Abstract
Escherichia coli W uses the aromatic compound 4-hydroxyphenylacetate (4-HPA) as a sole source of carbon and energy for growth. The monooxygenase which converts 4-HPA into 3,4-dihydroxyphenylacetate, the first intermediate of the pathway, consists of two components, HpaB (58.7 kDa) and HpaC (18.6 kDa), encoded by the hpaB and hpaC genes, respectively, that form a single transcription unit. Overproduction of the small HpaC component in E. coli K-12 cells has facilitated the purification of the protein, which was revealed to be a homodimer that catalyzes the reduction of free flavins by NADH in preference to NADPH. Subsequently, the reduced flavins diffuse to the large HpaB component or to other electron acceptors such as cytochrome c and ferric ion. Amino acid sequence comparisons revealed that the HpaC reductase could be considered the prototype of a new subfamily of flavin:NAD(P)H reductases. The construction of a fusion protein between the large HpaB oxygenase component and the choline-binding domain of the major autolysin of Streptococcus pneumoniae allowed us to develop a rapid method to efficiently purify this highly unstable enzyme as a chimeric CH-HpaB protein, which exhibited a 4-HPA hydroxylating activity only when it was supplemented with the HpaC reductase. These results suggest the 4-HPA 3-monooxygenase of E. coli W as a representative member of a novel two-component flavin-diffusible monooxygenase (TC-FDM) family. Relevant features on the evolution and structure-function relationships of these TC-FDM proteins are discussed.
Oxygenases are the enzymes that catalyze the initial reactions of aerobic catabolic pathways for aromatic compounds by incorporating either two atoms of molecular oxygen (dioxygenases) or a single oxygen atom (monooxygenases) (14, 15). For the monooxygenases that require the NAD(P)H cofactor, the reaction is separated into two steps, i.e., the oxidation of NAD(P)H to generate two reducing equivalents and the hydroxylation of substrates. Most of the monooxygenases catalyzing the hydroxylation of the aromatic ring are flavoprotein enzymes that carry out the two reactions on a single polypeptide chain (14, 15). However, multicomponent monoxygenases where NAD(P)H oxidation and the hydroxylation reaction are catalyzed by separate polypeptides linked by an electron transport chain have been also described (14, 15). The most complex monooxygenases described so far are the six-component proteins for the hydroxylation of aromatic compounds, such as phenol, benzene, and toluene (5, 14, 15).
Different two-component monooxygenases that hydroxylate aromatic compounds have been reported and they can be classified in two main categories according to the nature of the oxygenase component, that is, as heme and nonheme enzymes. Within the heme monooxygenase group, a flavoprotein constitutes the electron transfer component, and cytochrome c or cytochrome P450 is usually found as the oxygenase component (14, 15). The nonheme two-component monooxygenases can use either pteridines or flavins as cofactors (15). In turn, the flavin-dependent nonheme two-component monooxygenases can be grouped in two major families according to their electron transfer component. One family comprises those enzymes whose electron transfer component involves a ferredoxin-NAD(P) domain, e.g., the diiron XylMA (toluene-xylene monooxygenase) or the mononuclear iron TsaMB (p-toluenesulfonate monooxygenase) (19). The other family, referred to hereafter as the two-component nonheme flavin-diffusible monooxygenase (TC-FDM) family, comprises several enzymes of uncertain classification reported in the literature whose reductase component uses NAD(P)H to catalyze the reduction of a flavin that diffuses to the oxygenase component for oxidation of the substrate by molecular oxygen.
The 4-hydroxyphenylacetate (4-HPA) 3-monooxygenase from Escherichia coli W is a two-component enzyme encoded by the hpaB and hpaC genes and catalyzes the initial reaction in the degradation of 4-HPA, i.e., the introduction of a second hydroxyl group into the benzene nucleus at a position ortho to the existing hydroxyl group, giving rise to 3,4-dihydroxyphenylacetate (3,4-DHPA) (32, 33). This monooxygenase shows a broad substrate range, hydroxylating phenol derivatives (32, 33). While the HpaB protein (58.7 kDa) of 4-HPA 3-monooxygenase was shown to be the oxygenase component, HpaC (18.6 kDa) was assumed to be a coupling protein that enhanced the activity of HpaB and could prevent the wasteful oxidation of NADH in the absence of substrate (32). In this regard, a coupling protein enhancing the activity of an aromatic monooxygenase had been also described for the 4-HPA hydroxylase from Pseudomonas putida (2, 3). However, in the past four years several TC-FDM enzymes whose amino acid sequences have revealed significant similarities with those of the HpaB and HpaC proteins have been reported in different bacteria, suggesting that HpaB and HpaC could be considered as the representative oxygenase and reductase components, respectively, of this new TC-FDM family (13, 16, 21, 36, 41).
In this work, we provide the experimental demonstration that HpaC is the flavin:NADH oxidoreductase component of the 4-HPA 3-monooxygenase from E. coli W. Thus, this enzyme becomes the first sequenced protein in the aromatic TC-FDM family. A comparative analysis of different members of the rapidly growing TC-FDM family reveals interesting features on the evolution and the structure-function relationships of these proteins.
MATERIALS AND METHODS
Materials.
Restriction endonucleases and phenyl-Sepharose CL4B, Sephadex G-100, and Superose 12 HR columns were from Pharmacia Fine Chemicals. Flavin adenine dinucleotide (FAD), flavin mononucleotide (FMN), NADH, NADPH, 4-HPA, 3,4-DHPA, riboflavin, cytochrome c, ferrozine, glucose dehydrogenase, Blue Sepharose and DEAE-cellulose columns, and marker proteins for gel filtration were purchased from Sigma. Hydroxyapatite-HTP columns and marker proteins for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis were purchased from Bio-Rad. Catalase was purchased from Boehringer Mannheim. Culture media were obtained from Difco. All other chemicals were of the highest grade available and were purchased from Sigma or Merck.
Strains, plasmids, media, and growth conditions.
The E. coli K-12 strains used were DH1 (34) and TG1 (Amersham Pharmacia). Bacteria were grown in Luria-Bertani medium (34) at 37°C with shaking. The plasmids (and relevant genotype) used were pUC19 (34), pAJ27 (hpaB) (32), pAJ28 (hpaC) (32), pAJ22 (hpaB hpaC) (32), and pCE17 (c-lytA) (35). It is worth noting that the hpaC and hpaB genes were expressed in E. coli K-12 strains which lack the 4-HPA degradative cluster in their genomes (33).
DNA manipulations and transformation.
Plasmid preparation and isolation of DNA fragments were carried out by standard procedures (34). Restriction endonucleases were used according to the manufacturer's instructions. Transformations of E. coli cells were carried out by the RbCl method (34). Nucleotide sequences were determined directly from plasmids by using an ABI-377 automated DNA sequencer (Applied Biosystems, Inc., Foster City, Calif.).
Computer analyses.
Protein sequence similarity searches were made by using the BLASTP and BLASTX programs (1) via the National Institute for Biotechnology Information server (http://www.ncbi.nlm.nih.gov/cgi-bin/blast). Protein secondary structure predictions were performed with the GORI program (11) via the ExPASy server (http://expasy.hcuge.ch/www/tools.html). Pairwise and multiple protein sequence alignments were made with ALIGN (43) and CLUSTAL W (39) programs, respectively, at the Baylor College of Medicine-Human Genome Center server (http://kiwi.imgen.bcm.tmc.edu:8088/search-launcher/launcher.html). The E. coli database collection ECDC (25) was accessed via the Internet (http://susi.bio.uni-giessen.de/ecdc.html).
Purification of the reductase component HpaC.
E. coli DH1 cells harboring plasmid pAJ28 were cultured overnight at 37°C in 2 liters of Luria-Bertani medium containing 0.1 mg of ampicillin per ml. Cells were harvested by centrifugation, washed, and suspended in 100 ml of 50 mM HEPES buffer (pH 7.8). Cells were broken by passage through a French press (Aminco Corp.) operated at a pressure of 20,000 lb/in2, and the resulting cell extract was clarified by centrifugation at 26,000 × g for 30 min. Proteins contained in the clear supernatant fluid were precipitated with 60% ammonium sulfate at 4°C. The pellet was recovered by centrifugation and dialyzed against 50 mM Tris-HCl buffer, pH 8.0 (buffer A). The soluble protein was loaded on a DEAE-cellulose column (50-ml bed volume) equilibrated with buffer A. Proteins were eluted at a rate of 1 ml/min with 250 ml of ammonium sulfate in an increasing gradient from 0 to 0.5 M in buffer A with Bio-Rad Econo-System equipment. The fractions showing flavin reductase activity were pooled and loaded onto a phenyl-Sepharose CL-4B column (12-ml bed volume) equilibrated with buffer A plus 0.3 M ammonium sulfate. Proteins were eluted at a rate of 0.16 ml/min with 100 ml of a gradient with a decreasing concentration of ammonium sulfate (0.3 to 0 M) in buffer A. Under these conditions, the reductase eluted at the end of the gradient after the column was washed with 20 ml of buffer A. The fractions showing the highest reductase activity were pooled, concentrated by centrifugation with a Centricon 10 filter (Amicon) at 11,000 × g for 30 min, and loaded on a hydroxyapatite-HTP column (5 ml of bed volume) equilibrated with 10 mM Na-phosphate buffer, pH 7.0. The unbound protein containing the reductase activity was recovered by washing the column with 10 ml of the equilibration buffer. Fractions with reductase activity were concentrated with a Centricon 10 filter and loaded on a column of Sephadex G-100 (20 by 0.6 cm) that was equilibrated and eluted with buffer A at a flow rate of 0.2 ml/min. The reductase activity that was recovered as a sharp peak in the void volume was stored at −20°C. The yield and fold purification of HpaC were 40% and 166, respectively.
Purification of the oxygenase component HpaB.
The native oxygenase component HpaB was partially purified from extracts of E. coli DH1(pAJ27) cells by Blue Sepharose columns as described elsewhere (32). The chimeric CH-HpaB oxygenase was purified from E. coli TG1(pAJ31) cells by affinity on DEAE-cellulose through a single chromatographic step as previously described (35). In short, cells were broken by using a French press, and the resulting cell extract was clarified by centrifugation and loaded on a DEAE-cellulose column (10-ml bed volume) equilibrated with 20 mM sodium phosphate buffer, pH 6.9 (buffer B). The column was washed with 20 volumes of buffer B containing 1.5 M NaCl. The chimeric protein was eluted with 2 volumes of buffer B containing 1.5 M NaCl plus 140 mM choline. Fractions showing oxygenase activity were pooled, dialyzed against 2 liters of buffer A, and stored at −20°C after addition of 10% glycerol. The yield and fold purification of CH-HpaB were 47% and 12.5, respectively.
Molecular mass determination.
Fractions containing enzyme activity were tested for purity by SDS-PAGE (26) with 12.5% polyacrylamide gels and a molecular mass marker kit for determination of the subunit molecular mass. Polyacrylamide gels were stained with Coomassie brilliant blue. The molecular mass of the native protein was determined by gel filtration analysis on a Superose 12 HR 10/30 column equilibrated with 50 mM sodium phosphate buffer, pH 8.0, with Gilson high-performance liquid chromatography (HPLC) equipment. The standards used to calibrate the column were ferritin (480,000 Da), catalase (240,000 Da), alcohol dehydrogenase (150,000 Da), bovine serum albumin (67,000 Da), ovalbumin (45,000 Da), and chymotrypsinogen A (25,000 Da).
HPLC analysis of 4-HPA consumption and 3,4-DHPA production.
The production of 3,4-DHPA was analyzed with Gilson HPLC equipment with a Lichrosphere 5 RP-8 column (150 by 4.6 mm) after a guard column (mobile phase, 20% methanol-water containing 0.1% [vol/vol] trifluoracetic acid; flow rate, 1 ml/min). The detection was carried out spectrophotometrically at 280 nm. Metabolites were identified by comparison of their retention times with those of pure substances.
Enzyme assays.
Flavin:NAD(P)H reductase activity was detected by a spectrophotometric assay measuring the disappearance of the yellow color due to the reduction of FMN by NADH at 450 nm (ɛ = 12,200 M−1 cm−1) (18). The assay cuvette contained 0.06 mM FMN and 5 mM NADH in 50 mM HEPES buffer (pH 7.8), in a final volume of 0.5 ml. After the addition of the enzyme, the assay was run at 22°C during a controlled period of time. To determine the enzyme specificity, FAD (ɛ450nm = 11,300 M−1 cm−1) (18), riboflavin (ɛ450nm = 12,200 M−1 cm−1) (18), and NADPH were also tested as substrates.
Flavin:NADH oxidase activity was assayed at 22°C by monitoring the decrease in absorption of NADH at 340 nm (ɛ = 6,220 M−1 cm−1) (10) in 50 mM HEPES buffer, pH 7.8, containing 0.2 mM NADH and 0.01 mM flavin. Assays were initiated by the addition of the enzyme. Steady-state kinetic measurements were performed with a 1-cm light path cuvette in a final volume of 0.5 ml with a Shimadzu UV-160 spectrophotometer. This assay was used to determine the Km values for flavins.
NADH:cytochrome c reductase activity was assayed by recording the NADH-dependent reduction of cytochrome c at 550 nm (ɛ = 21,100 M−1 cm−1) (4) with 50 mM HEPES buffer, pH 7.8, at 22°C; the reaction mixture contained 0.04 mM cytochrome c, 0.2 mM NADH, and 0.03 mM FMN or FAD.
NADH:iron(III) reductase activity was determined with ferrozine as the iron (II) trap. The reaction was followed by recording the absorbance at 562 nm of the ferrozine-iron(II) chelate (ɛ = 28,000 M−1 cm−1) (10). The assay was performed at 22°C in 50 mM HEPES buffer, pH 7.8, containing 0.2 mM ferric citrate, 1 mM ferrozine, 3 mM NADH, and 0.02 mM FMN.
Oxygenase assays were performed at 22°C in 50 mM HEPES buffer, pH 8.0, containing 4 mM 4-HPA, 3 mM NADH, 0.01 mM FAD, 50 mM glucose, 120 U of catalase/ml, and 0.5 U of glucose dehydrogenase/ml. The mixture was gently stirred. Catalase was added to the reaction mixture to avoid accumulation of H2O2 produced due to substrate-independent oxygen consumption. Glucose dehydrogenase was added to regenerate the NADH. The reaction was stopped at different times with 5% trichloroacetic acid (wt/vol) and the samples were centrifuged at 30,000 × g for 10 min before the production of 3,4-DHPA was analyzed by HPLC.
Oxygenase activity was also determined by a two-compartment reaction assay. In this case, a solution (1 ml) carrying the HpaC reductase (0.6 μg of purified protein) in 50 mM HEPES buffer was placed inside a dialysis bag (6 mm in diameter; molecular weight cutoff, 12 to 14 kDa; Dialysis SERVA Visking) that was immersed into a solution (3 ml) of 50 mM HEPES buffer, pH 8.0, containing 4 mM 4-HPA, 3 mM NADH, 0.01 mM FAD, 50 mM glucose, 120 U of catalase/ml, 0.5 U of glucose dehydrogenase/ml, and the CH-HpaB oxygenase (30 μg of purified protein). The reaction was performed at 22°C with shaking, and the resulting 3,4-DHPA was analyzed by HPLC at different time intervals from 10 to 300 min. A control experiment was carried out under identical conditions but with the same amount of HpaC placed outside the dialysis bag.
N-terminal amino acid sequencing.
The N-terminal amino acid sequence was determined by Edman degradation with an Applied Biosystems model 470A gas phase sequencer fitted with an online PTH analytical system.
Protein determination.
Protein was determined by the method of Bradford (7) with bovine serum albumin as a standard.
RESULTS AND DISCUSSION
Purification and characterization of the small component (HpaC) of 4-HPA 3-monooxygenase from E. coli W.
We had observed that the 4-HPA 3-monooxygenase activity of HpaB was significantly increased after the addition of extracts of E. coli DH1(pAJ28) cells overexpressing the HpaC protein (32). Moreover, in vitro analyses demonstrated that the hydroxylating activity of HpaB was NADH and FAD dependent (32). Although the purified HpaB protein did not show the characteristic absorption bands of flavin enzymes, we assumed that the FAD and/or NADH binding sites of the 4-HPA 3-monooxygenase should be located in HpaB, since it can be specifically eluted by NADH from a Blue Sepharose column, whereas the HpaC protein did not bind to this matrix (32). Assuming that the behavior of HpaC resembled that of the coupling protein of the 4-HPA 3-hydroxylase from P. putida (2, 3), a similar role was tentatively ascribed to this protein (32). However, the possibility that the hpaC gene could encode a reductase instead of a coupling protein was not envisioned until a similar protein, the ORF6 of the actinorhodin cluster from Streptomyces coelicolor (hereafter named the ActVB protein), was shown to behave as a flavin:NADH oxidoreductase (21). According to this observation, analyses carried out with extracts of E. coli DH1(pAJ28) revealed the presence of a high level of flavin:NADH oxidoreductase activity compared with control extracts of E. coli DH1 cells harboring plasmid pUC18 (see below). To ascertain that FMN reduction in the presence of NADH was carried out specifically by the HpaC protein and not by another enzyme induced in the host cell as a consequence of the overexpression of the hpaC gene, we decided to purify the putative HpaC oxidoreductase enzyme.
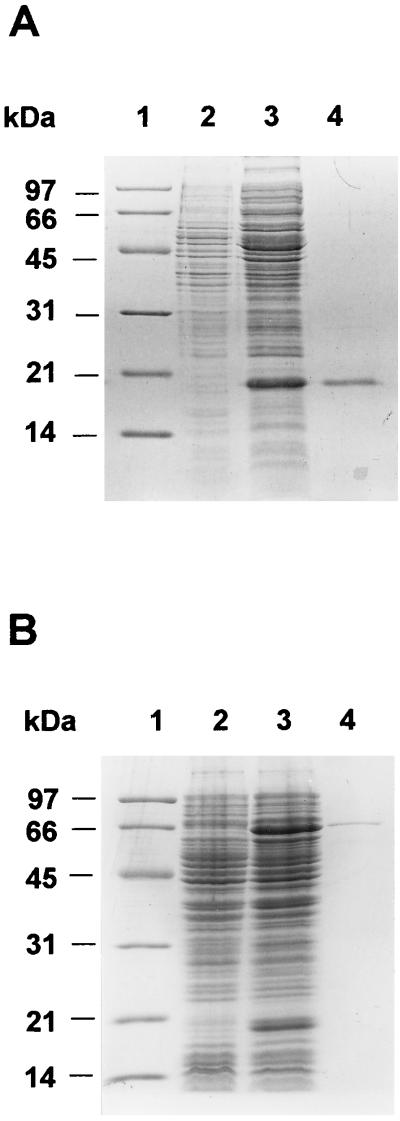
The purification of flavin:NADH oxidoreductase activity from crude extracts of E. coli DH1(pAJ28) cells rendered a protein of at least 95% homogeneity, as judged by denaturing gel electrophoresis (Fig. 1A). The purified enzyme showed an apparent molecular weight on SDS-PAGE of 20,000, which was in agreement with the predicted molecular mass for the HpaC protein (32). Moreover, its N-terminal amino acid sequence analysis revealed a sequence, Met-Gln-Leu-Asp-Glu, which was identical to that deduced from the nucleotide sequence of the hpaC gene (32). These findings strongly supported the assumption that the reductase activity observed in crude extracts of E. coli DH1(pAJ28) corresponded to that of the HpaC protein. The purified HpaC protein was very stable at −20°C, since no significant loss of activity was observed during 2 months of storage at this temperature.
FIG. 1.
SDS-PAGE analysis of the overproduction and purification of the two components of the 4-HPA 3-monooxygenase from E. coli W. (A) Lane 1, molecular mass markers; lane 2, soluble control extract from E. coli DH1(pUC18); lane 3, soluble crude extract from E. coli TG1(pAJ28); lane 4, purified HpaC reductase. (B) Lane 1, molecular mass markers; lane 2, soluble control extract from E. coli TG1(pUC18); lane 3, soluble crude extract from E. coli TG1(pAJ31); lane 4, purified CH-HpaB protein. The molecular mass marker proteins are indicated in kilodaltons.
Gel permeation chromatography on a Superose 12 HR column of crude extracts from E. coli DH1(pAJ28) indicated that HpaC formed dimers. However, purified HpaC eluted as a high molecular weight protein, which indicates the formation of soluble aggregates. A similar behavior has been also observed with the ActVB reductase from S. coelicolor, which is a dimer with a high tendency to form aggregates when purified (21).
Biochemical properties of the HpaC oxidoreductase.
The purified HpaC oxidoreductase was colorless, and the UV-visible spectrum showed no evidence for any chromogenic cofactor (data not shown). The reductase activity of HpaC depended on both NADH and flavin being added to the assay. No requirement for any other cofactors was apparent. In particular, these experiments showed that 4-HPA had no influence in this reaction. The most effective substrates were NADH and FMN, but FAD and riboflavin could also be turned over by the enzyme with similar Km values (see below). When the reductase assay was performed without shaking, FMN was reduced completely by an excess of NADH, and the absorption band at 450 nm corresponding to FMN was completely bleached. After shaking, reduced flavin mononucleotide (FMNH2) was recycled by reaction with oxygen to form H2O2, and the absorption at 450 nm returned. When FMN was added in a 200-fold molar excess of the HpaC protein, it became completely reduced (data not shown), suggesting that the flavin dissociated from the protein and behaved as a true substrate rather than as a tightly bound cofactor.
The HpaC reductase showed optimal activity at pH 7.0, but it maintained more than 80% of activity between pH values of 6.5 to 8. The Km values for NADH and different flavins (Table 1) were similar to those observed for other flavin reductases, that is, enzymes that generate free reduced flavins (10, 18, 21, 38). It is worth noting the high Km values for FMN when compared to the Km values in the nanomolar range that have been determined for other monooxygenases, an observation that reinforces the idea that FMN acts as a substrate rather than as a prosthetic group. The specific activity of HpaC on different flavins using NADH as an electron donor (Table 1) was similar to that reported for the SnaC reductase (38) but 10 times higher than that reported for ActVB reductase (21), the other two HpaC-like reductases (see below) that have been purified so far. Other flavin reductases that do not show sequence similarity to HpaC, like the Fre reductase from E. coli (10) and the major flavin reductase (FRase I) from Vibrio fischeri (18), also showed specific activities of around 100 μmol min−1 mg−1. Although the HpaC enzyme can also use NADPH as a substrate, its specific activities on FMN, FAD, and riboflavin were more than 2 orders of magnitude lower than those observed in the presence of NADH (Table 1). This behavior appears to be typical of flavin reductases that do not contain a flavin as a prosthetic group, since they reduce FMN, FAD, and riboflavin with similar efficiencies but present a higher selectivity for NADH or NADPH (10, 18, 21, 38). The low specificity could be ascribed to the fact that in these reductases, the flavin behaves as a real substrate and not as a tightly bound prosthetic group, as is the case in the majority of flavin enzymes (12).
TABLE 1.
Kinetic properties of HpaC
| Kinetic parameter and substrate | Value |
|---|---|
| Km (μM) | |
| FMNa | 2.1 |
| Riboflavina | 2.6 |
| FADa | 3.1 |
| NADHb | 40 |
| Vmax (μmol/min/mg) | |
| FMNa | 70 |
| Riboflavina | 59 |
| FADa | 38 |
| FMNc | 0.5 |
| Riboflavinc | 0.4 |
| FADc | 0.1 |
| Cytochrome cd | 1.8 |
| Fe3+d | 12.4 |
Km and Vmax were obtained by using 150 μM NADH as the electron donor.
Km was obtained by using 60 μM FMN as the electron acceptor.
Vmax was obtained by using 150 μM NADPH as the electron donor.
Vmax was obtained by using 150 μM NADH and 60 μM FMN.
The HpaC oxidoreductase can also reduce cytochrome c and iron(III) at a high velocity (Table 1). Although cytochrome c reductase activity has also been reported for the reductase components of pyrrole-2-carboxylate monooxygenase (4) and the chlorophenol 4-monooxygenase (16), two putative members of the TC-FDM family (see below), ferric reductase activity had not been detected so far in any other reductase component of this family of monooxygenases. It is worth noting that the ferric and cytochrome c reductase activities had been considered a peculiar characteristic of Fre reductase from E. coli (10) or of the major flavin reductase (FRase I) from V. fischeri (18). Therefore, in all these cases, the electron transfer from NADH to other electron acceptors appears to be mediated through FMNH2 generated by a reductase.
Analysis of the oxygenase-reductase interactions in the 4-HPA 3-monooxygenase.
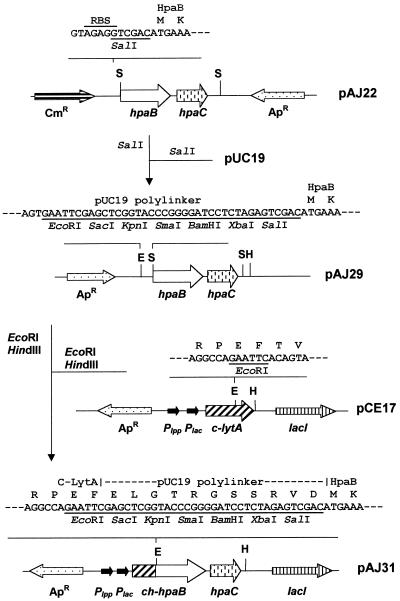
Although we have observed that HpaC was able to produce FMNH2 and reduced flavin adenine dinucleotide (FADH2) in vitro in the absence of 4-HPA, it was necessary to investigate whether such activity could be affected by the presence of the oxygenase component HpaB. In spite of the fact that the HpaB protein had been purified, it lost most of its original activity due to its low stability and to the time consumed by the complex purification procedure (32). When we tried to purify the HpaB protein by a single Blue Sepharose chromatography, the partially purified enzyme represented about 17% of the total protein, as determined by SDS-PAGE, and showed an activity of 140 nmol min−1 mg−1 in the presence of saturating concentrations of NADH and HpaC reductase (data not shown). Although this HpaB preparation presented a high level of activity, the possibility that some host reductase(s) could be retained in the Blue Sepharose column and thereafter coeluted with HpaB could not be ruled out. In fact, we have detected a low FAD-dependent reductase activity (5 nmol min−1 mg−1) in HpaB preparations. Therefore, to purify the HpaB enzyme by a faster and more selective procedure, we constructed a chimeric tagged HpaB protein (CH-HpaB) by fusing the choline-binding domain of the major autolysin of Streptococcus pneumoniae (35) to the N terminus of HpaB (Fig. 2). The CH-HpaB protein can be purified free of reductase-contaminating activities in a single step by affinity chromatography with DEAE-cellulose, a choline analogue-containing matrix (35). E. coli TG1 cells harboring plasmid pAJ31 produced large amounts of the CH-HpaB fusion both as soluble and insoluble (inclusion bodies) protein. The soluble protein was recovered by centrifugation and loaded on a DEAE-cellulose column. Figure 1B shows that the CH-HpaB protein eluted in the choline fraction is nearly pure. Interestingly, the purified CH-HpaB protein did not show a contaminating flavin reductase activity (data not shown) but presented a high level of oxygenase activity in the presence of saturating concentrations of NADH and HpaC reductase (see below).
FIG. 2.
Schematic representation of the construction of the gene encoding the CH-HpaB fusion protein. Abbreviations: Apr, ampicillin resistance; Cmr, chloramphenicol resistance; E, EcoRI; H, HindIII; S, SalI; Plac, lac promoter; Plpp, lpp promoter; RBS, ribosome binding site. Amino acids are indicated by their standard single letter codes.
To investigate the oxygenase activity of CH-HpaB in the absence or presence of HpaC, we determined the rate of synthesis of 3,4-DHPA from 4-HPA in vitro. Since the reaction must be carried out under aerobic conditions, we included an NADH-regenerating system and catalase in the assay to keep the concentration of NADH constant and to avoid the accumulation of H2O2, respectively. Interestingly, as we have avoided the contamination of the purified CH-HpaB protein with any flavin reductase, the chimeric enzyme was unable to hydroxylate 4-HPA in the absence of HpaC reductase. As expected, the addition of HpaC to CH-HpaB reaction mixtures (1:5 molar ratio) led to the production of 3,4-DHPA with activity levels of 460 nmol min−1 mg−1. This hydroxylating activity was NADH and FAD dependent, and neither FMN nor riboflavin could replace FAD in the reaction. This result is in agreement with the previous finding that hydroxylation of 4-HPA was stimulated by the addition of FAD to the assay (32). Although we were able to detect a very low level of hydroxylating activity when NADH was replaced by NADPH (data not shown), most probably this observation does not have any physiological relevance. The specific activity of the chimeric CH-HpaB enzyme was close to the theoretical value (800 nmol min−1 mg−1) that can be deduced from the activity of the wild-type HpaB enzyme partially purified on a Blue Sepharose column and the SDS-PAGE densitometric determination of the HpaB content in this preparation (see above). Taking into account all these results, it can be concluded that the fusion of the choline-binding domain to the N terminus of HpaB does not significantly affect its enzymatic activity but facilitates a rapid purification of the protein free from contaminant reductase activities. The use of this choline-binding domain offers, therefore, a suitable alternative for investigating the overexpression and easy purification of other oxygenases.
The observation that CH-HpaB activity was absolutely dependent on the presence of HpaC supports the hypothesis that both components are required for 4-HPA hydroxylation. Therefore, the low level of HpaC-independent oxygenase activity (0.5 nmol min−1 mg−1) detected with the native HpaB purified by the Blue Sepharose method should be ascribed to a contaminant flavin:NADH reductase from the host (see above) that generates the FADH2 required for 4-HPA hydroxylation.
To determine whether a physical interaction between the two components of the 4-HPA 3-monooxygenase is indispensable to catalyze the hydroxylation of 4-HPA, the HpaB and HpaC components were placed in two different compartments separated by a membrane permeable to compounds smaller than 14 kDa (see Materials and Methods). By this two-compartment reaction assay, 4-HPA was efficiently transformed into 3,4-DHPA at a rate of 138 nmol min−1 mg−1. This value is in the same order of magnitude of that obtained in a control experiment (345 nmol min−1 mg−1) carried out under the same conditions but with the reductase and oxygenase components placed in the same compartment. This result indicated that a physical interaction between HpaB and HpaC is not required for the hydroxylation of 4-HPA, although we cannot discard the idea that a direct interaction between HpaB and HpaC could enhance the hydroxylation reaction. Interestingly, the presence or absence of the CH-HpaB component did not affect the levels of FMN reduction by the HpaC reductase. All these data suggest that HpaC reduces FAD to FADH2, which then dissociates from the enzyme and diffuses to the medium, where it is captured by HpaB to catalyze the hydroxylation of 4-HPA. Since the HpaB oxygenase component does not require a direct interaction with the HpaC oxidoreductase to hydroxylate 4-HPA, any flavin reductase present in the host cell that is able to release FADH2 into the cytoplasm would supplant the role of HpaC. This finding explains the puzzling result observed in E. coli DH1(pAJ271), a strain that expressed the oxygenase HpaB component alone but showed a significant level of 4-HPA hydroxylating activity (32).
Structural and evolutionary analyses of the TC-FDM family.
Table 2 shows a compilation of the monooxygenases described so far that might be tentatively classified as members of the TC-FDM family. This family can be defined according to the following properties. (i) The reductase and the oxygenase components of the monooxygenase are encoded by two different genes. (ii) The reductase component uses NAD(P)H to catalyze the reduction of a flavin that diffuses to the oxygenase component for oxidation of the substrate (aromatics or nonaromatic compounds) by molecular oxygen. (iii) Both components are not flavoproteins, i.e., they do not contain any flavin prosthetic group and lack typical ferredoxin and/or flavin:NAD(P)H binding motifs. Interestingly, no three-dimensional structure is known for any of these proteins. It is worth noting that the pristinamycin IIA synthase was not included in Table 2 because although SnaC reductase shows a significant similarity with the HpaC protein, the oxygenase component is certainly an αβ heterodimer (6). Similarly, in spite of the fact that bacterial luciferase is probably the best-understood system in which the oxygenase component uses the FMNH2 produced by a reductase component (18), it was not included in Table 2 because its oxygenase component is also an αβ heterodimer. Furthermore, none of the luciferase components show a significant similarity with the reductase and oxygenase components of any member of the TC-FDM family. Finally, the two-component 4-HPA hydroxylase of P. putida was also not included in Table 2 since, apparently, it does not involve a flavin reductase component (2, 3).
TABLE 2.
Monooxygenases of the TC-FDM family
| Enzyme | Cofactors | Reductase (kDa) | Oxygenase (kDa) | Organism | Accession number or reference |
|---|---|---|---|---|---|
| 4-HPA 3-monooxygenase | FAD, NADH | HpaC (19) | HpaB (59) | E. coli | Z29081 |
| 4-HPA 3-monooxygenase | FAD, NADH | HpaH (19) | HpaA (59) | K. pneumoniae | L41068 |
| Chlorophenol 4-hydroxylase | FAD, NADH | HadB (21) | HadA (59) | B. pickettii | D86544 |
| D50690 | |||||
| Chlorophenol 4-hydroxylase | FAD, NADH | TftC (22) | TftD (58) | B. cepacia | U83405 |
| Styrene monooxygenase | FAD, NADH | StyB (18) | StyA (47) | Pseudomonas sp. strain Y2 | AJ000330 |
| Phenol hydroxylase | Unknown | Unknown | PheA (60) | B. thermoleovorans | AF031325 |
| p-Nitrophenol monooxygenase | FAD, NAD(P)H | Component A | Component B | B. sphaericus JS905 | 20 |
| Dibenzothiophene 5-5′ dioxide monooxygenase | FMN, NADH | DszD (21) | DszA (50) | R. erythropolis | AF048979 |
| L37363 | |||||
| Dibenzothiophene-5-oxide monooxygenase | FMN, NADH | DszD (21) | DszC (45) | R. erythropolis | AF048979 |
| L37363 | |||||
| Pyrrole-2-carboxylate monooxygenase | FAD, NADH | Small subunit (19) | Large subunit (54) | Rhodococcus sp. | 4 |
| Actinorhodin biosynthesis | FMN, NADH | ActVB (18) | ActVA-5 (40) | S. coelicolor | X58833 |
| Frenolicin biosynthesis | Unknown | FrnH (18) | FrnT (40) | S. roseofulvus | AF058302 |
| Granaticin biosynthesis | Unknown | Orf34 (19) | Orf21 (42) | S. violaceoruber | AJ011500 |
| Valanimycin biosynthesis (isobutylamine hydroxylase) | FAD, NADPH | VlmR (21) | VlmH (40) | S. viridifaciens | U93606 |
| U76606 | |||||
| Nitrilotriacetate monooxygenase | FMN, NADH | NtaB (35) | NtaA (51) | C. heintzii | U39411 |
| Nitrilotriacetate monooxygenase | FMN, NADH | NmoB (35) | NmoA (51) | C. heintzii | L49438 |
| EDTA-monooxygenase | FMN, NADH | cB′ (25) | cA′ (44) | Strain DSM9103 | 44 |
| EDTA-monooxygenase | FMN, NADH | Unknown | Oxygenase (45) | Strain BNC1 | 30 |
| 2,5-Diketocamphane 1,2-monooxygenase |
FMN, NADH | Oxidase component (36) | Oxygenase component (36) | P. putida ATCC 17453 | 37 |
| Aliphatic sulfonate monooxygenase | Unknown | Unknown | SsuD (41) | B. subtilis | Z93102 |
| Aliphatic sulfonate monooxygenase | Unknown | SsuE (21) | SsuD (42) | E. coli | AJ237695 |
| Methanesulfonate sulfonatase | FMN, NADH | MsuE (20) | MsuD (42) | P. aeruginosa | AF026067 |
The oxygenase components of the members of the TC-FDM family show marked differences in their primary structures, which might reflect the fact that the substrate specificity of these enzymes resides in these components (20). Interestingly, amino acid sequence comparisons among the oxygenase components of several aromatic hydroxylases of the TC-FDM family have revealed the existence of a conserved region (Fig. 3). Remarkably, a 54% identity has been observed between the HpaB oxygenase component of 4-HPA 3-monooxygenase from E. coli W and the phenol hydroxylase (PheA) from Bacillus thermoleovorans (9). This high amino acid sequence identity agrees with the observation that phenol is also a satisfactory substrate for HpaB (31, 32), suggesting that both enzymes might have evolved from a common ancestor able to hydroxylate phenol derivatives. Although no biochemical data on the cofactor requirements of PheA activity are available (9), the significant similarity observed between HpaB and PheA suggests that the latter does not contain a flavin:NAD(P)H reductase center, and therefore, it is likely to require an independent reductase enzyme which could provide this activity. On the other hand, it is worth noting that the central region of the HpaB-like oxygenases displays a significant similarity with a central region of the medium-chain acyl-coenzyme A dehydrogenases (data not shown) that is in close contact with the flavin nucleotide (23). Nevertheless, whether the central region of HpaB-like oxygenases has a role in the interaction with the reduced flavin provided by the cognate reductase component remains to be checked.
FIG. 3.
Multiple sequence alignment of the oxygenase components of several members of the TC-FDM family. Numbers in parentheses indicate the position of the residues in the complete amino acid sequence of the protein. A consensus sequence was deduced for positions where the residues were identical in more than half of the sequences. AF-HpaA1, putative 4-HPA 3-monooxygenase from Archaeoglobus fulgidus (AE001081); AF-HpaA2, putative 4-HPA 3-monooxygenase from A. fulgidus (AE001043); AF-HpaA3, putative 4-HPA 3-monooxygenase from A. fulgidus (AE001032); PA-PvcC putative hydroxylase of pyoverdine chromophore biosynthesis in Pseudomonas aeruginosa (AF002222); PL-HpaB, putative 4-HPA 3-monooxygenase from Pseudomonas luminescens (AF021839); KP-HpaA, 4-HPA 3-monooxygenase from Klebsiella pneumoniae (L41068); EC-HpaB, 4-HPA 3-monooxygenase from E. coli (Z29081); SD-HpaB, putative 4-HPA 3-monooxygenase from S. dublin (AF144422); BT-PheA, phenol hydroxylase from B. thermoleovorans (AF031325); BS-Yoal, putative 4-HPA 3-monooxygenase from Bacillus subtilis (Z99114); BP-HadA; chlorophenol 4-hydroxylase from B. pickettii (D86544); BC-TftD, chlorophenol 4-hydroxylase from B. cepacia (U83405); RE-DszC, dibenzothiophene monooxygenase from R. erythropolis (L37363); PP-MsuC, putative monooxygenase from P. aeruginosa (AF026067).
In contrast with the oxygenase components, we have observed an extended similarity among the reductase components of the TC-FDM proteins (Fig. 4). The only exceptions were the MsuE and the related SsuE reductases (Table 2) that lack similarity with any other described reductases (22). The similarities among the reductases correlate with the fact that all of them use the same substrates, i.e., FAD-FMN and NAD(P)H. Although most of the HpaC-like reductase components are colorless proteins, suggesting that flavin is not tightly bound to the enzyme, there are some exceptions, such as the NtaB reductase component of the nitrilotriacetate monooxygenase from Chelatobacter heintzii that shows a typical FMN spectrum although the flavin is not strongly bound to the enzyme (40). A similar behavior was also observed with the cB′ reductase component of the EDTA-monooxygenase (44).
FIG. 4.
Multiple sequence alignment of HpaC-like proteins. A comparison of the amino acid sequences of HpaC and other proteins of the databases that present a significant similarity is shown. Numbers in parentheses indicate the position of the residues in the complete amino acid sequence of the protein. A consensus sequence was deduced for positions where the residues were identical in more than half of the sequences. SC-ActVB, flavin reductase involved in the biosynthesis of actinorhodin in S. coelicolor (X58833); SR-FrnH, putative flavin reductase involved in frenolicin biosynthesis in Streptomyces roseofulvus (AF058302); SV-Orf34, putative flavin reductase involved in granaticin biosynthesis in Streptomyces violaceoruber (AJ011500); SV-VlmR, putative flavin reductase involved in valanimycin biosynthesis in Streptomyces viridifaciens (U93606); SP-SnaC, flavin reductase involved in pristimamycin IIA biosynthesis in Streptomyces pristinaespiralis (P54994); SA-Orf, putative reductase involved in chlortetracycline biosynthesis in Streptomyces aureofaciens (D38215); CH-NtaB, flavin reductase component of the nitrilotriacetate monooxygenase from C. heintzii (U39411); CH-NmoB, flavin reductase component of the nitrilotriacetate monooxygenase from C. heintzii (L49438); RE-Bph61, putative reductase involved in the metabolism of biphenyl derivatives in R. erythropolis (D88018); MT-14c, putative reductase from Mycobacterium tuberculosis (Z92774); PF-StyB, flavin reductase component of the styrene monooxygenase from Pseudomonas fluorescens (Z92524); PY-StyB, flavin reductase component of the styrene monooxygenase from Pseudomonas sp. strain Y2 (AJ000330); PS-StyB flavin reductase component of the styrene monooxygenase from Pseudomonas sp. strain VLB120 (AF031161); MT-11, putative reductase from M. tuberculosis (AL021929); ML-23, putative reductase from Mycobacterium leprae (AL022486); MT-25c, putative reductase from M. tuberculosis (Z84498); RE-DszD, flavin reductase involved in dibenzothiophene desulfurization from R. erythropolis (AF048979); PL-HpaC, HpaC-like reductase from P. luminescens (AF021838); KP-HpaH, putative reductase component of 4-HPA 3-monooxygenase from K. pneumoniae (L41068); EC-HpaC, reductase component of 4-HPA 3-monooxygenase from E. coli (Z29081); EC-F152, putative HpaC-like reductase from E. coli (AE000202); SD-HpaC, putative HpaC reductase from S. dublin (AF144422); BC-TftC, reductase component of chlorophenol 4-hydroxylase from B. cepacia (U83405); AF-HpaCL, putative reductase from A. fulgidus (AE001047); SS-HpaC-1, HpaC-like protein from Synechococcus sp. (L19521); SS-HpaC-2, HpaC-like protein from Synechococcus sp. (D64000); SS-F594, flavoprotein from Synechocystis sp. (D90900); SS-F578 flavoprotein from Synechocystis sp. (M96929); SS-F597 flavoprotein from Synechocystis sp. (D90914); SS-F573, flavoprotein from Synechocystis sp. (D64003).
Organisms have evolved a great variety of enzymes that catalyze the reduction of flavins by NAD(P)H. These flavin:NAD(P)H oxidoreductases can be classified within several families and subfamilies according to their sequence similarities and biochemical properties. One of these families is constituted by the flavoprotein reductases that contain a tightly bound flavin as a prosthetic group, e.g., the Frp reductase from Vibrio harveyi (27) and the sulfite reductase from E. coli (8), which are representative members of two different subfamilies. Another family is represented by those enzymes that do not contain a flavin as a prosthetic group and thus cannot be considered flavoproteins. Instead, they use flavins as substrates, with a rather broad substrate specificity. Based on sequence comparisons, at least two subfamilies were identified, one constituted by the Fre reductase from E. coli as well as the Fre-like and LuxG reductases of luminous bacteria (17), and the other constituted by the FRase I from V. fischeri (18). Interestingly, amino acid sequence comparison analyses revealed that the HpaC-like reductases are not similar to other nonflavoprotein flavin reductases described so far and therefore they appear to constitute a novel subfamily.
Multiple sequence alignment of flavin reductases of the HpaC subfamily revealed several conserved residues (Fig. 4). Thus, a residue of Ser (Thr or Cys) located before a pair of conserved proline residues is highly conserved at the N termini of the proteins (Fig. 4). Since Gly, Asp, and His residues at the C-terminal region of Fre are involved in NAD(P)H binding (17), it is tempting to assume that the highly conserved GDH motif found in the C-terminal regions of members of the HpaC family could play a role in NAD(P)H interaction.
Only two flavin reductases have been described so far in E. coli, the Fre reductase (17) and the sulfite reductase (8). However, the findings reported here demonstrate the existence in E. coli W of another highly active reductase, the HpaC reductase, that can be considered a prototype of a new subfamily of nonflavoprotein flavin reductases. The existence of several reductases capable of producing free reduced flavins in a microorganism, and the apparent functional interchangeability between them (29) poses some questions. A potential adaptive significance of this redundancy is to provide a readily available backup if an enzyme is lost by a mutational event. For instance, although the Fre reductase of E. coli appears to provide reduced flavins for specific purposes, it has been shown that the sulfite reductase can replace its activity in Fre-deficient mutants (8). Similarly, as it has been pointed out above, the existence of Fre or other flavin reductases could explain the residual 4-HPA 3-monooxygenase activity observed in E. coli K-12 cells expressing only the HpaB oxygenase component (32). Nevertheless, since the amount of FADH2 provided by the host reductases is not enough to achieve an optimal 4-HPA 3-monooxygenase activity, the acquisition of the hpaC reductase gene cotranscribed (coregulated) with the hpaB oxygenase gene might represent an evolutionary advantage for the development of a highly efficient 4-HPA catabolic pathway.
The genes encoding the two components of the enzymes of the TC-FDM family are located in the same operon or can be found very close in the chromosome (32), sometimes divergently oriented (24). Such an arrangement could favor a coordinated regulation of both genes and might facilitate the horizontal transfer of the monooxygenase activity. Nevertheless, there are several exceptions to this arrangement and thus the dszD gene encoding the reductase component involved in the S oxidation of dibenzothiophene is located on the genome far from the genes encoding the DszA and DszC oxygenase components (45). This genetic arrangement and the observed functional exchangeability among the flavin reductases (29) suggest that the genes encoding both components of the enzymes of the TC-FDM family might have evolved independently but frequently became physically associated in the genome.
A further step in the evolution of the HpaC-like reductases can be inferred from their amino acid sequence similarities to the C-terminal extension of four A-type flavoproteins from Synechocystis (Fig. 4) (42). It was suggested that these flavoproteins, which bind FAD and FMN at the same time in equimolecular amounts, might have evolved by the fusion of two flavin-binding domains located in the N- and C-terminus regions of the protein, showing different activities and functions (42). The existence of a HpaC-like FMN binding domain in A-type flavoproteins suggests that a fusion between the reductase and oxygenase components of the enzymes of the TC-FDM family might already exist in nature or, at least, it would be feasible to design in vitro such monocomponent monooxygenase by protein engineering (unpublished data). An evolutionary mechanism similar to that proposed here has been postulated to explain the origin of phthalate dioxygenase reductase (17).
The comparative analyses presented here also provide valuable information for ascribing functions to several still-unclassified genes found in recently sequenced genomes. Some putative HpaC-like flavin reductases that have been found in different microorganisms are included in the multisequence alignment shown in Fig. 4. Interestingly, a comparative analysis of the E. coli genome has revealed two open reading frames (ORFs), orf152 and orf196 (AE000202), that may encode two proteins showing 41 and 53% amino acid sequence identity with HpaC and HadB, respectively. orf152 is located at 23 min in the E. coli K-12 linkage map, immediately downstream of orf196 and within the same putative transcription unit that also contains five additional ORFs of unknown function, which are oriented opposite an ORF encoding a putative regulatory protein (25). Since one of these ORFs (orf382) displays a significant similarity with the DszA monooxygenase from Rhodococcus erythropolis (Table 2), it is reasonable to assume that this E. coli gene cluster might encode an oxygenolytic pathway whose physiological role is still unknown. Interestingly, in the Salmonella dublin genome there are two contiguous genes, hpaB and hpaC (AF144422), whose putative products share 92 and 79% identity with HpaB and HpaC from E. coli, respectively, thus suggesting the existence of a 4-HPA 3-monooxygenase of the TC-FDM family in this enteric bacteria.
It is surprising that whereas the HadA oxygenase component of the chlorophenol 4-hydroxylase from Burkholderia pickettii is homologous to the TftD oxygenase component of the chlorophenol 4-hydroxylase from Burkholderia cepacia (Fig. 3), the HadB component did not show any similarity with the corresponding TftC reductase or with any other HpaC-like flavin reductases. In fact, HadB shows significant similarity only with the H2O2-forming NADH oxidase from Thermus thermophilus HB8 (28). Interestingly, we have identified just at the 5′ end of the hadAB operon of B. pickettii (36), a partially sequenced ORF that might form part of the same operon and that encodes a protein showing 23% amino acid identity with the HpaC reductase (data not shown). Whether the protein encoded by this ORF could be the real reductase component of the chlorophenol 4-hydroxylase from B. pickettii instead of the proposed HadB protein (Table 2) requires further research.
In summary, the results presented above demonstrate that HpaC is a flavin:NADH oxidoreductase involved in the hydroxylation of 4-HPA by the HpaB oxygenase component of the 4-HPA 3-monooxygenase and therefore allow us to complete the functional characterization of all the genes of the 4-HPA catabolic cluster in E. coli W (31). Since the 4-HPA 3-monooxygenase from E. coli W was the first member of the new TC-FDM family whose primary structure was elucidated (32), this enzyme can be considered the archetype of this family, with the HpaC reductase being, in turn, the prototype of a new subfamily of flavin reductases.
ACKNOWLEDGMENTS
We thank J. Varela, A. Díaz, S. Carbajo, and G. Porras for their help with protein and DNA sequencing. We are indebted to M. Carrasco and E. Cano for their technical assistance. M. A. Prieto was a recipient of a Contrato de Incorporación de Doctores del Ministerio de Educación y Cultura.
This work was supported by grant AMB97-0630-C02-02 from the CICYT.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment searching tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Arunachalam U, Massey V, Miller S M. Mechanism of hydroxyphenylacetate-3-hydroxylase. A two-protein enzyme. J Biol Chem. 1994;269:150–155. [PubMed] [Google Scholar]
- 3.Arunachalam U, Massey V, Vaidayanathan C S. p-Hydroxyphenylactate-3-hydroxylase. A two-protein component enzyme. J Biol Chem. 1992;267:25848–25855. [PubMed] [Google Scholar]
- 4.Becker D, Schräder T, Andreesen J R. Two-component flavin-dependent pyrrole-2-carboxylate monooxygenase from Rhodococcus sp. Eur J Biochem. 1997;249:739–747. doi: 10.1111/j.1432-1033.1997.t01-1-00739.x. [DOI] [PubMed] [Google Scholar]
- 5.Bertoni G, Martino M, Galli E, Barbieri P. Analysis of the gene cluster encoding toluene/o-xylene monooxygenase from Pseudomonas stutzeri OX1. Appl Environ Microbiol. 1998;64:3626–3632. doi: 10.1128/aem.64.10.3626-3632.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanc V, Lagneaux D, Didier P, Gil P, Lacroix P, Crouzet J. Cloning and analysis of structural genes from Streptomyces pristinaespiralis encoding enzymes involved in the conversion of pristinamycin IIB to pristinamycin IIA (PIIA): PIIA synthase and NADH:riboflavin 5′-phosphate oxidoreductase. J Bacteriol. 1995;177:5206–5214. doi: 10.1128/jb.177.18.5206-5214.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Covès J, Nivière V, Eschenbrenner M, Fontecave M. NADPH-sulfite reductase from Escherichia coli. A flavin reductase participating in the generation of the free radical of the ribonucleotide reductase. J Biol Chem. 1993;268:18604–18609. [PubMed] [Google Scholar]
- 9.Duffner F M, Müller R. A novel phenol hydroxylase and catechol 2,3-dioxygenase from the thermophilic Bacillus thermoleovorans strain A2: nucleotide sequence and analysis of the genes. FEMS Microbiol Lett. 1998;161:37–45. doi: 10.1111/j.1574-6968.1998.tb12926.x. [DOI] [PubMed] [Google Scholar]
- 10.Fontecave M, Eliasson R, Reichard P. NAD(P)H:flavin oxidoreductase of Escherichia coli. A ferric iron reductase participating in the generation of the free radical of ribonucleotide reductase. J Biol Chem. 1987;262:12325–12331. [PubMed] [Google Scholar]
- 11.Garnier J, Osguthorpe D J, Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 12.Ghisla S, Massey V. Mechanism of flavoprotein-catalyzed reactions. Eur J Biochem. 1989;181:1–17. doi: 10.1111/j.1432-1033.1989.tb14688.x. [DOI] [PubMed] [Google Scholar]
- 13.Gibello A, Suárez M, Allende J L, Martin M. Molecular cloning and analysis of the genes encoding the 4-hydroxyphenylacetate hydroxylase from Klebsiella pneumoniae. Arch Microbiol. 1997;167:160–166. [PubMed] [Google Scholar]
- 14.Harayama S, Kok M, Neidle E L. Functional and evolutionary relationships among diverse oxygenases. Annu Rev Microbiol. 1992;46:565–601. doi: 10.1146/annurev.mi.46.100192.003025. [DOI] [PubMed] [Google Scholar]
- 15.Harayama S, Timmis K N. Aerobic biodegradation of aromatic hydrocarbons by bacteria. In: Siegel H, Siegel A, editors. Metal ions in biological systems. New York, N.Y: Marcel Dekker, Inc.; 1992. pp. 99–155. [Google Scholar]
- 16.Hübner A, Danganan C E, Xun L, Chakrabarty A M, Hendrickson W. Genes for 2,4,5-trichlorophenoxyacetic acid metabolism in Burkholderia cepacia AC100: characterization of the tftC and tftD genes and locations of the tft operon on multiple replicons. Appl Environ Microbiol. 1998;64:2086–2093. doi: 10.1128/aem.64.6.2086-2093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ingelman M, Ramaswamy S, Nivière V, Fontecave M, Eklund H. Crystal structure of NAD(P)H:flavin oxidoreductase from Escherichia coli. Biochemistry. 1999;38:7040–7049. doi: 10.1021/bi982849m. [DOI] [PubMed] [Google Scholar]
- 18.Inouye S. NAD(P)H-flavin oxidoreductase from the bioluminescent bacterium, Vibrio fischeri ATCC 7744, is a flavoprotein. FEBS Lett. 1994;347:163–168. doi: 10.1016/0014-5793(94)00528-1. [DOI] [PubMed] [Google Scholar]
- 19.Junker F, Kiewitz R, Cook A M. Characterization of the p-toluenesulfonate operon tsaMBCD and tsaR in Comamonas testosteroni T-2. J Bacteriol. 1997;179:919–927. doi: 10.1128/jb.179.3.919-927.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadiyala V, Spain J C. A two-component monooxygenase catalyzes both the hydroxylation of p-nitrophenol and the oxidative release of nitrite from 4-nitrocatechol in Bacillus sphaericus JS905. Appl Environ Microbiol. 1998;64:2479–2484. doi: 10.1128/aem.64.7.2479-2484.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kendrew S G, Harding S E, Hopwood D A, Marsh E N G. Identification of a flavin:NADH oxidoreductase involved in the biosynthesis of actinorhodin. Purification and characterization of the recombinant enzyme. J Biol Chem. 1995;270:17339–17343. doi: 10.1074/jbc.270.29.17339. [DOI] [PubMed] [Google Scholar]
- 22.Kertesz M A, Schmidt-Larbig K, Wüest T. A novel reduced flavin mononucleotide-dependent methanesulfonate sulfonatase encoded by the sulfur-regulated msu operon of Pseudomonas aeruginosa. J Bacteriol. 1999;181:1464–1473. doi: 10.1128/jb.181.5.1464-1473.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J-J P, Wu J. Structure of the medium-chain acyl-CoA dehydrogenase from pig liver mitochondria at 3-Å resolution. Proc Natl Acad Sci USA. 1988;85:6677–6681. doi: 10.1073/pnas.85.18.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knobel H-R, Egli T, Van der Meer J R. Cloning and characterization of the genes encoding nitrilotriacetate monooxygenase of Chelatobacter heintzii ATCC 29600. J Bacteriol. 1996;178:6123–6132. doi: 10.1128/jb.178.21.6123-6132.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kröger M, Wahl R. Compilation of DNA sequences of Escherichia coli K12: description of the interactive databases ECD and ECDC (update 1996) Nucleic Acids Res. 1997;25:39–42. doi: 10.1093/nar/25.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Lei B, Tu S-C. Mechanism of reduced flavin transfer from Vibrio harveyi NADPH-FMN oxidoreductase to luciferase. Biochemistry. 1998;37:14623–14629. doi: 10.1021/bi981841+. [DOI] [PubMed] [Google Scholar]
- 28.Park H J, Kreutzer R, Reiser C O, Sprinzl M. Molecular cloning and nucleotide sequence of the gene encoding a H2O2-forming NADH oxidase from the extreme thermophilic Thermus thermophilus HB8 and its expression in Escherichia coli. Eur J Biochem. 1992;205:875–879. doi: 10.1111/j.1432-1033.1992.tb16852.x. [DOI] [PubMed] [Google Scholar]
- 29.Parry R J, Wenying L. An NADPH:FAD oxidoreductase from the valanimycin producer Streptomyces viridifaciens. Arch Biochem Biophys. 1997;339:47–54. doi: 10.1006/abbi.1996.9857. [DOI] [PubMed] [Google Scholar]
- 30.Payne J W, Bolton H, Jr, Campbell J A, Xun L. Purification and characterization of EDTA monooxygenase from the EDTA-degrading bacterium BNC1. J Bacteriol. 1998;180:3823–3827. doi: 10.1128/jb.180.15.3823-3827.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prieto M A, Díaz E, García J L. Molecular characterization of the 4-hydroxyphenylacetate catabolic pathway of Escherichia coli W: engineering a mobile aromatic degradative cluster. J Bacteriol. 1996;176:111–120. doi: 10.1128/jb.178.1.111-120.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prieto M A, García J L. Molecular characterization of 4-hydroxyphenylacetate 3-hydroxylase of Escherichia coli. A two-protein component system. J Biol Chem. 1994;269:22823–22829. [PubMed] [Google Scholar]
- 33.Prieto M A, Pérez-Aranda A, García J L. Characterization of an Escherichia coli aromatic hydroxylase with a broad substrate range. J Bacteriol. 1993;175:2162–2167. doi: 10.1128/jb.175.7.2162-2167.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 35.Sánchez-Puelles J M, Sanz J M, García J L, García E. Immobilization and single-step purification of proteins using DEAE-cellulose. Eur J Biochem. 1992;203:153–159. doi: 10.1111/j.1432-1033.1992.tb19840.x. [DOI] [PubMed] [Google Scholar]
- 36.Takizawa N, Yokoyama H, Yanagihara K, Hatta T, Kiyohara H. A locus of Pseudomonas pickettii DTP0602, had, that encodes 2,4,6-trichlorophenol-4-dechlorinase with hydroxylase activity, and hydroxylation of various chlorophenols by the enzyme. J Ferment Bioeng. 1995;80:318–326. [Google Scholar]
- 37.Taylor D G, Trudgill P W. Camphor revisited: studies of 2,5-diketocamphane 1,2-monooxygenase from Pseudomonas putida ATCC 17453. J Bacteriol. 1986;165:489–497. doi: 10.1128/jb.165.2.489-497.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thibaut D, Tatet N, Bisch D, Faucher D, Debussche L, Blanche F. Purification of the two-enzyme system catalyzing the oxidation of the d-proline residue of pristinamycin IIB during the last step of pristinamycin IIA biosynthesis. J Bacteriol. 1995;177:5199–5205. doi: 10.1128/jb.177.18.5199-5205.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uetz T, Schneider R, Snozzi M, Egli T. Purification and characterization of a two-component monooxygenase that hydroxylates nitrilotriacetate from “Chelatobacter” strain ATCC29600. J Bacteriol. 1992;174:1179–1188. doi: 10.1128/jb.174.4.1179-1188.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Velasco A, Alonso S, García J L, Perera J, Díaz E. Genetic and functional analysis of the styrene catabolic cluster of Pseudomonas sp. strain Y2. J Bacteriol. 1998;180:1063–1071. doi: 10.1128/jb.180.5.1063-1071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wasserfallen A, Ragettli S, Jouanneau Y, Leisinger T. A family of proteins in the domains Archaea and Bacteria. Eur J Biochem. 1998;254:325–332. doi: 10.1046/j.1432-1327.1998.2540325.x. [DOI] [PubMed] [Google Scholar]
- 43.Wilbur W J, Lipman D J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci USA. 1983;80:726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Witschel M, Nagel S, Egli T. Identification and characterization of the two-enzyme system catalyzing oxidation of EDTA in the EDTA-degrading bacterial strain DSM 9103. J Bacteriol. 1997;179:6937–6943. doi: 10.1128/jb.179.22.6937-6943.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xi L, Squires C H, Monticello D J, Childs J D. A flavin reductase stimulates DszA and DszC proteins of Rhodococcus erythropolis IGTS8 in vitro. Biochem Biophys Res Commun. 1997;230:73–75. doi: 10.1006/bbrc.1996.5878. [DOI] [PubMed] [Google Scholar]