Abstract
Ralstonia eutropha (formerly Alcaligenes eutrophus) TF93 is pleiotropically affected in the translocation of redox enzymes synthesized with an N-terminal signal peptide bearing a twin arginine (S/T-R-R-X-F-L-K) motif. Immunoblot analyses showed that the catalytic subunits of the membrane-bound [NiFe] hydrogenase (MBH) and the molybdenum cofactor-binding periplasmic nitrate reductase (Nap) are mislocalized to the cytoplasm and to the inner membrane, respectively. Moreover, physiological studies showed that the copper-containing nitrous oxide reductase (NosZ) was also not translocated to the periplasm in strain TF93. The cellular localization of enzymes exported by the general secretion system was unaffected. The translocation-arrested MBH and Nap proteins were enzymatically active, suggesting that twin-arginine signal peptide-dependent redox enzymes may have their cofactors inserted prior to transmembrane export. The periplasmic destination of MBH, Nap, and NosZ was restored by heterologous expression of Azotobacter chroococcum tatA mobilized into TF93. tatA encodes a bacterial Hcf106-like protein, a component of a novel protein transport system that has been characterized in thylakoids and shown to translocate folded proteins across the membrane.
Periplasmic enzymes binding redox cofactors play a central role in alternative energy metabolism of gram-negative bacteria. In contrast to cytochrome c-type proteins, various periplasmic enzymes binding constituents, such as the molybdenum cofactor, a [NiFe] site, copper centers, or iron-sulfur clusters, contain a conserved, positively charged -S/T-RRXFLK- (twin-arginine) element within their N-terminal signal peptides, pointing to a special translocation pathway (2). Very recently, a system responsible for the membrane targeting and translocation of [NiFe] hydrogenases, as well as the molybdenum enzymes dimethylsulfoxide reductase, trimethylamine N-oxide reductase, and periplasmic nitrate reductase, has been characterized for Escherichia coli (32, 43). Mutant analyses suggested that the translocation of the twin-arginine signal peptide-bearing enzymes proceeds independently of the general secretion machinery of the cell (Sec), presumably via intermediates which have their cofactors inserted (31).
Ralstonia eutropha (formerly Alcaligenes eutrophus [7]) is the host of at least three periplasmic cofactor-containing enzymes synthesized with an N-terminal twin-arginine signal peptide: the membrane-bound hydrogenase (MBH) (involved in energy generation from H2), the periplasmic nitrate reductase (Nap) (reduces nitrate to nitrite), and the nitrous oxide reductase (NosZ) (a component of the denitrification pathway). All three enzymes are encoded in R. eutropha H16 by megaplasmid-borne genes.
The MBH of R. eutropha is a member of the [NiFe] hydrogenases (15) composed of a heterodimer (HoxKG) attached to the periplasmic surface of the inner membrane by a cytochrome b-type anchor protein (HoxZ) (4, 13). Hydrogen is activated at the [NiFe] site of HoxG (62 kDa), and the electrons are transferred via three iron-sulfur clusters within HoxK (35 kDa) to the physiological electron acceptor HoxZ (4). The small subunit HoxK contains a twin-arginine signal peptide. A deletion in this region blocks the membrane targeting of the MBH dimer and leads to the accumulation of inactive HoxG protein in the cytoplasm (3). The N-terminal amino acid sequence of the mature HoxG protein is colinear with the sequence predicted from the nucleotide sequence (17), indicating the absence of an export-triggering signal peptide at the N terminus. In contrast, HoxG contains a peptide extension of 15 amino acids (aa) at the C terminus, which is removed by a specific protease during [NiFe] cofactor assembly and plays a role in metal insertion (3). From these results, it was concluded that the two MBH subunits are cotranslocated in a tandem fashion and that this process is directed by the twin-arginine signal peptide-bearing small subunit. This conclusion gained support by a recent report on hydrogenase-2 of E. coli (27).
The periplasmic nitrate reductase, Nap, belongs to a large family of respiratory nitrate reductases, which appears to participate in denitrification, at least in some organisms (1, 28). Nap has been isolated from R. eutropha as a heterodimeric enzyme consisting of a 90-kDa subunit (NapA) and a 17-kDa subunit (NapB). NapA carries the catalytic site and exhibits sequence similarity with molybdopterin guanine dinucleotide (MGD) binding polypeptides of bacterial assimilatory nitrate reductases and formate dehydrogenases, both of which bind an iron-sulfur cluster at the N terminus (35). In fact, crystal structure analyses of NapA from Desulfovibrio desulfuricans show two MGD moieties per polypeptide and a [4Fe-4S] cluster (10). Comparison of the N-terminal amino acid sequence of the mature R. eutropha NapA subunit, as determined by Edman degradation, with the predicted primary structure identified a 29-aa twin-arginine signal peptide in NapA. NapB, which contains two binding sites for heme c, is synthesized with an N-terminal signal peptide resembling those required for translocation by Sec (35).
The periplasmic, copper-dependent nitrous oxide reductase, NosZ, is a key enzyme of denitrification which converts nitrous oxide to molecular dinitrogen. The primary structures of NosZ from various bacteria, including R. eutropha, are highly conserved and are all characterized by an unusually long N-terminal twin-arginine signal peptide of 45 to 56 aa (11, 47).
In this study, we reexamined the H2-oxidizing R. eutropha TF93 (ATCC 17697) strain, which had been reported in early studies to form a membrane-type of hydrogenase which occurred in the soluble fraction of the cell (14). We show that due to a missing or nonfunctional chromosomal factor, the strain is affected not only in membrane targeting of the twin-arginine signal peptide-bearing [NiFe] hydrogenase but also in the translocation of the periplasmic nitrate reductase and the nitrous oxide reductase. The mislocalized MBH and Nap proteins were active with artificial electron acceptors and donors, supporting the interpretation that these metalloenzymes have their cofactors inserted prior to translocation.
MATERIALS AND METHODS
Strains and plasmids.
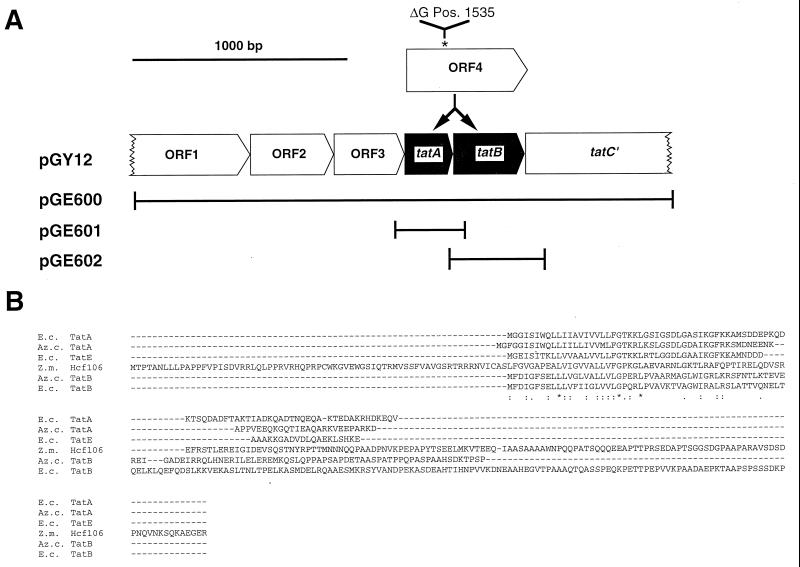
Bacterial strains and plasmids are listed in Table 1. Plasmid pGE600 is a derivative of the mobilizable vector pGE151, carrying the 2.7-kb BamHI/HindIII fragment of pGY12. DNA fragments extending from nucleotides 1268 to 1620 and from 1555 to 2006 of the Azotobacter chroococcum tat region (GenBank accession no. ACU48408) were synthesized by PCR, using plasmid pGY12 as a template. Amplification of tatA with primer pair 5′-ACCCAAGCTTGCGGAAAAGGCCGCACGGCC-3′ and 5′-CGGGATCCAGCAGGAGTTCGCTGAAGCCG-3′ produced a 366-bp fragment containing an additional HindIII and BamHI site. All the PCR amplifications were carried out with Pfu DNA polymerase (Stratagene). The resulting fragment was cut with HindIII and BamHI and ligated into pBluescript SK(+) (pCH800). In the same way, the primers 5′-ACCCAAGCTTGCAAGGTCGAGGAACCGGCCAGG-3′ and 5′-CGGGATCCCTGCGCAGCAGGCGCGAGCGC-3′ were used for amplification of a 468-bp fragment containing tatB and ligated into pBluescript SK(+) (pCH801). The identity of the subcloned PCR fragments was verified by nucleotide sequence determination. The HindIII/BamHI fragments containing either tatA or tatB were excised from the pBluescript derivatives and cloned into plasmid pGE151, yielding plasmids pGE601 and pGE602, respectively. For nucleotide sequence determination of the fusion in the tat region (see Fig. 6A), the native 1,638-bp SphI fragment of pGY12 was cloned into pUC18, yielding pCH802. E. coli phoA was PCR amplified on a 1.4-kb fragment with the primers 5′-TACATATGAAACAAAGCACTATTGCACTGG-3′ and 5′-TATCTAGATTATTTCAGCCCCAGAGCGGC-3′, with total DNA as the template. The PCR product was cut by NdeI and XbaI and cloned downstream of the strong R. eutropha promoter of the soluble hydrogenase into the broad-host-range plasmid pEDY309 as previously described (23), yielding pGE603.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| R. eutropha | ||
| H16 | Wild type, pHG1 | ATCC 17699 |
| HF345 | ΔhoxM; isogenic mutant of H16 | 3 |
| HF359 | ΔhoxG; isogenic mutant of H16 | 3 |
| HF405 | ΔhoxZ; isogenic mutant of H16 | 3 |
| TF93 | Wild type, pHG2 | ATCC 17697 |
| TF100 | Megaplasmid-free derivative of TF93; Nap negative | 14 |
| TF140 | Derivative of TF93 harboring pHG1 instead of pHG2 | B. Friedrich (Humboldt-University Berlin) |
| E. coli | ||
| S17-1 | Tra+; recA pro thi hsdR chr::RP4-2 | 36 |
| NovaBlue(DE3) | endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 lac[F′ proA+B+ lac1qZΔM15::Tn10] | Novagen |
| Plasmids | ||
| pBluescript II SK(+) | Apr, lacZ′, T7φ10 promoter, f1 ori | Stratagene |
| pEDY309 | Tcr, pRK290 replicon with multiple cloning site | 23 |
| pGE144 | Derivative of pVK101 harboring the nap gene region | 35 |
| pGE151 | Derivative of pRK404, containing Plac | 17 |
| pGY12 | 2.7-kb HindIII/BamHI fragment harboring tatABC′ of A. chroococcum | 45 |
| pGE600 | 2.7-kb HindIII/BamHI fragment of pGY12 in pGE151 | This work |
| pGE601 | 366-bp HindIII/BamHI fragment with tatA from pGY12 in pGE151 | This work |
| pGE602 | 468-bp HindIII/BamHI fragment with tatB from pGY12 in pGE151 | This work |
| pGE603 | 1.4-kb NdeI/XbaI fragment with E. coli phoA in pEDY309 | This work |
| pCH800 | 366-bp HindIII/BamHI fragment with tatA from pGY12 in pBluescript SK(+) | This work |
| pCH801 | 468-bp HindII/BamHI fragment with tatB from pGY12 in pBluescript SK(+) | This work |
| pCH802 | 1.6-kb SphI fragment of pGY12 in pUC18 | This work |
FIG. 6.
The tat locus on pGY12 of A. chroococcum according to GenBank accession no. ACU48404, with the nucleotide sequence correction. (A) The newly annotated genes are in black. The fragments used for complementation analysis are shown below with bars, and the corresponding plasmid designations are given at the left. (B) Alignment of the deduced A. chroococcum TatA and TatB amino acid sequences, with Hcf106 and Hcf106 analogs from E. coli. Az. c., A. chroococcum; E. c., E. coli; Z. m., Zea mays. Perfect matches are indicated by asterisks and high and low similarities are indicated by double and single dots, respectively.
Media and growth conditions.
Lithotrophic cultures of R. eutropha strains were grown in mineral salts medium under an atmosphere of hydrogen, carbon dioxide, and oxygen (8:1:1 [vol/vol/vol]) supplemented with 0.8 μM NiCl2 in place of the standard trace element mixture SL6 (12). Synthetic media for aerobic heterotrophic growth, in order to optimally express Nap, contained 0.4% (wt/vol) gluconate or 0.4% (wt/vol) fructose and SL6 as described previously (42). Anaerobic heterotrophic growth, at the expense of 0.2% (vol/vol) nitrate (denitrification), was performed in medium containing fructose as previously described (30). Strains of E. coli were grown in Luria-Bertani medium (20). Solid media contained 1.5% (wt/vol) agar. Antibiotics were added as appropriate for R. eutropha (tetracycline, 12.5 μg/ml) and for E. coli (tetracycline, 12.5 μg/ml; ampicillin, 80 μg/ml).
Conjugative plasmid transfer.
Mobilizable plasmids were transferred from E. coli S17-1 to R. eutropha by a spot mating technique (36). Transconjugants were selected on mineral medium plates containing the appropriate antibiotic under lithotrophic growth conditions.
DNA techniques.
Standard techniques were used in this study (29). Plasmid DNA isolation was carried out by the alkaline lysis procedure and ion-exchange chromatography according to the manufacturer's instructions (QIAGEN Inc.). DNA and PCR fragments used in plasmid constructions were isolated from agarose gels by QiaEx (QIAGEN Inc.). Nucleotide sequence determination of pCH802 was done by the dideoxy chain termination method and by cycle sequencing with sequence-derived fluorescence-labelled primers and the thermostable sequenase kit (Amersham Pharmacia Biotech) in an automatic sequencing device, as recommended by the manufacturer (LICOR).
In vivo expression of tatAB gene products.
Expression of tatA and tatB from plasmids pCH800 and pCH801 was under control of the phage T7 φ10 promoter. The plasmids were transformed into strain NovaBlue(DE3), which carries a chromosomally encoded T7 polymerase. Synthesis of the tat-encoded gene products was induced by IPTG (isopropyl-β-d-thiogalactopyranoside) and labelling with [35S]methionine followed the procedure previously described (38).
Isolation of subcellular fractions.
Subcellular fractions were prepared according to a method described by Bernhard et al. (4), with modifications. R. eutropha cells (100 ml) were grown in the presence of oxygen either under lithotrophic conditions for 36 h or heterotrophically on gluconate for 24 to 48 h and then harvested by centrifugation (4,000 × g, 4°C). Cells were washed with 10 ml of 10 mM Tris-HCl, pH 7.5 (4,000 × g, 4°C), and resuspended in 5 ml of 10 mM Tris-HCl, pH 7.8, containing 0.5 M sucrose. After a 10-min incubation at 30°C in the presence of 1 mM EDTA, the incubation was continued at room temperature for 30 min with lysozyme (10 mg/g [wet weight] of cells). The suspension was centrifuged (4,000 × g; 20 min; 4°C), and the supernatant contained the periplasmic fraction. The spheroplasts were washed and lysed by osmotic shock with 5 ml of 10 mM Tris-HCl, pH 7.8. Cell debris was removed (5,000 × g, 15 min, 4°C), and the membrane and cytoplasmic fractions were obtained by ultracentrifugation (88,000 × g, 45 min, 4°C). Membranes used for the detection of the MBH were washed three times and suspended in 50 mM KPO4, pH 7.0. Membranes used for the detection of NapA were resuspended in 250 mM NaKPO4, pH 6.5. The periplasmic, cytoplasmic, and membrane fractions were used directly or stored at −20°C until used for enzyme assays, activity staining, and immunoblotting analysis. The quality of the subcellular fractions of the lithotrophically grown cells was monitored by examining the distribution of the cytoplasmic, NAD-reducing hydrogenase activity of the soluble hydrogenase (SH) of R. eutropha and immunoblotting with SH-specific antibodies, as previously described (4). The purity of subcellular fractions of heterotrophically grown cells was controlled by immunoblotting with antiserum raised against the cytoplasmic marker flavohemoglobin of R. eutropha (9).
Immunoblot analysis.
Proteins were resolved by electrophoresis in sodium dodecyl sulfate (SDS)–10% or 12% (wt/vol) polyacrylamide gels and were transferred to nitrocellulose membranes, and the immunoblot analysis was done according to a standard protocol (41). Specific proteins were detected with polyclonal rabbit antisera and an alkaline phosphatase-labelled goat anti-rabbit immunoglobulin G (Jackson Immuno Research Laboratories). The proteins were applied at the following dilutions: anti-flavohemoglobin (1:5,000), anti-HoxH (1:10,000), anti-HoxG (1:2,000), anti-PhoA (1:1,000; 5 Prime → 3 Prime, Inc.), anti-nitrite reductase (1:10,000) (30), and anti-NapA (1:1,000).
Analytical procedures.
SH (hydrogen:NAD+ oxidoreductase [EC 1.12.1.2]) activity was assayed by spectrophotometric determination of H2-dependent NAD reduction (14). MBH (ferredoxin:H+ oxidoreductase [EC 1.18.99.1]) activity was determined according to a previously described method (37), with modifications. Hydrogenase activities of membranes were determined in N2-saturated 50 mM KPO4, pH 7.0. Soluble extracts at pH 5.5 were measured with 0.5 mM methylene blue and 86 μM H2. One unit of hydrogenase activity was the amount of enzyme which catalyzed the consumption of 1 μmol of substrate per min. Nap reductase activity was determined in subcellular fractions obtained from aerobically grown cells (42) exploiting the formate-dependent nitrate reduction (35). One unit of Nap activity was the amount of enzyme which catalyzed the formation of 1 μmol of nitrite per min. Nitrite concentration was determined colorometrically at 546 nm (18).
In-gel chromogenic detection of hydrogenase activity after native polyacrylamide gel electrophoresis was done according to a previously described method (4).
c-Type cytochromes were detected after SDS-polyacrylamide gel electrophoresis of subcellular fractions and specific staining for covalently attached heme, as previously described (39).
Protein determination of cells and subcellular fractions was done by the method of Lowry et al. (19).
Determination of nitrous oxide and dinitrogen by gas chromatography was done as previously described (9).
RESULTS AND DISCUSSION
The membrane-bound hydrogenase of R. eutropha TF93 is mislocalized to the cytoplasm.
The majority of R. eutropha strains form two enzymes for energy conservation from molecular hydrogen: a cytoplasmic NAD-dependent SH and an MBH. Both proteins have been shown to be also present in the natural isolate R. eutropha TF93; however, the MBH was identified in the soluble fraction instead of in the membrane. Mislocalization of the enzyme could not be restored by a megaplasmid exchange using a donor which synthesized the MBH properly attached to the membrane (14).
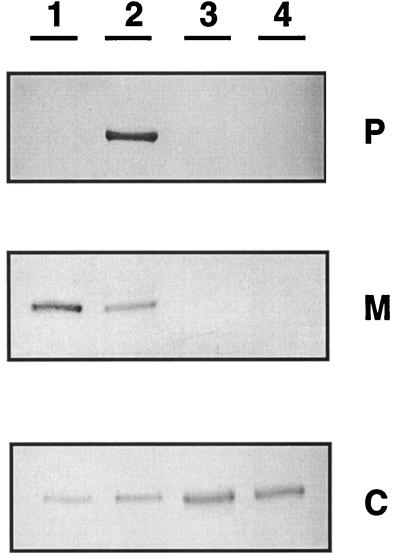
To determine the precise cellular localization of the MBH dimer, we examined periplasmic, membrane, and cytoplasmic fractions of autotrophically grown TF93 cells by immunoblot analysis using antiserum raised against the large MBH subunit (HoxG) of R. eutropha H16. HoxG of TF93 was exclusively found in the cytoplasm (Fig. 1, lane 3), unlike the MBH of strain H16, which occurred predominantly in the membrane fraction and only in trace amounts in the cytoplasm (Fig. 1, lane 1). Export of the MBH to the periplasm of TF93 can be excluded on the basis of the release of the MBH into the periplasm by a mutant of H16 impaired in the membrane anchor HoxZ (Fig. 1, lane 2) (4). These results unambiguously show that the MBH of TF93 is restricted to the cytoplasm, which was also the case in a transconjugant, TF140, harboring the megaplasmid pHG1 of R. eutropha H16 (Fig. 1, lane 4). This confirmed that no mutations in megaplasmid genes are responsible for the mislocalization of the MBH in TF93, but pointed to a defective or missing chromosomally encoded factor which is required for the proper targeting of the MBH to the membrane. Since pHG1 of strain H16 is better characterized than the native plasmid pHG2 of TF93, subsequent experiments were done with TF140 (Table 1).
FIG. 1.
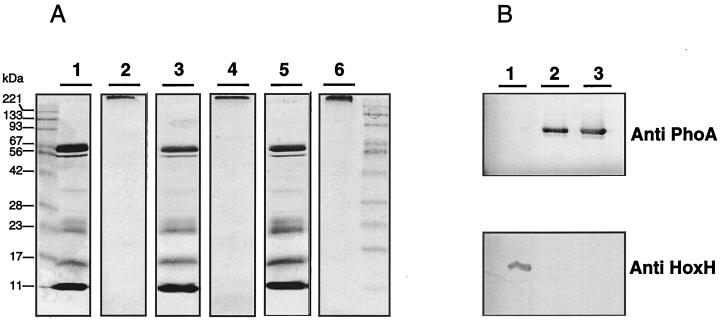
Cellular localization of the [NiFe] center-bearing subunit (HoxG) of MBH in strains of R. eutropha. Equal amounts of protein (25 μg) were loaded onto the gel except for the periplasmic fractions (100 μl each), because they contained high concentrations of lysozyme protein. The subcellular fractions are indicated: P, periplasm; M, membrane; C, cytoplasm. The strains tested are as follows: lane 1, H16 (harboring pHG1); lane 2, HF405, carrying ΔhoxZ in pHG1; lane 3, TF93 (harboring pHG2); lane 4, TF140 (harboring pHG1).
R. eutropha TF93 is pleiotropically deficient in translocation of redox enzymes.
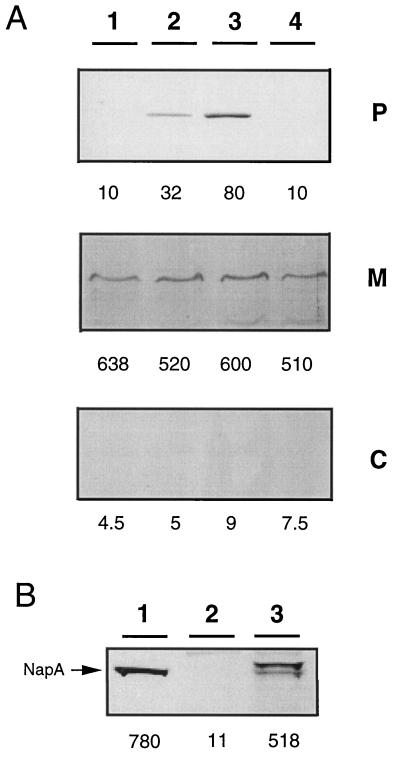
To examine whether the deficiency of TF93 plays a more general role in the export of cofactor-containing enzymes, we investigated the localization of Nap and of NosZ in comparison to proteins which are translocated via the general secretion system. Subcellular fractions from TF140 cells grown heterotrophically in synthetic medium in presence of oxygen were analyzed for Nap. Immunoblots with polyclonal NapA antiserum showed that the catalytic subunit NapA was trapped in the membrane, whereas both the periplasm and the cytoplasm were almost free of any cross-reacting material (Fig. 2, lane 1). This clearly pointed to a pleiotropic nature of the export deficiency in TF140. Identical results were obtained with the parental TF93 strain (data not shown).
FIG. 2.
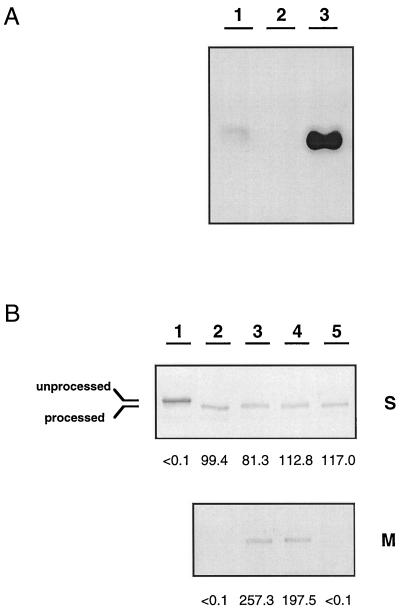
Mislocalization of NapA in TF140 and restoration of NapA export by tat genes (A) and specificity of the NapA detection system (B). The subcellular fractions are indicated: P, periplasm; M, membrane; C, cytoplasm. Nap activity (milliunits per milligram of protein) is given below the immunoblot. Note, the Nap activity in the periplasm is given as milliunits per gram (wet weight) of cells. (A) Lane 1, TF140; lane 2, TF140 harboring tatAB on pGE600; lane 3, TF140 harboring tatA on pGE601; lane 4, TF140 harboring tatB on pGE602. (B) Lane 1, partially purified NapA (arrow); lane 2, membrane of TF100 cells; lane 3, membrane of TF100 cells harboring the complete nap genes on pGE144 (35).
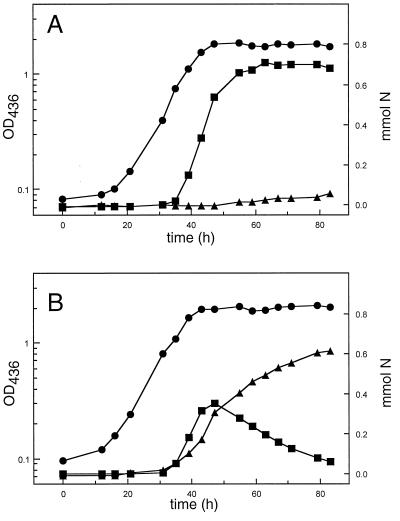
The second twin-arginine signal peptide-bearing redox enzyme, NosZ, was investigated on the physiological level. Cells of TF140 were grown anaerobically under denitrifying conditions with nitrate as the electron acceptor. Nitrous oxide production and its conversion to molecular dinitrogen were monitored by gas chromatography. Figure 3A shows that TF140 accumulated nitrous oxide as the final product, indicating that NosZ was incapable of converting nitrous oxide. Physiologically active periplasmic NosZ of R. eutropha, however, readily converts nitrous oxide, which only transiently accumulates in the gas phase (30). The result is compatible with the conclusion that the mislocalization of NosZ in TF140 impairs its physiological function. Indeed, mutational analysis of the NosZ system from Pseudomonas stutzeri has shown that the export to the periplasm is a prerequisite of the protein to be physiologically active (11). Due to the lack of an appropriate NosZ antibody, it was not possible to investigate the cellular localization of the enzyme. Attempts to use heterologous antiserum raised against NosZ by P. stutzeri (47) were not successful. Nevertheless, the results provide an excellent explanation for the failure of R. eutropha TF93 to produce dinitrogen gas during anaerobic nitrate respiration (44). Furthermore, the data demonstrate that the export deficiency in TF93 has an enormous physiological impact on the cells since it blocks the function of at least three independent redox systems.
FIG. 3.
Blocked nitrous oxide reduction in R. eutropha TF140 (A), which was restored by the expression of A. chroococcum tatA on pGE601 (B). Growth was performed anaerobically on fructose containing 10 mM nitrate, and the gaseous denitrification products were monitored by gas chromatography. ●, growth of R. eutropha culture at 436 nm; ■, nitrous oxide; ▴, dinitrogen. OD, optical density.
To definitely show that the translocation of twin-arginine signal peptide-dependent redox enzymes is specifically blocked and that the translocation of proteins across the cytoplasmic membrane is not generally impaired in R. eutropha TF93, the distribution of cytochrome c-type proteins of this strain was tested. Periplasmic and cytoplasmic fractions obtained from cells of TF140 grown aerobically and anaerobically were subjected to heme c staining (Fig. 4). Heme c-type proteins were identified exclusively in the periplasm; they were absent in the cytoplasmic fraction (Fig. 4A, lanes 1 and 2). Furthermore, immunoblot analyses confirmed the periplasmic localization of the heme cd1-containing nitrite reductase (data not shown) and also of the alkaline phosphatase (PhoA), which is a well-known substrate of the general secretory pathway (Sec) in E. coli (25). When heterologously expressed from plasmid pGE603, E. coli PhoA is exported to the periplasmic space of TF140 in the same manner as in an H16 derivative, as expected (Fig. 4B, lanes 2 and 3, top panel). Purity controls eliminated the possibility of a contamination by cytoplasmic proteins (Fig. 4B, bottom panel).
FIG. 4.
Periplasmic c-type cytochromes (A) and export of heterologously expressed E. coli PhoA to the periplasm (B) in R. eutropha strains. (A) Cytochrome c was visualized by heme staining of periplasmic (lanes 1, 3, and 5) and cytoplasmic (lanes 2, 4, and 6; loaded with 100 μg of protein each) fractions of TF140 cells grown anaerobically on nitrate. The proteins were separated on SDS–15% (vol/vol) polyacrylamide gels. Lanes 1 and 2, TF140; lanes 3 and 4, TF140 harboring tatAB on pGE600; lanes 5 and 6, TF140 harboring tatA on pGE601. Prestained protein markers are on the left and right of the stain. The nature of the staining at the border of the stacking and the running gel in the cytoplasmic fractions (lanes 2, 4, and 6) is unclear. (B) Immunoblot analysis of the periplasmic fractions of autotrophically grown R. eutropha strains with antiserum directed against E. coli PhoA (upper panel) and a purity control with antiserum raised against the SH protein (anti-HoxH) as a cytoplasmic marker (4) (lower panel). Lane 1, 1 μg of purified SH; lane 2, periplasm of TF140 expressing a copy of E. coli phoA on pGE603; lane 3, periplasm of H16 derivative (HF405) expressing a copy of E. coli phoA on pGE603.
MBH and Nap are enzymatically active in the translocation-arrested state.
In the process of cytochrome c maturation, the polypeptide and the prosthetic group heme c are translocated separately (reference 40 and references therein). Thus, neither preapocytochromes nor holocytochrome c containing the cofactor essential for the catalytic activity is detectable when export is blocked. Immunochemical studies showed that cofactor-free apoforms are unstable (21, 22).
The detection of export-blocked MBH and Nap prompted us to examine whether both are also enzymatically active. MBH activity was visualized by in-gel H2-dependent phenazine methosulfate reduction and showed a strong activity staining in the cytoplasmic compartment of TF140 (Fig. 5A, lane 3). For H16, only a faint stain corresponding to traces of anti-MBH material detected in the cytoplasm was observed (Fig. 1 and 5A, lane 1). No staining appeared in an H16 derivative which is devoid of the [NiFe]-containing subunit (HF359; ΔhoxG) (Fig. 5A, lane 2). This demonstrated that translocation-arrested MBH accumulates in its catalytically active form in the cytoplasm of TF140. Further immunoblot analyses showed that the MBH is processed in TF140, compared to a mutant in which the protease gene had been deleted (HF345; ΔhoxM) (Fig. 5B, compare lanes 1 and 2). This confirms that for metal center assembly, the mislocalized MBH undergoes the same proteolytic processing at the C terminus of the [NiFe]-containing subunit in TF140 as that documented for the H16 enzyme (4). These results are in agreement with the previous finding of the partially purified active MBH from the soluble fraction of R. eutropha TF93 (24). Nitrate reductase assays showed that the NapA protein trapped in the membrane of TF140 is also catalytically active (Fig. 2, lane 1). Only basal nitrate-reducing activities were found in the periplasmic and cytoplasmic fractions (Fig. 2, lane 1). The nitrate reductase activity measured is exclusively due to Nap and not due to the respiratory membrane-bound nitrate reductase, which is known to be expressed anaerobically only when nitrate serves as the alternative electron acceptor (42). Figure 2B shows that a Nap-negative variant of TF93 formed no anti-NapA reacting material, and in consequence, no Nap activity was trapped in the membrane of the aerobically grown cells. However, NapA-specific protein and the corresponding enzyme activity were detectable again upon complementation of the mutant nap strain by the nap genes residing on pGE144 (Fig. 2B, compare lanes 2 and 3). In summary, the results demonstrate that MBH and NapA have their respective cofactors incorporated in an identical manner appropriate for the physiological situation despite their translocation being blocked.
FIG. 5.
In-gel detection of MBH-dependent hydrogenase activity in the cytoplasmic fraction of TF140 (A) and the restoration of MBH targeting to the membrane by tat genes (B). (A) Lane 1, H16; lane 2, HF359, defective in the [NiFe]-containing subunit of MBH (ΔhoxG in pHG1); lane 3, TF140. (B) Analysis of restoration of MBH translocation was carried out by immunoblot analysis. S, the soluble extract consisting of the periplasmic and cytoplasmic fractions; M, the membrane fraction. MBH activity (milliunits per milligram of protein) is given below the blot. Lane 1, HF345, defective in the MBH-specific protease (ΔhoxM in pHG1); lane 2, TF140; lane 3, TF140 with tatAB (pGE600); lane 4, TF140 with tatA (pGE601); lane 5, TF140 with tatB (pGE602).
The pretranslocational fashion of cofactor insertion accompanied with folding of the preproteins precludes translocation by the Sec system (25). In fact, it has been shown recently that the twin-arginine signal peptide-dependent translocation of the trimethylamine N-oxide reductase proceeds independently of the Sec pathway in E. coli (31).
A bacterial analog of the thylakoid Hcf106 restores translocation of mislocalized redox enzymes in R. eutropha.
Interestingly, a class of plant proteins shares the twin-arginine element (2, 5, 8), which can direct these proteins in the folded state across the thylakoid membrane in a Sec-independent manner (16). The import to the lumen of thylakoids is equivalent to export across the inner membrane of bacterial cells. A component of this twin-arginine-protein transport system, Hcf106 (30 kDa), was first identified in maize, and proteins with local similarities have been deduced also from gene data banks and from sequenced genomes of bacteria and archaea (33, 34). First attempts to identify and clone a homologous gene of R. eutropha by virtue of the local similarities of the primary structures of bacterial Hcf106-like proteins were unsuccessful.
Hence, we used the orf4 gene from A. chroococcum (45), whose product had been determined to contain local similarities to Hcf106 of maize (34) for heterologous complementation. A. chroococcum contains a membrane-bound H2-oxidizing enzyme which is highly related to the R. eutropha system (15). It has been shown that orf4 is able to complement an A. chroococcum mutant with a membrane-bound hydrogenase activity misassembled to the soluble fraction, but the molecular basis remains to be elucidated (45). To express the A. chroococcum orf4 gene we cloned the 2.7-kb BamHI/HindIII fragment of pGY12 into the broad-host-range vector pGE151, yielding pGE600. In the course of our studies, we uncovered (by sequence alignment studies) that the orf4 gene product represents a fusion of two Hcf106-like proteins (Fig. 6). Nucleotide sequence determination of orf4 on plasmid pGY12 (Fig. 6A) resulted in the elimination of a guanine at position 1535 of the sequence (GenBank accession no. ACU48404), yielding two new orf genes, tatA and tatB (Fig. 6), which encode separate proteins as confirmed by Tabor expression (data not shown). The primary structure of both products showed local similarities to Hcf106 (Fig. 6B). Moreover, the newly annotated A. chroococcum gene products showed significant similarities to the products of the tat system, recently identified in the E. coli genome (32, 43). Since the product of the incomplete orf5 gene, immediately downstream of tatA and tatB, also showed significant primary sequence identity to a product of the tat system of E. coli, the respective A. chroococcum gene was designated tatC′ (Fig. 6). tatA and tatB were subcloned individually into pGE151 under the control of the lac promoter, resulting in plasmids pGE601 and pGE602, respectively. Upon mobilization of the cloned A. chroococcum genes tatAB (pGE600), tatA (pGE601), and tatB (pGE602) into strain TF140, the subcellular fractions of the resulting transconjugants were tested for the localization of MBH and of NapA. Figure 5B shows that the tatAB- and tatA-harboring derivatives targeted the MBH correctly to the membrane and restored hydrogenase activity in this fraction. Likewise, NapA was correctly exported in the tatAB- and tatA-harboring cells, accompanied by the occurrence of Nap activity in the periplasm (Fig. 2). Moreover, expression of A. chroococcum tatA (on pGE600 and pGE601) restored nitrous oxide reduction in TF140; the resulting transconjugants accumulated dinitrogen in the gas phase, as illustrated representatively for TF140 harboring a copy of tatA (Fig. 3B). Transfer of A. chroococcum tatB alone (on pGE602) did not restore any deficiency observed in TF140 (Fig. 2, 3, and 5). In contrast, the expression of the A. chroococcum tat genes had no effect at all on the export of cytochrome c-type proteins (Fig. 4A). These results strongly point to a lesion in a TatA-like protein in R. eutropha TF93 which appears to be required for the translocation of folded twin-arginine signal peptide-bearing redox enzymes.
Although we cannot exclude the possibility that the folding constraints for MBH and Nap in export-blocked TF93 are different from the export-competent cells, the results presented could be interpreted in favor of a cytoplasmic assembly rather than a periplasmic assembly pathway for twin-arginine signal peptide-bearing redox enzymes, as previously proposed (2). This interpretation also gains support by studies indicating that metal insertion has to take place before the molybdenum-dependent dimethylsulfoxide reductase from Rhodobacter sphaeroides f. sp. denitrificans (46) and hydrogenase-2 from E. coli (26) are translocated. Both enzymes are examples of bacterial redox proteins carrying the unusual signal peptide (2).
Hcf106-like proteins in targeting and translocation of bacterial enzymes binding different redox cofactors.
Very recently it was shown that E. coli contains a Sec-independent transport apparatus required for the membrane targeting and translocation of twin-arginine signal peptide-bearing enzymes (31, 32, 43). The novel system, Tat (twin-arginine translocator, formerly called Mtt [membrane targeting and translocation]), is encoded by the tatABCD operon and the unlinked tatE gene (32). Although controversial, it is now apparent that the E. coli Tat system comprises at least three gene products regarded as Hcf106-like proteins, designated TatA (11 kDa), TatB (18 kDa), and TatE (13 kDa) (Fig. 6) (32, 33; A. Chanal, C. Santini, and L. F. Wu, Letter, Mol. Microbiol. 30:674–676, 1998). Like the corresponding gene products identified in A. chroococcum, which are 8 (TatA) and 12 (TatB) kDa in size, they are predicted to have a very similar N-terminal membrane-spanning domain, followed by an amphipathic helix (Fig. 6B). The fourth component identified in E. coli is predicted to constitute a membrane-integral protein, TatC (29 kDa), and forms the essential core component of the translocation system (6). Although we have not yet identified any of the corresponding genes in R. eutropha, successful transcomplementation with a copy of tatA suggests that a homolog or analog to TatC could be functional in this organism. We report that A. chroococcum TatA promoted the translocation of three basically different twin-arginine signal peptide-proteins carrying the cofactors [NiFe], MGD, and polynuclear copper sites. Our observation that TatA recognizes a broad range of proteins is consistent with the very first analysis of bacterial Hcf106-like proteins in E. coli. Mutational analysis has suggested that they can compensate for each other in the translocation of molybdoenzymes and [NiFe] hydrogenases to a certain extent, depending on the enzyme studied (32, 43). One may speculate that the Hcf106 analogs function in concert as a membrane-bound receptor complex and thus respond to variations in the tertiary and oligomeric structures of the different Tat substrates. The question remains whether the Hcf106 analogs are sufficient to select, proofread, and guide the various metalloproteins through a core component. From this point of view, it is noteworthy that the formation of physiologically active Tat-dependent enzymes requires individual sets of auxiliary proteins. We have shown that eight accessory genes are involved in the energy generation by the MBH in R. eutropha. However, the functions of six of these gene products are still unknown (3). Even much less is known about accessory genes of the nap cluster and the nos locus of R. eutropha. Work is in progress to elucidate whether those accessory gene products assist the coordination of cofactor insertion and Tat-mediated translocation.
ACKNOWLEDGMENTS
We gratefully thank Geoffrey Yates for plasmid pGY12. The work of Hubert Schröder to provide NapA-specific antiserum is highly acknowledged. We thank Ursula Stegert and Christine Reinemann for technical assistance and Edward Schwartz for critically reading the manuscript.
The work was supported by grants from the Deutsche Forschungsgemeinschaft (to R.A.S. and B.F.) and the Fonds der Chemischen Industrie (to B.F.).
REFERENCES
- 1.Bedzyk L, Wang T, Ye R W. The periplasmic nitrate reductase in Pseudomonas sp. strain G-179 catalyzes the first step of denitrification. J Bacteriol. 1999;181:2802–2806. doi: 10.1128/jb.181.9.2802-2806.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berks B C. A common export pathway for proteins binding complex redox cofactors? Mol Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 3.Bernhard M, Schwartz E, Rietdorf J, Friedrich B. The Alcaligenes eutrophus membrane-bound hydrogenase gene locus encodes functions involved in maturation and electron transport coupling. J Bacteriol. 1996;178:4522–4529. doi: 10.1128/jb.178.15.4522-4529.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernhard M, Benelli B, Hochkoeppler A, Zannoni D, Friedrich B. Functional and structural role of the cytochrome b subunit of the membrane-bound hydrogenase complex of Alcaligenes eutrophus H16. Eur J Biochem. 1997;248:179–186. doi: 10.1111/j.1432-1033.1997.00179.x. [DOI] [PubMed] [Google Scholar]
- 5.Bogsch E, Brink S, Robinson C. Pathway specificity for a ΔpH-dependent precursor thylakoid lumen protein is governed by a ‘Sec-avoidance’ motif in the transfer peptide and a ‘Sec-incompatible’ mature protein. EMBO J. 1997;16:3851–3859. doi: 10.1093/emboj/16.13.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogsch E G, Sargent F, Stanley N R, Berks B C, Robinson C, Palmer T. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J Biol Chem. 1998;273:18003–18006. doi: 10.1074/jbc.273.29.18003. [DOI] [PubMed] [Google Scholar]
- 7.Brim H, Heyndrickx M, de Vos P, Wilmotte A, Springael D, Schlegel H G, Mergeay M. Amplified rDNA restriction analysis and further genotypic characterisation of metal-resistant soil bacteria and related facultative hydrogenotrophs. Syst Appl Microbiol. 1999;22:258–268. doi: 10.1016/S0723-2020(99)80073-3. [DOI] [PubMed] [Google Scholar]
- 8.Chaddock A M, Mant A, Karnauchov I, Brink S, Herrmann R G, Klösgen R B, Robinson C. A new type of signal peptide: central role of a twin-arginine motif in transfer signals for the ΔpH-dependent thylakoidal protein translocase. EMBO J. 1995;14:2715–2722. doi: 10.1002/j.1460-2075.1995.tb07272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cramm R, Siddiqui R A, Friedrich B. Primary sequence and evidence for a physiological function of the flavohemoprotein of Alcaligenes eutrophus. J Biol Chem. 1994;269:7349–7354. [PubMed] [Google Scholar]
- 10.Dias J M, Than M E, Humm A, Huber R, Bourenkov G P, Bartunik H D, Bursakov S, Calvete J, Caldeira J, Carneiro C, Moura J J, Moura I, Romao M J. Crystal structure of the first dissimilatory nitrate reductase at 1.9 Å solved by MAD methods. Structure. 1999;7:65–79. doi: 10.1016/s0969-2126(99)80010-0. [DOI] [PubMed] [Google Scholar]
- 11.Dreusch A, Burgisser D M, Heizmann C W, Zumft W G. Lack of copper insertion into unprocessed cytoplasmic nitrous oxide reductase generated by an R20D substitution in the arginine consensus motif of the signal peptide. Biochim Biophys Acta. 1997;1319:311–318. doi: 10.1016/s0005-2728(96)00174-0. [DOI] [PubMed] [Google Scholar]
- 12.Eberz G, Friedrich B. Three trans-acting regulatory functions control hydrogenase synthesis in Alcaligenes eutrophus. J Bacteriol. 1991;173:1845–1854. doi: 10.1128/jb.173.6.1845-1854.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eismann K, Mlejnek K, Zipprich D, Hoppert M, Gerberding H, Mayer F. Antigenic determinants of the membrane-bound hydrogenase in Alcaligenes eutrophus are exposed toward the periplasm. J Bacteriol. 1995;177:6309–6312. doi: 10.1128/jb.177.21.6309-6312.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedrich B, Hogrefe C, Schlegel H G. Naturally occurring genetic transfer of hydrogen-oxidizing ability between strains of Alcaligenes eutrophus. J Bacteriol. 1981;147:198–205. doi: 10.1128/jb.147.1.198-205.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedrich B, Schwartz E. Molecular biology of hydrogen utilization in aerobic chemolithotrophs. Annu Rev Microbiol. 1993;47:351–383. doi: 10.1146/annurev.mi.47.100193.002031. [DOI] [PubMed] [Google Scholar]
- 16.Hynds P J, Robinson D, Robinson C. The Sec-independent twin-arginine translocation system can transport both tightly folded and malfolded proteins across the thylakoid membrane. J Biol Chem. 1998;273:34868–34874. doi: 10.1074/jbc.273.52.34868. [DOI] [PubMed] [Google Scholar]
- 17.Kortlüke C, Horstmann K, Schwartz E, Rohde M, Binsack R, Friedrich B. A gene complex coding for the membrane-bound hydrogenase of Alcaligenes eutrophus H16. J Bacteriol. 1992;174:6277–6289. doi: 10.1128/jb.174.19.6277-6289.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowe R H, Evans H J. Preparation and some properties of soluble nitrate reductase from Rhizobium japonicum. Biochim Biophys Acta. 1964;85:377–389. doi: 10.1016/0926-6569(64)90301-3. [DOI] [PubMed] [Google Scholar]
- 19.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 20.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 21.Page M D, Ferguson S J. A bacterial c-type cytochrome can be translocated to the periplasm as an apo form; the biosynthesis of cytochrome cd1 (nitrite reductase) from Paracoccus denitrificans. Mol Microbiol. 1989;3:653–661. doi: 10.1111/j.1365-2958.1989.tb00213.x. [DOI] [PubMed] [Google Scholar]
- 22.Page M D, Ferguson S J. Apo forms of cytochrome c550 and cytochrome cd1 are translocated to the periplasm of Paracoccus denitrificans in the absence of haem incorporation caused either mutation or inhibition of haem synthesis. Mol Microbiol. 1990;4:1181–1192. doi: 10.1111/j.1365-2958.1990.tb00693.x. [DOI] [PubMed] [Google Scholar]
- 23.Pierik A, Schmelz M, Lenz O, Friedrich B, Albracht S P J. Characterization of the active site of a hydrogen sensor from Alcaligenes eutrophus. FEBS Lett. 1998;438:231–235. doi: 10.1016/s0014-5793(98)01306-4. [DOI] [PubMed] [Google Scholar]
- 24.Podzuweit H G, Schneider K, Knüttel H. Comparison of the membrane-bound hydrogenases from Alcaligenes eutrophus H16 and Alcaligenes eutrophus type strain. Biochim Biophys Acta. 1987;905:435–446. doi: 10.1016/0005-2736(87)90473-1. [DOI] [PubMed] [Google Scholar]
- 25.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodrigue A, Boxer D H, Mandrand-Berthelot M A, Wu L-F. Requirement for nickel of the transmembrane translocation of NiFe-hydrogenase 2 in Escherichia coli. FEBS Lett. 1996;392:81–86. doi: 10.1016/0014-5793(96)00788-0. [DOI] [PubMed] [Google Scholar]
- 27.Rodrigue A, Chanal A, Beck K, Müller M, Wu L-F. Co-translocation of a periplasmic enzyme complex by a hitchhiker mechanism through the bacterial tat pathway. J Biol Chem. 1999;274:13223–13228. doi: 10.1074/jbc.274.19.13223. [DOI] [PubMed] [Google Scholar]
- 28.Sabaty M, Gagnon J, Vermeglio A. Induction by nitrate of cytoplasmic and periplasmic proteins in the photodenitrifier Rhodobacter sphaeroides forma sp. denitrificans under anaerobic or aerobic condition. Arch Microbiol. 1994;162:335–343. doi: 10.1007/BF00263781. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Sann R, Kostka S, Friedrich B. A cytochrome cd1-type nitrite reductase mediates the first step of denitrification in Alcaligenes eutrophus. Arch Microbiol. 1994;161:453–459. doi: 10.1007/BF00307765. [DOI] [PubMed] [Google Scholar]
- 31.Santini C L, Ize B, Chanal A, Müller M, Giordano G, Wu L-F. A novel Sec-independent periplasmic protein translocation pathway in Escherichia coli. EMBO J. 1998;17:101–112. doi: 10.1093/emboj/17.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sargent F, Bogsch E G, Stanley N R, Wexler M, Robinson C, Berks B C, Palmer T. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 1998;17:3640–3650. doi: 10.1093/emboj/17.13.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Settles A M, Martienssen R. Old and new pathways of protein export in chloroplasts and bacteria. Trends Cell Biol. 1998;8:494–501. doi: 10.1016/s0962-8924(98)01387-7. [DOI] [PubMed] [Google Scholar]
- 34.Settles A M, Yonetani A, Baron A, Bush D R, Cline K, Martienssen R. Sec-independent protein translocation by the maize Hcf106 protein. Science. 1997;278:1467–1470. doi: 10.1126/science.278.5342.1467. [DOI] [PubMed] [Google Scholar]
- 35.Siddiqui R A, Warnecke-Eberz U, Hengsberger A, Schneider B, Kostka S, Friedrich B. Structure and function of a periplasmic nitrate reductase in Alcaligenes eutrophus H16. J Bacteriol. 1993;175:5867–5876. doi: 10.1128/jb.175.18.5867-5876.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:748–791. [Google Scholar]
- 37.Sweet W J, Houchins J P, Rosen P R, Arp D J. Polarographic measurement of H2 in aqueous solutions. Anal Biochem. 1980;107:337–340. doi: 10.1016/0003-2697(80)90393-0. [DOI] [PubMed] [Google Scholar]
- 38.Tabor S, Richardson C C. A bacteriophage T7 polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas P E, Ryan D, Levin W. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal Biochem. 1976;75:168–176. doi: 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- 40.Thöny-Meyer L. Biogenesis of respiratory cytochromes in bacteria. Microbiol Mol Biol Rev. 1997;61:337–376. doi: 10.1128/mmbr.61.3.337-376.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warnecke-Eberz U, Friedrich B. Three nitrate reductase activities in Alcaligenes eutrophus. Arch Microbiol. 1993;159:405–409. [Google Scholar]
- 43.Weiner J H, Bilous P T, Shaw G M, Lubitz S P, Frost L, Thomas G H, Cole J A, Turner R J. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell. 1998;93:93–101. doi: 10.1016/s0092-8674(00)81149-6. [DOI] [PubMed] [Google Scholar]
- 44.Yabuuchi E, Kosako Y, Yano I, Hotta H, Nishiuchi Y. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal of Ralstonia pickettii (Ralston, Palleroni and Doudoroff 1973) comb. nov., Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol Immunol. 1995;39:897–904. doi: 10.1111/j.1348-0421.1995.tb03275.x. [DOI] [PubMed] [Google Scholar]
- 45.Yates M G, De Souza E M, Kahindi J H. Oxygen, hydrogen and nitrogen fixation in Azotobacter. Soil Biol Biochem. 1997;29:863–869. [Google Scholar]
- 46.Yoshida Y, Takai M, Satoh T, Takami S. Molybdenum requirement for translocation to the periplasmic space in a photodenitrifier, Rhodobacter sphaeroides f. sp. denitrificans. J Bacteriol. 1991;173:3277–3281. doi: 10.1128/jb.173.11.3277-3281.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zumft W G, Dreusch A, Löchelt S, Cuypers H, Friedrich B, Schneider B. Derived amino acid sequences of the nosZ gene (respiratory N2O reductase) from Alcaligenes eutrophus, Pseudomonas aeruginosa and Pseudomonas stutzeri reveal potential copper-binding residues. Implications for the CuA site of N2O reductase and cytochrome-c oxidase. Eur J Biochem. 1992;208:31–40. doi: 10.1111/j.1432-1033.1992.tb17156.x. [DOI] [PubMed] [Google Scholar]