ABSTRACT
Increasing infections caused by blaNDM-carrying Klebsiella pneumoniae (NDM-KP) are an urgent threat to children with weakened immunity and limited antibiotic use. Preventing and intervening in NDM-KP infections requires a clear understanding of the pathogen’s molecular and epidemiological characteristics. We investigated the prevalence and characteristics of NDM-KP in six children’s hospitals from five Chinese provinces/municipalities. We collected 111 NDM-KP strains (40 NDM-1, one NDM-4 and 70 NDM-5) from neonatal intensive care units (NICUs) and pediatric intensive care units (PICUs) from June 2017 to June 2018; these strains accounted for 31.62% of all carbapenem-resistant K. pneumoniae (CR-KP). Although NDM-KP isolates exhibited high resistance to all carbapenems, including ertapenem (MIC: ≥32 mg/L, 96.4%), imipenem (MIC: ≥16 mg/L, 90.1%) and meropenem (MIC: ≥16 mg/L, 99.1%), they were fully sensitive to amikacin, tigecycline and polymyxin B, and presented low resistance to levofloxacin (9.9%) and gentamicin (15.3%). Whole-genome sequencing was conducted to gain insight into the molecular characterizations of NDM-KP isolates. The NDM-KP isolates belonged to 20 sequence types (STs), and ST2407 (n = 45) dominated in one hospital from Chengdu. ST2407 isolates with fewer single-nucleotide polymorphisms (SNP < 38) were found either in the same hospital or different hospitals. Most blaNDM (81.1%, 90/111), including all blaNDM-5 and blaNDM-4 and 47.5% (19/40) of blaNDM-1, in NDM-KP isolates with 13 STs were associated with the IncX3 plasmid. Our results indicated that both explosive clonal transmission and horizontal transmission of blaNDM occur among NDM-KP strains in children's hospitals. These data provide a basis for preventing and controlling NDM-KP-associated infectious diseases in hospitalized children, especially in neonates.
IMPORTANCE The blaNDM gene is playing an increasingly important role in infections caused by CR-KP, especially in children. However, systematic detection and bioinformatics analysis of NDM-KP in children's hospitals are lacking in China. In this study, a total of 111 NDM-positive K. pneumoniae isolates were selected from the China Antimicrobial Surveillance Network for further investigation. The isolates were further characterized using state-of-the-art molecular techniques. Our findings suggested the clonal and horizontal transmission of blaNDM in K. pneumoniae in NICUs/PICUs. Key plasmids (IncX3) and ST diversity contribute to the spread of blaNDM. In addition, our findings provided recommendations for pediatric clinicians to use antibiotics to treat NDM-KP infections. Our current large-scale epidemiological survey would support further infection intervention strategies of NDM-KP in NICU/PICU of children's hospitals.
KEYWORDS: carbapenem, bla NDM , Klebsiella pneumoniae, children, intensive care unit
INTRODUCTION
Klebsiella pneumoniae is one of the most common and important pathogens causing community-acquired and hospital-acquired infectious bacterial diseases. Particularly in hospital-acquired infections, K. pneumoniae causes various infectious diseases, including sepsis, urinary tract infections, pneumonia and soft tissue infections, especially in immunocompromised hosts such as newborns (1). The emergence and rapid spread of carbapenem-resistant K. pneumoniae (CR-KP) in recent decades led to failed infection treatments, making it an urgent threat to global public health (2). The World Health Organization listed carbapenem-resistant Enterobacterales, including CR-KP, as priority drug-resistant bacteria requiring novel treatment (3). Although it emerged later than other carbapenem resistance genes, such as blaKPC, blaIMP, blaVIM and blaOXA-48 (4), the blaNDM gene has been rapidly identified in clinical K. pneumoniae isolates worldwide (5–8).
The situation in China is grim as well. Data from the China Antimicrobial Surveillance Network (CHINET) revealed that CR-KP increased from 2.9% in 2005 to 27.1% in 2021 (http://www.chinets.com/). In addition to blaKPC-carrying K. pneumoniae (KPC-KP) (9), NDM-KP has begun to enter neonatal intensive care units (NICUs) and pediatric intensive care units (PICUs), with a handful of outbreaks among child patients in many areas of China (10–15). Notably, the isolation rate of NDM-KP in children's hospitals is increasing annually (14, 16). Considering the limited antibiotic selection for children, the characteristics of NDM-KP isolated from children need to be investigated.
Previous reports described NDM-KP outbreaks based mostly on fragmented surveillance of individual children’s hospitals and lacked systematic comparisons of the molecular epidemiology and strain characteristics between hospitals. To elucidate the epidemiology and molecular characteristics of NDM-KP in the NICUs/PICUs of different children’s hospitals, we collected NDM-KP clinical isolates from six children’s hospitals from five Chinese provinces or municipalities for 1 year (June 2017 to June 2018). Whole-genome sequencing and analysis were undertaken to gain insight into characterizing the isolates.
RESULTS
Phenotypes and genotypes of NDM-KP in six children’s hospitals.
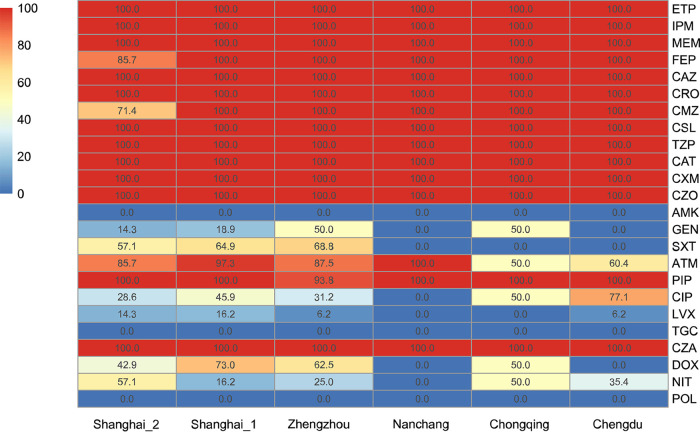
The blaNDM gene was detected in 111 CR-KP isolates originating from six children’s hospitals and accounted for 31.62% (111/351, 95% confidence interval [CI]: 26.8%–36.8%) of all CR-KP isolates in Chengdu (48/49, 97.96%), Chongqing (2/34, 5.88%), Nanchang (1/5, 20%), Zhengzhou (16/130, 12.31%), Shanghai_1 (37/111, 33.33%), and Shanghai_2 (7/22, 31.82%). All CR-KP isolates showed high resistance to carbapenems, including ertapenem (MIC: 96.4%, ≥32 mg/L), imipenem (MIC: 90.1%, ≥16 mg/L) and meropenem (MIC: 99.1%, ≥16 mg/L; Fig. 1). These isolates also exhibited high resistance rates to ceftazidime-avibactam (100%), piperacillin-tazobactam (100%), cefoperazone-sulbactam (100%), ceftolozane-tazobactam (100%), and other β-lactams (piperacillin, cefepime, ceftazidime, ceftriaxone, cefmetazole, cefuroxime, and cefazolin; 98.2%–100%); moderate resistance to doxycycline (n = 42; 37.8%), ciprofloxacin (n = 62; 55.9%) and aztreonam (n = 87; 78.4%); low resistance to gentamicin (n = 17; 15.3%), trimethoprim-sulfamethoxazole (n = 39; 35.1%), nitrofurantoin (n = 32; 28.8%) and levofloxacin (n = 11; 9.9%); and sensitivity to amikacin, tigecycline and polymyxin B (Fig. 1). No strains showed the hypermucoviscous phenotype.
FIG 1.
Antimicrobial-resistance profiles of 24 antimicrobial agents against NDM-KP. The number represents the antimicrobial resistance rate of the NDM-KP isolates. ETP: ertapenem; IPM: imipenem; MEM: meropenem; FEP: cefepime; CAZ: ceftazidime; CRO: ceftriaxone; CMZ: cefmetazole; CSL: cefoperazone-sulbactam; TZP: piperacillin-tazobactam, CAT: ceftolozane-tazobactam; CXM: cefuroxime; CZO: cefazolin; AMK: amikacin; GEN: gentamicin; SXT: trimethoprim-sulfamethoxazole; ATM: aztreonam; PIP: piperacillin; CIP: ciprofloxacin; LVX: levofloxacin; TGC: tigecyclin; CZA: ceftazidime-avibactam; DOX: doxycycline; NIT: nitrofurantoin; POL: polymyxin B.
Antibiotic-resistance and virulence-associated genes in the 111 NDM-KP isolates were identified from WGS data (Fig. 2A, Table S1). Seventy NDM-KP (63%) harbored blaNDM-5, 40 (36%) harbored blaNDM-1, and one (1%) harbored blaNDM-4. One ST37 strain (A96) carried two CR genes, blaNDM-1 and blaIMP-4. Some NDM-KP isolates also harbored the ESBL genes, blaCTX-M-14 (47; 42.3%), blaCTX-M-15 (19; 17.2%), blaCTX-M-3 (21; 18.9%) and blaSFO-1 (14; 12.6%); the tetracycline-resistance genes, tet(A) (19; 17.1%), tet(B) (2; 1.8%) and tet(D) (20; 18%); and the fluroquinolone-resistance genes, oqxAB (94; 84.7%), qnrB (46; 41.4%) and qnrS1 (24; 21.6%). For the genes associated with hypervirulent K. pneumoniae, the categorical virulence scores ranged from 0–5; 24.3% (27/111) of the K. pneumoniae isolates scored 0, and the remaining K. pneumoniae (75.7%, 84/111) isolates scored 1, suggesting that the NDM-KP isolates carried few genes mediating high virulence. Strains scoring 1 carried the siderophore yersiniabactin virulence loci. Only one strain, A156, carried the hypermucoid locus, rmpA, and the salmochelin loci, iroB, iroC, iroD and iroN. Additionally, 34.2% (38/111) of the K. pneumoniae isolates had different degrees of deletions in the outer membrane protein, OmpK35.
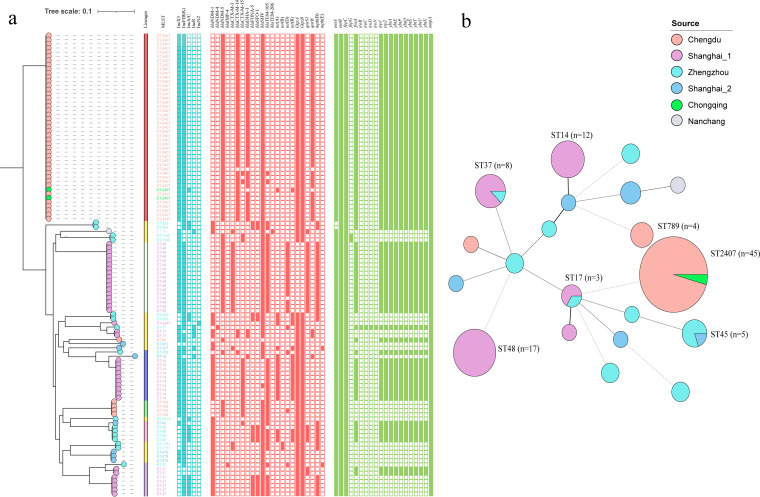
FIG 2.
Population structure of NDM-KP isolates. (a) Phylogeny of core-genome SNPs in 111 NDM-KP isolates. K. pneumoniae ST2407 S6, sequenced on MinION, was used as a reference strain. Sources of the isolates are differentiated by six colors. Inc-type plasmid (bule square), antimicrobial-resistance genes (red square), and virulence-associated genes (green square) among the isolates are denoted by filled squares for presence and empty squares for absence. (b) Minimum-spanning tree of NDM-KP isolates based on a core-genome MLST (cgMLST).
Nosocomial blaNDM outbreak and transmission caused by K. pneumoniae diversity.
The 111 NDM-KP isolates were assigned to 20 MLST types (Table S1). The minimum-spanning tree indicated the clonal transmission characteristics of some NDM-KP isolates in the NICUs/PICUs (Fig. 2B). For example, clone transmission of ST2407 (n = 45) occurred in a children's hospital from Chengdu. The same ST, like ST2407, ST37, ST45, and ST17, could exist in two children's hospitals that were geographically far apart. Additionally, 10 and nine STs occurred in Shanghai (two hospitals) and Zhengzhou (one hospital in Henan), respectively. Like the STs, the NDM-KP serotypes also exhibited diverse profiles. Identification of capsule synthesis loci revealed 20 different types. Each ST, except for ST37, corresponded to only one capsule synthesis locus. ST37 had three capsular synthesis loci: K8, K15 and K38. All K. pneumoniae ST14 serotypes were K2, which has been associated with high pathogenicity. Five O antigen (LPS) serotypes (O1–O5) were predicted (Table S1). The data showed that blaNDM can exist in various K. pneumoniae strains, showing its widespread spread at cloning levels in children's hospitals.
Phylogenetics of NDM-KP in six children's hospitals.
Phylogenetic analysis of the core-genome single-nucleotide polymorphisms (SNPs) revealed seven distinct population structure lineages among the 111 NDM-KP isolates (Fig. 2A). Shallow branching and scattered population structure also occurred: the same K. pneumoniae STs (except ST37) in the same hospital exhibited little branching and few SNPs (5–38), suggesting an outbreak of NDM-KP in one hospital. Notably, all NDM-KP ST2407 isolates were clustered as the significant phylogroup with limited SNP (5–29) divergences. The core genomes of two isolates from Chongqing and 43 isolates from Chengdu were nearly identical, with <25 SNPs (Table S2). These two hospitals are geographically distant (straight-line distance >280 km), indicating the spread of blaNDM through clonal transfer. In contrast, on the three branches for ST17, ST37, and ST45, the core genomes of the same STs from two children's hospitals differed significantly.
Epidemiology of the blaNDM-IncX3 plasmid in children's hospitals.
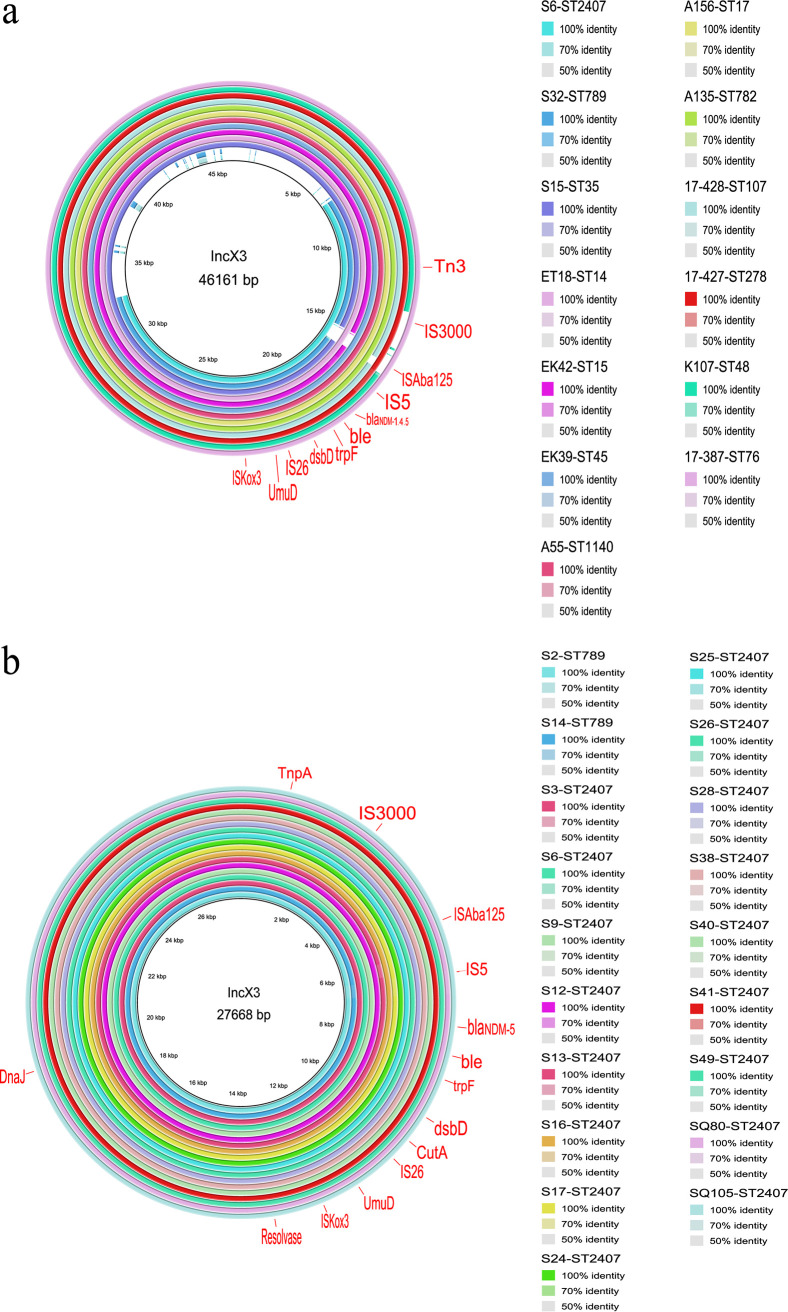
Plasmids are likely the most common carriers for blaNDM gene (17). WGS data detected major Inc-type plasmids, including IncX3, IncFIB(K), Inc(A/C), IncR and IncN2. A complete high prevalence 46.1 kb IncX3 plasmid (GenBank accession number: CP049352) carrying blaNDM-5 isolated from E. coli in China was used as a reference, then the BLAST Ring Image Generator (BRIG) was used to compare it with the WGS of different strains, revealing that the blaNDM-IncX3 plasmid presented in 13 different NDM-KP STs (Fig. 3A) and in 81.1% of the total NDM-KP (90/111), which is highly homologous to the reference plasmid. By BLAST analysis of larger contigs (>34 kb), we identified 18 isolates carrying the blaNDM-IncX3 plasmid. The Bandage association analysis revealed a high probability connection between IncX3 plasmids and blaNDM in the remaining 72 isolates. However, circularization of plasmids (i.e., use of Illumina plus Nanopore) in these isolates would prove this suggestion. IncX3 plasmid presented in all blaNDM-4 and blaNDM-5 isolates and 45.7% (19/40) of blaNDM-1 isolates, and this plasmid could be identified from isolates of the same lineage or different lineages in the same hospital, suggesting the spread of blaNDM through horizontal transfer (Fig. 2A). Notably, BRIG analysis showed that 49 IncX3 plasmids carried by ST2407 (n = 45) and ST789 (n = 4) isolates from the Sichuan and Chongqing hospitals was smaller than the original IncX3 plasmid (46.1 kb), and the conjugation-associated type IV secretion system (T4SS) play as the missing part. Therefore, we randomly selected one ST2407 isolate S6 carrying the smaller IncX3 plasmid for MinIon sequencing and confirmed the 27.7 kb size of the plasmid with T4SS deletion (Table S3). Further, the IncX3 plasmids in these NDM-KP ST2407 (n = 17) and ST789 (n = 2) strains had higher homology with this 27.7 kb IncX3 plasmid (Fig. 3B).
FIG 3.
Schematic diagram of mobile genetic elements integrated in NDM-KP isolates. (a) IncX3 plasmid carrying blaNDM was present in 13 K. pneumoniae STs. (b) Evolved IncX3 plasmid carrying blaNDM in ST789 and ST2407 K. pneumoniae.
Mobile genetic element arrangements in spreading blaNDM in children's hospitals.
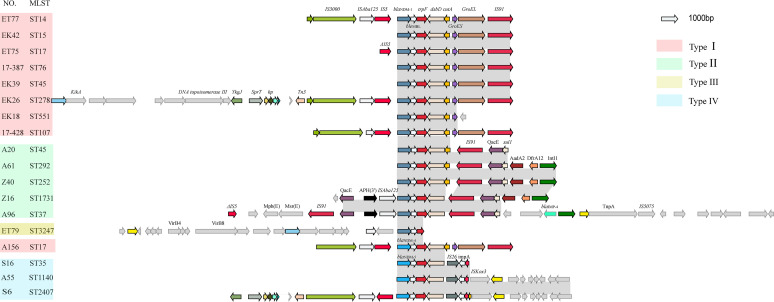
To further analyze the transfer of blaNDM via mobile genetic elements (MGEs), comparison of the blaNDM genetic environment in all 20 NDM-KP STs illustrated a conservative and complex combination of multiple genetic vehicles in spreading blaNDM (Fig. S2 and Fig. 4). The genetic environments of blaNDM were clustered into four types according to homologous regions, ST and blaNDM genotypes, showing a relatively conserved architecture in the variable context surrounding blaNDM. All four types retained the conserved sequence blaNDM-bleMBL-trpF-dsbD. All blaNDM-5 belonged to type 4 (n = 70), blaNDM-4 to type 1 (n = 1), and blaNDM-1 to types 1 (n = 19), 2 (n = 20) and 3 (n = 1). Compared with the conservative flanking region of blaNDM-5, the surrounding environments of blaNDM-1 exhibited more diverse, likely owing to the different Inc types of plasmids carrying blaNDM-1. Both types 1 and 4 were associated with the IncX3 backbone, with differences in the heat shock protein-related GroES and GroEL, which only appear downstream of blaNDM in type 1. The upstream IS3000, IS5, and the downstream IS91 of the segment blaNDM-bleMBL-trpF-dsbD are conserved. Type 2 belonged to the plasmid backbone of IncFIB(K) (contigs > 137 kb), in which the strain A96 of ST37 coexisted with blaIMP-4 at the 8921-bp downstream position of blaNDM-1. Additionally, the sulfanilamide-resistance gene, sul1, also coexisted with the downstream blaNDM-1. The blaNDM-1-carrying type 3 belonged to the plasmid backbone of IncN2 (contigs > 41 kb), in which the upstream virB4 and virB8 were associated with plasmid conjugation transfer, suggesting that blaNDM-1 can be transferred horizontally by plasmids other than IncX3.
FIG 4.
Genetic environment of blaNDM in K. pneumoniae. The four different types are represented by four colors. Open reading frames are designated by arrows indicating the direction of transcription and colored based on their predicted gene functions. Dark gray shading indicates homologous regions.
DISCUSSION
Clinical CR-KP in China is an increasing concern, and its prevalence is higher in children than in adults (China Antimicrobial Resistance Surveillance System, http://www.carss.cn/). Compared with KPC-KP, the notorious pathogen dominant in both children's and adult hospitals, NDM-KP has caused a higher proportion of CR-KP in children’s hospitals than in adult hospitals in recent years (18). As the possible transmission link for NDM-E. coli between humans and animals has been established (19–21), we proposed that the increasing trend for NDM-KP in child patients in this study might be associated with animals for the following four reasons. First, different from the ST11-dominant KPC-KP in China (9, 22, 23), the NDM-KP isolates collected from children’s hospitals exhibited diverse profiles (20 STs) and were not limited to one superior clone. Most clones, including ST45, ST48, and ST37, have been detected and are being transferred in livestock and poultry production chain (24–26). Second, the blaNDM-IncX3 dominant in children NDM-KP was proved to be a successful plasmid among various commercial farm animals (pigs and chicken), backyard animals (pigs, chickens, cattle, pets), and other animals (flies and birds). The high conjugation transfer frequency (27), low fitness cost (28) and favorable stability of the IncX3 plasmid in many Enterobacteriaceae may facilitate the circulation of blaNDM-IncX3 plasmids in bacteria of both animal and human origin. Third, unlike blaKPC, which disseminates via stable association with a lineage of K. pneumoniae, the spread of blaNDM is mediated by transient associations of diverse plasmids (including IncX3) with multiple lineages of K. pneumoniae (17). Notably, blaNDM, but not blaKPC, was frequently detected in high abundances in farm-animal feces and in high prevalence in bacteria of farm-animal origin (29). Finally, children are generally considered more susceptible to infection because of the immaturity of their immune and intestinal systems (30). CR-KP can emerge from the intestinal lumen and invade the bloodstream of vulnerable patients, causing disseminated infection (31). Thus, NDM-KP may be transmitted directly (via the food chain) or indirectly (in the environment) from animals to children, leading to colonization and infection. Therefore, the increasing NDM-KP in children’s hospitals suggests that the patterns and extent of the impact of non-human factors on NDM-KP infections in children must be investigated.
Our results for both antimicrobial-resistance genes and phenotypes indicated the presence of multidrug-resistant (MDR) K. pneumoniae in children's hospitals. The use of ß-lactams can co-select resistance determinants for different antibiotics harbored in the same plasmids as blaNDM, such as sulfonamides and even quinolones, which are not used in common children's treatment. These MGEs likely contribute to the prevalence of blaNDM and increase the threat to children’s health with further limited spectrum antibiotics. Fortunately, all NDM-KP are completely sensitive to amikacin, tigecycline and polymyxin B and highly sensitive to levofloxacin and gentamicin, which helps fight against NDM-KP. Gentamicin and polymyxin B are more suitable for clinical children, while amikacin (32), levofloxacin (33, 34) and tigecycline (35) are used with caution due to unclear safety concerns. Clinicians may consider these antibiotics as alternative options for treating NDM-KP infections. Additionally, these MDR K. pneumoniae are not highly virulent, which thus reduces the risk of severe infections in children. Nevertheless, these virulence-related genes and plasmids should not be ignored. Previous studies have predicted that the rate at which MDR K. pneumoniae acquires virulence plasmids far exceeds the rate at which hypervirulent K. pneumoniae (36) acquires MDR plasmids.
This work had several limitations. First, the amount of data collected from the six hospitals was inconsistent, which may have led to bias in the analysis. Second, we only collected the strain characteristic information during the study, and the individual medical history information (e.g., demographics [sex and age], antibiotics exposure, disease types) of the patients that the strains were isolated from was mismatched or absent due to the improper storage, thus we could not conduct a correlational study between epidemiological data and strain genetic information. Finally, we have not determined the mechanism causing the increased NDM-KP in children and its relevance to NDM-KP in animals; this will be a focus in our subsequent studies.
Our study revealed the clonal and horizontal transmission of blaNDM in K. pneumoniae in NICUs/PICUs. Key plasmids (IncX3) and ST diversity accelerate the spread of blaNDM. Considering these results, clinicians should include screening for NDM-KP when diagnosing infectious diseases in children, especially newborns, to avoid treatment time delays and antibiotic failure. Monitoring and data collection on blaNDM in children should be strengthened to better understand its epidemic patterns. Our results also suggest that there may be more confounding factors for prevalence of blaNDM in children, therefore, the “One Health” perspective is needed to address this increasing threat.
MATERIALS AND METHODS
Bacterial isolation and identification.
We collected 351 nonduplicate CR-KP clinical isolates from six children's hospitals in five provinces or municipalities across China (member units of the CHINET Study Group; Fig. S1) from June 2017 to June 2018. All CR-KP strains were identified using Vitek matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS, bioMérieux, Marcyl’Étoile, France) and selected by the Kirby-Bauer antimicrobial susceptibility method during the routine daily work of the microbiology laboratory in each children hospital. PCR (37) was used to screen the target blaNDM-positive strains for follow-up study. The NDM-KP isolates were obtained from sputum (n = 91), bronchoalveolar lavage fluid (n = 7), urine (n = 2) and blood (n = 11), collected from the NICU (n = 83) or PICU (n = 28) at six children's hospitals (Table S1).
Antimicrobial susceptibility testing.
The MICs for 24 antimicrobial agents were determined for all blaNDM-positive isolates using the microdilution broth method following the Clinical and Laboratory Standards Institute (CLSI) guidelines (38). The MIC results were interpreted according to CLSI documents M100-ED3015 (38) and European Committee on Antimicrobial Susceptibility Testing breakpoints (39). Escherichia coli ATCC 25922 and K. pneumoniae ATCC 13883 were used as quality-control standards.
Whole-genome sequencing and bioinformatics analysis.
All blaNDM-positive isolates were selected for whole-genome sequencing (WGS). Total DNA was extracted using a Magen Genomic DNA purification kit (Magen, Guangzhou, China) as per the manufacturer’s instructions. Indexed DNA libraries were prepared using a KAPA Hyper Prep Kit and sequenced on the Illumina HiSeq X 10 platform (Annoroad, Beijing, China). All draft genomes were assembled using SPAdes, version 3.9.0 (40). A K. pneumoniae strain S6 carrying blaNDM-IncX3 plasmid was selected and sequenced on the MinION platform (Oxford Nanopore Technologies, Oxford, UK). We used Unicycler v.0.4.8-beta (41) to generate genome assemblies combining the Illumina and MinION sequences. Minimum-spanning trees of all blaNDM-positive isolates were generated in BioNumerics. A phylogenetic tree was produced using snippy (https://github.com/tseemann/snippy) and gubbins (42) using core-genome alignments and was visualized using the online tool, iTOL (43). Phylogenetic trees for K. pneumoniae based on the core-genome sequences of the isolates were structured using Harvest, version 1.1.2 (44). Multilocus sequence typing (MLST), antimicrobial-resistance genes, virulence-associated genes and K (capsule) and O antigen (LPS) serotype prediction were identified using Kleborate (45). Plasmid types of the blaNDM-carrying contigs were identified using abricate (https://github.com/tseemann/abricate). All contigs from the WGS and MinION sequencing analyses were annotated using prokka (46). Genetic contexts in the different blaNDM-carrying plasmids were compared using BLAST Ring Image Generator (BRIG) (47). Gcluster (48) was used to visualize and compare the blaNDM genome contexts for all genomes. Bandage (49) was used to predicate and visualize the connection of contigs in which IncX3 plasmids located and contigs in which blaNDM located.
Data availability.
All raw data in this study were deposited in GenBank under Bioproject accession number PRJNA750893.
ACKNOWLEDGMENTS
This work was supported in part by the National Natural Science Foundation of China (81861138051, 81861138052, 81871690 and 81902101), and the CHINET Antimicrobial Surveillance Network (2020QD049). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Y.W. and F.H. designed the study. B.F, D.Y., R.B. performed isolates identified and sequenced. B.F, D.Y., C.S., Y.S, D.L. analyzed and interpreted the data. Y.W., F.H., B.F, D.Y., R.Z. and J.S. wrote and revised the report. All authors reviewed and approved the final report.
Footnotes
Supplemental material is available online only.
Contributor Information
Fupin Hu, Email: hufupin@fudan.edu.cn.
Yang Wang, Email: wangyang@cau.edu.cn.
N. Esther Babady, Memorial Sloan Kettering Cancer Center
REFERENCES
- 1.Gupta A. 2002. Hospital-acquired infections in the neonatal intensive care unit-Klebsiella pneumoniae. Semin Perinatol 26:340–345. doi: 10.1053/sper.2002.36267. [DOI] [PubMed] [Google Scholar]
- 2.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, Aboderin AO, Al-Abri SS, Jalil NA, Benzonana N, Bhattacharya S, Brink AJ, Burkert FR, Cars O, Cornaglia G, Dyar OJ, Friedrich AW, Gales AC, Gandra S, Giske CG, Goff DA, Goossens H, Gottlieb T, Blanco MG, Hryniewicz W, Kattula D, Jinks T, Kanj SS, Kerr L, Kieny M-P, Kim YS, Kozlov RS, Labarca J, Laxminarayan R, Leder K, Group WPPLW , et al. 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 4.Poirel L, Pitout JD, Nordmann P. 2007. Carbapenemases: molecular diversity and clinical consequences. Future Microbiol 2:501–512. doi: 10.2217/17460913.2.5.501. [DOI] [PubMed] [Google Scholar]
- 5.Poirel L, Revathi G, Bernabeu S, Nordmann P. 2011. Detection of NDM-1-Producing Klebsiella pneumoniae in Kenya. Antimicrob Agents Chemother 55:934–936. doi: 10.1128/AAC.01247-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Struelens MJ, Monnet DL, Magiorakos AP, Santos O’Connor F, Giesecke J, The European Ndm-1 Survey Participants C, Participants C the EN-1 S . 2010. New Delhi metallo-beta-lactamase 1–producing Enterobacteriaceae: emergence and response in Europe. Eurosurveillance 15. doi: 10.2807/ese.15.46.19716-en. [DOI] [PubMed] [Google Scholar]
- 7.Zou M-X, Wu J-M, Li J, Dou Q-Y, Zhou R-R, Huang Y, Liu W-E. 2012. NDM-1-producing Klebsiella pneumoniae in mainland China Zhongguo Dang Dai Er Ke Za Zhi Chin J Contemp Pediatrics 14:616–621. [PubMed] [Google Scholar]
- 8.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new Metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India ▿. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. 2011. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother 66:307–312. doi: 10.1093/jac/dkq431. [DOI] [PubMed] [Google Scholar]
- 10.Zhu J, Sun L, Ding B, Yang Y, Xu X, Liu W, Zhu D, Yang F, Zhang H, Hu F. 2016. Outbreak of NDM-1-producing Klebsiella pneumoniae ST76 and ST37 isolates in neonates. Eur J Clin Microbiol Infect Dis 35:611–618. doi: 10.1007/s10096-016-2578-z. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Chen D, Xu G, Huang W, Wang X. 2018. Molecular epidemiology and drug resistant mechanism in carbapenem-resistant Klebsiella pneumoniae isolated from pediatric patients in Shanghai, China. PLoS One 13:e0194000. doi: 10.1371/journal.pone.0194000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin Y, Shao C, Li J, Fan H, Bai Y, Wang Y. 2015. Outbreak of multidrug resistant NDM-1-producing Klebsiella pneumoniae from a neonatal unit in Shandong Province, China. PLoS One 10:e0119571. doi: 10.1371/journal.pone.0119571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng R, Zhang Q, Guo Y, Feng Y, Liu L, Zhang A, Zhao Y, Yang X, Xia X. 2016. Outbreak of plasmid-mediated NDM-1-producing Klebsiella pneumoniae ST105 among neonatal patients in Yunnan. China Ann Clin Microb Anti 15:10. doi: 10.1186/s12941-016-0124-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong F, Lu J, Wang Y, Shi J, Zhen JH, Chu P, Zhen Y, Han SJ, Guo YL, Song WQ. 2017. A Five-year surveillance of carbapenemase-producing Klebsiella pneumoniae in a pediatric hospital in China reveals increased predominance of NDM-1. Biomed Environ Sci 30:562–569. [DOI] [PubMed] [Google Scholar]
- 15.Li J, Hu X, Yang L, Lin Y, Liu Y, Li P, Wang K, Qiu S, Li P, Song H. 2019. New Delhi Metallo-β-Lactamase 1-Producing Klebsiella pneumoniae ST719 isolated from a neonate in China. Microb Drug Resist doi: 10.1089/mdr.2019.0058. [DOI] [PubMed] [Google Scholar]
- 16.Liu X, Geng S, Chan EW-C, Chen S. 2019. Increased prevalence of Escherichia coli strains from food carrying blaNDM and mcr-1-bearing plasmids that structurally resemble those of clinical strains, China, 2015 to 2017. Eurosurveillance 24:1800113. doi: 10.2807/1560-7917.ES.2019.24.13.1800113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.David S, Cohen V, Reuter S, Sheppard AE, Giani T, Parkhill J, Rossolini GM, Feil EJ, Grundmann H, Aanensen DM, Group the ES of C-PE (EuSCAPE) W, (ESGEM) the ESG for EM . 2020. Integrated chromosomal and plasmid sequence analyses reveal diverse modes of carbapenemase gene spread among Klebsiella pneumoniae. Proc Natl Acad Sci USA 117:25043–25054. doi: 10.1073/pnas.2003407117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han R, Shi Q, Wu S, Yin D, Peng M, Dong D, Zheng Y, Guo Y, Zhang R, Hu F, Group CASN (CHINET) S . 2020. Dissemination of carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) among carbapenem-resistant Enterobacteriaceae isolated from adult and children patients in China. Front Cell Infect Mi 10:314. doi: 10.3389/fcimb.2020.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Zhang R, Li J, Wu Z, Yin W, Schwarz S, Tyrrell JM, Zheng Y, Wang S, Shen Z, Liu Z, Liu J, Lei L, Li M, Zhang Q, Wu C, Zhang Q, Wu Y, Walsh TR, Shen J. 2017. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat Microbiol 2. doi: 10.1038/nmicrobiol.2016.260. [DOI] [PubMed] [Google Scholar]
- 20.Li J, Bi Z, Ma S, Chen B, Cai C, He J, Schwarz S, Sun C, Zhou Y, Yin J, Hulth A, Wang Y, Shen Z, Wang S, Wu C, Nilsson LE, Walsh TR, Börjesson S, Shen J, Sun Q, Wang Y. 2019. Inter-host transmission of carbapenemase-producing Escherichia coli among humans and backyard animals. Environ Health Perspect 127:107009. doi: 10.1289/EHP5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen Y, Hu F, Wang Y, Yin D, Yang L, Chen Y, Xu C, Li J, Jiang J, Wang X, Fu Y, Shao D, Liu D, Ma T, Cai C, Shen Z, Wang S, Li J, Zhang R, Ke Y, Wu C, Shen J, Walsh TR, Wang Y. 2022. Transmission of carbapenem resistance between human and animal NDM-positive Escherichia coli strains. Engineering-london doi: 10.1016/j.eng.2021.07.030. [DOI] [Google Scholar]
- 22.Dong N, Yang X, Zhang R, Chan EW-C, Chen S. 2018. Tracking microevolution events among ST11 carbapenemase-producing hypervirulent Klebsiella pneumoniae outbreak strains. Emerg Microbes Infec 7:1–8. doi: 10.1038/s41426-018-0146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li P, Liang Q, Liu W, Zheng B, Liu L, Wang W, Xu Z, Huang M, Feng Y. 2021. Convergence of carbapenem resistance and hypervirulence in a highly-transmissible ST11 clone of K. pneumoniae: An epidemiological, genomic and functional study. Virulence 12:377–388. doi: 10.1080/21505594.2020.1867468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang R, Li J, Wang Y, Shen J, Shen Z, Wang S. 2019. Presence of NDM in non-E. coli Enterobacteriaceae in the poultry production environment. J Antimicrob Chemoth 74:2209–2213. doi: 10.1093/jac/dkz193. [DOI] [PubMed] [Google Scholar]
- 25.Zhai R, Fu B, Shi X, Sun C, Liu Z, Wang S, Shen Z, Walsh TR, Cai C, Wang Y, Wu C. 2020. Contaminated in-house environment contributes to the persistence and transmission of NDM-producing bacteria in a Chinese poultry farm. Environ Int 139:105715. doi: 10.1016/j.envint.2020.105715. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Q, Lv L, Huang X, Huang Y, Zhuang Z, Lu J, Liu E, Wan M, Xun H, Zhang Z, Huang J, Song Q, Zhuo C, Liu J-H. 2019. Rapid increase in carbapenemase-producing Enterobacteriaceae in retail meat driven by the spread of the blaNDM-5-carrying IncX3 plasmid in China from 2016 to 2018. Antimicrob Agents 63:e00573-19. doi: 10.1128/AAC.00573-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Tong M-K, Chow K-H, Cheng VC-C, Tse CW-S, Wu AK-L, Lai RW-M, Luk W-K, Tsang DN-C, Ho P-L. 2018. Occurrence of highly conjugative IncX3 epidemic plasmid carrying blaNDM in Enterobacteriaceae isolates in geographically widespread areas. Front Microbiol 9:2272. doi: 10.3389/fmicb.2018.02272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma T, Fu J, Xie N, Ma S, Lei L, Zhai W, Shen Y, Sun C, Wang S, Shen Z, Wang Y, Walsh TR, Shen J. 2020. Fitness cost of blaNDM-5-carrying p3R-IncX3 plasmids in wild-type NDM-free Enterobacteriaceae. Microorg 8:377. doi: 10.3390/microorganisms8030377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Köck R, Daniels-Haardt I, Becker K, Mellmann A, Friedrich AW, Mevius D, Schwarz S, Jurke A. 2018. Carbapenem-resistant Enterobacteriaceae in wildlife, food-producing and companion animals–a systematic review. Clin Microbiol Infect 24:1241–1250. doi: 10.1016/j.cmi.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 30.Travier L, Alonso M, Andronico A, Hafner L, Disson O, Lledo P-M, Cauchemez S, Lecuit M. 2021. Neonatal susceptibility to meningitis results from the immaturity of epithelial barriers and gut microbiota. Cell Rep 35:109319. doi: 10.1016/j.celrep.2021.109319. [DOI] [PubMed] [Google Scholar]
- 31.Keith JW, Pamer EG. 2019. Enlisting commensal microbes to resist antibiotic-resistant pathogens. J Exp Med 216:10–19. doi: 10.1084/jem.20180399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCracken GH. 1986. Arninoglycoside toxicity in infants and children. Am J Medicine 80:172–178. doi: 10.1016/0002-9343(86)90497-3. [DOI] [PubMed] [Google Scholar]
- 33.Noel GJ, Bradley JS, Kauffman RE, Duffy CM, Gerbino PG, Arguedas A, Bagchi P, Balis DA, Blumer JL. 2007. Comparative safety profile of Levofloxacin in 2523 children with a focus on four specific musculoskeletal disorders. Pediatric Infect Dis J 26:879–891. doi: 10.1097/INF.0b013e3180cbd382. [DOI] [PubMed] [Google Scholar]
- 34.Bradley JS, Byington CL, Shah SS, Alverson B, Carter ER, Harrison C, Kaplan SL, Mace SE, McCracken GH, Moore MR, Peter SDS, Stockwell JA, Swanson JT, America PIDS and the IDS of . 2011. The management of community-acquired pneumonia in infants and children older than 3 months of age: clinical practice guidelines by the Pediatric Infectious Diseases Society and the Infectious Diseases Society of America. Clin Infect Dis 53:e25–e76. doi: 10.1093/cid/cir531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sánchez AR, Rogers RS, Sheridan PJ. 2004. Tetracycline and other tetracycline‐derivative staining of the teeth and oral cavity. Int J Dermatol 43:709–715. doi: 10.1111/j.1365-4632.2004.02108.x. [DOI] [PubMed] [Google Scholar]
- 36.Wyres KL, Wick RR, Judd LM, Froumine R, Tokolyi A, Gorrie CL, Lam MMC, Duchêne S, Jenney A, Holt KE. 2019. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet 15:e1008114. doi: 10.1371/journal.pgen.1008114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nordmann P, Poirel L, Carrër A, Toleman MA, Walsh TR. 2011. How to detect NDM-1 producers. J Clin Microbiol 49:718–721. doi: 10.1128/JCM.01773-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clinical and Laboratory Standards Institute (CLSI). 2020. Performance standards for antimicrobial susceptibility testing. CLSI supplement M100S. 30th ed. CLSI. Wayne, PA. [Google Scholar]
- 39.The European Committee on Antimicrobial Susceptibility Testing. 2020. Breakpoint tables for interpretation of MICs and zone diameters. Version 10.0. EUCAST. Växjö, Sweden. [Google Scholar]
- 40.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Letunic I, Bork P. 2021. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. 2020. Genomic surveillance framework and global population structure for Klebsiella pneumoniae. Biorxiv. doi: 10.1101/2020.12.14.422303. [DOI] [PMC free article] [PubMed]
- 46.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 47.Alikhan N-F, Petty NK, Zakour NLB, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. Bmc Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X, Chen F, Chen Y. 2020. Gcluster: a simple-to-use tool for visualizing and comparing genome contexts for numerous genomes. Bioinformatics 36:3871–3873. doi: 10.1093/bioinformatics/btaa212. [DOI] [PubMed] [Google Scholar]
- 49.Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.01574-21-s0001.pdf, PDF file, 0.6 MB (631KB, pdf)
Supplemental material. Download spectrum.01574-21-s0002.xlsx, XLSX file, 0.2 MB (155.2KB, xlsx)
Data Availability Statement
All raw data in this study were deposited in GenBank under Bioproject accession number PRJNA750893.