Abstract
Wild-type Escherichia coli K-12 ferments glucose to a mixture of ethanol and acetic, lactic, formic, and succinic acids. In anoxic chemostat culture at four dilution rates and two different oxidoreduction potentials (ORP), this strain generated a spectrum of products which depended on ORP. Whatever the dilution rate tested, in low reducing conditions (−100 mV), the production of formate, acetate, ethanol, and lactate was in molar proportions of approximately 2.5:1:1:0.3, and in high reducing conditions (−320 mV), the production was in molar proportions of 2:0.6:1:2. The modification of metabolic fluxes was due to an ORP effect on the synthesis or stability of some fermentation enzymes; thus, in high reducing conditions, lactate dehydrogenase-specific activity increased by a factor of 3 to 6. Those modifications were concomitant with a threefold decrease in acetyl-coenzyme A (CoA) needed for biomass synthesis and a 0.5- to 5-fold decrease in formate flux. Calculations of carbon and cofactor balances have shown that fermentation was balanced and that extracellular ORP did not modify the oxidoreduction state of cofactors. From this, it was concluded that extracellular ORP could regulate both some specific enzyme activities and the acetyl-CoA needed for biomass synthesis, which modifies metabolic fluxes and ATP yield, leading to variation in biomass synthesis.
A wealth of information is available on the response of Escherichia coli cellular metabolism to pH, water activity, or temperature variations, but little is known about the action of extracellular oxidoreduction potentials (ORP) on metabolism, although numerous reactions and regulations are of the oxidoreduction type. Previous studies have shown that substrates with different oxidation states yield a specific product spectrum. Thus, with glucose (oxidation state = 0), the spectrum of main end products (formate:acetate:ethanol:lactate) is equal to 2:1:1:2, with glucitol (oxidation state = −1), it is equal to 2:1:6:0.5, and with glucuronate (oxidation state = 2), it is equal to 1:5:1:1, with small amounts of succinate also being produced in each case (1). Those metabolic flux modifications are also observed when the nature of external electron acceptors varies (7). In the same way, the NADH/NAD+ ratio, which is responsible for regulation of enzymes (8) or genes (16), can be influenced by the oxidation level of the substrate (36) or by the availability and nature of electron acceptors (7). In addition, protein folding and disulfide bond formation are regulated and modified by different oxidoreductase enzymes and oxidoreduction couples (glutathione, thioredoxin) (26). Recently, Taylor and Zhulin in their review have shown that ORP influences or could influence numerous regulations of cell functions controlled via Per-Arnt-Sim (PAS)-containing receptors (signaling modules that monitor changes in light, ORP, oxygen, and the overall energy level of a cell), transducers, and regulators (34). Thus, E. coli senses the medium ORP and swims to a preferred ORP niche by redox taxis, involving a change in proton motive force as a hypothetical sensor (3).
Few experimental studies have explored the extracellular ORP effect on particular metabolite production. Kwong and Rao have shown that reducing conditions can increase productions of homoserine and lysine by Clostridium glutamicum (13). In the same way, for Clostridium acetobutylicum, a decrease from −300 to −370 mV increases the production level of butanol and decreases those of butyrate and acetate (24). These results show that extracellular ORP can modify metabolic fluxes.
To our knowledge, however, a more global study of ORP action on metabolism has not been realized, although Gill et al. have shown that extracellular ORP could act on the oxidoreduction state of cytoplasmic molecules, due to the low ORP buffer strength of cytoplasm (9). Furthermore, from a biotechnological point of view, if this parameter can modify metabolic fluxes, it might be an additional physicochemical parameter to take into account for the optimization of processes.
In this paper, we describe the effect of ORP on modifications of the metabolic end product spectrum (lactate, acetate, ethanol, formate, succinate, and CO2), enzyme activities (alcohol dehydrogenase [ADH], lactate dehydrogenase [LDH], acetate kinase [AK], pyruvate kinase [PK], and phosphoenolpyruvate carboxylase [PEPC]), intermediary metabolites (acetyl-coenzyme A [CoA], oxaloacetic acid [OAA], and phosphoenolpyruvate [PEP]), oxidized and reduced forms of NAD in E. coli, and ORP modified fermentation pathways, leading to changes in end product spectrum, growth yield, and oxidation balance.
MATERIALS AND METHODS
Batch cultivation: bacterial strain and culture conditions.
E. coli K-12 wild-type EMG2 (CGSC 4401) was grown in Trypticase soy broth sterilized by filtration, in anaerobic conditions. Cells were cultivated in an 800-ml bioreactor (Biostat Q, Braun, Germany). Temperature, pH, and agitation were maintained at 37°C, 7.0 (with NaOH as neutralizer), and 200 rpm respectively. ORP was measured with a redox combined electrode (Pt 4805-DXK; Mettler-Toledo S.A.R.L., Paris, France) connected to a redox controller (P507 Consort; Bioblock Scientific, Illkirch, France), and the values were corrected according to the reference electrode value (210 mV at 37°C). Before inoculation, the medium was sparged with nitrogen to eliminate oxygen. Metabolic end products were measured at the beginning of the stationary phase.
Chemostat cultivation: culture conditions.
The strain was grown in minimal M9 medium with glucose (20 g/liter) as the carbon source, sterilized by filtration. An anaerobic cultivation condition was chosen to avoid the uncertainties associated with the P/O ratio of oxidative phosphorylation for ATP yield calculations. In anaerobic growth without external electron acceptors such as nitrate or fumarate, energy generation occurs only through substrate level phosphorylation. Chemostat cultivation was chosen over batch cultivation to provide a better quality of specific rate data.
The chemostat cultivation was performed in a 2.0-liter bioreactor (Setric G.I., Toulouse, France) equipped with a weight control for maintaining a constant working volume of 1.5 liters. After inoculation of the fermentor, it was run in batch mode overnight and then put on chemostat condition. Four dilution rates (0.025, 0.05, 0.1, and 0.2 h−1) were studied. In these conditions, glucose was not a limiting nutrient. The continuous culture reached a steady state after 5 to 6 residence times. The pH, temperature, and agitation were maintained at 6.0 (with NaOH as a neutralizer), 37°C, and 200 rpm, respectively. Two distinct sets of ORP conditions were maintained, the first set, named low reducing conditions (LRC), was obtained by a sparging nitrogen gas flow which dispelled the hydrogen produced naturally by the bacteria (mean ORP ± standard deviation, −90 ± 50 mV, depending on the dilution rate). The second set, named high reducing conditions (HRC), was obtained by a low sparging nitrogen gas flow, with, if necessary, hydrogen added to maintain a mean ORP of −320 mV (standard deviation, 20 mV, depending on the dilution rate). To limit the end product evaporation loss (especially ethanol) by gas flow, the fermentor was equipped with an exhaust gas cooler, maintained at 0°C by a refrigerated circulating bath. ORP was measured with a redox combined electrode. To ensure that all oxygen traces were removed from the feeding medium, it was sparged with oxygen-free nitrogen before use.
Analytical technique.
Cell density was monitored at 620 nm in a spectrophotometer (Novaspec 4049; LKB Products) and reported as cellular dry weight. For substrate and fermentation end product measures, a 10-ml sample was centrifuged twice (20,000 × g, 10 min at 4°C) and the supernatant was frozen at −80°C until analysis. Glucose, acetate, ethanol, succinate, formate, and lactate were quantified using a high-pressure liquid chromatography system (Shimadzu) equipped with a cation-exchange column (HPX-87H; Bio-Rad Labs), a differential refractive index detector, and a UV detector. A mobile phase pH of 1.32 (H2SO4) at a 0.5-ml/min flow rate was used, and the column was operated at 60°C. CO2 production was measured by passing the effluent gas from the fermentor through a CO2 analyzer (infrared absorption technology; Leybold Heraeus).
For enzyme activity determination, a 200-ml sample was centrifuged (6,000 × g, 10 min at 4°C), washed twice with 50 mM potassium phosphate buffer (pH 7.5), resuspended in an equal protein volume at a concentration of 10 g · liter−1 and frozen for no more than 2 days at −20°C (15). To obtain the cell extracts, cells were sonicated three times for 3 min using cycles of 1 s of sonication (power 7) and 1 s of rest (Vibra cell disruptor; Sonics Materials, Danburry, Conn.) and cooled in an ice-water bath. The cell debris was removed by centrifugation (20,000 × g, 10 min at 4°C). The supernatant was used for the measurements of the activities of the five enzymes. All enzyme assays were realized in the linear response range (determined beforehand) of each enzyme by adequately diluting the sample.
LDH was assayed by the method described by Racker (25), in a reaction mixture containing potassium phosphate buffer (100 mM, pH 7.5), NADH (0.33 mM), and sodium pyruvate (30 mM). The reaction was initiated by the addition of pyruvate.
AK was assayed by the method described by Nakajima et al. (21), in a reaction mixture consisting of imidazole-HCl buffer (50 mM, pH 7.3), MgCl2 (10 mM), glucose (10 mM), NADP (1.6 mM), acetyl phosphate (12 mM), ADP (5 mM), hexokinase (56 U/ml), and glucose 6-phosphate-dehydrogenase (1.5 U/ml). The reaction was initiated by the addition of ADP.
ADH was assayed by the method described by Clark and Cronan, Jr. (5), in a reaction mixture containing sodium phosphate buffer (12 mM, pH 8.5), ethanol (300 mM), and NAD (0.3 mM). The reaction was initiated by the addition of ethanol.
PEPC was assayed by the method described by McAlister et al. (18), in a reaction mixture containing Tris-HCl buffer (100 mM, pH 8.5), PEP (6 mM), MgSO4 (5 mM), KHCO3 (10 mM), acetyl-CoA (1 mM), NADH (0.3 mM), and malate dehydrogenase (10 U/ml). The reaction was initiated by the addition of acetyl-CoA.
PK was assayed by the method described by Garrigues et al. (8), in a reaction mixture containing Tris-HCl buffer (100 mM, pH 7.2), MnSO4 (5 mM), KCl (10 mM), ADP, (1 mM), NADH (0.3 mM), LDH (10 U/ml), and PEP (2 mM). The reaction was initiated by the addition of PEP.
Protein concentration was determined by the method of Lowry et al. (17); bovine serum albumin was used as the standard. One unit of enzyme is defined as the amount of enzyme required to produce 1 μmol of product per min.
In vitro activity and stability of LDH have been measured in the two sets of ORP conditions, LRC and HRC, generated by a sparging gas flow of nitrogen and hydrogen, respectively, which created ORP conditions as close as possible to those utilized during batch cultivation. Cell extracts containing LDH were obtained from a batch cultivation of E. coli in M9 medium harvested at a culture pH of 6.0 and lysed as described above. LDH activity was measured with the LDH assay described above. In the case of in vitro LDH activity measurement, the two ORP conditions were generated with a gas sparging into the different solutions and maintained during the assay with a continued gas flow in the spectrometer cuvette. In the case of the in vitro LDH stability measurement (pH 7.5, 50°C), the two sets ORP conditions were applied to the cell extracts with gas sparging before and during heat treatment, and then LDH activity was determined.
Intracellular metabolite and coenzyme concentrations were measured by using an in vitro procedure based on rapid inactivation of metabolism followed by metabolite extraction directly from the cell sample. Cell samples were removed from the culture, and metabolite extraction was performed immediately. Methods of metabolite extraction (acidic or basic) and assays were based on those developed by Lebloas (15). A variable volume of either HCl (6 N) or KOH (10 N) was added to yield a final pH of 1.2 or 12.5, respectively. Most metabolites and coenzymes were extracted by incubating the HCl-treated culture at 50°C for 10 min before neutralizing it to a pH of 6.5 to 7 by KOH during vigorous agitation. After a 10-min centrifugation (6,000 × g at 4°C), the supernatant was immediately used for metabolite concentration measurements. Acid-labile coenzymes, like NADH, were extracted by incubating the KOH-treated culture (pH 12.5) for 10 min at room temperature (25°C). After centrifugation (6,000 × g, 10 min at 4°C), the supernatant was immediately tested for NADH. Metabolites were measured by coupling appropriate enzyme assays with fluorimetric determination of the coenzyme NADH or NADPH. Emission was measured at 460 nm after excitation at 350 nm with a spectrofluorometer (Hitachi F-4500; Hitachi Instrument Co., Tokyo, Japan).
Pyruvate was assayed in a reaction mixture containing potassium phosphate buffer (100 mM, pH 7), MgCl2 (2.5 mM), NADH (33 μM), acid extract, and LDH (20 U · ml−1) to initiate pyruvate consumption (6).
PEP was determined after complete depletion of the pyruvate present in the extract by using a mixture which contained potassium phosphate buffer (100 mM, pH 7), MgCl2 (2.5 mM), ADP (2 mM), a molar concentration of NADH sufficient to complete pyruvate elimination, acid extract, and LDH (20 U · ml−1). After all the pyruvate had been consumed, the level of remaining NADH was checked and adjusted to obtain the PEP measure and pyruvate kinase (5 U · ml−1) was added to initiate PEP assay (6).
Acetyl-CoA was assayed in a reaction mixture containing Tris-HCl buffer (200 mM, pH 8.1), malate (5 mM), NAD+ (33 μM), acid extract (first reading), malate dehydrogenase (1 U · ml−1) (second reading), and citrate synthase (0.08 U · ml−1) (third reading) (20).
Oxaloacetate was assayed in a reaction mixture containing potassium phosphate buffer (100 mM, pH 7), MgCl2 (2.5 mM), NADH (33 μM), acid extract, and MDH (120 U · ml−1) to initiate oxaloacetate consumption (procedure was adapted from that described in reference 18).
NAD+ was assayed in a reaction mixture containing pyrophosphate buffer (50 mM, pH 8.8), semicarbazide (2.5 g · liter−1), absolute ethanol (80 mM), acid extract, and alcohol dehydrogenase (0.5 U · ml−1) (12). The NADH present in the alkali extract was measured in a reaction mixture containing triethanolamine buffer (200 mM, pH 7), pyruvate (5 mM), alkaline extract, and LDH (20 U · ml−1) to initiate the pyruvate-dependent oxidation of NADH (11).
All metabolite concentrations obtained were relative to cellular dry weight but were expressed as aqueous molar values, using an average intracellular concentration of 2.3 ml · g (dry weight)−1 (10).
RESULTS
Batch cultivation.
The carbon recovery levels were close to 97% in the three experiments (Table 1). When the ORP increased from −360 to −280 mV, lactate production decreased from 0.15 to 0.01 mol per mol of consumed substrate, carbon dioxide decreased from 0.71 to 0.39 mol per mol of consumed substrate, and formate increased from 0.88 to 1.31 mol per mol of consumed substrate. In fact, the total formate production, equal to the sum of ethanol and acetate (Fig. 1), did not vary with ORP values, but CO2 produced from formate decreased when the ORP increased. The three other products, acetate, ethanol, and succinate, did not seem to be affected by ORP variations. Concerning the final biomass, the fermentation with 1 g of dithiothreitol (DTT) per liter presented a decrease of 30% compared to normal growth. The weight of dry biomass produced per ATP mole (YATP) varied from 5.6 to 7.9 g (dry weight) · mol of ATP−1.
TABLE 1.
Effect of ORP on metabolic end product distribution for anaerobic batch cultivation in Trypticase soy broth medium maintained at a pH of 7.0
| Reducing compound added (concn [g/liter]) | NaBH4 (0.2) | None | DTT (1) |
|---|---|---|---|
| ORP during growth phase | −360 | −325 | −280 |
| Final biomass (g [dry weight]/liter) | 1.02 ± 0.05 | 1.15 ± 0.08 | 0.77 ± 0.03 |
| Formatea | 0.88 ± 0.08 | 1.16 ± 0.10 | 1.31 ± 0.08 |
| Ethanola | 0.87 ± 0.08 | 0.89 ± 0.12 | 0.94 ± 0.15 |
| Acetatea | 0.81 ± 0.05 | 0.88 ± 0.09 | 0.87 ± 0.05 |
| Succinatea | 0.09 ± 0.02 | 0.09 ± 0.02 | 0.11 ± 0.02 |
| Lactatea | 0.15 ± 0.02 | 0.07 ± 0.01 | 0.01 ± 0.01 |
| CO2b | 0.71 | 0.58 | 0.39 |
| Carbon recovery (%) | 96 | 98 | 97 |
| YATP (g [dry weight]/mole of ATP) | 7.0 | 7.9 | 5.6 |
Values (in moles per mole of consumed substrate) are averages (± standard deviations) from two different experiments.
Calculated using theoretical flux (CO2 = ethanol + acetate − formate − CO2 consumed by succinate production).
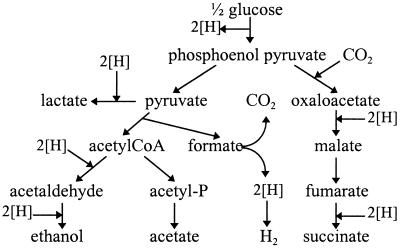
FIG. 1.
Fermentation pathways of E. coli. The flow of reducing equivalents ([H]) is shown.
Chemostat cultivation. (i) Effect of the ORP on growth.
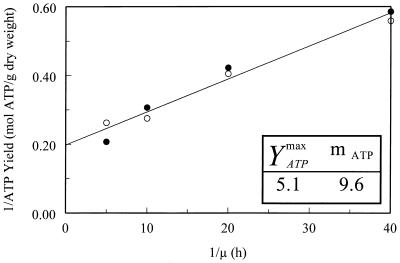
The biomass was affected by the ORP value and by the dilution rate (μ). When the μ increased from 0.025 to 0.2 h−1, in LRC, biomass decreased from 0.49 to 0.20 g · liter−1, and in HRC biomass decreased from 0.2 to 0.12 g · liter−1. Whatever the tested dilution rate was, a 250-mV ORP decrease led to a twofold decrease in biomass production (Table 2). The maximum YATP (YATPmax) and maintenance coefficient (mATP, in mmoles of ATP · g [dry weight]−1 · h−1) values were identical under the two sets of ORP conditions (Fig. 2). The glucose consumption was on average two times higher in LRC than in HRC (Table 2). As for the average ATP yield, there was a slight decrease from 2.8 mol of ATP · mol of consumed glucose−1 in LRC to 2.3 mol of ATP · mol of consumed glucose−1 in HRC (Table 2).
TABLE 2.
Results of anaerobic chemostat cultivation in minimum media with glucose as the carbon source maintained at a pH of 6.0
| Condition or activity | LRC and a dilution rate (h−1) of:
|
HRC and a dilution rate (h−1) of:
|
||||||
|---|---|---|---|---|---|---|---|---|
| 0.025 | 0.05 | 0.1 | 0.2 | 0.025 | 0.05 | 0.1 | 0.2 | |
| ORP (mV) | −140 | −80 | −50 | −40 | −340 | −320 | −320 | −315 |
| Biomass (g [dry weight]/liter)a | 0.49 ± 0.02 | 0.45 ± 0.02 | 0.42 ± 0.02 | 0.20 ± 0.01 | 0.20 ± 0.01 | 0.19 ± 0.01 | 0.15 ± 0.01 | 0.12 ± 0.01 |
| Glucose consumed (%)b | 96 ± 4 | 67 ± 3 | 46 ± 2 | 16 ± 1 | 51 ± 2 | 33 ± 2 | 18 ± 1 | 13 ± 1 |
| Carbon recovery (%)c | 97 | 92 | 92 | 90 | 96 | 97 | 98 | 96 |
| NADH recovery (%)d | 120 | 118 | 116 | 120 | 119 | 113 | 109 | 105 |
| Oxidation state balancee | 0.64 | 0.46 | 0.30 | 0.41 | −0.12 | −0.27 | −0.18 | −0.15 |
| ATP yieldf | 2.99 | 2.80 | 2.76 | 2.73 | 2.20 | 2.27 | 2.35 | 2.44 |
Values are averages (± standard deviations) of three measurements.
Values are percentages of the initial concentration (20 g · liter−1) ± standard deviations.
Biomass was taken into account to calculate carbon recovery, with a 50% carbon content.
The NADH recovery was calculated as the sum of pathways producing NADH versus those consuming NADH (NADH recovery = (2 × glucose + CO2 + CO2 consumed by succinate production)/(2 × ethanol + 2 × succinate + lactate) × 100.
Oxidation state balance was calculated as the sum of products with positive oxidation states and those with negative oxidation states per mole of consumed glucose (for more details, see Materials and Methods).
ATP yield values are in moles of ATP produced (lactate + acetate + formate + CO2 + CO2 consumed by succinate production) per mole of consumed glucose.
FIG. 2.
Effect of the dilution rate and ORP conditions (LRC [filled symbols] and HRC [open symbols]) on YATP of E. coli K-12 when it was grown in continuous culture.
(ii) Metabolic fluxes and balances.
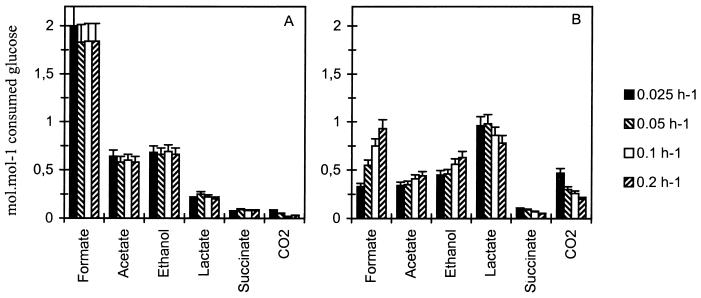
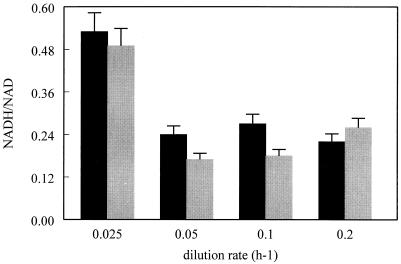
The carbon recovery level was between 90 and 98%, measured independently of the dilution rate and ORP conditions (Table 2). The distribution of the consumed glucose carbon between the different products of glucose metabolism was calculated for the eight conditions (Fig. 3). The elemental composition C4.2H8O1.25N0.68P0.1 was used to estimate the amount of assimilated carbon recovered in biomass (22). In the case of LRC, the product distribution, expressed in moles per mole of consumed substrate, was identical for all dilution rates. The fermentation produced formate (formate and CO2), acetate, ethanol, and lactate in proportions of approximately 2.5:1:1:0.3 and a small amount of succinate and CO2. With the HRC, this proportion was radically modified by increased production of lactate and CO2 and decreased production of acetate (Fig. 3), and the fermentation spectrum was on average 2:0.6:1:2, with a small amount of succinate. The lactate percentage increased from 10 to 12% of the total carbon in LRC to 35 to 47% of the total carbon in HRC. As for the ethanol/acetate ratio, it increased from 1.07 to 1.15 in LRC to 1.33 to 1.42 in HRC. The total carbon dioxide synthesis (CO2 gas measured and the part used in succinate synthesis) was about 2% in LRC and between 3.5 and 9% (depending on the dilution rate) in HRC. The NADH/NAD+ ratio was independent of ORP conditions but varied with the dilution rate from 0.2 in HRC to 0.5 in LRC (Fig. 4). The NADH recovery, was on average equal to 115%, independent of the dilution rate and ORP (Table 2).
FIG. 3.
Production rate of formate, acetate, ethanol, lactate, succinate, and CO2 in anaerobic glucose chemostat culture of E. coli K-12 regulated at a pH of 6.0, depending on dilution rate, and in LRC (range, −40 to −140 mV) (A) and HRC (range, −315 to −340 mV) (B). Each value is the average of two measurements. The standard deviation is ±10%.
FIG. 4.
NADH/NAD+ ratio versus dilution rate and ORP conditions (LRC [black] and HRC [gray]). The values are the means ± standard deviations (error bars) of at least three measurements.
(iii) Oxidation balance.
Each molecule has its own oxidation state (glucose, 0; acetate, 0; lactate, 0; formate, +1; succinate, +1; carbon dioxide, +2; hydrogen, −1; and ethanol, −2). Using the average composition formula of E. coli, C4.2H8O1.25N0.68P0.1 and 4% for trace elements (≈100 g/mol) (22), the oxidation state of the biomass is equal to −1.5. The sum of oxidized and reduced products should equal the oxidation state of the substrate (4). Whereas for HRC, there was an excess of reducing equivalents of 0.12 to 0.27 mol per mol of glucose, for LRC, the excess of oxidized equivalents was between 0.30 and 0.64 mol per mol of glucose (Table 2).
(iv) Enzyme synthesis.
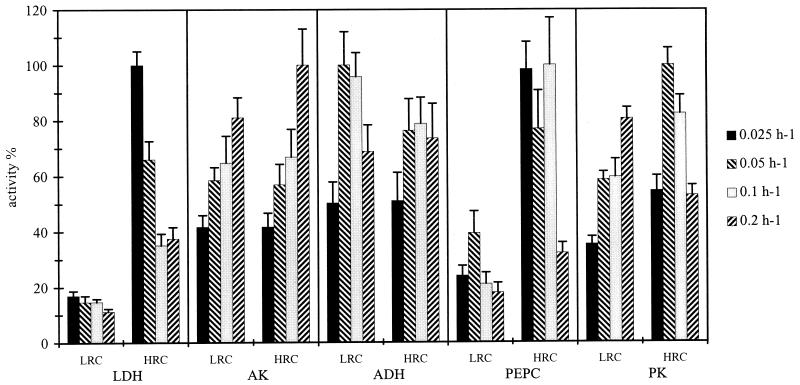
The activities of the PEPC, ADH, and PK were constant whatever the dilution rate, whereas the activity of LDH decreased and that of AK increased with an increase in the dilution rate, for the two sets of ORP conditions (Fig. 5). As for ORP, it especially affected the LDH and PEPC activities. The former increased three- to sixfold and the latter increased two- to fivefold in HRC compared to LRC.
FIG. 5.
Effect of dilution rate and the two sets of ORP conditions (LRC and HRC) on in vitro measurements of LDH, AK, ADH, PEPC, and PK in E. coli K-12 cell extract. For each enzyme 100% was equal to the highest values obtained whatever the dilution rate and ORP condition. Each value is the average ± standard deviation (error bar) of at least three measurements.
(v) Intracellular intermediary metabolites.
PEP, acetyl-CoA, OAA, and pyruvate are situated at four major branch points of glycolysis and fermentation pathways which intervene in numerous enzyme regulations. Due to a high extracellular concentration of pyruvate, the difference between intra- and extracytoplasmic concentrations was less than the assay error, and consequently, the intracellular concentration of pyruvate was not determined. But it was interesting to note that the extracellular concentration of pyruvate was on average 10-fold higher in HRC than in LRC (Table 3).
TABLE 3.
Intracellular concentrations of three intermediary metabolites and of extracellular pyruvate, depending on dilution rate and ORP conditionsa
| Metabolite or acid | LRC and a dilution rate (h−1) of:
|
HRC and a dilution rate (h−1) of:
|
||||||
|---|---|---|---|---|---|---|---|---|
| 0.025 | 0.05 | 0.1 | 0.2 | 0.025 | 0.05 | 0.1 | 0.2 | |
| PEP (mM) | 0.10 ± 0.02 | 0.16 ± 0.02 | 0.25 ± 0.04 | 0.63 ± 0.15 | 0.10 ± 0.01 | 0.23 ± 0.02 | ND | 0.65 ± 0.21 |
| Acetyl-CoA (mM) | 0.07 ± 0.03 | 0.15 ± 0.05 | 0.16 ± 0.06 | 0.32 ± 0.14 | 0.17 ± 0.05 | 0.21 ± 0.08 | 0.57 ± 0.18 | ND |
| OAA (mM) | 0.11 ± 0.02 | 0.26 ± 0.05 | 0.49 ± 0.20 | 1.00 ± 0.4 | 0.12 ± 0.03 | 0.48 ± 0.15 | 0.51 ± 0.20 | 0.86 ± 0.35 |
| Pyruvate (μM) | 87 ± 4 | 50 ± 3 | 35 ± 2 | 27 ± 2 | 705 ± 35 | 600 ± 20 | 350 ± 10 | ND |
Intracellular concentrations have been calculated with the correlation factor of 2.35 μl · mg of dry cells−1. Values are the averages ± standard deviations of at least three measurements. ND, not determined.
PEP, acetyl-CoA, and OAA concentrations increased five to tenfold when the dilution rate increased from 0.025 to 0.2 h−1. Concerning ORP effect, there was no significant difference for PEP and OAA, but acetyl-CoA concentration increased two- to eightfold in HRC (Table 3).
(vi) Steady-state fluxes of end products.
The glucose flux (qglucose) slightly increased in HRC (Table 4). In the cases of acetate flux (qacetate), ethanol flux (qethanol), and succinate flux (qsuccinate), the ORP had no significant effect. On the other hand, in HRC, lactate flux (qlactate) increased three- to sixfold, the CO2 flux (qCO2) increased 8- to 10-fold and the formate flux (qformate) decreased from 0.5- to 5-fold. Thus, pyruvate was preferentially used for lactate synthesis in HRC and for acetyl-CoA and formate production in LRC. The difference between total formate flux (qformate plus qCO2 plus qsuccinate) and the sum of qethanol and qacetate showed that acetyl-CoA and derived compounds were used for biomass synthesis. This need varied on average from 38% of total acetyl-CoA flux in LRC to 12% of total acetyl-CoA flux in LRC.
TABLE 4.
Steady-state fluxes in chemostat culture of E. coli K-12, depending on dilution rate and ORP conditions
| Flux measureda | LRC and a dilution rate (h−1) of:
|
HRC and a dilution rate of:
|
||||||
|---|---|---|---|---|---|---|---|---|
| 0.025 | 0.05 | 0.1 | 0.2 | 0.025 | 0.05 | 0.1 | 0.2 | |
| qglucose | −5.0 | −7.4 | −11.0 | −15.2 | −6.4 | −8.9 | −11.7 | −21.5 |
| qlactate | 1.0 | 1.8 | 2.8 | 3.0 | 6.2 | 8.7 | 10.1 | 16.8 |
| qacetate | 3.2 | 4.3 | 6.3 | 8.8 | 2.2 | 3.1 | 4.8 | 9.5 |
| qethanol | 3.4 | 4.9 | 7.3 | 10.0 | 2.9 | 4.1 | 6.6 | 13.5 |
| qformate | 10.1 | 13.6 | 20.2 | 28.0 | 2.1 | 4.9 | 8.7 | 20.8 |
| qsuccinate | 0.4 | 0.7 | 1.0 | 1.2 | 0.6 | 0.8 | 0.8 | 1.0 |
| qCO2 | 0.4 | 0.4 | 0.3 | 0.4 | 3.0 | 2.6 | 3.1 | 4.3 |
Steady-state fluxes are expressed in mmol · h−1 · g (dry weight)−1 and were determined with an average accuracy of ±10%.
(vii) In vitro activity and stability of LDH.
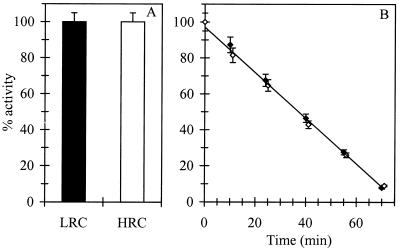
In vitro activity and stability at 50°C of LDH were identical in the two ORP conditions tested (Fig. 6).
FIG. 6.
Effect of ORP (LRC [filled symbols] and HRC [open symbols]) on in vitro activity (A) and stability (B) of LDH. Activity of 100% corresponds to the activity of LDH at a pH of 7.5 in LRC. The values are the averages ± standard deviations of two independent measurements.
DISCUSSION
The study realized in batch culture shows that ORP affects metabolic fluxes. The main results were a decrease in lactate and CO2 production and an increase in formate production when the ORP increased (Table 1). These results were obtained in batch culture using DTT and sodium borohydride to modify the ORP. Because of the known toxicity of DTT to E. coli (2) and because of the instability of sodium borohydride in solution, another reducing compound, hydrogen, recognized for its absence of toxicity, has been chosen for further experiments. The results obtained with chemostat cultivation confirm results obtained in the preliminary study and show clearly that changes in ORP affect metabolic fluxes of E. coli, but without modification of YATPmax and mATP values.
In HRC, a decrease in the level of biomass synthesis was observed, since ATP yield decreased from 2.8 to 2.3 mol of ATP · mol of consumed glucose−1 and substrate consumption was divided by two (Table 2). When ORP decreased, lactate production increased essentially to the detriment of formate and in a lower proportion of acetate. Thus, for an average ORP of −100 mV, 10% of carbon was transformed into lactate, and this percentage increased to 34 to 46% for an average ORP of −320 mV, for all tested dilution rates. Furthermore, for HRC and LRC respectively, acetyl-CoA used for biomass synthesis represented between 11 and 36% of acetyl-CoA flux. Decreases in acetyl-CoA need and in formate flux resulted in acetyl-CoA and pyruvate accumulation.
Measurements in HRC of the specific activities of LDH were in agreement with those of lactate flux, increasing by a factor 3.5 to 6 for LDH activity (Fig. 5) and 3.5 to 5.0 for lactate flux (Table 4), compared to values obtained in LRC. The similarity of variation between LDH-specific activity and lactate production shows that the apparent activity of LDH is independent of extracellular ORP conditions. Concerning the LDH activity, it could be assumed that its activity was not ORP sensitive, based on the knowledge that E. coli cytoplasm is not well ORP buffered, as has been demonstrated already (9). Measurement of in vitro activity of LDH confirmed the assumption (Fig. 6). To our knowledge, only a few enzyme activities of E. coli are classified as sensitive to ORP: the phosphotransferase system (PTS) (30), the sugar-proton and amino acid-proton symports (12, 29), and, as was more recently demonstrated, the glutathione-gated K+ channels (19).
Thus, modifications of metabolic fluxes are linked to variations of specific enzyme activities, principally an increase of LDH and probably of formate dehydrogenase (FDH), as observed by the carbon dioxide production in HRC, which was six to seven times higher than that in LRC (Fig. 3). These two enzymes are known to be overexpressed in acidic conditions (4), but chemostat is pH regulated and the higher activity observed cannot be due to extracellular pH variations. However, we have shown recently that on resting cells of E. coli, reducing conditions permeabilized the cytoplasmic membrane mainly to protons (28).
It should thereby be assumed that if the cytoplasmic membrane is permeabilized, the intracellular pH is lower than in less reducing conditions. The cell has different methods for detecting internal and external pH (27, 33), but the mechanisms of internal pH detection are not clearly understood. Although to our knowledge only a few genes show pH-dependent induction or overexpression, like the multiple drug resistance regulon mar (31), the ferric iron uptake regulation gene fur, and others identified or not (14), the regulation of other genes could be sensitive like LDH, and perhaps FDH, to an acidification of cytoplasm without external pH variation, as our results seem to show.
Another explanation may be that the enzyme has an ORP-dependent expression and/or protein stability. This last hypothesis has been tested, but no difference between results for LRC and HRC could be shown (Fig. 6). An ORP-dependent expression, as has been already demonstrated for the fumarate nitrate reductase (FNR) system (35) and for the adenosyl-cobalamin biosynthetic (Ado-CBL) operon responsible for coenzyme B12 synthesis (23), seems to be the right hypothesis to explain this modification of LDH-specific activity.
Concerning the ethanol/acetate ratio, in HRC, on average it was equal to 1.36, and it decreased to 1.12 at −100 mV. On the basis of NADH recovery, a normal ratio would be equal to 1. It has been shown that hydrogen produced by the formate hydrogenlyase (FHL) complex could be recycled by the hydrogenases Hyd-1 and/or Hyd-2 in fermentation conditions to increase the reductive pathway with ethanol formation (1, 32). This is what we observed in our study, and due to the nearly identical NADH recovery in the two ORP conditions, it can be supposed that extra reducing equivalents are directly transferred to ethanol synthesis, which slightly reduces acetate production in HRC. The redox balance, which presents a molar excess of 0.12 to 0.27 mol of reducing equivalents per mol of consumed glucose, tends to demonstrate this. On the contrary, in LRC, the redox balance presents an excess of oxidant equivalents (0.30 to 0.64 mol of oxidant equivalents per mol of consumed glucose), which confirms that extracellular ORP can modify the oxidation balance of metabolism.
The NAD cofactor ratio and NADH recovery are not modified by extracellular ORP, which illustrates the necessity of a balance in the rates of oxidation and reduction of these nucleotides to ensure the continuation of both catabolism and anabolism for a given carbon source in E. coli.
From this study, it can be concluded that manipulating the ORP changes the proportion of metabolic end products. The two assumed main causes involved in such a shift are alterations of the specific enzyme activities of LDH and FDH and of the acetyl-CoA requirement for biomass synthesis.
In fact, ORP modifies the biomass synthesis, affecting the ratio of glucose consumed while the YATPmax and the mATP values remain constant. Such a modification might be explained by an overall activation or inhibition of the microbial physiology in different oxidoreduction conditions.
ACKNOWLEDGMENTS
This work was supported by the program Aliment Demain contract R97/04 of the Ministère de l'Agriculture, de la Pêche et de l'Alimentation. C. Riondet holds doctorate fellowships from the Conseil Régional de Bourgogne.
REFERENCES
- 1.Alam K Y, Clark D P. Anaerobic fermentation balance of Escherichia coli as observed by in vivo NMR spectroscopy. J Bacteriol. 1989;171:6213–6217. doi: 10.1128/jb.171.11.6213-6217.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen C L, Mattey-Dupraz A, Missiakas D, Raina S. A new Escherichia coli gene, dsbG, encodes a periplasmic protein involved in disulphide bond formation, required for recycling DsbA/DsbB and DsbC redox proteins. Mol Microbiol. 1997;26:121–132. doi: 10.1046/j.1365-2958.1997.5581925.x. [DOI] [PubMed] [Google Scholar]
- 3.Bespalov V A, Zhulin I B, Taylor B L. Behavioral responses of Escherichia coli to changes in redox potential. Proc Natl Acad Sci USA. 1996;93:10084–10089. doi: 10.1073/pnas.93.19.10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Böck A, Sawers G. Fermentation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 262–282. [Google Scholar]
- 5.Clark D, Cronan J E., Jr Escherichia coli mutants with altered control of alcohol dehydrogenase and nitrate reductase. J Bacteriol. 1980;141:177–183. doi: 10.1128/jb.141.1.177-183.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Czok R, Lamprecht W. Pyruvate, phosphoenolpyruvate and d-glycerate-2-phosphate. In: Bergmeyer H U, editor. Methods of enzymatic analysis. Vol. 3. New York, N.Y: Academic Press, Inc.; 1974. pp. 1446–1451. [Google Scholar]
- 7.De Graef M R, Alexeeva S, Snoep J L, Joost Teixeira de Mattos M. The steady-state internal redox state (NADH/NAD) reflects the external redox state and is correlated with catabolic adaptation in Escherichia coli. J Bacteriol. 1999;181:2351–2357. doi: 10.1128/jb.181.8.2351-2357.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrigues C, Loubiere P, Lindley N D, Cocaign-Bosquet C. Control of the shift from homolactic acid to mixed-acid fermentation in Lactococcus lactis: predominant role of the NADH/NAD+ ratio. J Bacteriol. 1997;179:5282–5287. doi: 10.1128/jb.179.17.5282-5287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gill R T, Cha H J, Jain A, Rao G, Bentley W E. Generating controlled reducing environments in aerobic recombinant Escherichia coli fermentations: effects on cell growth, oxygen uptake, heat shock protein expression, and in vivo CAT activity. Biotechnol Bioeng. 1998;59:248–259. [PubMed] [Google Scholar]
- 10.Hickey E W, Hirshfield I N. Low-pH-induced effects on patterns of protein synthesis and on internal pH in Escherichia coli and Salmonella typhimurium. Appl Environ Microbiol. 1990;56:1038–1045. doi: 10.1128/aem.56.4.1038-1045.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klingenberg M. Nicotinamide-adenine dinucleotides (NAD, NADP, NADH, NADPH). Spectrophotometric and fluorimetric methods. In: Bergmeyer H U, editor. Methods of enzymatic analysis. Vol. 4. New York, N.Y: Academic Press, Inc.; 1974. pp. 2045–2059. [Google Scholar]
- 12.Konings W N, Robillard G T. Physical mechanism for regulation of proton solute symport in Escherichia coli. Proc Natl Acad Sci USA. 1982;79:5480–5484. doi: 10.1073/pnas.79.18.5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwong S C W, Rao G. Utility of culture redox potential for identifying state changes in amino acid fermentation. Biotechnol Bioeng. 1991;38:1034–1040. doi: 10.1002/bit.260380912. [DOI] [PubMed] [Google Scholar]
- 14.Lambert L A, Abshire K, Blankenhorn D, Slonczewski J L. Proteins induced in Escherichia coli by benzoic acid. J Bacteriol. 1997;179:7595–7599. doi: 10.1128/jb.179.23.7595-7599.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lebloas P. Etude des limitations et des régulations du métabolisme central de Eubacterium limosium. Ph.D. thesis. Toulouse, France: Institut National des Sciences Appliquées de Toulouse; 1992. [Google Scholar]
- 16.Leonardo M R, Dailly Y, Clark D P. Role of NAD in regulating the adhE gene of Escherichia coli. J Bacteriol. 1996;178:6013–6018. doi: 10.1128/jb.178.20.6013-6018.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowry O H, Rosebrough N J, Farr A L, Randell R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 18.McAlister L E, Evans E L, Smith T E. Properties of a mutant Escherichia coli phosphoenolpyruvate carboxylase deficient in coregulation by intermediary metabolites. J Bacteriol. 1981;146:200–208. doi: 10.1128/jb.146.1.200-208.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meury J, Robin A. Glutathione-gated K+ channels of Escherichia coli carry out K+ efflux controlled by the redox state of the cell. Arch Microbiol. 1990;154:475–482. doi: 10.1007/BF00245231. [DOI] [PubMed] [Google Scholar]
- 20.Michal G, Bergmeyer H U. Coenzyme A. In: Bergmeyer H U, editor. Methods of enzymatic analysis. Vol. 4. New York, N.Y: Academic Press; 1974. p. 2302. [Google Scholar]
- 21.Nakajima H, Suzuki K, Imahori K. Purification and properties of acetate kinase from Bacillus stearothermophilus. J Biochem. 1978;84:193–203. doi: 10.1093/oxfordjournals.jbchem.a132108. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen J, Villadsen J. Cellular growth reactions, bioreaction engineering principles. Vol. 1. New York, N.Y: Plenum Press; 1994. [Google Scholar]
- 23.O'Toole G A, Rondon M R, Trzebiatowski J R, Suh S, Escalante-Semerena J C. Biosynthesis and utilization of adenosyl-cobalamin (coenzyme B12) In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 710–720. [Google Scholar]
- 24.Peguin S, Soucaille P. Modulation of metabolism of Clostridium acetobutylicum grown in chemostat culture in a three-electrode potentiostatic system with methyl viologen as electron carrier. Biotechnol Bioeng. 1996;51:342–348. doi: 10.1002/(SICI)1097-0290(19960805)51:3<342::AID-BIT9>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 25.Racker E. Alcohol dehydrogenase from bakers yeast. Methods Enzymol. 1955;1:500–503. [Google Scholar]
- 26.Raina S, Missiakas D. Making and breaking disulfide bounds. Annu Rev Microbiol. 1997;51:179–202. doi: 10.1146/annurev.micro.51.1.179. [DOI] [PubMed] [Google Scholar]
- 27.Repaske D R, Adler J. Change in intracellular pH of Escherichia coli mediates the chemostatic response to certain attractants and repellents. J Bacteriol. 1981;145:1196–1208. doi: 10.1128/jb.145.3.1196-1208.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riondet C, Cachon R, Waché Y, Alcaraz G, Divies C. Changes in the proton motive force in Escherichia coli in response to external oxidoreduction potential. Eur J Biochem. 1999;262:595–599. doi: 10.1046/j.1432-1327.1999.00429.x. [DOI] [PubMed] [Google Scholar]
- 29.Robillard G T, Konings W N. A hypothesis for the role of dithiol-disulfide interchange in solute transport and energy-transducing processes. Eur J Biochem. 1982;127:597–604. doi: 10.1111/j.1432-1033.1982.tb06914.x. [DOI] [PubMed] [Google Scholar]
- 30.Robillard G T, Konings W N. Physical mechanism for regulation of phosphoenolpyruvate-dependent glucose transport activity in Escherichia coli. Biochemistry. 1981;20:5025–5032. doi: 10.1021/bi00520a032. [DOI] [PubMed] [Google Scholar]
- 31.Rosner J L, Slonczewski J L. Dual regulation of inaA by the multiple antibiotic resistance (Mar) and superoxide (SoxRS) stress response systems of Escherichia coli. J Bacteriol. 1994;176:6262–6269. doi: 10.1128/jb.176.20.6262-6269.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawers G. The hydrogenases and formate dehydrogenases of Escherichia coli. Antonie Leeuwenhoek. 1994;66:57–88. doi: 10.1007/BF00871633. [DOI] [PubMed] [Google Scholar]
- 33.Slonczewski J L, Macnab R M, Alger J R, Castle A M. Effects of pH and repellent tactic stimuli on protein methylation levels in Escherichia coli. J Bacteriol. 1982;152:384–399. doi: 10.1128/jb.152.1.384-399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor B L, Zhulin I B. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Unden G, Trageser M, Duchêne A. Effect of positive redox potentials (greater than +400 mV) on the expression of anaerobic respiratory enzymes in Escherichia coli. Mol Microbiol. 1990;4:315–319. doi: 10.1111/j.1365-2958.1990.tb00598.x. [DOI] [PubMed] [Google Scholar]
- 36.Vasconselos I, Girbal L, Soucaille P. Regulation of carbon and electron flow in Clostridium acetobutylicum grown in chemostat culture at neutral pH on mixtures of glucose and glycerol. J Bacteriol. 1994;176:1443–1450. doi: 10.1128/jb.176.5.1443-1450.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]