Abstract
We have previously shown that both ends of the Tn3 family transposon Tn4652 contain integration host factor (IHF) binding sites and that IHF positively regulates expression of the Tn4652 transposase gene tnpA in Pseudomonas putida (R. Hõrak, and M. Kivisaar, J. Bacteriol. 180:2822–2829, 1998). Tn4652 can activate silent genes by creating fusion promoters during the transposition. The promoters are created as fusions between the −35 hexamer provided by the terminal inverted repeats of Tn4652 and the −10 hexamers in the target DNA. Two fusion promoters, PRA1 and PLA1, that contain sequences of the right and left termini of Tn4652, respectively, were chosen for the study of mechanisms of transcription activation. Gel mobility shift analysis using crude extracts from P. putida cells allowed us to detect specific binding of P. putida IHF to the ends of the transposon Tn4652. We found that the rate of transcription from the fusion promoter PRA1 is enhanced by IHF. Notably, the positive effect of IHF on transcription from the promoter PRA1 appeared only when cells of P. putida reached the stationary growth phase. We speculate that the intracellular concentration of IHF might be critical for the in vivo effect of IHF on transcription from the fusion promoters in P. putida. In the case of PLA1, the mechanism of transcription modulation by IHF is different than that observed for PRA1. Our results demonstrate that transcription of neighboring genes from outwardly directed promoters at the ends of a mobile DNA element could be influenced by the same factors that control transposition of the element.
Integration host factor (IHF) induces a sharp bend of DNA with an overall angle of 160 to 180° at its binding site (14, 41). This small heterodimeric protein is involved in a diverse set of cellular functions, including the transposition of mobile DNA elements and the regulation of gene expression (16). In many of these processes, IHF has been shown to act as an architectural element that facilitates the establishment of DNA-protein complexes. IHF influences the transcription of genes both positively and negatively (15, 21). No uniform mechanism for IHF action on IHF-regulated promoters has been found. In the activation of ς54 promoters, IHF bends DNA to bring together RNA polymerase bound to the core promoter and activator protein bound to a distal enhancer (reviewed in references 21 and 38). The architectural role of IHF in gene expression is also demonstrated in the activation of ς70-dependent promoters. For example, in the case of the ilvpG promoter, IHF activates transcription by forming a higher order protein-DNA complex in the upstream region that structurally alters the DNA helix in a way that facilitates open complex formation at the downstream promoter site (37). In the lambda pL promoter and the Mu Pe promoter, IHF was shown to have a direct role in promoting binding of the carboxy-terminal domain of the α-subunit (α-CTD) of RNA polymerase to the UP-element-like sequence (18, 19, 44).
Several mobile DNA elements possess functional outwardly directed −35 hexamers of ς70 promoters in terminal inverted repeats, and when transposed at the correct distance from a resident −10 hexamer, new promoters capable of activating transcription of neighboring genes can be created (reviewed in reference 31). Galas and Chandler (17) have suggested that the formation of fusion promoters is a relatively common event associated with transposition. As shown for the 2,4,5-trichlorophenoxyacetic acid catabolic genes of Burkholderia cepacia, creation of a constitutive promoter would be especially important in the early steps of evolution of catabolic pathways and in horizontal transfer of genes when operon regulation may not function (27).
To reduce the deleterious mutagenic effect of high transposition activity on the host cell, transposition activity in cells is generally maintained at a low level and is controlled by host factors (3, 31). In addition to binding sites for the element-encoded transposase, binding sites for host-specified proteins are also often found within or close to terminal inverted repeats of mobile DNA elements. This indicates that transcription of neighboring genes from outwardly directed promoters at the ends of mobile DNA elements could be influenced by the same factors that control transposition of the element.
Transposon Tn4652 is a 17-kb-long DNA element derived from TOL plasmid pWW0, and it locates chromosomally in Pseudomonas putida PaW85 (42). Tn4652 belongs to the Tn3 family of transposons (43). We have previously shown that expression of the transposase gene tnpA of Tn4652 is positively affected by IHF (26). Because both ends of Tn4652 contain IHF binding sites, we have suggested that besides activation of the tnpA promoter, IHF may also participate in transposition of Tn4652 (26). Our recent studies demonstrated that promoters for the transcription of initially promoterless phenol degradation genes pheBA were created as a result of base substitutions, deletions, and transposition of Tn4652 when cells of P. putida PaW85 were selected for growth on phenol minimal plates (28, 36). Sequence analysis and mapping of the transcription start point of the pheBA operon in hybrid plasmids containing insertions of Tn4652 from the chromosome of PaW85 revealed that fusions between the −10 sequences present in the pheBA operon and the −35 sequence located in the terminal repeats of Tn4652 had generated functional promoters (36). Five of the six different fusion promoters identified were created at the junctions of the right terminus of Tn4652 and the target DNA, and in only one particular case was the left end sequence of the transposon involved. In this study, we investigated mechanisms of transcriptional activation from the fusion promoters. Two promoters, PLA1 and PRA1, containing sequences of the left and right termini of Tn4652, respectively, were chosen for more detailed examination (Fig. 1). We showed IHF-dependent positive effect on transcription from these promoters. Binding of P. putida IHF to Tn4652 terminal sequences was demonstrated by in vitro experiments using the gel shift assay. The possible mechanisms for IHF-mediated modulation of transcription at the ends of Tn4652 will be discussed.
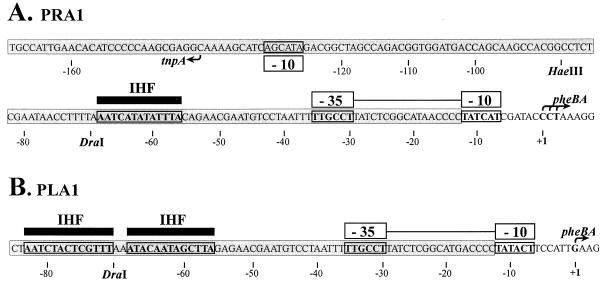
FIG. 1.
Nucleotide sequence of the fusion promoters PRA1 (A) and PLA1 (B). Fusions between −35 hexamer provided by the inverted repeats of Tn4652 and −10 hexamers found in the target DNA upstream of the pheBA genes created these promoters (36). The sequences of Tn4652 are shaded. Locations of −10 and −35 hexamers of the fusion promoters are indicated above the sequence. The upstream region of the promoter PRA1 overlaps with the oppositely directed promoter region of the Tn4652 transposase gene tnpA. Location of −10 hexamer of the tnpA promoter is shown below the sequence of the right end of Tn4652. Transcription start sites for the promoters, determined by primer extension experiments, are indicated by arrows. The potential IHF binding sites at the ends of Tn4652 resembling the E. coli IHF binding consensus sequence WATCAANNNNTTR are indicated above the sequences of the promoters PRA1 and PLA1 by black bars. Restriction sites relevant to the experiments presented in this paper are shown.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are described in Table 1. Escherichia coli TG1 (7) was used as a host for DNA cloning. The construction of plasmids pRA1, pRA1-7, pRA1-12, pLA1, pLA1-4, pLA1-5, and pLA1-12 is depicted in Fig. 2. For the construction of P. putida RT31 carrying the P. putida genes ihfA and ihfB under the control of Ptac promoter and lacIq repressor in its chromosome, the following DNA manipulations were performed. The ihfA gene was amplified by PCR from the chromosomal DNA of P. putida PaW85 (2) by using the primers pihfA1 5′-ACAAAGCTT(HindIII)GAACACCAACGTTAAGGAAAT-3′, complementary to nucleotides (nt) −35 to −7 relative to the ATG start codon of the ihfA, and pihfA2 5′-TCCGAATTC(EcoRI)CGCAGCACGTGTGGCTTTAC-3′, complementary to nt 67 to 95 downstream of the TAA stop codon of the ihfA. The amplified DNA fragment was cloned into EcoRI and HindIII sites in pBluescript SK vector. The gene ihfB, originating from plasmid pHip (6), was cloned as a 600-bp XbaI-SmaI fragment downstream of the ihfA gene. Thereafter, the ihfA and ihfB genes were inserted as a XbaI-HindIII fragment into plasmid pMMB208 (34) containing Ptac promoter and lacIq gene. Subsequently, a Ptac-ihfAB-lacIq cassette was inserted into pUC18 Not. The cleavage of pUT mini-Tn5 luxAB (11) by NotI enabled us to delete the luxAB genes from this plasmid and to clone the Ptac-ihfAB-lacIq cassette as NotI DNA fragment into mini-Tn5 to obtain plasmid pUTtetPF. Mating between E. coli S17-1 λpir (33) carrying the plasmid pUTtetPF and the ihfA knockout mutant P. putida A8759 (6), and selection of kanamycin-tetracycline-resistant P. putida transconjugants containing chromosomal insertion of Ptac-ihfAB-lacIq cassette plus the lacIq gene was carried out as previously described (10). The presence of this cassette in the chromosome of P. putida RT31 was verified by PCR by using the primers specific to the 5′ end of the ihfA and the 3′ end of the ihfB.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or construction | Source or reference |
|---|---|---|
| E. coli | ||
| TG1 | supE hsdΔ thiΔ (lac-proAB) F′ | 7 |
| WM2015 | subE thi Δ(lac-pro) | 30 |
| WM2017 | WM2015 himA::TcrhimD::Cmr | 30 |
| S17-1 λ pir | Tpr SmrrecA thi pro (r− m+) RP4::2 Tc::Mu::Km Tn7 λ pir | 33 |
| P. putida | ||
| PaW85 | Tn4652 | 2 |
| KT2442 | Tn4652 xylRS Pu-lacZ Rifr Smr | 6 |
| A8759 | KT2442 Tn4652 ihfA::Kmr Pu-lacZ Rifr Smr | 6 |
| RT31 | A8759 P. putida ihfA and ihfB under control of Ptac promoter and lacIq repressor Rifr Smr Tcr | This work |
| Plasmids | ||
| pBluescript SK | Cloning vector (Apr) | Stratagene |
| pEST1332 | Plasmid pAYC32 carrying promoterless pheBA operon | 29 |
| pRA1 | 64-bp DraI-ClaI DNA fragment containing PRA1 promoter region cloned into pEST1332 | 36, Fig. 2 |
| pRA1-7 | 82-bp HaeIII-ClaI DNA fragment containing PRA1 promoter region cloned into pEST1332 | This work, Fig. 2 |
| pRA1-12 | 288-bp Eco47III-ClaI DNA fragment containing PRA1 promoter region cloned into pEST1332 | This work, Fig. 2 |
| pLA1 | 110-bp DraI-ClaI DNA fragment containing PLA1 promoter region cloned into pEST1332 | 36, Fig. 2 |
| pLA1-4 | 152-bp BstUI-ClaI fragment containing PLA1 promoter region cloned into pEST1332 | This work, Fig. 2 |
| pLA1-5 | 200-bp EcoRV-ClaI fragment containing PLA1 promoter region cloned into pEST1332 | This work, Fig. 2 |
| pLA1-12 | 387-bp HaeIII-ClaI fragment containing PLA1 promoter region cloned into pEST1332 | This work, Fig. 2 |
| pHip | Plasmid carrying E. coli ihfB | 6 |
| pMMB208 | lacIq/Ptac-based broad-host-range expression plasmid (Kmr) | 34 |
| pUC18 Not | Identical to pUC18 but with NotI sites in multicloning region (Apr) | 25 |
| pUTmini-Tn5luxAB | Delivery plasmid for mini-Tn5 luxAB (Apr Tcr) | 11 |
| pUTtetPF | pUTmini-Tn5luxAB carrying P. putida ihfA and ihfB Ptac promoter and lacIq | This work |
| pUCPu130 | 129-bp DpnI fragment of Pu promoter region of xyl genes in TOL plasmid cloned into pUC18 | 26 |
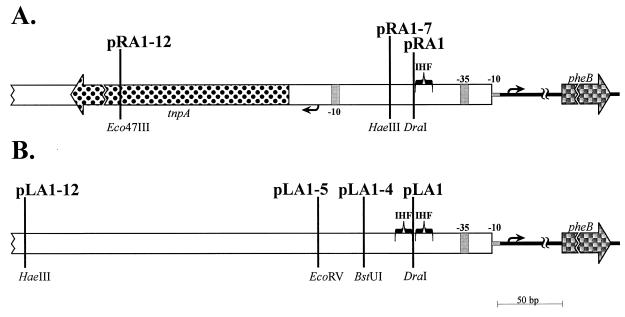
FIG. 2.
Schematic representation of PRA1 and PLA1 promoter constructs used in this study. Plasmids pRA1, pRA1-7, and pRA1-12 (A) carry the sequences of the right end of Tn4652 extending to the DraI, HaeIII, and Eco47III sites, respectively. Plasmids pLA1, pLA1-4, pLA1-5, and pLA1-12 (B) contain the sequences of the left end of Tn4652 sequences upstream of the fusion promoter PLA1 extending to the DraI, BstUI, EcoRV, and HaeIII sites, respectively. All the plasmids are promoter-pheBA fusions in pEST1332 (29). Plasmids pRA1 and PLA1 were constructed as in our previous study (36). The promoters were initially cloned into pBluescript SK, and multicloning sites SacI and ClaI were used in order to reclone them upstream of the pheBA genes in plasmid pEST1332. The large spotted arrow represents the tnpA gene of Tn4652, and the checked arrows represent the pheB gene. Open boxes designate noncoding sequences of Tn4652, and thick lines show noncoding sequences of plasmid pEST1332 locating between the reporter gene pheB and promoters PRA1 or PLA1. Grey regions indicate the locations of −10 and −35 elements of the promoters. The small arrows denote transcription start sites of the pheB gene and the tnpA gene. The IHF binding consensus-resembling sequences at the ends of Tn4652 are indicated by brackets.
Bacteria were grown on Luria-Bertani (LB) medium (32). P. putida was incubated at indicated final concentrations of antibiotics: carbenicillin, 1,500 μg/ml; kanamycin, 50 μg/ml; and tetracycline, 15 μg/ml. For E. coli, ampicillin at 100 μg/ml, kanamycin at 50 μg/ml, and tetracycline at 15 μg/ml were used. P. putida was incubated at 30°C, and E. coli was incubated at 37°C. E. coli was transformed with plasmid DNA as described by Hanahan (23), and P. putida was electroporated by using the protocol of Sharma and Schimke (41).
Enzyme assays.
Cells of P. putida strains harboring different plasmids were grown overnight in LB medium supplemented with appropriate antibiotics. Then the cultures were diluted into fresh LB medium to obtain the optical density at 580 nm of 0.01. Exponentially growing cells were sampled after 6 h, and stationary-phase cells were sampled after 16 h of cultivation. When necessary, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to growth media at indicated concentrations. The catechol 1,2-dioxygenase (C12O) assay was carried out as described by Hegeman (24). Protein concentration in cell lysates was measured by the Bradford method (4).
Gel mobility shift assay.
Cell lysates used in gel shift assays were prepared either from exponential- or stationary-phase 30-ml cultures. The cells were pelleted and sonicated in 1× binding buffer (25 mM Tris-HCl [pH 7.5]; 5 mM EDTA; 50 mM KCl; 25 mM NaCl; 5 mM dithiothreitol; 5% glycerol). The DNA fragments used in gel shift binding assays were generated by PCR as follows: (i) the primers TnR, 5′-CGTATCGATCAGCATAGACGGCTAGCCAG-3′, and TnotsSac, 5′-CGTGAGCTCGGGGTTATGCCGAGATAAGGC-3′ (complementary to nt 100 to 124 and to nt 1 to 21 relative to the right terminus of Tn4652), were used for amplification of the 140-bp DNA fragment containing the IHF binding site at the right end of Tn4652; (ii) the primers TnL, 5′-CGTAAGCTTCCTCAATGGATGGCTGAAG-3′ (complementary to nt 111 to 132 relative to the left terminus of Tn4652), and TnotsSac were used for amplification of the 140-bp DNA fragment containing the IHF binding site at the left end of Tn4652; and (iii) two primers, pAYC32, 5′-CTCGACCTTTGAGCCAAATG-3′, and ABC, 5′-GGGTATGCTTGGCAGTCGT-3′, complementary to sequences locating upstream and downstream of the SacI and ClaI cloning sites in plasmid pEST1332, respectively, were used to amplify the DNA regions of the fusion promoters lacking the A-T-rich sequences upstream of the IHF binding core sequences in the plasmids pRA1 and pLA1. The same primers were used to amplify the DNA regions of PRA1 and PLA1 in the presence of A-T-rich sequences upstream of these promoters in the plasmids pRA1-7 and pLA1-4. The DNA fragments were labeled with [γ-32P]dATP by using T4 polynucleotide kinase. The radiolabeled DNA fragments were purified by polyacrylamide gel electrophoresis. The binding reaction was carried out in a volume of 20 μl. DNA probes (500 cpm) were incubated at 23°C for 30 min with different cell lysates in 1× binding buffer containing 1 μg of bovine serum albumin and 2 μg of salmon sperm DNA. The specific nonlabeled competitor DNA containing the IHF binding site upstream of the Pu promoter of xyl genes in TOL plasmid was created by amplification of the 129-bp DpnI fragment of Pu promoter region cloned into pUC18 (26) by using pUC18 forward and reverse primers. When the specific competitor DNA was used, cell lysate was added last to the binding reaction. After incubation, the reaction mixture was loaded onto a 20-min-prerun 5% nondenaturing polyacrylamide gel. We wish to stress that gel electrophoresis conditions are critical for the detection of P. putida IHF complex in a gel. In our previous report, we could demonstrate binding of E. coli IHF to the ends of Tn4652 but failed to detect P. putida-IHF-dependent shift (26). In this study, we lowered the pH of the gel electrophoresis buffer as well as the gel running temperature. Electrophoresis was carried out at 4°C in 0.5× Tris-borate-EDTA buffer (pH 7.5) at 10 V/cm for 3 h. The gel was exposed to a phosphoimager screen. The IHF-bound DNA was quantified relative to the unbound DNA by using Phosphoimager (ImageQuant 4.2a software; Molecular Dynamics).
RESULTS
Level of transcription from the fusion promoter PRA1 is enhanced by DNA sequences of the right end of Tn4652 located upstream of the −35 hexamer of this promoter.
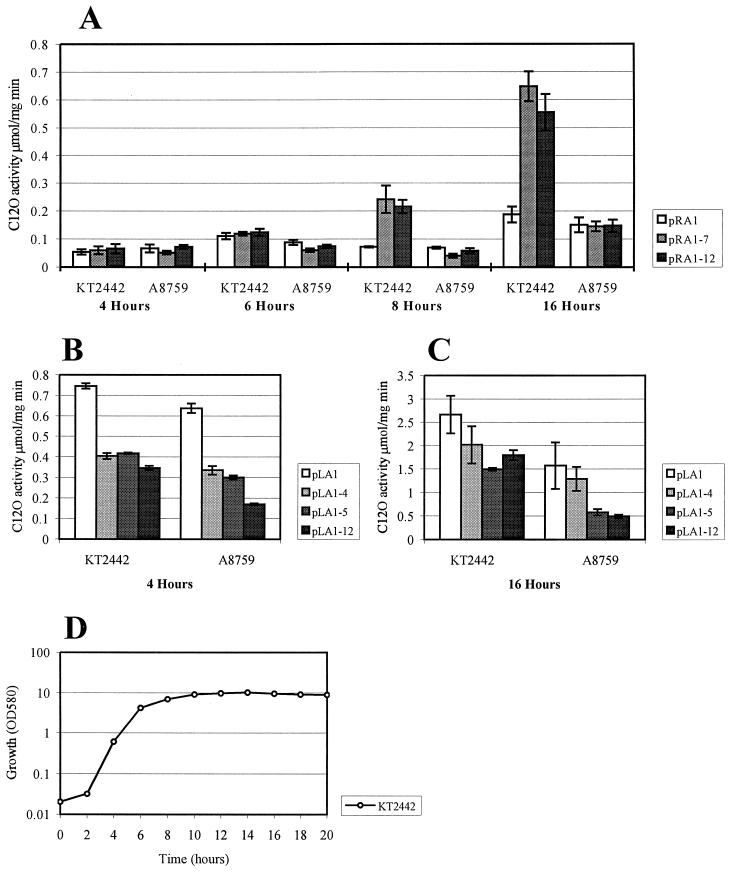
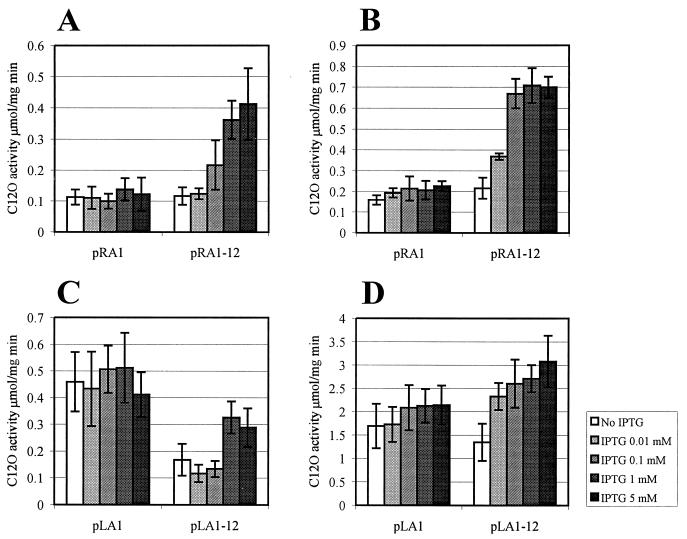
Phenol-degrading (Phe+) mutant clones carrying plasmids with functional promoters created upstream of the pheBA genes were picked up from phenol minimal plates (36). Interestingly, plasmid pRA1 containing the fusion promoter sequence PRA1 cloned from the original hybrid plasmid pEST1354 upstream of the pheBA genes in plasmid pEST1332 (36) did not provide growth of P. putida PaW85 cells on minimal medium containing phenol as the sole carbon and energy source. The plasmid pRA1 contained 57 nt of the right-end sequence of Tn4652, extending to the DraI cleavage site (up to nt −69 relative to the transcription start point of the promoter PRA1 [Fig. 1A and 2A]). This raised the question of whether, on phenol plates, the growth-supporting level of expression of the pheBA genes in the hybrid plasmid pEST1354 carrying the entire transposon (if compared to the plasmid pRA1) needs the presence of DNA sequences flanking the DraI site at the Tn4652 right end. To test the possible effect of upstream sequences on transcription from the fusion promoter PRA1, plasmids pRA1-12 and pRA1-7 were constructed (Fig. 2A). The plasmid pRA1-12 contained the transposon DNA extending to the Eco47III cleavage site located at position −292 upstream from the transcription start point of the promoter PRA1. Another construct, pRA1-7, contained the Tn4652 DNA extending to the HaeIII cleavage site located at nt −87 relative to the transcription initiation site of the promoter PRA1. We compared the level of expression of the pheB gene in P. putida KT2442 in the plasmids pRA1, pRA1-7, and pRA1-12. The level of expression of C12O activity in plasmids pRA1-7 and pRA1-12 was the same as in pEST1354 (data not shown). It turned out that both pRA1-7 and pRA1-12 expressed C12O activities at approximately threefold higher levels than pRA1 (Fig. 3A). Notably, the positive effect of the upstream sequences of the promoter PRA1 was dependent on the growth phase: it appeared when bacteria reached the stationary growth phase. The effect of growth phase of bacteria on transcription from the promoter PRA1 also became evident in the case of plasmid pRA1: the expression of C12O in stationary phase cells was 3.5-fold higher than in exponential cells (Fig. 3A).
FIG. 3.
Study of the effects of upstream sequences and IHF on transcription from the fusion promoters PRA1 (A) and PLA1 (B and C) by comparison of the levels of expression of C12O activity in the wild-type strain P. putida KT2442 and in its ihfA knockout derivative A8759. C12O activity was measured at different time points either in exponentially growing or stationary-phase cells of P. putida KT2442. Data (means ± standard deviations) of at least four independent experiments are presented. The growth curve of P. putida KT2442 in LB is shown (D). The growth rate of the strain A8759 is similar to that of KT2442 (not shown).
Analysis of the nucleotide sequence upstream of the fusion promoter PRA1 in the plasmids pRA1-7 and pRA1-12 did not reveal any sequence motifs exhibiting similarity to the ς70-recognized promoter consensus TTGACA-N16–18TATAAT. Also, no additional upstream-located transcriptional start sites of the reporter gene pheB were detected in primer extension analysis of the 5′ end of the pheB mRNA in P. putida PaW85 carrying pRA1-12 (data not shown). This indicated that the plasmids pRA1-7 and pRA1-12 do not contain any additional promoter sequences responsible for the higher level of transcription of the pheBA genes compared to that in the plasmid pRA1. Therefore, we speculated that the upstream-located sequences of Tn4652 could play a positive role in transcription enhancement due to binding some cellular factors and this suggested that protein-DNA interactions at this DNA region might influence transcription activation from the fusion promoter PRA1.
Study of the effect of IHF on transcriptional activation from the promoter PRA1.
In our earlier study, the IHF binding sites were localized by us at both ends of the transposon Tn4652 (26). The sequence AATCATATATTTA, which resembles the E. coli IHF binding consensus sequence WATCAANNNNTTR (where W = A/T and R = A/G) (8), locates at nt −68 to −56 relative to the transcription start point of the promoter PRA1 (Fig. 1A). Usually, the sequences that flank the 5′ end of this consensus are A-T rich. Several studies of IHF have shown that interruption of the sequence flanking the IHF binding motif strongly affects the binding of this protein (15). The DraI cleavage site in the Tn4652 right end locates just between the sequence resembling the IHF binding consensus and the A-T-rich DNA region. Since the presence of the Tn4652 right-end DNA sequence upstream from the DraI site was required for the transcription enhancement from the fusion promoter PRA1, the involvement of IHF in this effect was examined. P. putida A8759 is an ihfA knockout mutant of the strain P. putida KT2442 (6). The data in Fig. 3A show that the C12O activities measured in stationary-phase cells of the wild-type strain P. putida KT2442 bacteria harboring either the plasmid pRA1-7 or pRA1-12 were approximately three- to fourfold higher than in those containing pRA1. At the same time, differences were recorded when the ihfA knockout derivative P. putida A8759 was used as a host. Thus, the positive effect of the upstream sequences of the promoter PRA1 on transcription became evident only in P. putida cells containing the functional IHF.
Study of the effect of the upstream sequences on transcription from the promoter PLA1 that is formed at the junction between the left end of Tn4652 and the target DNA.
Two potential IHF binding sites locate at positions −83 to −71 and −68 to −56 from the transcriptional start point of PLA1 (Fig. 1B). We intended to study whether the effect of IHF on PLA1 could be similar to that found by us in the case of the promoter PRA1. The expression of the reporter gene pheB downstream from the fusion promoter PLA1 was measured in two plasmids, pLA1 and pLA1-12, that contained different lengths of the transposon DNA. pLA1 contained the Tn4652 left-end DNA up to the DraI cleavage site located at nt −71 relative to the transcription start site of the promoter (Fig. 1B and 2B). pLA12 included the left-end DNA of Tn4652 extending to the HaeIII site at nt −349 from the transcription start point of the promoter PLA1 (Fig. 2B). One of the potential IHF binding sites locating distantly from the promoter was deleted in the plasmid pLA1. pLA1 also lacked part of the A-T-rich DNA sequence upstream of the proximal potential IHF binding site. Results of the C12O activity measurements shown in Fig. 3B and C demonstrate the effect of the growth phase of bacteria on transcription of the reporter gene pheB from the promoter PLA1. The C12O activities measured in stationary-phase cells of P. putida KT2442 carrying either the plasmid pLA1 or pLA1-12 were approximately fourfold higher if compared to those observed in exponentially growing cells. In both cases, pLA1-12 exhibited a slightly lower level of expression of the pheBA genes than did pLA1. Moreover, pLA1-12 exhibited two- and 3.5-fold lower levels of C12O activity in IHF-deficient P. putida KT2442 derivative A8759 than in the wild-type strain, respectively, both in exponentially growing and stationary-phase cells (Fig. 3B and C). This indicated that IHF may somehow sequester the negative effect of upstream sequences on transcription from PLA1 in the case of pLA1-12. However, in stationary-phase cells, the level of expression from the promoter PLA1 in the plasmid pLA1 was also slightly lower in strain A8759 compared to wild-type strain KT2442 (Fig. 3C).
In order to localize the DNA regions responsible for negative effects on transcription from PLA1 more exactly, deletion analysis of the upstream region of this promoter was carried out. The plasmid pLA1-4 contained the upstream sequences extending to the BstUI cleavage site located at nt −113 relative to the transcription initiation site of the promoter PLA1. Another construct, pLA1-5, contained the Tn4652 DNA extending to the EcoRV site located at position nt −144 upstream from the transcription start point of the promoter PLA1 (Fig. 2). However, the results obtained with these plasmids indicated that modulation of the transcription from PLA1 in the presence of sequences located upstream of the DraI site seems to be more complex than that from PRA1. In the case of PRA1, the expression patterns of the reporter genes in pRA1-7 and pRA1-12 were identical, which indicates that the presence of Tn4652 sequences located upstream of the Eco47III site in plasmid pRA1-12 has no effect on transcription from this promoter. At the same time, the effects observed either in exponentially growing or in stationary-phase cells of the wild-type strain KT2442 and of the IHF-negative strain A8759 carrying either plasmids pLA1-4 or pLA1-5 were different. Although in stationary-phase A8759, pLA1-5 behaved similarly to pLA1-12 and pLA1-4 behaved similarly to pLA1, in exponentially grown cells, both pLA1-4 and pLA1-5 exhibited equal levels of expression of C12O. This expression level was higher than that of pLA1-12 but lower than that of pLA1 (Fig. 3B and C).
Binding of P. putida IHF to the ends of Tn4652 in vitro.
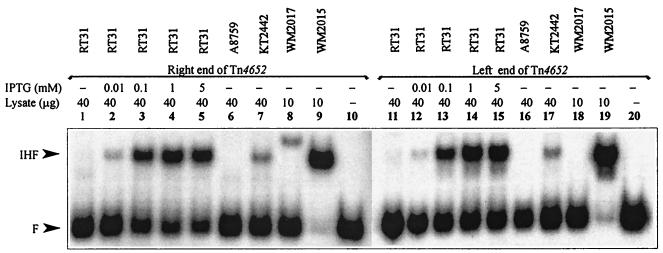
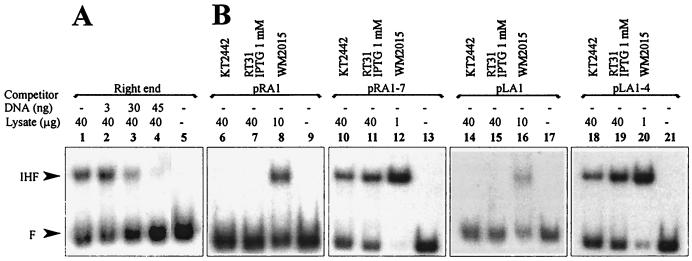
We have previously demonstrated by using gel shift assay that both ends of the transposon Tn4652 bind IHF from the cell lysate of E. coli (26). However, the experimental conditions used in in vitro binding assay did not enable detection of the IHF-dependent shift when cell extract of P. putida PaW85 was used (26). In this study, we have established conditions that enabled us to detect P. putida-IHF-dependent retardation of DNA containing the IHF binding sites (see Materials and Methods). The results of the gel shift assay with the DNA probes prepared either from the Tn4652 right or left end demonstrated the specific complex formation in the cell lysate from P. putida KT2442 but not in the cell lysate from ihfA-deficient P. putida A8759 (Fig. 4, lanes 6, 7, 16 and 17). This complex had the same mobility as the complex containing E. coli IHF (Fig. 4, lanes 7, 9, 17, and 19). To verify the specific binding of P. putida IHF to the ends of Tn4652, competition experiments with a Pu promoter region containing the binding site for IHF (1, 12) were carried out. Addition of the nonlabeled DNA probe from Pu promoter region to the binding reaction suppressed the formation of the proposed IHF-specific complex in crude lysate of P. putida with both ends of Tn4652 (only results with the right-end DNA are shown [Fig. 5A, lanes 1 to 4]).
FIG. 4.
Gel shift assay of in vitro binding of IHF from cell lysates of P. putida and E. coli to the ends of Tn4652. Cell lysates used were from P. putida wild-type strain KT2442 (lanes 7 and 17), P. putida A8759 defective in the ihfA gene (lanes 6 and 16), P. putida RT31 carrying the ihfA and ihfB genes under the control of Ptac promoter and lacIq repressor (lanes 1 to 5 and 11 to 15), E. coli wild-type strain WM2015 (lanes 9 and 19), and E. coli WM2017 defective in ihfA and ihfB genes (lanes 8 and 18). No cell lysate was added to the reaction mixture in the case of lanes 10 and 20. The protein-DNA complex visible on lane 8 is of unknown origin. All lysates were prepared from stationary-phase cells.
FIG. 5.
(A) Gel shift assay demonstrating suppression of the formation of the P. putida IHF complex with the right end of Tn4652 by nonlabeled DNA fragment containing Pu promoter region. Cell lysates used were from P. putida KT2442. (B) Gel shift assay of in vitro binding of P. putida and E. coli IHF to the ends of Tn4652 lacking A-T-rich regions upstream of the IHF binding core sequence (DNA probes from plasmids pRA1 and pLA1) and in the presence of A-T-rich regions (DNA probes from plasmids pRA1-7 and pLA1-4). Lysates used were from E. coli WM2015 (lanes 8, 12, 16, and 20) and P. putida KT2442 (lanes 6, 10, 14, and 18) and RT31 grown in the presence of 1 mM IPTG (lanes 7, 11, 15, and 19). No cell lysate was added to the reaction mixture in lanes 9, 13, 17, or 21. All lysates were prepared from stationary-phase cells.
Binding of the P. putida IHF to the ends of Tn4652 was detectable only in the case when A-T-rich regions flanked the IHF-binding core sequence at the 5′ side (plasmids pRA1-7 and pLA1-4 in Fig. 5B, lanes 10, 11, 18 and 19). No P. putida IHF-dependent complexes could be detected when the DNA fragments containing either the Tn4652 right-end or left-end sequences extending to the DraI site were used in binding reactions (plasmids pRA1 and pLA1 in Fig. 5B, lanes 6, 7, 14, and 15). However, lack of the A-T-rich DNA region still enabled binding of E. coli IHF to the Tn4652 right- and left-end sequences (Fig. 5B, lanes 8 and 16).
Effect of intracellular IHF concentration on transcription from the fusion promoters.
In order to study the effect of intracellular concentration of IHF on transcription from the promoters PRA1 and PLA1, P. putida RT31 carrying the P. putida ihfA and ihfB genes in the chromosome under the control of Ptac promoter and lacIq repressor was constructed (for details, see Materials and Methods). The level of expression of the genes encoding IHF could be modified in this strain by the addition of different concentrations of IPTG to the bacterial growth medium.
Figure 6 represents the results of C12O assay in P. putida RT31 cells from either exponential- or stationary-phase cells grown in the presence of different concentrations of IPTG. We found that if the intracellular concentration of IHF was artificially increased, the positive effect of this protein on the transcription from the promoter PRA1 could be detected in the exponentially growing cells as well. An approximately fourfold-elevated level of transcription in exponential phase cells of P. putida RT31 was recorded for the fusion promoter PRA1 in plasmid pRA1-12, but not in plasmid pRA1 when 1 to 5 mM IPTG was added to the growth medium (Fig. 6A). Similar results were recorded for the stationary-phase cells of RT31 carrying pRA1 and pRA1-12. In this case, we could observe 3.5 times the positive effect on transcription of the presence of 0.1 to 5 mM IPTG in the medium (Fig. 6B).
FIG. 6.
Study of the effect of different intracellular concentrations of IHF on transcription from the promoter PRA1 in plasmids pRA1 and pRA1-12 (A, exponentially growing cells; B, stationary-phase cells) and from the promoter PLA1 in plasmids pLA1 and pLA1-12 (C, exponentially growing cells; D, stationary-phase cells) containing different lengths of upstream sequences. P. putida RT31 carrying the ihfA and ihfB genes under the control of Ptac promoter and lacIq repressor was grown in LB in the presence or absence of inducer IPTG. Exponentially growing cells were sampled at 6 h, and stationary-phase cells were sampled at 16 h. Data (means ± standard deviations) of at least four independent experiments are presented.
The level of transcription from the promoter PLA1 in exponentially growing cells of P. putida RT31 was 1.5- to twofold lower in plasmid pLA1-12 than in the plasmid pLA1 either in the absence of IPTG or in the presence of 0.01 to 0.1 mM IPTG (Fig. 6C). In the presence of 1 to 5 mM IPTG, the level of expression of the promoter PLA1 in the plasmid pLA1-12 became closer to that measured in pLA1 (Fig. 6C). The level of transcription from the promoter PLA1 in plasmid pLA1-12 was increased to approximately 1.5- to twofold that of the plasmid pLA1 in the presence of 0.01 to 5 mM IPTG in the growth medium of the stationary-phase cells of P. putida RT31 (Fig. 6D).
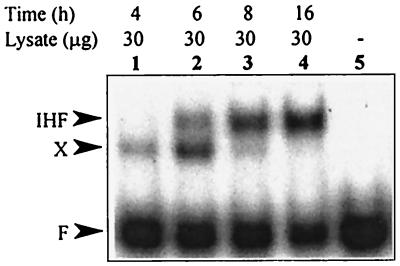
The efficiency of IHF-dependent protein-DNA complex formation in cell extracts prepared from P. putida RT31 grown at different concentrations of IPTG was investigated (Fig. 4). The IHF-dependent probe retardation was only slightly detectable in the cell lysates prepared from P. putida RT31 grown without IPTG (Fig. 4, lanes 1 and 11). The amount of IHF-specific complex detected with respect to the unbound DNA increased when 0.01 to 5 mM IPTG was added to the growth medium (Fig. 4, lanes 2 to 5 and 12 to 15). This indicated that the gel retardation assay revealing binding of IHF to the ends of Tn4652 would be an adequately sensitive method for the detection of substantial differences in intracellular concentrations of IHF. In order to find out whether the amount of P. putida IHF could be different in exponentially growing and stationary-phase cells, we performed the gel shift assay by using cell lysates prepared from cells sampled at different time points from P. putida KT2442 culture. According to the gel shift assay, the crude extracts prepared from stationary-phase cells of P. putida (sampled at 16 h) exhibited an approximately sevenfold-higher amount of the IHF-specific complex than did extracts prepared from exponentially growing bacteria (sampled at 4 h) (Fig. 7, lanes 1 and 4). In addition to the IHF-specific complex, another, faster moving, complex was detectable in the gel shift assay. The relative amount of this complex (detected with respect to the unbound DNA) was higher when cell extracts were prepared from exponentially growing cultures (Fig. 7, lanes 1 and 2), and it was only slightly detectable when the samples were taken either from the late-exponential or the stationary-phase cultures (Fig. 7, lanes 3 and 4).
FIG. 7.
Gel shift assay of IHF binding to the right end of Tn4652 in cell extracts prepared from different growth phases of cells of P. putida KT2442. No cell lysate was added to the reaction mixture in lane 5. Another complex moving faster than the IHF-specific complex is designated as X. The growth curve of this bacterium is shown in Fig. 3.
DISCUSSION
In this study, we have demonstrated that the activation of transcription from the fusion promoters PRA1 and PLA1 (formed at the junctions of the target DNA with the right end and left end of Tn4652, respectively) is positively affected by IHF in P. putida. The involvement of IHF in the modulation of transcription from the fusion promoters PRA1 and PLA1 was demonstrated by comparing the levels of expression from the fusion promoters in the wild type and in the IHF-negative background of P. putida (Fig. 3). Construction of the P. putida RT31 carrying the genes coding for P. putida IHF under the control of the Ptac promoter and the lacIq repressor in its chromosome allowed us to modify the level of IHF expression by changing the concentration of IPTG in the growth medium. The amount of the IHF-specific complex detected in the gel shift assay increased in crude extracts of P. putida RT31 cells grown at higher concentrations of IPTG in the growth medium inducing IHF expression (Fig. 4). The elevated level of transcription from the fusion promoters became apparent in P. putida RT31 only in the presence of IPTG in growth media, due to the result of induced expression of the genes for IHF (Fig. 6). However, the influence of IHF on transcription from the promoter PLA1 was not so clearly defined as that from PRA1, and the mechanism of modulation of transcription from PLA1 by upstream sequences seems to be more complex.
The presence of the A-T-rich element 5′ to the IHF-binding consensus sequence WATCAANNNNTTR has been shown to be important for the binding of IHF in several cases (20, 22, 46). We have found that the binding of IHF to the ends of Tn4652 is also dependent on the sequences flanking the core IHF binding site. This was confirmed in gel shift assays by using the DNA probes lacking the A-T-rich sequences upstream from the DraI site at the Tn4652 ends. In this case, we failed to detect the P. putida IHF-dependent shift (Fig. 5B). These results are in good accordance with the results obtained in in vivo experiments. Study of the expression of the fusion promoter PRA1 in plasmids carrying different lengths of DNA sequences upstream of the core IHF binding site revealed the IHF-mediated effect on transcription only when the A-T-rich DNA sequence upstream of the DraI site was present (Fig. 5B). This indicates that the IHF binding core consensus sequence together with the A-T-rich sequence flanking the DraI site is required for an enhanced level of transcription from the promoter PRA1 in P. putida.
The positive effect of IHF on transcription from the fusion promoter PRA1 investigated in this study appeared only in stationary-phase P. putida (Fig. 3). The results of a study by Ditto et al. (13) indicate that the abundance of E. coli IHF is increased five- to 10-fold during the transition from steady-state exponential growth to the late stationary phase. Although the level of IHF is already high in exponential-phase cultures, the occupancy of the IHF binding sites increases in stationary-phase cells (5, 13, 35). The IHF low-affinity sites are only partially occupied, and strong binding sites reach semisaturation in exponentially growing E. coli (35). The IHF content of P. aeruginosa is also increased more than 10-fold when cells enter the stationary phase (9). Prior to now, there have been no published data about direct measurements of concentration of IHF in P. putida cells. Our results indicate that the intracellular concentration of IHF in P. putida could be increased during transition to the stationary phase as well.
In Fig. 7, we compare the amount of IHF-dependent protein-DNA complex formation in cell extracts prepared either from exponentially growing or stationary-phase cultures of P. putida KT2442. The IHF-bound DNA was quantified relative to the unbound DNA by using Phosphoimager. An approximately sevenfold increase in the IHF-dependent DNA-binding activity was observed in the cell extracts from stationary phase bacteria if compared to that in crude extracts of exponentially grown cells sampled after 4 h. This indicates that the intracellular content of IHF could increase when P. putida enters the stationary growth phase, and the IHF concentration in exponential-phase cells might be insufficient to occupy IHF binding sites at the ends of Tn4652.
Interestingly, another complex (designated by us as complex X) migrating in the gel faster than the IHF-specific complex was dominant in crude extracts prepared from exponentially growing cells (Fig. 7). Meanwhile, the effect of transcription enhancement from the promoter PRA1 became evident just in the late-exponential growth phase of bacteria (at 8 h) when the amount of IHF-specific complex was increased and the amount of complex X was decreased (Fig. 3A and 7). It is possible that IHF and the unidentified factor X compete for the overlapping binding sites, if the cellular amount of IHF increases then the factor X is outcompeted and the positive effect of IHF appears. This hypothesis is supported by the results of experiments in which the level of IHF in cells was artificially increased by IPTG in exponentially growing cells of P. putida RT31 carrying pRA1-12. In this case, we observed the elevated level of transcription from PRA1 similar to that observed in the cells entering the stationary phase of growth (Fig. 6A). Because of the fact that IHF can bind upstream to the promoter PRA1 only in the presence of A-T-rich sequences at the 5′ side of the IHF binding consensus sequence, one could also speculate that the binding of IHF to this DNA might eliminate the negative effect of the factor X on transcription from PRA1 in plasmids pRA1-7 and pRA1-12. The positive role of IHF on transcription by competing with negatively acting factor for the binding site has also been described in other published reports. For example, IHF can counteract inhibition of transcription by H-NS at the Mu phage Pe promoter (45) and at the virB promoter of Shigella flexneri virulence genes (39). Similarly, our previous studies on the regulation of the expression of the tnpA promoter of Tn4652 indicate that IHF not only enhances transcription from this promoter but also alleviates the negative effect of the terminal sequences of this transposon on the promoter activity (26). The promoter of the transposase gene tnpA of Tn4652, localized by us to the right end of the element, and the fusion promoter PRA1 are oppositely directed (Fig. 1A). The DNA region of the Tn4652 right end that is involved in transcription enhancement from the promoter PRA1 overlaps with the promoter upstream region of the tnpA gene. Therefore, in addition to IHF, some other factors that regulate transcription from the tnpA promoter could also influence transcription from the promoter PRA1. We can hypothesize that factor X, which forms a complex with Tn4652 right end and interferes with the regulation of the fusion promoter, could be a regulator of the tnpA promoter. However, there is no effect of the tnpA promoter itself on the transcription from the promoter PRA1 because pRA1-7 lacking the tnpA promoter and pRA1-12 including this DNA element exhibited similar expression patterns.
Notably, the stronger negative effect of the upstream sequences on transcription from the fusion promoter PLA1 appeared in the IHF-negative strain P. putida A8759 carrying the plasmid pLA1-12 (Fig. 3B and C). This indicates that, in the case of the promoter PLA1, IHF could also partially eliminate the negative effect, but the DNA sequences involved in this IHF-mediated transcription modulation locate further upstream. The level of expression of PLA1 in pLA1-12 was also slightly decreased in the wild-type strain KT2442 compared with the expression of this promoter in pLA1. When IHF was overexpressed in RT31 stationary-phase cells, the positive effect of the presence of upstream sequences in pLA1-12 became evident. Our attempts to map the DNA regions involved in transcription modulation from PLA1 led to results that are not easily interpreted (Fig. 3B and C). The plasmids pLA1-4 and pLA1-5 revealed different expression patterns in KT2442 and in its ihfA-defective derivative strain A8759 when transcription from PLA1 in exponentially growing and stationary-phase cells was compared. This indicates that the regulation of transcription from the left end of Tn4652 could be different and more complex than that from the right end of the transposon.
We have also observed the effect of growth phase of P. putida cells on the level of transcription from the fusion promoters in the case of plasmids pRA1 and pLA1 (Fig. 3). The mechanism for stationary-phase-induced transcription from the fusion promoters still remains unclear. However, experiments carried out in our laboratory have revealed stationary-phase ς factor ςS-dependent transcription from PLA1 but not from PRA1 (E. Ojangu and A. Tover, unpublished results).
Summarizing the data presented in this report, we can conclude that IHF positively influences transcription from the fusion promoters generated at the junctions between the target DNA and the terminal sequences of the transposon Tn4652 in P. putida. The binding sites for the proteins that are involved in regulation of transposition usually overlap with DNA sequences containing outwardly directed promoter elements. We have previously shown that transcription of the Tn4652 transposase gene tnpA is positively affected by IHF (26). The presence of binding sites for IHF in the both ends of Tn4652 suggests the possible role of IHF in regulation of transposition of Tn4652. Thus, our results illustrate how regulation of two distinct processes, transposition of the DNA element and transcriptional activation of neighboring genes by this element, may be connected to each other.
ACKNOWLEDGMENTS
We thank V. de Lorenzo for kindly providing P. putida KT2442 and A8759, E. coli S17-1 λ pir, and plasmids pUC18 Not and pUT mini-Tn5 luxAB; V. Morales for providing plasmid pMMB208; and A. Oppenheim for providing plasmid pHip. We also thank T. Alamäe and L. Kasak for critically reading the manuscript and for their helpful discussions.
This work was supported by grants from the Estonian Science Foundation, by a grant from International Foundation for Science (Salen Foundation), and by grant no. LKH100 from the Joint Program of the Government of Estonia and the International Science Foundation.
REFERENCES
- 1.Abril M A, Buck M, Ramos J L. Activation of the Pseudomonas TOL plasmid upper pathway operon. Identification of binding sites for the positive regulator XylR and for integration host factor protein. J Biol Chem. 1991;266:15832–15838. [PubMed] [Google Scholar]
- 2.Bayley S A, Duggleby C J, Worsey M J, Williams P A, Hardy K G, Broda P. Two modes of loss of the TOL function from Pseudomonas putida mt-2. Mol Gen Genet. 1977;154:203–204. doi: 10.1007/BF00330838. [DOI] [PubMed] [Google Scholar]
- 3.Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Bushman W, Thompson J F, Vargas L, Landy A. Control of directionality in lambda site specific recombination. Science. 1985;230:906–911. doi: 10.1126/science.2932798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calb R, Davidovitch A, Koby S, Giladi H, Goldenberg D, Margalit H, Holtel A, Timmis K, Sanchez-Romero J M, de Lorenzo V, Oppenheim A B. Structure and function of the Pseudomonas putida integration host factor. J Bacteriol. 1996;178:6319–6326. doi: 10.1128/jb.178.21.6319-6326.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter P, Bedouelle H, Winter G. Improved oligonucleotide site-directed mutagenesis using M13 vectors. Nucleic Acids Res. 1985;13:4431–4443. doi: 10.1093/nar/13.12.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craig N L, Nash H A. E. coli integration host factor binds to specific sites in DNA. Cell. 1984;39:707–716. doi: 10.1016/0092-8674(84)90478-1. [DOI] [PubMed] [Google Scholar]
- 9.Delic-Attree I, Toussaint B, Froger A, Willison J C, Vignais P M. Isolation of an IHF-deficient mutant of a Pseudomonas aeruginosa mucoid isolate and evaluation of the role of IHF in algD gene expression. Microbiology. 1996;142:2785–2793. doi: 10.1099/13500872-142-10-2785. [DOI] [PubMed] [Google Scholar]
- 10.de Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 11.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative Eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Lorenzo V, Herrero M, Metzke M, Timmis K N. An upstream XylR- and IHF-induced nucleoprotein complex regulates the sigma 54-dependent Pu promoter of TOL plasmid. EMBO J. 1991;10:1159–1167. doi: 10.1002/j.1460-2075.1991.tb08056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ditto M D, Roberts D, Weisberg R A. Growth phase variation of integration host factor level in Escherichia coli. J Bacteriol. 1994;176:3738–3748. doi: 10.1128/jb.176.12.3738-3748.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelhorn M, Geiselmann J. Maximal transcriptional activation by the IHF protein of Escherichia coli depends on optimal DNA bending by the activator. Mol Microbiol. 1998;30:431–441. doi: 10.1046/j.1365-2958.1998.01078.x. [DOI] [PubMed] [Google Scholar]
- 15.Freundlich M, Ramani N, Mathew E, Sirko A, Tsui P. The role of integration host factor in gene expression in Escherichia coli. Mol Microbiol. 1992;6:2557–2563. doi: 10.1111/j.1365-2958.1992.tb01432.x. [DOI] [PubMed] [Google Scholar]
- 16.Friedman D I. Integration host factor: a protein for all reasons. Cell. 1988;55:545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- 17.Galas D J, Chandler M. Bacterial insertion sequences. In: Berg D E, Howe M H, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 109–162. [Google Scholar]
- 18.Giladi H, Kobu S, Prag G, Engelhorn M, Geiselmann J, Oppenheim A B. Participation of IHF and a distant UP element in the stimulation of the phage λ PL promoter. Mol Microbiol. 1998;30:443–451. doi: 10.1046/j.1365-2958.1998.01079.x. [DOI] [PubMed] [Google Scholar]
- 19.Giladi H, Murakami K, Ishihama A, Oppenheim A B. Identification of an UP element within the IHF binding site at the PL1-PL2 tandem promoter of bacteriophage λ. J Mol Biol. 1996;260:484–491. doi: 10.1006/jmbi.1996.0416. [DOI] [PubMed] [Google Scholar]
- 20.Goodrich J A, Schwartz M L, McClure W R. Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF) Nucleic Acids Res. 1990;18:4993–5000. doi: 10.1093/nar/18.17.4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goosen N, van de Putte P. The regulation of transcription initiation by integration host factor. Mol Microbiol. 1995;16:1–7. doi: 10.1111/j.1365-2958.1995.tb02386.x. [DOI] [PubMed] [Google Scholar]
- 22.Hales L M, Gumport R I, Gardner J F. Examining the contribution of a dA + dT element to the conformation of Escherichia coli integration host factor-DNA complexes. Nucleic Acids Res. 1996;24:1780–1786. doi: 10.1093/nar/24.9.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanahan D. Studies on the transformation of E. coli with plasmids. J Mol Biol. 1983;166:577–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 24.Hegeman D. Synthesis of the enzymes of the mandelate pathway by Pseudomonas putida. I. Synthesis of the enzymes by wild type. J Bacteriol. 1966;91:1140–1154. doi: 10.1128/jb.91.3.1140-1154.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hõrak R, Kivisaar M. Expression of the transposase gene tnpA of Tn4652 is positively affected by integration host factor. J Bacteriol. 1998;180:2822–2829. doi: 10.1128/jb.180.11.2822-2829.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hübner A, Hendrickson W. A fusion promoter created by a new insertion sequence, IS1490, activates transcription of 2,4,5-trichlorophenoxyacetic acid catabolic genes in Burkholderia cepacia AC1100. J Bacteriol. 1997;179:2717–2723. doi: 10.1128/jb.179.8.2717-2723.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kasak L, Hõrak R, Kivisaar M. Promoter-creating mutations in Pseudomonas putida: a model system for the study of mutation in starving bacteria. Proc Natl Acad Sci USA. 1997;94:3134–3139. doi: 10.1073/pnas.94.7.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kivisaar M, Hõrak R, Kasak L, Heinaru A, Habicht J. Selection of independent plasmids determining phenol degradation in Pseudomonas putida and the cloning and expression of genes encoding phenol monooxygenase and catechol 1,2-dioxygenase. Plasmid. 1990;24:25–36. doi: 10.1016/0147-619x(90)90022-5. [DOI] [PubMed] [Google Scholar]
- 30.Langer U, Richter S, Roth A, Weigel C, Messer W. A comprehensive set of DnaA-box mutations in the replication origin, oriC, of Escherichia coli. Mol Microbiol. 1996;21:301–311. doi: 10.1046/j.1365-2958.1996.6481362.x. [DOI] [PubMed] [Google Scholar]
- 31.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 33.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morales V, Bäckman A, Bagdasarian M. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene. 1991;97:39–47. doi: 10.1016/0378-1119(91)90007-x. [DOI] [PubMed] [Google Scholar]
- 35.Murtin C, Engelhorn M, Geiselmann J, Boccard F. A quantitative UV laser footprinting analysis of the interaction of IHF with specific binding sites: reevaluation of the effective concentration of IHF in the cell. J Mol Biol. 1998;284:949–961. doi: 10.1006/jmbi.1998.2256. [DOI] [PubMed] [Google Scholar]
- 36.Nurk A, Tamm A, Hõrak R, Kivisaar M. In-vivo-generated fusion promoters in Pseudomonas putida. Gene. 1993;127:23–29. doi: 10.1016/0378-1119(93)90612-7. [DOI] [PubMed] [Google Scholar]
- 37.Parekh B S, Hatfield G W. Transcriptional activation by protein-induced DNA bending: evidence for a DNA structural transmission model. Proc Natl Acad Sci USA. 1996;93:1173–1177. doi: 10.1073/pnas.93.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez-Martin J, de Lorenzo V. Clues and consequences of DNA bending in transcription. Annu Rev Microbiol. 1997;51:593–628. doi: 10.1146/annurev.micro.51.1.593. [DOI] [PubMed] [Google Scholar]
- 39.Porter M E, Dorman C J. Positive regulation of Shigella flexneri virulence genes by integration host factor. J Bacteriol. 1997;179:6537–6550. doi: 10.1128/jb.179.21.6537-6550.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rice P A, Yang S W, Mizuuchi K, Nash H A. Crustal structure of IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 41.Sharma R C, Schimke R T. Preparation of electro-competent E. coli using salt-free growth medium. BioTechniques. 1996;20:42–44. doi: 10.2144/96201bm08. [DOI] [PubMed] [Google Scholar]
- 42.Tsuda M, Iino T. Genetic analysis of a transposon carrying toluene degrading genes on a TOL plasmid pWWO. Mol Gen Genet. 1987;210:270–276. doi: 10.1007/BF00325693. [DOI] [PubMed] [Google Scholar]
- 43.Tsuda M, Minegishi K-I, Iino T. Toluene transposons Tn4651 and Tn4653 are class II transposons. J Bacteriol. 1989;171:1386–1393. doi: 10.1128/jb.171.3.1386-1393.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Ulsen P, Hillebrand M, Kainz M, Collard R, Zulianello L, van de Putte P. Function of the C-terminal domain of the alpha subunit of Escherichia coli RNA polymerase in basal expression and integration host factor mediated activation of the early promoter of bacteriophage Mu. J Bacteriol. 1997;179:530–537. doi: 10.1128/jb.179.2.530-537.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Ulsen P, Hillebrand M, Zulianello L, van de Putte P, Goosen N. Integration host factor alleviates the H-NS-mediated repression of the early promoter of bacteriophage Mu. Mol Microbiol. 1996;21:567–578. doi: 10.1111/j.1365-2958.1996.tb02565.x. [DOI] [PubMed] [Google Scholar]
- 46.Yang S W, Nash H A. Comparison of protein binding to DNA in vivo and in vitro defining an effective intracellular target. EMBO J. 1995;14:6292–6300. doi: 10.1002/j.1460-2075.1995.tb00319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]