Abstract
Escherichia coli cells contain potential division sites at midcell and adjacent to the cell poles. Selection of the correct division site at midcell is controlled by three proteins: MinC, MinD, and MinE. It has previously been shown (D. Raskin and P. de Boer, Cell 91:685–694, 1997) that MinE-Gfp localizes to the midcell site in an MinD-dependent manner. We use here Gfp-MinD to show that MinD associates with the membrane around the entire periphery of the cell in the absence of the other Min proteins and that MinE is capable of altering the membrane distribution pattern of Gfp-MinD. Studies with the isolated N-terminal and C-terminal MinE domains indicated different roles for the two MinE domains in the redistribution of membrane-associated MinD.
Cell division in Escherichia coli and most other rod-shaped bacteria occurs by formation of a division septum at the midpoint of the cell, thus ensuring the equipartition of cytoplasmic components into the two daughter cells. To ensure that division occurs only at the midcell site, the cell must select this site in preference to other potential division sites that are located adjacent to the cell poles. If the polar sites are used to support septum formation, small chromosomeless minicells are formed that are incapable of undergoing further divisions. In E. coli, the selective suppression of the polar division sites is accomplished by the cooperative action of the three gene products of the minCDE locus: MinC, MinD, and MinE.
Previous studies (4) have shown that the three Min proteins act in the following way. MinC and MinD act in concert to form a nonspecific inhibitor of septation. In this process, MinD functions to activate the latent division inhibitory activity of MinC (6). The MinCD division inhibitor lacks site specificity, as shown by the observation that expression of minC and minD in the absence of minE leads to a block in septation at all potential division sites, leading to formation of long nonseptate filaments. Filamentation is suppressed by MinE, which acts as a topological specificity factor to prevent the division inhibitor from acting at the midcell site while permitting it to block septation at polar division sites.
The ability of MinE to counteract the MinCD division inhibitor and the ability to impart topological specificity to the system reside in different domains of the 88-amino-acid MinE protein. The N-terminal MinE domain is responsible for the anti-MinCD function, as shown by the ability of MinE1–22 (20) or MinE1–34 (13) to prevent MinCD-induced filamentation. (Superscripts [e.g., minE1–53] refer to amino acid positions within MinE or within the minE gene product.) The C-terminal domain of MinE is thought to encode the MinE topological specificity function that is responsible for limiting its action to the division site at midcell. It has been proposed that topological specificity is accomplished by the binding of the C-terminal topological specificity domain to a putative topological target molecule at the new division site at midcell (17). The sequestration of MinE at midcell would permit it to interfere with the division inhibitor at this site without interfering with the division inhibition activity at the unwanted polar division sites. As a result, normal cell division would occur and polar divisions would be prevented. Excess MinE has been shown to induce minicell formation, suggesting that once the topological target is saturated, the excess MinE molecules are free to counteract the division inhibitor elsewhere in the cell.
Consistent with the ability of MinE to specifically counteract the division inhibitor at midcell, a MinE-green fluorescent protein chimera (MinE-Gfp) localizes to a ring-like structure at sites adjacent to the midcell, and this localization pattern requires the simultaneous expression of minD (14). This implicates MinD in the process that leads to localization of MinE at midcell.
MinD plays several roles in the Min system. First, it activates the MinC division inhibitor. Second, it is required to make the division inhibitor sensitive to MinE (5, 10). Third, MinD is required to localize MinE at midcell (14). After the initial submission of the present study for publication, it was shown in a related study that MinD localizes to the cell pole in a MinE-dependent fashion and undergoes a rapid oscillation from pole to pole (16).
We have also examined the cellular localization of the MinD protein and its membrane interactions with MinE by using a green fluorescent protein-MinD fusion protein (Gfp-MinD) to monitor the cellular distribution of MinD. In this report we describe the membrane rearrangements of Gfp-MinD that are induced by coexpression with MinE, and we define a specific role for the N-terminal MinE domain in this process.
MATERIALS AND METHODS
Bacterial strains.
E. coli PB114 (λDB217) was derived from PB114 (ΔminCDE) (4) and λDB217 (Plac-minC). λDB217 was constructed by in vivo recombination of plasmid pDB217 (6) with λNT5 (3). E. coli RC1 [ΔminCDE Δ(araAB01C-leu)] was constructed by P1-mediated transduction of Δmin from PB114 into E. coli MC1000.
Plasmids.
pSY1057 (Plac-minE22–88), a pUC derivative, has been previously described (19). pJPB262 (Plac-minE1–32) was a gift from Jean-Pierre Bouché (13); this pUC8-derived plasmid contains a termination codon immediately after minE1–32 and the minE33–88 portion of minE has been deleted.
For pSY1053 (Plac-minE1–53), the HindIII/XbaI fragment from pZC20 (20), in which a translational stop codon follows minE1–53, was subcloned into pBluescript KS (Stratagene). For pDB188 (Plac-minE), the HindIII/EcoRI fragment from pDB156 (4) was inserted into HindIII/EcoRI-digested pBluescript KS. For pAS72, NdeI/HindIII-digested pET21a (Novagen) was ligated to a fragment containing the minD gene from pDB175 (4), prepared by PCR; the fragment lacks any minE sequences. For pAS74 (Plac-minD), the XbaI/HindIII minD fragment from pAS72 was inserted into XbaI/HindIII-digested pBluescript SK. For pFX1 (Plac-minE1–53::gfp), pDB175 (4) was used as template in a PCR reaction to generate a minE fragment coding for amino acids 1 to 53, preceded by the terminal region of minD that includes the ribosome-binding site for minE translation; PCR was also used to generate a fragment from pKENgfpmut2 (1) that includes the entire gfpmut2 gene, immediately preceded by a BamHI site. The two fragments were cleaved with EcoRI/BamHI and BamHI/HindIII, respectively, and the fragments were ligated into EcoRI/HindIII-digested pBluescript. The resulting pFX1 plasmid contains minE1–53 cloned in frame to gfpmut2, with an intervening four amino acid linker (Gly-Ser-Glu-Phe). For pFX11 (Plac-gfp::minD minE1–53), a minE1–53 fragment with a TAG termination codon immediately after codon 53 was generated by PCR with pSLR23 as a template. The PCR fragment was cleaved with EcoRI/HindIII and ligated into EcoRI/HindIII-digested pMLB1113 (4). The minE1–53-gfp fusion was verified by sequencing. pYW3 (Plac-minC gfp::minD minE) was constructed by ligating the 1.9-kbp HindIII (Klenow enzyme-treated)/NheI fragment from pSLR23, which contains gfp::minD minE, into pDB217 (Plac-minC) (6) that had been digested with XbaI (Klenow-treated) and SpeI.
The following plasmids were derived from the pACYC-derived plasmid pBAD33 (8) or pSJ4, a plasmid (kindly provided by S. Justice) that was derived from pBAD33 by site-directed mutagenesis that destroyed the EcoRI site in the cat gene without inactivating chloramphenicol resistance. pAS73 (Para-minD) encodes minD preceded by the ribosome-binding site and leader sequence of pET21a (Novagen). The plasmid was constructed by subcloning the XbaI/HindIII fragment from pAS72 into XbaI/HindIII-digested pBAD33. pAS86 (Para-minE::gfp) contains minE fused in frame to gfpmut2; the minE::gfpmut2 fragment was originally prepared by PCR from pKEN1gfpmut2 (1) and was transferred to SmaI/HindIII-digested pBAD33 as an XmnI/HindIII fragment from the intermediate plasmid pAS82 (Para-minD minE::gfpmut2). pSLR21 (Para-gfpmut2) encodes the gfpmut2 gene, preceded by the ribosome-binding site and leader sequence of pET21a (Novagen). The plasmid was constructed by ligating a PCR-derived gfpmut2-containing fragment to EcoRI/HindIII-digested pSJ4, thereby placing gfpmut2 under Para control. The fragment was derived by PCR with a pKEN derivative encoding the gfpmut2 gene (1) as a template. pSLR22 (Para-gfpmut2::minD) encodes the gfpmut2 gene fused in frame to the 5′ end of the minD gene and preceded by the ribosome-binding site and leader sequence of pET21a. The stop codon of gfp was replaced with a tyrosine codon. In the protein product, Gfp is linked to MinD by a Gly-Ser-Arg-Phe linker which also encodes an XbaI site, and Met-1 of MinD has been replaced by Ile. This protein can be produced at a high concentration without the formation of inclusion bodies. The plasmid was constructed by ligating EcoRI/HindIII-digested pSJ4 to an EcoRI/XbaI-digested gfpmut2-containing fragment and an XbaI/HindIII-digested minD-containing fragment. The gfpmut2 gene was obtained via PCR by using the same primers and template as for pSLR21; the minD gene was obtained by PCR with pDB175 as a template. The entire fusion gene was sequenced to ensure it was correct. pSLR23 (Para-gfpmut2::minD minE) encodes the same gfpmut2::minD fusion as pSLR22, upstream of the minE gene; the DNA sequence between minD and minE is as found on the E. coli chromosome. The plasmid was constructed by ligating EcoRI/HindIII-digested pSJ4 to an EcoRI/XbaI-digested PCR product encoding gfpmut2 and to an XbaI/HindIII-digested minD minE PCR product derived from pDB175. The primers and template for gfpmut2 were the same as for pSLR22 (above). The entire PCR-generated insert was sequenced to ensure it was correct. pSLR24 (Para-minD minE) encodes the wild-type minD and minE genes in tandem preceded by the ribosome-binding site and leader sequence of pET 21a. It was constructed by ligating EcoRI/HindIII-digested pSJ4 to an EcoRI/HindIII-digested PCR product encoding minD minE, derived from plasmid pDB175 (4).
Growth conditions.
Strains were grown overnight on L agar plates containing 1% glucose and antibiotics needed for plasmid maintenance (50 or 100 μg of ampicillin per ml for pUC or pBluescript-derived plasmids and 20 μg of chloramphenicol per ml for pBAD33-derived plasmids) and were inoculated the next morning into L broth containing the same additives. After 3 to 4 h of growth at 37°C, the cells were collected by centrifugation, washed twice with L broth, and suspended at an A600 of 0.04 in fresh L broth lacking glucose but containing antibiotics and inducer, where indicated. Unless otherwise stated, genes under Para control were expressed by growth in 0.001 to 0.005% arabinose, while genes under Plac control were expressed at basal levels by growth in the absence of IPTG (isopropyl-β-d-thiogalactopyranoside). The cultures were incubated with shaking at 30°C for 4 to 4.5 h, and cells were sampled directly from the culture for microscopy (unfixed) or were fixed by addition to the culture of 1.7% formaldehyde and 0.17% glutaraldehyde (final concentrations), followed by incubation at room temperature for 45 min. The fixed cells were collected by centrifugation, resuspended in 0.9% saline, and stored at 4°C until microscopic examination. Western blot analysis showed that the concentrations of Gfp-MinD in ΔminCDE cells containing plasmid pSLR22 (Para-gfp::minD), in cells grown for 4.5 h in the presence of 0.001 or 0.0025% arabinose, were 0.96- and 1.78-fold the concentration of MinD in the wild-type strain, respectively. In ΔminCDE cells containing plasmid pSLR23 (Para-gfp::minD minE) and grown in the presence of 0.001 or 0.0025% arabinose, the concentrations of Gfp-MinD were 1.17- and 2.1-fold and the concentrations of MinE were 5.0- and 9.5-fold, respectively, their concentrations in the wild-type strain.
Gel electrophoresis and immunoblotting.
Cells for Western blot analysis were harvested by centrifugation and frozen immediately. Frozen samples were thawed by resuspension in sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer, boiled for 5 min, and subjected to sodium dodecyl sulfate-gel electrophoresis (11). Immunoblots were performed as previously described by using antibodies directed against MinE2–19 to compare the concentrations of MinE and N-terminal MinE fragments and antibody directed against a mixture of MinE29–38 and MinE70–88 to detect MinE+ or to compare the concentrations of MinE and of MinE C-terminal fragments (19, 20). Antibody raised against whole wild-type MinD protein was used to detect MinD and Gfp-MinD. Bands in Western blots were quantitated by using the ImageQuant program (Molecular Dynamics) on digitized images by using concentrations of cell extracts where the band intensity was proportional to amount of the sample applied to the gel.
Microscopy and data analysis.
Samples were examined by phase contrast, Nomarski, and fluorescence microscopy. A minicell phenotype was defined by the presence of moderate to large numbers of spherical minicells, polar septa, and cells with a length ranging from the wild-type length to short filaments of approximately four to six cell lengths. A filamenting phenotype was defined by the virtual to complete absence of minicells and by the presence of large numbers of filaments, ranging from 6 to 100 cell lengths. Fluorescence images were collected by using an integrating charge-coupled device camera. Measurements of cell lengths and positions of cell landmarks and data analysis were done by using the public domain NIH Image program (http://rsb.info.nih.gov/nih-image/) and in-house software.
RESULTS
Phenotypic effects of a Gfp-MinD fusion protein.
To study the cellular localization of MinD, we constructed a gfp::minD fusion that codes for a protein (Gfp-MinD) in which the high quantum yield Gfpmut2 (1) (called Gfp in the remainder of this study) is fused to the N-terminal end of MinD. The gfp::minD fusion was expressed under control of the arabinose-inducible BAD promoter to permit coexpression of other relevant genes under Plac control in the same cell.
MinD normally performs two functions that affect formation of the division septum and that can be assayed in vivo: (i) MinD is an activator of the MinC division inhibitor and (ii) MinD makes the division inhibitor sensitive to suppression by MinE. Evidence that the Gfp-MinD fusion protein retained these two functions was obtained by comparing the effects of MinD and Gfp-MinD on the division pattern of a Δmin strain in the presence of MinC and/or MinE.
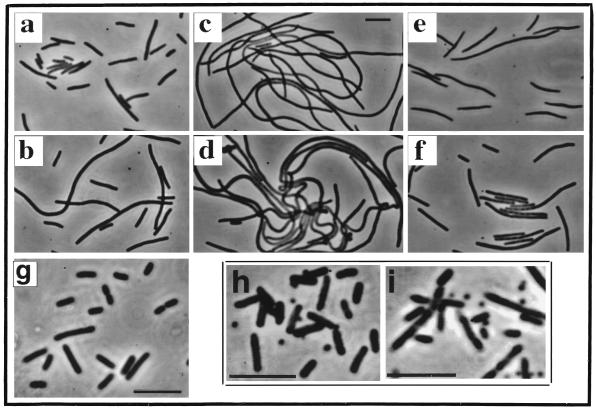
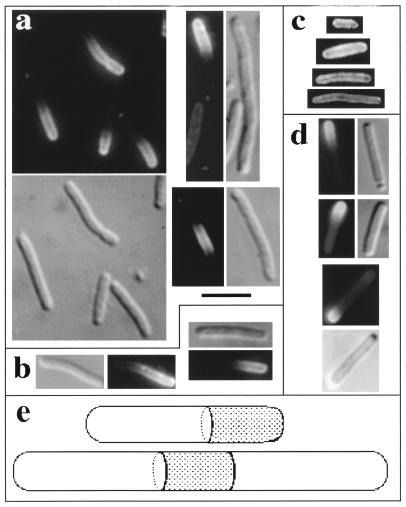
As previously reported (4), induction of minC from an integrated Plac-minC prophage in the absence of MinD failed to block division in a ΔminCDE strain (Fig. 1a), whereas coexpression of minC and minD led to the formation of nonseptate filaments (Fig. 1b and c). Coexpression of minC and gfp::minD also induced filamentation when Plac-minC and Para-gfp::minD were coinduced by growth in the presence of IPTG and arabinose (Fig. 1d). We conclude that Gfp-MinD retained the MinC activation effect of MinD.
FIG. 1.
Phenotypic effects of Gfp-MinD, MinD, and MinE1–53-Gfp. (Panels a to f) Strain PB114 (λDB217) (ΔminCDE [Plac-minC]) containing the indicated plasmids was grown at 30° for 3.5 h in L broth containing 1 mM IPTG and 0.01% arabinose unless otherwise indicated and then examined by phase-contrast microscopy. Panels: a, plasmid pBAD33 (vector); b, plasmid pSLR22 (Para-gfp::minD) (arabinose was omitted); c, plasmid pAS73 (Para-minD); d, plasmid pSLR22 (Para-gfp::minD); e, plasmid pSLR24 (Para-minD minE); f, plasmid pSLR23 (Para-gfp::minD minE); g, strain RC1/pYW3 (Plac-minC gfp::minD minE) was grown at 30° for 4.5 h in L broth containing 10 μM IPTG; h and i, strain DH5α/pFX1 (minC+D+E+/Plac-minE1–53::gfp) was grown for 4 h at 30° in L broth containing 0.05 mM IPTG (h) or 1 mM IPTG (i). Bars, 5 μm.
MinD is required to make MinC-mediated division inhibition sensitive to suppression by MinE (6). To ask whether Gfp-MinD retained this aspect of MinD function, minD or gfp::minD was expressed in cis with minE in cells that expressed minC from a Plac-minC λ prophage. As previously described, the expression of minE suppressed the filamentation that occurred when minC and minD were coexpressed in the absence of minE (Fig. 1e). Similarly, filamentation was suppressed when minD was replaced by gfp::minD (in pSLR23, Fig. 1f). Thus, Gfp-MinD behaves similarly to MinD in making the activated MinC division inhibitor sensitive to MinE.
Raskin and de Boer (16) have shown that Gfp-MinD can correct the minicelling phenotype of a minD1 mutant, in which the mutation is located near the carboxy terminus of MinD (10). This showed that the Gfp-MinD protein retains the MinD function that is lost due to the minD1 mutation. In the present study, evidence that Gfp-MinD could provide all of the functions of MinD was obtained by expressing minC, gfp::minD, and minE in a ΔminCDE strain (from PB114/pYW3 [ΔminCDE/Plac-minC gfp::minD minE]). As illustrated in Fig. 1g, low-level derepression of Plac by growth of PB114/pYW3 in 0.005% or 0% glucose or 10 μM IPTG led to almost complete disappearance of the minicell phenotype. Similar results were obtained with wild-type MinD, expressed from pDB170 (Plac-minC minD minE) (data not shown) (4). In this case, minicelling was corrected at even lower levels of derepression, obtained by growth in 0.05 or 0.01% glucose. We conclude that Gfp-MinD has retained all of the important functions of MinD, although the Gfp moiety may interfere somewhat with MinD efficiency or stability.
Pattern of membrane-associated Gfp-MinD in the absence of MinE.
It has previously been shown that MinE-Gfp can localize to sites near midcell and that this localization requires the presence of the MinD protein (14). We therefore studied the localization pattern of Gfp-MinD in the presence or absence of MinE. Experiments were performed in the Δmin strain PB114. Because of the Δmin background, the cells showed a minicell phenotype consisting of minicells plus cells ranging from wild-type cell length to short filaments. Cells were examined after low-level induction of Gfp-MinD from Para-gfp::minD by growth in the presence of 0.001 to 0.005% arabinose. Under these conditions the phenotype of the Δmin host was unchanged.
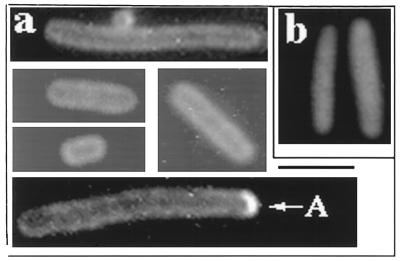
Most fluorescent cells in the population showed a peripheral pattern of fluorescence that extended entirely around the cell (Fig. 2a). The pattern was seen at all levels of arabinose induction that were tested. The peripheral pattern was not an optical artifact since expression of Gfp alone, in the absence of the MinD moiety, showed only diffuse cellular fluorescence without evidence of peripheral localization (Fig. 2b). We conclude that MinD can associate with the cell membrane around the entire periphery of the cell in the absence of other Min proteins, an idea consistent with the results of Raskin and de Boer (16).
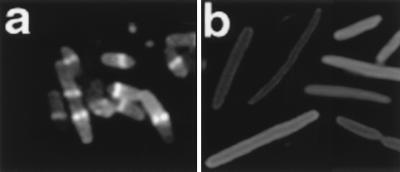
FIG. 2.
Distribution of Gfp-MinD in the absence of other Min proteins. Strain PB114/pSLR22 (ΔminCDE/Para-gfp::minD) was grown in the presence of 0.005% arabinose for 4 h at 30°C prior to fluorescence microscopy. (a) Fixed cells showing the peripheral pattern of Gfp-MinD fluorescence. A, polar arc. (b) Fixed cells; pSLR22 was replaced by pSLR21(Para-gfp).
In addition to the peripheral localization pattern, approximately 10% of the cells contained a short fluorescent polar arc (“A” in Fig. 2a). The polar arcs were usually present at only one of the two cell poles.
Effect of MinE on the Gfp-MinD distribution pattern: polar zones.
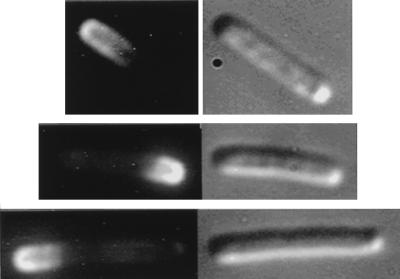
Evidence that MinE can induce changes in the distribution pattern of membrane-associated MinD came from experiments in which gfp::minD was coexpressed with minE in a Δmin strain (RC1/pSLR22/pDB188 [Δmin/Para-gfp::minD/Plac-minE]) by growth in the presence of 0.001 to 0.005% arabinose. Under these conditions many cells in the population contained a long fluorescent zone at one end of the cell (Fig. 3). In unfixed cells, the polar zones of Gfp-MinD oscillated from pole-to-pole (Fig. 4) as has previously been described (16).
FIG. 3.
Distribution of Gfp-MinD in the presence of MinE. Strain PB114/pSLR22/pDB188 (ΔminCDE/Para-gfp::minD/Plac-minE) was grown in the presence of 0.0025% arabinose for 4 h at 30°C. Fluorescence micrographs are shown on the left; Nomarski micrographs are shown on the right. Membrane-associated Gfp-MinD fluorescence is almost exclusively present in the polar zones.
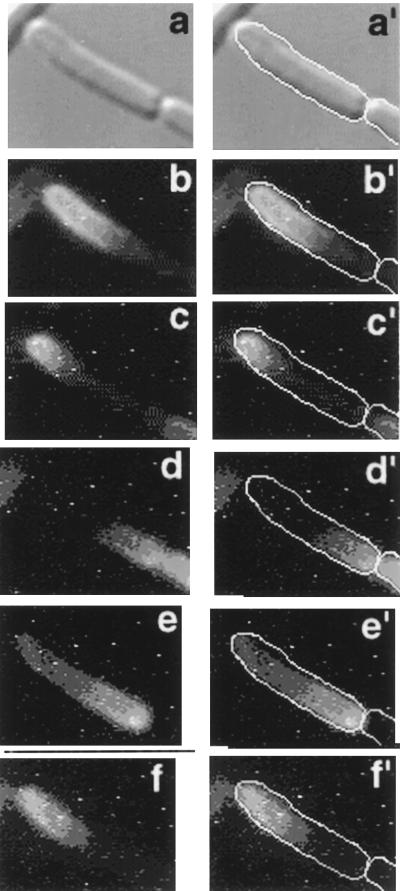
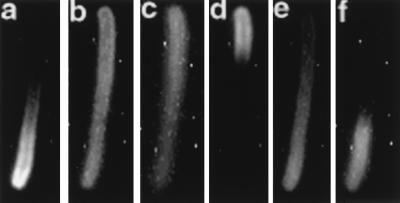
FIG. 4.
Pole-to-pole movement of Gfp-MinD in the presence of MinE. Strain PB114 (λDB156)/pSLR22 [ΔminCDE (Plac-minE)/Para-gfp::minD] was grown in 0.0025% arabinose and 10 μM IPTG for 4 h at 30°C. Unfixed cells were examined. A single field is shown. Panels: a, Nomarski differential interference micrograph; b and b′ to f and f′, fluorescence images were collected at 2-min intervals. In panels b′ to f′ a white outline has been added to indicate the position of the cell. A zone of polar fluorescence at the upper left pole can be seen to move to the opposite pole (d and e) and then to return to the original pole (f).
Quantitative Western blots, made by using anti-MinD antibody, were done to exclude the possibility that the redistribution of Gfp-MinD might have reflected secondary increases in Gfp-MinD concentration rather than a positive effect of MinE on MinD distribution. This showed no significant difference in the concentration of immunoreactive Gfp-MinD between cells that did not express MinE and those that coexpressed MinE and Gfp-MinD (data not shown). This excludes the possibility that the effect of MinE was due to a secondary change in MinD concentration.
Effect of MinE domains on localization of Gfp-MinD.
Previous work has indicated that the topological specificity domain of MinE is located within the C-terminal region (MinE36–88) of the 88-amino-acid MinE protein and that the anti-MinCD domain that is capable of suppressing the action of the MinCD division inhibitor is located in the N-terminal region of the protein (MinE1–22 or MinE1–34) (13, 20). It has been suggested that the topological specificity domain interacts with a topological target at midcell to direct MinE localization at midcell, whereas the anti-MinCD domain of MinE is thought to interact with the MinCD system via MinD (17).
We therefore asked whether the ability of MinE to cause Gfp-MinD to coalesce into an extended membrane-associated structure at the cell pole required both of the MinE domains. In these experiments gfp::minD was expressed in a Δmin host from pSLR22 (Para-gfp::minD) and C-terminal or N-terminal MinE fragments were expressed under Plac control.
Coexpression of Gfp-MinD with a MinE fragment that contained the topological specificity domain but lacked the anti-MinCD domain (fragment MinE22–88, in pSY1057) did not significantly alter the pattern that was present in the absence of MinE (Fig. 5c). Fluorescence was predominantly peripheral along the length of the cell, and no localized fluorescent zones were observed. Western blots with antibody directed against the C-terminal region of MinE showed that the concentration of MinE22–88 was severalfold higher than the concentration of full-length MinE (MinE1–88) in the experiments in which Plac-minE1–88 was coexpressed with gfp::minD (data not shown). Therefore, the failure of the C-terminal MinE domain to affect the Gfp-MinD pattern was not due to a lower cellular concentration of the fragment compared with the concentration of MinE1–88 in the parallel experiments.
FIG. 5.
Distribution of Gfp-MinD in the presence of MinE fragments. Cells were grown for 4 h at 30°C in the presence of 0.005% arabinose. Fluorescence and Nomarski micrographs are shown side by side to show the complete outline of the cells. Panels: a, PB114/pSLR22/pSY1053 (ΔminCDE/Para-gfp::minD/Plac-minE1–53), fixed cells; b, PB114/pSLR22/pJPB262 (ΔminCDE/Para-gfp::minD/Plac-minE1–32), fixed cells; c, PB114/pSLR22/pSY1057 (ΔminCDE/Para-gfp::minD/Plac-minE22–88), fixed cells; d, PB114/pSLR22/pSY1053 (ΔminCDE/Para-gfp::minD/Plac-minE1–53), unfixed cells; and e, diagrammatic representation of Gfp-MinD zones. Black bar, 3 μm.
In contrast to the failure of the C-terminal MinE fragment to change the Gfp-MinD distribution pattern, N-terminal fragments MinE1–32 and MinE1–53, which contain the anti-MinCD domain but lack the topological specificity domain, did significantly alter the distribution pattern of Gfp-MinD. In these cases, instead of the pattern of peripheral fluorescence that was characteristic of cells that expressed gfp::minD in the absence of MinE, cells that coexpressed Gfp-MinD with MinE1–32 or MinE1–53 contained long fluorescent zones at the cell pole (Fig. 5a and b). These were present in most cells in the population and were similar in appearance to the polar zones in cells that coexpressed gfp::minD and full-length minE (Fig. 3). The pattern was the same when expression of the N-terminal MinE fragments was varied over a wide range by growth at IPTG concentrations ranging from 0 to 2 mM.
The pattern was the same in fixed and unfixed cells (Fig. 5a and d), indicating the polar zones of fluorescence were not fixation artifacts and were not artifacts induced by UV irradiation during examination or by the attachment of unfixed cells to the glass slide. In cells of normal length (≤5.0-μm cell length), the zones were always located at one end of the cell. In some longer cells, fluorescent zones were present at both poles and were sometimes also present elsewhere along the length of the cell. The polar fluorescence was predominantly peripheral, indicating that it represented membrane-associated Gfp-MinD. Strikingly, whenever polar zones were present, the remainder of the cell periphery was essentially nonfluorescent (Fig. 5a, b, and d). This was in sharp contrast to cells that expressed Gfp-MinD in the absence of MinE, where the fluorescence was distributed around the entire cell periphery. This implies that the MinE fragments were responsible for the recruitment of essentially all of the membrane-associated Gfp-MinD into the polar zones.
Studies of unfixed cells showed that the N-terminal portion of MinE induced both the segregation of Gfp-MinD to one pole and its subsequent back-and-forth movement to the opposite pole (Fig. 6). The relatively slow oscillation may reflect lower-than-optimal ratios of MinE to MinD, which can significantly affect the rate of oscillation (16). The presence of oscillatory pole-to-pole movement of Gfp-MinD in the presence of MinE1–53 indicates that the mechanisms for the MinE-dependent polar localization and for transpolar movement of MinD do not require the topological specificity domain of MinE.
FIG. 6.
Pole-to-pole movement of Gfp-MinD in the presence of MinE1–53. Strain PB114/pFX11 (ΔminCDE/Plac-gfp::minD minE1–53) was grown for 4 h at 30°C in the presence of 25 μM IPTG. Unfixed cells were examined and images were collected at 2-min intervals. One cell is followed in panels a to f. A zone of polar fluorescence can be seen to move from the lower pole (panel a) to the upper pole (panel d) and back to the lower pole (panel f).
Localization of MinE-Gfp and MinE1–53-Gfp in the presence of MinD.
Full-length MinE (as MinE-Gfp) forms a ring at midcell when expressed in the presence of MinD (14). This led to the suggestion that the midcell MinE ring might be responsible for sequestering Gfp-MinD to one of the ends of the cell to form the polar MinD zones and might also play a role in inducing disassembly of Gfp-MinD from the polar membrane binding site and subsequent pole-to-pole movement (16).
It has been previously shown that MinE1–33-Gfp fails to form midcell MinE rings (14) although, as shown in the present work, MinE1–32 was capable of inducing formation of polar zones of MinD-Gfp. Because MinE1–53 also induced the formation of polar MinD zones, we used MinE1–53-Gfp to determine whether MinE1–53 was capable of forming a MinE ring at midcell. Localization studies in cells where MinE1–53-Gfp was coexpressed with MinD failed to show the characteristic MinE rings that are seen with full-length MinE-Gfp (Fig. 7). This confirms that the N-terminal MinE domain is unable to localize at midcell in the absence of the C-terminal topological specificity domain, although it is capable of inducing the redistribution of MinD into polar zones. These results suggest that a MinE ring is not required for the formation of polar zones of MinD.
FIG. 7.
Localization of MinE-Gfp and MinE1–53-Gfp in the presence of MinD. (a) Strain PB114/pAS86/pAS74 (ΔminCDE/Para-minE::gfp/Plac-minD) was grown in 0.0125% arabinose for 4 h. (b) Strain PB114/pAS73/pFX1 (ΔminCDE/Para-minD/Plac-minE1–53-gfp) was grown in 0.005% arabinose–30 μM IPTG for 4 h. Similar results were obtained when arabinose was varied between 0 and 0.005% and IPTG was varied between 0 and 30 μM and when the same experiments were performed on strain DH5α/pFX1 (minC+D+E+/Plac-minE1–53-gfp).
It has not been directly shown that MinE-Gfp, and MinE1–33-Gfp and MinE1–53-Gfp, behave similarly to the non-Gfp proteins in the ability to induce formation of polar MinD zones. However, wild-type MinE-Gfp can restore a wild-type phenotype to MinE-deficient cells (reference 14 and unpublished results). Similarly, MinE1–53-Gfp behaved like underivatized MinE1–53 (20) in its ability to induce minicell formation in wild-type cells (Fig. 1h, i), presumably by interacting with MinD. These observations suggest that the presence of the Gfp components does not significantly interfere with function of full-length MinE or the N-terminal MinE domain.
DISCUSSION
Proper placement of the E. coli division septum requires that MinD and MinE function cooperatively to modulate the division potential of cellular sites that are located at midcell and at the cell poles. A MinD-MinE interaction in this process is implied by the fact that localization of MinE at midcell requires MinD, that MinD is required to make the MinC division inhibitor sensitive to suppression by MinE, and that MinE is required for the formation of MinD zones at the cell pole.
Gfp-MinD is capable of associating with the membrane around the entire periphery of the cell in the absence of MinE and MinC, as shown by Raskin and de Boer (16) and as confirmed in the present study. In contrast, the membrane association of MinE (14) and MinC (9, 15) both require the presence of MinD. These observations suggest that the membrane attachment of MinD is the initial step in the membrane assembly of the Min proteins. The MinD sequence does not include an apparent membrane-spanning domain, and cell fractionation and immunoelectronmicroscopic studies suggest that MinD is a peripheral membrane protein (2). This suggests that MinD is likely to interact with another membrane component that anchors it to the membrane surface.
MinE dramatically changes the membrane distribution of MinD so that essentially all of the membrane-associated Gfp-MinD is recruited into a broad zone at one cell pole (reference 16 and this study). Previous studies with Gfp-MinE (14) have shown that MinE forms a ring near midcell under the same conditions that lead to formation of the polar zones of MinD, raising the possibility (16) that the midcell MinE ring plays a role in the observed redistribution of membrane-associated MinD. The present observations suggest that the two events, i.e., the formation of the midcell MinE ring and the formation of polar zones of MinD, are unrelated phenomena. Thus, in the present study the N-terminal domain of MinE, which did not form a midcell MinE ring in studies of MinE1–53-Gfp (this study) and MinE1–33-Gfp (14), was capable of inducing formation of polar zones of Gfp-MinD with high efficiency. This argues against models in which the MinE ring at midcell acts as a gasket to sequester Gfp-MinD to one end of the cell and/or to provoke release of Gfp-MinD from one pole so that it can move to the opposite pole (16).
Because MinE1–53 and MinE1–34 fragments retain their ability to counteract the division-inhibitory action of MinCD in a MinD-dependent fashion (13, 20), it is likely that the N-terminal MinE domain is the domain that interacts with MinD. It is this interaction that presumably provokes the redistribution of membrane-associated MinD to the cell pole.
We suggest the following sequence of events to explain the cooperative actions of MinE and MinD, based on the idea that formation of the MinE ring at midcell and formation of the MinD zone at the cell pole both result from the lateral movement of MinD within the two-dimensional membrane matrix. First, MinD associates with the inner surface of the cytoplasmic membrane around the entire periphery of the cell. Second, the N-terminal domain of MinE interacts with the membrane-associated MinD. This recruits MinE to the membrane. We speculate that the MinD-MinE interaction may alter MinD or its membrane attachment to permit MinD molecules to diffuse laterally within the two-dimensional membrane matrix, possibly in association with its putative membrane anchor. Alternatively, MinD may always be laterally mobile within the membrane. In this case, MinE could modify MinD to increase its affinity for polar sites (discussed below). This alternative is perhaps less likely since the fact that Gfp-MinD forms polar arcs in the absence of MinE (Fig. 2a) implies that MinD has affinity for the cell pole independently of MinE. In either case, the laterally mobile MinD molecules are suggested to be responsible both for the formation of the MinE ring at midcell and for the formation of the MinD polar zones. Third, when the laterally mobile MinD-MinE complex encounters the putative topological target for MinE at midcell, the midcell target interacts with the C-terminal topological specificity domain of MinE, thereby anchoring MinE as a ring structure at midcell and releasing it from its MinD carrier. This finding is consistent with the observation that the topological specificity domain is required for formation of the midcell MinE ring. Fourth, unrelated to formation of the MinE ring, the collision of laterally mobile MinD molecules with a hypothetical membrane-associated nucleation site adjacent to a cell pole leads to formation of a side-by-side array of MinD molecules (the polar zone) whose assembly depends on collisional interactions within the membrane matrix. The two-dimensional MinD lattice would be expected to grow and coalesce until most or all of the mobile membrane-associated MinD molecules were captured by collision with the polar lattice. This would explain the striking observation that only a single Gfp-MinD zone was present in most cells, with no visible fluorescence elsewhere in the membrane.
The fact that the MinD zone is apparently formed at only one pole might reflect a rapid MinD assembly process following the initial interaction with one of the polar nucleation sites, a process similar to the cooperative assembly process suggested by Raskin and de Boer (16). The forces that capture and retain MinD molecules within the polar zone have yet to be defined. The lattice could be based on direct interactions between MinD molecules or could involve the noncovalent cross-linking of MinD molecules or oligomers by another component that would also be part of the polar lattice structure. The suggested model invokes lateral diffusion of MinD molecules as the key event in the formation of the MinD polar zones and in the MinD-facilitated formation of the midcell MinE ring. The possibility also exists that the lateral translocation event might in part be an active process in which MinE modifies MinD into a form that can be actively translocated along the membrane.
A precedent for the capture of mobile membrane molecules into a single structure exists in the well-established ability of antibody molecules or lectins to induce the redistribution of eucaryotic membrane-associated surface proteins by noncovalently crosslinking them into “patches” and “caps” that are reminiscent of the Gfp-MinD structures described here (7, 12, 18). Ultimately, all of the proteins are captured into a single large domain, one analogous to the Gfp-MinD zones that are formed at the cell pole.
In an entirely different type of model, MinD would move directly from the cytoplasm to the polar membrane sites after its interaction with MinE. This cannot be excluded although it would require a second mechanism to explain the requirement for MinD in formation of the midcell MinE ring. Further work will be needed to distinguish between these and other possible models.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health to L.I.R. (GM41978 and GM53276). S.L.R. was a Long Term Fellow of the Human Frontiers Science Program.
REFERENCES
- 1.Cormack B P, Valdivia R H, Falkow S. FACS-optimised mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 2.de Boer P A J, Crossley R E, Hand A R, Rothfield L I. The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J. 1991;10:4371–4380. doi: 10.1002/j.1460-2075.1991.tb05015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Boer P A J, Crossley R E, Rothfield L I. Isolation and properties of minB, a complex genetic locus involved in correct placement of the division site in Escherichia coli. J Bacteriol. 1988;170:2106–2112. doi: 10.1128/jb.170.5.2106-2112.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Boer P A J, Crossley R E, Rothfield L I. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 5.de Boer P A J, Crossley R E, Rothfield L I. Central role for the Escherichia coli minC gene product in two different cell division-inhibition systems. Proc Natl Acad Sci USA. 1990;87:1129–1133. doi: 10.1073/pnas.87.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Boer P A J, Crossley R E, Rothfield L I. Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J Bacteriol. 1992;174:63–70. doi: 10.1128/jb.174.1.63-70.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edelman G. Some new views of the cell surface. J Biochem. 1976;79:1P–12P. doi: 10.1093/oxfordjournals.jbchem.a131116. [DOI] [PubMed] [Google Scholar]
- 8.Guzman L, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Z, Lutkenhaus J. Topological regulation in Escherichia coli involves rapid pole-to-pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol Microbiol. 1999;34:82–90. doi: 10.1046/j.1365-2958.1999.01575.x. [DOI] [PubMed] [Google Scholar]
- 10.Labie C, Bouché F, Bouché J-P. Minicell-forming mutants of Escherichia coli: suppression of both DicB- and MinD-dependent division inhibition by inactivation of the minC gene product. J Bacteriol. 1990;172:5852–5855. doi: 10.1128/jb.172.10.5852-5855.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 12.Nicolson G. The interactions of lectins with animal cell surfaces. Int Rev Cytol. 1974;39:89–190. doi: 10.1016/s0074-7696(08)60939-0. [DOI] [PubMed] [Google Scholar]
- 13.Pichoff S, Vollrath B, Touriol C, Bouché J-P. Deletion analysis of gene minE which encodes the topological specificity factor of cell division in Escherichia coli. Mol Microbiol. 1995;18:321–329. doi: 10.1111/j.1365-2958.1995.mmi_18020321.x. [DOI] [PubMed] [Google Scholar]
- 14.Raskin D, de Boer P. The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell. 1997;91:685–694. doi: 10.1016/s0092-8674(00)80455-9. [DOI] [PubMed] [Google Scholar]
- 15.Raskin D, de Boer P. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J Bacteriol. 1999;181:6419–6424. doi: 10.1128/jb.181.20.6419-6424.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raskin D, de Boer P. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothfield L I, Zhao C-R. How do bacteria decide where to divide? Cell. 1996;84:183–186. doi: 10.1016/s0092-8674(00)80971-x. [DOI] [PubMed] [Google Scholar]
- 18.Taylor R B, Duffus W P H, Raff M C, de Petris S. Redistribution and pinocytosis of lymphocyte surface immunoglobulin molecules induced by anti-immunoglobulin antibody. Nat New Biol. 1971;233:225–229. doi: 10.1038/newbio233225a0. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Rowland S, King G, Rothfield L. Relation of the oligomeric structure of MinE to its topological specificity function. Mol Microbiol. 1998;30:265–273. doi: 10.1046/j.1365-2958.1998.01059.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhao C-R, de Boer P, Rothfield L. Proper placement of the E. coli division site requires two functions that are associated with different domains of the MinE protein. Proc Natl Acad Sci USA. 1995;92:4313–4317. doi: 10.1073/pnas.92.10.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]