The recent enormous expansion of the microbial DNA databases has made it profitable to search for homologues and paralogues (homologues within species) of interesting genes. In combination with genetic, biochemical, and physiological investigations, such analyses may yield new, valuable information with impact on entire research fields. Here I present one such example, the combined description and database analyses of toxin-antitoxin (TA) loci from prokaryotes.
Naturally occurring plasmids are genetically stable. In most cases, stable plasmid inheritance is due to the presence of gene cassettes that actively prevent plasmid loss at cell division. These cassettes can be divided into three classes: (i) centromere-like systems that actively secure ordered segregation of replicons prior to cell division (31, 39, 40, 97), (ii) site-specific recombination systems that actively resolve tandem plasmid multimers into monomers (81, 85), and (iii) cassettes that mediate killing of newborn, plasmid-free cells resulting from failure of the first two systems to secure plasmid maintenance. This latter, paradoxical type of cell differentiation has been termed postsegregational killing (PSK) (24). Two types of PSK mechanisms have been described in detail at the molecular level. In both cases, the killing of plasmid-free progeny relies on stable toxins whose action or expression is counteracted by metabolically unstable regulators. The instability of the regulators results in activation of the toxins in cells that have lost the toxin-encoding plasmid. In one type of PSK mechanism, the regulators are unstable antisense RNAs that inhibit the translation of stable, toxin-encoding mRNAs (i.e., the hok mRNAs). The instability of the antisense RNAs leads to activation of translation of the toxin-encoding mRNAs specifically in plasmid-free cells, thereby leading to their elimination. The complex posttranscriptional regulation of the hok genes has been reviewed previously (26) and will not be discussed further here.
The other type of PSK mechanism relies on stable toxins whose action is prevented by cognate protein antitoxins (reviewed previously in references 35, 38, and 42). Again, the indigenous instability of the antitoxins (also called antidotes by some researchers) leads to activation of the toxins in plasmid-free cells. The PSK phenotype results in increased plasmid maintenance, since plasmid-free progeny have a much lower chance of survival than the plasmid-bearing cells. Accordingly, the plasmid-encoded TA loci have also been called plasmid addiction modules and proteic plasmid stabilization systems, terms that should be used exclusively for the plasmid-encoded loci. Here I present an overview combined with a database analysis of prokaryotic TA loci, with emphasis on recent findings. The ubiquity of the TA loci in prokaryotic chromosomes indicates that they have function(s) unrelated to plasmid maintenance. Two such potential alternative functions are discussed here. The general phenomenon of programmed cell death in bacteria has been reviewed in detail elsewhere (34, 35, 98).
PLASMID-ENCODED TA LOCI
General properties.
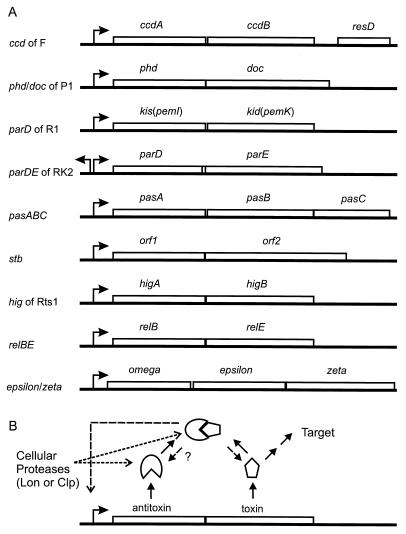
The genetic organization of the known plasmid-encoded TA loci are shown in Fig. 1A, and Table 1 gives an overview of their components. In general, the TA loci are organized into operons in which the first cistron encodes the antitoxin and the second cistron encodes the toxin. One exception to this rule is the hig locus of Rts1 in which the upstream cistron codes for the toxin (90). A second peculiarity of the hig locus is that the toxin is smaller than the antitoxin, whereas the reverse is the case for all other systems known (Table 1). Even though the genes in general do not exhibit sequence similarity, the genetic structures and functions of the components of the TA loci are quite similar, thus favoring the suggestion that they arose from a common ancestral gene. This conjecture is supported by the finding that the antitoxins of ccd of F and pem/parD of R100/R1 exhibit weak sequence similarity (74).
FIG. 1.
(A) Genetic organization of plasmid-encoded TA loci. In all cases but one (the hig locus), the antitoxins are encoded by the upstream gene of the TA operons. Arrows pointing right indicate promoters upstream of the genes. The arrow pointing left indicates a divergent promoter that promotes transcription into the parABC genes of RK2. The resD gene downstream of the ccd genes in F encodes a site-specific resolvase that resolves F multimers into monomers (43, 66). The derivation of gene designations follows: ccd, coupled cell division; phd, prevention of host death; doc, death on curing; kis, killing suppression; kid, killing determinant; pem, plasmid emergency maintenance; pas and stb, plasmid stability; hig, host inhibition of growth; rel, relaxed control of stable RNA synthesis. The pas locus is from the T. ferrooxidans plasmid pTF-CF2; stb is from the S. flexneri plasmid pMYSH6000; ω-ɛ-ζ is from pSM19025 of S. pyogenes; relBE homologues are present on plasmid P307 of E. coli, plasmid pJK2 of A. europaeus, plasmid R485 of M. morganii, and plasmid pRJF2 of B. fibrisolvens. Genes were not drawn to scale. (B) General genetic and functional setup of the TA loci. The antitoxins neutralize the toxins by forming tight complexes with them. The TA complexes bind to operators in the promoter regions and repress transcription (shown by broken arrow pointing to the promoter region). Cellular proteases (Lon or Clp) degrade the antitoxins, thereby leading to activation of the toxins in plasmid-free cells and perhaps during other, as yet unknown, conditions. The question mark indicates that it is not yet known if the antitoxins are degraded when complexed with the toxins or if the toxins and antitoxins dissociate before the antitoxins are degraded.
TABLE 1.
Properties of plasmid-encoded TA loci
| Locusa | Organism | Toxin (no. of aa) | Target | Antitoxin (no. of aa) | Protease | PSK | Reference(s) |
|---|---|---|---|---|---|---|---|
| ccd of F | E. coli | CcdB (101) | DNA gyrase | CcdA (72) | Lon | Yes | 57, 58, 67, 95, 96 |
| pem/parD of R100/R1 | E. coli | PemK/Kid (110) | DnaB? | PemI/Kis (84) | Lon | Yes | 10, 77, 91, 92 |
| phd-doc of P1 | E. coli | Doc (126) | Unknown | Phd (73) | ClpXP | Yes | 46, 47 |
| parDE of RK2 | BHRb | ParE (103) | Unknown | ParD (83) | Unknown | Yes | 72 |
| relBE/pasABC | T. ferrooxidans | PasB (90) | Unknown | PasA (74) | Lon | Yes | 82, 83, 84 |
| stb of pMYSH6000 | S. flexneri | Orf2 (133) | Unknown | Orf1 (75) | Unknown | NDc | 69 |
| hig of Rts1 | BHR | HigB (92) | Unknown | HigA (104) | Unknown | Yes | 90 |
| ɛ/ζ of pSM19035 | BHR | ζ (287) | Unknown | ɛ (90) | Unknown | Yes | 13 |
| relBE of P307 | E. coli | RelE (95) | Translation | RelB (83) | Lon | Yes | 30 |
| relBE/stbDE of R485 | M. morganii | RelE/StbE (93) | Unknown | RelB/StbD (83) | Unknown | Yes | 32 |
| relBE of pJK21 | A. europaeus | RelE (93) | Unknown | RelB (87) | Unknown | ND | This work |
| relBE of pRJF2 | B. fibrisolvens | RelE (93) | Unknown | RelB (83) | Unknown | ND | This work |
| relBE of pB171 | E. coli | RelE (95) | Unknown | RelB (83) | Unknown | ND | This work |
| relBE of p11184 | P. shigelloides | RelE (94) | Unknown | RelB (80) | Unknown | ND | This work |
Plasmid R485 is from M. morganii, pMYSH6000 is from Shigella flexneri, pJK21 from Acetobacter europaeus, pasABC is from the T. ferrooxidans plasmid pTF-CF2, ω-ɛ-ζ of pSM19035 is from S. pyogenes, and plasmid pRJF2 is from B. fibrisolvens; all other loci are from E. coli plasmids or broad-host-range plasmids (RK2/RP4, Rts1 and pSM19035). Broad-host-range plasmid RK2/RP4 is from gram-negative bacteria, pSM19035 is from gram-positive bacteria.
BHR, broad-host-range plasmid.
ND, not determined.
The toxins are very potent, and their artificial overproduction leads to rapid and massive cell killing, in most cases corresponding to several orders of magnitude in reduction of viable-cell counts. Cloning of a toxin-encoding gene in expression vectors can be difficult, usually due to a fortuitous low transcription rate of the toxin-encoding gene even without inducer present (e.g., isopropyl-β-d-thiogalactopyranoside [IPTG] for LacI-regulated promoters and arabinose for AraC-regulated promoters). Several solutions to this problem exist. For instance, my lab has developed expression vectors with large reductions in leakiness (29, 30, 68). Another solution relies on the fact that the replacement of the AUG start codon of the toxin gene with GUG reduces the level of gene expression 5- to 10-fold (28). Such mutations are easily introduced into a toxin gene of interest by recombinant PCR (29). A third solution relies on the use of an antitoxin-producing strain as the recipient in the cloning procedure.
The protein antitoxins counteract their cognate toxins by forming tight complexes with them. This has been shown directly in the cases of CcdA/CcdB of F (3, 87, 96), Kis/Kid of R1 (76), Phd/Doc of P1 (23, 51), and ParD/ParE of RK2 (41). The antitoxins, which are usually found in greater concentrations than those of the toxins, are degraded by cellular proteases Lon and Clp (Table 1), whereas the toxins are generally stable. The instability of the antitoxins is the basis for activation of the toxins in plasmid-free segregants (38). Thus, newborn, plasmid-free cells inherit a pool of TA complexes plus a pool of free antitoxin. By inference, the cellular proteases recognize the antitoxins both when the antitoxins are free in solution and when they are complexed with the cognate toxin. Alternatively, the observed activation of toxin activity is caused by dissociation of the toxin and antitoxins at a rate sufficiently high as to allow for the observed killing of plasmid-free cells.
The antitoxins autoregulate transcription of the TA operons via binding to operator sites upstream of or overlapping with the operon promoters (Fig. 1B). In many cases, the toxins act as corepressors of transcription, indicating that a TA complex is bound to the operator sites. Binding of such TA complexes to the promoter regions has been shown in several cases (17, 50, 51, 71, 75, 78, 87, 93) and inferred in others (29, 30, 83, 88). With respect to transcriptional regulation, ccd of F (Fig. 1A) poses a special case, since the CcdA antitoxin has no repressor activity by itself, as transcriptional regulation of the ccd promoter requires both CcdA and CcdB (17, 78, 87).
In a physiological study, my group compared the efficiency of the TA systems with respect to PSK using isogenic host-vector systems (38). We found that ccd of F and parD of R1 stabilized R1 plasmids only marginally (5- to 10-fold), whereas parDE of RK2 was considerably more efficient (1,000-fold stabilization). Similarly, phd-doc stabilized P1 sevenfold and relBE from P307 (relBEP307) stabilizes P307 only fivefold (30, 46). Thus, in general, it seems as if TA loci stabilize plasmids considerably less efficiently than true centromere-like systems, which yield 100- to 10,000-fold stabilization (9).
The instability of the antitoxins is due to degradation by the cellular proteases Lon and Clp (Table 1). Thus, CcdA, PemI, RelB of P307, and PasA of pTF-CF2 are degraded by Lon, whereas Phd of P1 is degraded by ClpXP (Table 1). In those cases investigated, degradation is relatively slow, as indicated by the long half-lives of the antitoxins in vivo (30 to 60 min). Thus, in steady-state cell growth, the antitoxins are slowly turned over. It is reasonable to assume that a higher turnover rate of the antitoxins would lead to more-efficient plasmid stabilization. Thus, in the context of plasmid stabilization, it does not seem appropriate with antitoxins whose half-lives are too long. TA systems have been identified on many low-copy-number plasmids replicating in gram-negative bacteria. Many features of the TA loci appear similar, and the following is a description of the well-characterized systems.
The ccd locus of F.
Originally, ccd was described as a system that couples cell division to plasmid replication by inhibiting division of cells with fewer than two plasmid copies (56, 57, 67). Later physiological analyses showed that ccd mediates plasmid maintenance by PSK (33, 36). Using a novel plasmid replication arrest system, we confirmed the latter conclusion (37). Lon degrades CcdA in vivo, and in vitro Lon degrades CcdA in an ATP-dependent fashion (95, 96). However, perhaps counterintuitively, CcdB protects CcdA from degradation by Lon in vitro. If this is also the case in vivo, then the activation of CcdB is not a simple consequence of Lon degrading CcdA in the CcdAB complex. One possibility is that the CcdAB complex dissociates at a rate that allows activation of CcdB in plasmid-free cells. Alternatively, factors (yet to be defined) present in vivo may modulate the interaction between CcdA and CcdB.
Selection of mutations that rendered host cells resistant to the toxic activity of CcdB showed that CcdB inhibits Escherichia coli DNA gyrase (6, 58). In a number of elegant analyses, Martine Couturier's lab investigated the mechanism of action of CcdB, its interaction with DNA gyrase subunit A and with CcdA, and the tertiary structure of CcdB (3, 7, 49, 78). The CcdB protein traps DNA gyrase in an inactive complex with DNA (7). Thus, the observed inhibition of cell division by CcdB is probably due, at least in part, to the trapped DNA gyrase (i.e., induction of the SOS response). CcdB-mediated poisoning of DNA gyrase in vitro was reversed by the addition of excess CcdA antitoxin (3). The observation that the CcdB-mediated gyrase poisoning is reversible suggests that cell killing is an extreme case of CcdB action observed when CcdB is overproduced. Thus, it is possible that CcdB during other, more-physiological conditions, inhibits DNA replication without concomitant killing of the host cell.
The pem/parD loci of plasmids R100/R1.
The parD and pem (for plasmid emergency maintenance) loci of R1 and R100, which are identical, code for the toxins Kid (for killing determinant)/PemK and the antitoxins Kis (for killing suppressor)/PemI (10, 91). The wild-type parD locus is functionally inactive or poorly effective, yielding a 2- to 10-fold stabilization of mini-R1 plasmids (37, 76). Surprisingly, and not yet explained, mutations in the repA gene that reduced plasmid copy number activated the wild-type parD locus. In this case, the mutant R1 plasmid derivative was stabilized highly efficiently by parD (10, 76). Thus, under certain circumstances, the parD killer locus is very efficient. The PemI/Kis protein is degraded by Lon, which is the basis for activation of PemK/Kid in plasmid-free cells (39, 92).
Ramon Diaz-Orejas' group showed that Kid/PemK inhibits initiation of replication in vitro (76). The target of Kid/PemK is probably the E. coli DnaB helicase, since multicopy plasmids carrying the dnaB gene suppress the lethal action of Kid/PemK in vivo. The same study showed that Kis and Kid form a complex in vitro. The pem/parD operon is autoregulated by the concerted action of the toxin and antitoxin, presumably via binding of the Kis-Kid complex to the promoter region (76, 93).
The phd-doc locus of P1.
Plasmid P1 codes for a TA system, phd-doc, that stabilizes P1 approximately sevenfold (46). Expression of Doc (for death on curing) is lethal in the absence of PhD (for prevention of host cell death). The Phd antidote is degraded by the ClpXP protease (47). Assuming that Doc is stable, then the instability of Phd is the cause of plasmid stabilization (by PSK). Phd autoregulates the phd-doc operon, and Doc acts as a corepressor of transcription (50). Phd binds cooperatively as a tetramer to inverted repeats in the region between the −10 box and the start site of transcription upstream of phd (22, 50, 51). Gel shift analyses indicated that Doc stimulates the cooperative binding of Phd to the promoter region (50, 51). In solution, Phd exists predominantly in an unfolded conformation, and DNA binding stabilizes the native Phd fold (22). A nontoxic mutant version of Doc interacted physically with Phd in a Phd2D trimeric complex and this interaction is probably the molecular basis for the antitoxic effect of Phd (23, 51). Using fluorescence resonance energy transfer, Gazit and Sauer (23) determined the in vitro half-life of the trimeric complex to be less than 1 s, perhaps indicating the presence of small amounts of free Doc protein in vivo. Furthermore, such a high dissociation rate may be the basis for activation of Doc, since it is perhaps difficult to reconcile how a protease can degrade the antitoxin in a TA complex without also degrading the toxin. This line of thinking is consistent with the finding that CcdB protects CcdA in the CcdAB complex from Lon in vitro (96). Interestingly, functional chromosomal homologues of phd-doc are located upstream of enterobacterial type IC restriction-modification systems within P1-like sequence contexts (94).
The parDE locus of broad-host-range plasmid RK2/RP4.
The parDE operon of RP4/RK2 encodes a potent PSK system that stabilizes mini-R1 and other types of replicons very efficiently (27, 37, 70, 72). The ParD and ParE proteins are dimers in solution, and the ParDE proteins form a tetrameric ParD2ParE2 complex in vitro (41). This tetrameric complex binds to the parDE promoter in vitro and autoregulates transcription of parDE (41). However, ParD protein alone is sufficient for autoregulation of the parDE operon (16, 19), and a ParD dimer binds to the promoter in vitro (71). It is not known if ParE participates in autoregulation of the operon. Database searching revealed ParDE homologues on Yersinia pestis plasmids pCD1 and pYVe227 and on the chromosomes of Vibrio cholerae, Yersinia enterocolitica, and Mycobacterium tuberculosis. This gene family will not be analyzed further here.
The pas (for plasmid stability) locus of pTF-CF2.
The pas locus of plasmid pTF-CF2 from Thiobacillus ferrooxidans constitutes a special TA locus since it codes for three genes, pasABC, organized in an operon (Fig. 1A) (82). The first two cistrons encode a TA couple, PasAB, whereas pasC apparently codes for a protein factor that modulates the interaction between PasA and PasB. Thus, it seems that the presence of PasC enhances the antitoxic effect of PasA towards PasB. The molecular mechanism behind this phenomenon is not yet known but could be due to the formation of a triple PasABC complex. The PasA antitoxin represses the pas promoter, and PasB acts as a corepressor of transcription (83). As shown in a number of other cases, PasA antitoxin is degraded by Lon (84). Interestingly, BLAST analyses revealed that pasAB belongs to the relBE family (see below). However, no obvious pasC homologues have been identified.
The ω-ɛ-ζ operon of the broad-host-range plasmid pSM19035 from gram-positive bacteria.
Plasmid pSM19035 is an inc18 broad-host-range replicon originally isolated from Streptococcus pyogenes, and unlike most other plasmids from gram-positive bacteria, it replicates via a θ-like mechanism (11). Despite its low copy number, pSM19035 is structurally and segregationally stable. Ceglowski et al. (13) showed that the presence of a region encoding genes ω-ɛ-ζ is required for plasmid stability. More-recent analyses showed that ζ encodes a cytotoxin, while ɛ encodes an antitoxin that combines in vivo with ζ (P. Ceglowski, personal communication). However, the TA locus of pSM19035 is unusual in that it also contains gene ω, which encodes an autorepressor of the ω-ɛ-ζ operon. Thus, the proteins encoded by genes ɛ and ζ are not involved in transcriptional regulation. Furthermore, the toxin encoded by ζ (287 amino acids [aa]) is much larger than the toxins of the other TA loci described here. Thus, the evolutionary origin of the ɛ-ζ couple is not clear.
The stb locus of pMYSH6000.
The stb locus of the Shigella flexneri virulence plasmid pMYSH6000 stabilizes plasmids in E. coli (69). stb encodes two small juxtaposed open reading frames designated STBORF1 (75 codons) and STBORF2 (133 codons). The mechanism of plasmid stabilization by stb is not yet known, but its genetic organization suggests that it could be a PSK system. Curiously enough, TRAORF1 and TRAORF2 of plasmid F are highly similar (98.7 and 98.5% identity) to STBORF1 and STBORF2, but the F genes apparently do not mediate plasmid stabilization (69). Using BLAST, additional homologues of stb were identified on a plasmid from Salmonella dublin and on the chromosomes of Haemophilus influenzae (20), Dichelobacter nodosus, Agrobacterium tumefaciens, and the photosynthetic bacteria Synechococcus and Synechocystis.
The relBE locus of P307.
We showed recently that the enteropathogenic plasmid P307 of E. coli codes for a TA locus that is homologous to relBE of E. coli K-12 (30). Here, further relBE loci were identified on plasmids from E. coli (pB171), Plesiomonas shigelloides (belongs to the family Vibrionaceae), Acetobacter europaeus (pJK21) and Butyrivibrio fibrisolvens (pRJF2) (see Fig. 2 and 3). As mentioned above, the pasAB genes from T. ferrooxidans also belong to the relBE family. The plasmid-encoded relBE loci are discussed below.
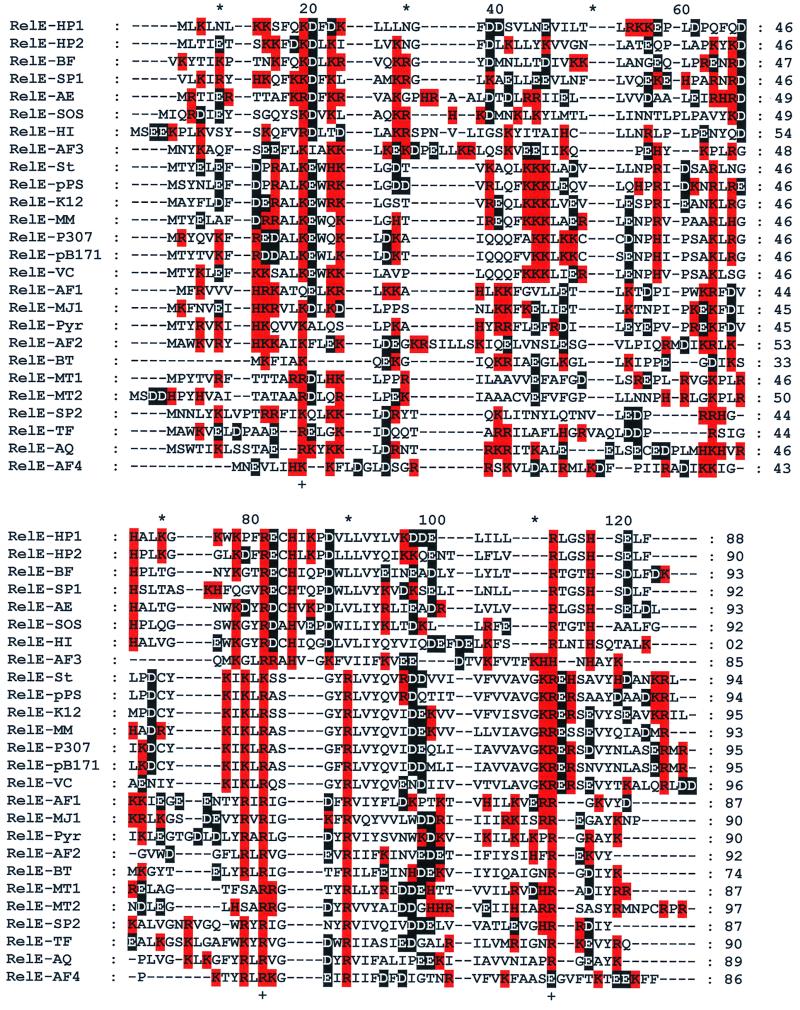
FIG. 2.
Multiple-sequence alignment of 27 RelE proteins from gram-negative and gram-positive bacteria and from archaea. Only RelE proteins with an upstream RelB partner were included in the alignment shown. The characteristics of the corresponding RelB partner proteins are listed in Table 3. Positively charged amino acids are shown in red, and negatively charged amino acids are shown in black. Note the conserved arginine at +82, and the positively charged amino acid at +112 (lysine or arginine). The primary alignment were accomplished by using the Wisconsin GCG package version 8.1.0(a). The multiple-sequence alignment file (msf) was transferred to ClustalX, and the final alignment was edited by eye, using the program Genedoc. The different species and plasmids from which the RelE homologues were derived are indicated by the following letters and numbers after the RelE- suffix: HP1 and HP2, H. pylori homologues 1 and 2, respectively; BF, B. fibrisolvens plasmid pRJF2; SP1, S. pneumoniae homologue 1; AE, A. europaeus plasmid pJK21; SOS, E. coli K-12 homologue 2; HI, H. influenzae; AF3, A. fulgidus homologue 3; St, S. enterica serovar Typhi; pPS, P. shigelloides plasmid; K12, E. coli K-12 homologue 1; MM, M. morganii plasmid R485; P307, E. coli plasmid P307; pB171, E. coli plasmid pB171; VC, V. cholerae; AF1, A. fulgidus homologue 1; MJ1, M. jannaschii homologue 1; Pyr, P. horikoshii OT3; AF2, A. fulgidus homologue 2; BT, B. thuringiensis; MT1 and MT2, M. tuberculosis homologues 1 and 2, respectively; TF, T. ferrooxidans plasmid TF-CF2, AQ, A. aeolicus; AF4, A. fulgidus homologue 4. For simplicity, the irregular RelE homologue of Synechocystis (120 amino acids) was omitted from the alignment. After completion of the database searches, RelE homologues in the unfinished genomes of Salmonella enterica serovars Typhimurium and Paratyphi and Klebsiella pneumoniae were identified. Gaps introduced to maximize alignment are indicated by the dashes.
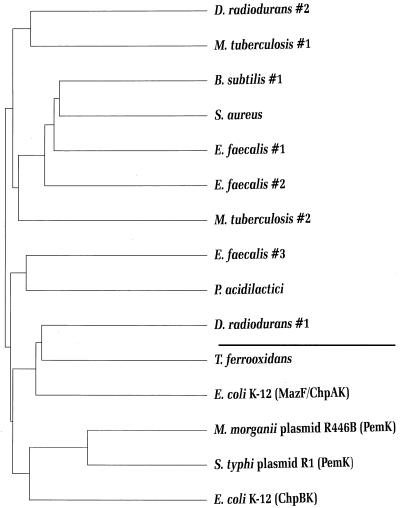
FIG. 3.

Chladogram (unrooted evolutionary tree) of RelE homologues in prokaryotes. The tree was calculated by PILEUP in the Wisconsin GCG package version 8.1.0(a). The aligned sequences shown in Fig. 2 were used as input. The lengths of horizontal lines indicate relative evolutionary distances, whereas the lengths of the vertical bars are arbitrary. The gram-negative bacterial species are shown in blue, the gram-positive bacterial species are shown in red, and the archaeal species are shown in green. For clarity, the deep-branching organism Aquifex aeolicus was grouped with the gram-negative bacteria.
CHROMOSOME-ENCODED TA LOCI
The relBE loci constitute a large gene family in prokaryotes.
Unexpectedly, we found recently that the two first cistrons of the relBEF operon of E. coli K-12 encode a TA locus (29). Furthermore, the E. coli plasmid P307 encodes a locus that is homologous with relBE of E. coli K-12 and which stabilizes mini-P307 replicons (30). The properties of these two relBE loci are strikingly similar: the relE genes encode cytotoxins whose lethal effect is counteracted by relB-encoded antitoxins; the antitoxins are degraded by Lon; the antitoxins repress transcription of the operons, presumably via binding to the cognate promoter regions, and the toxins act as corepressors, such that the promoters are very efficiently repressed during steady-state cell growth. The relBE promoters are very strong, and the degree of repression, presumably caused by binding of the RelBE complexes to the promoter regions, is in both cases on the order of 3 magnitudes. Because of these striking similarities, we also tested if the chromosomal relBE locus could mediate plasmid stabilization. This was indeed the case, and the fold stabilization was similar to that mediated by relBE of P307 (i.e., fourfold) (29, 30). Thus, although encoded by the chromosome, the relBE locus of E. coli appears to stabilize plasmids by PSK.
The third gene of the E. coli relBEF operon (also called hokD) codes for a cytotoxin that belongs to the Hok family of proteins (25, 26, 73). The function of relF/hokD is not known, but the relF/hokD cistron is not translated during steady-state cell growth and does not contribute to plasmid stabilization (29).
In a screening for plasmid stabilization cassettes, Finbarr Hayes identified a second plasmid-encoded relBE-homologous locus on the Morganella morganii plasmid R485 (denoted stbDE) (32). It is reasonable to suggest that the plasmid stabilization phenotype mediated by stbDE/relBER485 is a consequence of PSK. Curiously, the N-terminal two-thirds of the StbD/RelB protein of R485 exhibits similarity with E. coli DnaT protein. The significance of this is not known but may give a hint in the search for host-encoded interaction partners.
Using BLAST (2), I searched the entire DNA databases for homologues of the cytotoxins RelE, PemK/Kid, CcdB, Doc, and ParE (see below). Only toxin genes with a closely linked upstream antitoxin gene were included in the compilations described below. A small number of toxin homologues without such a closely linked putative antitoxin-encoding gene were identified. In principle, a partner antitoxin could be encoded anywhere on a chromosome, but the low degree of similarity between the antitoxins makes their identification by homology searches difficult. To simplify the analyses, toxin homologues without a linked antitoxin partner gene were discarded. To cover the whole spectrum of possible positive scores, every new toxin included in a gene family was used in a new round of searches in the databases.
At the time of writing, 27 TA loci belonging to the relBE gene family were identified. An alignment of the RelE toxin sequences is shown in Fig. 2, and their properties are listed in Table 2. Sixteen of the 27 relBE loci are from gram-negative bacteria, 5 are from gram-positive bacteria, and most surprisingly, 6 are from the Archaea domain. The RelE proteins are small and basic, with pIs of approximately 10, except for RelE from H. influenzae (Table 2). In contrast, all partner antitoxins are acidic except for the two homologues from Helicobacter pylori (Table 3), consistent with the proposal that the toxins and antitoxins interact physically. The RelB homologues are quite diverse, and a global alignment of all sequences did not yield meaningful information. The alignment of the subgroups of RelB homologues will be presented elsewhere.
TABLE 2.
relE homologues
| Bacterial species | relE genea | Characteristic of the RelE protein
|
||
|---|---|---|---|---|
| No. of aa | Molecular mass (kDa)b | pIb | ||
| Gram-negative bacteria | ||||
| E. coli K-12 | relEK-12 | 95 | 11.2 | 9.7 |
| E. coli K-12 | relESOSc | 92 | 10.8 | 9.5 |
| E. coli plasmid P307 | relEP307 | 95 | 11.2 | 9.9 |
| E. coli plasmid pB171 | relEpB171 | 95 | 11.1 | 10.7 |
| Salmonella enteria serovar Typhi | relEStyphi1 | 94 | 10.9 | 10.4 |
| P. shigelloides plasmid p11184 | relEPs | 94 | 11.2 | 10.7 |
| Haemophilus influenzae | relEHi | 102 | 11.9 | 6.7 |
| Vibrio cholerae | relEVc | 96 | 11.2 | 9.9 |
| Helicobacter pylori | relEHp1 | 88 | 10.4 | 7.9 |
| H. pylori | relEHp2 | 90 | 10.4 | 10.2 |
| Butyrivibrio fibrisolvens | relEBf | 93 | 11.0 | 9.7 |
| Acetobacter europaeus | relEAe | 93 | 10.9 | 10.0 |
| Morganella morganii | relEMm | 93 | 11.0 | 10.9 |
| Thiobacillus ferrooxidans | relETf | 90 | 10.3 | 10.8 |
| Aquifex aeolicusd | relEAq | 89 | 10.4 | 10.8 |
| Synechosystisd | relESy | 120 | 13.7 | 7.9 |
| Gram-positive bacteria | ||||
| Bacillus thuringiensis | relEBt | 74 | 8.6 | 9.7 |
| Mycobacterium tuberculosis | relEMt1 | 87 | 10.2 | 11.0 |
| M. tuberculosis | relEMt2 | 97 | 11.1 | 9.5 |
| Streptococcus pneumoniae | relESp1 | 92 | 10.9 | 10.3 |
| S. pneumoniae | relESp2 | 87 | 10.4 | 11.1 |
| Archaea | ||||
| Methanococcus jannaschii | relEMj1 | 90 | 11.0 | 10.2 |
| Archaeoglobus fulgidus | relEAf1 | 87 | 10.6 | 10.3 |
| A. fulgidus | relEAf2 | 92 | 11.0 | 9.9 |
| A. fulgidus | relEAf3 | 85 | 10.0 | 10.0 |
| A. fulgidus | relEAf4 | 86 | 10.2 | 9.9 |
| P. horikoshii OT3 | relEPyr | 90 | 10.9 | 10.9 |
Subscripts refer to the species in which the relE genes were identified (see the species shown in the leftmost column).
Molecular masses and isoelectric points were calculated by using the Wisconsin GCG package.
The relBESOS locus (dinJ - yafQ) of E. coli K-12 contains a LexA binding site in the promoter region, thus belonging to the SOS regulon (48).
For clarity, A. aeolicus and Synechosystis were grouped with the gram-negative bacteria.
TABLE 3.
relB homologues
| Bacterial species | relB genea | Characteristic of the RelB protein
|
||
|---|---|---|---|---|
| No. of aa | Molecular mass (kDa)b | pIb | ||
| Gram-negative bacteria | ||||
| E. coli K-12 | relBK-12 | 79 | 9.1 | 4.8 |
| E. coli K-12 | relBSOSc | 86 | 9.4 | 5.2 |
| E. coli plasmid P307 | relBP307 | 83 | 9.2 | 4.4 |
| E. coli plasmid pB171 | relBpB171 | 83 | 9.2 | 4.1 |
| S. enterica serovar Typhi | relBSt1 | 80 | 9.0 | 5.2 |
| P. shigelloides plasmid p11184 | relBPs | 83 | 9.2 | 4.1 |
| H. influenzae | relBHi | 98 | 11.0 | 4.7 |
| V. cholerae | relBVc | 82 | 8.9 | 4.4 |
| H. pylori | relBHp1 | 95 | 11.4 | 9.8 |
| H. pylori | relBHp2 | 95 | 11.4 | 10.6 |
| B. fibrisolvens | relBBf | 83 | 9.6 | 6.6 |
| A. europaeus | relBAe | 87 | 9.5 | 4.6 |
| M. morganii | relBMm | 83 | 9.2 | 4.3 |
| T. ferrooxidans | relBTf | 74 | 8.5 | 4.7 |
| A. aeolicus | relBAq | 80 | 9.3 | 5.4 |
| Synechosystis | relBSy | 86 | 9.9 | 4.7 |
| Gram-positive bacteria | ||||
| B. thuringiensis | relBBt | 85 | 10.1 | 4.5 |
| M. tuberculosis | relBMt1 | 93 | 10.2 | 4.6 |
| M. tuberculosis | relBMt2 | 89 | 9.8 | 5.1 |
| S. pneumoniae | relBSp1 | 87 | 10.0 | 4.4 |
| S. pneumoniae | relBSp2 | 80 | 9.2 | 4.2 |
| Archaea | ||||
| M. jannaschii | relBMj1 | 82 | 9.6 | 4.5 |
| A. fulgidus | relBAf1 | 65 | 7.8 | 4.8 |
| A. fulgidus | relBAf2 | 62 | 7.4 | 4.3 |
| A. fulgidus | relBAf3 | 72 | 8.5 | 4.5 |
| A. fulgidus | relBAf4 | 57 | 6.7 | 4.1 |
| P. horikoshii OT3 | relBPyr | 67 | 8.0 | 3.8 |
Subscripts refer to the species in which the relB genes were identified (see the species shown in the leftmost column).
Molecular masses and isoelectric points were calculated by using the Wisconsin GCG package.
The relBESOS system of E. coli K-12 contains a LexA binding site in the promoter region and may belong to the SOS regulon (48).
In contrast, even though the RelE proteins are quite diverse, they show significant similarities as revealed by the alignment in Fig. 2: +20 is positive (Arg or Lys), +82 is an invariable Arg (except for Lys in one case), and +112 is also a positively charged amino acid. Hence, the evolutionary kinship of the RelE proteins is clear. Furthermore, the asserted relationship is strongly supported by the fact that all genes encoding the RelE proteins (Fig. 2) have closely linked relB genes (Table 3). However, the sequence alignment shows that the RelE proteins are surprisingly diverse, given that their overall characteristics are conserved, as indicated by invariant sizes and pIs (Table 3) and similar genetic contexts. The interesting question of whether the RelE homologues have common cellular targets awaits further experiments, but preliminary analyses indicate that this is true at least in some cases.
The genetic organization of the relBE loci from these very diverse organisms are strikingly similar and concur with the general structure shown in Fig. 1A: putative promoter elements are present upstream of the relB homologs, the relB and relE reading frames are in all cases closely linked, and in many cases the stop codon of relB overlaps with the first one or two codons of relE. Such close linkage of the relB and relE genes may indicate translational coupling. This conjecture is consistent with the finding that the relE genes from E. coli K-12 and P307 are expressed at considerably lower levels than those of the cognate relB partner genes (29, 30).
Figure 3 shows a phylogram deduced from the 27 RelE protein sequences. This calculation revealed four major RelE groups: (i) RelE proteins from enteric bacteria or closely related bacteria, (ii) RelEs from other gram-negative bacteria also including one member from Streptococcus pneumoniae and one member from Archaeoglobus fulgidus, (iii) RelEs from gram-positive bacteria, including two homologues from gram-negative bacteria, and finally (iv) one group from Archaea which includes a homologue from Bacillus thuringiensis. Two homologues did not fit into any of the four groups (RelEs from Synechocystis and A. fulgidus homologue 4).
This grouping is largely consistent with the evolutionary division of prokaryotic organisms based on 16S rRNA sequences. Furthermore, the evolutionary relationship within the subgroups agrees well with the evolutionary grouping of the corresponding organisms (Fig. 3). However, the significant number of exceptions to regular grouping could reflect lateral gene transfer. Lateral interkingdom gene transfer is consistent with the finding that the deep-branching members of the domain Bacteria such as Aquifex aeolicus and Thermotoga maritima contain many genes like archaeal genes (16 and 24%, respectively) (61). Alternatively, irregular grouping of the homologues could reflect statistical fluctuations caused by the small size of the RelE proteins, rather than a true evolutionary relationship.
One striking finding is that several chromosomes contain two or more relBE homologues (paralogues). The complex relationship between multiple paralogues and orthologues as described by Tatusov et al. (89) is not considered here. However, E. coli K-12 contains two relBE paralogues called relBEK12 and relBESOS here (Table 2). Genes relBESOS were previously identified as dinJ and yafQ, respectively (8, 48). The promoter upstream of dinJ was mapped and contains a known LexA binding site at a proper location (48). Thus, transcription of relBESOS may be induced during the SOS response and be part of the stress response elicited by DNA damage. E. coli contains a third relB homologue called yafN (8). yafN has no apparent downstream relE homologue. The genome of Salmonella typhi contains two relE homologues, but only one of them has a closely linked upstream relB gene (relESt1 in Fig. 2). The gram-positive organism S. pneumoniae contains two complete relBE homologues, and the relatively small chromosome of the archaeon Archaeoglobus fulgidus contains four relBE homologues. The reason for this apparent redundancy is not known. Of the 27 relBE homologues described here, seven are located on plasmids from gram-negative bacteria. However, the majority of the relBE genes are located on prokaryotic chromosomes, arguing for functions other than plasmid stabilization by PSK (discussed below). Two of the chromosomal relBE loci are located on mobile genetic elements: relBE of V. cholerae is located within a mega-integron (15, 21, 54), and relBE of B. thuringiensis is located on transposon Tn504 (4). The localization of relBE genes on mobile genetic elements such as plasmids and transposable elements may increase their horizontal spread and may also accelerate their rate of evolution.
Prokaryotic cells respond rapidly and efficiently to stress situations such as amino acid or carbon source starvation by altering gene expression such that the harsh environmental conditions can be efficiently coped with. Carbon source starvation of E. coli cells leads to altered expression rates of a large number of genes (12, 62), and amino acid starvation leads to arrest of synthesis of stable RNA (rRNA and tRNA). This so-called stringent response is elicited by the increased rate of synthesis of the alarmone (p)ppGpp. In this case, (p)ppGpp synthesis is due to activated RelA protein (12). RelA, also called (p)ppGpp synthetase I, is activated by binding of uncharged tRNA to vacant ribosomal A-sites, and RelA-deficient cells fail to accumulate (p)ppGpp during amino acid starvation and other carbon source limitations. Consequently, such cells do not shut down stable RNA synthesis after amino acid starvation (12) and are said to have a relaxed phenotype with respect to stable RNA synthesis. During the induction of the stringent response, many proteins exhibit a reduced rate of synthesis. However, the reverse is also true: a large number of proteins exhibit an increased rate of synthesis (12). Among the latter are enzymes encoded by the amino acid synthetic operons. This makes sense, since the cells try to ameliorate the lack of amino acids. Lon and perhaps other cellular proteases are also activated during the stringent response (12, 14). This also makes sense, since endogenous building blocks must be generated for de novo protein synthesis.
The E. coli relB gene was discovered in a screen for mutations that abolish the stringent response without affecting the relA gene. Three different selection procedures were devised, and they all resulted in point mutations in the relB gene. The mutations yielded a phenotype called the delayed relaxed response (18, 44, 45, 59, 60). Delayed relaxed cells resume synthesis of stable RNA approximately 10 min after the onset of amino acid starvation. This is in contrast to relaxed mutants (defective in relA) in which stable RNA synthesis continues after amino acid starvation without any lag. Cells exhibiting the delayed relaxed phenotype contain point mutations in relB that partially inactivate RelB protein (5). The relB mutants were found to recover very slowly after amino acid starvation in that virtually no growth took place for about 3 h after release from amino acid starvation. This inhibition of cell growth was attributed to the accumulation of a factor, most probably a protein that inhibited translation (45). The molecular basis of the delayed relaxed response is not yet understood. However, from indirect experiments, Bech et al. (5) suggested that relB did not encode the translational inhibitor itself but rather a negative regulator of the inhibitor. Recently, we proposed that this protein synthesis inhibitor might be RelE, since that would provide a reasonable explanation for the delayed relaxed response exhibited by relB mutants: after amino acid starvation, the reduced activity of antagonist RelB would lead to activation of RelE. If RelE were to inhibit translation, this postulated inhibition, in turn, would lead to a reduced drain on tRNA and thereby reduce the number of vacant ribosomal A-sites bound to uncharged tRNA. Consequently, such cells would shut down (p)ppGpp synthetase I and resumption of stable RNA synthesis would follow and thereby elicit the delayed relaxed response (29). At present, we do not exclude other explanations for the delayed relaxed response. However, the clear effect on the stringent response suggests that the function of the E. coli relBE locus is to protect the cells from detrimental effects of stress rather than being suicide modules.
The chp loci constitute a novel gene family in the domain Bacteria.
DNA sequence analyses revealed that E. coli K-12 contains two loci (chpA and chpB) that are homologous to pem/parD of R100/R1 (52, 55). The chpA locus encodes two polypeptides, ChpAI and ChpAK, that are structurally and functionally similar to PemI/Kis and PemK/Kid of R100/R1, respectively. The chpA locus, which is located downstream of and adjacent to the relA gene, has also been called mazEF (1, 55). The chpB locus at 100 min on the E. coli K-12 chromosome encodes ChpBI and ChpBK (I for inhibitor; K for cell killing) that are also structurally and functionally related to the proteins encoded by the pem/parD locus. In both cases, it has been shown that the toxin homologues are toxic and that the upstream partners are antitoxic (1, 52). Deletion of the two chp loci, either alone or in combination, had no apparent effect on cell growth or viability (52, 53). Curiously, high concentrations of the ChpAI (MazE) and ChpBI antitoxins counteract PemK/Kid-mediated cell killing, indicating that there is cross talk between plasmid- and chromosome-encoded components of the TA loci (79, 80).
Evidence was obtained that MazE (ChpAI) and MazF (ChpAK) interact physically and that the antitoxin MazE is degraded by ClpAP in vivo (1). These researchers further suggested that the toxic effect of MazF is induced during the stringent response. This conjecture was based on the observation that the mazEF promoter was inhibited during (p)ppGpp overproduction via induction of a truncated RelA protein (RelA*), which synthesizes (p)ppGpp without the requirement for vacant ribosomal A-sites, combined with the observation that induction of (p)ppGpp synthesis at 42°C (but not at lower temperatures) resulted in mazEF-dependent cell death. The researchers reasoned that the induction of cell killing was consistent with the finding that the mazEF promoter was inhibited by (p)ppGpp, since that would lead to arrest of synthesis of mazEF mRNA, depletion of MazE antitoxin, and consequently, activation of MazF toxin. In many respects, overproduction of (p)ppGpp via overproduction of RelA* mimics the stringent response and has the advantage that the cells can be investigated without the need for carbon source or amino acid limitation (12). However, such artificial induction of (p)ppGpp synthesis is inevitably prone to result in erroneous conclusions if results from other ways of inducing stringent starvation are not contemplated as well (12). Using the same strain (MC4100) and growth conditions (media, temperature), my lab has not been able to reproduce the results reported by Aizenman et al. (1), and we do not observe cell killing during stringent starvation at 37°C (induced by the addition of serine hydroxamate or valine to growing cells). A complicating factor is that strain MC4100, which Aizenman et al. (1) used, carries the relA1 allele (an IS2 element between codons 85 and 86 of relA). How could the postulated programmed cell death or altruistic suicide during carbon source limitation be an advantage for a bacterial population? As argued by Nyström (65), such behavior would be detrimental to cells encountering stasis at low cell densities. Thus, to accommodate a reasonably realistic altruistic suicide theory, one has to postulate a regulatory network signaling cell density to the TA systems (e.g., by quorum sensing).
By database searching (BLAST), additional pem/chp/mazEF loci were identified (Fig. 4). Only toxins with a closely linked putative antitoxin-encoding gene were included in the alignment shown in Fig. 4. A new pem (parD)-homologous locus was identified on the enterobacterial plasmid R466B. Furthermore, chromosomal chp (mazEF) loci were identified in the gram-negative organism T. ferrooxidans and in the gram-positive organisms Bacillus subtilis (one complete TA locus and a ChpK toxin gene without an obvious partner), Enterococcus faecalis (three complete TA loci), M. tuberculosis (two loci), Staphylococcus aureus (one locus), Deinococcus radiodurans (two loci), and Pediococcus acidilactici (one locus). No ChpK homologues were identified in Archaea. Figure 4 shows an alignment of the putative 15 homologous ChpK proteins with upstream partners. As seen, the proteins are quite diverse yet clearly related. Their N termini contain conserved proline (+36 and +48) and arginine (+47) residues.
FIG. 4.
Multiple-sequence alignment of 15 ChpK proteins from gram-negative and gram-positive bacteria. Only ChpK proteins with identified putative antitoxin partners were included in the alignment shown. Amino acids with >80% conservation are shown by the black background. Note the fully conserved proline at position 37 and the fully conserved RP motif at positions +47 and +48. The different species and plasmids from which the ChpK (or PemK) homologues were derived are indicated by the following letters and numbers after the ChpK- (or PemK-) suffix: Dr2, D. radiodurans homologue 2; Mt1, M. tuberculosis homologue 1; Bsu1, B. subtilis homologue 1; Sa, S. aureus; Ef and Ef2, E. faecalis homologues 1 and 2, respectively; Mt2, M. tuberculosis homologue 2; Ef3, E. faecalis homologue 3; Pa, P. acidilactici; Dr1, D. radiodurans homologue 1; Tf, T. ferrooxidans; K12, E. coli K-12; R466B, M. morganii plasmid R446B. Gaps introduced to maximize alignment are indicated by the dashes.
A chladogram of the ChpK homologues is shown in Fig. 5. The distribution of the homologues into two major groups is consistent with the division of bacteria into gram-negative and gram-positive bacteria (Fig. 5). One of the two ChpK homologues of D. radiodurans appears to be more closely related to the gram-negative group than to the gram-positive group. As in the case of the RelE homologues, the presence of such a large number of chromosomal chp-homologous loci points to functions other than plasmid stabilization by PSK. The cellular target(s) of the ChpK proteins is not yet known. However, the database analyses presented here show that TA systems are surprisingly abundant in prokaryotic organisms.
FIG. 5.
Chladogram of the ChpK homologues from bacteria. The tree was calculated by PILEUP in the Wisconsin GCG package version 8.1.0(a). The aligned sequences shown in Fig. 4 were used as input. The organisms above the thick line in the figure are gram-positive bacteria, while those below the line are gram-negative bacteria.
Functions of the chromosome-encoded TA loci: cell killing functions or stress response elements?
The specific molecular targets of the toxins are known in only two cases. CcdB inhibits DNA gyrase, and PemK/Kid inhibits DNA replication, presumably via interaction with DnaB helicase (7, 77). The RelE proteins of E. coli K-12 and P307 presumably inhibit translation (29, 30), but their specific target(s) within the translation machinery is not yet known. It is reasonable to assume that at least some of the other RelE and ChpK proteins have similar, if not identical, cellular targets. Thus, in some or even many of the cases of the two largest TA gene families of toxins, RelE and ChpK, activation of the toxins may mediate inhibition of translation and DNA replication, respectively. What is common to these two processes? Clearly, both translation and DNA replication are highly expensive for the cell in terms of energy consumption (in the form of ATP). Thus, I note here the possibility that the chromosomal TA loci may be part of the global cellular response to environmental stress such as amino acid and/or glucose limitation, rather than being cell-killing modules. According to this hypothesis, the main function of the TA loci is to regulate the synthesis of macromolecules (i.e., proteins and DNA) at rates compatible with the external supply of nutrients. Thus, activation of RelE would reduce the rate of translation, while activation of ChpK would reduce the rate of replication, thus saving energy and/or building blocks vital for maintenance functions (64). Conceptually, this is very similar to the way (p)ppGpp works: starvation for amino acids or carbon source limitation provokes (p)ppGpp-dependent inhibition of transcription (inhibition of stable RNA promoters and reduced elongation rates) and thereby the shutdown of rRNA synthesis. In turn, this has an indirect effect on the rate of translation. It has also been suggested that (p)ppGpp might have a direct effect on translation in vivo (reference 86 and unpublished observations). However, it has not been possible to inhibit translation in vitro by the addition of (p)ppGpp. Thus, a primary function of (p)ppGpp is probably to inhibit transcription immediately after a nutritional downshift. After a shift to amino acid or carbon source starvation, translation continues at a high rate for a short period (5 to 10 min) before the new poststimulus steady-state level is reached (86). Clearly, the cell needs to respond rapidly and to regulate coordinately the rates of macromolecular synthesis during nutritional shift scenarios such that one parameter of macromolecular synthesis does not run wild. A simple and testable hypothesis then is that the toxins of the TA loci are induced after amino acid and glucose starvation and coordinately reduce DNA replication and translation, while accumulation of (p)ppGpp reduces transcription. Hence, as an alternative to the altruistic suicide theory proposed by Aizenman and coworkers (1), I suggest here the possibility that the TA loci are beneficial to cell survival by being part of the global stress response. My lab is currently testing this compelling hypothesis. If this hypothesis is true, why have the TA loci been mainly described as plasmid stabilization cassettes? Plasmids have been used extensively as model systems, and many of their components have been studied in detail. One explanation is that the PSK phenotype elicited by the plasmid-encoded TA loci is a fortuitous consequence of their genetic setup. However, it is also obvious that plasmids do evolve mechanisms that lead to their genetic stabilization, such as the efficient centromere systems found in F, P1, and R1. It is possible that the TA loci in parallel with their effect on cell metabolism provide an advantage to plasmids, either as stabilization cassettes or as stress response modules that increase host survival or both. Furthermore, TA loci may exploit plasmids (and transposable genetic elements) as vehicles for their rapid transfer and evolution.
The widespread occurrence of the relE and pem/parD/chp loci stands in contrast to the much narrower occurrence of ccd, which is present only on F and closely related plasmids. Although somewhat speculative, the tripartite physiological stress response hypothesis described above (i.e., inhibition of replication, transcription, and translation during severe stress) may reflect the contour of how evolution works: Since the target of CcdB is DNA gyrase and that of PemK/Kid/ChpK is DNA helicase, both toxins inhibit DNA replication. Thus, if the stress response theory is valid, then ccd and chp affect the same cellular parameter and are thus complementing each other. In other words, cells equipped with a chp locus might not obtain a high advantage by acquiring a ccd-like system and visa versa. The prevalence of chp loci as compared to ccd may indicate that chp is for some reason more advantageous and therefore has had a higher degree of evolutionary success. Another question relates to the abundant presence of relBE loci in Archaea, whereas none of the other TA systems have been identified in that domain. This suggests that the target of the RelE toxins is evolutionarily better conserved than those of the other toxins. It will be interesting to learn if RelE homologues from Archaea are active in E. coli and visa versa.
Concluding remarks.
In this minireview, I have presented data showing that the loci known as plasmid addiction modules or proteic plasmid stabilization systems are much more abundant than recognized previously. The presence of the TA loci on prokaryotic chromosomes, often in multiple copies, points to functions other than plasmid stabilization by PSK. Based on some indirect evidence and some logical speculation, I find it reasonable to suggest that the TA loci are beneficial to host cells, perhaps by functioning as stress response elements. If this is true, this idea may change the way TA loci are analyzed in the future.
ACKNOWLEDGMENTS
I thank Kenneth Rudd, Kim Pedersen, and Hugo Grønlund for valuable comments on the manuscript.
This work was supported in part by the Center for Interaction, Structure, Function and Engineering of Macromolecules (CISFEM).
REFERENCES
- 1.Aizenman E, Engelberg-Kulka H, Glaser G. An Escherichia coli chromosomal “addiction module” regulated by 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc Natl Acad Sci USA. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahassi E M, O'Dea M H, Allali N, Messens J, Gellert M, Couturier M. Interactions of CcdB with DNA gyrase. Inactivation of GyrA, poisoning of the gyrase-DNA complex, and the antidote action of CcdA. J Biol Chem. 1999;274:10936–10944. doi: 10.1074/jbc.274.16.10936. [DOI] [PubMed] [Google Scholar]
- 4.Baum J. Tn5401, a new class II transposable element from Bacillus thuringiensis. J Bacteriol. 1994;176:2835–2845. doi: 10.1128/jb.176.10.2835-2845.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bech F W, Jørgensen S T, Diderichsen B, Karlström O H. Sequence of the relB transcription unit from Escherichia coli and identification of the relB gene. EMBO J. 1985;4:1059–1066. doi: 10.1002/j.1460-2075.1985.tb03739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernard P, Couturier M. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J Mol Biol. 1992;226:735–745. doi: 10.1016/0022-2836(92)90629-x. [DOI] [PubMed] [Google Scholar]
- 7.Bernard P, Kezdy K E, Van Melderen L, Steyaert J, Wyns L, Pato M L, Higgins P N, Couturier M. The F plasmid CcdB protein induces efficient ATP-dependent DNA cleavage by gyrase. J Mol Biol. 1993;234:534–541. doi: 10.1006/jmbi.1993.1609. [DOI] [PubMed] [Google Scholar]
- 8.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode R, Mayhew C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 9.Boe L, Gerdes K, Molin S. Effects of genes exerting growth inhibition and plasmid stability on plasmid maintenance. J Bacteriol. 1987;169:4646–4650. doi: 10.1128/jb.169.10.4646-4650.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bravo A, de Torrontegui G, Diaz R. Identification of components of a new stability system of plasmid R1, ParD, that is close to the origin of replication of this plasmid. Mol Gen Genet. 1987;210:101–110. doi: 10.1007/BF00337764. [DOI] [PubMed] [Google Scholar]
- 11.Bruand C, Ehrlich S D, Janniere L. Unidirectional theta replication of the structurally stable Enterococcus faecalis plasmid pAMβ1. EMBO J. 1991;10:2171–2177. doi: 10.1002/j.1460-2075.1991.tb07752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- 13.Ceglowski P, Boitsov A, Chai S, Alonso J C. Analysis of the stabilization system of pSM19035-derived plasmid pBT233 in Bacillus subtilis. Gene. 1993;136:1–12. doi: 10.1016/0378-1119(93)90441-5. [DOI] [PubMed] [Google Scholar]
- 14.Chung C H, Goldberg A L. The product of the lon (capR) gene in Escherichia coli is the ATP-dependent protease, protease La. Proc Natl Acad Sci USA. 1981;78:4931–4935. doi: 10.1073/pnas.78.8.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark C A, Purins L, Kaewrakon P, Manning P A. VCR repetitive sequence elements in the Vibrio cholerae chromosome constitute a mega-integron. Mol Microbiol. 1997;26:1137–1138. doi: 10.1046/j.1365-2958.1997.d01-5533.x. [DOI] [PubMed] [Google Scholar]
- 16.Davis T L, Helinski D R, Roberts R C. Transcription and autoregulation of the stabilizing functions of broad-host-range plasmid RK2 in Escherichia coli, Agrobacterium tumefaciens and Pseudomonas aeruginosa. Mol Microbiol. 1992;6:1981–1994. doi: 10.1111/j.1365-2958.1992.tb01371.x. [DOI] [PubMed] [Google Scholar]
- 17.de Feyter R, Wallace C, Lane D. Autoregulation of the ccd operon in the F plasmid. Mol Gen Genet. 1989;218:481–486. doi: 10.1007/BF00332413. [DOI] [PubMed] [Google Scholar]
- 18.Diderichsen B, Fiil N P, Lavallé R. Genetics of the relB locus in Escherichia coli. J Bacteriol. 1977;131:30–33. doi: 10.1128/jb.131.1.30-33.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eberl L, Givskov M, Schwab H. The divergent promoters mediating transcription of the par locus of plasmid RP4 are subject to autoregulation. Mol Microbiol. 1992;6:1969–1979. doi: 10.1111/j.1365-2958.1992.tb01370.x. [DOI] [PubMed] [Google Scholar]
- 20.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 21.Franzon V L, Barker A, Manning P A. Nucleotide sequence of the mannose-fucose-resistant hemagglutinin of Vibrio cholera O1 and construction of a mutant. Infect Immun. 1993;61:3032–3037. doi: 10.1128/iai.61.7.3032-3037.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gazit E, Sauer R T. Stability and DNA binding of the Phd protein of the phage P1 plasmid addiction system. J Biol Chem. 1999;274:2652–2657. doi: 10.1074/jbc.274.5.2652. [DOI] [PubMed] [Google Scholar]
- 23.Gazit E, Sauer R T. The Doc toxin and Phd antidote proteins of the bacteriophage P1 plasmid addiction system form a heterotrimeric complex. J Biol Chem. 1999;274:16813–16818. doi: 10.1074/jbc.274.24.16813. [DOI] [PubMed] [Google Scholar]
- 24.Gerdes K, Rasmussen P B, Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid free cells. Proc Natl Acad Sci USA. 1986;83:3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerdes K, Bech F W, Jørgensen S T, Løbner-Olesen A, Atlung T, Boe L, Karlström O, Molin S, von Meyenburg K. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 1986;5:2023–2029. doi: 10.1002/j.1460-2075.1986.tb04459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerdes K, Gultyaev A P, Franch T, Pedersen K, Mikkelsen N D. Antisense RNA regulated programmed cell death. Annu Rev Genet. 1997;31:1–31. doi: 10.1146/annurev.genet.31.1.1. [DOI] [PubMed] [Google Scholar]
- 27.Gerlitz M, Hrabak O, Schwab H. Partitioning of broad-host-range plasmid RP4 is a complex system involving site-specific recombination. J Bacteriol. 1990;172:6194–6203. doi: 10.1128/jb.172.11.6194-6203.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- 29.Gotfredsen M, Gerdes K. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol Microbiol. 1998;29:1065–1076. doi: 10.1046/j.1365-2958.1998.00993.x. [DOI] [PubMed] [Google Scholar]
- 30.Grønlund H, Gerdes K. Toxin-antitoxin systems homologous to relBE of Escherichia coli plasmid P307 are ubiquitous in prokaryotes. J Mol Biol. 1999;285:1401–1415. doi: 10.1006/jmbi.1998.2416. [DOI] [PubMed] [Google Scholar]
- 31.Harry E J. Illuminating the force: bacterial mitosis? Trends Microbiol. 1997;5:295–297. doi: 10.1016/S0966-842X(97)01091-3. [DOI] [PubMed] [Google Scholar]
- 32.Hayes F. A family of stability determinants in pathogenic bacteria. J Bacteriol. 1998;180:6415–6418. doi: 10.1128/jb.180.23.6415-6418.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hiraga S, Jaffé A, Ogura T, Mori H, Takahashi H. F plasmid ccd mechanism in Escherichia coli. J Bacteriol. 1986;166:100–104. doi: 10.1128/jb.166.1.100-104.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hochman A. Programmed cell death in prokaryotes. Crit Rev Microbiol. 1997;23:207–214. doi: 10.3109/10408419709115136. [DOI] [PubMed] [Google Scholar]
- 35.Holcik M, Iyer V N. Conditionally lethal genes associated with bacterial plasmids. Microbiology. 1997;143:3403–3416. doi: 10.1099/00221287-143-11-3403. [DOI] [PubMed] [Google Scholar]
- 36.Jaffé A, Ogura T, Hiraga S. Effects of the ccd function of the F plasmid on bacterial growth. J Bacteriol. 1985;163:841–849. doi: 10.1128/jb.163.3.841-849.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen R B, Grohmann E, Schwab H, Diaz R, Gerdes K. Comparison of ccd of F, parDE of RP4, and parD of R1 using a novel conditional replication control system of plasmid R1. Mol Microbiol. 1995;17:211–220. doi: 10.1111/j.1365-2958.1995.mmi_17020211.x. [DOI] [PubMed] [Google Scholar]
- 38.Jensen R B, Gerdes K. Programmed cell death in bacteria: proteic killer gene systems. Mol Microbiol. 1995;17:205–210. doi: 10.1111/j.1365-2958.1995.mmi_17020205.x. [DOI] [PubMed] [Google Scholar]
- 39.Jensen R B, Lurz R, Gerdes K. Mechanism of DNA segregation in prokaryotes: replicon pairing by parC of plasmid R1. Proc Natl Acad Sci USA. 1998;95:8550–8555. doi: 10.1073/pnas.95.15.8550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jensen R B, Gerdes K. Mechanism of DNA segregation in prokaryotes: ParM partitioning protein of plasmid R1 co-localizes with its replicon during the cell cycle. EMBO J. 1999;18:4076–4084. doi: 10.1093/emboj/18.14.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson E P, Ström A R, Helinski D R. Plasmid RK2 toxin protein ParE: purification and interaction with the ParD antitoxin protein. J Bacteriol. 1996;178:1420–1429. doi: 10.1128/jb.178.5.1420-1429.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kobayashi I. Selfishness and death: raison d'etre of restriction, recombination and mitochondria. Trends Genet. 1998;14:368–374. doi: 10.1016/s0168-9525(98)01532-7. [DOI] [PubMed] [Google Scholar]
- 43.Lane D, de Feyter R, Kennedy M, Phua S H, Semon D. D protein of mini-F plasmid acts as a repressor of transcription and as a site-specific resolvase. Nucleic Acids Res. 1986;14:9713–9728. [PMC free article] [PubMed] [Google Scholar]
- 44.Lavallé R. Nouveaux mutants de régulation de la synthèse de l'Arn. Bull Soc Chim Biol. 1965;47:1567–1570. [PubMed] [Google Scholar]
- 45.Lavallé R, Desmarez L, De Hauwer G. Natural messenger translation impairment in an E. coli mutant. In: Kjeldgaard N O, Maaløe O, editors. Control of ribosome synthesis. Copenhagen, Denmark: Munksgaard; 1976. pp. 408–418. [Google Scholar]
- 46.Lehnherr H, Maguin E, Jafri S, Yarmolinsky M B. Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J Mol Biol. 1993;233:414–428. doi: 10.1006/jmbi.1993.1521. [DOI] [PubMed] [Google Scholar]
- 47.Lehnherr H, Yarmolinsky M B. Addiction protein Phd of plasmid prophage P1 is a substrate of the ClpXP serine protease of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:3274–3277. doi: 10.1073/pnas.92.8.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewis L K, Harlow G R, Gregg-Jolly L A, Mount D W. Identification of high affinity binding sites for LexA which define new DNA damage-inducible genes in Escherichia coli. J Mol Biol. 1994;241:507–523. doi: 10.1006/jmbi.1994.1528. [DOI] [PubMed] [Google Scholar]
- 49.Loris R, Dao-Thi M H, Bahassi E M, Van Melderen L, Poortmans F, Liddington R, Couturier M, Wyns L. Crystal structure of CcdB, a topoisomerase poison from E. coli. J Mol Biol. 1999;285:1667–1677. doi: 10.1006/jmbi.1998.2395. [DOI] [PubMed] [Google Scholar]
- 50.Magnuson R, Lehnherr H, Mukhopadhyay G, Yarmolinsky M B. Autoregulation of the plasmid addiction operon of bacteriophage P1. J Biol Chem. 1996;271:18705–18710. doi: 10.1074/jbc.271.31.18705. [DOI] [PubMed] [Google Scholar]
- 51.Magnuson R, Yarmolinsky M B. Corepression of the P1 addiction operon by Phd and Doc. J Bacteriol. 1998;180:6342–6351. doi: 10.1128/jb.180.23.6342-6351.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Masuda Y, Miyakawa K, Nishimura Y, Ohtsubo E. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J Bacteriol. 1993;175:6850–6856. doi: 10.1128/jb.175.21.6850-6856.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Masuda Y, Ohtsubo E. Mapping and disruption of the chpB locus in Escherichia coli. J Bacteriol. 1994;176:5861–5863. doi: 10.1128/jb.176.18.5861-5863.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mazel D, Dychinco B, Webb V A, Davies J. A distinctive class of integron in the Vibrio cholerae genome. Science. 1998;280:605–608. doi: 10.1126/science.280.5363.605. [DOI] [PubMed] [Google Scholar]
- 55.Metzger S, Dror I B, Aizenman E, Schreiber G, Toone M, Friesen J D, Cashel M, Glaser G. The nucleotide sequence and characterization of the relA gene of Escherichia coli. J Biol Chem. 1988;263:15699–15704. [PubMed] [Google Scholar]
- 56.Miki T, Yoshioka K, Horiuchi T. Control of cell division by sex factor F in Escherichia coli. I. The 42.84–43.6 F segment couples cell division of the host bacteria with replication of plasmid DNA. J Mol Biol. 1984;174:605–625. doi: 10.1016/0022-2836(84)90086-x. [DOI] [PubMed] [Google Scholar]
- 57.Miki T, Chang Z T, Horiuchi T. Control of cell division by sex factor F in Escherichia coli. II. Identification of genes for inhibitor protein and trigger protein on the 42.84–43.6 F segment. J Mol Biol. 1984;174:627–646. doi: 10.1016/0022-2836(84)90087-1. [DOI] [PubMed] [Google Scholar]
- 58.Miki T, Park J A, Nagao K, Murayama N, Horiuchi T. Control of segregation of chromosomal DNA by sex factor F in Escherichia coli. Mutants of DNA gyrase subunit A suppress letD (ccdB) product growth inhibition. J Mol Biol. 1992;225:39–52. doi: 10.1016/0022-2836(92)91024-j. [DOI] [PubMed] [Google Scholar]
- 59.Mosteller R D, Kwan S F. Isolation of relaxed-control mutants of Escherichia coli K-12 which are sensitive to glucose starvation. Biochem Biophys Res Commun. 1976;69:325–332. doi: 10.1016/0006-291x(76)90525-8. [DOI] [PubMed] [Google Scholar]
- 60.Mosteller R D. Evidence that glucose starvation-sensitive mutants are altered in the relB locus. J Bacteriol. 1978;133:1034–1037. doi: 10.1128/jb.133.2.1034-1037.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nelson K E, Clayton R A, Gill S R, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Nelson W C, Ketchum K A, McDonald L, Utterback T R, Malek J A, Linher K D, Garrett M M, Stewart A M, Cotton M D, Pratt M S, Phillips C A, Richardson D, Heidelberg J, Sutton G G, Fleischmann R D, Eisen J A, Fraser C M, et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 62.Nyström T. The glucose-starvation stimulon of Escherichia coli: induced and repressed synthesis of enzymes of central metabolic pathways and role of acetyl phosphate in gene expression and starvation survival. Mol Microbiol. 1994;12:833–843. doi: 10.1111/j.1365-2958.1994.tb01069.x. [DOI] [PubMed] [Google Scholar]
- 63.Nyström T. The trials and tribulations of growth arrest. Trends Microbiol. 1995;3:131–136. doi: 10.1016/s0966-842x(00)88901-5. [DOI] [PubMed] [Google Scholar]
- 64.Nyström T, Gustavsson N. Maintenance energy requirement: what is required for stasis survival of Escherichia coli? Biochim Biophys Acta. 1998;1365:225–231. doi: 10.1016/s0005-2728(98)00072-3. [DOI] [PubMed] [Google Scholar]
- 65.Nyström T. To be or not to be: the ultimate decision of the growth-arrested cell. FEMS Microbiol Rev. 1998;21:283–290. [Google Scholar]
- 66.O'Connor M B, Kilbane J J, Malamy M H. Site-specific and illegitimate recombination in the oriV1 region of the F factor. DNA sequences involved in recombination and resolution. J Mol Biol. 1986;189:85–102. doi: 10.1016/0022-2836(86)90383-9. [DOI] [PubMed] [Google Scholar]
- 67.Ogura T, Hiraga S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc Natl Acad Sci USA. 1983;80:4784–4788. doi: 10.1073/pnas.80.15.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pedersen K, Gerdes K. Multiple hok genes on the chromosome of Escherichia coli. Mol Microbiol. 1999;32:1090–1102. doi: 10.1046/j.1365-2958.1999.01431.x. [DOI] [PubMed] [Google Scholar]
- 69.Radnedge L, Davis M A, Youngren B, Austin S J. Plasmid maintenance functions of the large virulence plasmid of Shigella flexneri. J Bacteriol. 1997;179:3670–3675. doi: 10.1128/jb.179.11.3670-3675.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roberts R C, Helinski D R. Definition of a minimal plasmid stabilization system from the broad-host-range plasmid RK2. J Bacteriol. 1992;174:8119–8132. doi: 10.1128/jb.174.24.8119-8132.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roberts R C, Spangler C, Helinski D R. Characteristics and significance of DNA binding activity of plasmid stabilization protein ParD from the broad host-range plasmid RK2. J Biol Chem. 1993;268:27109–27117. [PubMed] [Google Scholar]
- 72.Roberts R C, Ström A R, Helinski D R. The parDE operon of the broad-host-range plasmid RK2 specifies growth inhibition associated with plasmid loss. J Mol Biol. 1994;237:35–51. doi: 10.1006/jmbi.1994.1207. [DOI] [PubMed] [Google Scholar]
- 73.Rudd K E, Humphery-Smith I, Wasinger V C, Bairoch A. Low molecular weight proteins: a challenge for post-genomic research. Electrophoresis. 1998;19:536–544. doi: 10.1002/elps.1150190413. [DOI] [PubMed] [Google Scholar]
- 74.Ruiz-Echevarria M J, de Torrontegui G, Gimenez-Gallego G, Diaz-Orejas R. Structural and functional comparison between the stability systems ParD of plasmid R1 and Ccd of plasmid F. Mol Gen Genet. 1991;225:355–562. doi: 10.1007/BF00261674. [DOI] [PubMed] [Google Scholar]
- 75.Ruiz-Echevarria M J, Berzal-Herranz A, Gerdes K, Diaz-Orejas R. The kis and kid genes of the parD maintenance system of plasmid R1 form an operon that is autoregulated at the level of transcription by the co-ordinated action of the Kis and Kid proteins. Mol Microbiol. 1991;5:2685–2693. doi: 10.1111/j.1365-2958.1991.tb01977.x. [DOI] [PubMed] [Google Scholar]
- 76.Ruiz-Echevarria M J, de la Torre M A, Diaz-Orejas R. A mutation that decreases the efficiency of plasmid R1 replication leads to the activation of parD, a killer stability system of the plasmid. FEMS Microbiol Lett. 1995;130:129–135. doi: 10.1111/j.1574-6968.1995.tb07709.x. [DOI] [PubMed] [Google Scholar]
- 77.Ruiz-Echevarria M J, Gimenez-Gallego G, Sabariegos-Jareno R, Diaz-Orejas R. Kid, a small protein of the parD stability system of plasmid R1, is an inhibitor of DNA replication acting at the initiation of DNA synthesis. J Mol Biol. 1995;247:568–577. doi: 10.1006/jmbi.1995.0163. [DOI] [PubMed] [Google Scholar]
- 78.Salmon M A, Van Melderen L, Bernard P, Couturier M. The antidote and autoregulatory functions of the F plasmid CcdA protein: a genetic and biochemical survey. Mol Gen Genet. 1994;244:530–538. doi: 10.1007/BF00583904. [DOI] [PubMed] [Google Scholar]
- 79.Santos-Sierra S, Giraldo R, Diaz-Orejas R. Functional interactions between homologous conditional killer systems of plasmid and chromosomal origin. FEMS Microbiol Lett. 1997;152:51–56. doi: 10.1111/j.1574-6968.1997.tb10408.x. [DOI] [PubMed] [Google Scholar]
- 80.Santos-Sierra S, Giraldo R, Diaz-Orejas R. Functional interactions between chpB and parD, two homologous conditional killer systems found in the Escherichia coli chromosome and in plasmid R1. FEMS Microbiol Lett. 1998;168:51–58. doi: 10.1111/j.1574-6968.1998.tb13254.x. [DOI] [PubMed] [Google Scholar]
- 81.Sherratt D J, Arciszewska L K, Blakely G, Colloms S, Grant K, Leslie N, McCulloch R. Site-specific recombination and circular chromosome segregation. Philos Trans R Soc Lond Ser B. 1995;347:37–42. doi: 10.1098/rstb.1995.0006. [DOI] [PubMed] [Google Scholar]
- 82.Smith A S, Rawlings D E. The poison-antidote stability system of the broad-host-range Thiobacillus ferrooxidans plasmid pTF-FC2. Mol Microbiol. 1997;26:961–970. doi: 10.1046/j.1365-2958.1997.6332000.x. [DOI] [PubMed] [Google Scholar]
- 83.Smith A S G, Rawlings D E. Autoregulation of the pTF-FC2 proteic poison-antidote plasmid addiction system (pas) is essential for plasmid stabilization. J Bacteriol. 1998;180:5463–5465. doi: 10.1128/jb.180.20.5463-5465.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Smith A S G, Rawlings D E. Efficiency of the pTF-FC2 pas poison-antidote stability system in Escherichia coli is affected by the host strain, and antidote degradation requires the Lon protease. J Bacteriol. 1998;180:5458–5462. doi: 10.1128/jb.180.20.5458-5462.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Summers D. Timing, self-control and a sense of direction are the secrets of multicopy plasmid stability. Mol Microbiol. 1998;29:1137–1145. doi: 10.1046/j.1365-2958.1998.01012.x. [DOI] [PubMed] [Google Scholar]
- 86.Svitil A L, Cashel M, Zyskind J W. Guanosine tetraphosphate inhibits protein synthesis in vivo. A possible protective mechanism for starvation stress in Escherichia coli. J Biol Chem. 1993;268:2307–2311. [PubMed] [Google Scholar]
- 87.Tam J E, Kline B C. The F plasmid ccd autorepressor is a complex of CcdA and CcdB proteins. Mol Gen Genet. 1989;219:26–32. doi: 10.1007/BF00261153. [DOI] [PubMed] [Google Scholar]
- 88.Tam J E, Kline B C. Control of the ccd operon in plasmid F. J Bacteriol. 1989;171:2353–2360. doi: 10.1128/jb.171.5.2353-2360.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tatusov R L, Koonin E V, Lipman D J. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 90.Tian Q B, Ohnishi M, Tabuchi A, Terawaki Y. A new plasmid-encoded proteic killer gene system: cloning, sequencing, and analyzing hig locus of plasmid Rts1. Biochem Biophys Res Commun. 1996;220:280–284. doi: 10.1006/bbrc.1996.0396. [DOI] [PubMed] [Google Scholar]
- 91.Tsuchimoto S, Ohtsubo H, Ohtsubo E. Two genes, pemI and pemK, responsible for stable maintenance of resistance plasmid R100. J Bacteriol. 1988;170:1461–1466. doi: 10.1128/jb.170.4.1461-1466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tsuchimoto S, Nishimura Y, Ohtsubo E. The stable maintenance system pem of plasmid R100: degradation of PemI protein may allow PemK protein to inhibit cell growth. J Bacteriol. 1992;174:4205–4211. doi: 10.1128/jb.174.13.4205-4211.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tsuchimoto S, Ohtsubo E. Autoregulation by cooperative binding of the PemI and PemK proteins to the promoter region of the pem operon. Mol Gen Genet. 1993;237:81–88. doi: 10.1007/BF00282787. [DOI] [PubMed] [Google Scholar]
- 94.Tyndall C, Lehnherr H, Sandmeier U, Kulik E, Bickle T A. The type IC hsd loci of the enterobacteria are flanked by DNA with high homology to the phage P1 genome: implications for the evolution and spread of DNA restriction systems. Mol Microbiol. 1997;23:729–736. doi: 10.1046/j.1365-2958.1997.2531622.x. [DOI] [PubMed] [Google Scholar]
- 95.Van Melderen L, Bernard P, Couturier M. Lon-dependent proteolysis of CcdA is the key control for activation of CcdB in plasmid-free segregant bacteria. Mol Microbiol. 1994;11:1151–1157. doi: 10.1111/j.1365-2958.1994.tb00391.x. [DOI] [PubMed] [Google Scholar]
- 96.Van Melderen L, Thi M H D, Lecchi P, Gottesman S, Couturier M, Maurizi M R. ATP-dependent degradation of CcdA by Lon protease. Effects of secondary structure and heterologous subunit interactions. J Biol Chem. 1996;271:27730–27738. doi: 10.1074/jbc.271.44.27730. [DOI] [PubMed] [Google Scholar]
- 97.Wheeler R T, Shapiro L. Bacterial chromosome segregation: is there a mitotic apparatus? Cell. 1997;88:577–579. doi: 10.1016/s0092-8674(00)81898-x. [DOI] [PubMed] [Google Scholar]
- 98.Yarmolinsky M B. Programmed cell death in bacterial populations. Science. 1995;267:836–837. doi: 10.1126/science.7846528. [DOI] [PubMed] [Google Scholar]