ABSTRACT
We conducted a pilot whole genome sequencing (WGS) study to characterize the genotypes of nine methicillin resistant staphylococci (MRS) isolates recovered from goats and their farm environments in Eastern Province, Saudi Arabia, between November 2019 to August 2020. Seven out of nine isolates were methicillin resistant Staphylococcus aureus (MRSA), and two were methicillin resistant Staphylococcus epidermidis (MRSE). All MRSA isolates possessed genotypes previously identified to infect humans, including isolates harboring ST6-SCCmec IV-t304 (n = 4), ST5-SCCmec VI- t688 (n = 2) and ST5-SCCmec V-t311 (n = 1). 2 MRSA isolates possessed plasmids that were genetically similar to those identified in S. aureus isolates recovered from humans and poultry. In contrast, plasmids found in three MRSA isolates and one MRSE isolate were genetically similar to those recovered from humans. All MRSA isolates harbored the host innate modulate genes sak and scn previously associated with human infections. The genotypes of MRSE isolates were determined as ST35, a well-known zoonotic sequence type and ST153, which has been associated with humans. However, the MRSE isolates were untypeable due to extra ccr complexes identified in their SCCmec elements. Moreover, we identified in ST153 isolate SCCmec element also harbored the Arginine Catabolic Mobile Element (ACME) IV. All MRS isolates were phenotypically resistant to trimethoprim-sulfamethoxazole, an antibiotic for the decolonization of MRS. Three isolates carried antibiotic resistance genes in their SCCmec elements that were not previously described, including those encoding fusidic acid resistance (fusC) and trimethoprim resistance (dfrC) incorporated in the MRSA SCCmec VI.
IMPORTANCE Our findings demonstrate a possible cross-transmission of methicillin resistant staphylococci between goats and their local environments and between goats and humans. Due to ever increasing resistance to multiple antibiotics, the burden of MRS has a significant impact on livestock farming, public health, and the economy worldwide. This study highlights that implementing a holistic approach to whole genome sequencing surveillance in livestock and farm environments would aid our understanding of the transmission of methicillin resistant staphylococci and, most importantly, allow us to implement appropriate infection control and hygiene practices.
KEYWORDS: MRS, MRSA, MRSE, Saudi Arabia, environment, genotypes, goat
INTRODUCTION
Methicillin resistant staphylococci (MRS) have become a major threat to human and animal health due to the infections they cause and have become increasingly challenging to treat as these bacteria developed resistance to multiple antibiotics, predominantly due to the overuse of antibiotics in health care and agriculture settings (1–3). Many staphylococcal species are commensal to animals and humans but can act as opportunistic pathogens (4). Staphylococcus aureus and Staphylococcus epidermidis are common causes of staphylococcal infections in humans and animals and can transmit across host species barriers either via direct contact, the environment, or the food chain (5).
Methicillin resistance is derived from the mecA gene conferring resistance to all beta-lactam antibiotics, including carbapenems, cephalosporins, cephamycins, and monobactams. The mecA gene is believed to have originated from staphylococcal species of animal origin before successfully transferring across other staphylococci species via the SCCmec mobile genetic element (6, 7). The mecA gene has widely been associated with methicillin resistant S. aureus (MRSA); one of the leading causes of hospital-acquired infections in the world, as well as a common cause of infections in the community and livestock (3, 8, 9) and methicillin resistant S. epidermidis (MRSE), which, although lacking the arsenal of virulence factors of S. aureus, has been associated with nosocomial and medical device infections as well as with community and livestock infections (10–15).
Goats are common agricultural animals in many Middle Eastern countries and are used as a source of meat and milk. In Saudi Arabia, an estimated 3.7 million heads of goats in 2019 produced31,839 tons of meat and 68,694 tons of milk (16). Mastitis, the infection of the mammary gland, is common among goats and regularly caused by Staphylococcus aureus (17–19). Goat mastitis causes significant economic losses due to the reduced quality and quantity of milk and, in some cases, leads to the slaughter of the animal (20). The economic impact of mastitis in goats is further exacerbated by antibiotic resistance, making it harder to treat. A study published in 2015 reported that among the isolates recovered from the mastitis milk in the farms in Eastern Province, Saudi Arabia, 33.8% were S. aureus, of which 9.2% were MRSA compared to 4.2% and 0.6%, respectively, recovered for normal milk (21). In addition, the authors (21) found that 17.9% of the isolates recovered from nasal swabs of diseased animals were S. aureus, of which 2.6% were MRSA compared to 10.2% and 0.8%, respectively, of nasal swabs recovered from healthy animals. However, they provided no information on whether the genotypes of the MRSA isolates recovered from goats in Saudi Arabia were similar to those commonly found in livestock or associated with humans or the environment. In this study, we provide insights into the genotypes of MRS isolates recovered from goats and their farm environments in Saudi Arabia and analyze the genetic features to identify the possible source of transmission.
RESULTS
Speciation and genotyping of MRS isolates from goats and their environment.
In total, 57 staphylococci isolates were recovered from November 2019 to August 2020 from goats and their surrounding environments on a farm in Eastern Province, Saudi Arabia (Table S2). Nine out of 57 (15.7%) isolates that showed resistance to methicillin were sequenced (Table 1). Areas where MRS isolates were recovered from include goat's nasal swabs (n = 6), goat’s milk (n = 1), goat's drinking water (n = 1) and from the soil (n = 1). S. epidermidis (n = 2) was only isolated from the nasal swabs, whereas S. aureus (n = 7) was isolated from the nasal swabs (n = 4), goat milk (n = 1), drinking water (n = 1), and the soil (n = 1).
TABLE 1.
Speciation and genotyping of MRS isolates from goats and the surrounding environments
| Isolate no. | ID | Species | Sourcea | Isolation date | MLST | CC | Spa type | SCCmec |
|---|---|---|---|---|---|---|---|---|
| 1 | SE1 | S. epidermidis | GNS | 12/2019 | 153 | NAb | NA | mec class B with two ccr class 4 and one ccr class 2 |
| 2 | SE2 | S. epidermidis | GNS | 1/2020 | 35 | NA | NA | VI with an extra ccr class4 |
| 3 | SA1 | S. aureus | GM | 8/2020 | 5 | 5 | t311 | V |
| 4 | SA2 | S. aureus | GNS | 2/2020 | 5 | 5 | t688 | VI |
| 5 | SA3 | S. aureus | GNS | 1/2020 | 6 | 5 | t304 | IV |
| 6 | SA4 | S. aureus | DW | 1/2020 | 5 | 5 | t688 | VI |
| 7 | SA5 | S. aureus | GNS | 3/2020 | 6 | 5 | t304 | IV |
| 8 | SA6 | S. aureus | GNS | 11/2019 | 6 | 5 | t304 | IV |
| 9 | SA7 | S. aureus | Soil | 11/2019 | 6 | 5 | t304 | IV |
GNS = goat’s nasal swab, GM = goat’s milk, DR = drinking water.
NA means ‘not applicable’ as S. epidermidis does not have spa type of clonal clusters.
Whole genome sequencing analysis showed that all S. aureus isolates belonged to clonal complex (CC) 5. The most common S. aureus genotype isolated was ST6-MRSA-SCCmec IV-spa type t304 (ST6-MRSA-SCCmec IV-t304) (n = 4), which was identified in isolates recovered from nasal swabs (n = 3) and the soil (n = 1). Two isolates recovered from goat's nasal swab (n = 1) and drinking water (n = 1) possessed ST5-MRSA-SCCmec VI-t688 and one isolate recovered from goat's milk possessed ST5-MRSA-SCCmec VI - t311. In addition, S. epidermidis isolates belonged to ST153-MRSE-mec class B, with two ccr class 4 (ccrA4/B4 complex) and one ccr class 2 (ccrA2/B2 complex) (n = 1) and ST35-MRSE-SCCmec VI with an extra ccr class 4 (n = 1).
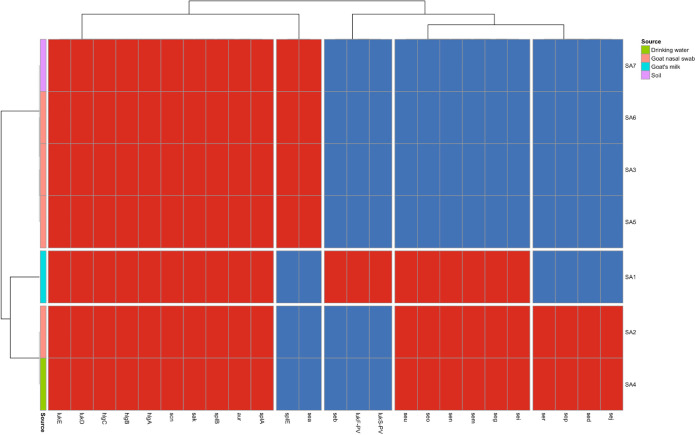
Determining the genetic similarities of MRS isolates recovered from goats and their environments.
To determine genetic similarities of MRS isolates recovered from goats and their surrounding environments, we performed digital DNA-DNA hybridization (dDDH), pairwise SNP distance of the core genome, and correlation between shared genes within the isolate's genome (Fig. 1). We found that the isolates cluster based on their species and MLST genotypes using all four methodologies. The dDDH scores of the MRSA isolates belonging to the same sequence type that has been recovered from goats and their surrounding environments were high (MRSA ST5 isolates dDDH range from 98.40 to 100% and MRSA ST6 isolates dDDH range from 99.4 to 100%) with small SNP differences (MRSA ST5 isolates SNP difference range 572-4 and MRSA ST6 isolates SNP difference range 162-1) and high correlation of genes (MRSA ST5 isolates gene correlation range r = 0.873-0.998 and MRSA ST6 isolates correlation range r = 0.950-1.0). Interestingly, we found that SA7 isolated from soil and SA6 from goat's nasal swab isolated at the same time (November 2019) were genetically identical by dDDH (dDDH = 100%), with a pairwise SNP distance of one and a correlation of r = 1 for genes they share. The SNP difference between SA7 and SA6 was a synonymous mutation within the Oxygen-dependent choline dehydrogenase gene betA. In addition, SA2 recovered from a goat nasal swab and SA4 from drinking water that were isolated 1 month apart (January 2020 and February 2020, respectively) were also shown to be genetically identical by dDDH (dDDH = 100%) with a pairwise SNP distance of 4 and a gene correlation of r = 0.998. Moreover, SA2 possessed four genes that had not previously been characterized and were not present in the SA2 genome and SA4 possessed one gene that had not been characterized and was not present in the SA2 genome.
FIG 1.
Hierarchy cluster of MRS isolates from goats and farm environments in Eastern Province, Saudi Arabia, showing genomic similarities between S. aureus isolates from different sources. (A) dDDH. (B) Pairwise SNP distance of the core genome containing 2,492,080 nucleotides for S. aureus isolates. (C) Pairwise SNP difference of the core genome containing 2,232,743 nucleotides for S. epidermidis isolates. (D) Gene correlation between isolates.
Antibiotic resistance genotyping and phenotyping.
All MRS isolates recovered from goats and surrounding environments tested against a panel of 14 antibiotics were resistant to benzylpenicillin, oxacillin and cefoxitin (all penicillin class of antibiotics); levofloxacin, moxifloxacin, and ciprofloxacin (all fluoroquinolones class of antibiotics) and trimethoprim-sulfamethoxazole (diaminopyrimidines- sulfonamide class of antibiotic) (Fig. 2A). However, all isolates were susceptible to rifampicin (antimycobacterial class of antibiotic), tigecycline (glycylcyclines class of antibiotic), quinupristin-dalfopristin (streptogramin class of antibiotics), gentamicin (aminoglycoside class of antibiotic), and clindamycin (macrolide class of antibiotic). We found that SA7 and SA6 differed in their resistance profile (SA7 was resistant to erythromycin [macrolide class of antibiotic] and tetracycline [tetracycline class of antibiotic], whereas SA6 was sensitive) despite being nearly genetically identical. All MRS isolates were sensitive to rifampicin, tigecycline, quinupristin-dalfopristin, gentamicin, and clindamycin. We found S. epidermidis isolates were resistant to 9 (SE2) and 8 (SE1), whereas S. aureus isolates were resistant to 9 (n = 2: SA1 and SA7) and 7 (n = 6: SA2, SA3, SA4, SA5, and SA6) antibiotics.
FIG 2.
Hierarchy cluster heatmap of MRS isolates from goats and surrounding farm environments in Eastern Province, Saudi Arabia. (A) Antibiotic resistance phenotype profiles. (B) Genotype resistance phenotypes. Red tiles presence of antibiotic resistance genotype/phenotype; blue tiles absence of antibiotic resistance phenotype/genotype.
Antibiotic resistance genotyping (Fig. 2B) showed that all MRS isolates possessed not only the mecA gene but also blaZ (encoding for beta-lactam resistance) and norA genes (encoding for fluoroquinolones resistance) and mgrA (genes responsible for regulating norA). In addition, lmrS (encoding for aminoglycoside, macrolide antibiotic, phenicol, diaminopyrimidine and oxazolidinone resistance), mepA and mepR (encoding for tetracycline and glycylcycline resistance), arlR, and arlS (genes responsible for regulating norA), and tet(38) (encoding for tetracycline resistance) were all present in all sequenced S. aureus isolates but were absent in S. epidermidis isolates. Other antibiotic resistance genes detected were mecR1 (regulator gene for mecA) in S. epidermidis (n = 2) and in S. aureus (n = 6); dfrC (diaminopyrimidine resistance) and fusC (fusidic acid resistance) in S. epidermidis (n = 2) and in S. aureus isolates (n = 3); fosB (fosfomycin resistance) in S. aureus isolates (n = 3); fexA (phenicol resistance) in S. aureus isolates (n = 2); tetM (tetracycline resistance) in S. aureus isolates (n = 2) and S. epidermidis isolates (n = 2); msrA (streptogramin, macrolide and streptogramin B resistance); mphC (macrolide resistance), fusB (fusidic acid resistance), APH(3′)-IIIa (aminoglycoside resistance), and tetK (tetracycline resistance) in an S. epidermidis isolate (n = 1), and dfrG (diaminopyrimidine resistance) in an S. aureus isolate (n = 1). Additionally, we detected antibiotic resistance conferring mutation parC S80F and gyrA S84L (fluoroquinolone resistances) in S. aureus (n = 2). S. epidermidis isolates had 12 (SE2) and 7 (SE1) genes/mutations that encode for antibiotic resistance, whereas S. aureus isolates had 16, (n = 2, SA2 and SA4), 15, (n = 1, SA1), 13 (n = 1, SA5), and 11 (n = 3, SA3, SA6, SA7) genes/mutations that encode for antibiotic resistance.
S. aureus virulence genotyping.
Twenty-five acquired virulence genes were detected in the sequenced MRSA isolates that were recovered from goats and the surrounding environments (Fig. 3). All isolates harbored toxin-producing genes, including hlgA, hlgB, hlgC, lukD, and lukE; exoenzyme genes aur, splA. and splB, and the host innate modulate genes sak and scn. Only SA1 isolate recovered from goat’s milk had the Panton-Valentine leucocidin toxin gene lukF-PV and lukS-PV and the Staphylococcus enterotoxin B gene seb. Overall, 20 (n = 2), 19 (n = 1) and 12 (n = 4) acquired virulence genes were detected S. aureus isolates.
FIG 3.
Acquired virulence genes of MRSA isolates recovered from goats and their surrounding farm environments in Eastern Province, Saudi Arabia.
Plasmids found in MRS isolates recovered from goats and the environment.
All isolates except for SA1 harbored a single plasmid (Fig. 4). Two different plasmids were identified in S. aureus isolates, and two different plasmids were identified in S. epidermidis isolates. Plasmid SA1 (pSA1) was detected in isolates (SA2 and SA4) carrying the ST5-MRSA-SCCmec VI -t688 genotype, whereas plasmid SA2 (pSA2) was found in all isolates (SA5, SA6, and SA7) with the ST5-MRSA-SCCmec VI -t688 genotype. Plasmid SE1 (pSE1) was identified in isolate SE1 and plasmid SE2 (pSE2) was identified in an isolate SE2 only.
FIG 4.
Maps of four plasmids isolated from MRS from goats and their surrounding farm environments in Eastern Province, Saudi Arabia.
Plasmid pSA1 and pSA2 (Fig. 4A and B) shared the Tn552 transposon; the beta-lactamase gene operon (blaZ, blaI, and blaR), cadmium metal resistance gene cadD, along with the metalloregulatory transcriptional repressors ArsR/SmtB. The only gene that pSE1 (Fig. 4C) shared with pSA1 and pSA2 was the Tn552 transposon. However, pSE did contain cadA and cadC genes responsible for cadmium resistance. We found that pSE2 did not share any genes with the other plasmids and that overall, the plasmid only contained three genes, of which one was hypothetical protein, the other two being the replication initiation protein and the general bacterial stress response protein csbD. Genes unique to pSA1 included the rep20 plasmid replication gene, three endotoxin genes (sed, ser, and sej), the marR family transcriptional regulator, putative replication-associate protein, recombinase gene binIII and the oxidoreductase gene. Genes that were unique to pSA2 were the two plasmid replication genes (rep16 and rep5a) and two recombinase genes, DNA invertase (hin), the lysR transcription factor, a helix-turn-helix transcriptional regulator gene and Lactococcin bacteriocin gene, the yxeA gene, which has not been charactered and the ABC transporter ATP binding protein. Genes unique to pSE1 were the rep19a plasmid replicon gene, the restriction-modification methylase Eco57I gene, a methylase gene, TIGR00730 family Rossman fold protein gene; the insertion sequence IS431mec, thyA gene involved in the biosynthesis of thymidylate, dfrC gene responsible for trimethoprim resistance, and the degV gene that binds long-chain fatty acids.
To investigate the novelty of plasmids found in our MRS isolates recovered from goats and the surrounding environment, we used PLSDB mash distance analysis (Tables S3, S4, S5 and S6) and blast (Fig. 5). We found that the plasmids found in MRSA isolates in this study have previously been reported in isolates recovered in different countries and sources. Plasmid pSA1 had 73 isolates with shared hashes of >900 (Table S3). Most of these plasmids were found in S. aureus isolates recovered from humans, including clinical samples (e.g., blood) in the USA isolated (accession no: CP030594); however, we did find pSA1 had hits with S. aureus isolates recovered from broiler chickens in Belgium (accession no: MH785250) and a bakery environment in the USA (accession no: CP045867) (Fig. 5A). Plasmid pSA2 had four hits with shared hashes of >900 with S. aureus isolates recovered from isolates from human and hospital wards in Denmark (accession no: CP047022), the USA (accession no: CP049373) and China (Fig. 5B) (Table S4). We also found that pSE2 from MRSE isolate SE2 had three hits with shared hashes >900 with S. epidermidis isolates recovered from France, Germany, and South Korea (Fig. 5D) (Table S6). Two of the isolates were recovered from humans (accession no: CP066372 and CP052957), whereas another isolated was recovered from a sofa in South Korea (accession no: CP069222). In addition, there were no isolates on the database with shared hashes >900 with the pSE1 found in MRSE SE1 in this study (Table S5). All 23 isolates which had hits for pSE1 were used for blast comparison to determine the genes they shared (Fig. 5C). We found that none of the plasmids shared the Eco57I, methylase gene, or the TIGR00730 family Rossman fold protein gene, along with three uncharacterized genes.
FIG 5.
BLAST analysis of plasmids that had hits from MASH distance analysis showing pSA1, pSA2 and pSA3 were not novel plasmids; however, pSE1 plasmid appeared to be novel due to low similarities to other plasmids on the database. (A) pSA1, (B) pSA2, (C) pSE1, and (D) pSE2.
SCCmec structure of MRS isolates from public settings.
Seven out of nine MRS isolates sequenced (recovered from goats and the surrounding farm environments) were assigned to the SCCmec typing scheme (Fig. 6). However, we found that S. epidermidis isolates (Fig. 6D and E) possessed an additional ccr complex in their SCCmec elements and SE1 harbored the Arginine catabolic mobile element (ACME) IV. In SE1, the ACME, ccr class 2 and class 4 complexes were separated by direct repeat (DR) sequences. Moreover, we found that SCCmec V and SCCmec with class B mec complex and two class 4 and one class 2 ccr complexes carried the fusidic acid resistance gene fusC (Fig. 6A and D); the SCCmec VI (SA2 and SA4) carried fusC and the trimethoprim resistance gene dfrC (Fig. 6B) and SCCmec IV with an extra class 4 ccr complex (SE2) carried fusC and downstream of the second DR sequence adjacent to a plasmid recombination gene are the tetracycline resistance gene tetK (Fig. 6E).
FIG 6.
SCCmec structure found in the MRS isolates recovered from goats and their surrounding farm environments in Eastern Province, Saudi Arabia. (A) SCCmec V found in SA1, (B) SCCmec VI found in SA2 and SA4, (C) SCCmec VI found in SA3, SA5, SA6, SA7. (D) SCCmec with class B mec complex and two class 4, one class 2 ccr complexes and ACME IV element found in SE1. (E) SCCmec IV with an extra class 4 ccr complex found in SE2. DR = direct repeat sequence of the SCCmec attachment sites.
DISCUSSION
MRS can be transmitted across to an animal host from the environment and act as an intermediate for intraspecies and interspecies transmission. This study found that MRSA isolates recovered from goats were genetically similar to those isolated from their local farm environments. It is plausible to hypothesize that these MRSA isolates were transmitted to goats from the farm environment or vice versa. It is also plausible to suggest that CC5 MRSA transmission occurred between goats indirectly through an environmental intermediate based on the low SNP diversity and high gene correlation between goat and environmental isolates. Previous studies have shown that MRSA isolates were recovered from farm environments, including Danish minks and pig farms and Italian dairy cattle herds (22–24). These studies showed that the MRSA isolates recovered from the farm environments were similar to lineages associated with pigs (22–24). However, in this study, we found that the MRSA genotype ST5- SCCmec VI- t688 has been previously recovered from a patient in a hospital in Kuwait; ST6- SCCmec IV- t304 was genetically similar to that found in patients in hospitals in Oman and Egypt and the genotype ST5-SCCmec V-spa type t311 to that found in patients in a hospital in Italy but to the best of our knowledge unreported in livestock and companion animals (25–28). Therefore, it is most likely that these isolates were originally cross transmitted from humans to goats. This is unsurprising as there have been multiple reports of cross-transmission of the CC5 S. aureus lineage between humans, companion animals and livestock (29–32). Another indicator of human to animal transmission is the presence of the Panton-Valentine leukocidin toxin genes lukF-PV and lukS-PV found in SA3 and the staphylokinase gene scn and staphylococcal complement inhibitor gene sak found in all MRSA isolates, which are strongly associated with human infections and sporadically have been found in livestock (33–35). Furthermore, these isolates carried genetically similar plasmids found in S. aureus recovered from humans (including isolates from clinical samples, e.g., blood) and their environments in Europe, the USA, and Asia. However, we observed that the pSA1 plasmid found in ST5-MRSA-SCCmec VI- t688 genotype was also genetically similar to a plasmid (pLUH02) found in S. aureus ST5, which has been transmitted to poultry from humans, suggesting that this genotype may have further occurrence in other animals (31, 36). These isolates most likely have been transmitted to goats from local farmers based on the fusC gene found in four of the five SCCmec elements, as there was a high prevalence of these SCCmec genotypes in the Arabian gulf isolated from humans and livestock compared to other regions of the world (37–40).
We found that MRSE isolates recovered from goat nasal swabs possessing ST35 genotype have previously also been recovered from a patient in Iraq, shared bicycle in China, birds of prey in Portugal, associated with farmers and hospital-associated isolates in Belgium, clinical isolates in Portugal, environmental sources from Germany as well as isolates recorded on pubMLST (accessed 18 November 2021) from a human source in Germany, South Korea, and Russia (41–47). Another genotype identified in our study, ST153 has been previously reported in isolates recovered from samples of catheter-related bloodstream infections in Belgium, nasal swabs, subgingival sites and oral rinse in Ireland and an isolate recovered from human samples in Ireland, the USA, and Latvia (44, 48, 49). We, therefore, hypothesize that these two S. epidermidis sequence types are not regularly reported. However, these reports suggested that ST35 is shown to be a zoonotic isolate. Further surveillance of S. epidermidis in animals is required to understand whether these genotypes are truly zoonotic or whether the isolates were originally transmitted by farmers. In addition, we also identified high homology between the plasmids found in S. aureus and S. epidermidis isolates recovered from goats to those recovered from humans, further indicating that the source of MRSE isolates in goats may have been humans.
Phenotypic resistance to fluoroquinolones class of antibiotics was found in the MRS isolates recovered from goat and their farm environments. The correlation of fluoroquinolone resistance with MRS is not unusual, as reports have shown that the use of fluoroquinolones is a significant risk factor for MRSA isolation from patients, companion animals and livestock (50–54). Moreover, our findings that all MRS isolates were phenotypically resistant to trimethoprim-sulfamethoxazole, an antibiotic used for the MRSA decolonization and treatment, is worrying (55, 56). Reports of high rates of trimethoprim-sulfamethoxazole resistance within hospitals and communities are not uncommon; however, the prevalence of trimethoprim-sulfamethoxazole resistance in livestock is generally low and has been reported in only one study that 68.4% of the MRSA isolates recovered from swine in the South of Italy were trimethoprim-sulfamethoxazole resistance (57–59).
We found that the majority of the resistance phenotypes identified in all MRS isolates correlate with a known resistance genotype (norA gene for fluoroquinolones class of antibiotics, mecA for penicillin class of antibiotics, LmrS in all S. aureus and dfrC in all S. epidermidis for trimethoprim-sulfamethoxazole; mgrA, in S. aureus and S. epidermidis; mepA, mepR and tet(38) in S. aureus and tet(K) in SE2 for tetracycline class of antibiotics and lmrS in SA1 and SA7 and mphC and msrA in SE2 for erythromycin) (6, 60–66). Interestingly we found that the ST6-MRSA-SCCmec IV-t304 isolates (SA6 and SA7), recovered from goat nasal swab and soil samples were genetically identical except for one synonymous mutation. However, they were phenotypically different in their antibiotic resistance profile, as one isolate was resistant to erythromycin and tetracycline, whereas the other was sensitive to these antibiotics. We hypothesize that this may be due to the phenomenon known as “bias portioning” described in E. coli AcrAB-TolC multidrug efflux pump, which is distributed asymmetrically on the poles of the cell; when the cell divides, the mother cell inherits old poles that are phenotypically more effective at pumping out the antimicrobial drugs then the daughter cells (67). This may be similar to the multidrug efflux pumps mgrA, which can actively pump out the tetracycline class of antibiotics from the cell and lmrS, which can actively pump out the macrolides class antibiotics (erythromycin) from the cell. However, no experimental data have been reported to show that such a phenomenon occurs in staphylococci.
Finally, we found certain additional features in the SCCmec elements in MRS isolates recovered from goats that may pose further challenges in treating infections caused by these bacteria. This included two S. epidermidis isolates that did not fit into the standard SCCmec typing scheme due to having additional ccr complexes (68–70). Previous reports have shown that these SCCmec elements with additional ccr complexes can be isolated from clinical settings, communities, and human public environments (71–75). In addition, S. epidermidis isolates from bovine mastitis have been reported to carry a SCCmec IV element with an additional ccr class 4 complex similar to that found in our study (SE2) (76). Moreover, in this study, we identified a SCCmec element found in an MRSE isolate (SE1) that possessed 3 ccr complex (1 class 2 and 2 class 4 complexes) and the ACME IV element associated with increased ability to colonize the skin and mucosa, which was originally identified in a S. epidermidis ST153 isolate (49). The class 2 ccr complex and ACME IV element were separated by DR sequences attachment site of the SCCmec element in the host chromosome suggesting these genomic regions were acquired in separate horizontal transfer events (77). Based on previous studies, it is plausible that these adaptions found in such untypeable SCCmec elements may be advantageous for bacteria as multiple ccr complexes have increased susceptibilities to oxacillin or cefoxitin (72). Our analysis showed inclusion of multiple antibiotic resistance genes including fusC and dfrC (SA2 and SA4) on the same SCCmec element (SE1’s SCCmec IV+ ccr4 class and SA2 and SA4’s SCCmec VI), which to the best of our knowledge, has not been reported previously. Furthermore, we found a possible inclusion of fusC and tetK (SE2 SCCmec) via the same SCCmec element; however, the integration of tetK within this element may have arisen separately via a recombinant plasmid as tetK was found adjacent to plasmid recombination protein. The inclusion of multiple antibiotic resistance genes on SCCmec elements may have a significant impact on public and animal health, making it more challenging to treat as well as increasing the potential of further dissemination of these antibiotic genes to other isolates via these mobile genetic elements.
The genetic similarities of MRS isolates recovered from goats and their farm environments indicate a possible transmission via the environment. Moreover, there is a strong indicator that these MRS isolates may have been initially transmitted from humans based on their molecular genotypes and plasmids possessed. These MRS isolates have also shown to be phenotypically resistant to multiple antibiotics, including trimethoprim-sulfamethoxazole used for decolonization patients with MRSA, which reduces options or treatment for animals and patients infected with these isolates. There were also isolates carrying two other antibiotic resistance genes apart from mecA (a SCCmec element with fusC and tetK, and another SCCmec element with fusC and dfrC) within the SCCmec element, which has the potential to transfer horizontally across to other staphylococci, making them multidrug resistant. Further large scale and structured surveillance studies are warranted to further our understanding of the human-livestock-environment cross transmission of these bacteria to improve hygiene practices in the farms.
MATERIALS AND METHODS
Sampling and bacterial isolation.
A total of 200 samples were collected from goats (nasal swabs; n = 130 and milk samples; n = 40) and their surrounding environments (soil; n = 15 and drinking water; n = 15) in one of the goat farms located in Eastern Province, Saudi Arabia, between November 2019 to August 2020. Nasal swabs were collected from nostrils after thorough cleaning and disinfection of the external nares. The collected swabs were kept in Amie’s transport medium (Difco, BD, Franklin Lakes, NJ, USA) and transported to the laboratory for microbiological examination. Milk samples were collected from lactating does in a sterile screw-cap tube, following the standard methods described by the National Mastitis Council (NMC, 1990). Environmental samples (soil and drinking water) were collected from herds in a sterile screw-cap container. All samples were labeled and transported cooled to the laboratory in an icebox (4°C). Samples were plated onto blood agar, Baird–Parker agar (Oxoid, Basingstoke, Hampshire, UK) supplemented with egg yolk tellurite and mannitol salt agar (Oxoid, Basingstoke, Hampshire, UK) and then incubated aerobically at 37°C for 24 h. A single morphologically typical staphylococcal isolate per sample was purified on 5% sheep blood agar (Oxoid, Basingstoke, Hampshire, UK) and further characterized by Gram staining, coagulase, and catalase test. Identification of staphylococci isolates were further confirmed to the species level by Vitek 2 COMPACT system (bioMérieux, Marcy l'Etoile, France) using Gram-positive cards, following the manufacturer guidelines.
Antibiotic susceptibility testing.
The phenotypic resistance profiles of all staphylococci isolates were determined by an automated Vitek 2 COMPACT system using AST-GP67 card (bioMérieux, Marcy l'Etoile, France), following the manufacturer guidelines. MIC values were interpreted according to the recommendations of EUCAST, 2021(78).
Whole genome sequencing and genomic assembly.
Nine isolates that showed resistance to methicillin were selected and submitted for whole genome sequencing (WGS) by MicrobeNG (Birmingham, UK) using Illumina sequencing platforms (San Diego, CA, USA). Short read files were deposited in European Nucleotide Archives (ENA) under the study PRJEB49547. The accession numbers for each isolate are included in the supplementary data (Table S1).
The quality of reads from sequencing was analyzed using the fastQC software, and reads were trimmed using the Trimmomatic software by setting the phred cutoff to Q20 (79, 80). Genomes and plasmids were assembled from the trimmed paired-end reads using SPades 3.15.3 (81).
Genome annotation and genetic typing.
Genomes and plasmids were initially annotated using Prokka 1.14.6 and then further annotated for antibiotic resistance genes/mutations using the Comprehensive Antibiotic Resistance Database; acquired virulence genes using the VirulenceFinder 2.0 webserver and insertion sequence using ISfinder webserver (82–85).
For genetic typing isolates, the python script mlst was used (https://github.com/tseemann/mlst) with the most up to date database obtained from pubMLST (accessed July 2021) to determine their multilocus sequence type (MLST); the SCCmecFinder 1.2 webserver for typing isolate SCCmec elements and spaTyper 1.0 webserver to type S. aureus spa gene (86, 87). SCCmec attachment sites were identified by blast searching against a list of known attachment sites direct repeats (DR) (88).
Genomic and plasmid comparison.
A digital DNA-DNA hybridization (dDDH) analysis using the TYGS webserver was conducted to determine the overall similarities between the genomes (89). To determine the pairwise SNP distance of the core genomes between the isolates, the Parsnp 1.5.6 alignment tool was employed using the reference genome DSM 20231 (accession no.: CP011526.1) for S. aureus and the reference genome ATCC 144990 (accession no.: CP035288.1) for S. epidermidis isolates. In addition, the analysis enabled recombination filtered, and the snp-dists tool (https://github.com/tseemann/snp-dists) was used to convert alignment to SNP distance matrix (90). For gene correlation, the pangenome pipeline roary was used to group genes that have a blastp minimum of 90% and the r package “corr” (https://cran.r-project.org/web/packages/corrr/index.html) to determine their Pearson correlation coefficient (91).
The plasmid novelty was determined using the mash dist function on the PLSDB webserver (92). A blast comparison of plasmids that had hits of similarities was conducted and visualized using the CGviewer webserver (93).
Hierarchy clustered heatmap.
Hierarchy clustered heatmap for genomic comparison was constructed using the r package “pheatmap.”
Ethical approval.
All experimental procedures used in the current study were approved by the guidelines of the Ethics Committee at King Faisal University, Saudi Arabia (Approval no: DSR-691).
ACKNOWLEDGMENTS
This work is supported by the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia (No. 23). This work is also supported through the Annual Funding track by the Deanship of Scientific Research, Vice Presidency for Graduate Studies and Scientific Research, King Faisal University, Saudi Arabia (Project No. AN000691).
We have no conflicts of interest to declare.
Funding Statement
This work is supported by the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia (No. 23). This work is also supported through the Annual Funding track by the Deanship of Scientific Research, Vice Presidency for Graduate Studies and Scientific Research, King Faisal University, Saudi Arabia [Project No. AN000691].
Footnotes
Supplemental material is available online only.
Contributor Information
Wael El-Deeb, Email: weldeeb@kfu.edu.sa.
Hermine V. Mkrtchyan, Email: hermine.mkrtchyan@uwl.ac.uk.
Kunyan Zhang, University of Calgary.
REFERENCES
- 1.Dadgostar P. 2019. Antimicrobial resistance: implications and costs. Infect Drug Resist 12:3903–3910. doi: 10.2147/IDR.S234610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nguyen TH, Park MD, Otto M. 2017. Host response to Staphylococcus epidermidis colonization and infections. Front Cell Infect Microbiol 7:90. doi: 10.3389/fcimb.2017.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner NA, Sharma-Kuinkel BK, Maskarinec SA, Eichenberger EM, Shah PP, Carugati M, Holland TL, Fowler VG. 2019. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol 17:203–218. doi: 10.1038/s41579-018-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker K, Heilmann C, Peters G. 2014. Coagulase-negative staphylococci. Clin Microbiol Rev 27:870–926. doi: 10.1128/CMR.00109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butaye P, Argudín MÁ, Threlfall J. 2015. Introduction to antimicrobial-resistant foodborne pathogens, p 1–17. In Chen C-Y, Yan X, Jackson CR (ed), Antimicrobial Resistance and Food Safety. Academic Press, San Diego. [Google Scholar]
- 6.Miragaia M. 2018. Factors contributing to the evolution of mecA-mediated β-lactam resistance in staphylococci: update and new insights from whole genome sequencing (WGS). Front Microbiol 9:2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. 2016. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anjum MF, Marco-Jimenez F, Duncan D, Marín C, Smith RP, Evans SJ. 2019. Livestock-associated methicillin-resistant Staphylococcus aureus from animals and animal products in the UK. Front Microbiol 10:2136. doi: 10.3389/fmicb.2019.02136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kluytmans J, Harbarth S. 2020. MRSA transmission in the community: emerging from under the radar. Lancet Infect Dis 20:147–149. doi: 10.1016/S1473-3099(19)30539-0. [DOI] [PubMed] [Google Scholar]
- 10.Chu VH, Miro JM, Hoen B, Cabell CH, Pappas PA, Jones P, Stryjewski ME, Anguera I, Braun S, Muñoz P, Commerford P, Tornos P, Francis J, Oyonarte M, Selton-Suty C, Morris AJ, Habib G, Almirante B, Sexton DJ, Corey GR, Fowler VG, International Collaboration on Endocarditis-Prospective Cohort Study Group . 2009. Coagulase-negative staphylococcal prosthetic valve endocarditis–a contemporary update based on the International Collaboration on Endocarditis: prospective cohort study. Heart 95:570–576. doi: 10.1136/hrt.2008.152975. [DOI] [PubMed] [Google Scholar]
- 11.Kanai H, Sato H, Takei Y. 2014. Community-acquired methicillin-resistant Staphylococcus epidermidis pyelonephritis in a child: a case report. J Med Case Rep 8:415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loncaric I, Künzel F, Klang A, Wagner R, Licka T, Grunert T, Feßler AT, Geier-Dömling D, Rosengarten R, Müller E, Reissig A, Spergser J, Schwarz S, Ehricht R, Monecke S. 2016. Carriage of meticillin-resistant staphylococci between humans and animals on a small farm. Vet Dermatol 27:191–e48. doi: 10.1111/vde.12321. [DOI] [PubMed] [Google Scholar]
- 13.Otto M. 2009. Staphylococcus epidermidis—the “accidental” pathogen. Nat Rev Microbiol 7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tong C, Wu Z, Zhao X, Xue H. 2018. Arginine catabolic mobile elements in livestock-associated methicillin-resistant staphylococcal isolates from bovine mastitic milk in China. Front Microbiol 9:1031. doi: 10.3389/fmicb.2018.01031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng Y, He L, Asiamah TK, Otto M. 2018. Colonization of Medical Devices by Staphylococci. Environ Microbiol 20:3141–3153. doi: 10.1111/1462-2920.14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.FAOSTAT. https://www.fao.org/faostat/en/#data/QCL.
- 17.Koop G, Collar CA, Toft N, Nielen M, van Werven T, Bacon D, Gardner IA. 2013. Risk factors for subclinical intramammary infection in dairy goats in two longitudinal field studies evaluated by Bayesian logistic regression. Prev Vet Med 108:304–312. doi: 10.1016/j.prevetmed.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 18.McDougall S, Malcolm D, Prosser C. 2014. Prevalence and incidence of intramammary infections in lactating dairy goats. N Z Vet J 62:136–145. doi: 10.1080/00480169.2013.865294. [DOI] [PubMed] [Google Scholar]
- 19.Schaeren W, Maurer J. 2006. [Prevalence of subclinical udder infections and individual somatic cell counts in three dairy goat herds during a full lactation]. Schweiz Arch Tierheilkd 148:641–648. doi: 10.1024/0036-7281.148.12.641. [DOI] [PubMed] [Google Scholar]
- 20.Novac CS, Andrei S. 2020. The Impact of mastitis on the biochemical parameters, oxidative and nitrosative stress markers in goat’s milk: a review. Pathogens 9:882. doi: 10.3390/pathogens9110882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El-Deeb W, Fayez M, Elmoslemany A, Kandeel M, Zidan K. 2018. Methicillin resistant Staphylococcus aureus among goat farms in Eastern province, Saudi Arabia: Prevalence and risk factors. Preventive Veterinary Medicine 156:84–90. doi: 10.1016/j.prevetmed.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 22.The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 11.0, 2021. http://www.eucast.org.
- 23.Babraham Bioinformatics. FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Retrieved 16 November 2021.
- 24.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. 2020. Using SPAdes de novo assembler. Curr Protoc Bioinformatics 70:e102. doi: 10.1002/cpbi.102. [DOI] [PubMed] [Google Scholar]
- 26.Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen A-LV, Cheng AA, Liu S, Min SY, Miroshnichenko A, Tran H-K, Werfalli RE, Nasir JA, Oloni M, Speicher DJ, Florescu A, Singh B, Faltyn M, Hernandez-Koutoucheva A, Sharma AN, Bordeleau E, Pawlowski AC, Zubyk HL, Dooley D, Griffiths E, Maguire F, Winsor GL, Beiko RG, Brinkman FSL, Hsiao WWL, Domselaar GV, McArthur AG. 2020. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48:D517–D525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleinheinz KA, Joensen KG, Larsen MV. 2014. Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage 4:e27943. doi: 10.4161/bact.27943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 29.Siguier P, Varani A, Perochon J, Chandler M. 2012. Exploring bacterial insertion sequences with ISfinder: objectives, uses, and future developments. Methods Mol Biol 859:91–103. doi: 10.1007/978-1-61779-603-6_5. [DOI] [PubMed] [Google Scholar]
- 30.Jolley KA, Bray JE, Maiden MCJ. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaya H, Hasman H, Larsen J, Stegger M, Johannesen TB, Allesøe RL, Lemvigh CK, Aarestrup FM, Lund O, Larsen AR. 2018. SCCmecFinder, a web-based tool for typing of Staphylococcal Cassette chromosome mec in Staphylococcus aureus using whole-genome sequence data. mSphere 3:e00612-17. doi: 10.1128/mSphere.00612-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arora S, Li X, Hillhouse A, Konganti K, Little SV, Lawhon SD, Threadgill D, Shelburne S, Hook M. 2020. Staphylococcus epidermidis MSCRAMM SesJ is encoded in composite islands. mBio 11:e02911-19. doi: 10.1128/mBio.02911-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meier-Kolthoff JP, Göker M. 2019. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun 10:2182. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Treangen T, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galata V, Fehlmann T, Backes C, Keller A. 2019. PLSDB: a resource of complete bacterial plasmids. Nucleic Acids Res 47:D195–D202. doi: 10.1093/nar/gky1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grant JR, Stothard P. 2008. The CGView server: a comparative genomics tool for circular genomes. Nucleic Acids Res 36:W181–W184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Espinosa-Gongora C, Larsen J, Moodley A, Nielsen JP, Skov RL, Andreasen M, Guardabassi L. 2012. Farm-specific lineages of methicillin-resistant Staphylococcus aureus clonal complex 398 in Danish pig farms. Epidemiol Infect 140:1794–1799. doi: 10.1017/S0950268811002391. [DOI] [PubMed] [Google Scholar]
- 39.Fertner M, Pedersen K, Jensen VF, Larsen G, Lindegaard M, Hansen JE, Chriél M. 2019. Within-farm prevalence and environmental distribution of livestock-associated methicillin-resistant Staphylococcus aureus in farmed mink (Neovison vison). Vet Microbiol 231:80–86. doi: 10.1016/j.vetmic.2019.02.032. [DOI] [PubMed] [Google Scholar]
- 40.Locatelli C, Cremonesi P, Caprioli A, Carfora V, Ianzano A, Barberio A, Morandi S, Casula A, Castiglioni B, Bronzo V, Moroni P. 2017. Occurrence of methicillin-resistant Staphylococcus aureus in dairy cattle herds, related swine farms, and humans in contact with herds. J Dairy Sci 100:608–619. doi: 10.3168/jds.2016-11797. [DOI] [PubMed] [Google Scholar]
- 41.Alseqely M, Newton-Foot M, Khalil A, El-Nakeeb M, Whitelaw A, Abouelfetouh A. 2021. Association between fluoroquinolone resistance and MRSA genotype in Alexandria, Egypt. Sci Rep 11:4253. doi: 10.1038/s41598-021-83578-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boswihi SS, Udo EE, Al-Sweih N. 2016. Shifts in the clonal distribution of methicillin-resistant Staphylococcus aureus in Kuwait hospitals: 1992–2010. PLoS One 11:e0162744. doi: 10.1371/journal.pone.0162744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lamanna O, Bongiorno D, Bertoncello L, Grandesso S, Mazzucato S, Pozzan GB, Cutrone M, Chirico M, Baesso F, Brugnaro P, Cafiso V, Stefani S, Campanile F. 2017. Rapid containment of nosocomial transmission of a rare community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) clone, responsible for the Staphylococcal Scalded Skin Syndrome (SSSS). Ital J Pediatr 43:5. doi: 10.1186/s13052-016-0323-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Udo EE, Al-Lawati BA-H, Al-Muharmi Z, Thukral SS. 2014. Genotyping of methicillin-resistant Staphylococcus aureus in the Sultan Qaboos University Hospital, Oman reveals the dominance of Panton–Valentine leucocidin-negative ST6-IV/t304 clone. New Microbes New Infect 2:100–105. doi: 10.1002/nmi2.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fan Y, Wang X, Li L, Yao Z, Chen S, Ye X. 2016. Potential relationship between phenotypic and molecular characteristics in revealing livestock-associated Staphylococcus aureus in Chinese humans without occupational livestock contact. Front Microbiol 7:1517. doi: 10.3389/fmicb.2016.01517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Köck R, Schaumburg F, Mellmann A, Köksal M, Jurke A, Becker K, Friedrich AW. 2013. Livestock-Associated Methicillin-Resistant Staphylococcus aureus (MRSA) as Causes of Human Infection and Colonization in Germany. PLoS One 8:e55040. doi: 10.1371/journal.pone.0055040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lowder BV, Guinane CM, Ben Zakour NL, Weinert LA, Conway-Morris A, Cartwright RA, Simpson AJ, Rambaut A, Nubel U, Fitzgerald JR. 2009. Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proc Natl Acad Sci USA 106:19545–19550. doi: 10.1073/pnas.0909285106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taniguchi Y, Koide S, Maeyama Y, Tamai K, Hayashi W, Tanaka H, Iimura M, Suzuki M, Nagano Y, Arakawa Y, Nagano N. 2020. Predominance of methicillin-resistant Staphylococcus aureus SCCmec type II-CC5 and SCCmec type IV-CC1/CC8 among companion animal clinical isolates in Japan: Findings from phylogenetic comparison with human clinical isolates. J Glob Antimicrob Resist 20:253–259. doi: 10.1016/j.jgar.2019.08.016. [DOI] [PubMed] [Google Scholar]
- 49.Chambers HF. 2005. Community-associated MRSA—resistance and virulence converge. N Engl J Med 352:1485–1487. doi: 10.1056/NEJMe058023. [DOI] [PubMed] [Google Scholar]
- 50.Egyir B, Hadjirin NF, Gupta S, Owusu F, Agbodzi B, Adogla-Bessa T, Addo KK, Stegger M, Larsen AR, Holmes MA. 2020. Whole-genome sequence profiling of antibiotic-resistant Staphylococcus aureus isolates from livestock and farm attendants in Ghana. J Glob Antimicrob Resist 22:527–532. doi: 10.1016/j.jgar.2020.03.029. [DOI] [PubMed] [Google Scholar]
- 51.Vandenesch F, Naimi T, Enright MC, Lina G, Nimmo GR, Heffernan H, Liassine N, Bes M, Greenland T, Reverdy M-E, Etienne J. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying panton-valentine leukocidin genes: worldwide emergence. Emerg Infect Dis 9:978–984. doi: 10.3201/eid0908.030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bukowski M, Piwowarczyk R, Madry A, Zagorski-Przybylo R, Hydzik M, Wladyka B. 2019. Prevalence of antibiotic and heavy metal resistance determinants and virulence-related genetic elements in plasmids of Staphylococcus aureus. Front Microbiol 10:805. doi: 10.3389/fmicb.2019.00805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Monecke S, Müller E, Braun SD, Armengol-Porta M, Bes M, Boswihi S, El-Ashker M, Engelmann I, Gawlik D, Gwida M, Hotzel H, Nassar R, Reissig A, Ruppelt-Lorz A, Senok A, Somily AM, Udo EE, Ehricht R. 2021. Characterisation of S. aureus/MRSA CC1153 and review of mobile genetic elements carrying the fusidic acid resistance gene fusC. Sci Rep 11:8128. doi: 10.1038/s41598-021-86273-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raji MA, Garaween G, Ehricht R, Monecke S, Shibl AM, Senok A. 2016. Genetic Characterization of Staphylococcus aureus Isolated from Retail Meat in Riyadh, Saudi Arabia. Front Microbiol 7:911. doi: 10.3389/fmicb.2016.00911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Senok A, Slickers P, Hotzel H, Boswihi S, Braun SD, Gawlik D, Müller E, Nabi A, Nassar R, Nitschke H, Reissig A, Ruppelt-Lorz A, Mafofo J, Somily AM, Udo E, Ehricht R, Monecke S. 2019. Characterisation of a novel SCCmec VI element harbouring fusC in an emerging Staphylococcus aureus strain from the Arabian Gulf region. PLoS One 14:e0223985. doi: 10.1371/journal.pone.0223985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Senok AC, Somily AM, Slickers P, Raji MA, Garaween G, Shibl A, Monecke S, Ehricht R. 2017. Investigating a rare methicillin-resistant Staphylococcus aureus strain: first description of genome sequencing and molecular characterization of CC15-MRSA. IDR 10:307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Argudín MA, Vanderhaeghen W, Vandendriessche S, Vandecandelaere I, André F-X, Denis O, Coenye T, Butaye P. 2015. Antimicrobial resistance and population structure of Staphylococcus epidermidis recovered from animals and humans. Vet Microbiol 178:105–113. doi: 10.1016/j.vetmic.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 58.Kozitskaya S, Olson ME, Fey PD, Witte W, Ohlsen K, Ziebuhr W. 2005. Clonal Analysis of Staphylococcus epidermidis Isolates Carrying or Lacking Biofilm-Mediating Genes by Multilocus Sequence Typing. J Clin Microbiol 43:4751–4757. doi: 10.1128/JCM.43.9.4751-4757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miragaia M, de Lencastre H, Perdreau-Remington F, Chambers HF, Higashi J, Sullam PM, Lin J, Wong KI, King KA, Otto M, Sensabaugh GF, Diep BA. 2009. Genetic diversity of arginine catabolic mobile element in Staphylococcus epidermidis. PLoS One 4:e7722. doi: 10.1371/journal.pone.0007722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.PubMLST. Public databases for molecular typing and microbial genome diversity. https://pubmlst.org/. Retrieved 18 November 2021.
- 61.Sousa M, Silva N, Igrejas G, Silva F, Sargo R, Alegria N, Benito D, Gómez P, Lozano C, Gómez-Sanz E, Torres C, Caniça M, Poeta P. 2014. Antimicrobial resistance determinants in Staphylococcus spp. recovered from birds of prey in Portugal. Vet Microbiol 171:436–440. doi: 10.1016/j.vetmic.2014.02.034. [DOI] [PubMed] [Google Scholar]
- 62.Talat A, Khalid S, Majeed HAR, Khan AU. 2020. Whole-genome sequence analysis of multidrug-resistant Staphylococcus epidermidis ST35 strain isolated from human ear infection of an Iraqi patient. J Glob Antimicrob Resist 21:318–320. doi: 10.1016/j.jgar.2020.03.020. [DOI] [PubMed] [Google Scholar]
- 63.Xu Z, Liu S, Chen L, Liu Y, Tan L, Shen J, Zhang W. 2019. Antimicrobial resistance and molecular characterization of methicillin-resistant coagulase-negative staphylococci from public shared bicycles in Tianjin, China. J Glob Antimicrob Resist 19:231–235. doi: 10.1016/j.jgar.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 64.Cherifi S, Byl B, Deplano A, Nagant C, Nonhoff C, Denis O, Hallin M. 2014. Genetic characteristics and antimicrobial resistance of Staphylococcus epidermidis isolates from patients with catheter-related bloodstream infections and from colonized healthcare workers in a Belgian hospital. Ann Clin Microbiol Antimicrob 13:20. doi: 10.1186/1476-0711-13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O'Connor AM, McManus BA, Coleman DC. 2018. First description of novel arginine catabolic mobile elements (ACMEs) types IV and V harboring a kdp operon in Staphylococcus epidermidis characterized by whole genome sequencing. Infect Genet Evol 61:60–66. doi: 10.1016/j.meegid.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 66.Feltrin F, Alba P, Kraushaar B, Ianzano A, Argudín MA, Di Matteo P, Porrero MC, Aarestrup FM, Butaye P, Franco A, Battisti A. 2016. A livestock-associated, multidrug-resistant, methicillin-resistant Staphylococcus aureus clonal complex 97 lineage spreading in dairy cattle and pigs in Italy. Appl Environ Microbiol 82:816–821. doi: 10.1128/AEM.02854-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hsueh P-R, Chen W-H, Teng L-J, Luh K-T. 2005. Nosocomial infections due to methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci at a university hospital in Taiwan from 1991 to 2003: resistance trends, antibiotic usage and in vitro activities of newer antimicrobial agents. Int J Antimicrob Agents 26:43–49. doi: 10.1016/j.ijantimicag.2005.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.MacDougall C, Powell JP, Johnson CK, Edmond MB, Polk RE. 2005. Hospital and community fluoroquinolone use and resistance in Staphylococcus aureus and Escherichia coli in 17 US hospitals. Clin Infect Dis 41:435–440. doi: 10.1086/432056. [DOI] [PubMed] [Google Scholar]
- 69.Weber SG, Gold HS, Hooper DC, Karchmer AW, Carmeli Y. 2003. Fluoroquinolones and the risk for methicillin-resistant Staphylococcus aureus in hospitalized patients1. Emerg Infect Dis 9:1415–1422. doi: 10.3201/eid0911.030284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wipf JR, Perreten V. 2016. Methicillin-resistant Staphylococcus aureus isolated from dogs and cats in Switzerland. Schweiz Arch Tierheilkd 158:443–450. doi: 10.17236/sat00070. [DOI] [PubMed] [Google Scholar]
- 71.Cadena J, Nair S, Henao-Martinez AF, Jorgensen JH, Patterson JE, Sreeramoju PV. 2011. Dose of trimethoprim-sulfamethoxazole to treat skin and skin structure infections caused by methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 55:5430–5432. doi: 10.1128/AAC.00706-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eliakim-Raz N, Hellerman M, Yahav D, Cohen J, Margalit I, Fisher S, Zusman O, Shaked H, Bishara J. 2017. Trimethoprim/sulfamethoxazole versus vancomycin in the treatment of healthcare/ventilator-associated MRSA pneumonia: a case-control study. J Antimicrob Chemother 72:882–887. doi: 10.1093/jac/dkw510. [DOI] [PubMed] [Google Scholar]
- 73.Pirolo M, Gioffrè A, Visaggio D, Gherardi M, Pavia G, Samele P, Ciambrone L, Di Natale R, Spatari G, Casalinuovo F, Visca P. 2019. Prevalence, molecular epidemiology, and antimicrobial resistance of methicillin-resistant Staphylococcus aureus from swine in southern Italy. BMC Microbiol 19:51. doi: 10.1186/s12866-019-1422-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sahibzada S, Pang S, Hernández-Jover M, Jordan D, Abraham S, O'Dea M, Heller J. 2020. Prevalence and antimicrobial resistance of MRSA across different pig age groups in an intensive pig production system in Australia. Zoonoses Public Health 67:576–586. doi: 10.1111/zph.12721. [DOI] [PubMed] [Google Scholar]
- 75.Schaumburg F, Idelevich EA, Peters G, Mellmann A, von Eiff C, Becker K, Study Group . 2014. Trends in antimicrobial non-susceptibility in methicillin-resistant Staphylococcus aureus from Germany (2004–2011). Clin Microbiol Infect 20:O554–557. doi: 10.1111/1469-0691.12519. [DOI] [PubMed] [Google Scholar]
- 76.Floyd JL, Smith KP, Kumar SH, Floyd JT, Varela MF. 2010. LmrS is a multidrug efflux pump of the major facilitator superfamily from Staphylococcus aureus. Antimicrob Agents Chemother 54:5406–5412. doi: 10.1128/AAC.00580-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Costa SS, Sobkowiak B, Parreira R, Edgeworth JD, Viveiros M, Clark TG, Couto I. 2019. Genetic diversity of norA, coding for a main efflux pump of Staphylococcus aureus. Front Genet 9. doi: 10.3389/fgene.2018.00710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dale GE, Broger C, Hartman PG, Langen H, Page MG, Then RL, Stüber D. 1995. Characterization of the gene for the chromosomal dihydrofolate reductase (DHFR) of Staphylococcus epidermidis ATCC 14990: the origin of the trimethoprim-resistant S1 DHFR from Staphylococcus aureus? J Bacteriol 177:2965–2970. doi: 10.1128/jb.177.11.2965-2970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matsuoka M, Inoue M, Endo Y, Nakajima Y. 2003. Characteristic expression of three genes, msr(A), mph(C) and erm(Y), that confer resistance to macrolide antibiotics on Staphylococcus aureus. FEMS Microbiol Lett 220:287–293. doi: 10.1016/S0378-1097(03)00134-4. [DOI] [PubMed] [Google Scholar]
- 80.Truong-Bolduc QC, Dunman PM, Strahilevitz J, Projan SJ, Hooper DC. 2005. MgrA is a Multiple regulator of two new efflux pumps in Staphylococcus aureus. J Bacteriol 187:2395–2405. doi: 10.1128/JB.187.7.2395-2405.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guay GG, Rothstein DM. 1993. Expression of the tetK gene from Staphylococcus aureus in Escherichia coli: comparison of substrate specificities of TetA(B), TetA(C), and TetK efflux proteins. Antimicrob Agents Chemother 37:191–198. doi: 10.1128/AAC.37.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McAleese F, Petersen P, Ruzin A, Dunman PM, Murphy E, Projan SJ, Bradford PA. 2005. A Novel MATE family efflux pump contributes to the reduced susceptibility of laboratory-derived Staphylococcus aureus mutants to tigecycline. Antimicrob Agents Chemother 49:1865–1871. doi: 10.1128/AAC.49.5.1865-1871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bergmiller T, Andersson AMC, Tomasek K, Balleza E, Kiviet DJ, Hauschild R, Tkačik G, Guet CC. 2017. Biased partitioning of the multidrug efflux pump AcrAB-TolC underlies long-lived phenotypic heterogeneity. Science 356:311–315. doi: 10.1126/science.aaf4762. [DOI] [PubMed] [Google Scholar]
- 84.2009. Classification of Staphylococcal cassette chromosome mec (SCC mec): guidelines for reporting novel SCC mec elements. Antimicrob Agents Chemother 53:4961–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu J, Chen D, Peters BM, Li L, Li B, Xu Z, Shirliff ME. 2016. Staphylococcal chromosomal cassettes mec (SCCmec): A mobile genetic element in methicillin-resistant Staphylococcus aureus. Microb Pathog 101:56–67. doi: 10.1016/j.micpath.2016.10.028. [DOI] [PubMed] [Google Scholar]
- 86.Saber H, Jasni AS, Jamaluddin TZMT, Ibrahim R. 2017. A review of Staphylococcal cassette chromosome mec (SCCmec) types in coagulase-negative Staphylococci (CoNS) species. Malays J Med Sci 24:7–18. doi: 10.21315/mjms2017.24.5.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cave R, Misra R, Chen J, Wang S, Mkrtchyan HV. 2019. Whole genome sequencing revealed new molecular characteristics in multidrug resistant staphylococci recovered from high frequency touched surfaces in London. Sci Rep 9:9637. doi: 10.1038/s41598-019-45886-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen X-P, Li W-G, Zheng H, Du H-Y, Zhang L, Zhang L, Che J, Wu Y, Liu S-M, Lu J-X. 2017. Extreme diversity and multiple SCCmec elements in coagulase-negative Staphylococcus found in the Clinic and Community in Beijing, China. Ann Clin Microbiol Antimicrob 16:57. doi: 10.1186/s12941-017-0231-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Singh-Moodley A, Strasheim W, Mogokotleng R, Ismail H, Perovic O. 2019. Unconventional SCCmec types and low prevalence of the Panton-Valentine Leukocidin exotoxin in South African blood culture Staphylococcus aureus surveillance isolates, 2013–2016. PLoS One 14:e0225726. doi: 10.1371/journal.pone.0225726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zong Z, Peng C, Lü X. 2011. Diversity of SCCmec elements in methicillin-resistant coagulase-negative staphylococci clinical isolates. PLoS One 6:e20191. doi: 10.1371/journal.pone.0020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xu Z, Mkrtchyan HV, Cutler RR. 2015. Antibiotic resistance and mecA characterization of coagulase-negative staphylococci isolated from three hotels in London. UK Front Microbiol 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fessler AT, Billerbeck C, Kadlec K, Schwarz S. 2010. Identification and characterization of methicillin-resistant coagulase-negative staphylococci from bovine mastitis. J Antimicrob Chemother 65:1576–1582. doi: 10.1093/jac/dkq172. [DOI] [PubMed] [Google Scholar]
- 93.Hiramatsu K, Ito T, Tsubakishita S, Sasaki T, Takeuchi F, Morimoto Y, Katayama Y, Matsuo M, Kuwahara-Arai K, Hishinuma T, Baba T. 2013. Genomic basis for methicillin resistance in Staphylococcus aureus. Infect Chemother 45:117–136. doi: 10.3947/ic.2013.45.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S6. Download spectrum.00387-22-s0001.xls, XLS file, 0.4 MB (448.5KB, xls)