photographed at various time points. Spore counts were performed after removing the cells from CF agar and resuspending them in water. Spore clumps were dispersed by sonication, and appropriate dilutions were placed in a Petroff-Hausser chamber for counting under magnification. Germination frequencies were determined by plating known spore numbers (from spore counts) onto CYE agar and incubating the plates for 4 or 5 days at 34°C.
That mutations in ald genes result in developmental defects in both M. xanthus and B. subtilis suggests that alanine dehydrogenase might play a similar role during development in both species. However, in B. subtilis, it has been suggested that the ald mutation results in reduced levels of pyruvate, thereby depleting energy levels in the cell. This explanation seems unlikely in the case of M. xanthus, as the cells are provided with significant amounts of pyruvate (9.1 mM sodium pyruvate) in CF fruiting medium (6). Pyruvate is included in CF medium because cells are considered unlikely to suddenly encounter absolute starvation conditions in the natural environment. On the other hand, exclusion of pyruvate from CF medium did not significantly alter the timing of developmental aggregation in either the wild-type or aldA mutant strains (data not shown). This suggests that it may be the increased pool of l-alanine in the cell which results in the delayed aggregation and other developmental defects seen in M. xanthus aldA mutants. We are currently unaware of the physiological significance of increased levels of a single amino acid on the cell, and alternative explanations for the aldA mutant phenotype are certainly possible. However, in E. coli, alanine, as well as leucine, has been shown to have a strong effector role by stimulating Lrp, the leucine response regulator (2, 19). The possibility that M. xanthus could also utilize a global regulator, similar to Lrp, during development would be an interesting area for future research.
Acknowledgments
Research in our laboratory is supported by Public Health Service grant GM20509 from the National Institutes of Health.
REFERENCES
- 1.Bretscher A P, Kaiser D. Nutrition of Myxococcus xanthus, a fruiting bacterium. J Bacteriol. 1978;133:763–768. doi: 10.1128/jb.133.2.763-768.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calvo J M, Matthews R G. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol Rev. 1994;58:466–490. doi: 10.1128/mr.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campos J M, Geisselsoder J, Zusman D R. Isolation of bacteriophage Mx4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978;119:167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- 4.Campos J M, Zusman D R. Regulation of development in Myxococcus xanthus: effect of 3′:5′ cyclic AMP, ADP and nutrition. Proc Natl Acad Sci USA. 1975;72:518–522. doi: 10.1073/pnas.72.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dworkin M. Nutritional requirements for vegetative growth of Myxococcus xanthus. J Bacteriol. 1962;84:250–257. doi: 10.1128/jb.84.2.250-257.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hagen D C, Bretscher A P, Kaiser D. Synergism between morphogenic mutants of Myxococcus xanthus. Dev Biol. 1978;64:284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- 7.Hemphill H E, Zahler S A. Nutritional induction and suppression of fruiting in Myxococcus xanthus FBa. J Bacteriol. 1968;95:1018–1023. doi: 10.1128/jb.95.3.1018-1023.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kashefi K, Hartzell P L. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF defect. Mol Microbiol. 1995;15:483–494. doi: 10.1111/j.1365-2958.1995.tb02262.x. [DOI] [PubMed] [Google Scholar]
- 9.Kottel R H, Orlowski M, White D, Grutsch J. Presence of amino acid dehydrogenases and transaminases in Myxococcus xanthus during vegetative growth and myxospore formation. J Bacteriol. 1974;119:650–651. doi: 10.1128/jb.119.2.650-651.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez J M, Dromerick A, Freese E. Response of guanosine 5′-triphosphate concentration to nutritional changes and its significance for Bacillus subtilis sporulation. J Bacteriol. 1981;146:605–613. doi: 10.1128/jb.146.2.605-613.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manoil C, Kaiser D. Accumulation of guanosine tetraphosphate and guanosine pentaphosphate in Myxococcus xanthus during starvation and myxospore formation. J Bacteriol. 1980;141:297–304. doi: 10.1128/jb.141.1.297-304.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi W, Zusman D R. The two motility systems of Myxococcus xanthus show different selective advantages on various surfaces. Proc Natl Acad Sci USA. 1993;90:3378–3382. doi: 10.1073/pnas.90.8.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimkets L J. The myxobacterial genome. In: Dworkin M, Kaiser D, editors. Myxobacteria II. Washington, D.C.: American Society for Microbiology; 1993. pp. 85–107. [Google Scholar]
- 14.Siranosian K J, Ireton K, Grossman A D. Alanine dehydrogenase (ald) is required for normal sporulation in Bacillus subtilis. J Bacteriol. 1993;175:6789–6796. doi: 10.1128/jb.175.21.6789-6796.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wall D, Kelenbrander P E, Kaiser D. The Myxococcus xanthus pilQ (sglA) gene encodes a secretin homolog required for the type IV pilus biogenesis, social motility, and development. J Bacteriol. 1999;181:24–33. doi: 10.1128/jb.181.1.24-33.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward M J, Lew H, Treuner-Lange A, Zusman D R. Regulation of motility behavior in Myxococcus xanthus may require an extracytoplasmic-function sigma factor. J Bacteriol. 1998;180:5668–5675. doi: 10.1128/jb.180.21.5668-5675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward M J, Zusman D R. Regulation of directed motility in Myxococcus xanthus. Mol Microbiol. 1997;24:885–893. doi: 10.1046/j.1365-2958.1997.4261783.x. [DOI] [PubMed] [Google Scholar]
- 18.Watson B F, Dworkin M. Comparative intermediary metabolism of vegetative cells and microcysts of Myxococcus xanthus. J Bacteriol. 1968;96:1465–1473. doi: 10.1128/jb.96.5.1465-1473.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhi J, Mathew E, Freundlich M. Lrp binds to two regions in the dadAX promoter region of Escherichia coli to repress and activate transcription directly. Mol Microbiol. 1999;32:29–40. doi: 10.1046/j.1365-2958.1999.01314.x. [DOI] [PubMed] [Google Scholar]