ABSTRACT
Methanogenic Archaea (methanogens) are a phylogenetically diverse group of microorganisms and are considered to be the most abundant archaeal representatives in the human gut. However, the gut methanogen diversity of human populations in many global regions remains poorly investigated. Here, we report the abundance and diversity of gut methanogenic Archaea in a multi-ethnic cohort of healthy Singaporeans by using a concerted approach of metagenomic sequencing, 16S rRNA gene amplicon sequencing, and quantitative PCR. Our results indicate a mutual exclusion of Methanobrevibacter species, i.e., the highly prevalent Methanobrevibacter smithii and the less prevalent Candidatus Methanobrevibacter intestini in more than 80% of the samples when using an amplicon sequencing-based approach. Leveraging on this finding, we were able to select a fecal sample to isolate a representative strain, TLL-48-HuF1, for Candidatus Methanobrevibacter intestini. The analyzed physiological parameters of M. smithii DSM 861T and strain TLL-48-HuF1 suggest high similarity of the two species. Comparative genome analysis and the mutual exclusion of the Methanobrevibacter species indicate potentially different niche adaptation strategies in the human host, which may support the designation of Candidatus M. intestini as a novel species.
IMPORTANCE Methanogens are important hydrogen consumers in the gut and are associated with differing host health. Here, we determine the prevalence and abundance of archaeal species in the guts of a multi-ethnic cohort of healthy Singapore residents. While Methanobrevibacter smithii is the most prevalent and abundant methanogen in the human gut of local subjects, the recently proposed Candidatus Methanobrevibacter intestini is the abundant methanogen in a minority of individuals that harbor them. The observed potential mutual exclusion of M. smithii and Ca. M. intestini provides further support to the proposal that the two physiologically similar strains may belong to different Methanobrevibacter species.
KEYWORDS: Archaea, human gut microbiota, Methanobrevibacter, methanogens
OBSERVATION
Methanogenic Archaea, also called methanogens, comprise a phylogenetically diverse group of microorganisms with methanogenesis as the exclusive metabolic pathway for energy conservation (1). The presence or absence of methanogens in the human gastrointestinal tract has been associated with different health phenotypes, such as body weight variations, periodontal disease, or cardiovascular disease (2–4). However, most studies aimed at characterizing the methanogen diversity have been performed in the United States or Europe (4, 5), while the gut methanogen diversity in other regions of the world has received little attention. This presents a potential knowledge gap in the understanding of human gut methanogen diversity as different ethnicities or cultural and dietary habits may potentially affect gut microbiota composition. In this regard, Southeast Asia is of particular interest as the gut microbiota of the rapidly growing and multi-ethnic populations in this region remains vastly understudied.
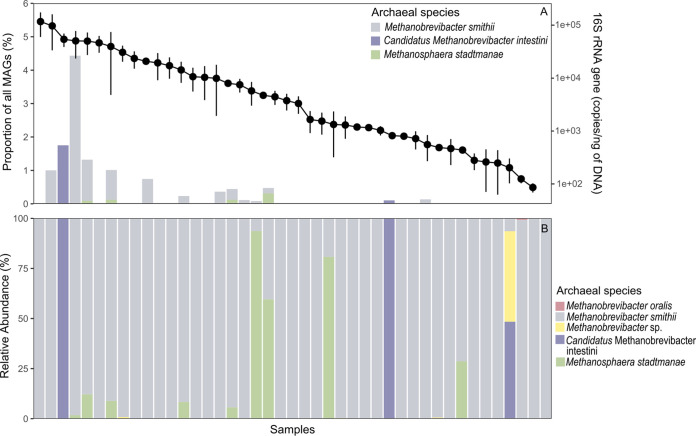
For this study, fecal samples were collected from 109 generally healthy Singapore residents aged 48 to 76 years old (median age = 60 years old) of Chinese (n = 53), Indian (n = 30), or Malay (n = 26) ethnicity (see Table S1 for metadata). The relative and total abundance of methanogens as well as the taxonomic composition in samples were determined by quantitative PCR (qPCR), metagenomic sequencing (MGS), and 16S rRNA gene amplicon sequencing (AS). By using methanogen specific primers for qPCR, methanogens were detected in 44 of 109 (40.3%) samples with a maximum of 1.16 × 105 ± 5.7 × 104 copies/ng of DNA (data variation is expressed as standard deviation throughout the manuscript), which is within the range of positive qPCR detections in studies involving healthy individuals (5, 6) (Fig. 1A). A hybrid-assembly approach using Nanopore long- and Illumina short-sequence reads was used to construct metagenome assembled genomes (MAGs). It was possible to assemble medium-quality (contamination <10%, >50% completeness), near-complete (contamination <5%, >90% completeness) and high-quality (near-complete MAG with a defined set of rRNA and tRNA genes) MAGs in 14 of 109 samples (12.8%) (see Table S2 for MAG statistics and quality). Twelve MAGs belonging to M. smithii, two to Candidatus Methanobrevibacter intestini, and five to Methanosphaera stadtmanae were detected. However, the observed methanogen population as a fraction of the total gut microbiota was low with a maximum relative abundance of 4.43% M. smithii, 1.75% Ca. Methanobrevibacter intestini, and 0.65% Msp. stadtmanae (Fig. 1A). Methanogens of other orders, e.g., Methanomassilicoccales, were not obtained even when low-quality (completeness <50%) MAGs were taken into consideration. A weak correlation (rho = 0.34; Spearman’s rank, P-value = 0.025) between the total abundance of methanogens (qPCR) 16S rRNA genes and relative abundance of methanogen MAGs in qPCR positive samples (n = 44) shows that reliable MAGs are not consistently obtained from samples of high total methanogen abundance. The cause for the MAG underrepresentation in some samples remains currently unclear, but this finding illustrates the importance of using a polyphasic sequencing approach to characterize the methanogen community.
FIG 1.
Methanogen diversity in fecal samples of multi-ethnic Singapore cohort. (A) Stacked barplot shows the relative abundance of methanogen species with MAGs (n = 19) of at least medium quality. The line-plot shows the abundance of archaeal 16S rRNA genes in methanogen positive samples (n = 43) via quantitative PCRs (qPCRs). Samples are arranged in the order of high to low abundance (left to right). One qPCR positive sample was not amplicon sequenced and was omitted from analysis. (B) Relative abundance (rarefied to 1,220 reads per sample) of methanogens in (n = 43) samples characterized using the same primers for qPCR via amplicon sequencing. Methanobrevibacter oralis is in a single sample at 0.8% relative abundance (amplicon sequencing).
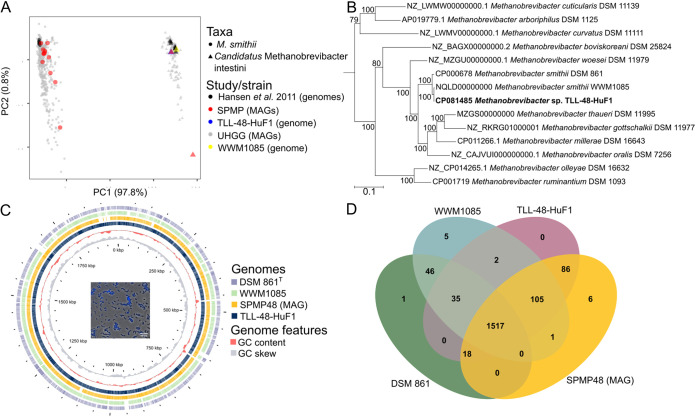
Recent reports suggest that the taxon, Methanobrevibacter smithii may comprise two species (7, 8). Metagenomic databases such as MGnify (9) and Genome Taxonomy Database (10) classified the M. smithii related sequence type as Methanobrevibacter_A smithii_A. A recent analysis of archaeal MAGs purports that M.smithii_A and M. smithii genomes are sufficiently distinct to meet the average nucleotide identity (ANI) species cut-off (>95%) for the former to be classified as ‘Candidatus Methanobrevibacter intestini’ (7, 11) (see Table S4 for ANI). We resolved the identities of previously isolated M. smithii strains to show that 13 out of the 20 isolates were Ca. Methanobrevibacter intestini strains (Fig. 2A) (5).
FIG 2.
Genomic and phylogenetic analysis of Candidatus Methanobrevibacter intestini and Methanobrevibacter strains. (A) A principal coordinate analysis plot of average nucleotide identities among Candidatus Methanobrevibacter intestini and Methanobrevibacter genomes. The variance for each principal coordinate (PC) is shown in parenthesis. The number of MAGs or genomes for each study is n = 729 Unified Human Gastrointestinal Genome (UHGG) collection (43), n = 14 Singapore Platinum Metagenomes Project (SPMP), n = 20 Hansen et al. (5), n = 1 TLL-48-HuF1 (this study) and n = 1 (WWM1085) (12). (B) A protein tree of concatenated alignment of 43 translated CDS. The tree consists of genomes of Methanobrevibacter type strains except for strains WWM1085 and TLL-48-HuF1 as representative strains of Candidatus Methanobrevibacter intestini. NCBI GenBank/RefSeq accession number for each taxon are shown. The tree is rooted to Saccharolobus solfataricus P1 (NCBI RefSeq NZ_LT549890) and is not shown. Scale bar represents 0.1 substitution per amino acid position. (C) Circular genomic map comparing strain TLL-48-HuF1 reference genome (complete) to metagenome-assembled genome of SPMP48 (incomplete), draft genome of strain WWM1085 and a complete genome of Methanobrevibacter smithii DSM 861T. Each ring shows the region of similarity (≥80%) to the reference genome detected by BLAST between translated CDS. CDS were predicted using RAST prior to genomic mapping. The insert is a merged fluorescence and phase contrast image of strain TLL-48-HuF1 viewed at 100 × magnification where blue cells indicate the presence of F420-autoflourescence. Scale bar represents 10 μm. (D) A Venn diagram showing the number of shared orthologous proteins among TLL-48-HuF1, metagenomics assembled genome (MAG) of SPMP48, Methanobrevibacter smithii DSM 861T and Ca. Methanobrevibacter intestini WWM1085.
The 16S rRNA genes of Ca. Methanobrevibacter intestini strains and M. smithii DSM 861T have been previously described as indistinguishable as they share a high sequence identity of >99% (7). Despite their close 16S rRNA gene identity, a few differences exist with the majority found in the V7 region. Specifically, a thymine to cytosine substitution at M. smithii DSM 861T position 1,056 (E. coli position 1,119) and deletions of one to two bases in a homopolymeric thymine-stretch at M. smithii DSM 861T positions 1,076 to 1,081 (E. coli position 1,135 to 1,138) can be observed in the Ca. Methanobrevibacter intestini 16S rRNA gene (see Table S3A for all the nucleotide differences). These differences were also detected in Ca. Methanobrevibacter intestini strain WWM1085 and 12 isolates from a pan-genomic study of gut M. smithii of related individuals (5, 12) (see Table S3B for nucleotide differences between Ca. Methanobrevibacter intestini strains). Due to these differences in the 16S rRNA gene, primer pair Ar915F/Ar1386R (13), which targets the V6-V8 region of the archaeal 16S rRNA gene, was used to characterize methanogen diversity and to distinguish between M. smithii and Ca. Methanobrevibacter intestini using AS.
Analysis of amplicon sequences confirmed the presence of Ca. Methanobrevibacter intestini in the two MGS samples concomitant with the apparent absence of other methanogen phylotypes (Fig. 1B). This contrasts with 26 samples that were solely of M. smithii phylotype. Co-existence of M. smithii and Ca. Methanobrevibacter intestini was detected in seven samples with the latter as a minor component of the methanogen community. In M. smithii positive samples (n = 64) the mean relative abundance was 84.1 ± 31.8%. Methanosphaera stadtmanae was detected in eight samples and had a mean relative abundance of 78% ± 17% in three samples that it dominated (Fig. 1B). The low abundance of Candidatus Methanobrevibacter intestini may have decreased the likelihood of obtaining more MAGs of at least medium quality from more samples. Overall, AS detected methanogens in 66 of 109 samples (60.5%) (see Fig. S2 for phylotypes in AS samples). Twenty-two out of the 66 samples had methanogen abundance below qPCR detection threshold of 30 copies/μL of sample. In contrast to MGS, these samples revealed the presence of methanogen species and orders, such as Methanomassilicoccales, of which some have been isolated from the human gut (14, 15). Methanogen abundance did not significantly differ among ethnicity (P = 0.471; Kruskal-Wallis) or gender (false discovery rate adjusted P = 0.29; Wilcoxon-rank sum test). No significant correlation between methanogen abundance with age was observed (rho = 0.027, p = 0.861, Spearman’s rank).
The apparent mutual exclusion of M. smithii and Ca. Methanobrevibacter intestini allowed the identification of two samples (SPMP39 and SPMP48) suitable as inoculum for the isolation of a Ca. Methanobrevibacter intestini strain. Strain TLL-48-HuF1 (a detailed description of the isolation is provided in the accompanying Materials and Methods section), was successful for one of the samples. Identification of the strain was confirmed by Sanger sequencing the 16S rRNA gene (99.8% of 1,105 bp identity to SPMP48; GenBank accession number OM535902) and the methyl-coenzyme reductase subunit A, mcrA gene (100% of 1,406 bp nucleotide identity to SPMP48; GenBank accession number OM642115). McrA and multilocus markers phylogenetic trees show a clear phylogenetic separation between Ca. Methanobrevibacter intestini and M. smithii DSM 861T compared to a 16S rRNA gene-based tree (Fig. 2B; Fig. S1) and that mcrA may be a more suitable marker gene to analyze methanogen and specifically Methanobrevibacter diversity in the gut.
Comparative genome analysis reveals differences between M. smithii and TLL-48-HuF1 (see Fig. 2D; Tables S5, S6, S7) but also a high degree of shared genome content. Notable exceptions include the absence of methyl-coenzyme M reductase isoenzymes (mrtABDG), some adhesins-like proteins (ALPs), molybdate transporter genes (modA and modB), with the latter two also being reported in other Ca. Methanobrevibacter intestini genomes (7). The lack of a molybdate transporter could potentially indicate a deficiency of molybdopterin metabolism in the cells, affecting enzymes involved in hydrogenotrophic methanogenesis, such as formyl-methanofuran dehydrogenases. Similar observations have been made for Msp. stadtmanae, which is deficient in molybdopterin cofactor (Moco) biosynthesis and thereby restricted to growth on hydrogen and methanol (16). Analysis of the TLL-48-HuF1 genome indicates that it encodes Moco biosynthesis genes and molybdopterin dependent enzyme. In addition, M. smithii DSM 861T and TLL-48-HuF1, showed very similar overall growth characteristics using hydrogen and carbon dioxide for growth (Table S8). While it cannot be completely ruled out that the gene annotation pipelines incorrectly annotated the transporter genes in independent studies, it could also indicate the presence of an alternative, yet to be characterized, molybdate transporter.
The absence of methyl-coenzyme M reductase isoenzyme (mrtABDG) as well as differences in ALP repertoire could hint to specific niche adaptations of Candidatus Methanobrevibacter intestini. Previous studies have shown that the gene expression of mcr and mrt genes may be regulated by hydrogen partial pressure (17). However, this has not been investigated in gut environments and genomes of gut methanogens. Adhesin-like proteins, which were originally discovered in the genome of Msp. stadtmanae, would potentially also contribute to niche adaptations as has been suggested before (16, 18). However, functional characterizations of ALPs remain poor and hypotheses regarding the purpose of the ALP repertoire in vivo remain speculative.
In summary, this study provides insights into the diversity of methanogenic Archaea in the human fecal microbiota of a tri-ethnic cohort in Southeast Asia. The analysis of the relative and absolute abundance of methanogens indicates mutual exclusion of the two species in most samples, which facilitated the isolation of a representative of Ca. Methanobrevibacter intestini. Additional phenotypic characterization of Ca. Methanobrevibacter intestini is required to determine if it is a novel species or a variant, e.g., a distant sequence type, of M. smithii. The ecological relationship of the two species and the temporal duration of the dominance of one sequence type within a subject remain to be elucidated. It is currently not clear if the dominance of Ca. Methanobrevibacter intestini in some subjects is temporary or if the abundance of either species may fluctuate over time. Reanalyzing data from a previous study indicates that heritability may also contribute to the distribution of either species among subjects (5), but this will need to be investigated in more detail with larger cohorts of twins.
MATERIALS AND METHODS
Sample collection and DNA extraction.
Feces from 109 individuals aged 48 to 76 years old of the Singapore Integrative Omics Study (SPMP) were collected in 2018 using a BioCollector (BioCollective) kit, according to the manufacturer’s instructions. Fecal samples were handled in a Coy anerobic chamber containing N2 (75%), CO2 (20%), and H2 (5%) gas mixture. Homogenized samples were transferred to 50 mL screw-cap tubes prior to storage at –80°C. The QIAamp Power Fecal Pro DNA kit was used to extract gDNA for genome (2 × 2 mL pure culture; OD600 = 0.17), metagenome (fecal material; ~0.5 g) sequencing, quantitative PCR, and 16S amplicon sequencing. DNA for genomic sequencing was further purified using a Qiagen Genomic Tip 20/g kit as described in the manufacturer’s protocol (Qiagen, Germany). Cells from cultures were concentrated at 10,000 × g for 15 min before DNA extraction. DNA was quantified using a Qubit 1.0 fluorometer with a broad range assay kit (Life Technologies) and a NanoDrop-2000 (Thermo Fisher Scientific).
Single marker gene-based analyses.
Using general primers for amplification of bacterial 16S rRNA genes and strain TLL-48-HuF1 mcrA-specific primers were used to test culture purity and enrichment of methanogen, respectively. Primer sequences and annealing temperatures are stated in Table S9. Each PCR contained 1 × GoTaq master mix (Promega), 0.2 μM final concentration per primer, and 1.7 ng/μL template. Thermal-cycler condition was generally 95°C for 3 min, 32 cycles of 95°C for 30 s, annealing temperature (Table S9) for 30 s, and 72°C for 30 s (amplicon ≤ 500 bp) followed by a final extension of 72°C for 10 min, unless stated otherwise. Primers designed in this study were checked for specificity using PRIMER-BLAST against NCBI nonredundant nucleotide database using default parameters (19). PCR products were separated by gel electrophoresis using 1.5% (≥1 kb) or 2% (<1 kb) agarose gel in 1 × TAE buffer (40 mM Tris, 20 mM acetic acid, 1 mM EDTA) stained with 1 × FloroSafe (1st Base). Primers Ar915F/Ar1386R were used to quantify methanogenic Archaeal 16S rRNA genes with SYBR-green based qPCR chemistry as previously described (13). The average quantity (copies/μL) of triplicate reactions was quantified as technical replicates and normalized by the amount (ng) of DNA extracted per sample. Standards used for qPCR were 10-fold dilutions of pGEM-T Easy vectors (Promega) ligated with M. smithii DSM 861T 16S rRNA gene 1.3 kb amplicons, which were amplified using Ar84F/Ar1386R. The standard curve ranged from 3 × 101 to 8 copies/μL of sample, with a slope of −3.6, y-intercept of 34.8, and R2 = 0.97. Postamplification melt curve analysis was used to check for reaction specificity based on the average melting temperature (Tm) of the 16S rRNA gene standard (87 ± 0.2°C). Primer pair mcrA-85F/mcrA-1649R was used to amplify a 1.5 kb fragment of the mcrA gene for Sangar sequencing.
Cultivation and isolation of strains.
Methanobrevibacter smithii PS (DSM 861T = ATCC 35061T) was purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH. Cultures were grown as batch cultures contained in 100 mL serum bottles with 20 mL sterile basal medium enclosed with butyl rubber stoppers and aluminum crimp caps, unless stated otherwise. Basal medium was also used to prepare semisolid agarose shakes (0.7% low melting agarose; Life Technologies) and 3% noble agar. The basal medium composition followed medium 1 described in Balch et al. (20) with less Trypticase peptone and yeast extract (1 g/L each) added. Na2S.9H2O 0.25% (wt/vol) and 1 g/L l-cysteine-HCl were added as reducing agents. Sodium bicarbonate 10% (wt/vol) and HEPES (1g/L) buffered the medium at pH 7. All chemicals were purchased from Sigma-Aldrich unless stated otherwise. Cultures were pressurized with H2:CO2 (4:1) at 1.8 bar unless stated otherwise. All cultures were incubated in the dark at 37°C with liquid cultures shaken horizontally at 150 rpm. Gases except hydrogen were purchased from Air Liquide (Singapore) at 99.9995% minimum purity. Hydrogen was generated using a LNI SwissGas HG PRO (Italy) hydrogen generator at 99.9999% purity. Growth was monitored spectrophotometrically at 600 nm optical density (OD600) in an Amersham Biosciences Ultrospec 2100 pro. For enrichment culture, 0.6 g (wet weight) fecal material from a 71-year-old (SGM-48) was added to basal medium amended with ampicillin (100 μg/mL), tetracycline (10 μg/mL), and vancomycin (25 μg/mL) to inhibit bacterial growth. After three consecutive weekly transfers (10%; vol/vol) of the parent enrichment culture, additional ampicillin (200 μg/mL), tetracycline (35 μg/mL), vancomycin (50 μg/mL), and norfloxacin (10 μg/mL) were added to a fourth transfer. Rumen fluid (5% vol/vol) and 0.5 g/L coenzyme M were also added to provide nutrients that might be lacking from the transfers. After 6 days of incubation, serial dilutions (10−1 to −9) and semisolid agarose shakes (0.7% wt/vol; Life Technologies) were prepared in 10-mL headspace glass vials (Agilent Technologies) filled with 3 mL basal medium. Colonies were picked using 1-mL syringes with 21 G 1.5-inch hypodermic needles (BD) and inoculated as batch cultures. To ensure purity, strain TLL-48-HuF1 was further streaked onto 3% (wt/vol) noble agar basal medium (12 mL) contained in agar bottle plates (Bellco Glass) with 1 bar of H2:CO2 (4:1). Substrate utilization tests were performed in triplicate batch cultures in basal media devoid of rumen fluid that contained sodium acetate (61 mM), sodium formate (74 mM), methanol (123.6 mM), or trimethylamine (52 mM). Cultures without hydrogen were filled with N2:CO2 (4:1) at 1.8 bar. OD600 was measured to monitor for growth.
Phase contrast and fluorescence microscopy.
Cultures (OD600 = ~0.4) were immobilized on 2% (wt/vol) agarose coated slides and visualized on an inverted microscope (Zeiss Axio Observer 7) equipped with a 100×/1.4 plan apochromat objective lens, a fluorescence filter (BP 365/12 excitation and LP 397 emission), and a Hamamatsu 2k × 2k CMOS camera. Phase contrast and fluorescent images were merged, and scale bars added using MetaMorph version 7.10.2.240 (Molecular Devices LLC). ImageJ version 1.53e was used to obtain the average length of 100 cells from five fields (21).
Methane analysis.
Methane was measured from culture headspace using a gas chromatograph equipped with a flame ionization detector (GC-FID; Agilent Technologies 6890). Headspace (50 μL) was manually injected using a 250 μL lockable gas-tight syringe (Trajan Scientific and Medical) into the GC-FID. The GC was equipped with a 30 m × 0.32 mm internal diameter GasPro columns (J&W Scientific). Methane was quantified from calibration curves of five-point standards (10 μmol, 100 μmol, 1 mmol, 10 mmol, 100 mmol) that were prepared in bottles under the same conditions as the batch cultures.
Sequencing library construction and DNA sequencing.
Library construct for 16S rRNA gene Illumina amplicon sequencing protocol followed that described for the bacterial 16S rRNA gene (22). The barcoded bacterial 515F/806R primers were replaced with methanogen specific primers Ar915F/Ar1386R (Table S9) while retaining the same sequences for Illumina adapters, Golay barcodes, primer pads, and linkers. PCR followed a previously described condition (13). The library preparation was sequenced on an Illumina MiSeq using 2 × 250 bp chemistry by an external vendor (Axil Scientific). The demultiplexed raw fastq reads were processed using Qiime 2 2021.4 release (23). Forward reads were denoised using DeBlur to obtain amplicon sequence variants (ASV) (24). ASVs fewer than four reads in total were removed to minimize spurious reads (25). ASVs were BLASTed against RIM-DB, a database specific to methanogens of human and animal gut at 80% identity threshold (26). The proportion of ASV per sample is based on a rarefied depth of 1,220 reads per sample using the “qiime diversity core-metrics-phylogenetic” command and default options. Full-length 16S rRNA gene sequence from SPMP48 (MAG) was manually added to RIM-DB prior to clustering at 99.5% using vsearch to remove identical sequences (27). The genome of strain TLL-48-HuF1 was sequenced on a Illumina NovaSeq 6000 Sequencing System and an Oxford Nanopore Technologies MinION equipped with a R9.4.1 flow cell to sequencing depths of 640 × and 208 ×, respectively. Library preparation for paired-end sequencing (2 × 150 bp) on NovaSeq PE150 flow cell was performed externally (NovogeneAIT). MinION sequencing for isolate genome and MAGs was performed using a ligation sequencing kit (SQK-LSK109) and base called using MinIT (Oxford Nanopore Technologies PLC) according to the manufacturer’s protocol. A hybrid genome assembly utilizing short and long reads was generated assembled using UniCycler version 0.4.8 using default parameters (28) (see Table S10 for sequencing coverage and statistics). The SPMP metagenomic library for short reads was constructed using NEBNext Ultra II FS DNA Library Prep Kit for Illumina (New England Biolabs) and paired-end sequenced (2 × 151 bp reads) on an Illumina HiSeq4K platform as previously described (29).
Comparative genome analysis.
Protein coding sequences (CDS) were predicted and annotated from genomes using RAST server (30). Genome statistics was calculated NCBI Prokaryote Annotation Pipeline (31). Circular genomes of CDS were generated using Gview using default parameters (32). Venn diagrams and Swiss-Prot annotations were obtained using OrthoVenn2 (e-value cutoff = 0.01) (33). BlastKoala was used to provide annotation against the Kyoto encyclopedia of genes and genomes (KEGG) database (34). FastANI v1.33 was used calculate ANI between genomes (11).
Phylogenetic analyses.
Maximum likelihood phylogenetic trees from 16S rRNA gene and McrA were generated using raxmlGUI 2.0.5 (35) and bootstrapped using 1,000 iterations each. CheckM v1.1.2 was used to obtain a concatenated amino acid sequence of 43 CDS from each of the 15 genomes compared, including strain WWM1085 (12, 36). A maximum likelihood tree from the 43 CDS was constructed using RAxML v7.0.3 based on the PROTGAMMAJTTF model and 1,000 bootstrap iterations (37).
Metagenomic sequencing assembly and analysis.
Methanogen MAGs are derived from the Singapore Platinum Metagenomes Project (SPMP) (29). Methanogen MAGs are hybrid assemblies of Illumina and MinION data using OPERA-MS v0.9.0 (38). Representative MAGs binned at species level cluster (ANI >95%, mash distances [39]) using sklearn v0.23.2 (40) were assigned taxonomic identities using MAG databases of GTDB-Tk v1.4.1 to Genome Taxonomic Database (GTDB; release 95) (41, 42), and Unified Human Gastrointestinal Genome (UHGG) (43). Relative abundances for MGS data were calculated using Kraken and Bracken with a custom database generated from MGS MAGs (44, 45). Genome quality was assessed for completeness and contamination using CheckM v1.04 (36), Trna and rRNA content using tRNA-scan SE v2.0.5 (46), and barrnap v0.9 (https://github.com/tseemann/barrnap) where threshold for quality MAGs followed the minimum information about a metagenome-assembled genome guidelines (47). MAGs from UHGG collection were downloaded from Mgnify (9). The principal coordinate analysis plot was drawn using the ggplot2 (48) package in R v.4.1.2 (49) based on pairwise fastANI v1.32 (11) distances between a set of genomes composed of both MGS MAGs and external genomes, including UHGG genomes with ANI > 95% (mash distances) and manually selected genomes.
NCBI accession numbers.
Raw AS fastq files have been deposited under BioProject number PRJNA780363. 16S rRNA gene and mcrA sequences from this study were deposited to GenBank with the respective accession numbers OM535902 and OM642115. The hybrid genome of strain TLL-48-HuF1 is assigned GenBank accession number CP081485. MGS short and long reads can be found under BioProject number PRJEB49168. MAGs are deposited at https://zenodo.org/record/6537609#.YnsqD4xByUk.
ACKNOWLEDGMENTS
We thank Jianzhong He for allowing us to use their GC-FID at the National University of Singapore. We extend our appreciation to Yang Fan and Melvin Wong for their help with microscopy at the Bioimaging facility at Temasek Life Sciences Laboratory.
This study was conducted in compliance with the Declaration of Helsinki and national and institutional standards. The collection of fecal samples for this study was approved under the National University of Singapore IRB code H-17-026. Samples were collected with informed consent of the subjects.
Footnotes
Supplemental material is available online only.
Contributor Information
Niranjan Nagarajan, Email: nagarajann@gis.a-star.edu.sg.
Henning Seedorf, Email: henning@tll.org.sg.
Zhenjiang Zech Xu, Nanchang University.
REFERENCES
- 1.Thauer RK, Kaster A-K, Seedorf H, Buckel W, Hedderich R. 2008. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol 6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- 2.Lepp PW, Brinig MM, Ouverney CC, Palm K, Armitage GC, Relman DA. 2004. Methanogenic Archaea and human periodontal disease. Proc Natl Acad Sci USA 101:6176–6181. doi: 10.1073/pnas.0308766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, Spector TD, Clark AG, Ley RE. 2014. Human genetics shape the gut microbiome. Cell 159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borrel G, McCann A, Deane J, Neto MC, Lynch DB, Brugère J-F, O'Toole PW. 2017. Genomics and metagenomics of trimethylamine-utilizing Archaea in the human gut microbiome. ISME J 11:2059–2074. doi: 10.1038/ismej.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen EE, Lozupone CA, Rey FE, Wu M, Guruge JL, Narra A, Goodfellow J, Zaneveld JR, McDonald DT, Goodrich JA, Heath AC, Knight R, Gordon JI. 2011. Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc Natl Acad Sci USA 108:4599–4606. doi: 10.1073/pnas.1000071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dridi B, Henry M, El Khechine A, Raoult D, Drancourt M. 2009. High prevalence of Methanobrevibacter smithii and Methanosphaera stadtmanae detected in the human gut using an improved DNA detection protocol. PLoS One 4:e7063. doi: 10.1371/journal.pone.0007063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chibani CM, Mahnert A, Borrel G, Almeida A, Werner A, Brugere JF, Gribaldo S, Finn RD, Schmitz RA, Moissl-Eichinger C. 2022. A catalogue of 1,167 genomes from the human gut archaeome. Nat Microbiol 7:48–61. doi: 10.1038/s41564-021-01020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasolli E, Asnicar F, Manara S, Zolfo M, Karcher N, Armanini F, Beghini F, Manghi P, Tett A, Ghensi P, Collado MC, Rice BL, DuLong C, Morgan XC, Golden CD, Quince C, Huttenhower C, Segata N. 2019. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 176:649–662. doi: 10.1016/j.cell.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell AL, Almeida A, Beracochea M, Boland M, Burgin J, Cochrane G, Crusoe MR, Kale V, Potter SC, Richardson LJ, Sakharova E, Scheremetjew M, Korobeynikov A, Shlemov A, Kunyavskaya O, Lapidus A, Finn RD. 2020. MGnify: the microbiome analysis resource in 2020. Nucleic Acids Res 48:D570–D578. doi: 10.1093/nar/gkz1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parks DH, Chuvochina M, Chaumeil PA, Rinke C, Mussig AJ, Hugenholtz P. 2020. Author correction: a complete domain-to-species taxonomy for bacteria and archaea. Nat Biotechnol 38:1098. doi: 10.1038/s41587-020-0539-7. [DOI] [PubMed] [Google Scholar]
- 11.Jain C, Rodriguez RL, Phillippy AM, Konstantinidis KT, Aluru S. 2018. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jennings ME, Chia N, Boardman LA, Metcalf WW. 2017. Draft genome sequence of Methanobrevibacter smithii Isolate WWM1085, obtained from a human stool sample. Genome Announc 5:e01055-17. doi: 10.1128/genomeA.01055-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kittelmann S, Seedorf H, Walters WA, Clemente JC, Knight R, Gordon JI, Janssen PH. 2013. Simultaneous amplicon sequencing to explore co-occurrence patterns of bacterial, archaeal and eukaryotic microorganisms in rumen microbial communities. PLoS One 8:e47879. doi: 10.1371/journal.pone.0047879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dridi B, Fardeau ML, Ollivier B, Raoult D, Drancourt M. 2012. Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int J Syst Evol Microbiol 62:1902–1907. doi: 10.1099/ijs.0.033712-0. [DOI] [PubMed] [Google Scholar]
- 15.Khelaifia S, Garibal M, Robert C, Raoult D, Drancourt M. 2014. Draft genome sequence of a human-associated isolate of Methanobrevibacter arboriphilicus, the lowest-G+C-content archaeon. Genome Announc 2:e01181-13. doi: 10.1128/genomeA.01181-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fricke WF, Seedorf H, Henne A, Kruer M, Liesegang H, Hedderich R, Gottschalk G, Thauer RK. 2006. The genome sequence of Methanosphaera stadtmanae reveals why this human intestinal archaeon is restricted to methanol and H2 for methane formation and ATP synthesis. J Bacteriol 188:642–658. doi: 10.1128/JB.188.2.642-658.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pennings JL, Keltjens JT, Vogels GD. 1998. Isolation and characterization of Methanobacterium thermoautotrophicum DeltaH mutants unable to grow under hydrogen-deprived conditions. J Bacteriol 180:2676–2681. doi: 10.1128/JB.180.10.2676-2681.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poehlein A, Daniel R, Seedorf H. 2017. The draft genome of the non-host-associated Methanobrevibacter arboriphilus strain dh1 encodes a large repertoire of adhesin-like proteins. Archaea 2017:4097425. doi: 10.1155/2017/4097425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform 13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. 1979. Methanogens: reevaluation of a unique biological group. Microbiol Rev 43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, Prill RJ, Tripathi A, Gibbons SM, Ackermann G, Navas-Molina JA, Janssen S, Kopylova E, Vazquez-Baeza Y, Gonzalez A, Morton JT, Mirarab S, Zech Xu Z, Jiang L, Haroon MF, Kanbar J, Zhu Q, Jin Song S, Kosciolek T, Bokulich NA, Lefler J, Brislawn CJ, Humphrey G, Owens SM, Hampton-Marcell J, Berg-Lyons D, McKenzie V, Fierer N, Fuhrman JA, Clauset A, Stevens RL, Shade A, Pollard KS, Goodwin KD, Jansson JK, Gilbert JA, Knight R, Earth Microbiome Project C . 2017. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551:457–463. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodriguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amir A, McDonald D, Navas-Molina JA, Kopylova E, Morton JT, Zech Xu Z, Kightley EP, Thompson LR, Hyde ER, Gonzalez A, Knight R. 2017. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2. doi: 10.1128/mSystems.00191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. 2013. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seedorf H, Kittelmann S, Henderson G, Janssen PH. 2014. RIM-DB: a taxonomic framework for community structure analysis of methanogenic archaea from the rumen and other intestinal environments. PeerJ 2:e494. doi: 10.7717/peerj.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gounot J-S, Chia M, Bertrand D, Saw W-Y, Ravikrishnan A, Low A, Ding Y, Ng A, Tan LWL, Teo Y-Y, Seedorf H, Nagarayan N. 2022. Genome-centric analysis of short and long read metagenomes reveals uncharacterized microbiome diversity in Southeast Asians. bioRxiv. [DOI] [PMC free article] [PubMed]
- 30.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W, O'Neill KR, Haft DH, DiCuccio M, Chetvernin V, Badretdin A, Coulouris G, Chitsaz F, Derbyshire MK, Durkin AS, Gonzales NR, Gwadz M, Lanczycki CJ, Song JS, Thanki N, Wang J, Yamashita RA, Yang M, Zheng C, Marchler-Bauer A, Thibaud-Nissen F. 2021. RefSeq: expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. Nucleic Acids Res 49:D1020–D1028. doi: 10.1093/nar/gkaa1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petkau A, Stuart-Edwards M, Stothard P, Van Domselaar G. 2010. Interactive microbial genome visualization with GView. Bioinformatics 26:3125–3126. doi: 10.1093/bioinformatics/btq588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu L, Dong Z, Fang L, Luo Y, Wei Z, Guo H, Zhang G, Gu YQ, Coleman-Derr D, Xia Q, Wang Y. 2019. OrthoVenn2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res 47:W52–W58. doi: 10.1093/nar/gkz333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M, Sato Y, Morishima K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 35.Edler D, Klein J, Antonelli A, Silvestro D. 2021. raxmlGUI 2.0: a graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol Evol 12:373–377. doi: 10.1111/2041-210X.13512. [DOI] [Google Scholar]
- 36.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 38.Bertrand D, Shaw J, Kalathiyappan M, Ng AHQ, Kumar MS, Li C, Dvornicic M, Soldo JP, Koh JY, Tong C, Ng OT, Barkham T, Young B, Marimuthu K, Chng KR, Sikic M, Nagarajan N. 2019. Hybrid metagenomic assembly enables high-resolution analysis of resistance determinants and mobile elements in human microbiomes. Nat Biotechnol 37:937–944. doi: 10.1038/s41587-019-0191-2. [DOI] [PubMed] [Google Scholar]
- 39.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. 2016. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 17:1–14. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V. 2011. Scikit-learn: machine learning in Python. J Mach Learn Res 12:2825–2830. [Google Scholar]
- 41.Parks DH, Chuvochina M, Rinke C, Mussig AJ, Chaumeil PA, Hugenholtz P. 2022. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res 50:D785–D794. doi: 10.1093/nar/gkab776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH. 2019. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Almeida A, Nayfach S, Boland M, Strozzi F, Beracochea M, Shi ZJ, Pollard KS, Sakharova E, Parks DH, Hugenholtz P, Segata N, Kyrpides NC, Finn RD. 2021. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat Biotechnol 39:105–114. doi: 10.1038/s41587-020-0603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu J, Breitwieser FP, Thielen P, Salzberg SL. 2017. Bracken: estimating species abundance in metagenomics data. PeerJ Comput Sci 3:e104. doi: 10.7717/peerj-cs.104. [DOI] [Google Scholar]
- 45.Wood DE, Lu J, Langmead B. 2019. Improved metagenomic analysis with Kraken 2. Genome Biol 20:1–13. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res 44:W54–W57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bowers RM, Kyrpides NC, Stepanauskas R, Harmon-Smith M, Doud D, Reddy TBK, Schulz F, Jarett J, Rivers AR, Eloe-Fadrosh EA, Tringe SG, Ivanova NN, Copeland A, Clum A, Becraft ED, Malmstrom RR, Birren B, Podar M, Bork P, Weinstock GM, Garrity GM, Dodsworth JA, Yooseph S, Sutton G, Glockner FO, Gilbert JA, Nelson WC, Hallam SJ, Jungbluth SP, Ettema TJG, Tighe S, Konstantinidis KT, Liu WT, Baker BJ, Rattei T, Eisen JA, Hedlund B, McMahon KD, Fierer N, Knight R, Finn R, Cochrane G, Karsch-Mizrachi I, Tyson GW, Rinke C, Genome Standards C, Lapidus A, Meyer F, Yilmaz P, Parks DH, Genome Standards Consortium, et al. 2017. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol 35:725–731. doi: 10.1038/nbt.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wickham H. 2011. ggplot2. Wiley interdisciplinary reviews: computational statistics. WIREs Comp Stat 3:180–185. doi: 10.1002/wics.147. [DOI] [Google Scholar]
- 49.R Core Team. 2021. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.00849-22-s0001.pdf, PDF file, 3.9 MB (4MB, pdf)