ABSTRACT
Chronic colonization by Pseudomonas aeruginosa is critical in cystic fibrosis (CF) and other chronic lung diseases, contributing to disease progression. Biofilm growth and a propensity to evolve multidrug resistance phenotypes drastically limit the available therapeutic options. In this perspective, there has been growing interest in evaluating combination therapies, especially for drugs that can be administered by nebulization, which allows high drug concentrations to be reached at the site of infections while limiting systemic toxicity. Here, we investigated the potential antibiofilm activity of N-acetylcysteine (NAC) alone and in combination with colistin against a panel of P. aeruginosa strains (most of which are from CF patients) and the transcriptomic response of a P. aeruginosa CF strain to NAC exposure. NAC alone (8,000 mg/L) showed a limited and strain-dependent antibiofilm activity. Nonetheless, a relevant antibiofilm synergism of NAC-colistin combinations (NAC at 8,000 mg/L plus colistin at 2 to 32 mg/L) was observed with all strains. Synergism was also confirmed with the artificial sputum medium model. RNA sequencing of NAC-exposed planktonic cultures revealed that NAC (8,000 mg/L) mainly induced (i) a Zn2+ starvation response (known to induce attenuation of P. aeruginosa virulence), (ii) downregulation of genes of the denitrification apparatus, and (iii) downregulation of flagellar biosynthesis pathway. NAC-mediated inhibition of P. aeruginosa denitrification pathway and flagellum-mediated motility were confirmed experimentally. These findings suggested that NAC-colistin combinations might contribute to the management of biofilm-associated P. aeruginosa lung infections. NAC might also have a role in reducing P. aeruginosa virulence, which could be relevant in the very early stages of lung colonization.
IMPORTANCE Pseudomonas aeruginosa biofilm-related chronic lung colonization contributes to cystic fibrosis (CF) disease progression. Colistin is often a last-resort antibiotic for the treatment of such P. aeruginosa infections, and it has been increasingly used in CF, especially by nebulization. N-acetylcysteine (NAC) is a mucolytic agent with antioxidant activity, commonly administered with antibiotics for the treatment of lower respiratory tract infections. Here, we show that NAC potentiated colistin activity against in vitro biofilms models of P. aeruginosa strains, with both drugs tested at the high concentrations achievable after nebulization. In addition, we report the first transcriptomic data on the P. aeruginosa response to NAC exposure.
KEYWORDS: N-acetylcysteine, Pseudomonas aeruginosa, biofilms, colistin, cystic fibrosis, synergism, transcriptomic response
INTRODUCTION
Pseudomonas aeruginosa is a leading pathogen infecting the airways of patients affected by cystic fibrosis (CF) and other chronic lung diseases (e.g., chronic obstructive pulmonary disease and non-CF bronchiectasis) (1). Once established in the CF airways, P. aeruginosa develops into chronic infections and generally persists indefinitely, contributing to frequent exacerbations, decline of pulmonary function, and higher rates of mortality (1, 2). Chronic infections by P. aeruginosa in CF lungs are associated with adaptive changes of the pathogen, such as conversion to a mucoid phenotype, switching to the biofilm mode of growth, and acquisition of antibiotic resistance (3). Cumulative exposure to antibiotics during treatment causes dissemination of multidrug-resistant (MDR) P. aeruginosa strains, leading to the ineffectiveness of the antibiotic therapy and consequently worse clinical outcomes (3).
Colistin is among the last-resort agents for the treatment of P. aeruginosa infections caused by MDR strains, with the advantage of being also administrable by nebulization, which allows the achieving of high lung concentrations while reducing systemic toxicity (4). In this perspective, inhaled colistin has been increasingly used for the treatment of difficult-to-treat respiratory tract infections, especially those related to biofilm formation (5).
N-acetylcysteine (NAC) is a mucolytic agent commonly administered with antibiotics for the treatment of lower respiratory tract infections, which has been demonstrated to exert also antimicrobial and antibiofilm activity against relevant respiratory pathogens (6–8). Recently, a potent in vitro antibiofilm synergism of NAC-colistin combinations was demonstrated against colistin-susceptible and colistin-resistant Acinetobacter baumannii and Stenotrophomonas maltophilia strains (9, 10).
NAC has been demonstrated to exert several heterogeneous biological activities (whose molecular bases have not always been clearly elucidated) and has recently been under extensive investigation for potential clinical applications beyond the approved therapeutic usage as an antidote in acetaminophen (paracetamol) overdose and as a mucolytic (11). Overall, NAC can act as a direct or indirect antioxidant, due to the ability of the free thiol group to react with reactive oxygen and nitrogen species and by constituting a precursor of intracellular glutathione (11). In addition, NAC can bind transition and heavy metal ions and act as a reducing agent of protein sulfhydryl groups involved in intracellular redox homeostasis (11). Despite several studies that have addressed the biological effects of NAC on planktonic and biofilm bacterial cultures (8), to the best of our knowledge, no data on bacterial transcriptomic response to NAC exposure have been reported so far.
In this study, we investigated the in vitro antibiofilm activities of NAC alone and in combination with colistin (at the high concentrations achievable by the inhalation route of administration) (8, 12) against a panel of P. aeruginosa strains (most of which are from CF patients) representative of different phenotypes (in terms of mucoidy, antimicrobial susceptibility pattern, and O type) and multilocus sequence type (MLST) genotypes. In addition, we provided original data on the transcriptomic response of P. aeruginosa planktonic cultures to NAC exposure.
RESULTS AND DISCUSSION
Activity of NAC alone against preformed biofilm.
The antibiofilm activity of NAC alone was tested with 17 P. aeruginosa strains (Table 1), of which 15 were from CF patients, using the Nunc-TSP lid system.
TABLE 1.
Features of the 17 P. aeruginosa strains included in this study
| Strain | yr of isolation | Phenotype | Origina | STb | O type | Resistance patternc | MIC (mg/L)d |
|
|---|---|---|---|---|---|---|---|---|
| CST | NAC | |||||||
| PAO1 | 1954 | Nonmucoid | Wound | ST549 | O5 | Wild type | 2 | 64,000 |
| Z33 | 2005 | Nonmucoid | CF | ST235 | O11 | CPr, FQr, AGr | 1 | 16,000 |
| Z34 | 2006 | Nonmucoid | CF | ST17 | O1 | CBr, CPr, FQr, AGr | 2 | 64,000 |
| Z35 | 2006 | Nonmucoid | CF | ST235 | O11 | 1 | 16,000 | |
| Z152 | 2013 | Mucoid | CF | ST155 | O6 | CBr, FQr, AGr | 2 | 8,000 |
| Z154 | 2016 | Mucoid | CF | ST412 | O6 | CPr, FQr, AGr | 2 | 16,000 |
| M1 | 2002 | Mucoid | CF | ST155 | O6 | CBr, CPr, FQr, AGr | 2 | 16,000 |
| M4 | 2005 | Mucoid | CF | ST155 | O6 | CBr, CPr, FQr, AGr | 2 | 32,000 |
| M7 | 2005 | Mucoid | CF | ST253 | O10 | AGr | 2 | 64,000 |
| M13 | 2000 | Mucoid | CF | ST274 | O3 | CBr, CPr, AGr | 1 | 32,000 |
| M19 | 2006 | Mucoid | CF | ST3509 | O7 | 1 | 64,000 | |
| M25 | 2002 | Mucoid | CF | ST235 | O11 | 2 | 16,000 | |
| M32 | 2006 | Mucoid | CF | ST235 | O11 | 2 | 16,000 | |
| M42 | 2007 | Mucoid | CF | ST2437 | O6 | CBr, CPr, FQr, AGr | 2 | 32,000 |
| FC237 | 2007 | Nonmucoid | CF | ST365 | O3 | CBr, FQr, AGr, CSTr | 512 | 64,000 |
| FC238 | 2007 | Nonmucoid | CF | ST910 | O6 | CBr, CSTr | 8 | 64,000 |
| FZ99 | 2018 | Nonmucoid | RTIICU | ST111 | O12 | CBr, CPr, FQr, AGr, CSTr | 4 | 64,000 |
CF, cystic fibrosis; RTIICU, respiratory tract infection in intensive care unit.
According to the MLST Pasteur scheme.
CBr, resistance to carbapenems (imipenem and meropenem); CPr, resistance to cephems (ceftazidime and cefepime); FQr, resistance to fluoroquinolones (ciprofloxacin); AGr, resistance to aminoglycosides (amikacin and gentamicin); CSTr, resistance to colistin.
CST, colistin; NAC, N-acetylcysteine.
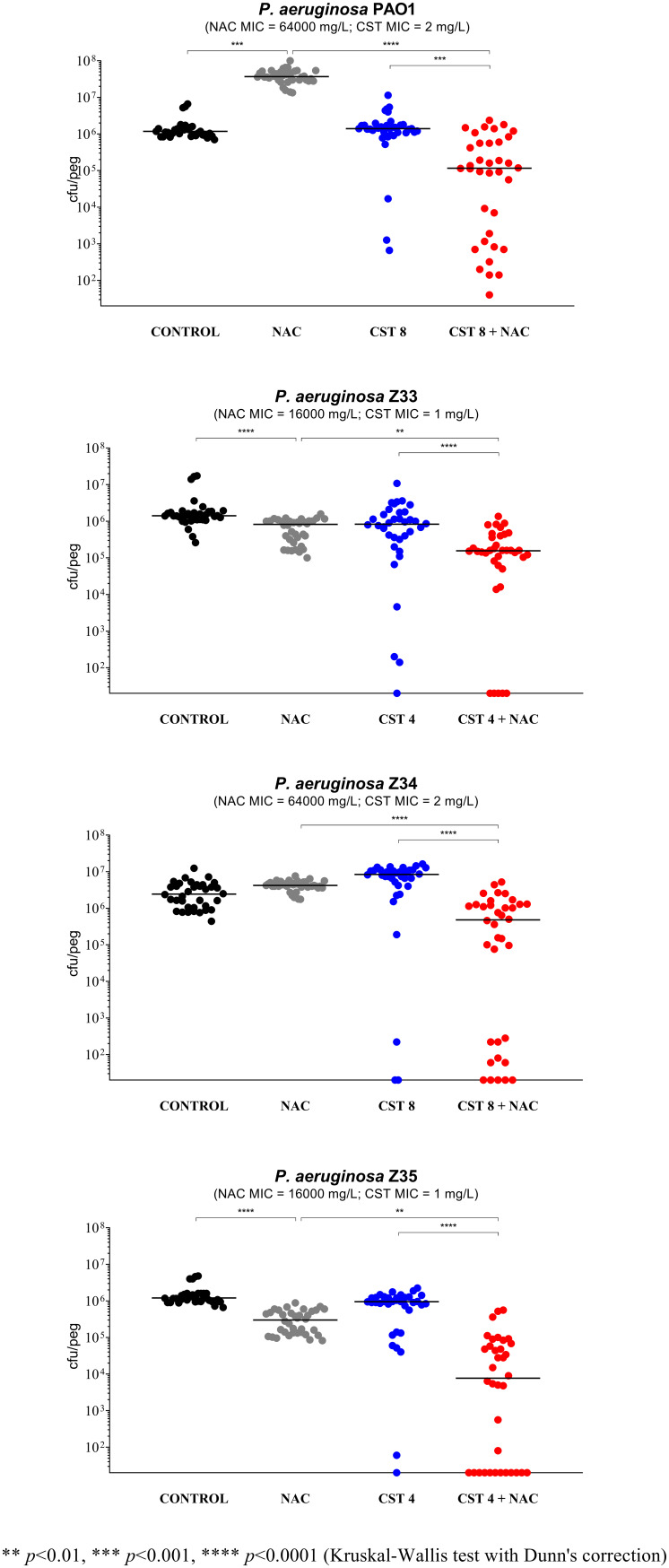
NAC at 8,000 mg/L (i.e., a high concentration achievable after inhalation) showed limited and strain-dependent activity (Fig. 1 to 4). In particular, major effects were observed with P. aeruginosa Z154 (i.e., decrease of >1 log CFU/peg compared to the control) (Fig. 1) and P. aeruginosa PAO1 (i.e., increase of >1 log CFU/peg compared to the control) (Fig. 2). With an additional 7 strains, a very slight but statistically significant activity was observed (i.e., <0.5 log CFU/peg compared to the control), resulting in biofilm reduction in six cases (i.e., P. aeruginosa Z33, Z35, Z152, M13, M19, and M25) and biofilm increase in the remaining one (i.e., P. aeruginosa M42) (Fig. 2 and 3).
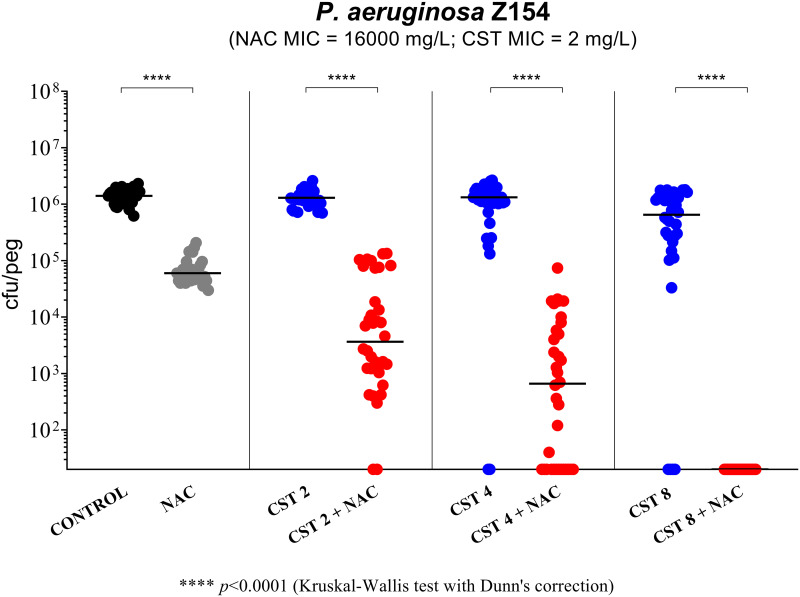
FIG 1.
Antibiofilm activity of N-acetylcysteine (NAC) at 8,000 mg/L, colistin (CST), and NAC-CST combinations against P. aeruginosa Z154 in the Nunc-TSP lid system. A relevant potentiation of colistin antibiofilm activity was observed with all NAC-CST combinations tested. CST 2, colistin at 2 mg/L; CST 4, colistin at 4 mg/L; CST 8, colistin at 8 mg/L. Biofilms not exposed to NAC or CST represent the control. Black lines indicate median values. The x axis is set at the limit of detection (20 CFU/peg).
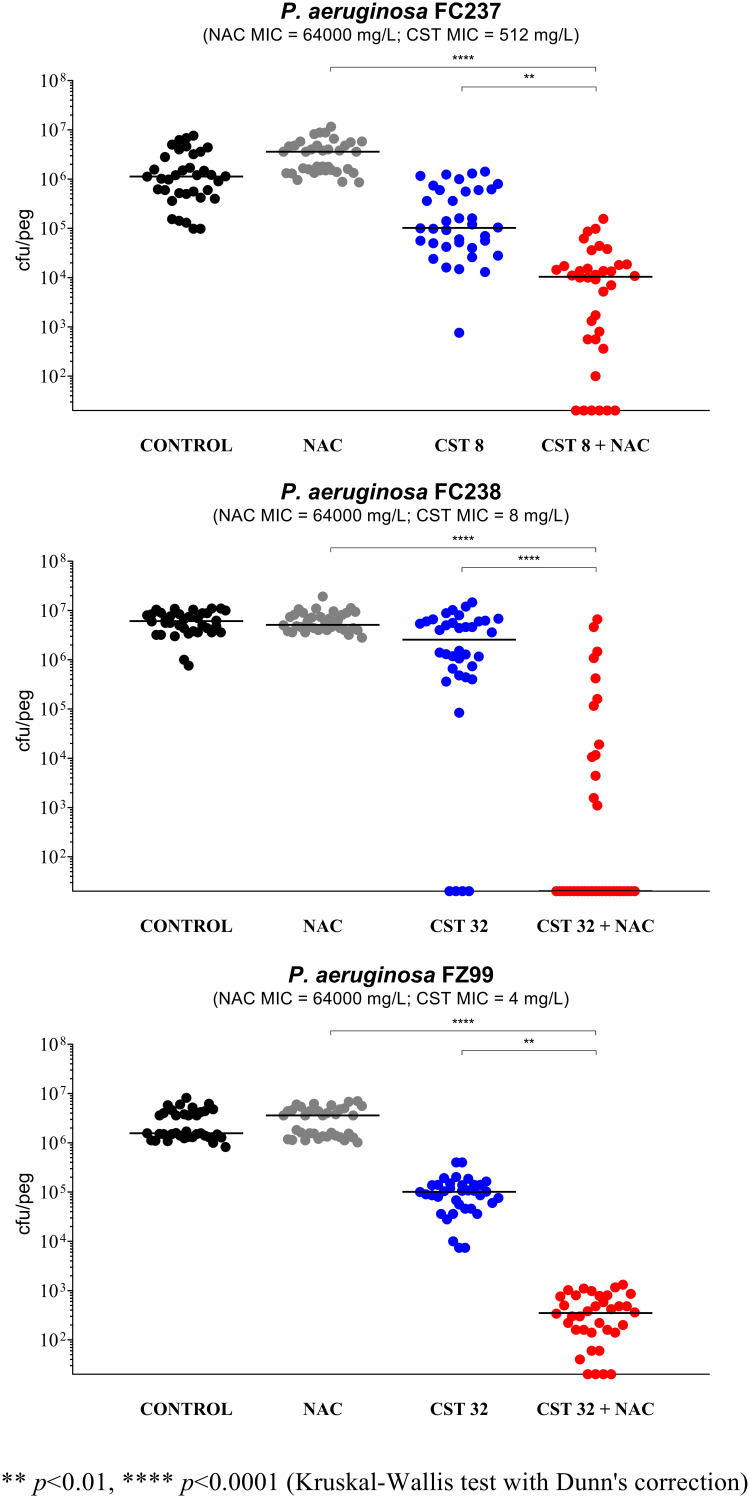
FIG 4.
Antibiofilm activity of N-acetylcysteine (NAC) at 8,000 mg/L, colistin (CST), and NAC-CST combinations against three colistin-resistant nonmucoid P. aeruginosa strains in the Nunc-TSP lid system. A potentiation by NAC of colistin antibiofilm activity was observed with all tested strains. CST 8, colistin at 8 mg/L; CST 32, colistin at 32 mg/L. Biofilms not exposed to NAC or CST represent the control. Black lines indicate median values. The x axis is set at the limit of detection (20 CFU/peg).
FIG 2.
Antibiofilm activity of N-acetylcysteine (NAC) at 8,000 mg/L, colistin (CST), and NAC-CST combinations against P. aeruginosa PAO1 and three colistin-susceptible nonmucoid strains in the Nunc-TSP lid system. A potentiation by NAC of colistin antibiofilm activity was observed with all tested strains. CST 4, colistin 4 mg/L; CST 8, colistin 8 mg/L. Biofilms not exposed to NAC or CST represented the control. Black lines indicate median values. The x axis is set at the limit of detection (20 CFU/peg).
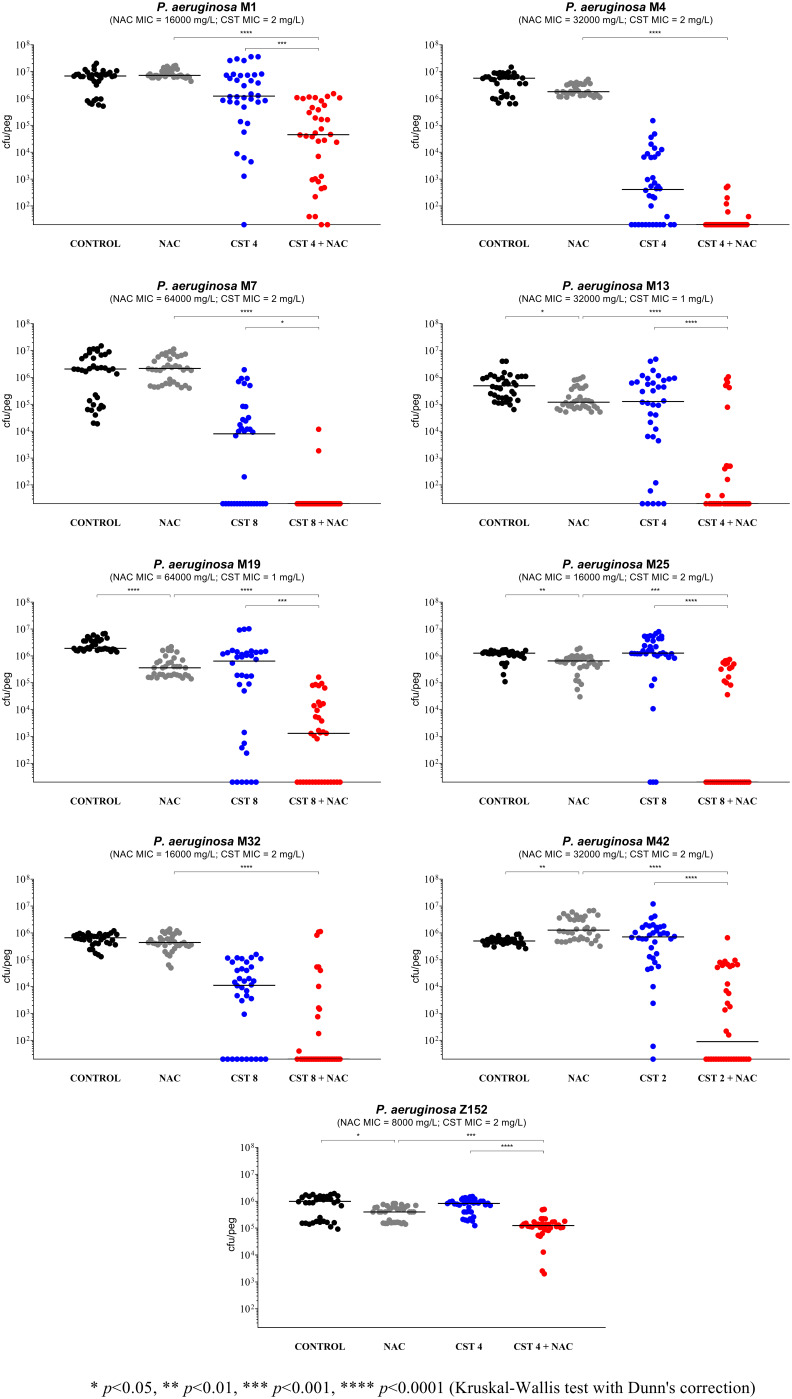
FIG 3.
Antibiofilm activity of N-acetylcysteine (NAC) at 8,000 mg/L, colistin (CST), and NAC-CST combinations against nine colistin-susceptible mucoid P. aeruginosa strains in the Nunc-TSP lid system. A potentiation by NAC of colistin antibiofilm activity was observed with all tested strains, although in two cases, statistical significance was not achieved (i.e., strains M4 and M32). CST 2, colistin at 2 mg/L; CST 4, colistin at 4 mg/L; CST 8, colistin at 8 mg/L. Biofilms not exposed to NAC or CST represent the control. Black lines indicate median values. The x axis is set at the limit of detection (20 CFU/peg).
Overall, these results indicated that inhaled NAC alone might not have major effects on P. aeruginosa biofilms already established in the lung and that the response to NAC was not related to phenotypic or genotypic features. The few previous studies that have addressed the activity of NAC against preformed P. aeruginosa biofilms have reported similar results (i.e., usually limited and strain-dependent effects), although a direct comparison of data is not straightforward due to different methodological approaches (e.g., different biofilm models and different NAC concentrations tested) and the low number of strains often tested in such studies (i.e., usually reference strains) (8, 13, 14). This study provided a wider picture on this topic by investigating a panel of characterized P. aeruginosa strains using a standardized in vitro biofilm model and in vivo achievable NAC concentrations. Interestingly, NAC alone (at the concentration used in this study and the same biofilm model) was recently shown to exert relevant activity against preformed biofilms of two relevant CF pathogens, namely, S. maltophilia and Burkholderia cepacia complex (BCC) (7). The reasons for such a diverse response of P. aeruginosa compared to S. maltophilia and BCC should deserve further attention, because they could possibly help identifying critical targets in the complex biofilm environments, to be used for the implementation of new antibiofilm strategies.
Activity of NAC-colistin combinations against preformed biofilms.
P. aeruginosa Z154 (a mucoid, MDR, colistin-susceptible CF strain) was first used to test the potential antibiofilm synergism of NAC at 8,000 mg/L plus diverse colistin concentrations. As shown in Fig. 1, a relevant synergism was observed already with colistin at 2 mg/L (i.e., the colistin MIC for the tested strain), with a dose-dependent effect at increasing colistin concentrations, and complete biofilm eradication was achieved with the combination of NAC at 8,000 mg/L plus colistin at 8 mg/L (Fig. 1).
The remaining 16 strains were initially tested with the combination of NAC at 8,000 mg/L plus colistin at 8 mg/L. In order to detect a potential synergism, the concentration of colistin was then modified for strains forming biofilms highly susceptible to colistin (n = 7) or particularly resistant (n = 2) (Fig. 2 to 4). Overall, a relevant synergism of NAC-colistin combinations was observed with all tested strains (including the three colistin-resistant ones), although in two cases (i.e., P. aeruginosa M4 and M32), statistical significance was not achieved (Fig. 2 to 4). These latter strains were also tested with lower colistin concentrations (i.e., 2 and 4 mg/L, respectively), but synergism was not observed (data not shown). Concerning the synergism observed with the three colistin-resistant strains (Fig. 4), it is interesting to note that with strain FC237 (nonmucoid, MDR), an important decrease in viable biofilm cells was observed with a combination including a colistin concentration much lower than the colistin MIC for this strain (i.e., 1/64 MIC) (Fig. 4).
Overall, these data demonstrated that NAC could potentiate colistin activity against preformed biofilms of colistin-susceptible and colistin-resistant P. aeruginosa strains, regardless of the mucoid/nonmucoid phenotype, the resistance pattern, and the ST and O type. Present findings are consistent with the previously observed antibiofilm synergism of NAC-colistin combinations against colistin-susceptible and colistin-resistant strains of A. baumannii and S. maltophilia (9, 10). Further studies with a higher number of P. aeruginosa clinical isolates, especially with a colistin-resistant phenotype, are encouraged.
Activity of NAC-colistin combinations in the ASM biofilm model.
Two P. aeruginosa CF strains exhibiting different phenotypes were selected for susceptibility assays with the artificial sputum medium (ASM) biofilm model: P. aeruginosa Z34 (nonmucoid, MDR, ST17, O1) and P. aeruginosa Z154 (mucoid, MDR, ST412, O6). Biofilms were grown in ASM, in order to mimic the P. aeruginosa biofilm environmental conditions experienced in the CF mucus. Preformed biofilms were then challenged in the same medium with NAC-colistin combinations.
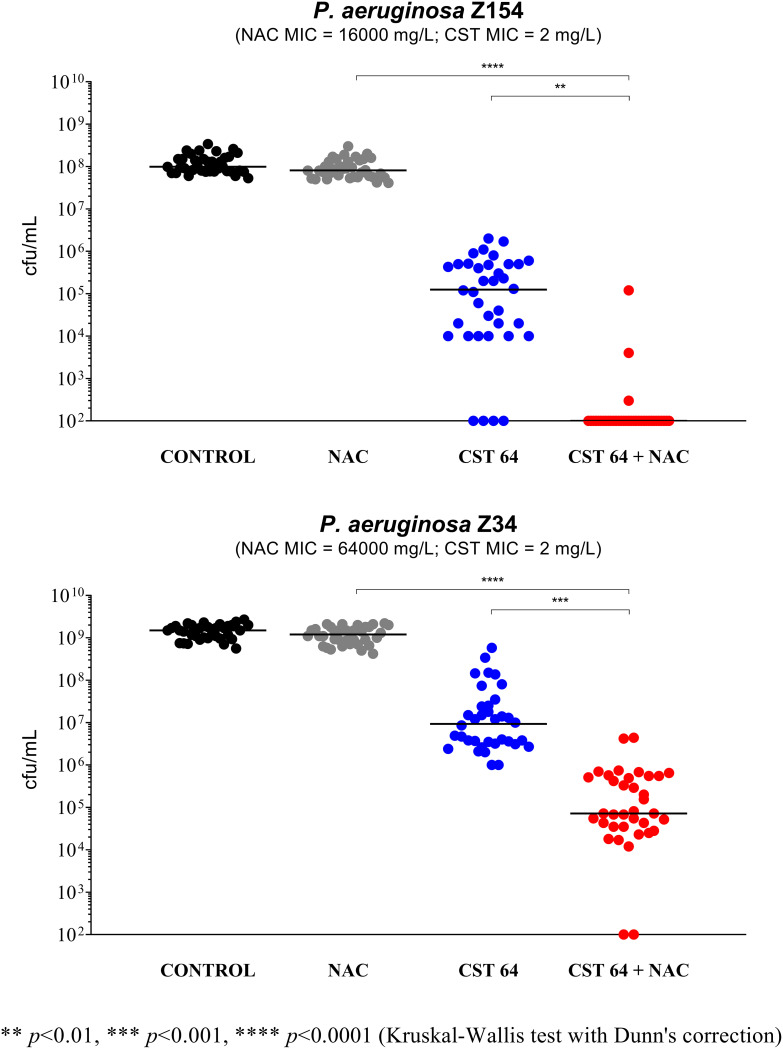
As shown in Fig. 5, a clear synergism of NAC at 8,000 mg/L in combination with colistin at 64 mg/L was observed with both strains (Fig. 5). Compared to the experiments performed with the Nunc-TSP lid system, the concentration of colistin that allowed observation of a synergism was much higher (i.e., 32× the MIC), possibly due to colistin strong ionic interactions with ASM components (e.g., extracellular DNA and mucin) (15). Indeed, preliminary experiments carried out with lower colistin concentrations did not show either colistin antibiofilm activity or synergism with NAC (data not shown). In addition, the antibiofilm activity of NAC alone observed against P. aeruginosa Z154 in the Nunc-TSP lid system was not observed in the ASM model (Fig. 5), confirming that the efficacy of NAC alone against preformed P. aeruginosa biofilms could be limited in vivo.
FIG 5.
Antibiofilm activity of N-acetylcysteine (NAC) at 8,000 mg/L, colistin at 64 mg/L (CST 64), and the NAC-CST combination against P. aeruginosa Z154 and P. aeruginosa Z34 in the ASM biofilm model. A potentiation by NAC of colistin antibiofilm activity was observed with both strains. Biofilms not exposed to NAC or CST represent the control. Black lines indicate median values. The x axis is set at the limit of detection (100 CFU/mL).
Overall, these data demonstrated that the antibiofilm synergism of NAC-colistin combinations against P. aeruginosa strains is preserved also under the environmental conditions mimicking the CF mucus, which is promising for clinical applications. Furthermore, the lower susceptibility to colistin of P. aeruginosa biofilms in the ASM model compared to biofilm susceptibility in standard media observed in this study is consistent with what was previously reported with P. aeruginosa (16).
Transcriptomic response of P. aeruginosa Z154 to NAC exposure.
P. aeruginosa Z154 (i.e., colistin-susceptible CF strain, mucoid, MDR, ST412, O6) was selected for investigating the transcriptome response of planktonic cultures to NAC exposure (i.e., NAC at 8,000 mg/L). A total of 66 differentially expressed genes (DEGs) were identified (adjusted P value of <0.05 with 99% confidence interval [CI]), of which 46 were upregulated and 20 downregulated compared to the control (Table 2).
TABLE 2.
DEGs in P. aeruginosa Z154 planktonic cultures exposed to 8,000 mg/L NAC compared to control
| DEG | Locus tag in P. aeruginosa strain |
Gene | Product (function)a | Zur regulon | Adjusted P value | Log2 fold change | ||
|---|---|---|---|---|---|---|---|---|
| Z154 | PAO1 | UCBPP-PA14 | ||||||
| Upregulated | IS492_10415 | PA0781 | PA14_54180 | znuD | TBDR ZnuD (zinc uptake) | + | 4.6E−36 | 1.9 |
| IS492_17070 | PA1922 | PA14_39650 | cirA | TBDR CirA (iron and zinc uptake) | + | 0.0E+00 | 2.4 | |
| IS492_17075 | PA1923 | PA14_39640 | Cobaltochelatase subunit CobN-like (cobalamin biosynthesis) | + | 7.9E−36 | 1.9 | ||
| IS492_17080 | PA1924 | PA14_39630 | exbD | ExbD proton channel family protein (energy support for TBDR, cotranscribed with PA1922) | + | 1.7E−03 | 0.6 | |
| IS492_17085 | PA1925 | PA14_39620 | Hypothetical protein (unknown function, DUF2149 domain-containing protein) | + | 7.5E−06 | 0.8 | ||
| IS492_19940 | PA2437 | PA14_33110 | HflC family modulator of membrane FtsH protease | + | 5.1E−06 | 0.8 | ||
| IS492_19945 | PA2438 | PA14_33080 | HflC modulator of membrane FtsH protease | + | 7.0E−03 | 0.6 | ||
| IS492_19950 | PA2439 | PA14_33070 | hflK | HflK family modulator of membrane FtsH protease | + | 6.5E−03 | 0.6 | |
| IS492_23615 | PA2911 | PA14_26420 | TBDR (possibly involved in zinc uptake) | + | 7.6E−03 | 0.6 | ||
| IS492_27310 | PA3600 | PA14_17710 | rpmJ2 | Zinc-independent paralog type B 50S ribosomal protein L36 | + | 2.0E−16 | 1.3 | |
| IS492_27315 | PA3601 | PA14_17700 | rpmE2 | Zinc-independent paralog type B 50S ribosomal protein L31 | + | 1.2E−04 | 0.7 | |
| IS492_29825 | PA4063 | PA14_11320 | Zinc SBP (zinc uptake) | + | 7.0E−41 | 2.0 | ||
| IS492_29830 | PA4064 | PA14_11310 | Zinc ABC transporter, ATP-binding protein (zinc uptake) | + | 4.2E−08 | 0.9 | ||
| IS492_29835 | PA4065 | PA14_11290 | Zinc ABC transporter, permease (zinc uptake) | + | 4.9E−13 | 1.2 | ||
| IS492_29840 | PA4066 | PA14_11280 | Zinc SBP (zinc uptake) | + | 8.5E−05 | 0.7 | ||
| IS492_06220 | PA4834 | PA14_63910 | cntI | Pseudopaline transport plasma membrane protein CntI (zinc uptake) | + | 6.1E−05 | 0.7 | |
| IS492_06215 | PA4835 | PA14_63920 | cntM | Pseudopaline biosynthesis dehydrogenase CntM (zinc uptake) | + | 8.1E−26 | 1.7 | |
| IS492_06210 | PA4836 | PA14_63940 | cntL | Pseudopaline biosynthesis enzyme CntL (zinc uptake) | + | 9.3E−39 | 2.0 | |
| IS492_06205 | PA4837 | PA14_63960 | cntO | Pseudopaline transport outer membrane protein CntO (zinc uptake) | + | 0.0E+00 | 2.5 | |
| IS492_06200 | PA4838 | PA14_63970 | Hypothetical membrane protein | + | 8.0E−04 | 0.7 | ||
| IS492_31595 | PA5498 | PA14_72550 | znuA | Zinc soluble binding protein ZnuA (zinc uptake) | + | 9.0E−08 | 0.9 | |
| IS492_31600 | PA5499 | PA14_72560 | zur | Transcriptional regulator for zinc homeostasis | + | 5.3E−10 | 1.0 | |
| IS492_31605 | PA5500 | PA14_72580 | znuC | Zinc ABC transporter, ATP-binding protein ZnuC (zinc uptake) | + | 1.2E−07 | 0.9 | |
| IS492_31610 | PA5501 | PA14_72590 | znuB | Zinc ABC transporter, ZnuB permease (zinc uptake) | + | 1.9E−03 | 0.6 | |
| IS492_31780 | PA5534 | PA14_73000 | Hypothetical protein (unknown function, DUF1826 domain-containing protein) | + | 9.8E−23 | 1.5 | ||
| IS492_31785 | PA5535 | PA14_73010 | zigA | Zinc metallochaperone GTPase ZigA | + | 5.9E−42 | 2.1 | |
| IS492_31790 | PA5536 | PA14_73020 | dksA2 | Zinc-independent paralog of RNA polymerase-binding protein DksA | + | 2.4E−23 | 1.5 | |
| IS492_31800 | PA5538 | PA14_73040 | amiA | N-acetylmuramoyl-l-alanine amidase (splitting of septal peptidoglycan during cell division) | + | 1.3E−08 | 1.0 | |
| IS492_31805 | PA5539 | PA14_73050 | folE2 | Zinc-independent paralog of GTP-cyclohydrolase FolE (folate biosynthesis) | + | 4.5E−28 | 1.7 | |
| IS492_31810 | PA5540 | PA14_73060 | cam | γ-Carbonic anhydrase (reversible hydration of carbon dioxide) | + | 1.5E−24 | 1.6 | |
| IS492_31815 | PA5541 | PA14_73070 | pyrC2 | Zinc-independent paralog of dihydroorotase PyrC (pyrimidine biosynthesis) | + | 3.1E−09 | 1.0 | |
| IS492_02205 | PA0433 | PA14_05630 | Hypothetical protein (unknown function, DUF2946 domain-containing protein) | 1.3E−03 | 0.7 | |||
| IS492_02210 | PA0434 | PA14_05640 | TBDR for which the siderophore has not been identified | 1.5E−28 | 1.7 | |||
| IS492_02430 | PA0478 | PA14_06250 | fiuC | GNAT family N-acetyltransferase (release of iron from desferrichrome in the cytoplasm) | 3.9E−06 | 0.8 | ||
| IS492_10765 | PA0848 | PA14_53300 | ahpB | AhpC-like alkylhydroperoxide reductase (oxidative stress response and cell redox homeostasis) | 3.9E−16 | 1.3 | ||
| IS492_17945 | PA2100 | NDb | mdrR2 | Transcriptional regulator, regulatory partner of MdrR1 (regulator of efflux systems) | 6.3E−05 | 0.7 | ||
| IS492_17950 | PA2101 | ND | Conserved hypothetical protein (EamA-like transporter family) | 1.7E−26 | 1.7 | |||
| IS492_17955 | PA2102 | ND | Hypothetical protein (unknown function, Mov34/MPN/PAD-1 family protein) | 5.7E−13 | 1.2 | |||
| IS492_17960 | PA2103 | ND | moeB | Probable molybdopterin biosynthesis protein MoeB (ubiquitin-like modifier-activating activity) | 7.5E−06 | 0.8 | ||
| IS492_25770 | PA3287 | PA14_21530 | Ankyrin repeat domain-containing protein (unknown function) | 1.9E−04 | 0.7 | |||
| IS492_27305 | PA3599 | PA14_17720 | Probable transcriptional regulator | 5.2E−12 | 1.1 | |||
| IS492_28275 | PA3784 | PA14_15130 | Hypothetical protein (unknown function) | 1.4E−05 | 0.8 | |||
| IS492_28280 | PA3785 | PA14_15120 | Copper chaperone PCu(A)C | 8.6E−07 | 0.9 | |||
| IS492_28305 | PA3790 | PA14_15070 | TBDR copper receptor OprC (copper uptake) | 1.0E−03 | 0.6 | |||
| IS492_06715 | PA4739 | PA14_62690 | Hypothetical protein (unknown function, BON domain-containing protein) | 9.8E−03 | 0.6 | |||
| IS492_31510 | PA5481 | PA14_72360 | Hypothetical periplasmic protein (inhibitor of vertebrate lysozyme) | 3.9E−04 | 0.7 | |||
| Downregulated | IS492_00850 | PA0164 | PA14_02050 | γ-Glutamyltransferase family protein | 8.0E−04 | −0.6 | ||
| IS492_02660 | PA0524 | PA14_06830 | norB | Nitric oxide reductase subunit NorB (denitrification) | 3.9E−03 | −0.6 | ||
| IS492_02685 | PA0529 | PA14_06890 | Hypothetical protein (unknown function, MOSC domain-containing protein) | 2.0E−05 | −0.7 | |||
| IS492_02690 | PA0530 | PA14_06900 | Probable class III pyridoxal phosphate-dependent aminotransferase (diverse metabolic pathways) | 5.7E−05 | −0.8 | |||
| IS492_02695 | PA0531 | PA14_06920 | Aspartate aminotransferase family protein | 4.7E−03 | −0.6 | |||
| IS492_12670 | PA1101 | PA14_50140 | fliF | Flagellar M-ring protein FliF (motility) | 5.7E−05 | −0.7 | ||
| IS492_12855 | PA1136 | PA14_49700 | Probable transcriptional regulator | 1.5E−12 | −1.1 | |||
| IS492_12860 | PA1137 | PA14_49690 | Oxidoreductase zinc-binding dehydrogenase family protein (protection from oxidative stress) | 0.0E+00 | −2.3 | |||
| IS492_14625 | PA1453 | PA14_45660 | flhF | Flagellar biosynthesis protein FlhF (motility) | 7.6E−03 | −0.6 | ||
| IS492_19230 | PA2298 | PA14_34900 | Probable oxidoreductase | 4.9E−05 | −0.7 | |||
| IS492_19235 | PA2299 | PA14_34880 | Probable transcriptional regulator | 3.2E−04 | −0.7 | |||
| IS492_26340 | PA3391 | PA14_20230 | nosR | Regulatory protein NosR (denitrification) | 3.2E−04 | −0.6 | ||
| IS492_26345 | PA3392 | PA14_20200 | nosZ | Nitrous oxide reductase (denitrification) | 4.1E−05 | −0.8 | ||
| IS492_26895 | PA3519 | PA14_18810 | Iron-containing redox enzyme family protein | 2.8E−05 | −0.3 | |||
| IS492_26920 | PA3523 | PA14_18760 | mexP | Resistance-nodulation-cell division (RND) efflux membrane fusion protein | 3.2E−03 | −0.2 | ||
| IS492_27180 | PA3574 | PA14_18080 | nalD | Transcriptional regulator NalD (second repressor of MexAB-OprM) | 1.5E−19 | −1.3 | ||
| IS492_27185 | PA3574a | PA14_18070 | copZ | Copper chaperone CopZ (copper efflux) | 9.1E−11 | −1.0 | ||
| IS492_27760 | PA3690 | PA14_16660 | Heavy metal-translocating P-type ATPase (efflux) | 1.1E−08 | −1.0 | |||
| IS492_28975 | PA3920 | PA14_13170 | copA | Copper-translocating P-type ATPase CopA1 (copper efflux) | 1.2E−27 | −1.2 | ||
| IS492_04870 | PA5100 | PA14_67350 | hutU | Urocanate hydratase (histidine catabolic process) | 4.0E−04 | −0.6 | ||
TBDR, TonB-dependent receptor; SBP, soluble binding protein; ABC, ATP-binding cassette. Protein functions were inferred from the literature and PseudoCAP (https://www.Pseudomonas.com/pseudocap).
ND, not determined.
Analysis of DEGs revealed that NAC mainly acted as Zn2+ chelator, inducing a strong Zn2+ starvation response. DEGs associated with such response were consistent with data reported in previous studies addressing zinc homeostasis in P. aeruginosa and other bacteria (Table 2) (17–22). In particular, 31 of the 46 upregulated DEGs belonged to the zur regulon and are known to be activated in response to Zn2+ starvation (Table 2) (17–22). Such genes mainly included operons involved in zinc uptake (e.g., the PA4063-PA4064-PA4065-PA4066 operon, cntOLMI operon, and znuABC operon) and genes encoding zinc-independent paralogs of cellular proteins (i.e., type B 50S ribosomal proteins L31 and L36, RNA polymerase-binding protein DksA2, adn GTP-cyclohydrolase FolE2) (Table 2) (17–23). Upregulated DEGs belonging to the zur regulon also included genes encoding an N-acetylmuramoyl-l-alanine amidase (AmiA, involved in splitting of septal peptidoglycan during cell division), a γ-carbonic anhydrase (Cam, involved in reversible hydration of carbon dioxide and important for growth under low-CO2 conditions), and three modulators of the membrane FtsH protease (i.e., HflC and HflK family modulators) (Table 2). The membrane FtsH zinc-dependent protease is required for the expression of diverse unrelated phenotypes (e.g., swimming and twitching motility, biofilm formation, autolysis, production of secondary metabolites, maintenance of plasma membrane integrity by degrading misfolded proteins), and it has been recently demonstrated to represent an important virulence factor in P. aeruginosa clone C (23). HflC and HflK family modulators interact with FtsH at the level of the plasma membrane, usually with an inhibitory effect (23). The NAC-mediated effects on the phenotypes related to FtsH would deserve further attention.
The remaining 15 upregulated DEGs included genes encoding a recently described transcriptional regulator, PA2100 (also named MdrR2) (24), an AhpC-like alkyl hydroperoxide reductase (involved in protection from oxidative stress) (25), and proteins possibly involved in copper and iron uptake (Table 2).
MdrR2, together with MdrR1, has been demonstrated to repress the mexAB-oprM operon (independently from the MexR repressor), activate the EmrAB efflux pump, and indirectly inhibit biofilm formation (Table 2) (24). The effect of NAC on the MdrR1-MdrR2 dual-regulation system should be further investigated. Nonetheless, a previous study aimed at investigating the potential antagonism of high NAC concentrations (i.e., as those tested in this study) on the activity of the major classes of antibiotics used in the clinical practice, did not show major effects (with the exception of carbapenems, due to a chemical instability of carbapenems in the presence of NAC) (26), suggesting that the activation of the EmrAB efflux could not be relevant or circumvented by compensatory mechanisms.
Analysis of downregulated DEGs identified genes involved in denitrification, in particular norB (encoding the nitric oxide reductase subunit NorB), nosR (encoding the regulatory protein NosR), and nosZ (encoding the nitrous oxide reductase NosZ) (Table 2). These data suggested that NAC might affect P. aeruginosa anaerobic respiration (which is crucial in the deeper biofilm layers and in the CF mucus) (27), because the nitric oxide reductase NorBC and the regulatory protein NosR have been recently demonstrated to constitute the nucleus of the denitrification protein network (28). NAC-mediated inhibition of the P. aeruginosa denitrification pathway might be implicated in the observed antibiofilm synergism of the NAC-colistin combination. Indeed, colistin has been demonstrated to exert increased antibiofilm activity against P. aeruginosa under anaerobic conditions, possibly due to a lower ability to implement the tolerance mechanism (e.g., lipopolysaccharide [LPS] modification) because of the low metabolism accompanying anaerobic growth (29). In this perspective, the inhibition of anaerobic respiration by NAC would further inhibit a P. aeruginosa adaptive response to colistin toxicity. This could be particularly relevant in P. aeruginosa biofilm in the CF mucus, where the anoxic conditions of biofilm cells are related not only to the position of the bacteria within the biofilm (i.e., anoxic conditions in the deeper layers), but also to the intense O2 depletion caused by polymorphonuclear leukocytes (PMNs), determining entire biofilm growth without aerobic respiration (29).
Downregulated DEGs also included the following: (i) two genes involved in flagellar biosynthesis (i.e., fliF, encoding the flagellar M-ring protein FliF, and flhF, encoding the flagellar biosynthesis protein FlhF); (ii) a NAD(P)H-quinone oxidoreductase protecting against ROS-induced oxidative stress, which was recently demonstrated to be part of the core biofilm transcriptome (PA1137) (30); and (iii) nalD, encoding a second repressor of the mexAB-oprM operon (31). Finally, consistent with previous studies on Pseudomonas response to zinc starvation, downregulation of copA and copZ, involved in copper efflux, was observed, suggesting interplay between zinc and copper homeostasis (Table 2) (32).
NAC-mediated inhibition of P. aeruginosa denitrification pathway.
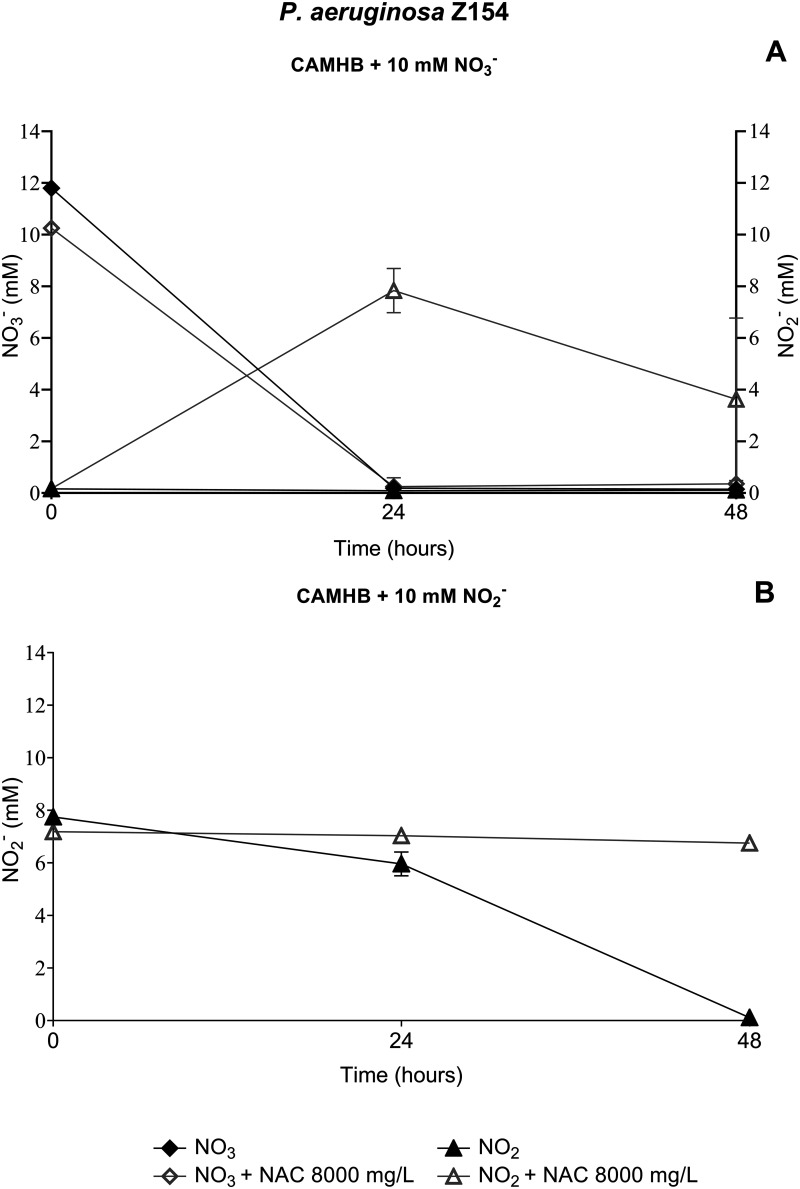
The role of NAC in the inhibition of the denitrification pathway was confirmed by measuring NO3− and NO2− concentrations during anaerobic growth of the P. aeruginosa Z154 strain (i.e., the strain used for transcriptomic analysis) in culture media supplemented with 10 mM NaNO3 or KNO2, in the presence or absence of NAC at 8,000 mg/L.
As expected from previous studies (33), in NaNO3-containing medium, the levels of NO3− and its reduction product, NO2−, fell below the detection limit after 24 h, in the absence of NAC (Fig. 6A). However, in the presence of NAC at 8,000 mg/L, the depletion of NO3− was followed by an accumulation of NO2− (evident at both 24 and 48 h), indicating that further reduction of NO2− was inhibited in the presence of NAC (Fig. 6A). In order to consolidate these data, the experiments were repeated using a medium supplemented with KNO2. In the absence on NAC, complete reduction of NO2 was observed after 48 h (Fig. 6B), as expected (33). On the contrary, in the presence of NAC at 8,000 mg/L, NO2 levels did not decrease (Fig. 6B).
FIG 6.
NAC-mediated inhibition of P. aeruginosa Z154 denitrification pathway. (A) NO3− and NO2− concentrations in anaerobic CAMHB supplemented with 10 mM NO3−, with or without NAC at 8,000 mg/L; (B) NO2− concentration in anaerobic CAMHB supplemented with 10 mM NO2−, with or without NAC at 8,000 mg/L. Data are plotted as the mean values of NO3− and/or NO2− levels detected at each time point.
These results were consistent with the transcriptomic data and showed that NAC was able to inhibit the denitrification pathway in anaerobic environments, such as those encountered in endobronchial CF mucus. This feature might contribute to the observed antibiofilm synergism of NAC-colistin combinations, as previously discussed.
Time-kill assays of the NAC-colistin combination against planktonic cultures grown under anaerobic and aerobic conditions.
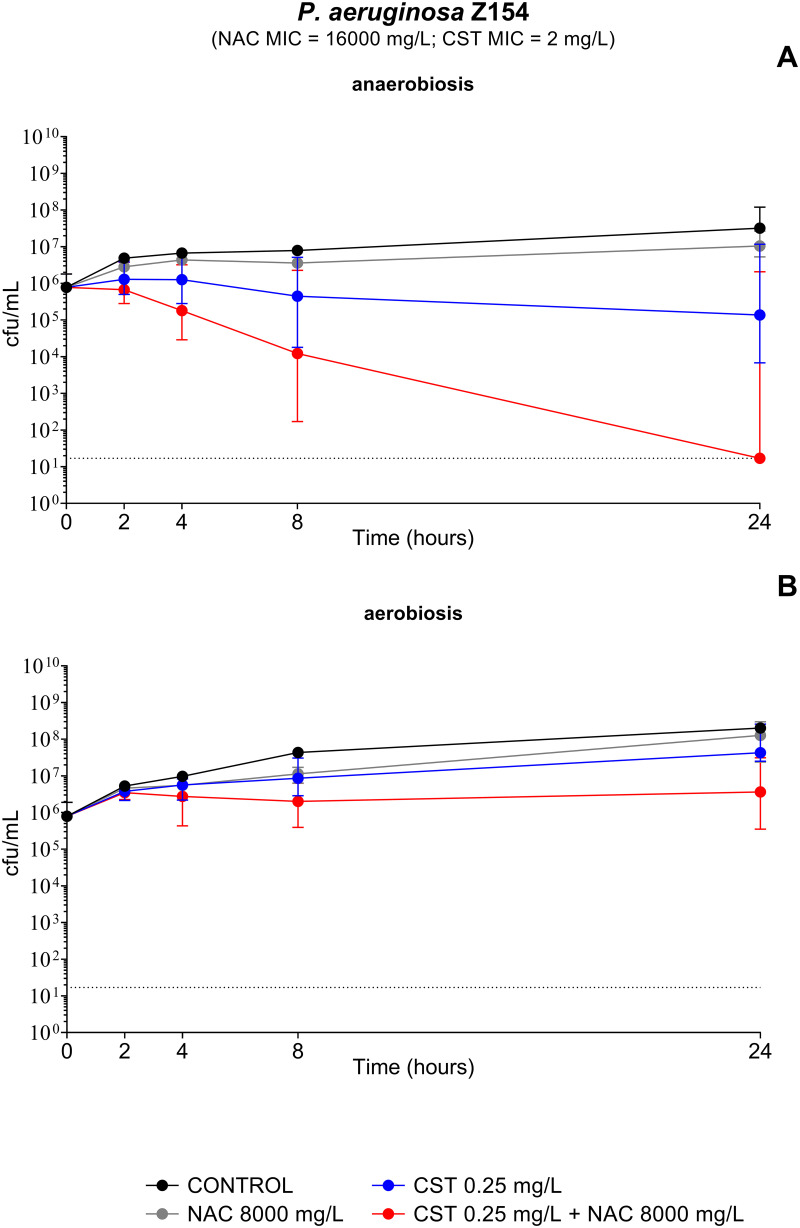
Transcriptomic and biological data from this study suggested a role of NAC in inhibiting the P. aeruginosa denitrification apparatus, which could contribute to the observed antibiofilm synergy of NAC-colistin combinations. In order to further investigate this issue, time-kill assays of the NAC-colistin combination were performed with P. aeruginosa Z154 (i.e., the strain used for transcriptomic analysis) planktonic cultures, under both anaerobic and aerobic conditions. Consistent with previous studies, anaerobic cultures were more susceptible to killing by colistin than aerobic cultures (34, 35) (Fig. 7A and B). Interestingly, a clear bactericidal effect of colistin at 0.25 mg/L (i.e., 1/8 MIC) in combination with NAC at 8,000 mg/L was observed in planktonic cultures grown under anaerobic conditions, with eradication achieved after 24 h of exposure (Fig. 7A). The wide error bars were due to the fact that in 2 out of 8 replicates (related to two independent experiments), no synergism was observed (Fig. 7A). This discrepancy was probably related to the low colistin concentration tested and the possible presence of heteroresistant subpopulations. On the contrary, cultures grown in the presence of oxygen were not affected by the NAC-colistin combination, demonstrating the influence of the growth conditions on the susceptibility of P. aeruginosa to such combination (Fig. 7B).
FIG 7.
Time-kill curves of P. aeruginosa Z154 planktonic cultures exposed to N-acetylcysteine (NAC) at 8,000 mg/L, colistin (CST) at 0.25 mg/L, and the NAC-CST combination under anaerobic (A) and aerobic (B) conditions. NAC potentiated the bactericidal activity of colistin only under anaerobic conditions. Data are plotted as the median values of CFU per milliliter for each time point. Dotted lines indicate the detection limit (17 CFU/mL).
These results supported the hypothesis that, under anoxic conditions like those present in the deeper biofilm layers and in CF mucus, NAC-mediated inhibition of anaerobic respiration would prevent an adaptive response of P. aeruginosa to protect from colistin toxicity.
NAC-mediated inhibition of P. aeruginosa swimming and swarming motility.
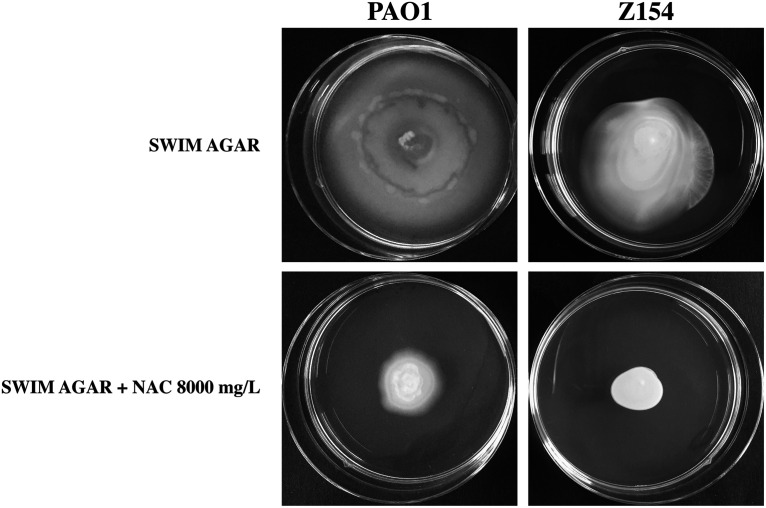
Transcriptomic results indicated that NAC downregulated two genes belonging to P. aeruginosa flagellar apparatus (i.e., fliF and flhF), which are necessary for the first step of flagellum assembly (36). In order to confirm the potential NAC-induced inhibition of flagellum-mediated motility, we performed classical swimming and swarming tests with the reference strain P. aeruginosa PAO1 and the CF strain P. aeruginosa Z154 (i.e., the strain used for transcriptomic analysis). P. aeruginosa Z154 was not capable of swarming motility under our laboratory conditions, so only the effect of NAC on swimming motility could be tested with this strain.
Overall, the results showed a clear inhibition of both swimming and swarming motility in the presence of NAC at 8,000 mg/L (Fig. 8 and 9). Such inhibition could be related to the downregulation of crucial genes of the flagellar apparatus and/or the induction of a zinc starvation response. Indeed, zinc starvation has been demonstrated to affect the ability of P. aeruginosa to express several virulence phenotypes, crucial for the ability of this pathogen to colonize CF lung, including motility, biofilm formation and siderophore synthesis (37).
FIG 8.
NAC-mediated inhibition of P. aeruginosa PAO1 and Z154 swimming motility. Assays were performed in at least three independent experiments (with three replicates per condition per experiment), and representative data are shown.
FIG 9.
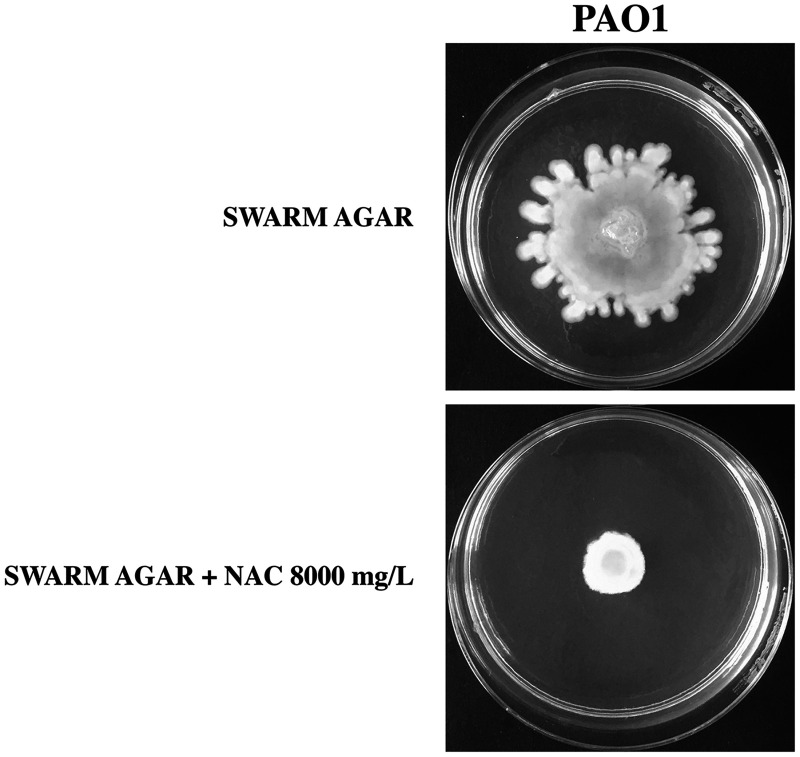
NAC-mediated inhibition of P. aeruginosa PAO1 swarming motility. Assays were performed in at least three independent experiments (with three replicates per condition per experiment), and representative data are shown.
Conclusions.
In conclusion, the results of this study demonstrated a relevant antibiofilm synergism of NAC-colistin combinations (at the high concentrations achievable by inhalation) against P. aeruginosa, which would deserve further investigation for potential clinical applications of inhaled formulations. Transcriptomic and biological experiments suggested that NAC inhibited P. aeruginosa anaerobic respiration, which could be relevant for the observed antibiofilm synergism with colistin.
In addition, although NAC alone was not demonstrated to be effective against preformed P. aeruginosa biofilms, transcriptomic analysis of NAC-exposed planktonic cultures revealed that NAC could attenuate P. aeruginosa virulence, mainly by inducing a zinc starvation response, affecting anaerobic respiration and inhibiting flagellum-mediated motility (with the last two features confirmed experimentally). In this perspective, NAC, at the high concentrations achievable by inhalation, might have beneficial effects in the very first steps of lung infection, possibly preventing biofilm formation and the establishment of a chronic colonization, which should be further investigated.
MATERIALS AND METHODS
Bacterial strains.
Seventeen strains were investigated, including 15 clinical isolates from CF patients, an MDR clinical isolate from a respiratory tract infection (RTI) from an intensive care unit (ICU), and the reference strain, P. aeruginosa PAO1 (Table 1). Identification was performed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Bruker, Shimadzu). Antimicrobial susceptibility was determined using the reference broth microdilution method (38). Whole-genome sequencing of clinical isolates was performed with the Illumina (San Diego, CA, USA) MiSeq platform, using a 2× 150-bp paired-end approach. Raw reads were assembled using SPAdes (39), and draft genomes were used to determine multilocus sequence types (MLSTs) and O types at the Oxford PubMLST site (https://pubmlst.org/) (40) and at the Center for Genomic Epidemiology site (https://cge.food.dtu.dk/services/PAst/) (41), respectively. The complete genome of P. aeruginosa Z154 was obtained by combining results from Illumina with those obtained using the Oxford Nanopore Technologies (Oxford, United Kingdom) MinION platform, and de novo assembly was generated using Unicycler v0.4.4 as previously described (42).
Preparation of culture media.
NAC stock solutions (100 g/L) were prepared immediately before use. NAC powder (Zambon, Bresso, Italy) was dissolved in sterile distilled water, the pH was adjusted to 6.5 to 6.8 with NaOH at 10 M, and the solution was filtered through a 0.22-μm-pore membrane filter. All experiments were performed in cation-adjusted Mueller-Hinton broth (CAMHB) (Becton Dickinson, Milan, Italy), unless otherwise specified, starting from an appropriately concentrated medium to avoid broth dilution when NAC solution was used. The artificial sputum medium (ASM) was also used in selected experiments and was prepared as previously described by Kirchner et al. (43).
In vitro biofilm susceptibility testing.
Biofilm susceptibility testing was first performed using the Nunc-TSP lid system (Thermo Fisher Scientific, Waltham, MA, USA), as described previously (44). Briefly, biofilms were grown for 24 h in CAMHB at 35°C under static conditions. Preformed biofilms were then exposed to NAC at 8,000 mg/L and colistin (colistin sulfate; Applichem, Darmstadt, Germany) at 2 to 32 mg/L, alone and in combination. The colistin concentration was selected according to preliminary results of antibiofilm susceptibility testing and the colistin MIC for each strain. After 24 h of exposure (i.e., 35°C, static conditions), biofilms were washed twice with 200 μL of phosphate-buffered saline (PBS) (Sigma-Aldrich, Milan, Italy) to remove loosely adherent bacteria, and sessile cells were removed from pegs by sonication for 30 min (Elma Transsonic T 460; Elma, Singen, Germany) in 200 μL of tryptic soy broth (TSB) (Oxoid, Milan, Italy) supplemented with 1% Tween 20 (Sigma-Aldrich) (i.e., the recovery medium). The median number of CFU per peg was then determined by plating 10 μL of appropriate dilutions of the recovery medium onto tryptic soy agar (TSA) (Oxoid) and incubating for 24 h at 35°C (detection limit, 20 CFU/peg). The colony count was also double-checked after 48 h of incubation.
The potential antibiofilm synergism of NAC-colistin combinations was further investigated using an in vitro ASM biofilm model (43) in order to mimic P. aeruginosa biofilm conditions within the CF mucus. The study was carried out with two selected CF strains (P. aeruginosa Z154 and Z34), exhibiting different features (i.e., mucoid/nonmucoid phenotype, antimicrobial susceptibility pattern, MLST, and O type) (Table 1). In brief, biofilms were grown in 2 mL ASM in 24-well plates (Sarstedt, Nümbrecht, Germany), for 72 h at 35°C under static conditions. Preformed biofilms were then exposed to NAC at 8,000 mg/L and colistin at 64 mg/L, alone and in combination. Preliminary experiments carried out with lower colistin concentrations (i.e., 2 to 32 mg/L) did not show evident synergistic antibiofilm activity, while higher colistin concentrations (i.e., >64 mg/L) led to eradication of the biofilm cultures even in the absence of NAC (data not shown). After 24 h of exposure (i.e., 35°C, static conditions), bacterial biofilms were disrupted by 30 min of sonication followed by manual pipetting, and the median number of CFU per milliliter was determined following the same protocol described for the Nunc-TSP lid assay.
Data from both biofilm models were obtained in at least three independent experiments, with at least 12 replicates per condition per experiment.
RNA-seq and transcriptomic analysis.
P. aeruginosa Z154 (i.e., colistin-susceptible CF strain, mucoid, MDR, ST412, O6) (Table 1) was selected for studies aimed at investigating the transcriptomic response of P. aeruginosa to NAC exposure. A CF strain, rather than a reference strain (such as P. aeruginosa PAO1), was selected for this analysis because of the known adaptive diversification of P. aeruginosa into “specialized” types during chronic/recurrent infections in CF patients (3).
Because these represented the first data on the transcriptomic response of P. aeruginosa to NAC exposure, and considering the complex and still largely unknown effects of NAC on microbial physiology, we decided to perform the experiments with planktonic cultures, which represent a more homogenous and better standardized model for transcriptomic studies.
Overnight cultures in CAMHB were diluted at 1:50 in the same medium and incubated at 35°C with agitation to achieve an optical density at 600 nm (OD600) of 1.0. The cells were then exposed to NAC at 8,000 mg/L for 30 min at 35°C under static conditions. Cultures treated in the same way but not exposed to NAC represented the control. Total RNA extraction was performed using the SV total RNA isolation system (Promega, Madison, WI, USA) following the manufacturer’s instructions. rRNA depletion, cDNA library construction, and Illumina HiSeq 4000 platform-based transcriptome sequencing (RNA-seq) were performed by Eurofins Genomics Europe Sequencing (Constance, Germany). The transcriptome libraries were single-end sequenced with 50-bp reads for a total of 10 million reads per sample. Bioinformatic analysis was performed using the SeqMan NGen v17.3 software tool (DNASTAR Lasergene, Madison, WI, USA), with default parameters. Reads were aligned using P. aeruginosa Z154 complete genome (n = 6,344 coding DNA sequences [CDSs]) as a reference. Differentially expressed genes (DEGs) of the NAC-exposed cultures compared to the control were analyzed considering false-discovery rate (FDR) adjusted P values of <0.05 from DeSeq2. DEGs with a 99% confidence interval (CI) were discussed. Results were obtained from two independent experiments. In order to favor comparison with data present in the literature, genes without a univocal name have been indicated as P. aeruginosa PAO1 locus tags throughout the text and reported in Table 2 also as P. aeruginosa UCBPP-PA14 locus tags.
NO3− and NO2− quantification.
NAC-mediated inhibition of the denitrification pathway was investigated by measuring the concentration of NO3− and NO2− in anaerobic cultures of P. aeruginosa Z154 (i.e., the strain used for transcriptomic analysis). For this purpose, the Griess nitrite/nitrate colorimetric assay (Cayman Chemicals, Ann Arbor, MI, USA) was used according to the manufacturer’s recommendations and as previously described, with some modification (33). CAMHB was supplemented with 10 mM NaNO3 or KNO2 and allowed to equilibrate for 3 days at 35°C in an anaerobic atmosphere by using the AnaeroGen kit (Oxoid). Overnight cultures were then diluted in 20 mL of each anoxic culture medium to reach a concentration of 106 CFU/mL and challenged with NAC at 8,000 mg/L. At times 0, 24, and 48 h of incubation under anoxic conditions at 35°C, supernatants were harvested and subjected to Griess colorimetric reaction in order to detect NO3− and NO2− levels. NAC-free cultures represented the control. Experiments were carried out in triplicate with one replicate per time point per condition.
Time-kill assays.
Time-kill assays were performed according to CLSI guidelines (45) with the colistin-susceptible strain P. aeruginosa Z154 (i.e., the strain used for transcriptomic analysis). Colistin at 0.25 mg/L was tested alone and in combination with NAC at 8,000 mg/L under both aerobic and anaerobic conditions. We decided to use this colistin concentration since a higher concentration led to eradication of the planktonic cultures (data not shown). The medium (CAMHB) used to obtain anoxic cultures was placed under an anaerobic atmosphere by using the AnaeroGen kit (Oxoid) for 3 days prior to use and during the whole experiment. The killing curves were carried out in borosilicate glass bottles with a final volume of 20 mL of CAMHB. At 0, 2, 4, 8, and 24 h of exposure, CFU per milliliter were determined by plating 60 µL of appropriate dilutions of each condition onto TSA and incubating for 24 h at 35°C (detection limit, 17 CFU/mL). Data were obtained from at least four independent experiments with two replicates per condition per experiment.
Motility tests.
NAC-induced inhibition of flagellum-mediated motility (i.e., both swimming and swarming motility) was investigated with the reference strain P. aeruginosa PAO1, which has been used for similar motility experiments in several previous studies (46), and P. aeruginosa Z154 (i.e., the strain used for transcriptomic analysis). P. aeruginosa Z154 was not capable of swarming motility under our laboratory conditions (perhaps due to the known reduction of flagellar expression in mucoid CF-adapted strains) (47), so only the effect of NAC on swimming motility could be tested with this strain. Swim plates consisted of Luria-Bertani (LB) broth (Oxoid) containing 0.3% agar (46). Swarm plates consisted of nutrient broth (Oxoid) with 0.5% glucose and 0.5% agar (46). Overnight cultures in CAMHB were diluted in the same medium to a final OD600 of 3.0, and 5 μL was spotted onto swim and swarm plates, with or without NAC at 8,000 mg/L. Results were observed after incubation at 35°C for 48 h. Assays were performed in at least three independent experiments with three replicates per condition per experiment.
Statistical analysis.
Statistical analysis of biofilm susceptibility assays was performed using GraphPad Prism version 8.0 (San Diego, CA, USA). Multiple-comparison tests were performed by the Kruskal-Wallis test with Dunn’s correction. A P value of ≤0.05 was considered significant. RNA-seq statistical analysis was performed using the SeqMan NGen v17.3 software tool.
Data availability.
The complete genome sequence of P. aeruginosa Z154 was deposited in GenBank under accession no. CP069177. RNA-seq data were also deposited in the NCBI Gene Expression Omnibus (GEO) database under accession no. GSE190946.
ACKNOWLEDGMENTS
This work was supported by a research grant from Zambon S.p.A. G.M.R. and L.P. have been Advisory Board members for Zambon S.p.A. and have participated in scientific events financed by Zambon. The remaining authors declare no conflict of interest.
P. aeruginosa strains Z154 and Z152 were kindly provided by Lisa Cariani, Cystic Fibrosis Microbiology Laboratory, IRCCS Fondazione Cà Granda, Ospedale Maggiore Policlinico, Milan, Italy.
Contributor Information
Lucia Pallecchi, Email: lucia.pallecchi@unisi.it.
Cezar M. Khursigara, University of Guelph
REFERENCES
- 1.Malhotra S, Hayes D, Wozniak DJ. 2019. Cystic fibrosis and Pseudomonas aeruginosa: the host-microbe interface. Clin Microbiol Rev 32:e00138-18. doi: 10.1128/CMR.00138-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkins MD, Somayaji R, Waters VJ. 2018. Epidemiology, biology, and impact of clonal Pseudomonas aeruginosa infections in cystic fibrosis. Clin Microbiol Rev 31:e00019-18. doi: 10.1128/CMR.00019-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossi E, La Rosa R, Bartell JA, Marvig RL, Haagensen JAJ, Sommer LM, Molin S, Johansen HK. 2021. Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis. Nat Rev Microbiol 19:331–342. doi: 10.1038/s41579-020-00477-5. [DOI] [PubMed] [Google Scholar]
- 4.Karaiskos I, Souli M, Galani I, Giamarellou H. 2017. Colistin: still a lifesaver for the 21st century? Expert Opin Drug Metab Toxicol 13:59–71. doi: 10.1080/17425255.2017.1230200. [DOI] [PubMed] [Google Scholar]
- 5.Ding L, Wang J, Cai S, Smyth H, Cui Z. 2021. Pulmonary biofilm-based chronic infections and inhaled treatment strategies. Int J Pharm 604:120768. doi: 10.1016/j.ijpharm.2021.120768. [DOI] [PubMed] [Google Scholar]
- 6.Manos J. 2021. Current and emerging therapies to combat cystic fibrosis lung infections. Microorganisms 9:1874. doi: 10.3390/microorganisms9091874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pollini S, Di Pilato V, Landini G, Di Maggio T, Cannatelli A, Sottotetti S, Cariani L, Aliberti S, Blasi F, Sergio F, Rossolini GM, Pallecchi L. 2018. In vitro activity of N-acetylcysteine against Stenotrophomonas maltophilia and Burkholderia cepacia complex grown in planktonic phase and biofilm. PLoS One 13:e0203941. doi: 10.1371/journal.pone.0203941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blasi F, Page C, Rossolini GM, Pallecchi L, Matera MG, Rogliani P, Cazzola M. 2016. The effect of N-acetylcysteine on biofilms: implications for the treatment of respiratory tract infections. Respir Med 117:190–197. doi: 10.1016/j.rmed.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 9.Pollini S, Boncompagni S, Di Maggio T, Di Pilato V, Spanu T, Fiori B, Blasi F, Aliberti S, Sergio F, Rossolini GM, Pallecchi L. 2018. In vitro synergism of colistin in combination with N-acetylcysteine against Acinetobacter baumannii grown in planktonic phase and in biofilms. J Antimicrob Chemother 73:2388–2395. doi: 10.1093/jac/dky185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciacci N, Boncompagni S, Valzano F, Cariani L, Aliberti S, Blasi F, Pollini S, Rossolini GM, Pallecchi L. 2019. In vitro synergism of colistin and N-acetylcysteine against Stenotrophomonas maltophilia. Antibiotics 8:101. doi: 10.3390/antibiotics8030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tenório MCDS, Graciliano NG, Moura FA, de Oliveira ACM, Goulart MOF. 2021. N-Acetylcysteine (NAC): impacts on human health. Antioxidants 10:967. doi: 10.3390/antiox10060967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boisson M, Jacobs M, Grégoire N, Gobin P, Marchand S, Couet W, Mimoz O. 2014. Comparison of intrapulmonary and systemic pharmacokinetics of colistin methanesulfonate (CMS) and colistin after aerosol delivery and intravenous administration of CMS in critically ill patients. Antimicrob Agents Chemother 58:7331–7339. doi: 10.1128/AAC.03510-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen Y, Li P, Chen X, Zou Y, Li H, Yuan G, Hu H. 2020. Activity of sodium lauryl sulfate, rhamnolipids, and N-acetylcysteine against biofilms of five common pathogens. Microb Drug Resist 26:290–299. doi: 10.1089/mdr.2018.0385. [DOI] [PubMed] [Google Scholar]
- 14.Belfield K, Bayston R, Hajduk N, Levell G, Birchall JP, Daniel M. 2017. Evaluation of combinations of putative anti-biofilm agents and antibiotics to eradicate biofilms of Staphylococcus aureus and Pseudomonas aeruginosa. J Antimicrob Chemother 72:2531–2538. doi: 10.1093/jac/dkx192. [DOI] [PubMed] [Google Scholar]
- 15.Huang JX, Blaskovich MAT, Pelingon R, Ramu S, Kavanagh A, Elliott AG, Butler MS, Montgomery AB, Cooper MA. 2015. Mucin binding reduces colistin antimicrobial activity. Antimicrob Agents Chemother 59:5925–5931. doi: 10.1128/AAC.00808-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diaz Iglesias Y, Van Bambeke F. 2020. Activity of antibiotics against Pseudomonas aeruginosa in an in vitro model of biofilms in the context of cystic fibrosis: influence of the culture medium. Antimicrob Agents Chemother 64:e02204-19. doi: 10.1128/AAC.02204-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kandari D, Joshi H, Bhatnagar R. 2021. Zur: zinc-sensing transcriptional regulator in a diverse set of bacterial species. Pathogens 10:344. doi: 10.3390/pathogens10030344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ducret V, Abdou M, Goncalves MC, Leoni S, Martin-Pelaud O, Sandoz A, Segovia CI, Tercier-Waeber M-L, Valentini M, Perron K. 2021. Global analysis of the zinc homeostasis network in Pseudomonas aeruginosa and its gene expression dynamics. Front Microbiol 12:739988. doi: 10.3389/fmicb.2021.739988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lhospice S, Gomez NO, Ouerdane L, Brutesco C, Ghssein G, Hajjar C, Liratni A, Wang S, Richaud P, Bleves S, Ball G, Borezée-Durant E, Lobinski R, Pignol D, Arnoux P, Voulhoux R. 2017. Pseudomonas aeruginosa zinc uptake in chelating environment is primarily mediated by the metallophore pseudopaline. Sci Rep 7:17132. doi: 10.1038/s41598-017-16765-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schalk IJ, Cunrath O. 2016. An overview of the biological metal uptake pathways in Pseudomonas aeruginosa. Environ Microbiol 18:3227–3246. doi: 10.1111/1462-2920.13525. [DOI] [PubMed] [Google Scholar]
- 21.Pederick VG, Eijkelkamp BA, Begg SL, Ween MP, McAllister LJ, Paton JC, McDevitt CA. 2015. ZnuA and zinc homeostasis in Pseudomonas aeruginosa. Sci Rep 5:13139. doi: 10.1038/srep13139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haas CE, Rodionov DA, Kropat J, Malasarn D, Merchant SS, de Crécy-Lagard V. 2009. A subset of the diverse COG0523 family of putative metal chaperones is linked to zinc homeostasis in all kingdoms of life. BMC Genomics 10:470. doi: 10.1186/1471-2164-10-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamal SM, Rybtke ML, Nimtz M, Sperlein S, Giske C, Trček J, Deschamps J, Briandet R, Dini L, Jänsch L, Tolker-Nielsen T, Lee C, Römling U. 2019. Two FtsH proteases contribute to fitness and adaptation of Pseudomonas aeruginosa clone C strains. Front Microbiol 10:1372. doi: 10.3389/fmicb.2019.01372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heacock-Kang Y, Sun Z, Zarzycki-Siek J, Poonsuk K, McMillan IA, Chuanchuen R, Hoang TT. 2018. Two regulators, PA3898 and PA2100, modulate the Pseudomonas aeruginosa multidrug resistance MexAB-OprM and EmrAB efflux pumps and biofilm formation. Antimicrob Agents Chemother 62:e01459-18. doi: 10.1128/AAC.01459-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hare NJ, Scott NE, Shin EHH, Connolly AM, Larsen MR, Palmisano G, Cordwell SJ. 2011. Proteomics of the oxidative stress response induced by hydrogen peroxide and paraquat reveals a novel AhpC-like protein in Pseudomonas aeruginosa. Proteomics 11:3056–3069. doi: 10.1002/pmic.201000807. [DOI] [PubMed] [Google Scholar]
- 26.Landini G, Di Maggio T, Sergio F, Docquier J-D, Rossolini GM, Pallecchi L. 2016. Effect of high N-acetylcysteine concentrations on antibiotic activity against a large collection of respiratory pathogens. Antimicrob Agents Chemother 60:7513–7517. doi: 10.1128/AAC.01334-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossi E, Falcone M, Molin S, Johansen HK. 2018. High-resolution in situ transcriptomics of Pseudomonas aeruginosa unveils genotype independent patho-phenotypes in cystic fibrosis lungs. Nat Commun 9:3459. doi: 10.1038/s41467-018-05944-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borrero-de Acuña JM, Rohde M, Wissing J, Jänsch L, Schobert M, Molinari G, Timmis KN, Jahn M, Jahn D. 2016. Protein network of the Pseudomonas aeruginosa denitrification apparatus. J Bacteriol 198:1401–1413. doi: 10.1128/JB.00055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolpen M, Appeldorff CF, Brandt S, Mousavi N, Kragh KN, Aydogan S, Uppal HA, Bjarnsholt T, Ciofu O, Høiby N, Jensen PØ. 2016. Increased bactericidal activity of colistin on Pseudomonas aeruginosa biofilms in anaerobic conditions. Pathog Dis 74:ftv086. doi: 10.1093/femspd/ftv086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thöming JG, Tomasch J, Preusse M, Koska M, Grahl N, Pohl S, Willger SD, Kaever V, Müsken M, Häussler S. 2020. Parallel evolutionary paths to produce more than one Pseudomonas aeruginosa biofilm phenotype. NPJ Biofilms Microbiomes 6:2. doi: 10.1038/s41522-019-0113-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morita Y, Cao L, Gould VC, Avison MB, Poole K. 2006. nalD encodes a second repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa. J Bacteriol 188:8649–8654. doi: 10.1128/JB.01342-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim CK, Hassan KA, Penesyan A, Loper JE, Paulsen IT. 2013. The effect of zinc limitation on the transcriptome of Pseudomonas protegens Pf-5. Environ Microbiol 15:702–715. doi: 10.1111/j.1462-2920.2012.02849.x. [DOI] [PubMed] [Google Scholar]
- 33.Kolpen M, Kragh KN, Bjarnsholt T, Line L, Hansen CR, Dalbøge CS, Hansen N, Kühl M, Høiby N, Jensen PØ. 2015. Denitrification by cystic fibrosis pathogens—Stenotrophomonas maltophilia is dormant in sputum. Int J Med Microbiol 305:1–10. doi: 10.1016/j.ijmm.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 34.Pompilio A, Crocetta V, Pomponio S, Fiscarelli E, Di Bonaventura G. 2015. In vitro activity of colistin against biofilm by Pseudomonas aeruginosa is significantly improved under “cystic fibrosis-like” physicochemical conditions. Diagn Microbiol Infect Dis 82:318–325. doi: 10.1016/j.diagmicrobio.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 35.Brochmann RP, Toft A, Ciofu O, Briales A, Kolpen M, Hempel C, Bjarnsholt T, Høiby N, Jensen PØ. 2014. Bactericidal effect of colistin on planktonic Pseudomonas aeruginosa is independent of hydroxyl radical formation. Int J Antimicrob Agents 43:140–147. doi: 10.1016/j.ijantimicag.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 36.Bouteiller M, Dupont C, Bourigault Y, Latour X, Barbey C, Konto-Ghiorghi Y, Merieau A. 2021. Pseudomonas flagella: generalities and specificities. Int J Mol Sci 22:3337. doi: 10.3390/ijms22073337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mastropasqua MC, Lamont I, Martin LW, Reid DW, D'Orazio M, Battistoni A. 2018. Efficient zinc uptake is critical for the ability of Pseudomonas aeruginosa to express virulence traits and colonize the human lung. J Trace Elem Med Biol 48:74–80. doi: 10.1016/j.jtemb.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 38.Clinical and Laboratory Standards Institute. 2018. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: M07-A1111. Committee for Clinical Laboratory Standards, Wayne, PA. [Google Scholar]
- 39.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jolley KA, Bray JE, Maiden MCJ. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thrane SW, Taylor VL, Lund O, Lam JS, Jelsbak L. 2016. Application of whole-genome sequencing data for O-specific antigen analysis and in silico serotyping of Pseudomonas aeruginosa isolates. J Clin Microbiol 54:1782–1788. doi: 10.1128/JCM.00349-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Di Pilato V, Aiezza N, Viaggi V, Antonelli A, Principe L, Giani T, Luzzaro F, Rossolini GM. 2020. KPC-53, a KPC-3 variant of clinical origin associated with reduced susceptibility to ceftazidime-avibactam. Antimicrob Agents Chemother 65:e01429-20. doi: 10.1128/AAC.01429-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirchner S, Fothergill JL, Wright EA, James CE, Mowat E, Winstanley C. 2012. Use of artificial sputum medium to test antibiotic efficacy against Pseudomonas aeruginosa in conditions more relevant to the cystic fibrosis lung. J Vis Exp doi: 10.3791/3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harrison JJ, Stremick CA, Turner RJ, Allan ND, Olson ME, Ceri H. 2010. Microtiter susceptibility testing of microbes growing on peg lids: a miniaturized biofilm model for high-throughput screening. Nat Protoc 5:1236–1254. doi: 10.1038/nprot.2010.71. [DOI] [PubMed] [Google Scholar]
- 45.National Committee for Clinical Laboratory Standards, Barry AL. 1999. Methods for determining bactericidal activity of antimicrobial agents: approved guideline. National Committee for Clinical Laboratory Standards, Wayne, PA. [Google Scholar]
- 46.Rashid MH, Kornberg A. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci USA 97:4885–4890. doi: 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Høiby N, Molin S. 2012. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol 10:841–851. doi: 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete genome sequence of P. aeruginosa Z154 was deposited in GenBank under accession no. CP069177. RNA-seq data were also deposited in the NCBI Gene Expression Omnibus (GEO) database under accession no. GSE190946.