Abstract
Background:
NF1-mutated tumors represent a small subset (10–15%) of melanomas, not sufficiently analyzed in large clinical cohorts. This study investigated the largest multicenter collection of NF1-mutated melanomas to date.
Methods:
This study analyzed a multicenter tumor tissue sample cohort from 266 patients with NF1-mutated melanoma. Targeted next-generation sequencing of the TERT promoter and 29 relevant melanoma genes was performed. Survival was compared with NF1-wild-type cohorts from the TRIM project (n = 432).
Results:
Most NF1-mutated melanoma arose in the head-and-neck region of patients > 60 years of age. NF1 alterations were frequently inactivating, primarily non-sense, less frequently truncating mutations. Non-inactivating NF1 mutations more frequently co-occurred with activating BRAF and RAS mutations. NF1-mutated tumors had higher numbers of gene mutations and UV-signature C>T and CC>TT transitions than BRAF, RAS and triple wild-type melanomas. NF1-mutated acral and mucosal melanomas harbored a different mutation signature and were frequent in females (69 and 83%, respectively), differing from non-acral cutaneous NF1-mutated melanomas (males 73%, females 27%). Overall survival in stage IV disease was comparable for patients with NF1-mutated or -wild-type melanoma. However, in patients receiving first-line immune checkpoint inhibitor treatment., better overall survival was observed for NF1-mutated than -wild-type tumors (mOS = not reached vs. mOS = 25.82, p = 0.0154, n = 80 and 432, respectively).
Conclusions:
Cutaneous, acral, and mucosal NF1-mutated melanomas vary in clinical and genetic characteristics and demonstrate a favorable outcome upon immune checkpoint inhibition therapy.
Keywords: NF1, BRAF, NRAS, melanoma, mutation profiling
Introduction
Cutaneous melanoma is a highly malignant tumor with a potential for distant metastasis 1,2. The prognosis for patients with metastatic disease remains poor despite significant recent improvements in therapeutic strategies 3.
The development of next-generation sequencing (NGS) technologies increasingly elucidated the genetic landscape of melanoma 4,5. The Cancer Genome Atlas (TCGA) suggested to classify melanomas into four main genetic subtypes: BRAF-mutated, NRAS-mutated, NF1-mutated or triple wild-type 6. Alterations in the V600 codon of BRAF and the Q61, G12 or G13 codons of RAS genes all lead to MAP Kinase activation 6. The NF1 gene product is a GTPase-activating protein downregulating RAS activity. NF1 inactivation thus leads to MAPK activation 7.
Recently introduced therapeutic approaches have significantly improved overall survival of patients with advanced or unresectable melanoma 3,8,9. These therapies can be classified into two groups, namely immune checkpoint inhibitors (ICI) targeting programmed death-1 and its ligand (PD-1/PD-L1) (nivolumab and pembrolizumab) or cytotoxic T lymphocyte antigen 4 (CTLA-4) (ipilimumab), and tyrosine kinase inhibitors (TKI) targeting the MAPK pathway, namely BRAF or MEK, which are applicable for patients with tumors harboring a BRAF V600 mutation. 10–13. The most potent combination immunotherapy of anti-PD-1 (nivolumab) and anti-CTLA-4 (ipilimumab) antibodies has achieved a 5-year-survival rate of 52%, accompanied by a high rate of toxicity 14,15. Besides these therapeutic regimens, treatment options for patients with advanced melanoma remain limited and targeted therapies for specific mutations in NRAS and NF1 genes are not available.
Melanoma has a variable mutation frequency, largely based on varying exposure to UV radiation 5,16. Melanomas arising in chronically sun-exposed skin harbor larger amounts of mutations including frequent mutations in the NF1 gene 17. A high mutational burden is associated with improved and more durable therapeutic responses to anti-CTLA-4 or anti-PD1 monotherapy in metastatic melanoma 18–20. However, mutations in NF1 also occur in tumors arising in anatomic sites with little or no UV exposure, such as acral and mucosal melanomas 17. Patient age at diagnosis is also associated with the mutational pattern of melanoma - BRAF mutations are more common in younger, NF1-mutations in older patients 17,21. NF1 mutations occur particularly frequently in desmoplastic melanoma 22. Loss-of-function mutations or deletions in NF1 have been linked to a decreased sensitivity to BRAF-inhibitors in BRAF-mutated melanomas 23. Enhanced sensitivity of NF1-mutated melanomas to MEK-inhibitors has been reported 7.
In the present study, we gathered the largest multi-center cohort of NF1-mutated melanomas investigated to date in order to further understand the NF1-mutated melanoma subtype and its implications on clinical course and possible therapeutic approaches in the respective patients.
Materials and Methods
Patients and Samples:
Screening 3837 NGS reports of melanoma patients, we identified 266 patients with NF1-mutated melanoma diagnosed between 2013 and 2020. Related data and tumor samples were obtained from the Westdeutsche Biobank Essen (WBE/SCABIO), University Hospital Essen (11-4715-BO, n=157) and from the multicenter prospective translational study “tissue registry in melanoma” (TRIM; 15-6566-BO, n=109). Tumors were classified according to the American Joint Committee on Cancer (AJCC 8th) staging system 24,25. NF1-wild-type cohort data was obtained from the TRIM cohort. This study was performed in accordance with the Declaration of Helsinki, was approved by the Ethics Committee of the Medical Faculty of the University of Duisburg-Essen (ethics approval no. 20-9606-BO) and followed the guidelines for good clinical practice (GCP).
A customized amplicon-based sequencing panel covering the NF1 gene as well as 29 additional genes known to harbor oncogenic mutations relevant for melanoma was used (genes list in Supplemental Table 4).
Targeted sequencing:
DNA was isolated from Formalin-fixed paraffin-embedded (FFPE) tumor tissue according to standard procedures as previously described26. A custom amplicon-based sequencing panel covering 29 genes (listed in Supplemental Table 4) known to be recurrently mutated in cutaneous and uveal melanoma was designed and prepared applying the GeneRead Library Prep Kit from QIAGEN® according to the manufacturer’s instructions. For adapter ligation and barcoding of individual samples, the NEBNext Ultra DNA Library Prep Mastermix Set and NEBNext Multiplex Oligos for Illumina from New England Biolabs were applied. Twelve samples were sequenced in parallel on an Illumina MiSeq next generation sequencer.
Sequencing analysis was performed applying the CLC Cancer Research Workbench from QIAGEN® (currently version 20.0.4). In brief, the following steps were applied. The workflow in CLC included adapter trimming and read pair merging before mapping to the human reference genome (hg19). InDels and Structural Variants were assessed allowing 3 maximum mismatches (unaligned end breakpoints). Single nucleotide variant detection, local realignment and primer trimming followed. Additional information was then obtained regarding potential mutation type, known single nucleotide polymorphisms and conservation scores by cross-referencing varying databases (COSMIC, ClinVar, dbSNP, 1000 Genomes Project, HAPMAP and PhastCons-Conservation_scores_hg19). After the CLC Cancer Research Workbench processing, resulting csv files were analyzed manually. Mutations affecting the protein coding portion of the gene were considered if predicted to result in non-synonymous amino acid changes. The functional consequences of mutations were predicted by later by performing an analysis on the server based SIFT27, PROVEAN28 and PolyPhen-227 assays. A detailed list of all mutations and database references is included in Supplemental Table 4. To eliminate questionable low frequency background mutation calls, mutations were reported only if ≥ 10 reads reported the mutated variant, coverage of the mutation site was ≥ 30 reads and the mutation frequency was ≥ 10%. The average read coverage of the targeted area achieved was 2607x. (A detailed listing of the individual settings applied in CLC cancer research workbench are listed in the Supplemental Material and Methods)
Statistical analysis:
Associations of tumor origin with clinical parameters were investigated using chi-squared tests or Fisher’s exact tests as indicated. Continuous variables are presented as means with standard deviation or as median with interquartile range, as appropriate. Categorical variables are presented as counts and percentages. Survival curves were drawn using the Kaplan-Meier method and the log-rank test was used for comparisons. OS was calculated from first date of stage IV diagnosis or start of ICI therapy until death or last patient contact (censored observation), respectively. Statistical analyses were performed using Microsoft Excel, GraphPad Prism (version 6), SPSS 27.0 (IBM Corp., Armonk NY, USA), R (R version 4.0.3 (2020-10-10)) and RStudio 29,30. A p-value < 0.05 was considered significant.
Results
Patient characteristics
Two hundred sixty-six patients (95 females and 171 males) diagnosed with NF1-mutated melanoma were included in this study. The clinical characteristics are shown in Table 1 (additional data for all patients are shown in Supplemental Table 1). Tumors of the head and neck region were the most common (44% of tumors with documented primary tumor localization) (Figure 1, B middle). Mucosal melanomas consisted of 8 vaginal, 3 anal, 2 nasal, 2 esophageal and 1 urethral mucosal cases. In two cases the localization was not documented. Of all 16 acral melanomas, 7 were located on the lower extremities, whereas 2 were on the upper extremities with information missing in 7 cases. An overview of the clinical characteristics of stage IV NF1-mutated melanomas in relation to available date from the NF1-wild-type cohort used for survival and treatment comparisons is shown in Supplemental Table 2.
Table 1 –
Clinical characteristics of non-acral, acral and mucosal melanomas
| Variable, n (%) | Non acral cutaneous (n = 197) | Mucosal (n = 18) | Acral (n = 16) | p-Value |
|---|---|---|---|---|
| Age at first diagnosis | .86 | |||
| Median (range) | 67 (16 – 94) | 67 (38 – 85) | 68,5 (41 – 80) | |
| ≤60 years | 67 (34.0) | 6 (33.3) | 7 (43.8) | |
| >60 years | 130 (66.0) | 12 (66.7) | 9 (56.3) | |
| Sex | < .0001 | |||
| Female | 55 (27.9) | 15 (83.3) | 11 (68.8) | |
| Male | 142 (72.1) | 3 (16.7) | 5 (31.3) | |
| Mutated oncogene | .39 | |||
| BRAF V600E | 39 (19.8) | 1 (5.6) | 2 (12.5) | |
| NRAS Q61 | 35 (17.8) | 1 (5.6) | 1 (6.3) | |
| NF1 | 197 (100) | 18 (100) | 16 (100) | |
| Invasion depth of primary | .69 | |||
| Mean ± SD | 4,61 ± 4,54 | 5,26 ± 4,29 | 3,73 ± 2,82 | |
| < 1 mm | 11 (5.6) | 0 (0) | 2 (12.5) | |
| 1 – 2 mm | 37 (18.8) | 3 (16.7) | 3 (18.8) | |
| 2 – 4 mm | 53 (26.9) | 0 (0) | 6 (37.5) | |
| > 4 mm | 72 (36.6) | 3 (16.7) | 5 (31.3) | |
| Unknown | 24 (12.2) | 12 (66.7) | 0 (0) | |
| Ulceration of primary | .26 | |||
| Present | 80 (40.6) | 2 (11.1) | 11 (68.8) | |
| Absent | 93 (47.2) | 3 (16.7) | 5 (31.3) | |
| Unknown | 24 (12.2) | 13 (72.2) | 0 (0) | |
| TERT promoter | ||||
| TERTp228 | 106 (49.8) | 0 (0) | 4 (25.0) | |
| TERTp242 | 15 (7.0) | 0 (0) | 1 (6.3) | |
| TERTp250 | 60 (28.2) | 2 (11.1) | 1 (6.3) | |
| PD-L1 | .33 | |||
| Positive | 72 (36.6) | 2 (11.1) | 5 (31.3) | |
| Negative | 93 (47.2) | 8 (44.4) | 6 (37.5) | |
| Not performed | 32 (16.2) | 8 (44.4) | 5 (31.3) | |
Figure 1 – Characteristics of NF1-mutated melanomas.
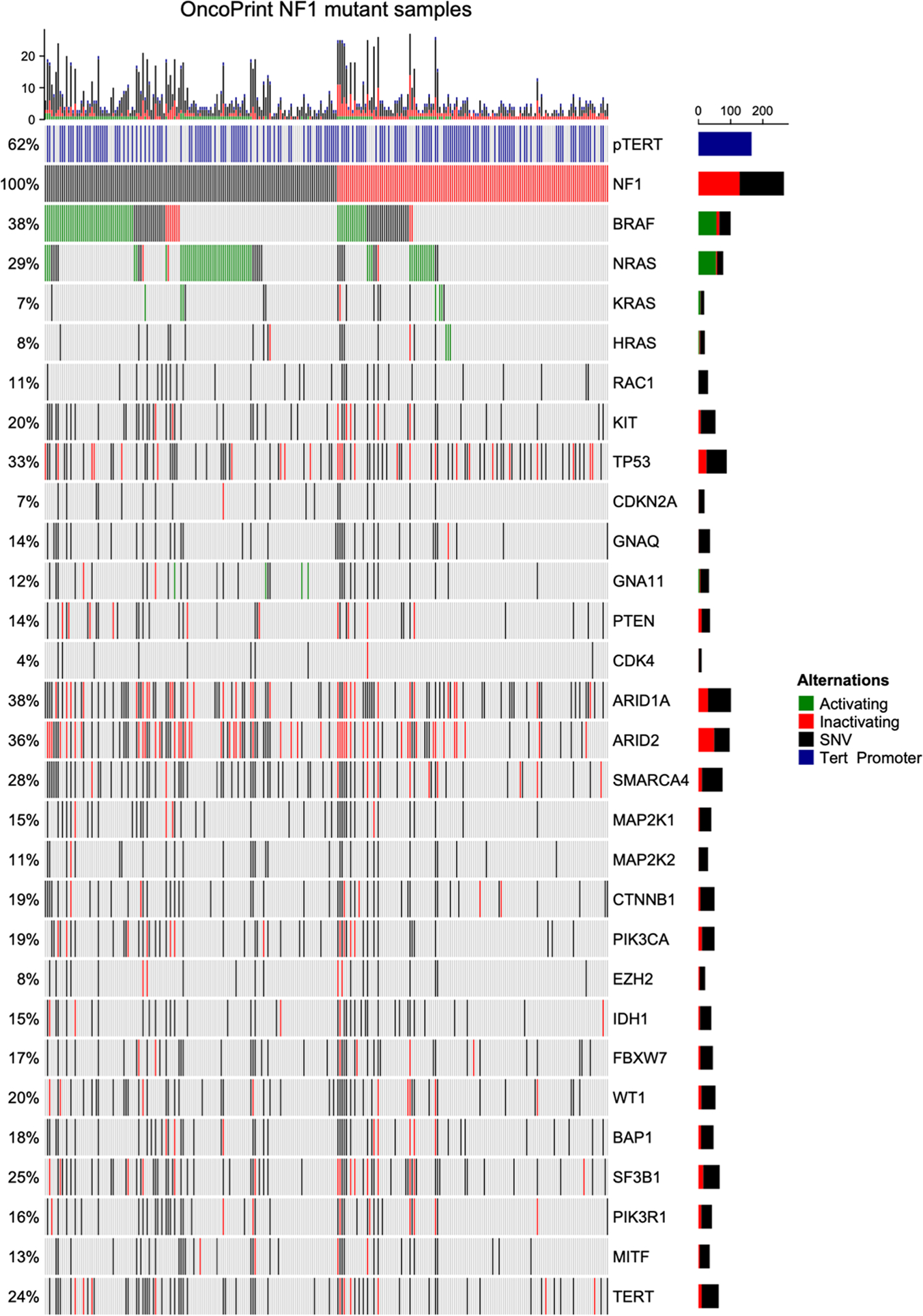
Non-acral cutaneous NF1-mutated melanoma harbored more frequent concurrent BRAF and NRAS mutations compared to both acral and mucosal melanoma (A). Left: Non-acral cutaneous melanoma shows a trend towards higher Breslow tumor thickness. Middle: Most non-acral cutaneous melanomas were located within the head and neck region. Right: Acral and non-acral cutaneous melanoma harbor significantly more mutations compared with mucosal melanoma (B). Left: NF1 mutated melanomas harbor more C>T substitutions compared to BRAF, NRAS or triple-wt melanomas. Right: NF1 mutated melanomas harbor the highest mutational burden among melanoma genetic subtypes. (C). Statistical tests performed are Mann-Whitney U test and Wilcoxon test. Data is shown as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Cutaneous melanomas harbor more mutations than mucosal melanomas
A subgroup analysis of the total 231 patients with known primary tumor localization for different melanoma subtypes revealed 197 (85.3%), 16 (6.9%) and 18 (7.8%) patients with cutaneous non-acral, acral and mucosal melanomas, respectively (Table 1). No age difference was noted (medians of 67, 68.5 and 67 years, respectively). A highly significant (p<0.0001) difference in sex distribution was noticed. In non-acral cutaneous melanomas, the majority of patients was male (72%), whereas both mucosal and acral melanomas arose more often in female patients (83% and 69%, respectively). BRAF, NRAS and TERT promoter mutations were more frequent in patients with non-acral cutaneous melanoma (Figure 1, A; Table 1). Mucosal melanomas had significantly fewer mutations than both acral and non-acral cutaneous melanoma (Figure 1, B right).
Distribution of UV-induced mutations amongst melanoma subtypes
Analysis of mutational patterns within the NF1-mutated tumors revealed a UV-signature. Single nucleotide variants were grouped into six mutation types, as previously described 31. We compared mutational patterns of single nucleotide variants of NF1-mutated melanomas to BRAF, NRAS and triple-wild-type melanomas. NF1-mutated melanomas revealed the highest number of UV-induced signature C>T substitutions and CC>TT substitutions (Figure 1, C and Supplemental Figure 1, B). Upon normalization per sample, mucosal melanomas showed a considerably lower number of UV-induced signature C>T transitions compared to other NF1-mutated melanomas (Supplemental Figure 1, A).
NF1-mutated melanoma exhibits the highest mutation number
Comparing the number of mutations identified in our sequencing panel (Supplemental Table 4, 0.88 megabase coverage) NF1-mutated melanomas were found to exhibit higher numbers of mutations per tumor (median 6) compared to BRAF-mutated (median 3), NRAS-mutated (median 4) and triple-WT (median 2) melanomas. (Figure 1, C).
NF1-mutated melanomas show a better response to ICI
Comparison of overall survival after first diagnosis of distant metastatic disease (AJCC stage IV) independently of treatment revealed no difference between patients with NF1-mutated (n = 128) or -wild-type (n = 387) melanomas (Supplemental Table 2) (mOS = 47 vs. 37 months, respectively [p = 0.35]) (Figure 2, A). However, a trend towards longer overall survival was noted in NF1-mutated melanoma patients receiving first line immune-checkpoint inhibitors (anti-PD1-monotherapy, or anti-PD1 + anti-CTLA-4 combination therapy) for stage IV disease compared to other first-line treatments (p = 0.32) (other treatments included targeted therapies, chemotherapy and tyrosine kinase inhibitors) (Figure 2, B). Further analysis of survival upon first-line immune checkpoint inhibitor treatment in advanced or metastatic disease, revealed a prolonged overall survival of NF1-mutated melanoma patients (n = 80) compared to a cohort of patients with NF1 wild-type melanoma (n = 432, TRIM cohort) (mOS = not reached vs. 25.15, respectively [p = 0.01]) (HR (95% CI) = 0.60 (0.40 to 0.90); p = 0.01). Combination nivolumab and ipilimumab, nivolumab or pembrolizumab treatment was given in 28, 31 and 21 cases, respectively in the NF1-mutated group and 132, 153 and 147 cases, respectively in the NF1 wild-type group (p = 0.39) (Figure 2, C).
Figure 2 – NF1-mutated melanomas exhibit favorable ICI therapy response.
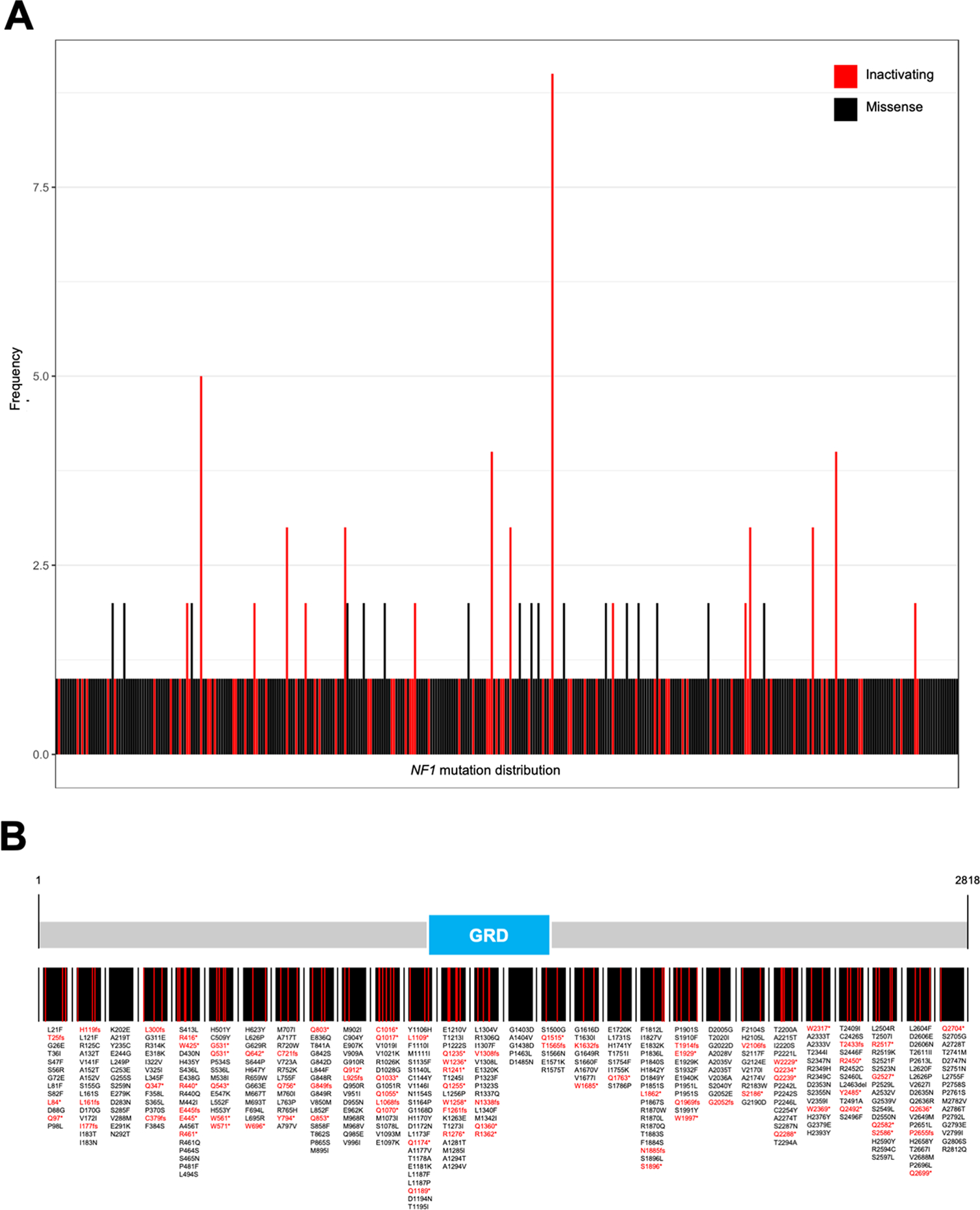
Patients with stage IV NF1-mutated melanoma (n = 128) do not show a difference in overall survival compared to patients with stage IV NF1-wild-type melanoma (n = 387) (A). Within the group of NF1 mutated melanoma, patients undergoing therapy with immune-checkpoint inhibitors exhibit a trend towards better overall survival compared to other therapies (B). Patients treated with first-line ICI therapies with NF1 mutated melanomas show a prolonged mOS compared to NF1-wild-type melanomas (C).
Targeted next generation sequencing
Within 266 study samples, 538 NF1 mutations were identified, with many harboring more than one mutation. BRAF mutations were frequent (n = 99, 38%) (Figure 3, Supplemental Table 3), with 45 V600E, 11 V600K, and 1 V600D activating mutation. NRAS mutations were found in 77 samples (29%), 56 of which were activating Q61/G12/G13 mutations. Q61 mutations included 4 Q61H, 9 Q61K, 16 Q61L and 17 Q61R (Supplemental Table 3). KRAS and HRAS mutations were less frequent, in 7% and 8% of samples, respectively (Figure 3). Activating TERT-promoter mutations were present in 166 samples (62%) (Supplemental Table 3). Other frequently mutated genes included TP53 (33%), ARID1A (38%), ARID2 (36%) and KIT (20%). Less frequent mutations were reported in RAC1, CDKN2A, GNAQ, GNA11, PTEN, CDK4, SMARCA4, MAP2K1, MAP2K2, CTNNB1, PIK3CA, EZH2, IDH1, FBXW7, WT1, BAP1, SF3B1, PIK3R1, MITF, and TERT.
Figure 3 – Mutation distribution in NF1-mutated melanomas.
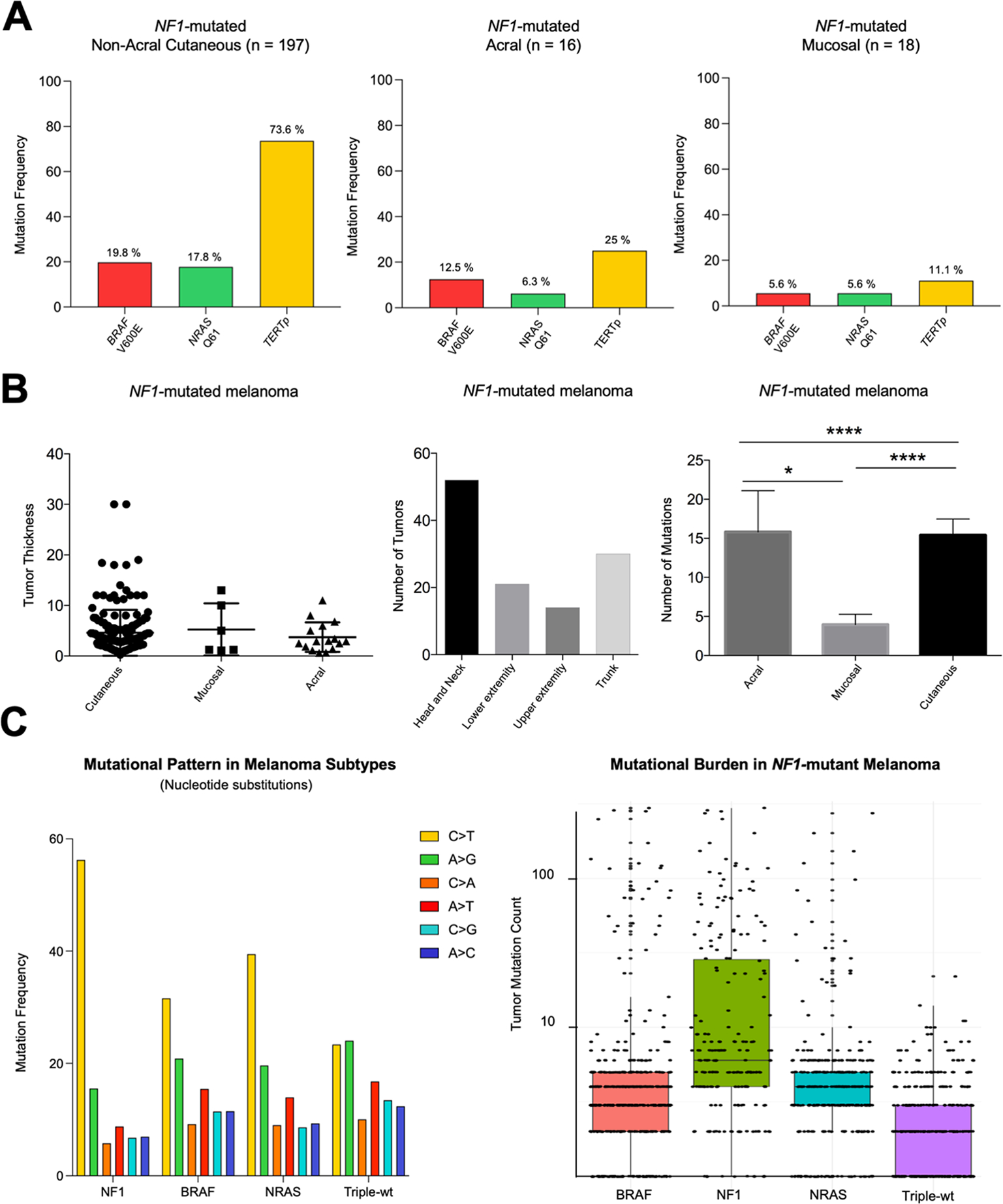
Green: mutations known or assumed to be activating. Red: loss of function mutations.
Blue: mutations in the TERT promoter region.
NF1 mutations are distributed throughout the neurofibromin protein, not sparing the GTPase-activating protein-related domain
NF1 mutations in the 2818 amino acid protein neurofibromin occured in a pattern typical of loss-of-function gene alterations, being evenly distributed across the entire protein with an enrichment of inactivating truncating mutations. (Figure 4). The well characterized Ras-GTPase-activating protein (GAP)-related domain (GRD), which down-regulates the Ras signaling pathway, harbored both missense and inactivating mutations.
Figure 4 – Distribution of NF1-mutations within neurofibromin.
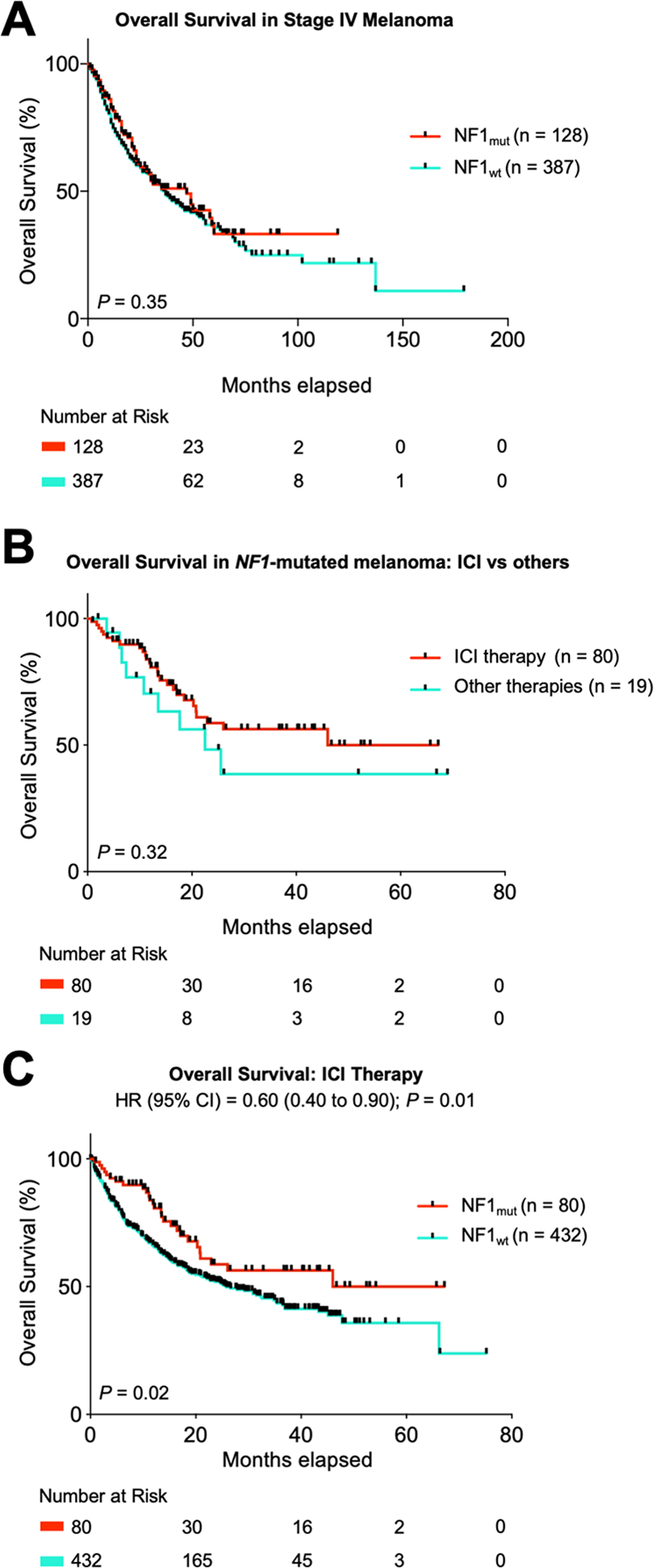
Analysis of mutations in NF1 mutated melanoma reveals no clustering or hotspot of mutations and no sparing of the GRD region on neurofibromin (A, B).
Discussion
To our knowledge, this study reports the largest cohort of NF1-mutated melanomas investigated to date. Analysis revealed a striking difference in the sex distribution pattern of NF1-mutated melanomas in favor of male patients - this distribution varied among different subgroups of NF1-mutated melanoma: 72% of patients with non-acral cutaneous melanoma were male, whereas 69% and 83% of patients with acral and mucosal melanoma were female, respectively. Mucosal melanoma being more frequent amongst female patients was documented previously 32,33. The reason for the strong male predominance in non-acral cutaneous NF1-mutated melanoma in our cohort is not apparent. The median age at diagnosis of NF1-mutated patients in our cohort (67 years), is significantly older compared to patients of other large melanoma cohort studies 34,35.
A large proportion of NF1-mutated cutaneous melanomas arose in the head and neck region. This observation is consistent with previous findings that NF1 mutations preferentially occur in sun-exposed areas as UV-radiation is the main driver for mutagenesis and represents a clustering specific for this subgroup 22,36.
The frequency of concurrent BRAFV600 mutations (17%) and NRASQ61 mutations (17%) was higher compared to previous studies of NF1-mutated melanomas 6,22,36. This discrepancy could be due to smaller numbers of NF1-mutated melanoma assessed in previous studies and differences in terms of tumor origin. Activating BRAF and NRAS mutations were less frequent in tumors harboring inactivating NF1 mutations (Table 2). This fits with missense or truncating NF1 mutations leading to a stronger activation of the MAPK pathway. Both mucosal and acral melanomas harbored comparatively fewer BRAF and NRAS mutations, fitting previous studies 33,37–40.
Table 2 –
Distribution of BRAF and NRAS mutations among NF1 mutated melanoma
| Variable, n (%) | Truncating / Frameshift (n = 115) | Other Mutations (n = 151) | p-Value |
|---|---|---|---|
| Mutation | p < 0.0001 | ||
| BRAF V600E | 10 (8.7) | 34 (22.5) | |
| NRAS Q61 | 9 (7.8) | 36 (23.8) | |
Patients with mucosal melanoma showed significantly fewer tumor mutations (Figure 1, B), a consequence of the lack of UV exposure, the main driver of mutations in cutaneous melanoma 41. Mucosal melanomas revealed, as expected, a significantly lower number of UV-signature C>T substitutions (Supplemental Figure 1).
Mutations in the TERT promoter region, usually exhibiting C>T transitions, may be UV-induced 42 but are also frequent in non-UV-induced tumors such as gliomas 43. TERT promoter mutations were rare in mucosal melanoma in our study, but were present in >30% of acral melanomas and >70% of non-acral cutaneous melanomas. We found that NF1-mutated melanoma exhibits more UV-induced C>T and CC>TT substitutions compared to other genetic subtypes. Further, NF1-mutated melanomas in our cohort harbor significantly more mutations compared to other melanoma subtypes, supporting previously published data 17.
Both missense and inactivating mutations in NF1 were distributed throughout the neurofibromin protein, typical of loss-of-function mutations 44. Whereas activating, gain-of-function, mutations have to occur at specific sites, inactivating alterations, in particular frame-shift and truncating mutations can generally occur throughout the gene. An enrichment of truncating mutations was identified in our cohort (Figure 4). We detected 96 truncating and 23 frameshift mutations. This is in line with findings from previous studies, in which an enrichment of truncating, but not frameshift mutations was noted 36. A limitation of our study is that our assay could not reliably detect losses of larger indels of the NF1 gene, meaning that likely some patients with bona fide NF1 alterations were not recognized45. Another generally difficult issue, is that NF1 is a very large gene (cDNA of circa 9000bp, depending on splice variant) and mutations can occur throughout the gene. Considering melanoma generally has a very high number of passenger mutations, these certainly do also occur in the NF1 gene. As demonstrated in other studies36,45,46, one can assume NF1 mutations are functionally more relevant when inactivating and occurring in tumors not harboring other known activating mutations (i.e. BRAF, NRAS, etc.). The predicted functional relevance of the individual NF1 mutations as determined by different algorithms (PROVEAN, SIFT and PolyPhen-2) is listed in Supplemental Table 6.
Survival analysis of stage IV melanoma patients showed no significant difference between the overall survival in NF1-mutated to -wild-type melanoma. The clinical data (Supplemental Table 1) showed a significant age difference with NF1-mutated melanomas occuring in older individuals. The prolonged OS observed in NF1-mutant tumor patients receiving first-line immunotherapy compared to patients with NF1 wild-type melanoma may be associated with tumor mutational burden. The size of our sequencing panel did not allow a valid estimation of tumor mutational burden (generally requiring sequencing of 1 megabase of DNA), however in our panel, we did observe a higher average number of mutations per sample in NF1-mutated melanoma than other subtypes (Figure 1). This supports existing data from patients with a high mutational burden compared to patients with a low mutational burden 18–20. The trend of NF1-mutated tumors for better OS of patients receiving ICI compared to other therapies including targeted therapies, tyrosine kinase inhibitors, and chemotherapies (Figure 2, B) did not reach statistical significance, probably due to the limited available patient numbers (n = 80 ICI vs. n = 19 other). Within the group of NF1-mutated ICI therapy treated patients, the only parameter determined to be significantly different in non-responders (SD/PD) versus responders (PR/CR) was the pre-treatment tumor PD-L1 status, which was more often positive (>= 5% of tumor cells stained) in the group of treatment responders (Supplemental Table 5). The prognosis of all NF1-mutated melanoma patients has been reported to be significantly worse than patients with other mutation patterns 17. This was not observed in our cohort, but this may be due to many of the patients having received immunotherapy. If validated in larger studies, the improved response to immune checkpoint inhibition therapy we observed, supports treating patients with NF1-mutated non-acral cutaneous melanoma with immunotherapy.
In the mucosal melanoma group, five patients received treatment with PD-1 agents. One patient exhibited a complete response, three progressive disease and one stable disease were noted. In acral melanoma group, of six patients received treatment with PD-1 agents, 5 patients exhibited progressive disease and one stable disease. This data suggests patients with NF1-mutated acral or mucosal melanoma may differ from cutaneous non-acral melanoma and a potential benefit from checkpoint-blockade therapy will need to be evaluated in larger studies.
Our data suggests that patients with NF1-mutated melanoma treated with immunotherapy exhibit better overall survival than those with NF1-wild-type melanoma. If validated in future studies, NF1 mutation status may be a biomarker to consider when selecting which melanoma patients may benefit from immunotherapy. However, given the retrospective nature of this study, survival analysis should be adjusted for factors such as LDH in future analyses. Unfortunately, we were unable to perform this analysis because of missing information in some cases of ICI-treated melanoma. Acral and mucosal NF1-mutated melanomas should be considered distinct subtypes of NF1-mutated melanomas with unique clinical characteristics and mutational patterns meriting further exploration in larger studies.
Conclusion:
Non-acral NF1-mutated melanoma occurs frequently in male patients and is clinically distinct from NF1-mutated acral and mucosal melanoma
NF1-mutated melanoma had a similar overall survival to NF1 wild-type melanoma when not stratifying for therapy received
NF1-mutated melanoma harbored more UV signature mutations than BRAF-mutated, NRAS-mutated or Triple-wild-type melanomas
NF1-mutated melanoma patients receiving immunotherapy had improved overall survival compared to NF1 wild-type melanoma patients
Supplementary Material
Supplemental Figure 1 – Mutational patterns in NF1-mutated melanomas
Non-Acral and acral NF1-mutated melanomas harbor higher numbers of C>T substitutions compared to mucosal NF1-mutated melanomas (A). NF1-mutated melanomas exhibit the greatest amount of CC>TT substitutions compared to BRAF, NRAS, or triple-wild-type melanomas (B).
Highlights.
NF1-mutated melanoma patients respond favorably to PD-1 based immunotherapy
NF1-mutated metastatatic melanoma had a similar overall survival to NF1 wild-type
Non-acral, acral and mucosal NF1-mutated melanoma are clinically distinct
NF1-mutated melanoma exhibit a large amount of UV signature mutations
Acknowledgments
The authors are indebted to all patients and their relatives.
Footnotes
Declaration of interests
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
C.M.T.: No relevant conflicts of interest.
E.C.: No relevant conflicts of interest.
J.M.: Declares travel support from Bristol Myers Squibb, Novartis and Sun Pharmaceutical Industries, outside the submitted work.
R.M.: No relevant conflicts of interest.
A.Z.: Declares travel support from Novartis, Sanofi Grenzyme, and Bristol-Myers Squibb, outside the submitted work.
G.L.: Declares travel support from Sun Pharma, outside the submitted work.
P.J.: No relevant conflicts of interest.
L.R.: No relevant conflicts of interest.
J.K.: No relevant conflicts of interest.
I.M.: No relevant conflicts of interest.
A.S.: No relevant conflicts of interest.
R.H.: Declares speakers and advisory board honoraria from BMS, MSD, Novartis, Pierre Fabre, Roche and SUN-Pharma, outside the submitted work.
P.T.: Declares honoraria from Bristol-Myers Squibb, Novartis, Merck Sharp & Dohme, Pierre-Fabre, CureVac, Merck Serono, Sanofi, Roche, Kyowa Kirin, Biofrontera; travel support from Bristol-Myers Squibb and Pierre-Fabre, outside the submitted work.
J.U.: Declares advisory board or has received honoraria and travel support from Amgen, Bristol Myers Squibb, GSK, LeoPharma, Merck Sharp and Dohme, Novartis, Pierre Fabre, Roche, Sanofi, outside the submitted work.
C.P.: served as consultant and/or has received honoraria from Bristol-Myers Squibb, Merck Sharp & Dohme, Novartis, Sanofi, Sunpharma, Pierre Fabre, AbbVie, Kyona Kirin and Amgen and received travel support from Amgen, Merck Sharp & Dohme, Bristol-Myers Squibb, Pierre Fabre, Sunpharma and Novartis, outside the submitted work.
J.Ul: Declares travel support: Medac, Sun Pharma; consulting: Bristol-MyersSquibb, Sun Pharma; lectures: Bristol-MyersSquibb, MSD, Merck, Novartis, Roche, Sanofi, Sun Pharma; grants: Novartis, outside the submitted work.
A.K.: No relevant conflicts of interest.
P.M.: Declares research support (to institution): Bristol-MyersSquibb, Novartis, MSD. Honoraria for lectures (personally): Roche Pharma, Bristol-MyersSquibb, Novartis, MSD, Almirall-Hermal, Amgen, Merck-Serono, Bayer, Pierre-Fabre, Sanofi. Honoraria for advisory boards: Bayersdorf, Roche Pharma, Bristol-MyersSquibb, Novartis, MSD, Almirall-Hermal, Amgen, Pierre-Fabre, Merck-Serono, SUN, Merck-Serono, Sanofi, outside the submitted work.
R.G.: Invited speaker: Roche, BMS, MSD, Novartis, Amgen, Merck Serono, Almirall Hermal, SUN, Sanofi, Pierre-Fabre. Advisory board: BMS, Roche, Novartis, Almirall Hermal, MSD, Amgen, SUN, Sanofi, Pierre-Fabre, 4SC, Bayer, MerckSerono, Pfizer, Immunocore. Research grants: Novartis, Pfizer, Johnson & Johnson, Amgen, Merck-Serono, SUN Pharma, Sanofi. Travel/meeting support: Roche, BMS, SUN, Merck-Serono, Pierre-Fabre, outside the submitted work.
F.M.: Declares travel support or/and speaker’s fees or/and advisor’s honoraria by Novartis, Roche, BMS, MSD and Pierre Fabre and research funding from Novartis and Roche, outside the submitted work.
E.D.: No relevant conflicts of interest.
M.W.: No relevant conflicts of interest.
A.P.: No relevant conflicts of interest.
E.L.: Served as consultant and/or has received honoraria from Bristol-Myers Squibb, Merck Sharp & Dohme, Novartis, Medac, Sanofi, Sunpharma and travel support from Amgen, Merck Sharp & Dohme, Bristol-Myers Squibb, Pierre Fabre, Sunpharma and Novartis, outside the submitted work.
LZ: served as consultant and/or has received honoraria from Bristol-Myers Squibb, Merck Sharp & Dohme, Novartis, Pierre-Fabre, Sunpharma and Sanofi; Research funding to institution: Novartis; travel support from Merck Sharp & Dohme, Bristol-Myers Squibb, Amgen, Pierre-Fabre, Sanofi, Sunpharma and Novartis, outside the submitted work.
D.S. reports personal fees and non-financial support from Roche/Genentech, grants, personal fees, non-financial support and other from BMS, personal fees from Merck Sharp & Dohme, personal fees and non-financial support from Merck Serono, grant, personal fees and non-financial support from Amgen, personal fees from Immunocore, personal fees from Incyte, personal fees from 4SC, personal fees from Pierre Fabre, personal fees and non-financial support from Sanofi/Regeneron, personal fees from Array BioPharma, personal fees from Pfizer, personal fees from Philogen, personal fees from Regeneron, personal fees from Nektar, personal fees from Sandoz, grants, personal fees and non-financial support from Novartis, personal fees and non-financial support from SunPharma, Replimune, Helsinn, OncoSec and InFlaRx outside the submitted work.
E.H.: No relevant conflicts of interest.
S.U. declares research support from Bristol Myers Squibb and Merck Serono; speakers and advisory board honoraria from Bristol Myers Squibb, Merck Sharp & Dohme, Merck Serono, Novartis and Roche, and travel support from Bristol Myers Squibb, and Merck Sharp & Dohme, outside the submitted work.
K.G.: No relevant conflicts of interest.
Financial disclosure:
CMT was supported as a Junior Clinician Scientist within the University Medicine Essen Academy (UMEA) funded by the Faculty of Medicine, University of Duisburg-Essen. This research was funded in part through the NIH/NCI Cancer Center Support Grant P30 CA008748. This work was partly funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) - SCHA 422/17-1 and HO 6389/2-1 (KFO 337). This work was in part supported by Bristol Myers Squibb for the multicenter translational study “Tissue Registry in Melanoma” (TRIM) within the framework of the skin cancer registry ADOREG of the German Dermatologic Cooperative Oncology Group (DeCOG).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ajithkumar T, Parkinson C, Fife K, Corrie P, Jefferies S. Evolving treatment options for melanoma brain metastases. The lancet oncology. 2015;16(13):e486–497. [DOI] [PubMed] [Google Scholar]
- 2.Schadendorf D, van Akkooi ACJ, Berking C, et al. Melanoma. Lancet. 2018;392(10151):971–984. [DOI] [PubMed] [Google Scholar]
- 3.Ugurel S, Rohmel J, Ascierto PA, et al. Survival of patients with advanced metastatic melanoma: The impact of MAP kinase pathway inhibition and immune checkpoint inhibition - Update 2019. Eur J Cancer. 2020;130:126–138. [DOI] [PubMed] [Google Scholar]
- 4.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. The New England journal of medicine. 2005;353(20):2135–2147. [DOI] [PubMed] [Google Scholar]
- 5.Krauthammer M, Kong Y, Ha BH, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nature genetics. 2012;44(9):1006–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas N Genomic Classification of Cutaneous Melanoma. Cell. 2015;161(7):1681–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nissan MH, Pratilas CA, Jones AM, et al. Loss of NF1 in cutaneous melanoma is associated with RAS activation and MEK dependence. Cancer research. 2014;74(8):2340–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donia M, Ellebaek E, Ollegaard TH, et al. The real-world impact of modern treatments on the survival of patients with metastatic melanoma. Eur J Cancer. 2019;108:25–32. [DOI] [PubMed] [Google Scholar]
- 9.Ugurel S, Rohmel J, Ascierto PA, et al. Survival of patients with advanced metastatic melanoma: the impact of novel therapies-update 2017. Eur J Cancer. 2017;83:247–257. [DOI] [PubMed] [Google Scholar]
- 10.Hodi FS, O’Day SJ, McDermott DF, et al. Improved survival with ipilimumab in patients with metastatic melanoma. The New England journal of medicine. 2010;363(8):711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McArthur GA, Chapman PB, Robert C, et al. Safety and efficacy of vemurafenib in BRAF(V600E) and BRAF(V600K) mutation-positive melanoma (BRIM-3): extended follow-up of a phase 3, randomised, open-label study. The lancet oncology. 2014;15(3):323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schadendorf D, Hodi FS, Robert C, et al. Pooled Analysis of Long-Term Survival Data From Phase II and Phase III Trials of Ipilimumab in Unresectable or Metastatic Melanoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015;33(17):1889–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolchok JD, Chiarion-Sileni V, Gonzalez R, et al. Overall Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. The New England journal of medicine. 2017;377(14):1345–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gellrich FF, Schmitz M, Beissert S, Meier F. Anti-PD-1 and Novel Combinations in the Treatment of Melanoma-An Update. J Clin Med. 2020;9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romanski NA, Holmstroem RB, Ellebaek E, Svane IM. Characterization of risk factors and efficacy of medical management of immune-related hepatotoxicity in real-world patients with metastatic melanoma treated with immune checkpoint inhibitors. Eur J Cancer. 2020;130:211–218. [DOI] [PubMed] [Google Scholar]
- 16.Sample A, He YY. Mechanisms and prevention of UV-induced melanoma. Photodermatol Photoimmunol Photomed. 2018;34(1):13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cirenajwis H, Lauss M, Ekedahl H, et al. NF1-mutated melanoma tumors harbor distinct clinical and biological characteristics. Mol Oncol. 2017;11(4):438–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372(26):2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351(6280):1463–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bauer J, Buttner P, Murali R, et al. BRAF mutations in cutaneous melanoma are independently associated with age, anatomic site of the primary tumor, and the degree of solar elastosis at the primary tumor site. Pigment cell & melanoma research. 2011;24(2):345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiesner T, Kiuru M, Scott SN, et al. NF1 Mutations Are Common in Desmoplastic Melanoma. The American journal of surgical pathology. 2015;39(10):1357–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whittaker SR, Theurillat JP, Van Allen E, et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer discovery. 2013;3(3):350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balch CM, Gershenwald JE, Soong SJ, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27(36):6199–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gershenwald JE, Scolyer RA. Melanoma Staging: American Joint Committee on Cancer (AJCC) 8th Edition and Beyond. Annals of surgical oncology. 2018;25(8):2105–2110. [DOI] [PubMed] [Google Scholar]
- 26.Griewank KG, Westekemper H, Murali R, et al. Conjunctival melanomas harbor BRAF and NRAS mutations and copy number changes similar to cutaneous and mucosal melanomas. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19(12):3143–3152. [DOI] [PubMed] [Google Scholar]
- 27.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31(16):2745–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. . 2020. [Google Scholar]
- 30.Team R. RStudio: Integrated Development for R. RStudio, Inc., Boston, MA. 2019. [Google Scholar]
- 31.Petljak M, Alexandrov LB. Understanding mutagenesis through delineation of mutational signatures in human cancer. Carcinogenesis. 2016;37(6):531–540. [DOI] [PubMed] [Google Scholar]
- 32.Cosgarea I, Ugurel S, Sucker A, et al. Targeted next generation sequencing of mucosal melanomas identifies frequent NF1 and RAS mutations. Oncotarget. 2017;8(25):40683–40692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heppt MV, Roesch A, Weide B, et al. Prognostic factors and treatment outcomes in 444 patients with mucosal melanoma. Eur J Cancer. 2017;81:36–44. [DOI] [PubMed] [Google Scholar]
- 34.Anderson WF, Pfeiffer RM, Tucker MA, Rosenberg PS. Divergent cancer pathways for early-onset and late-onset cutaneous malignant melanoma. Cancer. 2009;115(18):4176–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morgese F, Sampaolesi C, Torniai M, et al. Gender Differences and Outcomes in Melanoma Patients. Oncol Ther. 2020;8(1):103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krauthammer M, Kong Y, Bacchiocchi A, et al. Exome sequencing identifies recurrent mutations in NF1 and RASopathy genes in sun-exposed melanomas. Nature genetics. 2015;47(9):996–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abu-Abed S, Pennell N, Petrella T, Wright F, Seth A, Hanna W. KIT gene mutations and patterns of protein expression in mucosal and acral melanoma. J Cutan Med Surg. 2012;16(2):135–142. [DOI] [PubMed] [Google Scholar]
- 38.Beadling C, Jacobson-Dunlop E, Hodi FS, et al. KIT gene mutations and copy number in melanoma subtypes. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14(21):6821–6828. [DOI] [PubMed] [Google Scholar]
- 39.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24(26):4340–4346. [DOI] [PubMed] [Google Scholar]
- 40.Zaremba A, Murali R, Jansen P, et al. Clinical and genetic analysis of melanomas arising in acral sites. Eur J Cancer. 2019;119:66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newell F, Kong Y, Wilmott JS, et al. Whole-genome landscape of mucosal melanoma reveals diverse drivers and therapeutic targets. Nat Commun. 2019;10(1):3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339(6122):957–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGillicuddy LT, Fromm JA, Hollstein PE, et al. Proteasomal and genetic inactivation of the NF1 tumor suppressor in gliomagenesis. Cancer Cell. 2009;16(1):44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayward NK, Wilmott JS, Waddell N, et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545(7653):175–180. [DOI] [PubMed] [Google Scholar]
- 46.Shain AH, Garrido M, Botton T, et al. Exome sequencing of desmoplastic melanoma identifies recurrent NFKBIE promoter mutations and diverse activating mutations in the MAPK pathway. Nature genetics. 2015;47(10):1194–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1 – Mutational patterns in NF1-mutated melanomas
Non-Acral and acral NF1-mutated melanomas harbor higher numbers of C>T substitutions compared to mucosal NF1-mutated melanomas (A). NF1-mutated melanomas exhibit the greatest amount of CC>TT substitutions compared to BRAF, NRAS, or triple-wild-type melanomas (B).


