Abstract
Epigenetic processes enable environmental inputs such as diet, exercise, and health behaviors to reversibly tag DNA with chemical “marks” that increase or decrease the expression of an individual’s genetic template. Over time, epigenetic adaptations enable the effects of healthy or unhealthy stresses to become stably expressed in the tissue of an organism, with important consequences for health and disease. New research indicates that seemingly non-biological factors such as social stress, poverty, and childhood hardship initiate epigenetic adaptations in gene pathways that govern inflammation and immunity, two of the greatest contributors to chronic diseases such as diabetes and obesity. Epigenetic processes therefore provide a biological bridge between the genome—an individual’s genetic inheritance—and the Social Determinants of Health—the conditions in which they are born, grow, live, work, and age. This Perspective paper argues that physical therapy clinicians, researchers, and educators can use the theoretical framework provided by the International Classification of Functioning, Disability, and Health (ICF model) to harmonize new discoveries from both public health research and medically focused genomic research. The ICF model likewise captures the essential role played by physical activity and exercise, which initiate powerful and widespread epigenetic adaptations that promote health and functioning. In this proposed framework, epigenetic processes transduce the effects of the social determinants of health and behaviors such as exercise into stable biological adaptations that affect an individual’s daily activities and their participation in social roles. By harmonizing “nature” and “nurture,” physical therapists can approach patient care with a more integrated perspective, capitalizing on novel discoveries in precision medicine, rehabilitation science, and in population-level research. As the experts in physical activity and exercise, physical therapists are ideally positioned to drive progress in the new era of patient-centered population health care.
Keywords: DNA, Genomics, Precision Medicine, Precision Rehabilitation, RNA, Social Determinants of Health
Introduction
From its earliest days, physical therapy has endorsed a personalized, patient-centered approach. Astute clinicians have always understood that the patient’s emotional state, family dynamics, work circumstances, genetic predisposition, and daily sources of stress can either amplify or impede the efficacy of our interventions. Because our fundamental tool is movement (a behavioral intervention), we have always needed to attend to psychosocial factors that affect behavior and modulate pain. The movement strategies we prescribe require engagement of the patient as a whole person in all their complexity, creating an imperative for patient-centered care.
Public health research supports that our profession’s careful attention to the conditions in which people are born, grow, live, work, and age—the social determinants of health1—likely enhances our efficacy as clinicians. County-level studies indicate that socioeconomic factors and behaviors account for 30% to 50% of the variation observed in health outcomes compared with just 16% attributable to medical care.2 Other studies using this same population-level approach show strong associations between a community’s level of physical activity and the health outcomes they collectively experience.3 Although it is challenging to pinpoint the consequences of social determinants of health at the level of the individual, they are unquestionably part of the complete picture of physical therapy care.4 Moreover, the social determinants of health play a central role in the International Classification of Functioning, Disability, and Health (ICF) model,5 a framework for understanding health and disability used by health care researchers and practitioners the world over, including physical therapists (Fig. 1).
Figure 1.
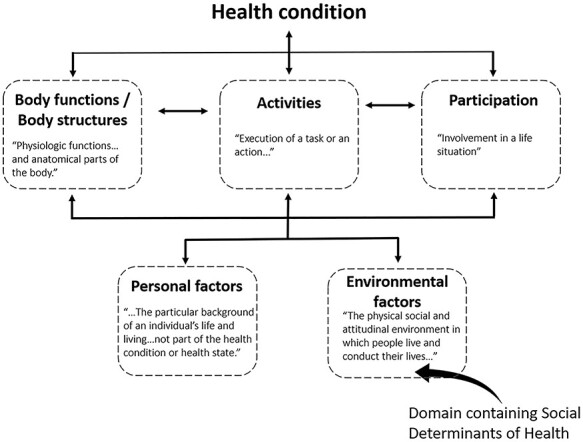
The ICF model. New discoveries in biology are helping to explain the ways that individual domains influence each other, culminating in function or disability.
Since the mid-20th century, medicine has rightfully interpreted that a person’s genetic makeup is fundamental to determining their individual health trajectory. The ICF model, on the other hand, does not explicitly conceptualize genetic inheritance as a contributor to function and disability. With the advent of “big data” informatics capabilities, the health care system has entered the precision health care era, in which providers may soon select individualized treatments (drugs) that are optimized for each patient’s molecular genetic signatures, social determinants of health, and disease state. Despite the potential for precision-based health care to impact population health, the pace of translation has been slow.6 Perspectives advocating for a “precision” approach have only recently appeared in the physical therapy literature.7–10
In addition to this lag in translation, the term “precision medicine” is occasionally confused with a related term, “personalized medicine.” Personalized medicine suggests that the keys to health and disease can be revealed by fine-scale analysis of individual genetic information without explicit attention to the social determinants of health.11 This runs contrary to public health research, which points to the relevance of complex environmental interactions that operate far above the level of the individual: across communities and even across cultures, with modifying influences from health care delivery systems and cost. A healthy tension currently exists between these 2 valid but seemingly dichotomous health care worldviews.12 This tension poses an important question: can a contemporary understanding of biology help explain the ways various ICF factors—including the social determinants of health and behaviors such as exercise—influence each other and thereby affect health? If environmental factors such as the social determinants of health can modify gene expression, then genomic and population health approaches represent a continuum, not a dichotomy.
The bridge between genetics and the social determinants of health is epigenetics, a collection of cellular processes that enable an organism’s environment to regulate the expression of its genetic template. Through the mediation of epigenetic processes, the genome, lifestyle, and the social determinants of health form a matrix whose interactions and mutual dependencies create a powerful etiologic framework for all health care practitioners, including physical therapists. The most important feature of epigenetic processes is that they are sensitive to the organism’s environment: the “nurture” half of the nature/nurture dyad. The term “exposome” is sometimes used to describe an organism’s environmental inputs, encompassing “…life-course environmental exposures (including lifestyle factors), from the prenatal period onwards.”13 The exposome includes environmental factors such as pollution and chemical exposure, physical activity and exercise, health behaviors such as smoking, nutrition, the social determinants of health (eg, poverty, social stress, poor health care access, discrimination), as well as factors within the body such as the gut microbiome (Fig. 2).14 The epigenome mediates the effects of all these factors on the genome, serving as an integrator by which the exposome regulates the output of the genome. Importantly, the epigenome acts in a cumulative fashion, amplifying and solidifying long-lasting exposome influences into stable phenotypic traits. In the context of rehabilitation care, movement interventions prescribed by a physical therapist can potentially yield epigenetic adaptations if they become long-term lifestyle behaviors.
Figure 2.
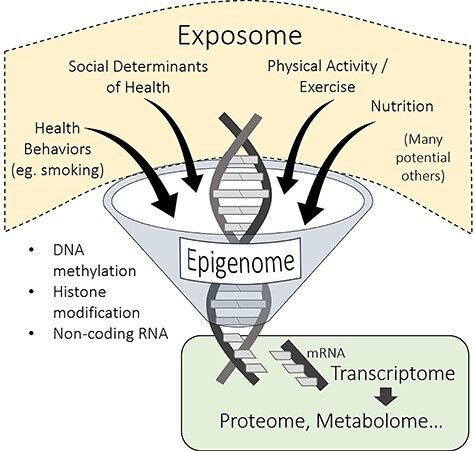
The 3 known epigenetic processes (lower left) modify the output of the genome (the transcriptome) in response to a wide range of inputs from the exposome. This has ramifications for the proteome, metabolome, and so on.
The unique power of epigenetic processes is that environmental stressors reversibly tag DNA with chemical “marks” that increase or decrease the expression of an individual’s genes. This does not underplay the importance of an acquired genetic pre-disposition. Epigenetic marks can be passed down with each successive mitotic cell duplication, enabling the effects of healthy or unhealthy stresses to become stably expressed in the tissue of an organism. But because they are reversible, epigenetic processes also create the potential for healthy stressors to mitigate the effects of earlier adverse events. As we will describe below, movement, physical activity, and exercise, fundamental tools used by physical therapists, are powerful epigenetic stressors that promote a healthful gene expression profile in multiple body systems. We predict that the progression of epigenetic research in the coming decades will reveal the specific biologic mechanisms that underlie the health-promoting effects of many activity-based interventions, including community-focused interventions that use behavioral approaches to improve the social determinants of health. We also believe that by providing a common biological currency between genomic and public health research, epigenetic research may synergize discovery in both fields.
For a physical therapist treating complex acquired non-communicable diseases such as obesity, diabetes, dementia, and heart disease, understanding the influence of lifestyle and environmental factors on health outcomes is essential to patient-centered care. Likewise, these same lifestyle and environmental factors will affect the progression of diseases with known genetic etiologies, such as muscular dystrophy and spina bifida. In both these scenarios, the therapeutic advantages to be gained through consideration of environmental factors are accompanied by the potential ethical peril of “detect[ing] adverse exposures and conditions that require resources beyond the scope of clinical care.”15 In addition, physical therapists who strive to use epigenetic mechanism-based reasoning16 to select interventions face a steep challenge to keep pace with the rapidly changing frontiers of “-omics,” including genomics, epigenomics, proteomics, and metabolomics.
The purpose of this Perspective paper is to describe a harmonizing framework by which physical therapy clinical care can incorporate new discoveries from both public health and genomic research, building on the foundation laid by the ICF model. Key to this discussion will be new scientific evidence that the social determinants of health, experienced by an individual as healthy or unhealthy stress, epigenetically modify the expression of an individual’s genome in ways that mediate health and disease states. Because physical therapists are equipped to intervene at any level of the ICF model—biological response, lifestyle behaviors, health systems, and society—they are uniquely suited to advance the frontiers of precision health care.
A Brief History of Human Genomic Research
Although genetic contributions to disease were long known from heredity studies, the first disease to be mapped to a human chromosome was Huntington Disease in 1983.17 It took a consortium of 58 research groups an additional 10 years to map the specific locus of the genetic defect in the huntingtin gene.18 By the late 1980s, it had become clear that more rapid progress could be achieved if a complete map of the entire human genome were available. On October 1, 1990, the National Institutes of Health and international partners launched the Human Genome Project, biology’s first large-scale, multi-national, collaborative research endeavor.19 The project’s goal was to sequence all 3 billion human base pairs within 15 years. “Working draft” maps of chromosome segments, combined with rapid technological advances, dramatically reduced the time and personnel required to find new disease genes. By 2001, a single laboratory using a “working draft” for chromosome 7 was able to identify the location of a tumor suppressor gene within a span of several weeks.20 Comparable work to isolate huntingtin, as mentioned above, took a decade of work by a multi-institution consortium.21
On April 14, 2003, the National Human Genome Research Institute (NHGRI) announced the completion of the human genome map a full 2 years ahead of schedule. Multi-national collaborative groups began to uncover the complex poly-genic basis of lifestyle-based diseases such as diabetes and cardiovascular disease.22 As it became apparent that medicine could someday incorporate genomic data into diagnosis and treatment of disease—the dawn of personalized medicine—NHGRI began to heavily invest in the development of technologies to reduce the cost of gene sequencing and to build the informatics capacity to manage large genomic datasets. In 2008, an international research consortium launched the 1000 Genomes Project, an ambitious effort to completely map the genomes of 1000 people worldwide. By 2015, technological progress had made possible what was once unthinkable: full genomic mapping of 1,000,000 individuals. In that year, NIH launched the “All of Us” research program, a precision medicine initiative that aimed first to enhance knowledge of the genetic basis of cancer and then to more widely study other diseases.23 A major objective of this ongoing project is to “reduce health care costs by matching the right person with the right treatment the first time.”24 By genotyping 1 million Americans and obtaining detailed information about their lifestyle, environment, and health history, this initiative is building the foundation for the era of precision health care.
The All of Us program and other large-scale genomic studies facilitate a population-scale approach that mines huge datasets for statistical correlations among diseases and discrete combinations of genes. This approach is called a Genome-Wide Association Study (GWAS). In complex disorders such as cardiovascular disease, “before the advent of GWAS, genetic analysis focused on individual variants in candidate genes and generated often poorly reproducible results. In contrast, GWAS can explore the genetic variability of the entire genome for association with any given phenotype.”25 For example, GWAS has identified nearly 200 gene loci with significant associations to coronary artery disease.25
As almost always occurs with scientific progress, new discoveries add new complexity to the next generation of research questions. It became clear early in the history of genomic research that “genes alone did not always determine destiny.” Evidence accumulated for individuals who beat the genetic odds, such as patients with a genetic predisposition for cardiovascular disease who never developed the disease phenotype. Or that the lifespan of a person with an acquired genetic disease, such as muscular dystrophy, was prolonged in some and not others. It soon became clear that the next frontier of discovery in health care would be the biological factors that control the expression of the pre-determined genetic template within each human being.
Epigenetics: Where DNA Meets the Environment
The Human Genome Project revealed the puzzling fact that only a tiny fraction (approximately 2%) of the human genome encodes proteins. These DNA regions are transcribed to messenger RNA (mRNA), which is then translated into the proteins that build and regulate every cell of every tissue (Fig. 3A). We now know that our DNA contains vast regions that do not code for proteins: 70,000 promoter regions that initiate the transcription of genes; 400,000 enhancer regions for non-contiguous genes; and copious nucleotide sequences that code for RNA molecules but not for proteins.26 Once considered “junk” DNA, these regions became prime candidates for sites that regulate how genes are activated or deactivated. The discovery that a damaged or healthy gene can either be epigenetically suppressed or promoted opens the door for new strategies to modulate our “predetermined” genetic destiny.
Figure 3.
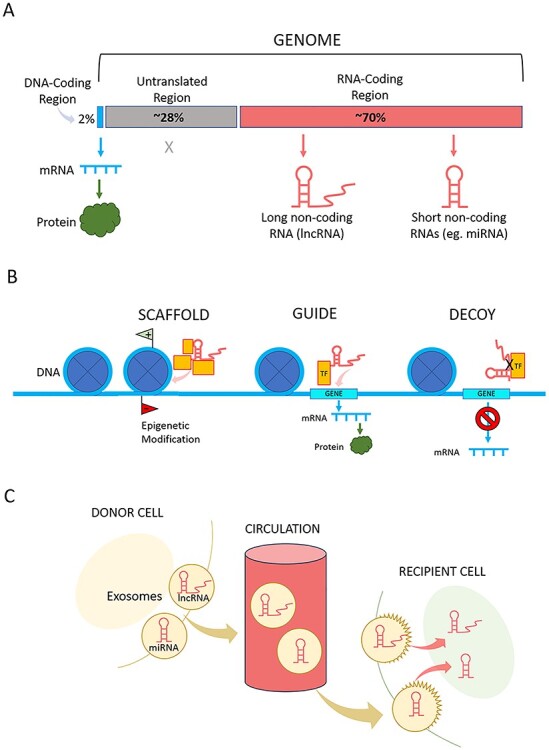
(A) The majority of the genome is untranslated (approximately 28%: promotor and enhancer regions) or encodes non-coding RNA (approximately 70%). Only approximately 2% encodes information for producing proteins. (B) A few of the newly discovered functions of non-coding RNA are “SCAFFOLD”: long non-coding RNA (lncRNA) serves as a scaffold on which 2 or more factors connect and carry out a shared function, such as epigenetic modification; “GUIDE”: lncRNA guides other elements to DNA to carry out a function such as gene transcription; and “DECOY”: lncRNA acts as a decoy to block molecular interactions, such as between transcription factors (TF) and their target genes. (C) Micro-RNAs (miRNA) and lncRNA are packaged into extracellular vesicles (exosomes) and released into the circulation where they travel to target cells in distant tissues. There, they exert regulatory effects on the genome and epigenome.
A long-term NHGRI initiative called ENCODE (the Encyclopedia of DNA Elements)27 has laid the groundwork for assigning molecular functions to non-protein coding DNA segments, the plentiful epigenetic regulators of the protein-coding genome. The project identified over 18,400 genes that are transcribed into non-coding RNA molecules (ncRNAs).28 Instead of moving to ribosomes to undergo translation into proteins, as mRNA does, ncRNAs engage in an ever-growing list of functions (Fig. 3B).29,30 They may serve as scaffolds on which other molecules transiently join in order to carry out shared functions, or they may guide other molecules to DNA sites that initiate gene transcription or place epigenetic marks. Some types of ncRNAs serve as decoys for transcription factors, preventing them from interacting with genes to initiate mRNA transcription. Importantly, some types of ncRNA can be released to systemic circulation via extracellular vesicles, enabling them to travel to distant target organs to carry out wide-ranging regulatory activities (Fig. 3C).
The ENCODE project mapped DNA regions that can be “tagged” (marked) by methyl chemical groups, which generally indicate that the associated genes have been silenced (not transcribed to mRNA). ENCODE also cataloged patterns of chemical modifications to histone proteins, the structural molecules that help package DNA into chromosomes. These histone modification sections signified DNA regions that had either been silenced or amplified, depending on the gene. These 3 features of DNA that are not themselves genes—ncRNA, methylation sites, and histone modifications—are the key factors that appear to regulate and coordinate DNA transcription/translation to proteins. Because these elements lie outside the genome, they have been termed the “epigenome.” The discovery of these controlling elements, the epigenome, in no way diminishes the importance of the human genome. The genome and epigenome are intricately entwined by bringing to the forefront the biological importance of “nature” and “nurture.”
Epigenetics initially gained public attention31 after rodent studies revealed surprising evidence that noxious stress, including social stress, experienced by both the dam and the sire can yield transgenerational epigenetic differences in offspring.32,33 This does not constitute a mutation, which is an alteration in the DNA base-pair sequence. Rather, these studies raised the thought-provoking possibility that environmental stressors, including what seemingly was construed as non-biological factors such as social stress, can yield biochemical adaptations that reverberate across generations. As we learn more about the non-coding regions of the human genome, we predict that the capacity to measure transgenerational epigenetic regulation will be enhanced. For example, recently a child and his mother showed maternally inherited epigenetic silencing of a damaged gene associated with Duchenne Muscular Dystrophy,34 opening the door for new therapeutics. Moreover, newborns of parents with obesity, considered an environmental stress, showed different methylation signatures than infants born to parents with normal weight.35 Transgenerational epigenetic research in humans is just beginning but may someday create powerful mechanistic connections between population-level public health research and precision health care.
For physical therapists practicing today, the most important epigenetic adaptations are those that yield gene regulation changes within the lifespan of an individual. Both healthful and noxious stresses experienced by an individual trigger epigenetic changes in their tissues, which, through mitotic cell division, are perpetuated in future generations of that individual’s cells, tissues, and organs. If these epigenetic changes are maintained long term, the phenotype of their tissues may skew toward healthful or diseased states. Just as importantly, the potential reversibility of some epigenetic adaptations opens the door for health-promoting interventions that can remove epigenetic marks applied by exposure to prior noxious stressors. An intense search is underway for epigenetically modifiable DNA loci that are responsible for the long-term adaptations to exercise and healthy lifestyle behaviors.
Epigenetics as a Transducer of Social Stress
Figure 4 illustrates the wide variety of environmental factors that comprise the social determinants of health.36 In the past decade, evidence has begun to accumulate that these factors, experienced by humans as positive or negative stress, epigenetically alter gene transcription pathways in ways that yield stable tissue and systems-level adaptations with direct consequences for health and disease.
Figure 4.
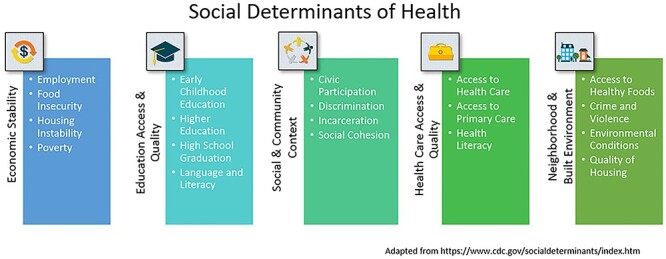
Centers for Disease Control categories and descriptors of the social determinants of health. Adapted from https://www.cdc.gov/socialdeterminants/index.htm.
In a large Italian cohort, childhood socio-economic status (SES) correlated with differential epigenetic tagging (methylation) of several genes involved in inflammation.37 Systemic inflammation plays a key role in many non-communicable diseases of adulthood, particularly cardiovascular disease, metabolic syndrome/diabetes, and osteoarthritis. Childhood epigenetic modification in response to social stress, if it did indeed promote inflammation in adulthood, could be a “smoking gun” helping to explain the differential rates of non-communicable diseases observed across the SES continuum. However, few studies have linked SES-related epigenetic methylation differences to clinical measures of inflammation, a key requirement to demonstrate any potential functional significance of epigenetic adaptations.
To date, the study that most convincingly addressed this question accessed detailed childhood SES data from a public health study in the Philippines.38 Researchers obtained new blood samples from the study participants, now all young adults, and determined that childhood environmental stressors (nutritional, microbial, psychosocial) yielded differential methylation of 10 DNA sites across 9 genes that regulate inflammation. To confirm that these epigenetic differences carried functional consequences, the researchers assayed several biomarkers of inflammation, determining that 4 of the methylation sites did indeed correlate with adulthood levels of inflammation. These results provide new evidence that the inflammation cascade may be a key system in humans that is epigenetically regulated by the social determinants of health. They are consistent with a growing body of work39–41 suggesting that infancy and childhood are a sensitive period during which social and environmental stressors can be transduced by epigenetic processes into persistent biological adaptations, often described as “biological embedding.”42,43 Early biological embedding of social stress also appears to underlie differential methylation of genes that regulate immune function in adults, with potential consequences for pathogen resistance and exacerbation of systemic inflammation.40,44 This insight is vital as we negotiate the challenges of the COVID-19 pandemic, which has demonstrated high mortality in adults with immune compromise45,46 and in groups commonly exposed to social stress.47
Additional “adaptive calibration of the DNA methylome”40 after early childhood social stress occurs within the hypothalamic–pituitary–adrenal (HPA) axis. The HPA axis governs the production of glucocorticoid (GC) stress hormones, which play key signaling roles in the physiologic processes that regulate both inflammation and immune function. Malfunction of the HPA axis is implicated in major depressive disorder, chronic pain syndromes, cardiovascular disease, metabolic syndrome, and many other non-communicable diseases.48–50 Primate studies using rhesus macaques, which have highly stratified social groups, showed that low-status individuals experienced epigenetic changes that favored transcription of inflammatory genes.51 High-status individuals showed different epigenetic changes that favored transcription of anti-inflammatory genes. Epigenetic differences between social classes diminished in animals receiving GC drugs, supporting that the effect was mediated by the HPA axis. Numerous clinical physiology studies have reported associations between social stress and elevated GC levels and/or GC resistance.52,53 Epigenetic response to social stress in humans is complex and appears to be dependent on genotype and to be subject to stochastic (random) effects.54 However, epigenetic regulation of genes governing the HPA axis could play a role in transducing social stress, as demonstrated in the primate study above,51 perhaps playing a role in diseases that are differentially borne across sociodemographic groups, such as the especially high rates of chronic pain55 and COVID-19 mortality47 among African Americans. Precision medicine of the future will more than likely focus on epigenetic adaptations to social stress in addition to the inter-individual differences in inherited genes.56 Epigenetics may empower us to discover the key relationships among the patient’s genome, environment, prenatal exposure, and disease risk in time to prevent disease development or secondary chronic co-morbidities.56
Movement, Physical Activity, and Exercise as Epigenetic Regulators
A key question facing precision health care is whether epigenetic adaptations that are biologically embedded in childhood can be reversed by health-promoting behaviors later in life. Fortunately, a wealth of new studies suggest that movement, physical activity, and exercise are powerful “epigenetic interventions” that can circumvent or prevent the development of chronic disease. For physical therapists, the coming decades will reveal many new ways that activity-based therapies can improve health and longevity by triggering epigenetic adaptations.
At present, we have a very limited understanding of the specific epigenetic adaptations that occur in response to unhealthy environmental stressors such as sedentary lifestyle, overfeeding, and social stress. It is therefore difficult to determine whether the salutary effects of movement, physical activity, and exercise remove these negative adaptations or whether they initiate a different set of healthful epigenetic responses. “Epigenome-wide association studies” are now underway to identify specific DNA segments whose epigenetic status differs in, for example, sedentary individuals versus those who habitually engage in physical activity.57 One such study examined 850,000 potential DNA methylation loci in human skeletal muscle, showing hypomethylation of >9000 sites after resistance training.58 (Removal of methyl groups would remove a “block” to DNA transcription and would therefore facilitate expression of these DNA sequences.) After engaging in prescribed resistance training that resulted in hypertrophy, participants ceased training and muscle mass returned to baseline levels. But on resumption of resistance training, 4 genes demonstrated even stronger hypomethylation and consequent gene up-regulation than had previously occurred during the initial training period. This provides the first compelling evidence for an epigenetic “memory” for training in human skeletal muscle. Although more investigation is needed, known functions of these 4 genes include skeletal muscle cell differentiation and repair, processes that would be essential in any adaptive response to exercise. Studies such as this have begun to reveal the fundamental biological processes that activity and exercise harness, underscoring the summative power of repeated, long-term exposure to activity-based stresses. Future discoveries could have prognostic value for physical therapy care. For instance, understanding a patient’s methylation status at key DNA loci that direct activity-based adaptations59 could help clinicians identify individuals who may be late responders or non-responders to exercise-based interventions.7 Armed with this knowledge, clinicians may craft precision rehabilitation plans that sidestep less fruitful interventions and prioritize alternatives with a greater potential to elicit desired adaptations.
Beyond skeletal muscle, new research is also revealing the broader ways that physical activity and exercise training elicit epigenetic adaptations in key body systems. Additional details about the various exercise protocols (mode, dose, intensity) can be found in several recent reviews.8,60–64 Briefly, evidence is amassing that the “gene–environment networks”61 that control glucose and energy homeostasis—and play a role in metabolic disease and type-2 diabetes—are epigenetically regulated by exercise. In skeletal muscle, exercise appears to epigenetically enhance the ability of insulin to “open the door” for glucose to enter the working cell. When activated by insulin, the insulin receptor at the muscle cell’s surface interacts with a chain of many other intra-cellular proteins (kinases) that create a channel through which glucose can be transported down its concentration gradient. Clinical studies are underway to determine what role epigenetic adaptations in insulin-mediated glucose transport play in the well-substantiated ability of physical activity and exercise to regulate glucose and insulin and prevent metabolic disease.
Exercise also appears to trigger other types of epigenetic changes in the systems that prevent metabolic disease. Exercise triggers widespread epigenetic changes (hypomethylation) across the skeletal muscle genome, notably in the gene PPARGC1A, which encodes a protein called PGC1α.65 PGC1α is a master transcription factor that coordinates metabolic adaptations to exercise by initiating the transcription of a host of downstream pathways, including some in the brain.66 Its widespread effects on genes in multiple physiologic systems makes PGC1α a critical gene for transducing physical exercise into organism-wide metabolic and cognitive health adaptations.66,67 As would be expected, PGC1α is hyper-methylated in type-2 diabetes, signifying that its transcription is repressed and its downstream effects are blunted in these individuals.65 We recently reported that even in people with spinal cord injury who cannot engage in lower extremity voluntary exercise, demethylation of PGC1α can be achieved by chronic doses of electrically induced muscle exercise.68 We are working to determine whether these epigenetic changes are accompanied by transcriptional adaptations in downstream pathways that regulate glucose and insulin homeostasis, and whether these adaptations yield improvements in clinically important biomarkers of metabolic health.
Exercise Dose and Epigenetic Adaptations
A key theme that we predict will emerge in future studies is that epigenetic change, while initiated rapidly by a single exercise bout, requires time and repeated exposures to “develop a memory.” Especially in human research, a major unknown variable is how long a behavioral intervention must be maintained before epigenetic adaptation becomes stable. For example, epigenetic changes induced by the brief negative environmental stress of a 5-day high-fat diet did not resolve even after 6 weeks of resumption of a healthy diet.69 This potential lag in reversing epigenetic adaptations increases the difficulty of measuring the time course for consolidation of epigenetic benefits of new activity or behavior-based interventions. We recently demonstrated that very long durations of electrically induced exercise were required to maximally promote genes in paralyzed muscle that had been repressed after long periods of chronic disuse.68 Although all signs indicate that movement, physical activity, and exercise are powerful epigenetic environmental regulators, we predict that future studies will unequivocally show that exercise must be recommended to clients as a lifestyle-based intervention, not as a 6- to 12-week quick fix. Although such short intervention durations are typical in exercise studies and clinical episodes of care, researchers and clinicians should have realistic expectations for the likely pace of epigenetic change.
ICF Model and Epigenetics
A central theme of this perspective is that epigenetics is a multimodal, temporal integrator of exposome stimuli that offers a biological bridge between nature and nurture, between an organism’s outer and inner worlds. From a broader health care perspective, epigenetic processes may also provide a mechanistic link among the many interrelated domains of the ICF. This theoretical framework and classification tool provides clinicians and researchers with a standard language to describe disability and function.5 Rather than the traditional “medical model,” which considers disability to be a linear consequence of biological disease processes, the ICF model facilitates a more coherent view of function and disability that places social, behavioral, environmental, and personal factors on an equal footing with biological change. In the ICF framework, the social determinants of health typically exert their effects on function via the domain “environmental factors” (Fig. 5). Although biological factors are typically presumed to exert their effects through the domain “body functions/structures,” the ICF classification system currently does not include coding designations for genetic or epigenetic signatures. We propose that inherited as well as acquired epigenetic signatures can be conceptualized as mediators of the interactions between several ICF domains. Figure 5 illustrates potential ways that epigenetic processes may mechanistically link environmental and behavioral ICF domains to affect an individual’s biological systems and overall expression of function or disability. This theoretical framework may be useful to academic physical therapy programs that emphasize the ICF model in their curricula but that are looking for ways to enhance content relating to medical genetics. In the future, precision rehabilitation will certainly respect the importance of genetic inheritance (a potential “personal factors” trait; Fig. 5) but need to understand the environmentally and behaviorally relevant epigenetic processes that affect body tissues.
Figure 5.
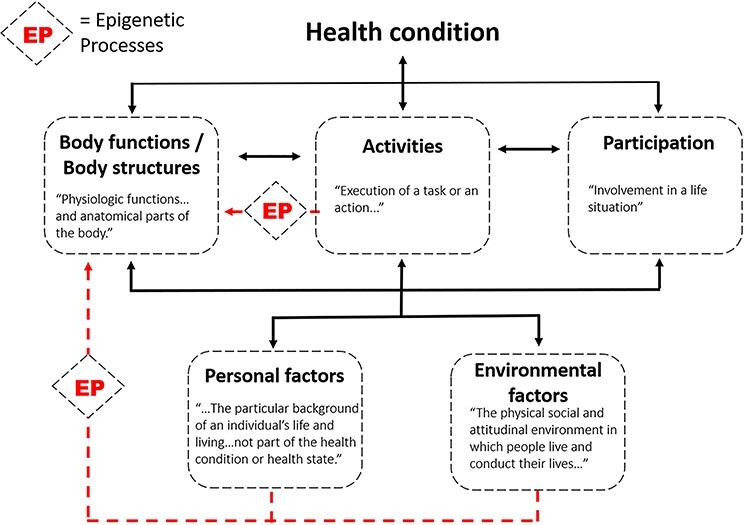
Epigenetic processes (EP) may biologically mediate the interrelationships among several International Classification of Functioning, Disability, and Health (ICF) domains. Activities (eg, exercise), environmental factors (eg, social determinants of health) and personal factors (eg, smoking, alcohol abuse) may yield epigenetic adaptations of tissues that culminate in positive or negative outcomes in body functions or structures.
Figure 6 depicts an example of how the ICF model can capture the ways that the social determinants of health (environmental factors), health-related personal factors, and regular exercise (activities) can all affect the experience of disability and functioning for a patient with type 2 diabetes. Epigenetic processes may transduce the effects of pervasive environmental and behavioral stressors and initiate biological adaptations such as impaired glucose metabolism (body functions and structures domain). These biological adaptations may exacerbate the underlying health condition and undermine an individual’s ability to carry out daily activities and participate in social roles.
Figure 6.
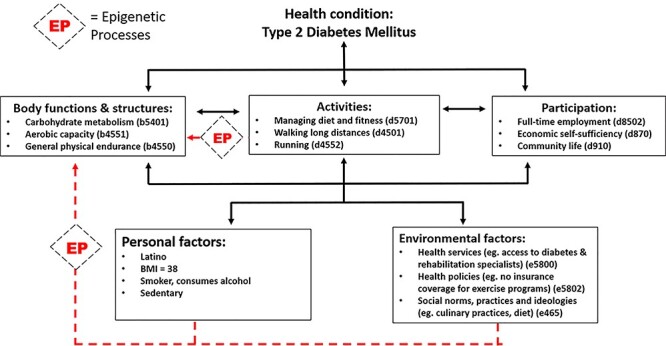
Example of International Classification of Functioning, Disability, and Health (ICF) qualifiers that can be used to characterize disability and functioning for a patient with Type 2 diabetes. Epigenetic processes (EP) likely transduce the effects of environmental, personal, and behavioral (activity) factors into tissue-level adaptations that culminate in changes in body structures and function. These changes may, in turn, exacerbate the underlying health condition and trigger further limitations in activities and participation.
Epigenetics and Patient-Centered Physical Therapy
In this Perspective, we link the burgeoning fields of genomics and epigenomics with the social determinants of health and exercise-based behaviors within the ICF model. Short term, we envision that embedding genetics and epigenetics within the ICF theoretical framework will promote a broader academic understanding of the complex interactions among personal predisposing factors, social/environmental factors, and biologic adaptation as it relates to physical therapist exercise-based interventions.
Long term, we envision that genomic/epigenomic, personal, and environmental factors will be predictive of responders or non-responders to our exercise interventions, assisting us in developing sound guidelines and more precise preventative treatments. We also envision that artificial intelligence algorithms will someday play a pivotal role in developing more precise practice guidelines by using personalized data about lifestyle as well as genetic/epigenetic molecular signatures, similar to drug selection, interaction, and delivery programs in medicine today.70
Several recent studies linking epigenetics to activity, behavior, and the environment can help to illustrate where future expansion may occur in physical therapy theory and practice. Four examples of recent papers and the implication to physical therapist practice are offered.
First, excessive alcohol consumption triggers epigenetic effects that are directly opposite to those observed after exercise.71 In light of this finding, do physical therapists routinely query clients about alcohol consumption and consider this social behavior when prescribing a dose of exercise?
Second, epigenetic regulation of 3 metabolic genes is affected by sleep duration and quality in a sex-dependent manner.72 Do physical therapists routinely consider sleep habits of a client before prescribing an exercise program, and do they consider how sleep may influence the probability of inducing a true behavioral change?
Third, in muscle cell culture, transcription of certain genes relating to metabolism and development are fine-tuned by epigenetic mechanisms that respond to an individual’s circadian rythmn.73 Do physical therapists optimize the time of day for a client to exercise, according to their own biological clocks, to promote adaptation, compliance, and ultimately a change in behavior?
Lastly, skeletal muscle has been shown to be a key epigenetic predictor of a person’s overall “true” biological age.74 Do physical therapists consider the biological age versus the chronological age of a client and adjust the prescribed exercise dose accordingly?
Although many physical therapists intuitively consider these factors as part of their routine clinical reasoning process, the underlying biological framework supporting their approach continues to evolve. Importantly, patient-centered care requires that we more comprehensively understand the social, environmental, and biological context for each client so that we can prevent disease and improve the probability that our treatment will be successful.
Conclusions
For physical therapy clinicians today, epigenetic discoveries about social determinants of health and lifestyle (exercise and physical activity) may seem far removed from immediate decisions concerning intervention selection and dose prescription. But more actionable discoveries are approaching quickly, and it is time for all health care professions to prepare to capitalize on this new science. We do not need to reinvent the wheel. The ICF model gains mechanistic power by expanding to include epigenetic processes, and it already emphasizes the social determinants of health, a factor that deserves greater formal integration into physical therapy care. Academic programs and clinicians can leverage the power of the ICF model to prepare the next generation of therapists to integrate genomic and epigenomic insights, attention to social factors, and an expert understanding of activity into precision rehabilitation care that yields enhanced patient outcomes and reduced costs. Precision medicine was once conceptually limited to genomics but now strives to incorporate environmental and behavioral factors,75 as shown by the All of Us research program.24 As the experts in physical activity and exercise, physical therapists must engage to drive progress in precision health care.
Contributor Information
Richard K Shields, Department of Physical Therapy and Rehabilitation Science, Roy J. and Lucille A. Carver College of Medicine, University of Iowa, Iowa City, Iowa, USA.
Shauna Dudley-Javoroski, Department of Physical Therapy and Rehabilitation Science, Roy J. and Lucille A. Carver College of Medicine, University of Iowa, Iowa City, Iowa, USA.
Author Contributions
Concept/idea/research design: R.K. Shields
Writing: R.K. Shields, S. Dudley-Javoroski
Project management: R.K. Shields
Fund procurement: R.K. Shields
Providing facilities/equipment: R.K. Shields
Clerical/secretarial support: R.K. Shields
Consultation (including review of manuscript before submitting): R.K. Shields
Funding
Research findings presented in this study were funded by National Center for Medical Rehabilitation Research grants R01HD-084645 and RO1HD-082109 to R.K. Shields. The funders played no role in the writing of this Perspective.
Disclosures
The authors completed the ICMJE Form for Disclosure of Potential Conflicts of Interest and reported no conflicts of interest.
References
- 1. World Health Organization . Social determinants of health. 2020. Accessed July 28, 2020. https://www.who.int/social_determinants/sdh_definition/en/.
- 2. Hood CM, Gennuso KP, Swain GR, Catlin BB. County health rankings: relationships between determinant factors and health outcomes. Am J Prev Med. 2016;50:129–135. [DOI] [PubMed] [Google Scholar]
- 3. Sato M, Du J, Inoue Y. Rate of physical activity and community health: evidence from U.S. counties. J Phys Act Health. 2016;13:640–648. [DOI] [PubMed] [Google Scholar]
- 4. Rethorn ZD, Cook C, Reneker JC. Social determinants of health: if you aren't measuring them, you aren't seeing the big picture. J Orthop Sports Phys Ther. 2019;49:872–874. [DOI] [PubMed] [Google Scholar]
- 5. World Health Organization . International Classification of Functioning, Disability and Health (ICF). 2020. Accessed July 28, 2020. https://www.who.int/classifications/icf/en/.
- 6. Allen CG, Peterson S, Khoury MJ, Brody LC, McBride CM. A scoping review of social and behavioral science research to translate genomic discoveries into population health impact. Transl Behav Med. 2021;11:901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shields RK. 48th Mary McMillan lecture: turning over the hourglass. Phys Ther. 2017;97:949–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Woelfel JR, Dudley-Javoroski S, Shields RK. Precision physical therapy: exercise, the epigenome, and the heritability of environmentally modified traits. Phys Ther. 2018;98:946–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chimenti RL, Frey-Law LA, Sluka KA. A mechanism-based approach to physical therapist management of pain. Phys Ther. 2018;98:302–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Magnusson DM, Eisenhart M, Gorman I, Kennedy VK, Davenport TE. Adopting population health frameworks in physical therapist practice, research, and education: the urgency of now. Phys Ther. 2019;99:1039–1047. [DOI] [PubMed] [Google Scholar]
- 11. Ferryman K, Pitcan M. What is Precision Medicine. Washington, DC, USA: Data and Society Research Institute; 2018. [Google Scholar]
- 12. Horesh Bergquist S, Lobelo F. The limits and potential future applications of personalized medicine to prevent complex chronic disease. Public Health Rep. 2018;133:519–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wild CP. Complementing the genome with an "exposome": the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev. 2005;14:1847–1850. [DOI] [PubMed] [Google Scholar]
- 14. Miller GW, Jones DP. The nature of nurture: refining the definition of the exposome. Toxicol Sci. 2014;137:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garg A, Boynton-Jarrett R, Dworkin PH. Avoiding the unintended consequences of screening for social determinants of health. JAMA. 2016;316:813–814. [DOI] [PubMed] [Google Scholar]
- 16. Polli A, Nijs J, Ickmans K, Velkeniers B, Godderis L. Linking lifestyle lactors to complex pain states: 3 reasons why understanding epigenetics may improve the delivery of patient-centered care. J Orthop Sports Phys Ther. 2019;49:683–687. [DOI] [PubMed] [Google Scholar]
- 17. Gusella JF, Wexler NS, Conneally PM, et al. A polymorphic DNA marker genetically linked to Huntington's disease. Nature. 1983;306:234–238. [DOI] [PubMed] [Google Scholar]
- 18. MacDonald ME, Ambrose CM, Duyao MP, et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993; 971–983. [DOI] [PubMed]
- 19. Green ED, Watson JD, Collins FS. Human genome project: twenty-five years of big biology. Nature. 2015;526:29–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zenklusen JC, Conti CJ, Green ED. Mutational and functional analyses reveal that ST7 is a highly conserved tumor-suppressor gene on human chromosome 7q31. Nat Genet. 2001;27:392–398. [DOI] [PubMed] [Google Scholar]
- 21. National Human Genome Research Institute . NHGRI history and timeline of events. Accessed July 29, 2020. https://www.genome.gov/about-nhgri/Brief-History-Timeline.
- 22. Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. National Institutues of Health All of Us Research Program . All of Us Research Program overview. 2020. Accessed July 29, 2020. https://allofus.nih.gov/about/all-us-research-program-overview.
- 25. Schunkert H, Samani NJ. Into the great wide open: 10 years of genome-wide association studies. Cardiovasc Res. 2018;114:1189–1191. [DOI] [PubMed] [Google Scholar]
- 26. Maher B. ENCODE: the human encyclopaedia. Nature. 2012;489:46–48. [DOI] [PubMed] [Google Scholar]
- 27. National Human Genome Research Institute . The Encyclopedia of DNA Elements (ENCODE). 2020. Accessed July 29, 2020. https://www.genome.gov/Funded-Programs-Projects/ENCODE-Project-ENCyclopedia-Of-DNA-Elements.
- 28. Pennisi E. Genomics. ENCODE project writes eulogy for junk DNA. Science. 2012;337:1159–1161. [DOI] [PubMed] [Google Scholar]
- 29. Balas MM, Johnson AM. Exploring the mechanisms behind long noncoding RNAs and cancer. Noncoding RNA Res. 2018;3:108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaikkonen MU, Lam MT, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res. 2011;90:430–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cloud J. Why DNA isn't your destiny. Time. 2010;18. [Google Scholar]
- 32. Blewitt M, Whitelaw E. The use of mouse models to study epigenetics. Cold Spring Harb Perspect Biol. 2013;5:a017939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cunningham AM, Walker DM, Nestler EJ. Paternal transgenerational epigenetic mechanisms mediating stress phenotypes of offspring. Eur J Neurosci. 2021;53:271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martone J, Lisi M, Castagnetti F, et al. Trans-generational epigenetic regulation associated with the amelioration of Duchenne muscular dystrophy. EMBO Mol Med. 2020;5:e12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Soubry A, Murphy SK, Wang F, et al. Newborns of obese parents have altered DNA methylation patterns at imprinted genes. Int J Obes. 2015;39:650–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Office of Disease Prevention and Health Promotion . Healthy people 2030 - social determinants of health. 2020. Accessed November 16, 2020. https://health.gov/healthypeople/objectives-and-data/social-determinants-health.
- 37. Stringhini S, Polidoro S, Sacerdote C, et al. Life-course socioeconomic status and DNA methylation of genes regulating inflammation. Int J Epidemiol. 2015;44:1320–1330. [DOI] [PubMed] [Google Scholar]
- 38. McDade TW, Ryan C, Jones MJ, et al. Social and physical environments early in development predict DNA methylation of inflammatory genes in young adulthood. Proc Natl Acad Sci U S A. 2017;114:7611–7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Alfano R, Guida F, Galobardes B, et al. Socioeconomic position during pregnancy and DNA methylation signatures at three stages across early life: epigenome-wide association studies in the ALSPAC birth cohort. Int J Epidemiol. 2019;48:30–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bush NR, Edgar RD, Park M, et al. The biological embedding of early-life socioeconomic status and family adversity in children's genome-wide DNA methylation. Epigenomics. 2018;10:1445–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Needham BL, Smith JA, Zhao W, et al. Life course socioeconomic status and DNA methylation in genes related to stress reactivity and inflammation: the multi-ethnic study of atherosclerosis. Epigenetics. 2015;10:958–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hertzman C. Putting the concept of biological embedding in historical perspective. Proc Natl Acad Sci U S A. 2012;109:17160–17167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Demetriou CA, van Veldhoven K, Relton C, Stringhini S, Kyriacou K, Vineis P. Biological embedding of early-life exposures and disease risk in humans: a role for DNA methylation. Eur J Clin Invest. 2015;45:303–332. [DOI] [PubMed] [Google Scholar]
- 44. Austin MK, Chen E, Ross KM, et al. Early-life socioeconomic disadvantage, not current, predicts accelerated epigenetic aging of monocytes. Psychoneuroendocrinology. 2018;97:131–134. [DOI] [PubMed] [Google Scholar]
- 45. Holuka C, Merz MP, Fernandes SB, et al. The COVID-19 pandemic: does our early life environment, life trajectory and socioeconomic status determine disease susceptibility and severity? Int J Mol Sci. 2020;21:5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Salimi S, Hamlyn JM. COVID-19 and crosstalk with the hallmarks of aging. J Gerontol A Biol Sci Med Sci. 2020;75:e34–e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maroko AR, Nash D, Pavilonis BT. COVID-19 and inequity: a comparative spatial analysis of New York City and Chicago hot spots. J Urban Health. 2020;97:461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cernackova A, Durackova Z, Trebaticka J, Mravec B. Neuroinflammation and depressive disorder: the role of the hypothalamus. J Clin Neurosci. 2020;75:5–10. [DOI] [PubMed] [Google Scholar]
- 49. Rahman MH, Bhusal A, Lee WH, Lee IK, Suk K. Hypothalamic inflammation and malfunctioning glia in the pathophysiology of obesity and diabetes: translational significance. Biochem Pharmacol. 2018;153:123–133. [DOI] [PubMed] [Google Scholar]
- 50. Nees F, Loffler M, Usai K, Flor H. Hypothalamic-pituitary-adrenal axis feedback sensitivity in different states of back pain. Psychoneuroendocrinology. 2019;101:60–66. [DOI] [PubMed] [Google Scholar]
- 51. Snyder-Mackler N, Sanz J, Kohn JN, et al. Social status alters chromatin accessibility and the gene regulatory response to glucocorticoid stimulation in rhesus macaques. Proc Natl Acad Sci U S A. 2019;116:1219–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Miller GE, Cohen S, Ritchey AK. Chronic psychological stress and the regulation of pro-inflammatory cytokines: a glucocorticoid-resistance model. Health Psychol. 2002;21:531–541. [DOI] [PubMed] [Google Scholar]
- 53. Walsh CP, Ewing LJ, Cleary JL, et al. Development of glucocorticoid resistance over one year among mothers of children newly diagnosed with cancer. Brain Behav Immun. 2018;69:364–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Matthews SG, McGowan PO. Developmental programming of the HPA axis and related behaviours: epigenetic mechanisms. J Endocrinol. 2019;242:T69–T79. [DOI] [PubMed] [Google Scholar]
- 55. Aroke EN, Joseph PV, Roy A, et al. Could epigenetics help explain racial disparities in chronic pain? J Pain Res. 2019;12:701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Feinberg AP. The key role of epigenetics in human disease prevention and mitigation. N Engl J Med. 2018;378:1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fernandez-Sanles A, Sayols-Baixeras S, Castro DEMM, et al. Physical activity and genome-wide DNA methylation: the REgistre GIroni del COR study. Med Sci Sports Exerc. 2020;52:589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Seaborne RA, Strauss J, Cocks M, et al. Human skeletal muscle possesses an epigenetic memory of hypertrophy. Sci Rep. 2018;8:1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hibler E, Huang L, Andrade J, Spring B. Impact of a diet and activity health promotion intervention on regional patterns of DNA methylation. Clin Epigenetics. 2019;11:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zimmer P, Bloch W. Physical exercise and epigenetic adaptations of the cardiovascular system. Herz. 2015;40:353–360. [DOI] [PubMed] [Google Scholar]
- 61. Barres R, Zierath JR. The role of diet and exercise in the transgenerational epigenetic landscape of T2DM. Nat Rev Endocrinol. 2016;12:441–451. [DOI] [PubMed] [Google Scholar]
- 62. Grazioli E, Dimauro I, Mercatelli N, et al. Physical activity in the prevention of human diseases: role of epigenetic modifications. BMC Genomics. 2017;18:802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fernandes J, Arida RM, Gomez-Pinilla F. Physical exercise as an epigenetic modulator of brain plasticity and cognition. Neurosci Biobehav Rev. 2017;80:443–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Beiter T, Niess AM, Moser D. Transcriptional memory in skeletal muscle. Don't forget (to) exercise. J Cell Physiol. 2020;235:5476–5489. [DOI] [PubMed] [Google Scholar]
- 65. Barres R, Yan J, Egan B, et al. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 2012;15:405–411. [DOI] [PubMed] [Google Scholar]
- 66. Wrann CD, White JP, Salogiannnis J, et al. Exercise induces hippocampal BDNF through a PGC-1alpha/FNDC5 pathway. Cell Metab. 2013;18:649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bonen A. PGC-1alpha-induced improvements in skeletal muscle metabolism and insulin sensitivity. Appl Physiol Nutr Metab. 2009;34:307–314. [DOI] [PubMed] [Google Scholar]
- 68. Petrie M, Sharma A, Taylor EB, Suneja M, Shields RK. Impact of short and long-term electrically induced muscle exercise on gene signaling pathways, gene expression, and PGC1a methylation in men with spinal cord injury. Physiol Genomics. 2020;52:71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jacobsen SC, Brons C, Bork-Jensen J, et al. Effects of short-term high-fat overfeeding on genome-wide DNA methylation in the skeletal muscle of healthy young men. Diabetologia. 2012;55:3341–3349. [DOI] [PubMed] [Google Scholar]
- 70. Voermans AM, Mewes JC, Broyles MR, LMG S. Cost-effectiveness analysis of a procalcitonin-guided decision algorithm for antibiotic stewardship using real-world U.S. hospital data. OMICS. 2019;23:508–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chen J, Hutchison KE, Bryan AD, et al. Opposite epigenetic associations with alcohol use and exercise intervention. Front Psych. 2018;9:594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jansen EC, Dolinoy DC, O'Brien LM, et al. Sleep duration and fragmentation in relation to leukocyte DNA methylation in adolescents. Sleep. 2019;42:zsz121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Altintas A, Laker RC, Garde C, Barres R, Zierath JR. Transcriptomic and epigenomics atlas of myotubes reveals insight into the circadian control of metabolism and development. Epigenomics. 2020;12:701–713. [DOI] [PubMed] [Google Scholar]
- 74. Voisin S, Eynon N, Yan X, Bishop DJ. Exercise training and DNA methylation in humans. Acta Physiol. 2015;213:39–59. [DOI] [PubMed] [Google Scholar]
- 75. McEwen BS. Integrative medicine: breaking down silos of knowledge and practice an epigenetic approach. Metabolism. 2017;69S:S21–S29. [DOI] [PubMed] [Google Scholar]


