ABSTRACT
Mobile colistin resistance (mcr) gene mcr-10.1 has been distributed widely since it was initially identified in 2020. The aim of this study was to report the first mcr-10.1 in Africa and the first mcr in Sierra Leone; furthermore, we presented diverse modular structures of mcr-10.1 loci. Here, the complete sequence of one mcr-10.1-carrying plasmid in one clinical Enterobacter cloacae isolate from Sierra Leone was determined. Detailed genetic dissection and comparison were applied to this plasmid, together with a homologous plasmid carrying mcr-10.1 from GenBank. Moreover, a genetic comparison of 19 mcr-10.1 loci was performed. In this study, mcr-10.1 was carried by an IncpA1763-KPC plasmid from one Enterobacter cloacae isolate. A total of 19 mcr-10.1 loci displayed diversification in modular structures through complex transposition and homologous recombination. A site-specific tyrosine recombinase XerC was located upstream of mcr-10.1, and at least one insertion sequence element was inserted adjacent to a conserved xerC-mcr-10.1-orf336-orf177 region. Integration of mcr-10.1 into a different gene context and carried by various Inc plasmids contributed to the wide distribution of mcr-10.1 and enhanced the ability of bacteria to survive under colistin selection pressure.
IMPORTANCE Colistin is used as one of the last available choices of antibiotics for patients infected by carbapenem-resistant bacterial strains, but the unrestricted use of colistin aggravated the acquisition and dissemination of mobile colistin resistance (mcr) genes. So far, 10 mcr genes have been reported in four continents around the world. This study presented one mcr-10.1-carrying Enterobacter cloacae isolate from Sierra Leone. The mcr-10.1 gene was identified on an IncpA1763-KPC plasmid. According to the results of genetic comparison of 19 mcr-10.1 loci, the mcr-10.1 gene was found to be located in a conserved xerC-mcr-10.1-orf336-orf177 region, and at least one insertion sequence element was inserted adjacent to this region. To our knowledge, this is the first report of identifying the mcr-10.1 gene in Africa and the mcr gene in Sierra Leone.
KEYWORDS: Enterobacter cloacae, colistin resistance, mcr-10.1, IncpA1763-KPC plasmid
INTRODUCTION
Colistin is one of the last choices of antibiotic to treat severe Gram-negative bacterial infections of humans, especially infections caused by bacteria with reduced susceptibility to carbapenem antibiotics, and it has been used in livestock for more than 60 years in most countries of the world (1). Morganellaceae, the Burkholderia cepacia complex, and Serratia marcescens are intrinsically resistant to colistin due to the presence of the cell wall that inhibits colistin binding with the susceptible lipid target site or the lipid A modification to reduce binding (2). Recently, the unrestricted use of colistin aggravated the acquisition and dissemination of mobile colistin resistance (mcr) genes in Enterobacteriaceae (3–5), Moraxellaceae (6), Morganellaceae (6, 7), Aeromonas (7), Alcaligenes (8), Cupriavidus (9), Pseudomonas (6), Serratia (6), Shewanella (6), and Vibrio (6). The mcr genes encode phosphoethanolamine (PEA) transferases that catalyze the combination of PEA with lipid A and thus modify the structure of lipid A to reduce the binding affinity to colistin (10). So far, 10 mcr genes, including mcr-1 to mcr-10 with different subvariants, have been reported in four continents around the world (11).
The mcr-10 gene was first identified in an IncFIA plasmid, pMCR10_090065, from Enterobacter roggenkampii in China in 2020 (11). Since then, mcr-10 has been found in IncFIB (12), IncFII:IncFIA (13), IncFII:IncFIB, and IncFIB:IncFIA plasmids from Asia, Europe, Oceania, and North America, but not from Africa, South America, and Antarctica (11).
In Africa, seven (except for mcr-6, mcr-7, and mcr-10) of the 10 mcr genes have been found in IncFIB, IncFII, IncHI, IncI, IncN, IncP, IncR, and IncX plasmids from Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Pseudomonas putida, Pseudomonas luteola, Enterobacter hormaechei, Acinetobacter baumannii, Citrobacter werkmanii, and Alcaligenes faecalis (8, 14). These mcr-carrying bacteria were isolated from human, animals, plants, and contaminated soil, water, and wildlife ecosystems. So far, none of mcr genes have been reported in Sierra Leone (8).
This study presented the complete sequence of one mcr-10.1-carrying plasmid in one sequenced clinical Enterobacter cloacae isolate from Sierra Leone. Detailed genetic dissection and comparison were applied to this plasmid, together with a plasmid carrying mcr-10.1 from GenBank. Moreover, a genetic comparison of 19 mcr-10.1 loci was performed to present diversification in modular structures of mcr-10.1. To our knowledge, this is the first report of identifying the mcr-10.1 gene in Africa and the mcr gene in Sierra Leone.
RESULTS
Identification and antimicrobial susceptibility of Enterobacter cloacae SL12517.
Strain SL12517 has a 98.74% average nucleotide identity (ANI) value with the reference strain Enterobacter cloacae ATCC 13047 (accession number CP001918). Multilocus sequencing typing (MLST) analysis revealed that strain SL12517 belonged to sequence type 850 (ST850).
Strain SL12517 was resistant to colistin (MIC, 8 μg/mL), cefazolin (MIC, ≥64 μg/mL), gentamicin (MIC, ≥16 μg/mL), and trimethoprim/sulfamethoxazole (MIC, ≥320 μg/mL), intermediate to piperacillin (MIC, 32 μg/mL), tobramycin (MIC, 8 μg/mL), and nitrofurantoin (MIC, 64 μg/mL), and susceptible to piperacillin/tazobactam (MIC, ≤4 μg/mL), cefuroxime (MIC, 4 μg/mL), ceftazidime (MIC, ≤1 μg/mL), ceftriaxone (MIC, ≤1 μg/mL), cefepime (MIC, ≤1 μg/mL), aztreonam (MIC, ≤1 μg/mL), imipenem (MIC, ≤1 μg/mL), meropenem (MIC, ≤0.25 μg/mL), amikacin (MIC, ≤2 μg/mL), ciprofloxacin (MIC, ≤0.5 μg/mL), and levofloxacin (MIC, ≤1 μg/mL).
Identification of resistance genes carried by strain SL12517.
Resistance genes carried by strain SL12517 were identified using the Comprehensive Antibiotic Resistance Database (CARD) and the ResFinder database. The chromosome of strain SL12517 carried the blaCMH-3 gene. An IncFII plasmid, pSL12517-TEM, carried blaTEM-1B and aacC2e genes. An IncpA1763-KPC plasmid, pSL12517-mcr10.1, contained mcr-10.1, aac2d, strA, strB, tetA(D), qnrS1, catA2, dfrA14b, tmrB, and sul2 genes. A ColRNAI plasmid, pSL12517-NR, harbored no resistance genes.
Sequence comparison of two IncpA1763-KPC plasmids.
A detailed sequence comparison was applied to two mcr-10.1-carrying IncpA1763-KPC plasmids; one was plasmid pSL12517-mcr10.1, which was isolated from strain SL12517, sequenced here, and the other one was pEC27-2 (15) from GenBank, which was recovered from one Enterobacter cloacae isolate in Vietnam in 2010. The plasmid pSL12517-mcr10.1 shared 99.94% nucleotide identity with pEC27-2 with 99% coverage. A total of 57 and 70 open reading frames (ORFs) were predicted in pSL12517-mcr10.1 (58.1 kb long; Fig. 1) and pEC27-2 (84.6 kb long; Fig. 2), respectively. At least 12 antimicrobial resistance genes, mcr-10.1, blaTEM-1, blaLAP-2, aac2d, strA, strB, tetA(D), qnrS1, catA2, dfrA14b, tmrB, and sul2, involved in resistance to 9 different categories of antimicrobials (colistin, β-lactams, aminoglycosides, tetracycline, quinolone, chloramphenicol, trimethoprim, tunicamycin, and sulfonamide), were identified in these two plasmids.
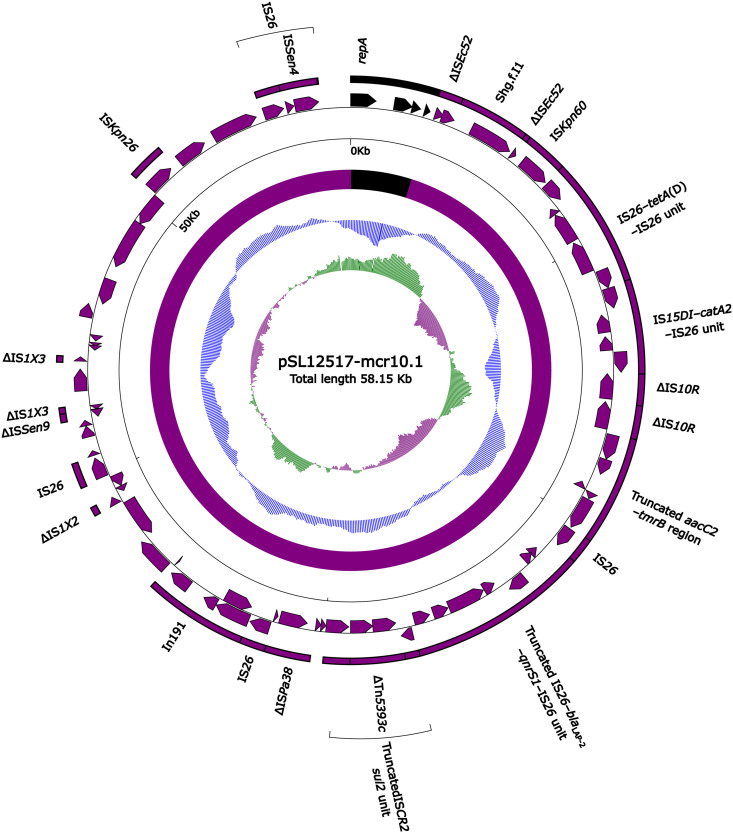
FIG 1.
Schematic map of plasmid pSL12517-mcr10.1. Genes are denoted by arrows, and the backbone and accessory module regions are highlighted in black and purple, respectively. The innermost circle presents GC-skew [(G–C)/(G+C)], with a window size of 500 bp and a step size of 20 bp. The next-to-innermost circle presents GC content.
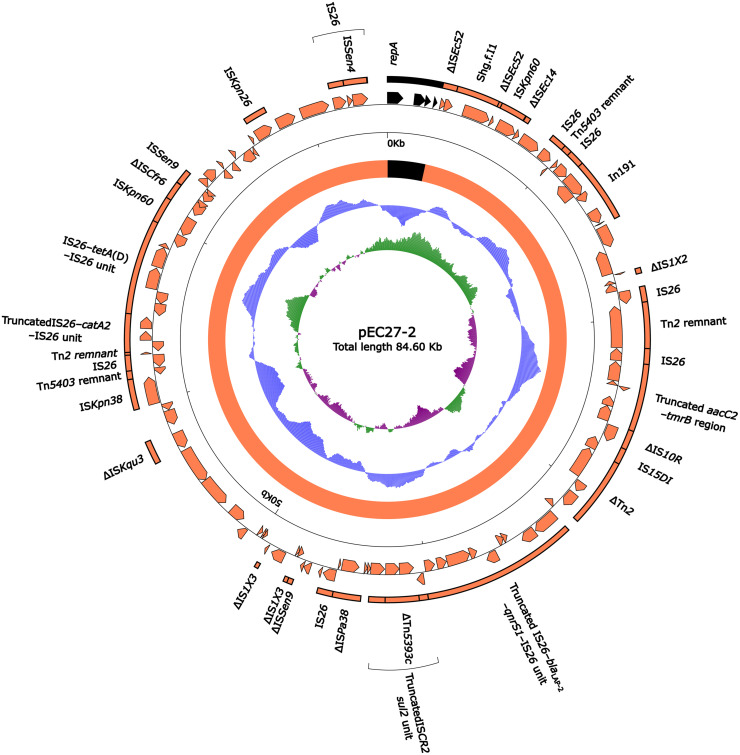
FIG 2.
Schematic map of plasmid pEC27-2. Genes are denoted by arrows, and the backbone and accessory module regions are highlighted in black and orange, respectively. The innermost circle presents GC-skew [(G–C)/(G+C)], with a window size of 500 bp and a step size of 20 bp. The next-to-innermost circle presents GC content.
The two plasmids shared a small backbone region (2.8 kb in length), including repIncpA1763-KPC, parA, and two undetermined genes (hypothetical proteins). Two multidrug resistance (MDR) regions (Fig. 3) MDRpSL1217-mcr10.1 (55.2 kb long) and MDRpEC27-2 (81.6 kb long) were integrated at the same site adjacent to the rep within the two plasmids, respectively.
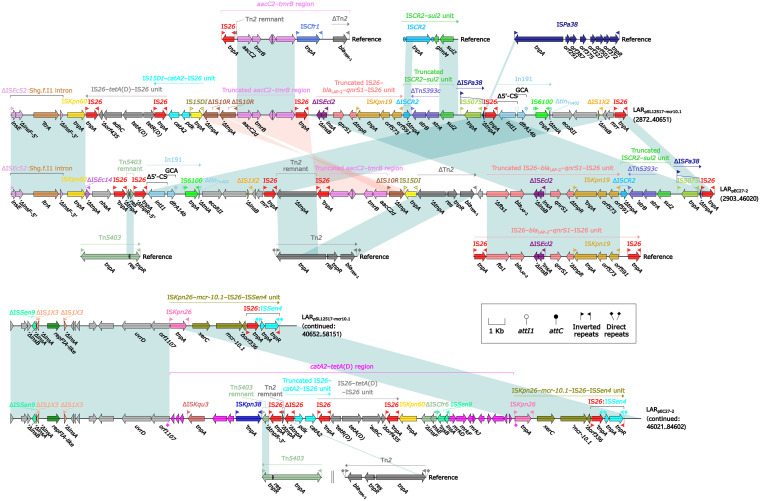
FIG 3.
Comparison of MDR regions from pSL12517-mcr10.1 and pEC27-2. Genes are denoted by arrows. Genes, accessory genetic elements (AGEs), and other features are colored based on their functional classification. Shading in light blue or light pink denotes regions of homology (nucleotide identity ≥95%). Numbers in brackets indicate nucleotide positions within plasmids pSL12517-mcr10.1 and pEC27-2. Accession numbers of Tn5403 (40), the aacC2-tmrB region (41), Tn2 (42), ISPa38, the ISCR2-sul2 unit (43), and the IS26 = blaLAP-2-qnrS1-IS26 unit (40) used as reference are KJ958926, JX101693, HM749967, CP003149, AE014073, and HF545433, respectively.
MDRpSL1217-mcr10.1 and MDRpEC27-2 shared a truncated aacC2-tmrB region, a truncated IS26-blaLAP-2-qnrS1-IS26 unit, a truncated ISCR2-sul2 unit (containing the strAB-carrying ΔTn5393c), a concise class 1 integron In191 with the gene cassette array (GCA) dfrA14b, and ISKpn26-mcr-10.1-IS26-ISSen4 unit, but each of them integrated two additional resistance loci: (i) the IS26-tetA(D)-IS26 unit and IS15DI-catA2-IS26 unit in MDRpSL1217-mcr10.1 and (ii) the ΔTn2 and catA2-tetA(D) region (bracketed by the same 4-bp direct repeats [DRs]; target site duplication signals for transposition) in MDRpEC27-2. Notably, 8 and 12 copies of IS26, IS15DI, and IS6100 were presented in MDRpSL1217-mcr10.1 and MDRpEC27-2, respectively, all of which belonged to the IS6 family and carried almost identical 14-bp inverted repeat (IR) sequences. It showed that these IS elements participate in complex homologous recombination events and promote the assembly of complex mosaic structures as observed in MDRpSL1217-mcr10.1 and MDRpEC27-2 (16).
Comparison of 19 mcr-10.1 loci from 19 plasmids.
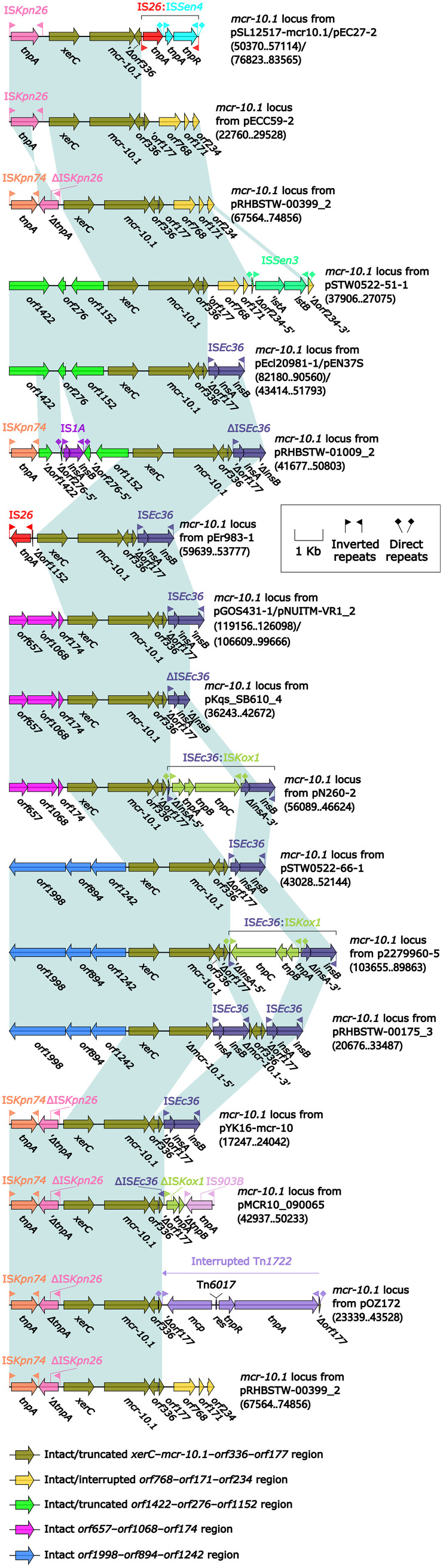
Detailed genetic dissection and sequence comparison were applied to 19 mcr-10.1 loci (Fig. 4) from 19 plasmids identified from GenBank as of 25 January 2022 (Table 1; see Table S1 in the supplemental material). Each mcr-10.1 loci carried an intact or truncated version of xerC (site-specific tyrosine recombinase)-mcr-10.1-orf336 (hypothetical protein)-orf177 (hypothetical protein) region. Various insertion sequence (IS) elements, unit transposons, and undetermined genes were present upstream or downstream of the xerC-mcr-10.1-orf336-orf177 region: (i) an intact ISKpn26 upstream of xerC in each mcr-10.1 loci from pECC59-2, pSL12517-mcr10.1, and pEC27-2; (ii) an orf1422-orf276-orf1152 region upstream of xerC in mcr-10.1 loci from pSTW0522-51-1, pEcl20981-1, and pEN37S, and a truncated orf1422-orf276-orf1152 region upstream of xerC in each mcr-10.1 loci from pRHBSTW-01009_2 and pEr983-1 (10); (iii) an orf657-orf1068-orf174 region upstream of xerC in mcr-10.1 loci from pGOS431-1, pNUITM-VR1_2, pKqs_SB610_4, and pN260-2 (12); (iv) an orf1998-orf894-orf1242 region upstream of xerC in mcr-10.1 loci from pSTW0522-66-1, p2279960-5, and pRHBSTW-00175_3; (v) an incomplete ISKpn26 truncated by ISKpn74 upstream of xerC in each mcr-10.1 loci from pRHBSTW-00399_2, pOZ172 (13), pMCR10_090065, and pYK16-mcr-10 (17); (vi) IS26, contributing to truncation of orf336 in mcr-10.1 loci from pSL12517-mcr10.1 and pEC27-2; (vii) an orf768-orf171-orf234 region downstream of orf177 in mcr-10.1 loci from pECC59-2 and pRHBSTW-00399_2, and a truncated orf768-orf171-orf234 region downstream of orf177 in the mcr-10.1 locus from pSTW0522-51-1; (viii) intact or truncated ISEc36, leading to truncation of orf177, in mcr-10.1 loci from 13 (except for pECC59-2, pRHBSTW-00399_2, pSTW0522-51-1, pOZ172, pEC27-2, and pSL12517-mcr10.1) of 19 plasmids; and (ix) an interrupted Tn1722, resulting in truncation of orf177, in the mcr-10.1 locus from pOZ172. These results indicated that the xerC-mcr-10.1-orf336-orf177 region might be the most conserved structure, and we could not determine which mcr-10.1 locus was the earliest among these 19 plasmids.
FIG 4.
Comparison of 19 mcr-10.1 loci from 19 plasmids. Genes are denoted by arrows. Genes, AGEs, and other features are colored based on their functional classification. Shading in light blue denotes regions of homology (nucleotide identity ≥95%). Numbers in brackets indicate nucleotide positions within the 19 plasmids.
TABLE 1.
General features of the 19 mcr-10.1-carrying plasmidsa
| Plasmid | GenBank accession no. | Total length (bp) | Location | Host bacterium | Reference or sourceb |
|---|---|---|---|---|---|
| pSL12517-mcr10.1 | MW048777 | 58,151 | Sierra Leone | Enterobacter cloacae SL12517 | This study |
| pEC27-2 | CP020091 | 84,602 | Vietnam | Enterobacter cloacae PIMB10EC27 | 15 |
| pECC59-2 | CP080472 | 64,293 | China | Enterobacter hormaechei ECC59 | NA |
| pRHBSTW-00399_2 | CP056561 | 137,623 | UK | Enterobacter cloacae RHBSTW-00399 | NA |
| pSTW0522-51-1 | AP022432 | 159,829 | Japan | Enterobacter kobei STW0522-51 | Not applicable |
| pEcl2098-1 | CP048651 | 161,986 | China | Enterobacter roggenkampii Ecl_20_981 | NA |
| pEN37S | AP024497 | 70,277 | Japan |
Enterobacter cloacae En37 |
NA |
| pRHBSTW-01009_2 | CP056127 | 70,650 | UK | Enterobacter asburiae RHBSTW-01009 | NA |
| pEr983-1 | CP060738 | 100,102 | China | Enterobacter roggenkampii Ecl-983 | 10 |
| pGOS431-1 | CP023893 | 231,294 | Canada | Raoultella ornithinolytica FDAARGOS_431 | NA |
| pNUITM-VR1_2 | AP025011 | 261,835 | Vietnam | Raoultella ornithinolytica NUITM-VR1 | NA |
| pKqs_SB610_4 | CP084774 | 124,980 | Netherlands | Klebsiella quasipneumoniae SB610 | NA |
| pN260-2 | AP023449 | 244,996 | Japan | Enterobacter roggenkampii OIPH-N260 | 12 |
| pSTW0522-66-1 | AP022466 | 324,199 | Japan | Enterobacter roggenkampii STW0522-66 | NA |
| p2279960-5 | LR890193 | 120,029 | Australia | K. pneumoniae INF133-sc-2279960 | NA |
| pRHBSTW-00175_3 | CP055932 | 68,715 | UK | Enterobacter sp. strain RHBSTW-00175 | NA |
| pYK16-mcr-10 | MT468575 | 117,855 | China | Enterobacter roggenkampii YK16 | 17 |
| pMCR10_090065 | CP045065 | 71,775 | China | Enterobacter roggenkampii WCHER090065 | 11 |
| pOZ172 | CP016763 | 127,005 | China | Citrobacter freundii B38 | 13 |
All the completely sequenced and nonredundant mcr-10.1-carrying plasmids available in GenBank (last accessed 25 January 2022) are included. Three unnamed plasmids from strain Ecl_20_981, FDAARGOS_431, and INF133-sc-2279960 were here named pEcl20981-1, pGOS431-1, and p2279960-5, respectively.
NA, not applicable.
Conjugation experiments.
We failed to obtain transconjugants containing mcr-10.1 no matter how many times the conjugation experiments were performed, which might be because the essential conjugal transfer genes, including rlx (relaxase), oriT (origin of conjugative replication), pri (DNA primase), cpl (coupling protein), and type IV secretion system (T4SS), were absent in pSL12517-mcr10.1.
DISCUSSION
Enterobacter cloacae is a vital nosocomial pathogen and is able to cause bacteremia and other infections in humans and animals (18). Due to the wide use of antibiotics, multidrug-resistant, especially carbapenem-resistant, Enterobacter cloacae emerged (19); therefore, colistin is used as one of the last available choices of antibiotics for patients infected by carbapenem-resistant strains (20). However, mcr-carrying Enterobacteriaceae have been identified all over the world recently (9, 21, 22). This study presented the complete sequence of one mcr-10.1-carrying IncpA1763-KPC plasmid in one sequenced Enterobacter cloacae isolate from Sierra Leone. Detailed genetic dissection and comparison were applied to this plasmid, together with a homologous IncpA1763-KPC plasmid carrying mcr-10.1 from GenBank. Moreover, a genetic comparison of 19 mcr-10.1 loci was performed to display diversification in modular structures of mcr-10.1.
The mcr-10.1 usually mediated low-level colistin resistance in early reports (10, 11), but strain SL12517 in this study displayed high-level colistin resistance with a MIC of 8 μg/mL. A previous study demonstrated that mcr-10.1 was able to cofunction with phoP (two-component system response regulator) and phoQ (two-component system sensor histidine kinase) to mediate the high-level colistin resistance (10). In this study, phoPQ was identified on the chromosome of strain SL12517, indicating that phoPQ might very likely participate in the high-level colistin resistance.
The IncpA1763-KPC plasmid carried an IncpA1763-KPC replicon, which was composed of repAIncpA1763-KPC and its iterons (23). The IncpA1763-KPC replicon (previously called RepBRep_3-family) was initially found in pK245 from K. pneumoniae in 2006 in Taiwan (24); since then, it has been frequently found in different plasmids in many K. pneumoniae isolates. In this study, two IncpA1763-KPC plasmids, pSL12517-mcr10.1 and pEC27-2 (15), were identified in Enterobacter cloacae recovered from Sierra Leone in 2018 and from Vietnam in 2010, respectively. Only two mcr-10.1-carrying IncpA1763-KPC plasmids (pSL12517-mcr10.1 and pEC27-2) have been identified until now, and no mcr-10.1-carrying IncpA1763-KPC plasmids were found in other species of bacteria. This result indicated that transfer of the IncpA1763-KPC plasmids without mcr-10.1 from K. pneumoniae to Enterobacter cloacae was prior to acquisition of mcr-10.1 by the IncpA1763-KPC plasmids. pEC27-2 was found earlier than pSL12517-mcr10.1, and colistin has not been used clinically in Sierra Leone (25); therefore, we speculate that pEC27-2 was possibly transferred from Vietnam to Sierra Leone through international food (animal- and plant-based) trade or travel (8).
According to detailed genetic dissection and comparison of 19 mcr-10.1 loci, the genetic organization xerC-mcr-10.1-orf336-orf177 might be the original modular structure of the mcr-10.1 locus. Various IS elements or transposons were inserted upstream or downstream of the xerC-mcr-10.1-orf336-orf177 region, which resulted in the truncation of orf177, but no truncation of xerC was found. Some mobile genetic elements (MGEs) integrated into the chromosomes using xerC-encoding tyrosine recombinases in Enterobacter cloacae (26, 27). This indicated that xerC could participate in mobilization of mcr-10.1 (10, 11). Diverse IS elements or transposons inserted upstream or downstream of the xerC-mcr-10.1-orf336-orf177 region suggest that the area surrounding this conserved region is the high-frequency region for insertion of MGEs (3).
In conclusion, this is the first report of identifying the mcr-10.1 gene in Africa and the mcr gene in Sierra Leone. The mcr-10.1 gene was able to rely on plasmids to accomplish intercellular transfer and on site-specific tyrosine recombinase to achieve intracellular transfer. Although mcr-10.1 was first identified in 2020, it showed the tendency of rapid propagation throughout the world due to uncontrolled colistin consumption. So far, mcr-10.1, which could be carried by Enterobacter cloacae, Enterobacter kobei, Enterobacter roggenkampii, Enterobacter asburiae, K. pneumoniae, Klebsiella quasipneumoniae, Raoultella ornithinolytica, and Citrobacter freundii, had been found in Sierra Leone, China, Japan, Vietnam, the United Kingdom, Netherlands, Canada, and Australia. It could be captured by various MGEs and integrated in diverse types of plasmids. Particularly, it should be noted that the high MIC value due to mcr-10.1 might enhance the ability of bacteria to survive under colistin selection pressure and aggravate the difficulty in treating infections caused by mcr-10.1-carrying bacteria, especially in low-income countries. Therefore, it is necessary to continuously monitor the spread of mcr-10.1 in the future.
MATERIALS AND METHODS
Bacterial isolation and identification.
Strain SL12517 was recovered from a public hospital in Sierra Leone in 2018 (28). The MIC of colistin was determined by the broth microdilution method according to Clinical and Laboratory Standards Institute (CLSI) guidelines (29). The breakpoint of colistin was defined by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (http://www.eucast.org). The Escherichia coli ATCC 25922 strain was used as a control. MICs of piperacillin, piperacillin/tazobactam, cefazolin, cefuroxime, ceftazidime, ceftriaxone, cefepime, aztreonam, imipenem, meropenem, amikacin, gentamicin, tobramycin, ciprofloxacin, levofloxacin, nitrofurantoin, and trimethoprim/sulfamethoxazole were tested using Vitek 2 and interpreted according to the CLSI guidelines (29).
Sequencing and sequence assembly.
Bacterial genomic DNA was isolated from strain SL12517 using the UltraClean microbial kit (Qiagen, North Rhine-Westphalia, Germany), and sequenced with a PacBio RS II sequencer (Pacific Biosciences, CA, USA). The reads were assembled de novo utilizing SMARTdenovo (http://github.com/ruanjue/smartdenovo).
Bacterial precise species identification and genotyping.
Bacterial precise species identification was performed using pairwise ANI analysis between strain SL12517 and the reference genome (http://www.ezbiocloud.net/tools/ani). A ≥95% ANI cutoff was used to define a bacterial species (30). Genotyping of strain SL12517 was performed by MLST at the online database PubMLST (http://pubmlst.org).
Sequence annotation and comparison.
RAST 2.0 (31) and blastp/blastn (32) searches were used to predicted ORFs. The online databases CARD (33), ResFinder (34), ISfinder (35), INTEGRALL (36), and Tn number registry (37) were used to find resistance genes and mobile elements. Pairwise sequence comparisons were carried out with blastn. Inkscape 1.0 was used to draw gene organization diagrams (http://inkscape.org/en/).
Conjugation experiments.
Conjugation experiments were performed with strain SL12517 used as a donor and rifampin-resistant Escherichia coli EC600 as a recipient (38, 39). Donor and recipient strains (3 mL each) were cultured overnight at 37°C and mixed together. The mixed cells were harvested by centrifugation for 3 min at 1,200 × g, washed with 3 mL of Luria-Bertani (LB) broth and resuspended in 150 μL of LB broth. The mixture was spotted on a 1-cm2 hydrophilic nylon membrane filter with a 0.45-μm pore size (Millipore), which was placed on an LB agar plate and then incubated for mating at 37°C for 6 h. The cells were recovered from the filter membrane and spotted on Muller-Hinton (MH) agar (BD Biosciences) plates containing 1,500 μg/mL rifampin and 4 μg/mL colistin for selecting an mcr-10.1-carrying transconjugant.
Data availability.
The complete sequence of plasmid pSL12517-mcr10.1 has been submitted to GenBank under the accession number MW048777.
ACKNOWLEDGMENTS
This research was funded by the National Key Research and Development Program of China, grant number 2016YFD0501305.
Conceptualization, X.G. and L. Zhu; investigation, J.G., L. Zheng, X.J., and Y.S.; formal analysis, J.G., G.L., Y.W., B.J., and J.L.; resources, S.L. and S.S.; writing–original draft preparation, J.G. and L.L.; writing–review and editing, X.G., L. Zhu, and S.L. All authors have read and agreed to the published version of the manuscript.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Lingwei Zhu, Email: lingweiz@126.com.
Xuejun Guo, Email: xuejung2021@163.com.
Daria Van Tyne, University of Pittsburgh School of Medicine.
REFERENCES
- 1.Sun J, Zhang H, Liu YH, Feng Y. 2018. Towards understanding MCR-like colistin resistance. Trends Microbiol 26:794–808. doi: 10.1016/j.tim.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Samonis G, Korbila IP, Maraki S, Michailidou I, Vardakas KZ, Kofteridis D, Dimopoulou D, Gkogkozotou VK, Falagas ME. 2014. Trends of isolation of intrinsically resistant to colistin Enterobacteriaceae and association with colistin use in a tertiary hospital. Eur J Clin Microbiol Infect Dis 33:1505–1510. doi: 10.1007/s10096-014-2097-8. [DOI] [PubMed] [Google Scholar]
- 3.Xu L, Wan F, Fu H, Tang B, Ruan Z, Xiao Y, Luo Q. 2022. Emergence of colistin resistance gene mcr-10 in Enterobacterales isolates recovered from fecal samples of chickens, slaughterhouse workers, and a nearby resident. Microbiol Spectr 10:e0041822. doi: 10.1128/spectrum.00418-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hmede Z, Sulaiman AAA, Jaafar H, Kassem II. 2019. Emergence of plasmid-borne colistin resistance gene mcr-1 in multidrug-resistant Escherichia coli isolated from irrigation water in Lebanon. Int J Antimicrob Agents 54:102–104. doi: 10.1016/j.ijantimicag.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Caselli E, D’Accolti M, Soffritti I, Piffanelli M, Mazzacane S. 2018. Spread of mcr-1-driven colistin resistance on hospital surfaces, Italy. Emerg Infect Dis 24:1752–1753. doi: 10.3201/eid2409.171386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khedher MB, Baron SA, Riziki T, Ruimy R, Raoult D, Diene SM, Rolain J-M. 2020. Massive analysis of 64,628 bacterial genomes to decipher water reservoir and origin of mobile colistin resistance genes: is there another role for these enzymes? Sci Rep 10:5970. doi: 10.1038/s41598-020-63167-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Zhai W, Li J, Liu D, Zhang Q, Shen Z, Wang S, Wang Y. 2018. Presence of an mcr-3 variant in Aeromonas caviae, Proteus mirabilis, and Escherichia coli from one domestic duck. Antimicrob Agents Chemother 62:e02106-17. doi: 10.1128/AAC.02106-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anyanwu MU, Okpala COR, Chah KF, Shoyinka VS. 2021. Prevalence and traits of mobile colistin resistance gene harbouring isolates from different ecosystems in Africa. Biomed Res Int 2021:6630379. doi: 10.1155/2021/6630379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El-Sayed Ahmed MAE-G, Zhong L-L, Shen C, Yang Y, Doi Y, Tian G-B. 2020. Colistin and its role in the Era of antibiotic resistance: an extended review (2000–2019). Emerg Microbes Infect 9:868–885. doi: 10.1080/22221751.2020.1754133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu T, Zhang C, Ji Y, Song J, Liu Y, Guo Y, Zhou K. 2021. Identification of mcr-10 carried by self-transmissible plasmids and chromosome in Enterobacter roggenkampii strains isolated from hospital sewage water. Environ Pollut 268:115706. doi: 10.1016/j.envpol.2020.115706. [DOI] [PubMed] [Google Scholar]
- 11.Wang C, Feng Y, Liu L, Wei L, Kang M, Zong Z. 2020. Identification of novel mobile colistin resistance gene mcr-10. Emerg Microbes Infect 9:508–516. doi: 10.1080/22221751.2020.1732231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Umeda K, Nakamura H, Fukuda A, Matsumoto Y, Motooka D, Nakamura S, Yasui Y, Yoshida H, Kawahara R. 2021. Genomic characterization of clinical Enterobacter roggenkampii co-harbouring blaIMP-1- and blaGES-5-encoding IncP6 and mcr-9-encoding IncHI2 plasmids isolated in Japan. J Glob Antimicrob Resist 24:220–227. doi: 10.1016/j.jgar.2020.11.028. [DOI] [PubMed] [Google Scholar]
- 13.Xiong J, Deraspe M, Iqbal N, Ma J, Jamieson FB, Wasserscheid J, Dewar K, Hawkey PM, Roy PH. 2016. Genome and plasmid analysis of blaIMP-4-carrying Citrobacter freundii B38. Antimicrob Agents Chemother 60:6719–6725. doi: 10.1128/AAC.00588-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snyman Y, Reuter S, Whitelaw AC, Stein L, Maloba MRB, Newton-Foot M. 2021. Characterisation of mcr-4.3 in a colistin-resistant Acinetobacter nosocomialis clinical isolate from Cape Town, South Africa. J Glob Antimicrob Resist 25:102–106. doi: 10.1016/j.jgar.2021.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Le-Ha TD, Le L, Le-Vo HN, Anda M, Motooka D, Nakamura S, Tran LK, Tran PT, Iida T, Cao V. 2019. Characterization of a carbapenem- and colistin-resistant Enterobacter cloacae carrying Tn6901 in blaNDM-1 genomic context. Infect Drug Resist 12:733–739. doi: 10.2147/IDR.S194495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Partridge SR, Kwong SM, Firth N, Jensen SO. 2018. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 31:e00088-17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lei CW, Zhang Y, Wang YT, Wang HN. 2020. Detection of mobile colistin resistance gene mcr-10.1 in a conjugative plasmid from Enterobacter roggenkampii of chicken origin in China. Antimicrob Agents Chemother 64:e01191-20. doi: 10.1128/AAC.01191-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders WE, Sanders CC. 1997. Enterobacter spp.: pathogens poised to flourish at the turn of the century. Clin Microbiol Rev 10:220–241. doi: 10.1128/CMR.10.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davin-Regli A, Lavigne JP, Pagès JM. 2019. Enterobacter spp.: update on taxonomy, clinical aspect, and emerging antimicrobial resistance. Clin Microbiol Rev 32:32. doi: 10.1128/CMR.00002-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stefaniuk EM, Tyski S. 2019. Colistin resistance in Enterobacterales strains: a current view. Pol J Microbiol 68:417–427. doi: 10.33073/pjm-2019-055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cuadrat RRC, Sorokina M, Andrade BG, Goris T, Dávila AMR. 2020. Global ocean resistome revealed: exploring antibiotic resistance gene abundance and distribution in TARA Oceans samples. GigaScience 9:giaa046. doi: 10.1093/gigascience/giaa046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martiny H-M, Munk P, Brinch C, Szarvas J, Aarestrup FM, Petersen TN. 2022. Global distribution of mcr gene variants in 214K metagenomic samples. mSystems 7:e0010522. doi: 10.1128/msystems.00105-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H, Zhan Z, Jiang X, Qing Y, Yin Z, Mei L, Zhou D, Ni B, Zhang Y. 2021. Comparative genomic analyses of IncpA1763-KPC plasmids. J Basic Microbiol 61:219–229. doi: 10.1002/jobm.202000668. [DOI] [PubMed] [Google Scholar]
- 24.Chen YT, Shu HY, Li LH, Liao TL, Wu KM, Shiau YR, Yan JJ, Su IJ, Tsai SF, Lauderdale TL. 2006. Complete nucleotide sequence of pK245, a 98-kilobase plasmid conferring quinolone resistance and extended-spectrum-β-lactamase activity in a clinical Klebsiella pneumoniae isolate. Antimicrob Agents Chemother 50:3861–3866. doi: 10.1128/AAC.00456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lakoh S, Adekanmbi O, Jiba DF, Deen GF, Gashau W, Sevalie S, Klein EY. 2020. Antibiotic use among hospitalized adult patients in a setting with limited laboratory infrastructure in Freetown Sierra Leone, 2017–2018. Int J Infect Dis 90:71–76. doi: 10.1016/j.ijid.2019.10.022. [DOI] [PubMed] [Google Scholar]
- 26.Antonelli A, D’Andrea MM, Di Pilato V, Viaggi B, Torricelli F, Rossolini GM. 2015. Characterization of a novel putative Xer-dependent integrative mobile element carrying the blaNMC-A carbapenemase gene, inserted into the chromosome of members of the Enterobacter cloacae complex. Antimicrob Agents Chemother 59:6620–6624. doi: 10.1128/AAC.01452-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castillo F, Benmohamed A, Szatmari G. 2017. Xer site specific recombination: double and single recombinase systems. Front Microbiol 8:453. doi: 10.3389/fmicb.2017.00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lakoh S, Li L, Sevalie S, Guo X, Adekanmbi O, Yang G, Adebayo O, Yi L, Coker JM, Wang S, Wang T, Sun W, Habib AG, Klein EY. 2020. Antibiotic resistance in patients with clinical features of healthcare-associated infections in an urban tertiary hospital in Sierra Leone: a cross-sectional study. Antimicrobial Resistance and Infection Control 9:38. doi: 10.1186/s13756-020-0701-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.CLSI. 2020. Performance standards for antimicrobial susceptibility testing, 30th ed. Supplement M100. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 30.Richter M, Rossello-Mora R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA III, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, Madden TL, Matten WT, McGinnis SD, Merezhuk Y, Raytselis Y, Sayers EW, Tao T, Ye J, Zaretskaya I. 2013. BLAST: a more efficient report with usability improvements. Nucleic Acids Res 41:W29–W33. doi: 10.1093/nar/gkt282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, Doshi S, Courtot M, Lo R, Williams LE, Frye JG, Elsayegh T, Sardar D, Westman EL, Pawlowski AC, Johnson TA, Brinkman FS, Wright GD, McArthur AG. 2017. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moura A, Soares M, Pereira C, Leitão N, Henriques I, Correia A. 2009. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25:1096–1098. doi: 10.1093/bioinformatics/btp105. [DOI] [PubMed] [Google Scholar]
- 37.Roberts AP, Chandler M, Courvalin P, Guédon G, Mullany P, Pembroke T, Rood JI, Smith CJ, Summers AO, Tsuda M, Berg DE. 2008. Revised nomenclature for transposable genetic elements. Plasmid 60:167–173. doi: 10.1016/j.plasmid.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qu D, Shen Y, Hu L, Jiang X, Yin Z, Gao B, Zhao Y, Yang W, Yang H, Han J, Zhou D. 2019. Comparative analysis of KPC-2-encoding chimera plasmids with multi-replicon IncR:IncpA1763-KPC:IncN1 or IncFIIpHN7A8:IncpA1763-KPC:IncN1. Infect Drug Resist 12:285–296. doi: 10.2147/IDR.S189168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carraro N, Poulin D, Burrus V. 2015. Replication and active partition of integrative and conjugative elements (ICEs) of the SXT/R391 family: the line between ICEs and conjugative plasmids is getting thinner. PLoS Genet 11:e1005298. doi: 10.1371/journal.pgen.1005298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang S, Dai E, Jiang X, Zeng L, Cheng Q, Jing Y, Hu L, Yin Z, Gao B, Wang J, Duan G, Cai X, Zhou D. 2019. Characterization of the plasmid of incompatibility groups IncFIIpKF727591 and IncpKPHS1 from Enterobacteriaceae species. Infect Drug Resist 12:2789–2797. doi: 10.2147/IDR.S212321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhan Z, Hu L, Jiang X, Zeng L, Feng J, Wu W, Chen W, Yang H, Yang W, Gao B, Yin Z, Zhou D. 2018. Plasmid and chromosomal integration of four novel blaIMP-carrying transposons from Pseudomonas aeruginosa, Klebsiella pneumoniae and an Enterobacter sp. J Antimicrob Chemother 73:3005–3015. doi: 10.1093/jac/dky288. [DOI] [PubMed] [Google Scholar]
- 42.Heffron F, Sublett R, Hedges RW, Jacob A, Falkow S. 1975. Origin of the TEM-β-lactamase gene found on plasmids. J Bacteriol 122:250–256. doi: 10.1128/jb.122.1.250-256.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo X, Yin Z, Zeng L, Hu L, Jiang X, Jing Y, Chen F, Wang D, Song Y, Yang H, Zhou D. 2021. Chromosomal integration of huge and complex blaNDM-carrying genetic elements in Enterobacteriaceae. Front Cell Infect Microbiol 11:690799. doi: 10.3389/fcimb.2021.690799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download spectrum.01127-22-s0001.pdf, PDF file, 0.07 MB (76.8KB, pdf)
Data Availability Statement
The complete sequence of plasmid pSL12517-mcr10.1 has been submitted to GenBank under the accession number MW048777.