ABSTRACT
A total of 38 isolates of carbapenem-resistant Klebsiella pneumoniae harboring blaNDM were obtained during surveillance of 10 hospitals in Myanmar. Of these 38 isolates, 19 (50%) harbored genes encoding 16S rRNA methylases, such as armA or rmtB. The K. pneumoniae strains tested belonged to 17 sequence types (STs), including the high-risk clonal lineages ST101 and ST147. The ST101 and ST147 isolates carried IncFII plasmids harboring blaNDM-5 and IncFIB(pQil) plasmids harboring blaNDM-1, respectively. These results indicate that IncFII plasmids harboring blaNDM-5 and IncFIB(pQil) plasmids harboring blaNDM-1 have been spreading in K. pneumoniae ST101 and ST147 isolates, respectively, in Myanmar.
IMPORTANCE The emergence of carbapenem-resistant K. pneumoniae has become a serious problem in medical settings worldwide. The present study demonstrated that carbapenem-resistant K. pneumoniae strains have been spreading in medical settings in Myanmar. In particular, plasmid genes encoding NDMs and 16S rRNA methylases have been spreading in K. pneumoniae high-risk clones.
KEYWORDS: carbapenemase-producing Enterobacteriaceae, Klebsiella pneumoniae, NDM-type metallo-β-lactamase, 16S rRNA methylase
INTRODUCTION
The emergence and spread of carbapenemase-producing Enterobacteriaceae (CPE) have become a serious medical problem worldwide (1). Several types of carbapenemases have been detected to date in Enterobacteriaceae, with New Delhi metallo-β-lactamase (NDM-type MBL) first being detected in Escherichia coli and Klebsiella pneumoniae isolates obtained from a patient in Sweden in 2008 (2). NDM-type MBLs subsequently spread rapidly worldwide (3), with 40 variants of NDM-type MBLs being detected to date [https://www.ncbi.nlm.nih.gov/pathogens/refgene/#gene_family:(blaNDM)].
NDM-type MBL-producing K. pneumoniae complex isolates have plasmids that carry the blaNDM gene (4). Many of these isolates have shown multidrug resistance and have been found to harbor genes, such as armA and rmtB, encoding 16S rRNA methylases that have been associated with aminoglycoside resistance (4). Plasmids carrying these genes, which have been associated with virulence and antibiotic resistance, belong to various incompatibility (Inc) types.
The present study describes the molecular epidemiology of clinical isolates of K. pneumoniae obtained from patients hospitalized in 10 hospitals in three regions of Myanmar from 2015 to 2017. All of these isolates were carbapenem resistant and produced NDM-type MBLs.
RESULTS
Clinical features of carbapenem-resistant K. pneumoniae complex isolates.
The whole genomes of 46 isolates of the K. pneumoniae complex were sequenced using MiSeq. Average nucleotide identity (ANI) and Type (Strain) Genome Sever (TYGS) analyses revealed that 38 were K. pneumoniae subsp. pneumoniae, 7 were K. quasipneumoniae subsp. similipneumoniae, and 1 was K. quasipneumoniae subsp. quasipneumoniae. The 46 carbapenem-resistant K. pneumoniae complex strains were isolated from clinical samples obtained from patients hospitalized at 10 hospitals in Myanmar from December 2015 to September 2017. Of the 46 isolates, 30 were from six hospitals in the Yangon region, 14 were from three hospitals in the Mandalay region, and 2 were from one hospital in Kachin State (see Fig. S1 in the supplemental material). The susceptibilities of these isolates to various antibiotics were tested by the microdilution method, according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (5). All 46 isolates were resistant to aztreonam (AZT), ceftazidime (CAZ), meropenem (MEM), and tigecycline (TGC); 43 (94%) were resistant to imipenem (IPM); 39 (85%) were resistant to ciprofloxacin (CIP); 27 (59%) were resistant to amikacin (AMK); and 1 (2%) was resistant to colistin (CST) (Table 1).
TABLE 1.
Drug susceptibility profiles of carbapenem-resistant K. pneumoniae complex isolates in 10 hospitals in Myanmar (n = 46)
| Antibiotic | Breakpoint for resistancea (μg/mL) |
% resistant isolates |
MIC (μg/mL) |
||
|---|---|---|---|---|---|
| Range | MIC50 | MIC90 | |||
| Amikacin | ≥64 | 50 | 1 to >1,024 | >1,024 | >1,024 |
| Aztreonam | ≥16 | 100 | 16 to >1,024 | 256 | 512 |
| Ceftazidime | ≥16 | 100 | 512 to >1,024 | >1,024 | >1,024 |
| Ciprofloxacin | ≥1 | 92 | 0.25 to >1,024 | 64 | 256 |
| Colistin | ≥4 | 3 | 0.0625 to 4 | 0.25 | 2 |
| Imipenem | ≥4 | 92 | 0.5 to 256 | 8 | 64 |
| Meropenem | ≥4 | 100 | 4 to 128 | 32 | 128 |
| Tigecycline | ≥0.5 | 100 | 0.5 to 4 | 1 | 2 |
The breakpoint for tigecycline was determined according to EUCAST guidelines.
Drug resistance genes of carbapenem-resistant K. pneumoniae complex isolates.
All 38 isolates of K. pneumoniae subsp. pneumoniae harbored blaNDM genes, with 22 harboring blaNDM-1, 1 harboring blaNDM-4, 11 harboring blaNDM-5, and 4 harboring blaNDM-7, as well as blaCTX-M genes, including blaCTX-M-15 or blaCTX-M-14. Nineteen (50%) isolates also harbored genes encoding 16S rRNA methylases, including armA or rmtB, making them highly resistant to aminoglycosides. Thirty-three isolates (87%) harbored aac(6′)-Ib-cr, which is the most common plasmid-mediated quinolone resistance gene (6). The 26 quinolone-resistant isolates (68%) with MICs of ≥1 μg/mL had point mutations at quinolone resistance-determining regions, including GyrA and ParC (Table 2). A summary of the characteristics of the 8 carbapenem-resistant Klebsiella species, including K. quasipneumoniae subsp. similipneumoniae and K. quasipneumoniae subsp. quasipneumoniae, is shown in Table S1. Of them, 6 isolates harbored blaNDM-1, blaCTX-M-15, and armA, and 2 harbored blaNDM-7 and blaCTX-M-15.
TABLE 2.
Summary of the characteristics of the 38 carbapenem-resistant K. pneumoniae strains, including MLST types and drug resistance genesa
| MLST type |
No. of isolates |
Hospital(s) | Carbapenemase genes(s) | Extended-spectrum β-lactamase-encoding gene(s) | 16S rRNA methylase gene(s) |
Aminoglycoside acetyltransferase-encoding gene(s) | Mutation(s) in DNA gyrase |
|
|---|---|---|---|---|---|---|---|---|
| GyrA | ParC | |||||||
| ST15 | 1 | A | bla NDM-1 | blaCTX-M-15, blaSHV-106 | armA | aac(6′)-Ib-cr, aac(3)-IId, aadA2, aadA16 | S83F, D87A | S80I |
| ST16 | 2 | B (1/2), F (1/2) | blaNDM-1 (1/2), blaNDM-5 (1/2) | blaCTX-M-15, blaSHV-26, blaSHV-78, blaSHV-98 | armA, rmtB (1/2) | aac(6′)-Ib-cr, aadA2, aac(6′)-Ib3 (1/2), aac(3)-Ild (1/2), aadA16 (1/2) | S83F, D87N | E84K |
| ST17 | 2 | G | bla NDM-1 | bla CTX-M-14 | —b | aac(3)-Iid, aac(6′)-Ib-cr, aac(6′)-Ib3 | — | — |
| ST36 | 1 | J | bla NDM-1 | blaCTX-M-15, blaSHV-11, blaSHV-13, blaSHV-70 | armA | aac(3)-Iid, aadA16 | — | — |
| ST42 | 1 | A | bla NDM-4 | blaCTX-M-15, blaSHV-26, blaSHV-78, blaSHV-98 | — | aac(3)-Iid, aac(6′)-Ib-cr, aadA16 | S83L, D87Y | S80I |
| ST101 | 11 | A (8/11),E (2/11),I(1/11) | blaNDM-1 (3/11), blaNDM-5 (8/11) | bla CTX-M-15 | armA(1/11), rmtB (9/11) | aac(3)-Ild (8/11), aac(6′)-Ib-cr (10/11), aadA1 (1/11), aadA2 (8/11), aadA16 (7/11) | S83Y, D87G (8/11) | S80I (8/11) |
| ST147 | 10 | A (8/10), C (1/10), G (1/10) | blaNDM-1 (9/10), blaNDM-5 (1/10) | blaCTX-M-15, blaSHV-11 | rmtB (1/10) | aac(3)-Ild (5/10), aac(6′)-Ib-cr (9/10), aac(6′)-Ib3, (2/10), aadA1 (9/10), aadA2 (4/10), aadA16 (5/10) | S83I (9/10), S83Y (1/10), D87A (1/10) | S80I |
| ST273 | 1 | H | bla NDM-7 | blaCTX-M-15, blaSHV-11 | — | aac(3)-Ild, aac(6′)-Ib-cr, aadA16, aph(3′)-I, aph(6)-Id | S83I | S80I |
| ST394 | 1 | C | bla NDM-5 | bla CTX-M-15 | rmtB | aadA2, aadA16 | — | — |
| ST401 | 1 | J | bla NDM-1 | bla CTX-M-15 | armA | aac(6′)-Ib-cr, aadA16 | — | — |
| ST420 | 1 | E | bla NDM-1 | blaCTX-M-15, blaSHV-75 | armA | aac(3)-IId, aadA16 | — | — |
| ST534 | 1 | A | bla NDM-7 | blaCTX-M-15, blaSHV-11, blaSHV-13, blaSHV-70, blaSHV-77, blaSHV-80 | — | aac(3)-Iid, aac(6′)-Ib-cr, aadA16 | S83I | S80I |
| ST1655 | 1 | G | bla NDM-1 | blaCTX-M-15, blaSHV-26, blaSHV-78, blaSHV-98 | armA | aac(3)-Iid, aac(6′)-Ib-cr, aadA16 | — | — |
| ST4029 | 2 | G | bla NDM-1 | blaCTX-M-15, blaSHV-40, blaSHV-56, blaSHV-79, blaSHV-85, blaSHV-89 | — | aac(6′)-Ib-cr, aadA1, aac(6′)-Ib | — | — |
| ST4030 | 1 | G | bla NDM-7 | blaCTX-M-15, blaSHV-187 | — | aac(3)-Iid, aac(6′)-Ib-cr, aadA16 | S83I | S80I |
| ST5912 | 1 | D | bla NDM-7 | blaCTX-M-15, blaSHV-26, blaSHV-78, blaSHV-98 | — | aac(6′)-Ib-cr, aadA16 | S83F, D87N | E84K |
Numbers in parenthesis indicate that the number of isolates with resistant genes or amino acid mutations per each ST isolate. No parenthesis indicate that all each ST isolate had resistant genes or amino acid mutations.
— means that the isolate has no mutation, i.e. S83S.
MLST and phylogenetic analyses of carbapenem-resistant K. pneumoniae complex isolates.
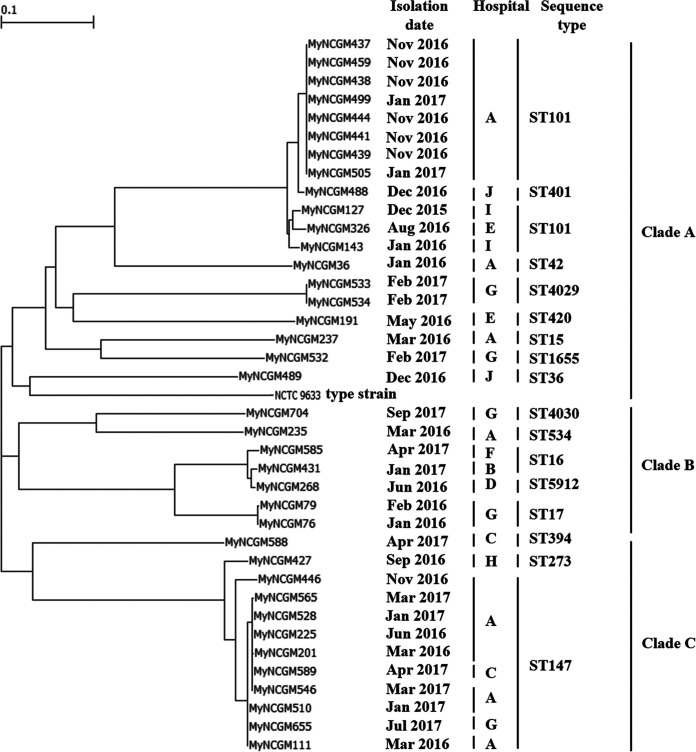
Multilocus sequence typing (MLST) analysis revealed that 10 isolates (26%) belonged to sequence type 147 (ST147); 11 (29%) belonged to ST101; 2 each (5%) belonged to ST16, ST17, and ST4029; and 1 each (3%) belonged to ST15, ST36, ST42, ST273, ST394, ST401, ST420, ST534, ST1655, ST4030, and ST5912. A phylogenetic tree revealed three clades, designated clades A, B, and C (Fig. 1). Clade A consisted of isolates belonging to ST15, ST36, ST42, ST101, ST401, ST420, ST1655, and ST4029 and the K. pneumoniae reference strain; clade B consisted of isolates belonging to ST16, ST17, ST534, ST4030, and ST5912; and clade C consisted of isolates belonging to ST147, ST273, and ST394. The high-risk clonal lineages ST101 and ST147 belonged to clades A and C, respectively. The other high-risk clonal lineage, ST15, of one isolate belonged to clade A (7). MLST showed that carbapenem-resistant K. quasipneumoniae subsp. similipneumoniae belonged to ST705, ST1473, ST3590, and ST5967, and K. quasipneumoniae subsp. quasipneumoniae belonged to ST3866 (Table S1).
FIG 1.
Phylogenetic tree of 38 carbapenem-resistant K. pneumoniae complex isolates obtained from clinical samples at 10 hospitals in Myanmar. The tree was constructed by the maximum likelihood method based on core-genome SNPs.
As shown in Fig. 1, isolates of the high-risk clonal lineage ST101 in clade A were from hospitals A, E, and I, whereas isolates of the high-risk clonal lineage ST147 in clade C were from hospitals A, C, and G. It is difficult to reveal the relationship between the other STs and hospitals.
Eight isolates belonging to ST101 from hospital A had numbers of single nucleotide polymorphisms (SNPs) ranging from 68 to 126, two ST4029 isolates from hospital G had 68 SNPs, two ST17 isolates from hospital G had 95 SNPs, and five ST147 isolates from hospital A had numbers of SNPs ranging from 93 to 29,684. Of the five ST147 isolates, three isolates (MyNCGM201, MyNCGM225, and MyNCGM528) had numbers of SNPs ranging from 90 to 93.
Plasmids carrying blaNDM.
All blaNDM genes, including blaNDM-1, blaNDM-4, blaNDM-5, and blaNDM-7, were located on plasmids ranging in size from 45,321 bp to 176,315 bp. These plasmids belonged to eight types of plasmid incompatibility complexes, including IncC (5 isolates), IncFII (6 isolates), IncFIB(pQil) (9 isolates), IncFIB(pQil)/IncFII(K) (3 isolates), IncFIB(K)/IncFII/IncFII(pKP91) (1 isolate), IncM2 (4 isolates), IncR (1 isolate), and IncX3 (5 isolates) (Table 3). The remaining four plasmids did not belong to any Inc type in Enterobacteriaceae (Table 3). The blaNDM-1 gene was located on IncC, IncFIB(pQil), IncFIB(pQil)/IncFII(K), IncM2, and IncR-type plasmids; blaNDM-4 was located on IncX3 plasmids; blaNDM-5 was located on IncFII and IncFIB(K)/IncFII/IncFII(pKP91) plasmids; and blaNDM-7 was located on IncX3 plasmids (Table 3).
TABLE 3.
Genetic characterization of carbapenemase resistance plasmids of 38 K. pneumoniae subsp. pneumoniae isolates harboring blaNDMa
| Inc type |
No. of isolates |
Hospital(s) | MLST type(s) | Plasmid size (bp) |
Carbapenemase- and ESBL-encoding gene(s) | Aminoglycoside resistance gene(s) |
|---|---|---|---|---|---|---|
| IncC | 5 | A (1/5), B (1/5), G (3/5) | ST15 (1/5), ST16 (1/5), ST17 (2/5), ST1655 (1/5) | 158,959–176,315 | bla NDM-1 | armA (3/5), aac(6′)-Ib-cr, aac(6′)-Ib3 (3/5), aadA2 (1/5) |
| IncFII | 6 | A (5/6), C (1/6) | ST101 (4/6), ST147 (1/6), ST394 (1/6) | 94,549–94,603 | bla NDM-5 | rmtB, aadA2 |
| IncFIB(pQil) | 9 | A (7/9), C (1/9), G (1/9) | ST147 | 51,716–87,316 | blaNDM-1, blaCTX-M-15 | aac(6′)-Ib-cr |
| IncFIB(pQil)/IncFII(K) | 3 | G (2/3), I (1/3) | ST101 (1/3), ST4029 (2/3) | 119,263–126,228 | blaNDM-1, blaCTX-M-15 | aac(6′)-Ib-cr, aac(6′)-Ib, aadA1 |
| IncFIB(K)/IncFII/IncFII(pKP91) | 1 | A | ST101 | 199,295 | blaNDM-5, blaCTX-M-15 | rmtB, aac(6′)-Ib-cr, aac(3)-Iid, aadA2, aadA16 |
| IncM2 | 4 | E (1/4), I (1/4), J (2/4) | ST36 (1/4), ST101 (1/4), ST401 (1/4), ST420 (1/4) | 80,663–80,798 | bla NDM-1 | armA |
| IncR | 1 | E | ST101 | 67,399 | bla NDM-1 | rmtB, aac(6′)-Ib-cr, aadA16 |
| IncX3 | 5 | A (2/5), D (1/5), H (1/5), G (1/5) | ST42 (1/5), ST273 (1/5), ST534 (1/5), ST4030 (1/5), ST5912 (1/5) | 45,321–46,161 | blaNDM-4 (1/5), blaNDM-7 (4/5) | — |
| —b | 4 | A (3/4), F (1/4) | ST16 (1/4), ST101 (3/4) | 10,494–122,000 | bla NDM-5 | aadA2 (3/4), aac(6′)-Ib-cr (1/4), aadA16 (1/4) |
Numbers in parenthesis indicate that the number of isolates with resistant genes or amino acid mutations per each ST isolate. No parenthesis indicate that all each ST isolate had resistant genes or amino acid mutations.
— means that the isolate has no mutation, i.e. S83S.
Of the 38 plasmids carrying blaNDM, 15 (39%) harbored genes encoding both NDMs and 16S rRNA methylases. These 15 plasmids included 7 that harbored armA on IncC- or IncM2-type plasmids and 8 that harbored rmtB on IncFII-, IncFIB(K)/IncFII/IncFII(pKP91)-, or IncR-type plasmids (Table 3).
The IncFIB(pQil)-type plasmids carrying blaNDM were detected in isolates from three regions in Myanmar, Kachin, Mandalay, and Yangon, whereas the IncC-type, IncFIB(pQil)/IncFII(K), IncM2, and IncX3-type plasmids carrying blaNDM were detected in isolates from urban areas, including the Mandalay and Yangon regions, in Myanmar (Table 3 and Fig. S1).
Genetic environments surrounding blaNDM and 16S rRNA methylases.
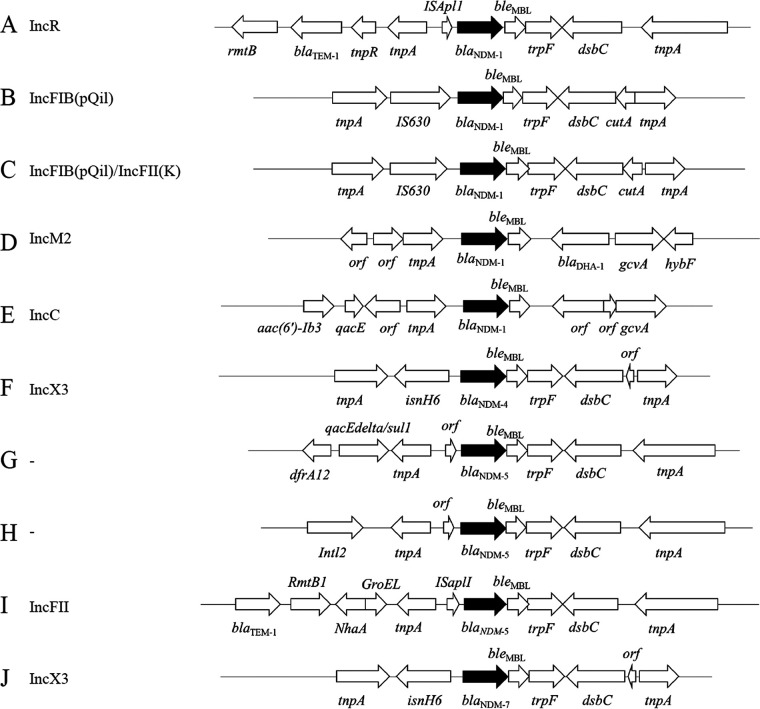
Assessment of the genomic environments surrounding blaNDM revealed 10 types of genetic structures, including blaNDM-1 (Fig. 2A to E), blaNDM-4 (Fig. 2F), bla NDM-5 (Fig. 2G to I), and blaNDM-7 (Fig. 2J).
FIG 2.
Genomic environments of blaNDM in K. pneumoniae complex strains isolated from various clinical samples obtained at 10 hospitals in Myanmar.
The genetic structure surrounding blaNDM-1 could be divided into five types (Fig. 2A to E). The structure of type A was rmtB-blaTEM-1-tnpR-tnpA-ISApll-blaNDM-1-bleMBL-trpF-dsbC-tnpA. The structure tnpA-ISApll-blaNDM-1-bleMBL-trpF-dsbC-tnpA was identical to those of plasmids in other types of Enterobacteriaceae, including Escherichia coli pC06114_1 (GenBank accession no. CP016035) detected in 2015 in Germany and K. pneumoniae pM941-NDM5 (GenBank accession no. AP023454) detected in 2018 in Myanmar. The structure of types B and C was tnpA-IS630-blaNDM-1-bleMBL-trpF-dsbC-cutA-tnpA, which was identical to the structures of plasmids of K. pneumoniae AATZP (GenBank accession no. CP014757) detected in 2014 in the United States; K. pneumoniae K66-45 (GenBank accession no. CP020902) detected in 2010 in Norway; and K. pneumoniae C435, C069, and C070 (GenBank accession no. LC521845, LC613144, and LC521839, respectively) detected in Thailand in 2016. The structure of type D was orf-orf-tnpA-blaNDM-1-bleMBL-blaDHA-1-gcvA-hybF, which was identical to the structures of plasmids of E. coli Es_ST2350_SE1 (GenBank accession no. CP031322), first detected in 2018 in the United Kingdom, and K. pneumoniae 3347689I (GenBank accession no. CP071086), first detected in 2020 in Switzerland. The structure of type E was aac(6′)-Ib3-qacE-orf-tnpA-blaNDM-1-bleMBL-orf-orf-gcvA, which was identical to the structure of a plasmid in E. coli Carbapenemase (NDM-1)_IncA/C2 (GenBank accession no. CP050162), first detected in 2012 in Hong Kong.
The genetic structure surrounding blaNDM-4 was tnpA-isnH6-blaNDM-4-bleMBL-trpF-dsbC-orf-tnpA (Fig. 2F), which was identical to those of plasmids of E. coli M2-16 (GenBank accession no. AP018146) in 2015 in Myanmar and E. coli TUM18530 (GenBank accession no. AP023194) in 2018 in Japan.
Three genetic structures were observed to surround blaNDM-5 (Fig. 2G to I). The structure of type G was dfrA12-qacEΔ/sul1-tnpA-orf-blaNDM-5-bleMBL-trpF-dsbC-tnpA, which was identical to those of plasmids of E. coli isolated from 2013 to 2019 in China, Malawi, Myanmar, South Korea, Thailand, and the United States. The structure of type H was Intl2-tnpA-orf-blaNDM-5-bleMBL-trpF-dsbC-tnpA, which was identical to that of a plasmid of Enterobacter hormaechei, p388, isolated in 2017 in the United States (GenBank accession no. CP021168). The structure of type I was blaTEM-1-rmtB1-nhaA-groEL-tnpA-ISApll-blaNDM-5-bleMBL-trpF-dsbC-tnpA, which was identical to those of E. coli plasmids pM214_FII and pM105_mF (GenBank accession no. AP018144 and AP018137, respectively), isolated in 2015 in Myanmar.
The genetic structure surrounding blaNDM-7 (Fig. 2J), tnpA-isnH6-blaNDM-7-bleMBL-trpF-dsbC-orf-tnpA, was similar to that surrounding blaNDM-4, with the latter being identical to those of plasmids of E. coli M2-16 (GenBank accession no. AP018146) in 2015 in Myanmar and E. coli TUM18530 (GenBank accession no. AP023194) in 2018 in Japan.
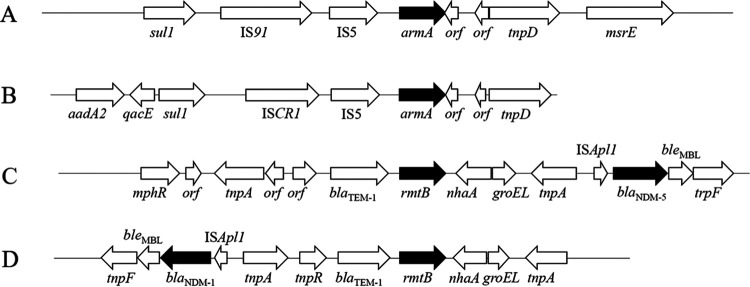
The structures of the genomic environments surrounding armA and rmtB are shown in Fig. 3. The structure surrounding armA of type A was detected in four of seven isolates and was identical to those in E. coli isolated from 2003 to 2018 in China, Hong Kong, India, Norway, and Poland (GenBank accession no. CP072463, HQ451074, CP030858, CP020902, and CP058363, respectively). The structure surrounding armA of type B was detected in three of seven isolates and was identical to those in K. pneumoniae isolated in Oman, Japan, and South Africa (GenBank accession no. JX988621, AB759690, and CP023488, respectively). The structure surrounding rmtB of type C was detected in 11 of 12 isolates and was identical to those in a strain of K. pneumoniae isolated in 2018 in the Czech Republic (GenBank accession no. CP050367) and strains of E. coli isolated from 2012 to 2019 in India, Italy, and Switzerland (GenBank accession no. CP033159, MN007141, and CP048368, respectively). The structure surrounding rmtB of type D was detected in 1 of the 12 isolates.
FIG 3.
Genomic environments of armA and rmtB in K. pneumoniae complex strains isolated from various clinical samples obtained at 10 hospitals in Myanmar.
Of the 38 isolates, 7 harbored both blaNDM-1 and armA on the same plasmids, including four IncM2 and three IncC plasmids; 6 harbored both blaNDM-5 and rmtB on the same plasmids, including plasmid type IncFII or IncFIB(K)/IncFII/IncFII (pKP91); and 1 harbored both blaNDM-1 and rmtB on the same plasmid belonging to IncR.
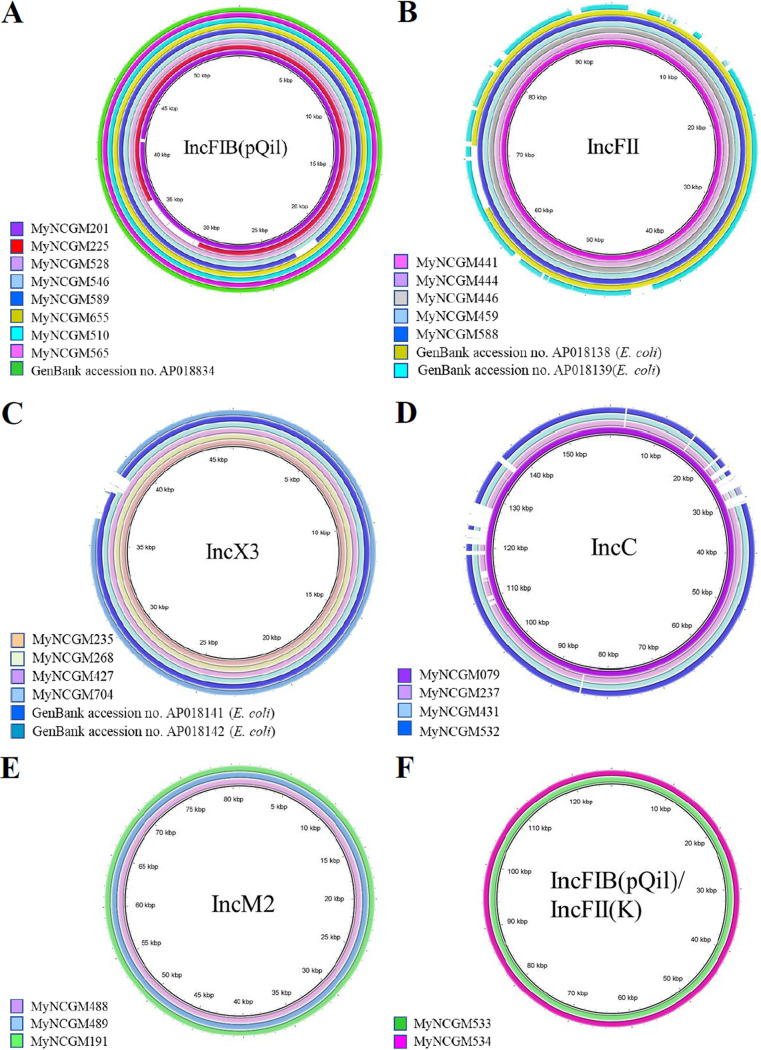
The plasmid structures belonging to IncFIB(pQil), IncFII, IncX3, IncC, IncM2, and IncFIB(pQil)/IncFII(K) are compared in Fig. 4. Five of nine plasmids belonging to IncFIB(pQil) had structures identical to that of a plasmid in K. pneumoniae in 2015 in Myanmar (GenBank accession no. AP018834). All six plasmids belonging to IncFII had structures 92% identical to that of a plasmid in E. coli isolated in 2015 in Myanmar (GenBank accession no. AP018138). Six of seven plasmids belonging to IncX3 had structures 97% identical to that of a plasmid in E. coli isolated in 2015 in Myanmar (GenBank accession no. AP018141). On the other hand, K. quasipneumoniae subsp. similipneumoniae harbored IncX3, IncC, or IncM2 plasmids, and K. quasipneumoniae subsp. quasipneumoniae harbored an IncX3 plasmid (Fig. 4).
FIG 4.
Comparison of the plasmid sequences of IncFIB(pQil), IncFII, IncX3, IncC, IncM2, and IncFIB(pQil)/IncFII(K). The images were generated using BLAST Ring Image Generator software (https://sourceforge.net/projects/brig/files/BRIG-0.95-dist.zip/download). Plasmid sequences belonging to each Inc type were compared with plasmids of MyNCGM111 for IncFIB(pQil) (A), MyNCGM439 for IncFII (B), MyNCGM036 for IncX3 (C), MyNCGM076 for IncC (D), MyNCGM127 for IncM2 (E), and MyNCGM143 for IncFIB(pQil)/IncFII(K) (F).
DISCUSSION
The present study suggests that K. pneumoniae ST101 isolates harboring blaNDM-5 on IncFII plasmids and ST147 isolates harboring blaNDM-1 on IncFIB(pQil) plasmids have spread in three regions in Myanmar in recent years. IncFII plasmids harboring blaNDM-5 in ST101 isolates and IncFIB(pQil) plasmids harboring blaNDM-1 in ST147 isolates seem to be horizontally spreading in hospital A. IncFII and IncFIB(pQil) plasmids harboring blaNDM were detected in E. coli ST354 and K. pneumoniae ST147 strains isolated in Myanmar (8), the United States (GenBank accession no. CP014757), Norway (GenBank accession no. CP020902), and Thailand (GenBank accession no. LC521839).
IncX3 plasmids harboring blaNDMs will be spreading among Enterobacteriaceae, including Citrobacter sp., Enterobacter sp., E. coli, and K. pneumoniae subspecies in Myanmar. In this study, we revealed that K. pneumoniae subsp. pneumoniae, K. quasipneumoniae subsp. similipneumoniae, and K. quasipneumoniae subsp. quasipneumoniae had IncX3 plasmids harboring blaNDM-4 or blaNDM-7 (Table 3; see also Table S1 in the supplemental material). Sugawara et al. reported that Citrobacter amalonaticus, Citrobacter freundii, Enterobacter asburiae, Enterobacter xiangfangensis, E. coli, Klebsiella pneumoniae, Klebsiella quasipneumoniae, Leclercia adecarboxylata, and Lelliottia nimipressuralis harbored IncX3 plasmids harboring blaNDM-4, blaNDM-5, or blaNDM-7 in Myanmar (8). Another study showed that IncX3 plasmids harboring blaNDM-5 have spread in K. pneumoniae isolates in China (9).
Carbapenem-resistant Enterobacteriaceae are a significant public health concern in Myanmar (8). K. pneumoniae ST101 and ST147 isolates producing NDM-1 caused outbreaks in Spain (10), and ST101 isolates producing extended-spectrum β-lactamases (ESBLs) were also reported in Tanzania (11). Previous studies in Myanmar revealed that K. pneumoniae ST101 and ST147 strains were detected in samples from patients in medical settings and environments, including foodstuff in the Yangon region (12, 13). In addition to ST101 and ST147, other high-risk clones, ST11, ST15, ST14, and ST48, were reported in Myanmar (14), Bangladesh (7), Saudi Arabia (15), and China (16), respectively.
The results of SNP analysis of closely related isolates (Fig. 1) suggested that eight ST101 isolates from hospital A, two ST4029 isolates from hospital G, two ST17 isolates from hospital G, and three ST147 isolates from hospital A represented outbreaks.
Strains of Enterobacteriaceae containing plasmids carrying genes encoding NDMs and 16S rRNA methylases, making these bacteria resistant to carbapenems and aminoglycosides, will likely spread throughout medical settings in Myanmar. Isolates of the Enterobacter cloacae complex coproducing NDM-1/4 and ArmA/RmtC/RmtE have been detected in five regions in Myanmar (17), and other species of Enterobacteriaceae, including E. coli and Citrobacter freundii, resistant to carbapenems and aminoglycosides and producing NDM-1/4/5 and ArmA/RmtB/RmtC/RmtE have been detected in environments as well as medical settings in Yangon, Myanmar (13). These findings emphasize the need to monitor Enterobacteriaceae in Myanmar for the presence of plasmid-borne genes encoding carbapenemases and 16S rRNA methylases.
In conclusion, this is the first report describing the molecular epidemiology of carbapenem-resistant K. pneumoniae isolates in medical settings in three regions of Myanmar. The incidence of multidrug-resistant (MDR) K. pneumoniae clinical isolates in hospitals differed regionally, being 57.9% in the Yangon region (8) but 39.5% in the three regions included in the present study. Epidemiological surveillance is required to prevent the emergence and spread in Myanmar of MDR Enterobacteriaceae harboring genes encoding enzymes associated with drug resistance.
MATERIALS AND METHODS
Bacterial strains.
Forty-six clinical isolates of the carbapenemase-resistant K. pneumoniae complex, defined as strains showing resistance to imipenem or meropenem (MIC, ≥4 μg/mL), were obtained between December 2015 and September 2017 from patients treated at 10 hospitals in Myanmar. Of these 46 isolates, 20, 1, 2, 1, 4, 1, 8, 5, 2, and 2 were from hospitals A through J, respectively. Bacteria were identified using the Vitek 2 system (bioMérieux, Marcy l’Etoile, France), with identities confirmed by sequencing of 16S rRNA. Of the 46 isolates, 22 were from blood, 10 were from tracheal aspirates and sputum, 7 were from pus and wounds, and 7 were from urine. As the situation in Myanmar has become increasingly uncertain in recent months, it is difficult to update the clinical information on the 46 K. pneumoniae isolates tested.
Drug susceptibility testing.
Drug susceptibility was tested according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (5). The ranges of antibiotic concentrations tested were 0.5 to 1,024 μg/mL amikacin (AMK), 0.5 to 1,024 μg/mL aztreonam (AZT), 0.5 to 1,024 μg/mL ceftazidime (CAZ), 0.25 to 1,024 μg/mL ciprofloxacin (CIP), 0.0625 to 8 μg/mL colistin (CST), 0.5 to 1,024 μg/mL imipenem (IPM), 0.5 to 1,024 μg/mL meropenem (MEM), and 0.5 to 1,024 μg/mL tigecycline (TGC) (Table 1). The MICs of each antimicrobial agent were determined by broth microdilution methods using Mueller-Hinton broth and 96-well microtiter plates (Kohjin Bio Co., Ltd., Saitama, Japan).
Whole-genome sequencing and genomic analysis.
Genomic DNAs of the 46 isolates were extracted using DNeasy blood and tissue kits (Qiagen, Tokyo, Japan) or 20-gauge genomic tips (Qiagen), and their complete genomes were sequenced using the MiSeq platform (Illumina, San Diego, CA) and MinION (Oxford Nanopore Technologies, Oxford, United Kingdom). Raw reads of each isolate were assembled using CLC Genomic Workbench version 10.0.1 (CLC Bio, Aarhus, Denmark). Species identities of these isolates were determined using an ANI calculator (18) or the Type (Strain) Genome Sever (TYGS) (https://tygs.dsmz.de). The sequences of drug resistance genes were determined using ResFinder 4.1, and plasmids were typed using Plasmid finder 2.1, both from the Center for Genomic Epidemiology (CGE) (https://www.genomicepidemiology.org/). The sequences of plasmids were annotated using the DDBJ Fast annotation and submission tool (https://dfast.ddbj.nig.ac.jp). Fluoroquinolone resistance has been associated with mutations in the quinolone resistance-determining region, which includes the gyrA and parC genes that encode DNA gyrase and topoisomerase IV, respectively. The gyrA and parC genes were detected in silico using CLC Genomics Workbench v.11.0.1 (CLC Bio, Denmark) (19). Comparative analysis of plasmid sequences surrounding blaNDM was performed using BLAST and visualized using in silico molecular cloning (In Silico Biology Inc., Kanagawa, Japan). Imaging of plasmid similarity was performed using the BLAST Ring Image Generator (https://sourceforge.net/projects/brig/files/BRIG-0.95-dist.zip/download).
MLST and phylogenetic analyses.
Multilocus sequence typing (MLST) was performed according to protocols of the MLST databases (https://bigsdb.pasteur.fr/). Phylogenetic trees were constructed using kSNP3.1 software (https://sourceforge.net/projects/ksnp/files/) (20) and visualized using FigTree v.1.4.4 (https://github.com/rambaut/figtree/releases). The type strain K. pneumoniae NCTC 9633 was used as the reference strain.
Accession number(s).
The whole-genome sequences of all 46 isolates have been deposited in GenBank under accession no. DRA009233.
ACKNOWLEDGMENTS
This study was supported by grants from the Research Program on Emerging and Re-emerging Infectious Diseases from the Japan Agency for Medical Research and Development (grant no. 22fk0108604h0702) and Asahi Group Holdings, Ltd. (grant no. AM227CH501 [S.W.]).
Footnotes
Supplemental material is available online only.
Contributor Information
Tatsuya Tada, Email: t-tada@juntendo.ac.jp.
Ana Paula D’Alincourt Carvalho-Assef, Instituto Oswaldo Cruz.
REFERENCES
- 1.Bush K. 2001. New beta-lactamases in Gram-negative bacteria: diversity and impact on the selection of antimicrobial therapy. Clin Infect Dis 32:1085–1089. doi: 10.1086/319610. [DOI] [PubMed] [Google Scholar]
- 2.Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. 2016. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol 7:895. doi: 10.3389/fmicb.2016.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pitout JD, Nordmann P, Poirel L. 2015. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother 59:5873–5884. doi: 10.1128/AAC.01019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2019. Performance standards for antimicrobial susceptibility testing, 29th ed. CLSI supplement M100. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 6.Aung MS, Win NC, San N, Hlaing MS, Myint YY, Thu PP, Aung MT, Yaa KT, Maw WW, Urushibara N, Kobayashi N. 2021. Prevalence of extended-spectrum β-lactamase/carbapenemase genes and quinolone-resistance determinants in Klebsiella pneumoniae clinical isolates from respiratory infections in Myanmar. Microb Drug Resist 27:36–43. doi: 10.1089/mdr.2019.0490. [DOI] [PubMed] [Google Scholar]
- 7.Farzana R, Jones LS, Barratt A, Rahman MA, Sands K, Portal E, Boostrom I, Espina L, Pervin M, Uddin A, Walsh TR. 2020. Emergence of mobile colistin resistance (mcr-8) in a highly successful Klebsiella pneumoniae sequence type 15 clone from clinical infections in Bangladesh. mSphere 5:e00023-20. doi: 10.1128/mSphere.00023-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugawara Y, Akeda Y, Hagiya H, Sakamoto N, Takeuchi D, Shanmugakani RK, Motooka D, Nishi I, Zin KN, Aye MM, Myint T, Tomono K, Hamada S. 2019. Spreading patterns of NDM-producing Enterobacteriaceae in clinical and environmental settings in Yangon, Myanmar. Antimicrob Agents Chemother 63:e01924-18. doi: 10.1128/AAC.01924-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu W, Wang X, Qin J, Liang W, Shen Z. 2020. Dissemination and stability of the blaNDM-5-carrying IncX3-type plasmid among multiclonal Klebsiella pneumoniae isolates. mSphere 5:e00917-20. doi: 10.1128/mSphere.00917-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez-Vazquez M, Sola Campoy PJ, Ortega A, Bautista V, Monzon S, Ruiz-Carrascoso G, Mingorance J, Gonzalez-Barbera EM, Gimeno C, Aracil B, Saez D, Lara N, Fernandez S, Gonzalez-Lopez JJ, Campos J, Kingsley RA, Dougan G, Oteo-Iglesias J, Spanish NDM Study Group . 2019. Emergence of NDM-producing Klebsiella pneumoniae and Escherichia coli in Spain: phylogeny, resistome, virulence and plasmids encoding blaNDM-like genes as determined by WGS. J Antimicrob Chemother 74:3489–3496. doi: 10.1093/jac/dkz366. [DOI] [PubMed] [Google Scholar]
- 11.Büdel T, Kuenzli E, Clément M, Bernasconi OJ, Fehr J, Mohammed AH, Hassan NK, Zinsstag J, Hatz C, Endimiani A. 2019. Polyclonal gut colonization with extended-spectrum cephalosporin- and/or colistin-resistant Enterobacteriaceae: a normal status for hotel employees on the island of Zanzibar, Tanzania. J Antimicrob Chemother 74:2880–2890. doi: 10.1093/jac/dkz296. [DOI] [PubMed] [Google Scholar]
- 12.Sugawara Y, Akeda Y, Hagiya H, Zin KN, Aye MM, Takeuchi D, Matsumoto Y, Motooka D, Nishi I, Tomono K, Hamada S. 2021. Characterization of blaNDM-5-harbouring Klebsiella pneumoniae sequence type 11 international high-risk clones isolated from clinical samples in Yangon General Hospital, a tertiary-care hospital in Myanmar. J Med Microbiol 70:1348. doi: 10.1099/jmm.0.001348. [DOI] [PubMed] [Google Scholar]
- 13.Sugawara Y, Hagiya H, Akeda Y, Aye MM, Myo Win HP, Sakamoto N, Shanmugakani RK, Takeuchi D, Nishi I, Ueda A, Htun MM, Tomono K, Hamada S. 2019. Dissemination of carbapenemase-producing Enterobacteriaceae harbouring blaNDM or blaIMI in local market foods of Yangon, Myanmar. Sci Rep 9:14455. doi: 10.1038/s41598-019-51002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakamoto N, Akeda Y, Sugawara Y, Takeuchi D, Motooka D, Yamamoto N, Laolerd W, Santanirand P, Hamada S. 2018. Genomic characterization of carbapenemase-producing Klebsiella pneumoniae with chromosomally carried blaNDM-1. Antimicrob Agents Chemother 62:e01520-18. doi: 10.1128/AAC.01520-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alghoribi MF, Alqurashi M, Okdah L, Alalwan B, AlHebaishi YS, Almalki A, Alzayer MA, Alswaji AA, Doumith M, Barry M. 2021. Successful treatment of infective endocarditis due to pandrug-resistant Klebsiella pneumoniae with ceftazidime-avibactam and aztreonam. Sci Rep 11:9684. doi: 10.1038/s41598-021-89255-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian D, Wang B, Zhang H, Pan F, Wang C, Shi Y, Sun Y. 2020. Dissemination of the blaNDM-5 gene via IncX3-type plasmid among Enterobacteriaceae in children. mSphere 5:e00699-19. doi: 10.1128/mSphere.00699-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oshiro S, Tada T, Watanabe S, Tohya M, Hishinuma T, Uchida H, Kuwahara-Arai K, Mya S, Zan KN, Kirikae T, Tin HH. 2020. Emergence and spread of carbapenem-resistant and aminoglycoside-panresistant Enterobacter cloacae complex isolates coproducing NDM-type metallo-β-lactamase and 16S rRNA methylase in Myanmar. mSphere 5:e00054-20. doi: 10.1128/mSphere.00054-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoon SH, Ha SM, Lim J, Kwon S, Chun J. 2017. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286. doi: 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
- 19.Deguchi T, Fukuoka A, Yasuda M, Nakano M, Ozeki S, Kanematsu E, Nishino Y, Ishihara S, Ban Y, Kawada Y. 1997. Alterations in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV in quinolone-resistant clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother 41:699–701. doi: 10.1128/AAC.41.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardner SN, Slezak T, Hall BG. 2015. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31:2877–2878. doi: 10.1093/bioinformatics/btv271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.00673-22-s0001.pdf, PDF file, 1.3 MB (1.3MB, pdf)