Abstract
Allergic rhinitis (AR) is an allergen-specific immunoglobulin E-mediated inflammatory disease. Both genetic and environmental factors could play a role in the pathophysiology of AR. 5-methylcytosine (5mC) can be converted to 5-hydroxymethylcytosine (5hmC) by the ten-eleven translocation (Tet) family of proteins as part of active deoxyribonucleic acid (DNA) demethylation pathway. 5hmC plays an important role in the regulation of gene expression and differentiation in immune cells. Here, we show that loss of Tet protein 2 (Tet2) could impact the severity of AR in the ovalbumin-induced mouse model. Genome-wide 5hmC profiling of both wild-type and Tet2 KO mice in response to AR revealed that the loss of Tet2 could lead to 5hmC alteration at specific immune response genes. Both partial loss and complete loss of Tet2 alters the 5hmC dynamic remodeling for the adaptive immune pathway as well as cytokines. Thus, our results reveal a new role of Tet2 in immunology, and Tet2 may serve as a promising target in regulating the level of immune response.
Introduction
Allergic sensitization, defined as the propensity to produce immunoglobulin E (IgE) antibodies in response to environmental and food antigens, could affect up to 50% of general population and is a common thread linking and predisposing to different manifestations of allergic diseases, including asthma, eczema and rhinitis (1). Allergic rhinitis (AR) is an allergen-specific IgE-mediated inflammatory disease of the nasal mucosa, characterized by sneezing, nasal itching, rhinorrhea and nasal congestion. Epidemiologic studies have revealed that the prevalence of AR is high globally and has increased progressively within the last 30 years, currently affecting up to 40% of the population worldwide (2).
Although multiple AR therapies are available, the precise molecular mechanisms underlying the pathogenesis of AR remain unclear. It has been proposed that the interaction between genetic and environmental factors could play an important role in AR pathophysiology (3). The inflammation progress and signal pathway of AR are complex and share similarity with allograft rejection and autoimmune diseases, which still lack effective treatment. Many previous studies have identified the altered DNA methylation associated with autoimmune diseases, hypersensitive diseases and anti-tumor adaptive immune response. Studies in murine asthma models have demonstrated a role of DNA methylation/demethylation in interferon regulation, T cell polarization, cytokine and human leukocyte antigen (HLA) production (4,5).
Methylation of cytosine (C) at the fifth carbon position [5-methylcytosine (5mC)] has long been regarded as a relatively stable DNA modification in mammalian genome. In 2009, it was discovered that 5mC can be step-wise oxidized to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) by the ten-eleven translocation (Tet) protein family (Tet1, 2 and 3), which constitute the endogenous active DNA demethylation pathway (6,7). It has been shown that different Tet family members have unique and non-overlapping functions, as highlighted by the phenotypes displayed by distinct Tet-deficient mice. Both 5hmC and Tet proteins could modulate the differentiation and behavior of human immune cells which in turn regulates the level of adaptive immune response. However, it is still unknown whether 5hmC- and Tet-mediated regulations are involved in the pathogenesis of AR.
To address this question, here, we employ ovalbumin (OVA), a classic AR-induction method, to investigate the role of 5hmC and Tet proteins in AR. We observed that mice without Tet2 could develop severe allergy symptoms; whereas, the mice without Tet1 only displayed a very mild response. We determined the genomic distribution and dynamic changes of 5hmC and correlated 5hmC changes with transcriptomic analysis to determine the functional significance of 5hmC modifications for transcriptional regulation of gene expression. Our results provide a genome-wide view of 5hmC remodeling during mouse hypersensitive response and reveal that the loss of Tet2 could lead to the up-regulation of MHC in nasal mucosa, thus promoting their immune response. Our study suggests the important roles of DNA methylation dynamics in regulating the expression of adaptive immune response and reveals a new role of Tet2 in AR, indicating that Tet2 may serve as a promising target in regulating the level of immune response in the future.
Results
Dynamic 5hmC changes induced by allergic exposure in an AR mouse model
An experimental OVA-induced allergic mouse model was previously established as an AR disease model (8). OVA exposure induces sneezing, nasal rubbing and nasal discharge. The phenotype is quantified using a phenotype score based on the number of sneezes, nasal rubbing and nasal discharge amount in the 15 min after last OVA exposure. We used the OVA-induced model to determine the 5hmC dynamics upon allergic exposure. To determine the change of 5hmC abundance upon OVA exposure, we first performed the immunostaining using a 5hmC-specific antibody and found that, during allergic inflammation, the AR nasal mucosa had less 5hmC when compared with the control group (Fig. 1A). We then performed a dot blot using a 5hmC-specific antibody (Fig. 1B) and observed a similar decrease of genome-wide 5hmC abundance in rhinitis.
Figure 1 .
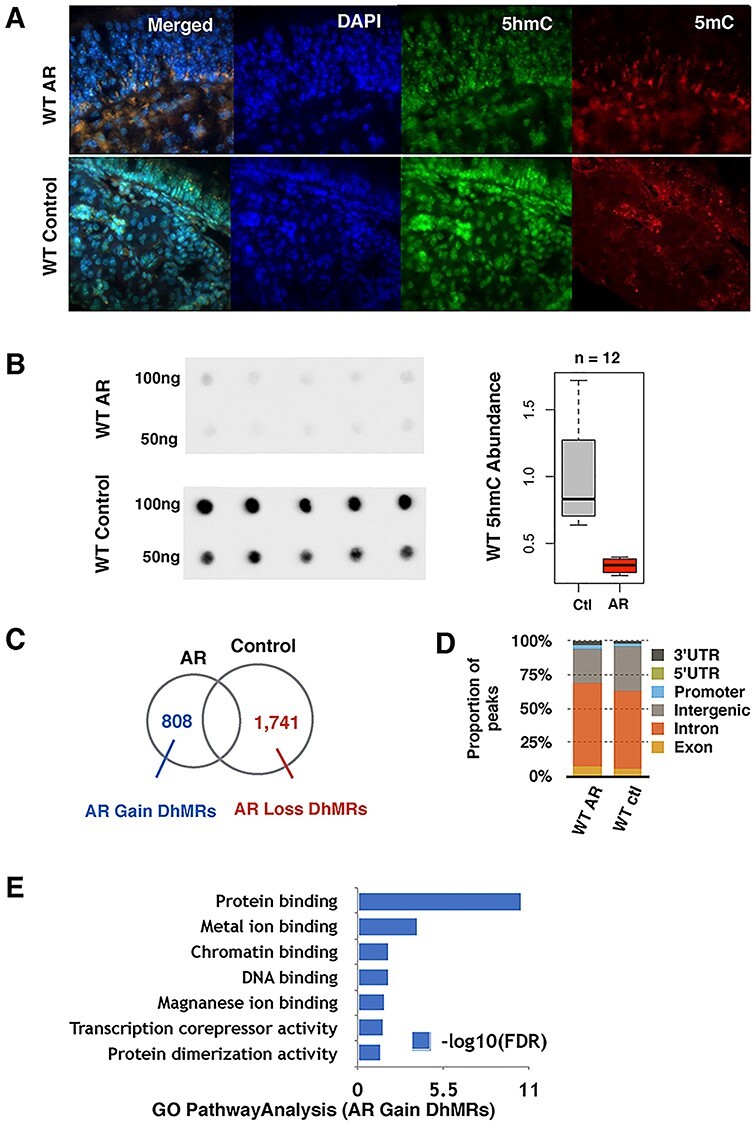
OVA-induced AR causes dynamic changes of 5hmC in WT mice nasal mucosa. (A) Immunofluorescence of the in vivo WT mice nasal mucosa 5hmC distribution is shown in green and 5mC in red. (B) Global 5hmC content of WT nasal mucosa purified 100 ng and 50 ng DNA measured by immuno-dot blot. Density analysis shows that 5hmC levels significantly decrease during the AR condition. (C) Identification of AR-induced dynamically hydroxymethylated gain-DhMRs and loss-DhMRs by DESeq2 (P-value < 0.05, absolute log 2 folds change >0.848). (D) Genomic annotation of DhMRs to demonstrate percentage change of each genomic region. WT AR mice gained coding region 5hmC and lost non-coding region 5hmC. (E) GO analysis functional annotation for the 808 WT-AR-gain-DhMRs.
To identify the genomic loci with altered 5hmC, we profiled the genome-wide 5hmC distribution by employing a previously established chemical labeling and affinity purification method, coupled with high-throughput sequencing technology (9). The specific AR dynamically hydroxymethylated regions (DhMRs) in wild-type (WT) mice (AR-DhMRs, P-value < 0.05) could be divided into two groups, AR-gain-DhMRs (fold change >1.8) and AR-loss-DhMRs (fold change <−1.8). In total, there are 808 WT-AR-gain-DhMRs and 1741 WT-AR-loss-DhMRs (Fig. 1C), which is consistent with our observations using the dot blot assay and indicates that AR could induce a global reduction of 5hmC in WT mice nasal mucosa. Analyses of genomic features associated with these WT-AR-DhMRs suggest that these DhMRs are enriched at intron, exon and promoter regions (Fig. 1D). We next performed gene ontology (GO) analysis and found that WT-AR-gain-DhMRs were significantly associated with the genes involved in protein and DNA binding, implicating that altered 5hmC could modulate the transcriptional activities of some AR-specific genes (Fig. 1E).
Loss of Tet1 and Tet2 leads to distinct responses in AR mouse model
Considering that the Tet protein family is responsible for generating 5hmC and subsequent demethylation processes (6,7), we examined the expression of each Tet family member upon OVA exposure, including Tet1, Tet2 and Tet3 (Fig. 2A). RNA extracted from both WT AR and control mice nasal mucosa was used for qRT-PCR to determine the expression levels. We did not observe significant changes for Tet1 and Tet3 expressions; however, the expression of Tet2 was significantly elevated in AR.
Figure 2 .
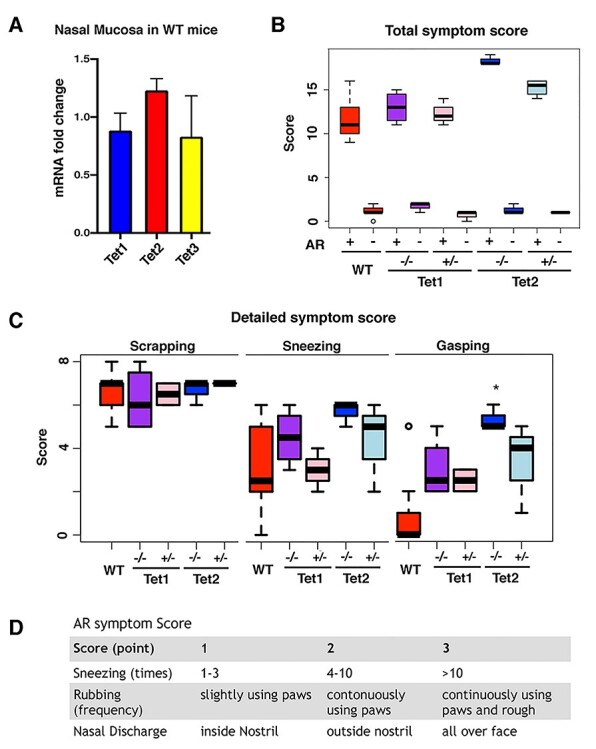
The loss of Tet2 leads to more severe symptoms. (A) Tet proteins fold changes compared with the control group in WT AR mice nasal mucosa using cDNA qRT-PCR. Tet2 is the only member that shows a significant difference (P-value < 0.05). (B) Total score of each group’s allergic symptoms for 15 min after the last OVA exposure. All AR models had severe allergic symptoms. (C) Phenotype scores for scraping, sneezing and gasping in AR mice. Tet2−/− mice have a significantly higher score than WT mice (P < 0.05) in gasping. *P-value < 0.05. (D) AR symptom score scale is shown.
To determine the role of the different Tet proteins in AR, we employed the previously established global knockout lines of Tet1 and Tet2 for the OVA-induced allergic paradigm. Both heterozygotes and homozygotes as well as their WT littermates were used for the analyses. Either partial or complete loss of Tet1 had no impact on the response to allergic exposure. However, the loss of Tet2 could significantly increase the severity of the phenotype, which was reflected by the higher symptom scores and increased gasping symptoms. Compared with WT littermates, both heterozygotes and homozygotes displayed stronger phenotypes, and the severity of phenotype correlated with the Tet2 dosage (Fig. 2B–D).
To further confirm the behavioral symptoms that we observed before, we performed an enzyme-linked immunosorbent assay (ELISA) to determine the serum levels of total IgE and OVA-specific IgE. As shown in Figure 3, the increased total IgE and OVA-specific IgE confirms that the AR group of mice had hypersensitive responses induced by OVA exposure (Fig. 3A and B). Interestingly, IL-17, an inflammatory cytokine production inducer, was highest in the Tet2 homozygous group, suggesting that the more severe hypersensitive response in Tet2 homozygote AR mice might be due the activation of the cytokine pathway (Fig. 3C). These molecular analyses are consistent with our observations in the behavioral analyses. These results together suggest that Tet2 expression level plays an important role(s) in regulating the response to allergic exposure.
Figure 3 .
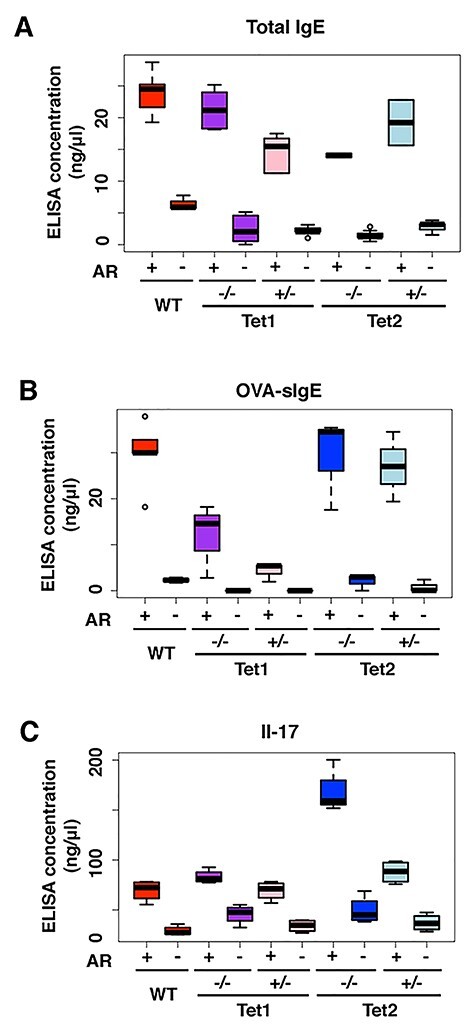
Loss of Tet1 and Tet2 leads to distinct inflammatory signal responses. (A–C) The IgE, OVA-IgE and Il-17 ELISA results of WT, Tet1−/−, Tet1+/−, Tet2−/− and Tet2+/− mice of AR and control groups, respectively. *, P-value < 0.05.
Tet2-mediated 5hmC changes in AR
To determine how 5hmC is altered in response to allergic exposure in the partial or complete loss of Tet1 or Tet2, we performed a dot blot assay using a 5hmC-specific antibody. We found that upon exposure to OVA, the loss of Tet2 leads to the accumulation of 5hmC (Fig. 4A), while the loss of Tet1 decreased 5hmC levels in response to the allergic exposure, similar to WT mice. Because the Tet2 protein is the only Tet family member with significantly altered expression levels during the AR disease process in WT mice (Fig. 2A), and the lack of Tet2 could induce the most severe phenotype (Fig. 2B and C) and highest inflammation response (Fig. 3C), we focused on the global Tet2 knockout (Tet2−/−) mice and heterozygous loss (Tet2+/−) mice in the OVA-induced AR model.
Figure 4 .
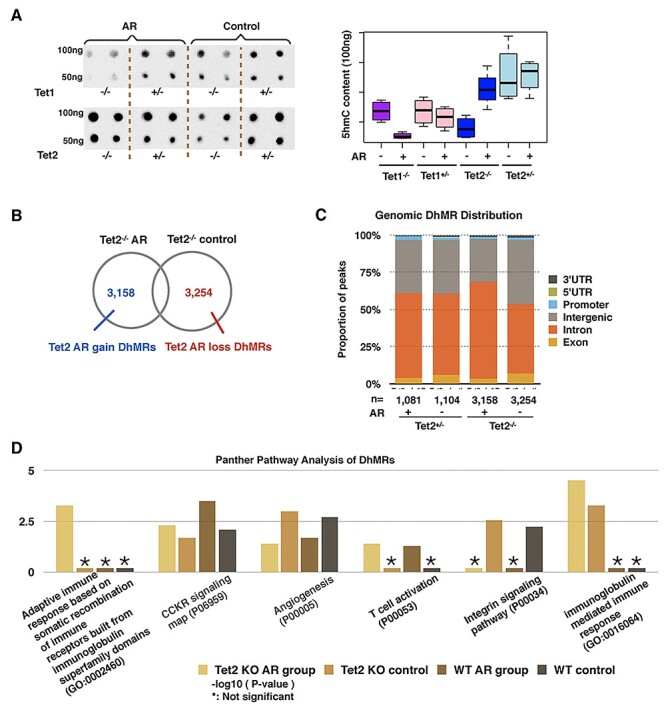
Loss of Tet2 mediated different immune response patterns in AR mice. (A) Global 5hmC content of nasal mucosa purified 100 and 50 ng DNA measured by immune-dot blot in Tet1−/−, Tet1+/−, Tet2−/− and Tet2+/− mice. Tet2−/− mice gained 5hmC content globally in nasal mucosa. The density measurement was performed by ImageJ. (B) Identification of Tet2−/− AR-induced dynamically hydroxymethylated gain-DhMRs and loss-DhMRs by DESeq2 (P-value < 0.05, absolute log 2 folds change >0.848). (C) Genomic annotation of DhMRs shows the percentage change of each genomic region. Tet2−/− AR-induced mice show a decrease in the intergenic percentage and an increase in the intronic region percentage. (D) Panther pathway analyses of DhMRs in both the wildtype and Tet2−/− groups.
We performed 5hmC profiling of Tet2−/− and Tet2+/− mice to determine 5hmC changes that occurred during the AR process. In response to AR exposure, the Tet2−/− group has 3158 AR-gain-DhMRs and 3254 AR-loss-DhMRs (Fig. 4B, with P-value < 0.05 and absolute fold change >1.8), while the Tet2+/− group had 1081 AR-gain-DhMRs and 1104 AR-loss-DhMRs (Fig. 4C). The genomic features associated with DhMRs in the Tet2+/− group displayed little change, while the Tet2−/− AR group had fewer DhMRs in intergenic regions but more in coding regions (Fig. 4C).
We next performed Panther pathway analyses and observed the enrichment of T cell activation in response to AR in both the WT and Tet2−/− groups (Fig. 4D). Interestingly, AR exposure could lead to a decrease in 5hmC at the genes involved in the integrin signaling pathway. We also observed Tet2−/−-specific 5hmC alterations in the pathway of immunoglobulin-mediated immune response, suggesting that 5hmC alterations at these genes could potentially explain the increased severity in AR mouse model. The pathway of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains was significantly changed in Tet2−/− mice under the AR condition.
The loss of Tet2 promotes the expression of MHC molecules
Given that the loss of Tet2 leads to the most severe AR phenotype, we were interested in how the loss of Tet2 could impact the transcriptional profile of the immune response genes. To determine this, we performed RNA-seq analyses on all six groups, including WT, Tet2+/− and Tet2−/− for both the AR and control groups. Through KEGG pathway analysis, we observed that the differential expressed genes of WT AR mice are enriched with olfactory receptor compensation pathway as well as various pro-inflammatory genes, like cytokines and chemokines (Fig. 5A). In the Tet2−/− AR group, the GO analysis revealed that the antigen processing and presentation pathway surpassed any other pathways (Fig. 5B).
Figure 5 .
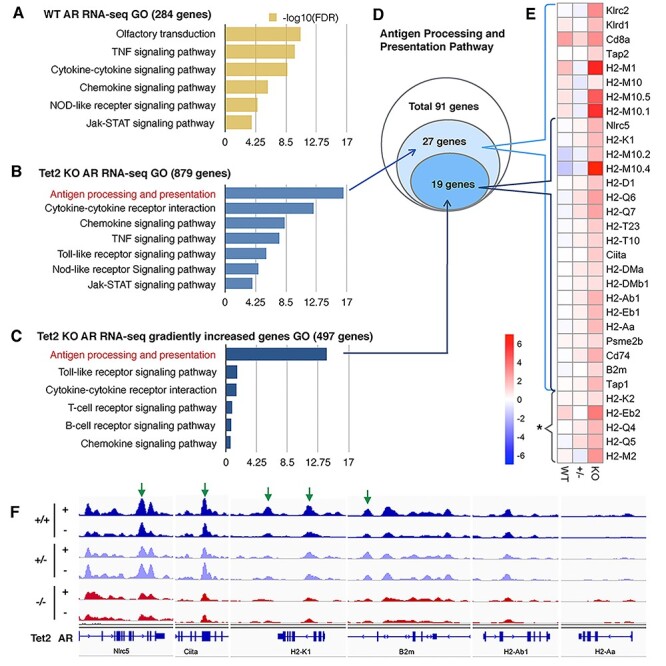
(A, B) GO analysis on pathways identified from up-regulated genes using RNA-seq on WT (284 genes) and Tet2−/− (879 genes) AR mice, respectively. (C) GO analysis results of Tet2−/− AR mice up-regulated genes from RNA-seq that showed a dosage effect with Tet2 (Tet2−/− > Tet2+/− > WT, 497 genes) suggest the significant involvement of the antigen processing and presentation pathway. (D) Of the 91 genes in the antigen processing and presentation pathway, 27 genes were captured in Tet2-KO AR mice [first pathway in (B)], with 19 of these genes showing a dosage effect with the loss of the Tet2 gene [first pathway in (C)]. (E) Heatmap of log 2 folds change of genes that involved in the first pathway of (B) and (C). *MHC molecules but do not belong to the antigen processing pathway. (F) IGV view of 5hmC dynamic changes in some MHC and linked genes. Green arrow, AR-gain-DhMRs loci (P-value > 0.05).
To highlight the dosage effect of Tet2, from the 879 up-regulated Tet2−/− AR RNA-seq genes, we filtered out the genes that showed gradually increased fold change (i.e. fold change Tet2−/− > Tet2+/− > WT from DESeq2 output). The 497 genes that showed a Tet2 allele dosage effect were associated with antigen processing and presentation pathway and pro-inflammatory genes (Fig. 5C). Of the 91 total genes of the antigen presenting KEGG pathway, Tet2−/− AR mucosa included 27 genes of which 19 genes showed a Tet2 dosage effect (Fig. 5D). The heatmap shows the RNA-seq fold change of all the MHC family molecules, including MHC class I and class II molecules, their regulator Nlrc5 and Ciita, respectively, and their associated proteins, such as β2m (Fig. 5E). The Tet2 KO AR group had the highest MHC expression levels overall.
We further examined the 5hmC marks at the MHC family genes. Multiple MHC molecules, including the MHC regulator Nlrc5 (for class I) and Ciita (for class II) displayed altered 5hmCs in the absence of Tet2 even without OVA exposure (Fig. 5F). Interestingly, both partial loss and complete loss of Tet2 could alter 5hmC modifications at these MHC loci. This suggests that the partial or complete loss of Tet2 could predispose the animals to stronger immune responses.
Discussion
There has been a rapid increase in AR prevalence over the past 30 years. Genetics cannot fully explain this rapid increase. Therefore, increasing attention has focused on the epigenetic regulation of AR. Many factors, such as antibiotics, microorganisms and environmental pollution, may eventually lead to epigenetic changes. Many epigenetic studies have linked DNA methylation/demethylation to inflammatory, oncogenesis, adaptive immune response and especially hypersensitive diseases (4,10–12). AR is an IgE-mediated allergic disease characterized by inflammation of the nasal mucosa and is caused by exposure to allergens. Accumulating evidence suggests that DNA methylation is closely related to AR. In particular, Tet2 has been shown to regulate the adaptive immune response. In our current study, we show that loss of Tet protein (Tet2) can impact the severity of AR in OVA-induced mouse model. Genome-wide 5hmC profiling of both WT and Tet2 KO mice in response to AR revealed that the loss of Tet2 led to 5hmC alteration at specific immune response genes. Both partial loss and complete loss of Tet2 could alter the 5hmC dynamic remodeling for the adaptive immune pathway as well as cytokines. Our data reveal a new role of Tet2 in AR and suggest that Tet2 could be a potential target in regulating the level of immune response.
Environment could affect the epigenetic regulation of the immune system. It was reported that particulate matter ≤2.5 μm (PM2.5) exposure could exacerbate AR in mice by increasing DNA methylation in the INF-γ gene promoter in CD4 + T cells via the extracellular signal-regulated protein kinase (ERK)-DNA methyltransferase (DNMT) pathway (13). Participants with grass pollen-induced AR underwent epigenetic changes within 3 h of exposure to grass pollen. Further, schlafen 12 (SLFN12) DNA methylation significantly correlated with symptoms, and the baseline DNA methylation pattern was found to be predictive of symptom severity upon grass allergen exposure (14). A recent study showed that increased histone deacetylase (HDAC) activity was a potential tissue injury mechanism responsible for dysregulated epithelial cell repair, leading to defective epithelial barriers in AR (15). In this study, we show that 5hmC, an oxidized product of 5mC, is very dynamic and could play an important role in AR disease pathogenesis. Tet family members (Tet1/2/3) are the key enzymes involved in the oxidation of 5mC and production of 5hmC as well as 5fC and 5caC, which have been shown to play important roles in various biological processes (16), including numerous immune components and pathways. Previous studies have shown that Tet1 and Tet2 have distinct roles in T cell differentiation (17–21). Interestingly, we observed that mice without Tet2 could develop severe allergy symptoms; whereas, mice without Tet1 displayed only a very mild response, indicating that Tet2 could play a unique role in AR.
Antigen presentation by MHC proteins is essential for adaptive immunity. Following exposure of nasal mucosa to allergens, allergens are captured and processed by dendritic cells (DCs). DCs present antigens to T cells in the context of MHC class II molecules, causing T cells to proliferate and differentiate, resulting in a series of immune responses. Some studies have reported a relationship between the human MHC gene (HLA) and allergic airway diseases. A genome-wide association study (GWAS) of asthma has confirmed that HLA-DR/DQ, part of the HLA class II region, is associated with asthma (22). MHC class II molecules, including HLA-DQ, are known for their roles in allergen binding and immune responses driven by T helper type 2 (Th2) cells. Waage et al. (23) identified the risk loci and genetic pathways underlying AR through genome-wide association and HLA fine-mapping studies, finding that the GWAS signal at this locus involved structural changes related to allergen binding properties. Morii et al. (24) studied the association of Japanese cedar (JC) pollinosis and sensitization with HLA-DPB1 in the Japanese adolescent, demonstrating that amino acid changes in the allergen-binding pocket of HLA-DPB1 could influence pollinosis or sensitization to the allergenic peptide of JC pollen and could determine the pollinosis risk. Zhao et al. (25) found that HLA-DRB1 and HLA-DQB1 were related to HDM-sensitive AR in China. Further, they demonstrated that HLA-DRB1 and HLA-DQB1 gene polymorphisms were associated with allergen-specific immunotherapy (AIT) efficacy in HDM-sensitive Chinese patients with AR (26). Here, we show that the loss of Tet2 can increase the severity in the OVA-induced AR mouse. Accordingly, we observed alterations of both MHC classes I and II molecule (e.g. H2-M10, H2-Ab1), their regulators (Ciita, Nlrc5 and STAT1) and ligands (e.g. β2m). Interestingly, most of the altered antigen-presenting pathway could be regulated by Nlrc5 (MHC class I cotransfactor) and Ciita (MHC class II cotransfactor).
Lastly, the nasal mucosa contains different types of cells, including epithelium cells, mesenchymal cells and immunocytes. Although we observed no significant gain of lymphocyte infiltration in Tet2−/− or Tet2+/− AR models, it remains to be determined whether the alteration at MHC loci occurred in immunocytes or non-immunocyte cells. In summary, our data revealed that the loss of Tet2 could predispose the host to the hyper-immune response, thus Tet2 could be a potential target in regulating the level of immune response.
Materials and Methods
Murine model of AR
C57BL/6 WT mice (The Jackson Lab, Maine, USA) were used for the initial AR experiment. To further explore the epigenetic mechanism involved in OVA-induced AR mice model, we used the previously generated global Tet1 KO (Tet1−/−), Tet2 KO (Tet2−/−), Tet1 heterozygous (Tet1+/−) and Tet2 heterozygous (Tet2+/−) mice. The Tet1 KO line was obtained from The Jackson Laboratory (stock number 017358, B6; 129S4-Tet1tm1.1dae/J) (27) and has been bred with C57BL/6 WT mice. The Tet2 KO line was previously generated in 129/sv background and has been maintained in 129/sv background (28). All animal procedures occurred according to protocols approved by Emory University Institutional Animal Care and Use Committee, and all mice used in this study were male and were 6–8 weeks old.
The AR model was established as previously described (8). Briefly, mice were sensitized by administration of an intraperitoneal injection of OVA (75 μg) in 200 μl of phosphate-buffered solution (PBS) containing 2 mg of aluminum hydroxide (Sigma Aldrich, St Louis, MO, USA) in a total volume of 200 μl on days 0, 7 and 14. After initial sensitization, mice were challenged with nasal instillation of OVA 500 μg in 20 μl of PBS into bilateral nasal cavities on days 16–25. The control group was challenged with PBS instead of OVA.
Behavioral test and scoring
After nasal challenged (day 25), mouse behavior was observed and recorded. The frequencies of sneezing and nasal rubbing were recorded for 15 min after final OVA challenge on day 25. Two blinded observers recorded the frequencies of sneezing and nasal rubbing in a 15-min interval with video. In a double-blind approach, two researchers observed and recorded the number and duration of symptoms in each mouse. Mild: 0–3, moderate: 4–6 and severe: 7–9. The mice were then euthanized 24 h after the last OVA challenge. Serum samples, nasal lavage and nasal mucosa from each mouse were also obtained and stored at −80°C for further analysis.
Histology and immunofluorescence staining
24 hours after the last challenge, mice were sacrificed under isoflurane anesthesia. Their decapitated heads were immersed in 4% PFA overnight, then immersed in 150 mM EDTA for 5–7 days, 30% sucrose for 24 h, then embedded in O.C.T. and cut into 5 μm sections. Sections were stained with hematoxylin and eosin (H&E) and immunofluorescence staining (5hmc and 5mc) according to the manufacturers’ recommendations.
ELISA
Serum was stored at −80°C. Serum levels of total IgE, OVA-specific IgE, IL-5, IL-6, IL-10, IL-13, IL-17 and TGF-β were measured by ELISA method according to the instructions of the kits (Thermo Fisher Scientific). The results were presented in relation to the total protein concentration for each sample.
Genomic DNA preparation
Genomic DNA was isolated from nasal tissues in 550 μl of digestion buffer (100 mM Tris-HCl, pH 8.5, 5 mM EDTA, 0.2% SDS, 200 mM NaCl), then treated with Proteinase K (Thermo) at 55°C overnight. The second day, 550 μl of phenol:chloroform:isoamyl alcohol (25:24:1 saturated with 10 mM Tris, pH 8.0, 1 mM EDTA) (P-3803, Sigma) was added to samples, mixed completely and centrifuged for 10 min at 12 000 rpm. The aqueous layer was transferred into a new Eppendorf tube and precipitated with 600 μl of isopropanol. The pellet was washed with 75% ethanol, air-dried and resuspended with Nuclease-Free Water (Ambion). DNA samples were treated extensively with an RNase cocktail (Ambion) to remove contaminating RNA.
5hmC dot blot
Dot blot was performed on a Bio-Dot Apparatus (cat#170-6545, BIO-RAD), as described previously (9), using rabbit antibody to 5hmC (1:10 000, cat#39769, Active Motif) as the primary antibody, incubated overnight at 4°C. Horseradish peroxidase–conjugated antibody to rabbit (1:5000, cat#A-0545, Sigma) was used as the secondary antibody and was incubated for 45 min at 20–25°C. Standard DNA templates were loaded (cat#D5405, ZYMO) for the quantification and to verify the specificity of antibodies. The density of each dot signal was quantified by ImageJ software (29).
RNA isolation and qRT-PCR
Nasal mucosa samples were homogenized in TRIzol (Invitrogen, cat# 15596026) and processed according to the manufacturer’s instructions. RNA was then reverse transcribed using a SuperScript III First-Strand Synthesis System (Thermo Fisher, cat# 18080051). cDNA was quantified by qPCR using TaqMan Universal PCR Master Mix (Thermo Fisher, cat# 4304437) and SYBR Green PCR Master Mix (Thermo Fisher, cat# 4309155). Each reaction was run in triplicate and analyzed following the ΔΔCT method using glyceralde-hyde-3-phosphate dehydrogenase (Gapdh) as normalization control. All experiments were repeated at least twice. The Tet1/2/3 primers are from TaqMan (Thermo Fisher, cat# 4304437). Detailed primer sequences are: 5-CTGGCGCCGTCTTGATAGT-3 for Gata3 forward and 5-GACGGTTGCTCTTCCGATCA-3 for Gata3 reverse; 5-ACTTCAGAGTCATGAGAAGGATGC-3 for Il-5 forward and 5-GCATTTCCACAGTACCCCCA-3 for Il-5 reverse; 5-GAGGTATTGAGGGTGGGTGTC-3 for Foxp3 forward and 5-CAGCATGGGTCTGTCTTCTCTAA-3 for Foxp3 backward; 5-CTGGGCTACCCTACTGAGGA-3 for Rorc forward and 5-GTGCAGGAGTAGGCCACATT-3 for Rorc reverse; 5-GCCCTTTGCTATGGTGTCCT-3 for Il-10 forward and 5-TAGGGGAACCCTCTGAGCTG-3 for Il-10 reverse; 5-AGTCTTTAACTCCCTTGGCG-3 for Il-17a forward and 5-ATCAGGGTCTTCATTGCGGT-3 for Il-17a reverse; 5-GAGACGTGGGGACTTCTTGG-3 for Tgf-b forward and 5-AATAGGGGCGTCTGAGGAAC-3 for Tgf-b reverse; 5-GGGGTGGGGAAGAGATTGTC-3 for Ifng forward and 5-CACTGCAGCTCTGAATGTTTCTT-3 for Ifng reverse; 5-GTACCACACCGTGCTACAGG-3 for Il-9 forward and 5-CACTGCAGCTCTGAATGTTTCTT-3 for Il-9 reverse; 5-GGGTCCCAGCTTAGGTTCAT-3 for Gaphd forward and 5-TACGGCCAAATCCGTTCACA-3 for Gapdh reverse.
RNA-seq
Three biological replicates were subjected to RNA-seq. RNA-seq library construction followed instructions of Illumina mRNA sample prep kit (cat# RS-100-0801). In brief, the poly-A-containing mRNA was purified using poly-T oligo-attached magnetic beads. The mRNA was then fragmented into small pieces using divalent cations under elevated temperature. The cleaved RNA fragments were copied into first-strand cDNA using reverse transcriptase and random primers. This was followed by second-strand cDNA synthesis using DNA polymerase I and RNaseH. These cDNA fragments went through an end repair process, the addition of a single ‘A’ base and then ligation of the adapters. These products were gel-purified and enriched with PCR to create the final cDNA libraries. The library constructs were run on the bioanalyzer to verify the size and concentration before sequencing on the Illumina HiSeq2000 machines. RNA-seq reads were aligned using STAR v2.7 (30), reference genome GRCm38 and the differential expression values were extracted using DESeq2 v1.26.0 (31). Genes with absolute fold change >1.8 (WT group absolute fold change >1.4) and P-value < 0.05 were defined as differentially expressed genes.
5hmC-specific chemical labeling, affinity purification and sequencing
5hmC enrichment was performed using a previously described procedure with an improved selective chemical labeling method (32). DNA libraries were generated following the Illumina protocol for ‘Preparing Samples for ChIP Sequencing of DNA’ (Part# 111257047 Rev. A) using 25–50 ng of input genomic DNA or 5hmC-captured DNA to initiate the protocol. All sequencing libraries were run on Illumina Hi-seq 2000 machines. FASTQ sequence files from biological replicates were concatenated and aligned to the Mus musculus reference genome (GRCm38/mm10) using Bowtie2 v2.3.5.1 (33), keeping unique non-duplicate genomic matches with no more than two mismatches within the first 25 bp.
Identification and annotation of DhMRs
A Poisson-based peak identification algorithm MACS2 v2.2.6 (34) was used to find the candidate peak location (tag size = 38, bandwidth = 300, P-value cutoff = 1.00 × 10E−5). All candidate peak loci were summarized and binned for the whole genome. We used R to count each sample’s read number for each candidate loci and then used DESeq2 v1.26.0 (31) to find DhMR (any peak loci which was P-value < 0.05 and absolute fold change >1.8). We used Hypergeometric Optimization of Motif EnRichment (HOMER) software (35) to annotate with known genomic features obtained from NCBI38v16/mm10. Genomic Regions Enrichment of Annotations Tool (GREAT, version 3.0) and The Database for Annotation, Visualization and Integrated Discovery (DAVID) were employed to perform GO analyses (36,37). HOMER Motif Analysis software performed known and de novo motif searches.
Read coverage and visualization
Genomic views of read coverage were generated using Integrated Genomics Viewer tools and browser (IGV 1.4.2, http://www. broadinstitute.org/igv/) (38) with a window size of 25 and a read extend of 270. We used ngsplot (39) for 5hmC remodeling visualization in specific gene regions.
Quantification and statistical analysis
All statistical analyses were performed in Prism 7.0 (GraphPad Software). Datasets were analyzed for significance using either unpaired Student’s two-tailed t-tests or ANOVA with multiple comparison post hoc tests; all data are presented as mean ± SEM. Samples and animal groups with P-value < 0.05 were considered to be statistically significant. Pearson’s chi-square tests with Yates’ continuity correction were performed in R software (http://www.r-project.org/). Sample sizes and descriptions of sample collection are provided in each figure legend.
Data and software availability
The sequencing data are archived at Gene Expression Omnibus under accession number GSE171005.
Acknowledgements
This work was supported in part by grants from the National Natural Science Foundation of China (81870701), the Program of Jilin Provincial Finance Department (2018scz042), Jilin Province Health Science and Technology Talent Project (2019scz004) and Jilin Provincial Natural Science Foundation (20200201411JC). Conflict of Interest statement: None declared.
Contributor Information
Cuida Meng, Department of Otolaryngology Head and Neck Surgery, China-Japan Union Hospital of Jilin University, Changchun, Jilin Province 130033, China; The Key Laboratory of Precise Diagnosis and Treatment of Upper Airway Allergic Diseases of Jilin Province, Changchun, Jilin Province 130033, China.
Lei Gu, Department of Ophthalmology, The Affiliated First hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang Province 310000, China.
Yujing Li, Department of Human Genetics, Emory University School of Medicine, Atlanta, GA 30322, USA.
Ronghua Li, Department of Human Genetics, Emory University School of Medicine, Atlanta, GA 30322, USA.
Yiqu Cao, Department of Human Genetics, Emory University School of Medicine, Atlanta, GA 30322, USA.
Ziyi Li, Department of Biostatistics, The University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Emily G Allen, Department of Human Genetics, Emory University School of Medicine, Atlanta, GA 30322, USA.
Dongdong Zhu, Department of Otolaryngology Head and Neck Surgery, China-Japan Union Hospital of Jilin University, Changchun, Jilin Province 130033, China; The Key Laboratory of Precise Diagnosis and Treatment of Upper Airway Allergic Diseases of Jilin Province, Changchun, Jilin Province 130033, China.
Peng Jin, Department of Human Genetics, Emory University School of Medicine, Atlanta, GA 30322, USA.
Funding
National Natural Science Foundation of China (81870701), the Program of Jilin Provincial Finance Department (2018scz042), Jilin Province Health Science and Technology Talent Project (2019scz004) and Jilin Provincial Natural Science Foundation (20200201411JC).
References
- 1. Ziyab, A.H. (2019) Prevalence of food allergy among schoolchildren in Kuwait and its association with the coexistence and severity of asthma, rhinitis, and eczema: a cross-sectional study. World Allergy Organ J, 12, 100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brożek, J.L., Bousquet, J., Agache, I., Agarwal, A., Bachert, C., Bosnic-Anticevich, S., Brignardello-Petersen, R., Canonica, G.W., Casale, T., Chavannes, N.H. et al. (2017) Allergic rhinitis and its impact on asthma (ARIA) guidelines-2016 revision. J. Allergy Clin. Immunol., 140, 950–958. [DOI] [PubMed] [Google Scholar]
- 3. Chiarella, S.E., Fernandez, R. and Avila, P.C. (2015) The genes and the environment in nasal allergy. Curr. Opin. Allergy Clin. Immunol., 15, 440–445. [DOI] [PubMed] [Google Scholar]
- 4. Burleson, J.D., Siniard, D., Yadagiri, V.K., Chen, X., Weirauch, M.T., Ruff, B.P., Brandt, E.B., Hershey, G.K.K. and Ji, H. (2019) TET1 contributes to allergic airway inflammation and regulates interferon and aryl hydrocarbon receptor signaling pathways in bronchial epithelial cells. Sci. Rep., 9, 7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yue, H., Yan, W., Ji, X., Gao, R., Ma, J., Rao, Z., Li, G. and Sang, N. (2017) Maternal exposure of BALB/c mice to indoor NO2 and allergic asthma syndrome in offspring at adulthood with evaluation of DNA methylation associated Th2 polarization. Environ. Health Perspect., 125, 097011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tahiliani, M., Koh, K.P., Shen, Y., Pastor, W.A., Bandukwala, H., Brudno, Y., Agarwal, S., Iyer, L.M., Liu, D.R., Aravind, L. et al. (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1) Science (New York, N.Y.), 324, 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ito, S., Shen, L., Dai, Q., Wu, S.C., Collins, L.B., Swenberg, J.A., He, C. and Zhang, Y. (2011) Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science (New York, N.Y.), 333, 1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang, N., Li, H., Jia, J. and He, M. (2015) Anti-inflammatory effect of curcumin on mast cell-mediated allergic responses in ovalbumin-induced allergic rhinitis mouse. Cell. Immunol., 298, 88–95. [DOI] [PubMed] [Google Scholar]
- 9. Song, C.X., Szulwach, K.E., Fu, Y., Dai, Q., Yi, C., Li, X., Li, Y., Chen, C.H., Zhang, W., Jian, X. et al. (2011) Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol., 29, 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bird, A. (2002) DNA methylation patterns and epigenetic memory. Genes Dev., 16, 6–21. [DOI] [PubMed] [Google Scholar]
- 11. Liang, G. and Weisenberger, D.J. (2017) DNA methylation aberrancies as a guide for surveillance and treatment of human cancers. Epigenetics, 12, 416–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kondilis-Mangum, H.D. and Wade, P.A. (2013) Epigenetics and the adaptive immune response. Mol. Asp. Med., 34, 813–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li, Y., Zhou, J., Rui, X., Zhou, L. and Mo, X. (2019) PM2.5 exposure exacerbates allergic rhinitis in mice by increasing DNA methylation in the IFN-γ gene promoter in CD4+T cells via the ERK-DNMT pathway. Toxicol. Lett., 301, 98–107. [DOI] [PubMed] [Google Scholar]
- 14. North, M.L., Jones, M.J., MacIsaac, J.L., Morin, A.M., Steacy, L.M., Gregor, A., Kobor, M.S. and Ellis, A.K. (2018) Blood and nasal epigenetics correlate with allergic rhinitis symptom development in the environmental exposure unit. Allergy, 73, 196–205. [DOI] [PubMed] [Google Scholar]
- 15. Steelant, B., Wawrzyniak, P., Martens, K., Jonckheere, A.C., Pugin, B., Schrijvers, R., Bullens, D.M., Vanoirbeek, J.A., Krawczyk, K., Dreher, A. et al. (2019) Blocking histone deacetylase activity as a novel target for epithelial barrier defects in patients with allergic rhinitis. J Allergy Clin. Immunol., 144, 1242–1253.e1247. [DOI] [PubMed] [Google Scholar]
- 16. Wu, H. and Zhang, Y. (2011) Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev., 25, 2436–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koh, K.P., Yabuuchi, A., Rao, S., Huang, Y., Cunniff, K., Nardone, J., Laiho, A., Tahiliani, M., Sommer, C.A., Mostoslavsky, G. et al. (2011) Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell, 8, 200–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li, X., Yao, B., Chen, L., Kang, Y., Li, Y., Cheng, Y., Li, L., Lin, L., Wang, Z., Wang, M. et al. (2017) Ten-eleven translocation 2 interacts with forkhead box O3 and regulates adult neurogenesis. Nat. Commun., 8, 15903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Piccolo, F.M., Bagci, H., Brown, K.E., Landeira, D., Soza-Ried, J., Feytout, A., Mooijman, D., Hajkova, P., Leitch, H.G., Tada, T. et al. (2013) Different roles for Tet1 and Tet2 proteins in reprogramming-mediated erasure of imprints induced by EGC fusion. Mol. Cell, 49, 1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang, R.R., Cui, Q.Y., Murai, K., Lim, Y.C., Smith, Z.D., Jin, S., Ye, P., Rosa, L., Lee, Y.K., Wu, H.P. et al. (2013) Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell, 13, 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang, R., Qu, C., Zhou, Y., Konkel, J.E., Shi, S., Liu, Y., Chen, C., Liu, S., Liu, D., Chen, Y. et al. (2015) Hydrogen sulfide promotes Tet1- and Tet2-mediated Foxp3 demethylation to drive regulatory T cell differentiation and maintain immune homeostasis. Immunity, 43, 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li, X., Howard, T.D., Zheng, S.L., Haselkorn, T., Peters, S.P., Meyers, D.A. and Bleecker, E.R. (2010) Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J. Allergy Clin. Immunol., 125, 328–335.e311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Waage, J., Standl, M., Curtin, J.A., Jessen, L.E., Thorsen, J., Tian, C., Schoettler, N., Flores, C., Abdellaoui, A., Ahluwalia, T.S. et al. (2018) Genome-wide association and HLA fine-mapping studies identify risk loci and genetic pathways underlying allergic rhinitis. Nat. Genet., 50, 1072–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Morii, W., Sakai, A., Ninomiya, T., Kidoguchi, M., Sumazaki, R., Fujieda, S. and Noguchi, E. (2018) Association of Japanese cedar pollinosis and sensitization with HLA-DPB1 in the Japanese adolescent. Allergol. Int., 67, 61–66. [DOI] [PubMed] [Google Scholar]
- 25. Zhao, Y., Zhao, Y., Li, J., Zhang, Y. and Zhang, L. (2016) HLA-DRB1*08:03:02 and HLA-DQB1*06:01:01 are associated with house dust mite-sensitive allergic rhinitis in Chinese subjects. Int. Forum Allergy Rhinol., 6, 854–861. [DOI] [PubMed] [Google Scholar]
- 26. Zhao, Y., Zhao, Y., Zhang, Y. and Zhang, L. (2019) HLA-II genes are associated with outcomes of specific immunotherapy for allergic rhinitis. Int. Forum Allergy Rhinol., 9, 1311–1317. [DOI] [PubMed] [Google Scholar]
- 27. Dawlaty, M.M., Ganz, K., Powell, B.E., Hu, Y.C., Markoulaki, S., Cheng, A.W., Gao, Q., Kim, J., Choi, S.W., Page, D.C. et al. (2011) Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell, 9, 166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li, Z., Cai, X., Cai, C.L., Wang, J., Zhang, W., Petersen, B.E., Yang, F.C. and Xu, M. (2011) Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood, 118, 4509–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schneider, C.A., Rasband, W.S. and Eliceiri, K.W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dobin, A., Davis, C.A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., Batut, P., Chaisson, M. and Gingeras, T.R. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England), 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Love, M.I., Huber, W. and Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol., 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Szulwach, K.E., Li, X., Li, Y., Song, C.X., Wu, H., Dai, Q., Irier, H., Upadhyay, A.K., Gearing, M., Levey, A.I. et al. (2011) 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci., 14, 1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Langmead, B. and Salzberg, S.L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods, 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang, Y., Liu, T., Meyer, C.A., Eeckhoute, J., Johnson, D.S., Bernstein, B.E., Nusbaum, C., Myers, R.M., Brown, M., Li, W. et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol., 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heinz, S., Benner, C., Spann, N., Bertolino, E., Lin, Y.C., Laslo, P., Cheng, J.X., Murre, C., Singh, H. and Glass, C.K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell, 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang da, W., Sherman, B.T. and Lempicki, R.A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc., 4, 44–57. [DOI] [PubMed] [Google Scholar]
- 37. McLean, C.Y., Bristor, D., Hiller, M., Clarke, S.L., Schaar, B.T., Lowe, C.B., Wenger, A.M. and Bejerano, G. (2010) GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol., 28, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Robinson, J.T., Thorvaldsdóttir, H., Winckler, W., Guttman, M., Lander, E.S., Getz, G. and Mesirov, J.P. (2011) Integrative genomics viewer. Nat. Biotechnol., 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shen, L., Shao, N., Liu, X. and Nestler, E. (2014) ngs.plot: quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics, 15, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequencing data are archived at Gene Expression Omnibus under accession number GSE171005.


