ABSTRACT
Intravenous drug users (IDUs) are a high-risk group for HIV-1, hepatitis C virus (HCV), and hepatitis B virus (HBV) infections, which are the leading causes of death in IDUs. However, the plasma virome of IDUs and how it is influenced by above viral infections remain unclear. Using viral metagenomics, we determined the plasma virome of IDUs and its association with HIV-1, HCV, and/or HBV infections. Compared with healthy individuals, IDUs especially those with major viral infections had higher viral abundance and diversity. Anelloviridae dominated plasma virome. Coinfections of multiple anelloviruses were common, and anelloviruses from the same genus tended to coexist together. In this study, 4,487 anellovirus ORF1 sequences were identified, including 1,620 (36.1%) with less than 69% identity to any known sequences, which tripled the current number. Compared with healthy controls (HC), more anellovirus sequences were observed in neg-IDUs, and HIV-1, HCV, and/or HBV infections further expanded the sequence number in IDUs, which was characterized by the emergence of novel divergent taxons and blooms of resident anelloviruses. Pegivirus was mainly identified in infected IDUs. Five main pegivirus transmission clusters (TCs) were identified by phylogenetic analysis, suggesting a transmission link. Similar anellovirus profiles were observed in IDUs within the same TC, suggesting transmission of anellome among IDUs. Our data suggested that IDUs suffered higher plasma viral burden especially anelloviruses, which was associated with HIV-1, HCV, and/or HBV infections. Blooms in abundance and unprecedented diversity of anellovirus highlighted active evolution and replication of this virus in blood circulation, and an uncharacterized role it may engage with the host.
IMPORTANCE Virome is associated with immune status and determines or influences disease progression through both pathogenic and resident viruses. Increased viral burden in IDUs especially those with major viral infections indicated the suboptimal immune status and high infection risks of these population. Blooms in abundance and unprecedented diversity of anellovirus highlighted its active evolution and replication in the blood circulation, and sensitive response to other viral infections. In addition, transmission cluster analysis revealed the transmission link of pegivirus among IDUs, and the individuals with transmission links shared similar anellome profiles. In-depth monitoring of the plasma virome in high-risk populations is not only needed for surveillance for emerging viruses and transmission networks of major and neglected bloodborne viruses, but also important for a better understanding of commensal viruses and their role it may engage with immune system.
KEYWORDS: plasma virome, IDUs, viral expansion, anellovirus, transmission cluster, HIV-1, HCV
INTRODUCTION
The virome is part of the microbiome, and it can be described as the collection of all the viruses, including prokaryotic viruses or bacteriophages, and eukaryotic viruses (1, 2). In the past decades, along with the development of viral metagenomics, substantial progress was achieved in the discovery of virosphere diversity and the uncovering of virus evolution (3–6), identification of viral etiologies responsible for specific symptoms or disease outbreaks (7–12), as well as the virome-host interactions and potential role it may engage with the physiology and disease of the host (13–18). In the context of humans, the unbiased viral metagenomics greatly improves our recognition of the viral sequences present in both healthy individuals and those with different disease status, and is increasingly accepted and used in clinical settings (19–21).
Previous studies of the human virome mainly focused on gut, respiratory tract, and skin (22–27). Blood virome consist of many commensal viruses, as well as pathogenic viruses (21). Certain primary viral infection leads to altered plasma virome. For example, an increased viral burden (e.g., anellovirus and human endogenous retrovirus or HERV) was associated with HIV-1 infection in certain populations (28, 29). The altered virome is associated with immune status and disease progression, which raises questions about whether resident plasma viruses could participate in immune response and influence health conditions (30). Among these resident plasma viruses, anelloviridae is the most abundant and widespread virus family. Anellovirus is a single-stranded circular DNA (ssDNA) virus, which was first reported in hepatitis patient. Subsequent studies revealed a commensal nature of anellovirus because the virus circulates in both health people and those with disease. Even though no association of anellovirus with specific disease was established, it was reported to engage with our host immune system, and could be used as surrogate marker for immune status (31, 32). Great diversities of anelloviruses have been reported in human blood. In particular, a recent study nearly tripled the whole anellovirus genomic sequence number. These data highlighted uncovered diversity and the mystery nature of these viruses (33).
Besides, characterizing the blood virome is of great importance for surveillance of pathogenic viruses and for transfusion safety. Unrecognized or neglected viral infections can be present without symptoms or with mild symptoms and spread to other individuals through blood transfusion or organ transplantation, which may lead to serious complications in the recipients (20, 34). Intravenous drug users (IDUs) are one of the major routes of acquiring main blood transmitted viral (HIV-1, HCV, and HBV) infections worldwide (35, 36) due to frequent needle sharing and unsafe syringe cleaning practices (37). Besides, as these population may live with risk behavior, poor health outcome or even suboptimal immune status, they may also tend to acquire other viral infections and even carry novel viruses.
The metagenomics era has unmasked unprecedented diversity of viruses, and makes it possible to study viruses in human health with a new ecological perspective (38–41). In this study, we used unbiased viral metagenomics to explore the plasma virome of individuals who are at high risk of potential exposure to viral infections through long-term injection drug use. We compared the virome of individuals who were previously identified to have HIV-1, HCV, and/or HBV infections to determine whether these major blood transmitted viruses would have any influence on their plasma viral composition and diversity, as well as revealing additional or novel viruses that may be neglected but are of potential risk to human health. Besides, by analyzing the diversity and evolution of the most abundant and widespread anellovirus in detail, we provided a comprehensive knowledge of their dynamics in the plasma virome of IDUs.
RESULTS
Participants.
Of 99 plasma samples from IDUs, 26, 11, and 10 were detected as HIV-1, HCV, and HBV positive, respectively, 29 were identified as positive for both HIV-1 and HCV, and other 23 were negative for all three viruses (neg-IDUs) by RT-qPCR assay (Table 1). The subjects had a median age around 30, and the vast majority (95%) of them are male. Only one person from HC had higher education experience, and all the IDUs had primary or middle school education experience. Ten out of 11 healthy individuals were married, while the majority (73%) of the IDUs were either single or divorced. Compared with neg-IDUs, IDUs positive for HIV-1, HCV, and/or HBV had longer history and higher frequency of drug use, higher rates of syringe sharing and repeated use, and poor cleaning and disinfection practice (Table 1).
TABLE 1.
Characteristics of healthy participants and IDUs in this study
| Sociodemographics | Healthy controla | Intravenous drug users (IDUs) |
||||
|---|---|---|---|---|---|---|
| Neg-IDUsd | HIV-1+ | HCV+ | HIV-1/hCV+ | HBV+ | ||
| No. of individuals | 11 | 23 | 26 | 11 | 29 | 10 |
| Age | 31 (19 to 60) | 30 (21 to 37) | 31 (22 to 53) | 29 (21 to 39) | 31 (18 to 43) | 30.5 (28 to 36) |
| Gender (male/female) | 11/0 | 23/0 | 20/0 | 10/1 | 25/4 | 11/0 |
| Higher education | 9.1% | 0 | 0 | 0 | 0 | 0 |
| Married | 10 | 8 | 7 | 4 | 6 | 2 |
| Single/divorced | 1 | 15 | 20 | 7 | 23 | 8 |
| Duration of drug use/y | 3.5 (1 to 16) | 8.0 (1 to 20) | 4.0 (1 to 13) | 10.0 (1 to 22) | 6.0 (2 to 15) | |
| Frequency of drug use (times/day) | 3.0 (0.3 to 7) | 3.0 (1.5 to 10) | 3.0 (1 to 3) | 3.5 (1 to 9) | 4.0 (1 to 5) | |
| Syringe sharing | 38.1% | 60.0% | 30.0% | 50.0% | 77.8% | |
| Times of repeated syringe use | 1.0 (1 to 10) | 2.0 (1 to 8) | 1.5 (1 to 5) | 2.0 (1 to 10) | 3.0 (1 to 5) | |
| Cleaning method | ||||||
| Tap water | 75% | 50% | 66.7% | 78.6% | 71.4% | |
| Boiled water | 25% | 28.6% | 16.7% | 7.1% | 14.3% | |
| Never | 0 | 21.4% | 16.7% | 14.3% | 14.3% | |
| STDb history | 10.0% | 5.9% | 33.3% | 0 | 22.2% | |
| Sexual partners | 3.0 (0 to 10) | 1.0 (0 to 6) | 2.0 (0 to 5) | 1.0 (0 to 5) | 3.0 (1 to 10) | |
| Ways of drug use | ||||||
| Intravenous | 33.3% | 23.8% | 70.0% | 65.0% | 44.4% | |
| Intravenous and oral | 66.7% | 76.2% | 30.0% | 35.0% | 55.6% | |
| Drug rehabilitation times | 2.0 (1 to 4) | 2.0 (1 to 15) | 2.0 (1 to 5) | 4.0 (2 to 15) | 2.5 (1 to 5) | |
| Other drug usec | 74.7% | 75.0% | 85.7% | 78.9% | 100% | |
Healthy control group (non-IDU and without main viral infections).
STD, sexually transmitted disease.
IDUs without HIV-1/HCV/HBV infections.
Other drugs include Diazepam and Triazolam.
Overview of the plasma virome of IDUs.
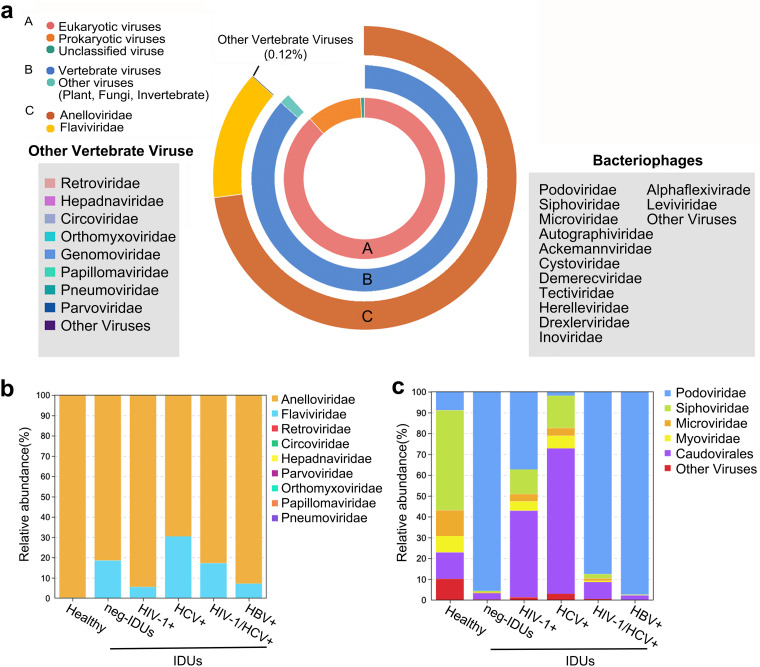
Unbiased viral metagenomic method was used to determine the viromes of all plasma samples, including centrifugation, filtration, nuclease digestion, followed by random amplification, library preparation, and sequencing (see Materials and Methods). In total, 1.12 billion paired-end clean reads were obtained with an average of 10 million reads for each sample. After removing host and bacterial sequences, 29.9% of the reads were classified as viral sequences. In general, the plasma virome of the HC and IDUs were dominated by eukaryotic viruses (88.6% of total viral reads), followed by prokaryotic viruses (bacteriophages, 10.7%) and unclassified viruses (0.7%) (Fig. 1a). Vertebrate viral reads accounted for the vast majority (98%) of eukaryotic viral reads, and the rest of the reads were from plant, fungi and invertebrate host origins. We identified the presence of main vertebrate viral families, including Anelloviridae, Flaviviridae, Retroviridae, Hepadnaviridae, Circoviridae, Orthomyxoviridae, Genomoviridae, Papillomaviridae, Pneumoviridae, Parvoviridae, and Kolmioviridae. Anelloviridae and Flaviviridae were the most prevalent viruses, and they accounted for 84% and 15.9% of all vertebrate viral abundance, respectively (Fig. 1a).
FIG 1.
Summary of the viral compositions detected in this study. The donut chart shows the distribution of all viral reads according to taxonomical ranks (a). Relative abundance of main vertebrate viruses (b) and prokaryotic viruses (bacteriophages) (c) in different groups.
We then compared the sensitivity of metagenomic sequencing with the qPCR results (Table S1). HIV-1 was detected in 24 out of 26 samples in HIV-1+ group, HCV in 11/11 for HCV+ group, HIV-1 and HCV in 28/29 for both HIV+ and HCV+ group, and HBV in 9/10 for HBV+ group. These data indicated that the metagenomic sequencing method had a good sensitivity for the detection of different viruses. Expect for HBV, no significant correlations between the number of viral reads and Ct values were observed for HIV-1 and HCV (Fig. S1a).
Plasma viromes of both healthy individuals and IDUs were dominated by anellovirus (Fig. 1b; Fig. S2a), and a significant correlation between the number of viral reads and Ct values was observed for anellovirus (Fig. S1b). Compared with HC, plasma virome of IDUs, regardless of infection or free for HIV-1, HCV and/or HBV, were more likely to carry other viruses, such as pegivirus (35 in IDUs, one in HC), parvoviridae (20 in IDUs, 0 in HC), and circoviridae (34 in IDUs, 3 in HC) (see supplemental material). The distribution of plasma bacteriophages was relatively even in healthy individuals, but was characterized by one or two dominant bacteriophage taxons in IDUs (Podoviridae and Caudovirale) (Fig. 1c; Fig. S2b).
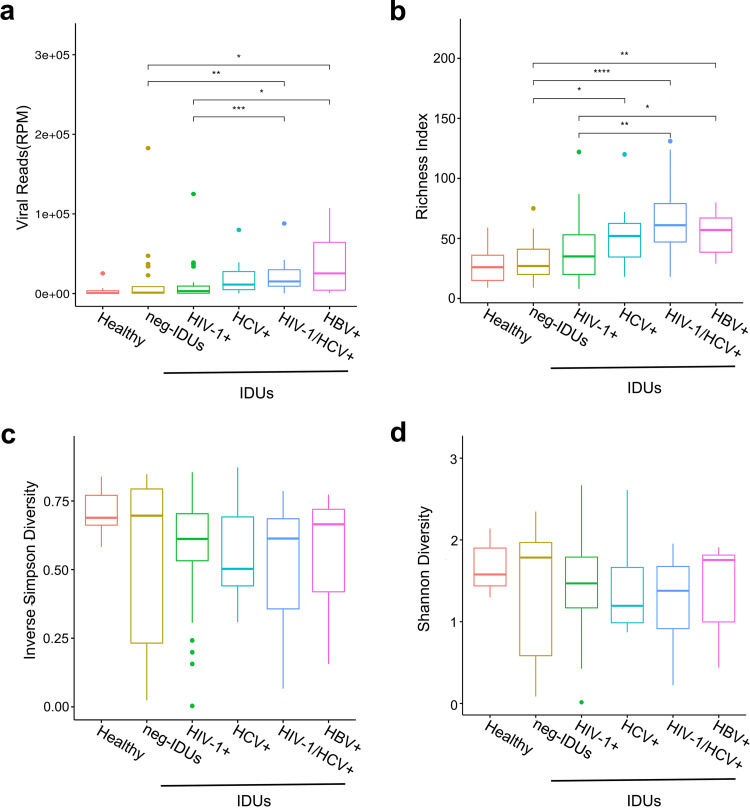
To investigate the plasma viral burden and complexity in HC and IDUs, we determined the viral abundance (reads per million, RPM) and viral diversity (richness, Simpson and Shannon indexes) of different groups. Compared with HC, the plasma of neg-IDUs had a relatively higher total viral abundance (P = 0.20), and IDUs with main viral infections appeared to have even higher plasma viral abundance (Fig. 2a). Besides, richness score showed a similar trend of the distribution of viral taxons. HC had the lowest numbers of plasma viral taxons, neg-IDUs, and those with main viral infections showed increased numbers (Fig. 2b). As for both Inverse Simpson and Shannon indexes, there were no apparent differences between HC and neg-IDUs, but IDUs with viral infections showed lower scores (Fig. 2c and d), which indicated unevenly distributed viral populations possibly due to the expansion of certain viruses. In addition, we observed significant positive correlations between duration of drug use and both viral abundance and richness (Fig. S3). There was no influence of age on the composition of plasma virome.
FIG 2.
Viral abundance and diversity among different groups. Viral abundance was shown as reads per million (RPM) (a). Viral diversity was shown for each group with richness score (number of annotated taxons) (b), Inverse Simpson index (c), and Shannon index (d).
Profile of the plasma Anelloviridae.
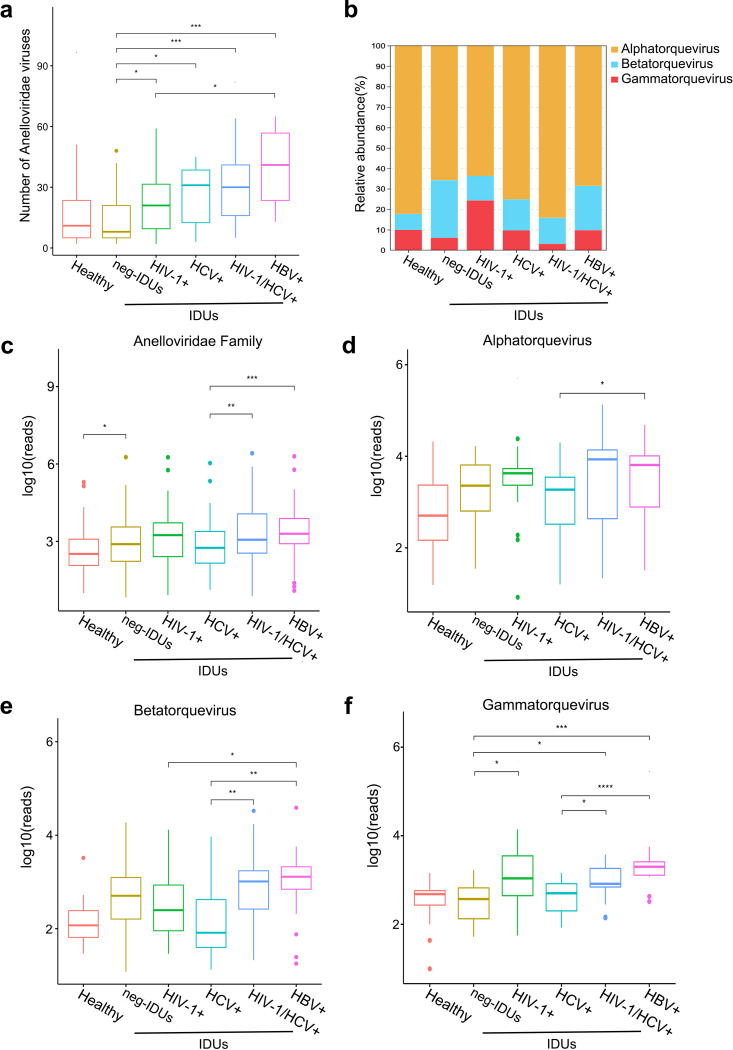
As the anelloviruses were the most abundant and prevalent eukaryotic viruses in plasma virome, we further characterized their diversity and distribution among different groups. Anelloviridae was present in all the plasma samples tested, and at least two different anellovirus taxons could be annotated in each sample. Over 71% of the samples had more than 10 anellovirus taxons, and 8.9% had over 50 different anelloviruses. Compared with HC, IDUs had more anelloviruses in individual and group levels. In particular, one sample from IDUs was found to carry 65 different anelloviruses (Fig. S4a, b). These results indicated a great diversity and individual variation of plasma anelloviruses. The number of anelloviruses identified among neg-IDUs did not significantly differ from that in HC, while infection with HIV-1, HCV, and/or HBV significantly increased the number of anelloviruses in IDUs (Fig. 3a). For the three main genera, alphatorquevirus was the most abundant genera among all samples. Compared with HC, a slight increase of relative abundance of betatorquevirus was observed in neg-IDUs (P > 0.05) (Fig. 3b), and similar increase were also observed for the absolute reads number of total anellovirus, alphatorquevirus, and betatorquevirus genera (Fig. 3c to e). Expect IDUs with HCV infections, higher abundance of not only total anellovirus, but also alphatorquevirus and gammatorquevirus genera was associated with HIV-1 and HBV infections, as well as HIV-1/HCV coinfections (Fig. 3c, d, and f). For betatorquevirus genera, its increase was only observed in IDUs with HIV-1 or HBV infections (Fig. 3e).
FIG 3.
Expansion and distribution of anelloviruses among different groups. Number of annotated anelloviruses (a). Relative abundance of three anellovirus genera in different groups (b). Abundance (log10 reads) of Anelloviridae (c), alpha-(d), beta-(e), and gammatorquevirus (f).
The enormous genetic diversity and expansion of Anelloviridae.
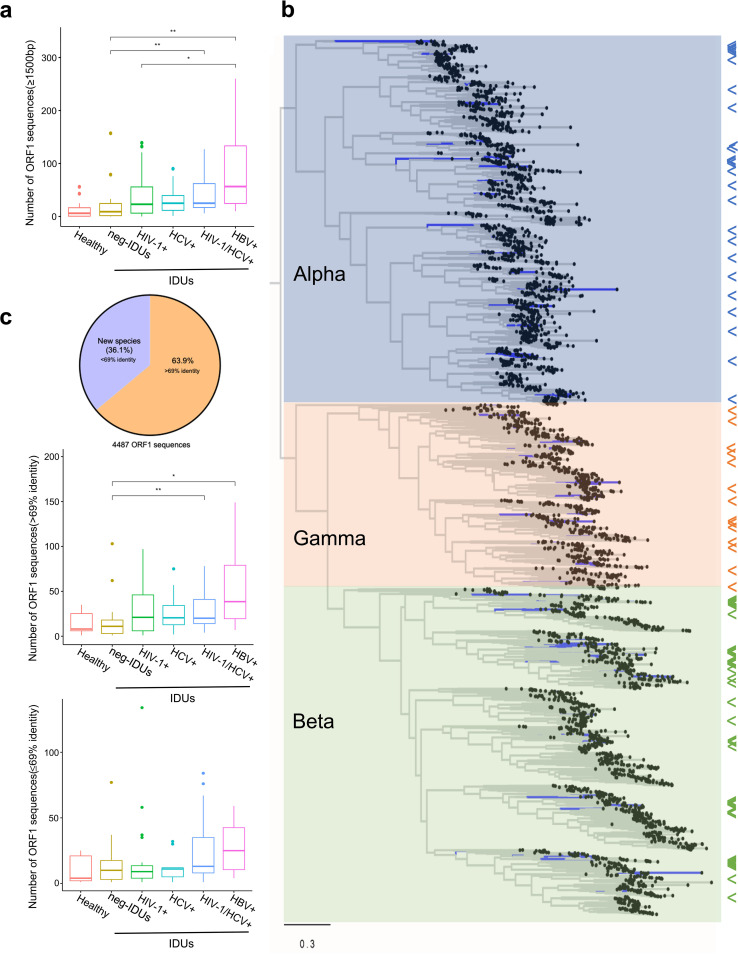
Due to the extremely high genetic diversity of anelloviruses, different subtypes/strains within the same species are common. In order to reflect the actual diversity and potential novel species of the anelloviridae, we extracted all the ORF1 sequences and analyzed their distribution and diversity among different groups. In total, we obtained 4,487 ORF1 sequences (over 1,500 bp) and found that ORF1 sequences heavily outnumbered annotated anellovirus taxons (Fig. 3a and 4a), which indicates a significant variability of anellovirus genomes. Compared with healthy individuals, neg-IDUs showed a higher number of ORF1 sequences, and the numbers were even higher in IDUs with HIV-1, HCV, and/or HBV infections, which are very similar to the observation in the distribution of anellovirus abundance (Fig. 3c). We then performed a phylogenic analysis with maximum-likelihood method using all the ORF1 sequences together with reference sequences from the three main genera, and found that all these sequences could cluster into the three main genera (Fig. 4b). Most sequences clustered closely with references; but the other sequences either did not cluster with known references, or formed independent phylogenetic clade, suggesting high genetic diversity of anellovirus and the presence of potential new species. Alignment analysis showed that 63.9% of the ORF1 had identities over 69% to all known anellovirus sequences in the database, and the rest (n = 1,620) were potential novel species with identities below 69% (Fig. 4c). Further analyses of all these ORF1 sequences showed an expansion of both novel and known anelloviruses in plasma of IDUs, especially those with HIV-1, HCV, and/or HBV infections (Fig. 4c).
FIG 4.
Unprecedented plasma anellovirus diversity. Number of anellovirus ORF1 sequences (>1,500 bp) in each group (a). Maximum-likelihood phylogenetic tree of ORF1 sequences. Arrows to the right of the tree indicate the positions of reference sequences (b). Distributions of new ORF1 sequences with over or below 69% identity to all currently known anelloviruses (c).
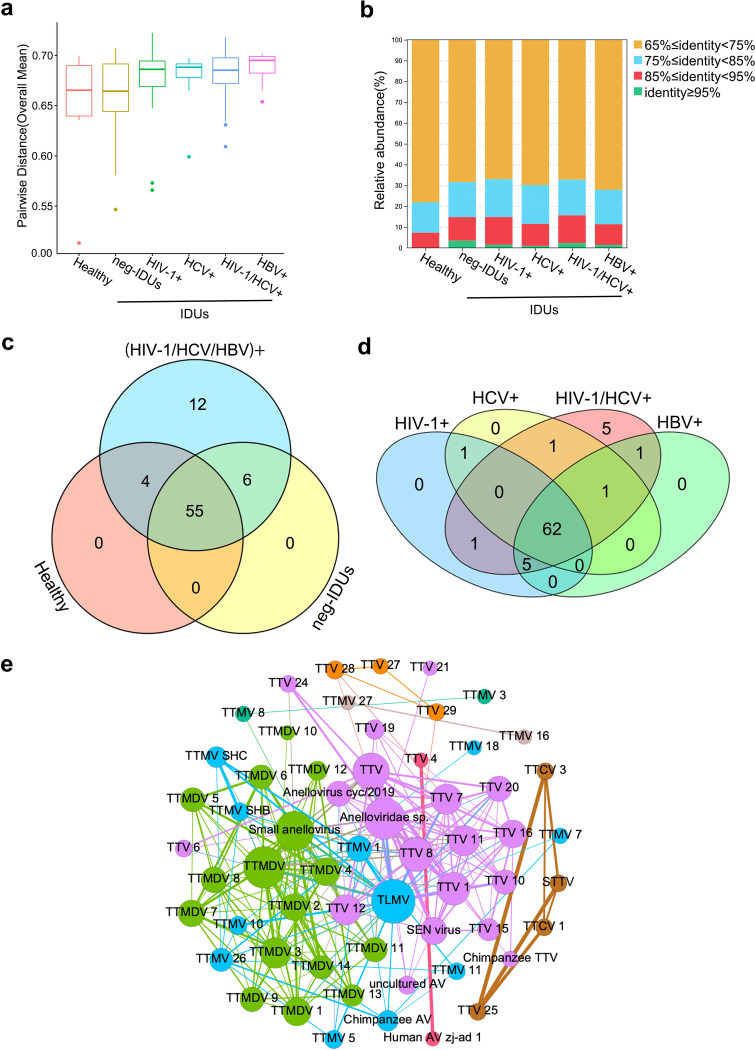
Next, we measured the pairwise distance of ORF1 sequences within each individual to determine genetic variation. Even though in HC whose anellovirus abundance is relatively low, the pairwise distance remained high (mean distance of 0.645). Neg-IDUs showed a slightly higher (0.658) pairwise distance than HC. Consistent with the abundance and number of anellovirus sequences, IDUs with HIV-1, HCV, and/or HBV infections had the highest plasma virus pairwise distance (mean distance from 0.673 to 0.688) (Fig. 5a). These data indicated the expansion and diversification of anelloviruses after certain viral infections in IDUs.
FIG 5.
Blooms and coexistence of certain anelloviruses. Comparison of within individual pairwise distances between different groups (a). Distributions of ORF1 sequence clusters that have different identities (b). Venn diagrams of different anellovirus taxons between healthy individuals, neg-IDUs, IDUs with HIV-1/HCV/HBV infections (c), and IDUs with different infection patterns (d). Co-occurrence network of different anelloviruses. Viruses in the same network were highlighted with same color. Bigger size of the circle means more viruses it interacts with, and the line between two circles indicates the frequency of the co-occurrence. TTV, Torque teno virus; TTMV, Torque teno mini virus; TTMDV, Torque teno midi virus; TLMV, TTV-like mini virus; STTV, Simian torque teno virus; AV, anellovirus.
To determine whether the virus expansion favors certain viruses and how different viruses coexist with each other, we further analyzed ORF1 sequence clusters according to their genetic identities. Healthy individuals had more sequences that fell into clusters with relatively low identities (<75%), while IDUs with or without main viral infections had more sequences belonging to higher identity clusters (≥85%), and all sequences in the clusters with ≥95% identity were found in IDUs (Fig. 5b). This result revealed that HIV-1, HCV, and/or HBV infections in IDUs may lead to the expansion of anellovirus and form many clusters with high genetic identities.
Even though most of the anelloviruses were shared by different individuals (more than 71%) (Fig. 5c), we found 12 anelloviruses were only detected in IDUs with major viral infections, and no anelloviruses were found to be specifically associated with certain viral infections (Fig. 5d). The 12 anelloviruses were mainly from alphatorquevirus and betatorquevirus, and different from the anelloviruses that fell into the >95% identity clusters (Fig. S4c; Table S2). These data further indicated the expansion of anellovirus after viral infection may lead to both the emergence of new anelloviruses and blooming of existing anelloviruses. Using co-occurrence network analysis, we found that the same anellovirus genera tended to form close networks and coexist together (Fig. 5e). The main hubs of the alphatorquevirus (purple) and gammatorquevirus (green) networks included Torque teno virus (TTV) 1, 7, 8, and 11 and Torque teno midi virus (TTMV) 2, 3, and 4, as well as two unclassified viruses in each network, respectively. TTV-like mini virus (TLMV) dominated the betatorquevirus (blue) networks. Eight of the 12 anelloviruses only detected in IDUs with major viral infections formed several independent clusters (Fig. S4d).
Transmission of pegivirus and anallovirus in IDUs.
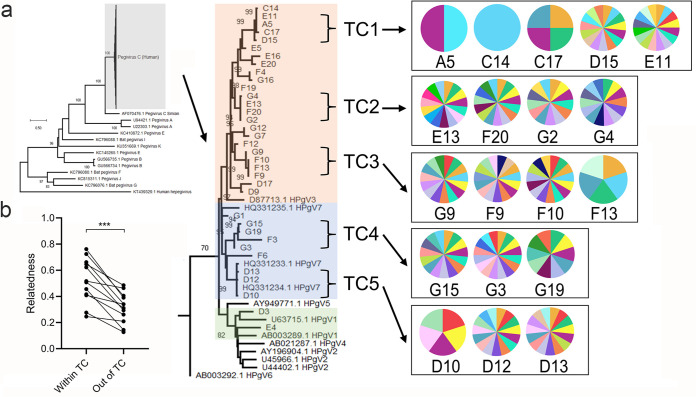
Pegivirus belongs to Flaviridae. Human pegivirus (HPgV) has high prevalence in HIV-1 and HCV infected individual, and shows a beneficial effect on HIV-1 infection (42). In this study, HPgV was detected in 37 individuals, and most of these individuals were IDUs with HIV-1, HCV and/or HBV infections (Fig. S5). Of all the pegivirus, 35 were HPgV-C, and two were HPgV-H (or HPgV-2). Only one healthy individual and five neg-IDUs were also positive for pegivirus. The detection rate of HPgV was well consistent with its prevalence in these populations (42). We previously reported significantly negative correlations of HPgV abundance with HIV-1 and anellovirus abundance in men who have sex with men (MSM) (30), while the correlations were not found in IDUs (P = 0.53 and 0.76) (Fig. S5). Only a weak positive correlation between pegivirus and HCV (P = 0.27) was detected (Fig. S5). Phylogenetic analysis of full-length NS5B sequences of human HPgV-C showed that most of them belonged to HPgV3/7 genotypes (Fig. 6a), which was consistent with the prevalence of both pegivirus genotypes in this region (43). HPgV-C genotype 1, which was rarely detected in China, was found in two individuals.
FIG 6.
Transmission of pegivirus and anellovirus among IDUs. (a) Maximum-likelihood phylogentic tree of human pegivirus C. Three human pegivirus genotypes are highlighted with orange, blue, and green colors. Five transmission clusters (TC1 to 5) are labeled. Pie charts to the right show the individual anellome profile within each TC (presence of each anellovirus), only top 15 abundant anelloviruses are shown. (b) Relatedness of anellovirus reads within TC versus unrelated individuals (also described in Table S3). Relatedness was compared with Wilcoxon matched-pairs signed rank test.
In order to determine whether HPgV identified among these IDUs had transmission links, we performed transmission cluster analysis based on genetic distances of closely related sequences. At least five transmission clusters that contained 3–5 HPgV variants with more than 99% identity were identified. In total, 54.3% (n = 19) of HPgV sequences formed TCs, and 18 of them were from IDUs. These data indicated that HPgV, like HIV and HCV, was mainly transmitted among IDUs in transmission cluster manner (Fig. 6a). In particular, TC5 contained two references that were isolated from IDUs in the same region of Yunnan, supporting the local transmission link of HPgV among IDUs.
As the transmission of anelloviruses was common in blood transfusion or organ transplantation (44), we asked whether anellome is transmissible among IDUs. Because the identification of TCs of HPgV allowed us to determine the IDUs who have potential transmission and epidemiological links, we further characterized and compared the anellovirus profile among each individual within the five TCs. Two to three IDUs in each TC shared highly similar anellovirus profiles that shared 67% to 99% of annotated anelloviruses. For example, two individuals in TCs 1, 4, and 5, and three IDUs in TCs 2 and 3 displayed highly similar anellovirus profiles (Fig. 6). Of particular importance is that two pairs of IDUs in TC3 shared 100% the same anelloviruses (Table S3). One healthy individual in TC1, and one neg-IDUs in TC1 and TC3 displayed relatively “simple” anellome, which was consistent with the relatively low diversity of anellovirus in general population and neg-IDUs (Fig. 2b and 3a). However, different individuals showed great variations in anellovirus abundance, even between those had similar anellome profiles (Fig. S6). As more than 36% of anelloviruses found in this study were potential novel species, and in order to reflect the transmission of anelloviruses more accurately among IDUs, we compared the relatedness of all anelloviruses (both annotated and undefined) within or outside the TCs defined by pegivirus. Compared with unrelated individuals outside each TC, individuals from the same TC that shared similar anellome profiles had more related anelloviruses (P = 0.0005) (Table S3; Fig. 6b). Both results from Fig. 6 and Table S3 indicated the transmission of anelloviruses along TCs in IDUs.
DISCUSSION
In this study, using viral metagenomics we explored the plasma virome of IDUs, and whether the changes of viral compositions circulating in these individuals was associated with HIV-1, HCV, and/or HBV infections. IDUs generally bear a high burden of the main bloodborne viruses (e.g. HIV-1/HCV/HBV), and injection drug use as well as other risk practices are major drivers of new infections and transmissions globally (45–48). Relatively low viral population diversity, high viral taxon number and viral abundance in IDUs, especially those with major viral infections, indicated higher viral burden of different viruses and probably the expansion of certain viruses in the blood of IDUs. We observed higher proportion of risk behaviors in IDUs with HIV-1, HCV, and/or HBV infections than healthy individuals, including longer duration of drug use, higher frequency of syringe sharing and repeated use, and worse syringe cleaning practices. In particular, longer duration of drug use was found to be associated with higher viral abundance and richness, indicating that long-time drug abuse would change the virome composition probably due to higher risk of exposure to various viral infections. Besides, all the IDUs were from the underdeveloped regions of Yunnan, near the China-Myanmar border, which is close to the “Golden Triangle,” where active trading, travelling, and syringe sharing between different risk populations would be bridges for further transmissions (49, 50).
Anelloviridae is the most ubiquitous and abundant viral family, and infections by anelloviruses are most probably asymptomatic (32, 51). Even though anelloviruses are generally known as a part of the commensal virome, numerous studies found the association of anellovirus with host immune status, and suggest anellovirus to be considered as a potential marker for immunocompetence in clinical organ transplantation (52–54). Anellovirus has also been associated with various liver diseases, such as acute or transfusion associated hepatitis, and chronic hepatitis B and C (55). Compared with healthy individuals, the increase of anellovirus abundance in IDUs might indicate a poor health condition, and therefore a higher risk of infection by various viruses. The infections with HIV-1, HCV and/or HBV may have impaired the immune system, and make the latter unable to control the replication of some resident viruses such as anelloviruses. This is particularly true as some previous studies have consistently found that increased anellovirus load was associated with HIV-1 infections, and the anellovirus load was inversely correlated with CD4+ T cell counts in MSM (30, 56).
Anellovirus is one of the most divergent viruses ever discovered, and has higher genetic diversity than HIV and papillomavirus. A recent study focusing on blood transfusion cohorts reported more than 1,600 anellovirus sequences, which nearly tripled the number of previously known anellovirus sequences (33). In this study, we obtained more than 4,450 ORF1 sequences, which further nearly tripled current anellome database. More than 36% of them have below 69% identity to known anelloviruses, unveiling the vast diversity of anellovirus. The results indicated that an expanded diversity and pairwise distance, as well as an increased abundance of anellovirus, was associated with injection drug use and major viral infections. Whether the blooms of anellovirus diversity and population size during HIV-1/HCV/HBV infections are a consequence of fast mutation rate and/or frequent recombination between different variants need further analyses (33, 57).
Because IDUs are the main population for transmission of many blood-borne viruses, we asked whether the anellome could be transmitted like other viruses. Even though viral abundance varied largely in different individuals, the similar anellovirus profiles in IDUs within the HPgV transmission clusters suggested that anelloviruses can be transmitted among these IDUs like other common virus (e.g., HIV-1, HPgV). However, not all the individuals within each transmission cluster showed the same anellome profile. There may have two explanations. First, the syringe sharing could happen before HPgV infections, and the “donor” may have a different anellome. Second, the anellome was similar between each individual at the beginning after transmission, but after a long period interaction with host’s immune system, it evolved to a different profile, as different individuals may favor certain virus types. Furthermore, these data also suggested a potential use of commensal viruses to trace transmission networks of main blood-borne viruses. For example, a recent study suggested that anellovirus could help to reveal drug use network, and early accumulation of anellovirus may predict the infection risk of HIV-1 and HCV (58).
We further confirmed the huge and yet to be discovered diversity of anelloviruses. The expansion and transmission of anelloviruses in these IDUs after main viral infections showed the ubiquity and active replication of these viruses within the host. We didn’t know whether the blooms of anellovirus would have any influence on either other viruses’ replication or disease progression, but their unique traits could provide opportunities for potential translational applications. For example, the sensitivity of anellovirus’s response to immune status or possible disease progression implied its diagnostic potential to support the development of personalized treatment strategies (51, 52). Besides, commensalism, transmission, and persistence of diverse anelloviruses imply less or no damage to the host, making them attractive vector candidates for delivering therapeutic strategies for different diseases. We could imagine more anelloviruses will be unveiled, and whether different species or genotypes of anelloviruses have different tissue or cell tropisms, and then influence host immunity and health condition need further studies.
HIV-1/HCV/HBV are globally screened, and the drugs and prevention measures against these viruses are available. However, many emerging and neglected viruses could be circulating in IDUs. These viruses may have potential disease outcomes or pose threats to others through blood transmission (55). In this study, several other blood-borne viruses that may associate with hepatitis were identified, including human pegivirus C (HPgV-1), more recently discovered pegivirus H (HPgV-2), and hepatitis delta virus (HDV) (Table S4). Even though no pathogenicity of HPgV is established, growing lines of evidences confirmed its high prevalence in persons with blood-borne or sexually transmitted infections. However, a lot of previous studies indicated a beneficial effect of HPgV during HIV-1 infection (42, 59). Whether the coinfections of HPgV with other viruses would impact the health outcome needs further investigation. HDV depends on HBV for propagation and may lead to the most severe form of viral hepatitis (60). The detection of HDV together with HPgV, HIV-1, and HCV in IDUs showed a complex circulation of blood-borne viruses and raised concerns about severe disease outcome and transmission risk among these populations. Besides, we were able to assemble a full circular viral genome (GenBank: ON226770) in one IDU sample, and this genome was most closely related to porcine circovirus 3 with only 70% identity, implying that it may be a novel circovirus specific to humans. The presence of these potential emerging viruses in this high-risk group highlighted the importance to screen and investigate their prevalence in IDUs and other high-risk cohorts, and to further determine their origin and transmission pattern (61).
There were several limitations of this study. First, the number of individuals in several groups was relatively small. Second, compared with many virome studies that focused on either RNA or DNA viruses, we employed an unbiased method targeting both RNA and DNA viruses. The strategy is time-saving, but the random amplification method used here may lead to further enrichment of circular genome sequences of Anelloviridae, which may disturb the relative viral abundance of some viruses. A methodology comparison between the methods with and without random amplification in performance will be very helpful in future study. Furthermore, even though the method used here was generally sensitive, as the virome composition may vary due to different populations, it is important to repeat a similar study and validate the findings in another cohort. Third, the lack of longitudinal samples limits us for further analysis of the virome dynamics within each individual.
Conclusions.
Using viral metagenomics, this study unveiled the vast diversity of plasma virome in IDUs, and the blooms of anelloviruses may be associated with certain viral infections. Complex expansion and transmission of anelloviruses raised concerns about whether they may influence the susceptibility of IDUs and other groups to the infection by other viruses and disease outcome. In-depth monitoring of plasma virome in high-risk populations might not only be needed for both surveillance for emerging viruses and transmission networks of major and neglected blood-borne viruses, but also be important for a better understanding of the commensal viruses and their role it may engage with our immune system.
MATERIALS AND METHODS
IDUs were from a previous transmitting viral infections surveillance program (2009 to 2012) in Yunnan Province, China. This study was approved by the Ethics Committees of Kunming Institute of Zoology, Chinese Academy of Sciences, and complied with all relevant ethical regulations (SWYX-2008010, SMKX-20180102-178). Blood samples from healthy non-IDUs of the same community were collected as controls (HC). Oral or written informed consents were obtained from all participants before sample collection. At least 2 mL of blood was collected from each participant (99 drug users and 11 healthy controls), and plasma was separated within 24 h upon sampling. All plasma samples were stored at −80°C until use. Initial detection of HIV-1/HCV/HBV was done by reverse transcription-real-time PCR (RT-qPCR; Sansure Biotech, China).
Sample processing, library construction, and sequencing.
Enrichment of encapsidated DNA and RNA viruses was performed as previously described (18, 62). Briefly, 200 μL plasma was thawed on ice and homogenized for 3 to 5min, and then the suspension was centrifuged at 12,000 × g at room temperature for 15 min. The supernatant was passed through a 0.45 μm sterile filter to reduce background materials (Costar Spin-X centrifuge tube filters, Corning, USA). Filtrates were incubated with a cocktail of nucleases, including 15U Turbo DNase (Invitrogen, USA), 20U Benzonase (Novagen, Germany), and 20U RNase I (Promega, USA) for 2 h at 37°C. The reaction was terminated with 30 mM EDTA at 65°C for 10 min. Total nucleic acids (including both DNA and RNA) were extracted using QIAamp MinElute virus kit (Qiagen, Germany), which were then amplified using a random-amplification approach (REPLI-g Single Cell WTA kit, Qiagen, Germany) according to the manufactures’ instructions. The amplified products were purified by QIAquick PCR purification kit (Qiagen, Germany). DNA libraries were prepared using NEBNext UltraII FS DNA library Prep Kit (Illumina, USA), quantified by Qubit3.0 (Invitrogen, USA) and sequenced on the Illumina Novaseq platform (Illumina, USA) with 2 × 150-bp paired reads.
Virome bioinformatic analyses.
Sequencing data were analyzed using an in-house pipeline. The NGS raw data were filtered by Cutadapt v.1.18 (63) and Trimmomatic v.0.38 (64) by removing Illumina sequencing adaptor and low-quality sequences. Human- and bacterium-derived sequences were subtracted from the data by Bowtie2 v.2.3.4.3 (65). The remaining high-quality reads were de novo assembled by Megahit v.1.1.3 (66). Assembled contigs, as well as singlets, were mapped against the viral nucleic acid and protein database using BLASTn (E < 10−10) and BLASTx (E < 10−5) (DIAMOND v.0.9.24) (67), respectively. All the viral hit candidates were then searched against the NCBI nonredundant nt and nr database to remove reads or contigs that have higher similarity to sequences related to host, bacteria, fungi, plasmids, vectors, and other nonviral sequences than to viral sequences (false positives). To reduce the risk of potential cross-library contamination due to index-hoping (68), viruses with a read count less than 0.1% of the highest count for that virus among the other libraries was removed for subsequent analyses. Viral abundance was calculated by reads per million (RPM).
Phylogenetic analyses.
The viral sequences of anellovirus and pegivirus were extracted from all viral contigs/reads, and were aligned to reference viral genomes to generate full or partial genomes using Geneious R11 program (69). ORF1 region of anellovirus was extracted using NCBI’s ORF Finder tool using the “any sense codon” option, which were then curated by aligning against anellovirus reference sequences. All the obtained ORF1 sequences of anellovirus were deduplicated with a threshold of 99% identity. ORF1 region of the reference anellovirus and NS5B region of the reference pegivirus were downloaded from the NCBI’s GenBank database. Viral nucleic acid sequences were first translated into amino acids and aligned using MAFFT (70). Phylogenetic trees were inferenced using the maximum likelihood method with IQ-Tree (71). Model test program was used to determine the best substitution model. Phylogenetic trees based on nucleotide sequences were generated using the bootstrap method (1,000 times) under a GTR+G model. According to previous studies of HIV-1 and HCV (72), we used a stringent threshold of 1% genetic distance for the detection of transmission clusters (TC) of pegiviruses.
Anellovirus diversity and distance.
Chao Richness Score was used to compute the number of anellovirus species in each group. The diversity of anellovirus community were indicated by both the Shannon Diversity Index and Inverse Simpson Index. Pairwise distances of ORF1 amino acid sequences within each group were calculated by MEGA 7.0 (73). All the ORF1 amino acid sequences were clustered into different groups based on their identities to each reference (65% to 75%, 75% to 85%, 85% to 95%, ≥95%) using CD-HIT (74), and only clusters that had at least 10 sequences were used for downstream analyses. In order to determine the potential transmission of Anelloviridae among IDUs, the general diversity and relatedness of Anelloviridae between individuals within or out of the TC (determined by pegivirus phylogeny) were compared (44, 58). Anellovirus reads were mapped to corresponding anellovirus contigs using highly stringent threshold with Bowtie 2 (44), and relatedness was shown as the proportion of recovered anellovirus reads within/out of TC relative to cognate anellovirus reads (anellovirus reads that were recovered from contigs of each individual).
Detection of anellovirus with qPCR.
Primers (forward: 5′-ACWKMCGAATGGCTGAGTTT-3′, reverse: 5′-CCCKWGCCCGARTTGCCCCT-3′) targeting the conserved UTR region of anelloviridae was used (52). A SYBR qPCR method was used for the detection. The reaction contained 10 μL of 2× SYBR Green Pro Taq HS Premix (AGBio, China), 1 μL of both primers (10 uM each), 3 μL template, and 5 μL H2O. qPCR programs were as follows: 95°C for 1 min and 30 s, 40 cycles at 95°C for 15 s, and 63°C for 1 min.
Statistics.
Differences between groups were determined by the nonparametric Kruskal-Wallis Test, correcting for multiple comparisons with Dunn’s procedure. A difference with P < 0.05 was considered to be statistically significant. Co-occurrence networks of different anelloviruses were illustrated by Gephi v.0.9.2. Relatedness of anellovirus reads within TC versus unrelated individuals was compared with Wilcoxon matched-pairs signed rank test. Spearman’s correlation was calculated to determine the correlation between the qPCR and metagenomic sequencing results (RPM). Influence of age and time of drug use on the blood virus composition was also determined by spearman’s correlations (age/time of drug use versus RPM, Richness, Shannon index). All statistical analyses were performed using RStudio v.3.8.
Availability of data and materials.
All the raw sequencing data were deposited in the CNSA (https://db.cngb.org/cnsa/) of CNGBdb (project number CNP0002340) under the accession numbers CNS0468254-CNS0468363. Anellovirus and pegivirus sequences generated in this study can be found in the same project under the accession numbers CNS0483741-CNS0483743.
Supplementary Material
ACKNOWLEDGMENTS
Thank for Kai Liu for the support and suggestions at the early stage of the project. Thank for Rui Yu for the support of the revision of the manuscript. We also thank Jian-Hua Wang at Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, for his very helpful discussion and suggestions on the draft.
This study was approved by the Ethics Committees of Kunming Institute of Zoology, Chinese Academy of Sciences, and complied with all relevant ethical regulations (SWYX-2008010, SMKX-20180102-178).
The authors declare no conflict of interests.
This study was supported by National Natural Science Foundation of China (32170147, U1302224, 81271892), Shanghai Science & Technology Innovation Action Program (20MC1920100), Yunnan Key Research and Development Program (202103AQ100001, 202102AA310055), and the Key Program of Chinese Academy of Sciences (KJZD-SW-L11-02).
C.Z., Y.-T.Z., and Y.L. participated in study design; M.Y., X.C., R.-R.T., and F.-L.L. collected samples; R.X., Y.M., and P.Z. performed the main experiments; L.C. and Y.L. performed bioinformatic analyses; Y.L., L.C., and C.Z. interpreted the data; Y.L. and C.Z. wrote the manuscript; Y.-T.Z. and Y.-Q.K. contributed to data interpretation and manuscript revision; C.Z. and Y.-T.Z. supervised the study. All authors read and approved the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Yong-Tang Zheng, Email: zhengyt@mail.kiz.ac.cn.
Chiyu Zhang, Email: zhangcy1999@hotmail.com.
Alison Sinclair, University of Sussex.
REFERENCES
- 1.Zárate S, Taboada B, Yocupicio-Monroy M, Arias CF. 2017. Human virome. Arch Med Res 48:701–716. doi: 10.1016/j.arcmed.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Virgin HW. 2014. The virome in mammalian physiology and disease. Cell 157:142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chow CE, Suttle CA. 2015. Biogeography of viruses in the sea. Annu Rev Virol 2:41–66. doi: 10.1146/annurev-virology-031413-085540. [DOI] [PubMed] [Google Scholar]
- 4.Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, Thomas AD, Huntemann M, Mikhailova N, Rubin E, Ivanova NN, Kyrpides NC. 2016. Uncovering Earth’s virome. Nature 536:425–430. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- 5.Zhang YZ, Shi M, Holmes EC. 2018. Using metagenomics to characterize an expanding virosphere. Cell 172:1168–1172. doi: 10.1016/j.cell.2018.02.043. [DOI] [PubMed] [Google Scholar]
- 6.Shi M, Lin XD, Chen X, Tian JH, Chen LJ, Li K, Wang W, Eden JS, Shen JJ, Liu L, Holmes EC, Zhang YZ. 2018. The evolutionary history of vertebrate RNA viruses. Nature 556:197–202. doi: 10.1038/s41586-018-0012-7. [DOI] [PubMed] [Google Scholar]
- 7.Delwart E. 2012. Animal virus discovery: improving animal health, understanding zoonoses, and opportunities for vaccine development. Curr Opin Virol 2:344–352. doi: 10.1016/j.coviro.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grard G, Fair JN, Lee D, Slikas E, Steffen I, Muyembe JJ, Sittler T, Veeraraghavan N, Ruby JG, Wang C, Makuwa M, Mulembakani P, Tesh RB, Mazet J, Rimoin AW, Taylor T, Schneider BS, Simmons G, Delwart E, Wolfe ND, Chiu CY, Leroy EM. 2012. A novel rhabdovirus associated with acute hemorrhagic fever in central Africa. PLoS Pathog 8:e1002924. doi: 10.1371/journal.ppat.1002924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edridge AWD, Deijs M, Namazzi R, Cristella C, Jebbink MF, Maurer I, Kootstra NA, Buluma LR, van Woensel JBM, de Jong MD, Idro R, Boele van Hensbroek M, van der Hoek L. 2019. Novel orthobunyavirus identified in the cerebrospinal fluid of a Ugandan child with severe encephalopathy. Clin Infect Dis 68:139–142. doi: 10.1093/cid/ciy486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma J, Lv XL, Zhang X, Han SZ, Wang ZD, Li L, Sun HT, Ma LX, Cheng ZL, Shao JW, Chen C, Zhao YH, Sui L, Liu LN, Qian J, Wang W, Liu Q. 2021. Identification of a new orthonairovirus associated with human febrile illness in China. Nat Med 27:434–439. doi: 10.1038/s41591-020-01228-y. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Li Y, He X, Ma J, Hong W, Hu F, Zhao L, Li Q, Zhang J, Zhang C, Zhang F. 2019. Gemykibivirus genome in lower respiratory tract of elderly woman with unexplained acute respiratory distress syndrome. Clin Infect Dis 69:861–864. doi: 10.1093/cid/ciz072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hannigan GD, Duhaime MB, Ruffin MT, Koumpouras CC, Schloss PD. 2018. Diagnostic potential and interactive dynamics of the colorectal cancer virome. mBio 9:e02248-18. doi: 10.1128/mBio.02248-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, Kambal A, Monaco CL, Zhao G, Fleshner P, Stappenbeck TS, McGovern DP, Keshavarzian A, Mutlu EA, Sauk J, Gevers D, Xavier RJ, Wang D, Parkes M, Virgin HW. 2015. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160:447–460. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Legoff J, Resche-Rigon M, Bouquet J, Robin M, Naccache SN, Mercier-Delarue S, Federman S, Samayoa E, Rousseau C, Piron P, Kapel N, Simon F, Socié G, Chiu CY. 2017. The eukaryotic gut virome in hematopoietic stem cell transplantation: new clues in enteric graft-versus-host disease. Nat Med 23:1080–1085. doi: 10.1038/nm.4380. [DOI] [PubMed] [Google Scholar]
- 16.Lang S, Demir M, Martin A, Jiang L, Zhang X, Duan Y, Gao B, Wisplinghoff H, Kasper P, Roderburg C, Tacke F, Steffen HM, Goeser T, Abraldes JG, Tu XM, Loomba R, Stärkel P, Pride D, Fouts DE, Schnabl B. 2020. Intestinal virome signature associated with severity of nonalcoholic fatty liver disease. Gastroenterology 159:1839–1852. doi: 10.1053/j.gastro.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao G, Vatanen T, Droit L, Park A, Kostic AD, Poon TW, Vlamakis H, Siljander H, Härkönen T, Hämäläinen AM, Peet A, Tillmann V, Ilonen J, Wang D, Knip M, Xavier RJ, Virgin HW. 2017. Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children. Proc Natl Acad Sci USA 114:E6166–E6175. doi: 10.1073/pnas.1706359114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Fu X, Ma J, Zhang J, Hu Y, Dong W, Wan Z, Li Q, Kuang YQ, Lan K, Jin X, Wang JH, Zhang C. 2019. Altered respiratory virome and serum cytokine profile associated with recurrent respiratory tract infections in children. Nat Commun 10:2288. doi: 10.1038/s41467-019-10294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson MR, Sample HA, Zorn KC, Arevalo S, Yu G, Neuhaus J, Federman S, Stryke D, Briggs B, Langelier C, Berger A, Douglas V, Josephson SA, Chow FC, Fulton BD, DeRisi JL, Gelfand JM, Naccache SN, Bender J, Dien Bard J, Murkey J, Carlson M, Vespa PM, Vijayan T, Allyn PR, Campeau S, Humphries RM, Klausner JD, Ganzon CD, Memar F, Ocampo NA, Zimmermann LL, Cohen SH, Polage CR, DeBiasi RL, Haller B, Dallas R, Maron G, Hayden R, Messacar K, Dominguez SR, Miller S, Chiu CY. 2019. Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N Engl J Med 380:2327–2340. doi: 10.1056/NEJMoa1803396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zanella MC, Cordey S, Laubscher F, Docquier M, Vieille G, Van Delden C, Braunersreuther V, Ta MK, Lobrinus JA, Masouridi-Levrat S, Chalandon Y, Kaiser L, Vu DL. 2021. Unmasking viral sequences by metagenomic next-generation sequencing in adult human blood samples during steroid-refractory/dependent graft-versus-host disease. Microbiome 9:28. doi: 10.1186/s40168-020-00953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moustafa A, Xie C, Kirkness E, Biggs W, Wong E, Turpaz Y, Bloom K, Delwart E, Nelson KE, Venter JC, Telenti A. 2017. The blood DNA virome in 8,000 humans. PLoS Pathog 13:e1006292. doi: 10.1371/journal.ppat.1006292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD. 2013. Rapid evolution of the human gut virome. Proc Natl Acad Sci USA 110:12450–12455. doi: 10.1073/pnas.1300833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nayfach S, Páez-Espino D, Call L, Low SJ, Sberro H, Ivanova NN, Proal AD, Fischbach MA, Bhatt AS, Hugenholtz P, Kyrpides NC. 2021. Metagenomic compendium of 189,680 DNA viruses from the human gut microbiome. Nat Microbiol 6:960–970. doi: 10.1038/s41564-021-00928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Handley SA. 2016. The virome: a missing component of biological interaction networks in health and disease. Genome Med 8:32. doi: 10.1186/s13073-016-0287-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi S, Sohn KH, Jung JW, Kang MG, Yang MS, Kim S, Choi JH, Cho SH, Kang HR, Yi H. 2021. Lung virome: new potential biomarkers for asthma severity and exacerbation. J Allergy Clin Immunol 148:1007–1015. doi: 10.1016/j.jaci.2021.03.017. [DOI] [PubMed] [Google Scholar]
- 26.Dodi G, Attanasi M, Di Filippo P, Di Pillo S, Chiarelli F. 2021. Virome in the lungs: the role of anelloviruses in childhood respiratory diseases. Microorganisms 9:1357. doi: 10.3390/microorganisms9071357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh J, Byrd AL, Park M, Kong HH, Segre JA, NISC Comparative Sequencing Program . 2016. Temporal stability of the human skin microbiome. Cell 165:854–866. doi: 10.1016/j.cell.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L, Deng X, Linsuwanon P, Bangsberg D, Bwana MB, Hunt P, Martin JN, Deeks SG, Delwart E. 2013. AIDS alters the commensal plasma virome. J Virol 87:10912–10915. doi: 10.1128/JVI.01839-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li SK, Leung RK, Guo HX, Wei JF, Wang JH, Kwong KT, Lee SS, Zhang C, Tsui SK. 2012. Detection and identification of plasma bacterial and viral elements in HIV/AIDS patients in comparison to healthy adults. Clin Microbiol Infect 18:1126–1133. doi: 10.1111/j.1469-0691.2011.03690.x. [DOI] [PubMed] [Google Scholar]
- 30.Liu K, Li Y, Xu R, Zhang Y, Zheng C, Wan Z, Li H, Yang Z, Jin X, Hao P, Zhao J, Zhang C. 2021. HIV-1 infection alters the viral composition of plasma in men who have sex with men. mSphere 6:e00081-21. doi: 10.1128/mSphere.00081-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaczorowska J, van der Hoek L. 2020. Human anelloviruses: diverse, omnipresent and commensal members of the virome. FEMS Microbiol Rev 44:305–313. doi: 10.1093/femsre/fuaa007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webb B, Rakibuzzaman A, Ramamoorthy S. 2020. Torque teno viruses in health and disease. Virus Res 285:198013. doi: 10.1016/j.virusres.2020.198013. [DOI] [PubMed] [Google Scholar]
- 33.Arze CA, Springer S, Dudas G, Patel S, Bhattacharyya A, Swaminathan H, Brugnara C, Delagrave S, Ong T, Kahvejian A, Echelard Y, Weinstein EG, Hajjar RJ, Andersen KG, Yozwiak NL. 2021. Global genome analysis reveals a vast and dynamic anellovirus landscape within the human virome. Cell Host Microbe 29:1305–1315.e1306. doi: 10.1016/j.chom.2021.07.001. [DOI] [PubMed] [Google Scholar]
- 34.Zanella MC, Cordey S, Kaiser L. 2020. Beyond cytomegalovirus and Epstein-Barr virus: a review of viruses composing the blood virome of solid organ transplant and hematopoietic stem cell transplant recipients. Clin Microbiol Rev 33:e00027-20. doi: 10.1128/CMR.00027-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kilonzo SB, Gunda DW, Majinge DC, Jaka H, Manyiri PM, Kalokola F, Mtui G, Shao ER, Bakshi FA, Stephano A. 2021. Seroprevalence of hepatitis B virus infection, anti-HCV antibodies and HIV and knowledge among people who use drugs attending methadone therapy clinic in Tanzania; a cross-sectional study. BMC Infect Dis 21:699. doi: 10.1186/s12879-021-06393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Degenhardt L, Peacock A, Colledge S, Leung J, Grebely J, Vickerman P, Stone J, Cunningham EB, Trickey A, Dumchev K, Lynskey M, Griffiths P, Mattick RP, Hickman M, Larney S. 2017. Global prevalence of injecting drug use and sociodemographic characteristics and prevalence of HIV, HBV, and HCV in people who inject drugs: a multistage systematic review. Lancet Glob Health 5:e1192–e1207. doi: 10.1016/S2214-109X(17)30375-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elkbuli A, Polcz V, Dowd B, McKenney M, Prado G. 2019. HIV prevention intervention for substance users: a review of the literature. Subst Abuse Treat Prev Policy 14:1. doi: 10.1186/s13011-018-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.French RK, Holmes EC. 2020. An ecosystems perspective on virus evolution and emergence. Trends Microbiol 28:165–175. doi: 10.1016/j.tim.2019.10.010. [DOI] [PubMed] [Google Scholar]
- 39.Hsu CL, Duan Y, Fouts DE, Schnabl B. 2021. Intestinal virome and therapeutic potential of bacteriophages in liver disease. J Hepatol 75:1465–1475. doi: 10.1016/j.jhep.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koonin EV, Dolja VV, Krupovic M. 2021. The healthy human virome: from virus-host symbiosis to disease. Curr Opin Virol 47:86–94. doi: 10.1016/j.coviro.2021.02.002. [DOI] [PubMed] [Google Scholar]
- 41.Liang G, Bushman FD. 2021. The human virome: assembly, composition and host interactions. Nat Rev Microbiol 19:514–527. doi: 10.1038/s41579-021-00536-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu Y, Wan Z, Wang JH, Yang X, Zhang C. 2022. Review of human pegivirus: prevalence, transmission, pathogenesis, and clinical implication. Virulence 13:324–341. doi: 10.1080/21505594.2022.2029328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miao Z, Gao L, Song Y, Yang M, Zhang M, Lou J, Zhao Y, Wang X, Feng Y, Dong X, Xia X. 2017. Prevalence and clinical impact of human pegivirus-1 infection in HIV-1-infected individuals in Yunnan, China. Viruses 9:28. doi: 10.3390/v9020028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abbas AA, Young JC, Clarke EL, Diamond JM, Imai I, Haas AR, Cantu E, Lederer DJ, Meyer K, Milewski RK, Olthoff KM, Shaked A, Christie JD, Bushman FD, Collman RG. 2019. Bidirectional transfer of Anelloviridae lineages between graft and host during lung transplantation. Am J Transplant 19:1086–1097. doi: 10.1111/ajt.15116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taherkhani R, Farshadpour F. 2017. Global elimination of hepatitis C virus infection: progresses and the remaining challenges. World J Hepatol 9:1239–1252. doi: 10.4254/wjh.v9.i33.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El-Bassel N, Wechsberg WM, Shaw SA. 2012. Dual HIV risk and vulnerabilities among women who use or inject drugs: no single prevention strategy is the answer. Curr Opin HIV AIDS 7:326–331. doi: 10.1097/COH.0b013e3283536ab2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grebely J, Tyndall MW. 2011. Management of HCV and HIV infections among people who inject drugs. Curr Opin HIV AIDS 6:501–507. doi: 10.1097/COH.0b013e32834bcb36. [DOI] [PubMed] [Google Scholar]
- 48.Nelson PK, Mathers BM, Cowie B, Hagan H, Des Jarlais D, Horyniak D, Degenhardt L. 2011. Global epidemiology of hepatitis B and hepatitis C in people who inject drugs: results of systematic reviews. Lancet 378:571–583. doi: 10.1016/S0140-6736(11)61097-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen X, Zhou YH, Ye M, Wang Y, Duo L, Pang W, Zhang C, Zheng YT. 2018. Burmese injecting drug users in Yunnan play a pivotal role in the cross-border transmission of HIV-1 in the China-Myanmar border region. Virulence 9:1195–1204. doi: 10.1080/21505594.2018.1496777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou YH, Liang YB, Pang W, Qin WH, Yao ZH, Chen X, Zhang C, Zheng YT. 2014. Diverse forms of HIV-1 among Burmese long-distance truck drivers imply their contribution to HIV-1 cross-border transmission. BMC Infect Dis 14:463. doi: 10.1186/1471-2334-14-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lolomadze EA, Rebrikov DV. 2020. Constant companion: clinical and developmental aspects of torque teno virus infections. Arch Virol 165:2749–2757. doi: 10.1007/s00705-020-04841-x. [DOI] [PubMed] [Google Scholar]
- 52.Thijssen M, Tacke F, Beller L, Deboutte W, Yinda KC, Nevens F, Laleman W, Van Ranst M, Pourkarim MR. 2020. Clinical relevance of plasma virome dynamics in liver transplant recipients. EBioMedicine 60:103009. doi: 10.1016/j.ebiom.2020.103009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jaksch P, Kundi M, Görzer I, Muraközy G, Lambers C, Benazzo A, Hoetzenecker K, Klepetko W, Puchhammer-Stöckl E. 2018. Torque teno virus as a novel biomarker targeting the efficacy of immunosuppression after lung transplantation. J Infect Dis 218:1922–1928. doi: 10.1093/infdis/jiy452. [DOI] [PubMed] [Google Scholar]
- 54.Focosi D, Maggi F. 2019. Torque teno virus monitoring in transplantation: the quest for standardization. Am J Transplant 19:1599–1601. doi: 10.1111/ajt.15194. [DOI] [PubMed] [Google Scholar]
- 55.Mrzljak A, Tabain I, Premac H, Bogdanic M, Barbic L, Savic V, Stevanovic V, Jelic A, Mikulic D, Vilibic-Cavlek T. 2019. The role of emerging and neglected viruses in the etiology of hepatitis. Curr Infect Dis Rep 21:51. doi: 10.1007/s11908-019-0709-2. [DOI] [PubMed] [Google Scholar]
- 56.Li Y, Altan E, Pilcher C, Hartogensis W, Hecht FM, Deng X, Delwart E. 2020. Semen virome of men with HIV on or off antiretroviral treatment. AIDS 34:827–832. doi: 10.1097/QAD.0000000000002497. [DOI] [PubMed] [Google Scholar]
- 57.Fahsbender E, Burns JM, Kim S, Kraberger S, Frankfurter G, Eilers AA, Shero MR, Beltran R, Kirkham A, McCorkell R, Berngartt RK, Male MF, Ballard G, Ainley DG, Breitbart M, Varsani A. 2017. Diverse and highly recombinant anelloviruses associated with Weddell seals in Antarctica. Virus Evol 3:vex017. doi: 10.1093/ve/vex017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kandathil AJ, Cox AL, Page K, Mohr D, Razaghi R, Ghanem KG, Tuddenham SA, Hsieh YH, Evans JL, Coller KE, Timp W, Celentano DD, Ray SC, Thomas DL. 2021. Plasma virome and the risk of blood-borne infection in persons with substance use disorder. Nat Commun 12:6909. doi: 10.1038/s41467-021-26980-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Williams CF, Klinzman D, Yamashita TE, Xiang J, Polgreen PM, Rinaldo C, Liu C, Phair J, Margolick JB, Zdunek D, Hess G, Stapleton JT. 2004. Persistent GB virus C infection and survival in HIV-infected men. N Engl J Med 350:981–990. doi: 10.1056/NEJMoa030107. [DOI] [PubMed] [Google Scholar]
- 60.Stockdale AJ, Kreuels B, Henrion MYR, Giorgi E, Kyomuhangi I, de Martel C, Hutin Y, Geretti AM. 2020. The global prevalence of hepatitis D virus infection: systematic review and meta-analysis. J Hepatol 73:523–532. doi: 10.1016/j.jhep.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Turlewicz-Podbielska H, Augustyniak A, Pomorska-Mól M. 2022. Novel porcine circoviruses in view of lessons learned from porcine circovirus type 2-epidemiology and threat to pigs and other species. Viruses 14:261. doi: 10.3390/v14020261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Y, Altan E, Reyes G, Halstead B, Deng X, Delwart E. 2021. Virome of bat guano from nine northern California roosts. J Virol 95:e01713-20. doi: 10.1128/JVI.01713-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kechin A, Boyarskikh U, Kel A, Filipenko M. 2017. cutPrimers: a new tool for accurate cutting of primers from reads of targeted next generation sequencing. J Comput Biol 24:1138–1143. doi: 10.1089/cmb.2017.0096. [DOI] [PubMed] [Google Scholar]
- 64.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li D, Liu CM, Luo R, Sadakane K, Lam TW. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 67.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 68.Geoghegan JL, Di Giallonardo F, Wille M, Ortiz-Baez AS, Costa VA, Ghaly T, Mifsud JCO, Turnbull OMH, Bellwood DR, Williamson JE, Holmes EC. 2021. Virome composition in marine fish revealed by meta-transcriptomics. Virus Evol 7:veab005. doi: 10.1093/ve/veab005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Clipman SJ, Mehta SH, Rodgers MA, Duggal P, Srikrishnan AK, Saravanan S, Balakrishnan P, Vasudevan CK, Ray SC, Kumar MS, Quinn TC, Cloherty GA, Lucas GM, Solomon SS. 2021. Spatiotemporal phylodynamics of hepatitis c among people who inject drugs in India. Hepatology 74:1782–1794. doi: 10.1002/hep.31912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.01447-22-s0001.pdf, PDF file, 1.2 MB (1.2MB, pdf)
Data Availability Statement
All the raw sequencing data were deposited in the CNSA (https://db.cngb.org/cnsa/) of CNGBdb (project number CNP0002340) under the accession numbers CNS0468254-CNS0468363. Anellovirus and pegivirus sequences generated in this study can be found in the same project under the accession numbers CNS0483741-CNS0483743.