Background and importance
mRNA-based host response signatures have been reported to improve sepsis diagnostics. Meanwhile, prognostic markers for the rapid and accurate prediction of severity in patients with suspected acute infections and sepsis remain an unmet need. IMX-SEV-2 is a 29-host-mRNA classifier designed to predict disease severity in patients with acute infection or sepsis.
Objective
Validation of the host-mRNA infection severity classifier IMX-SEV-2.
Design, settings and participants
Prospective, observational, convenience cohort of emergency department (ED) patients with suspected acute infections.
Outcome measures and analysis
Whole blood RNA tubes were analyzed using independently trained and validated composite target genes (IMX-SEV-2). IMX-SEV-2-generated risk scores for severity were compared to the patient outcomes in-hospital mortality and 72-h multiorgan failure.
Main results
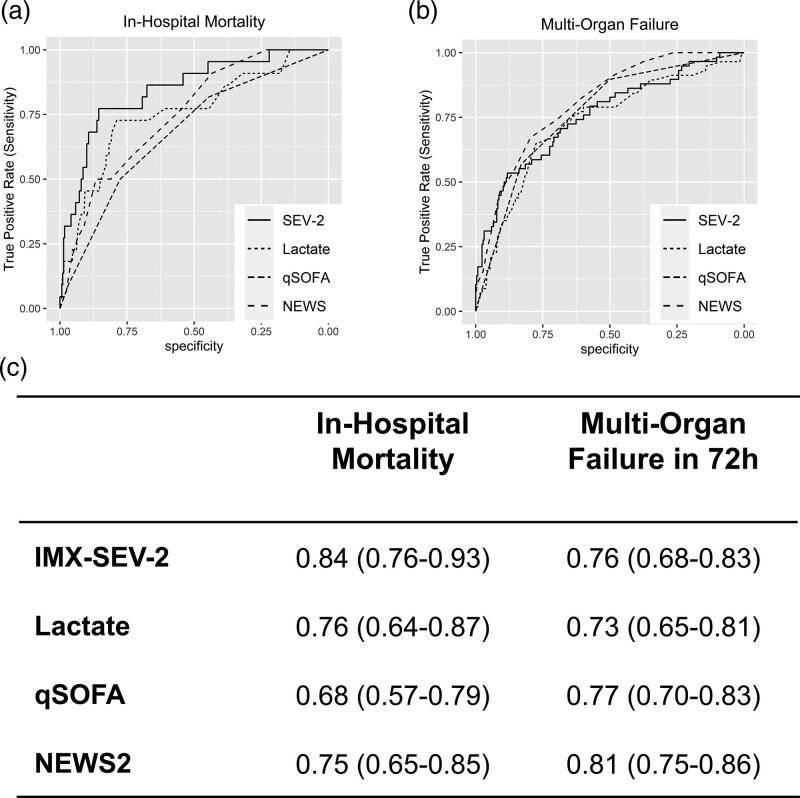
Of the 312 eligible patients, 22 (7.1%) died in hospital and 58 (18.6%) experienced multiorgan failure within 72 h of presentation. For predicting in-hospital mortality, IMX-SEV-2 had a significantly higher area under the receiver operating characteristic (AUROC) of 0.84 [95% confidence intervals (CI), 0.76–0.93] compared to 0.76 (0.64–0.87) for lactate, 0.68 (0.57–0.79) for quick Sequential Organ Failure Assessment (qSOFA) and 0.75 (0.65–0.85) for National Early Warning Score 2 (NEWS2), (P = 0.015, 0.001 and 0.013, respectively). For identifying and predicting 72-h multiorgan failure, the AUROC of IMX-SEV-2 was 0.76 (0.68–0.83), not significantly different from lactate (0.73, 0.65–0.81), qSOFA (0.77, 0.70–0.83) or NEWS2 (0.81, 0.75–0.86).
Conclusion
The IMX-SEV-2 classifier showed a superior prediction of in-hospital mortality compared to biomarkers and clinical scores among ED patients with suspected infections. No improvement for predicting multiorgan failure was found compared to established scores or biomarkers. Identifying patients with a high risk of mortality or multiorgan failure may improve patient outcomes, resource utilization and guide therapy decision-making.
Keywords: critical care, host-response, infection, messenger, RNA, sepsis, transcriptome
Introduction
Acute infections can lead to sepsis, which is still associated with estimated mortality rates between 10 and 35% [1]. Even clinically banal infections risk becoming septic and currently no test can quantify that risk [2]. Recognizing potential deterioration to sepsis is especially hindered in elderly patients and the immunosuppressed, as before decompensation they often lack the clinical signs of sepsis [3,4].
These clinically suspected septic patients are for the most part initially encountered in the emergency department (ED) where timely recognition and initiation of adequate therapy is challenging [5,6]. There is therefore a need for improved sepsis screening and risk-stratification tools to support physician decision-making [2]. Currently, decision-making depends on the treating physician’s experience, guided by clinical scores and a limited number of routine biomarkers.
The assessment of clinical scores, such as Sequential Organ Failure Assessment (SOFA), while highly accurate, comes with several difficulties in the ED due to time constraints and a diverse patient population [7,8].
The sepsis-3 definition advises the use of the quick-SOFA (qSOFA), a simpler alternative to SOFA and a more accurate alternative to systemic inflammatory response syndrome criteria for determining severity [9,10]. While helpful in recognizing acute organ dysfunction, qSOFA offers little benefit as a screening tool for sepsis, and current Surviving Sepsis Guidelines recommend against the use of qSOFA as a screening tool [11].
The National Early Warning Score 2 (NEWS2) has been suggested as a general severity indicator for a range of uses, settings and patient populations [12,13]. Although it lacks specificity for infections, NEWS2 has shown high accuracy for the prediction of severe outcomes including septic shock, organ failure and mortality in patients with acute infections [14].
Importantly, acute infections and sepsis exist on a continuum of severity [2]. ‘Severity’ covers a variety of complications associated with poor outcomes ranging from organ dysfunction to death.
A variety of biomarkers for infection severity have been proposed to complement clinical decision-making. Currently, lactate is the most commonly used biomarker for shock. Among patients with suspected infections, lactate is, therefore, a prognostic marker for sepsis. However, the prognostic accuracy is highly variable and lacks specificity for organ dysfunction caused by acute infections or sepsis [15]. Indeed, lactate cannot be used as a single, reliable marker for risk stratification in the ED [9]. Further prognostic markers have been studied to complement existing standard-of-care markers, including clinical scores, hematological markers, machine learning classifiers and multi-RNA transcriptomic signatures [13,16–18].
To address the unmet medical need described above, the classifier ‘Inflammatix Severity version 2’ (IMX-SEV-2, Inflammatix, Burlingame, California, USA) was developed for predicting disease severity. Objectively quantifying the risk of deterioration from a banal infection into sepsis would assist ED physicians in adequately assigning monitoring, therapy escalation and the application of local sepsis protocols to patients who will deteriorate. This study is a prospective validation of the IMX-SEV-2 severity score in a cohort of adult ED patients presenting to the ED with clinical suspicion of acute infection or sepsis.
Methods
Study design and participant selection
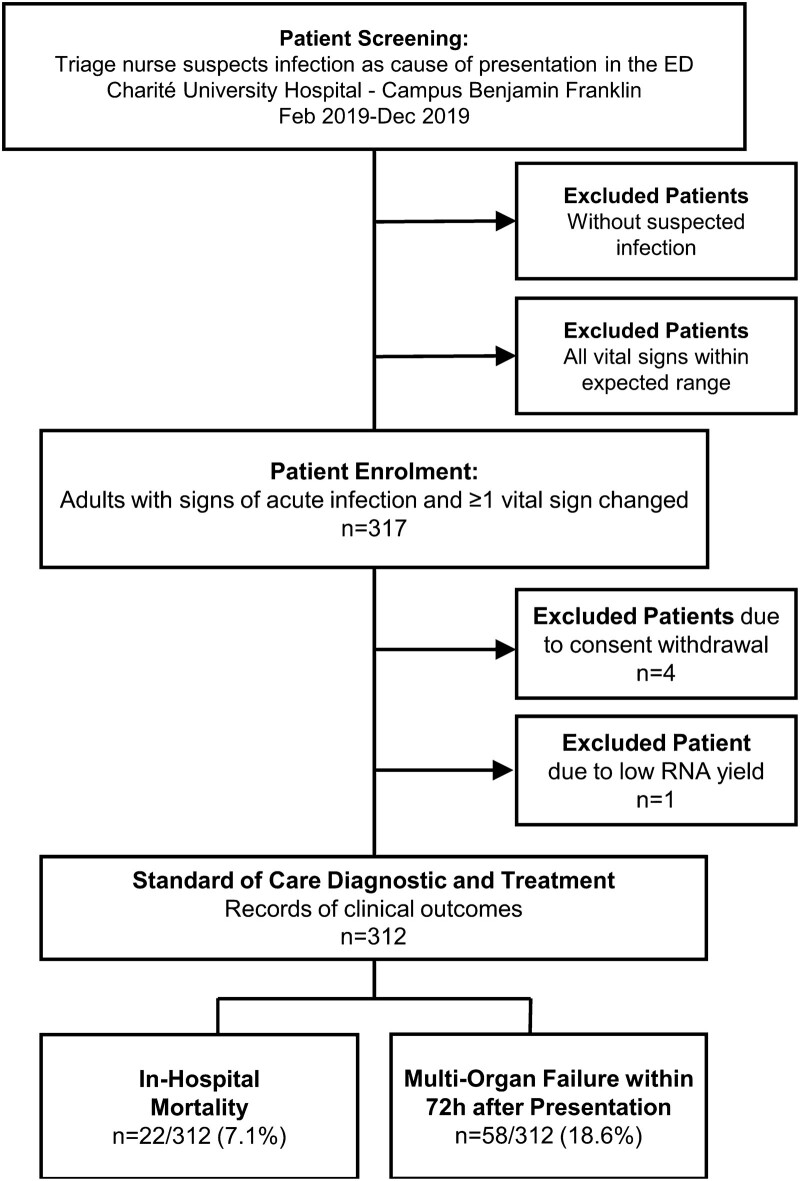
A prospective, observational study was conducted at the ED of the Charité-Universitätsmedizin Berlin, Campus Benjamin Franklin (Berlin, Germany) from February until December 2019. Enrollment criteria were (1) adult patients (≥18 years) presenting to the ED with clinical suspicion of acute infection, (2) at least one vital sign change. Samples were collected at the point of enrollment, and patients were then managed following local standard of care guidelines. Figure 1 shows the study design. For more details regarding enrollment, sample collection, and sample processing, see the supplemental methods (Supplementary Methods, Supplemental digital content 1, http://links.lww.com/EJEM/A334).
Fig. 1.
Flowchart of patient enrollment and outcomes. In total 312 of 317 patients enrolled met the inclusion criteria and were treated and diagnosed according to the standard of care by physicians blinded to the IMX-SEV-2 severity score results. Patient clinical outcomes were recorded from medical records.
The study was approved by the institutional review board and registered with the German Clinical Trials Register (DRKS-ID 0017395). We previously reported the performance of the bacterial versus viral infection diagnostic classifier Inflammatix Bacterial Viral Non-infected version 2 (IMX-BVN-2) in the same cohort [19].
Severity endpoints
The primary endpoint of this analysis was in-hospital mortality, with a secondary endpoint of 72-h multiorgan failure. Two senior physicians trained in Emergency and Internal Medicine retrospectively assessed each case, to determine whether patients experienced 72-h multiorgan failure, including organ failure already manifest at presentation, and defined as severe impairment of at least two SOFA score organ systems. The physicians were blinded to the IMX-SEV-2 results. For in-hospital mortality, to reduce the effects of unrelated mortality after prolonged inpatient care for chronic conditions, patients still hospitalized after 28 days were considered as not meeting the endpoint [20].
mRNA amplification and IMX-SEV-2 classifier
The development of mRNA classifiers which report a severity score for acute infections has been previously described [21–23]. In brief, the expression of 29 genes is read via a logistic regression classifier to compute a single severity output. The IMX-SEV-2 classifier was trained on 30-day mortality outcomes. To aid clinical decision-making across a range of severities, IMX-SEV-2 incorporates two preset cutoffs to generate three interpretation bands: low risk, moderate risk and high risk. Importantly, these cutoffs were developed on separate training datasets and formally locked before application to this patient cohort. For a more detailed explanation of the classifier development, see Supplementary Methods, Supplemental digital content 1, http://links.lww.com/EJEM/A334.
Statistical analysis and blinding
The primary outcome of this clinical validation study was the performance of IMX-SEV-2 for predicting in-hospital mortality, with the secondary outcome being performance for identifying and predicting 72-h multiorgan failure. Additional analyses included the performance of IMX-SEV-2 in patients with qSOFA ≥2 at presentation and a linear regression model for IMX-SEV-2.
Performance was assessed and compared (1) without predefined thresholds using the area under receiver operating characteristics (AUROCs) and (2) using predefined thresholds to show sensitivities, specificities and likelihood ratios. IMX-SEV-2 bands were analyzed and compared to the comparison markers using the routine thresholds. For NEWS2, three bands were created using the established thresholds [12] and combining ‘low-medium’ and ‘medium’ into one band, resulting in the following banding: low risk (0–4), medium risk (5–6, and/or ≥3 in any single category), high risk (≥7).
Further details on the statistical analysis are in Supplementary Methods, Supplemental digital content 1, http://links.lww.com/EJEM/A334. Raw clinical data and IMX-SEV-2 scores are listed in Supplemental Data, Supplemental digital content 1, http://links.lww.com/EJEM/A334.
Results
Characteristics of study subjects
The enrolled cohort consisted of 317 adult patients with signs of acute infection and ≥1 vital sign change at initial presentation to the ED. After excluding four patients due to consent withdrawal and one due to insufficient RNA yield, the final cohort included 312 patients. Patient characteristics segmented by in-hospital mortality are summarized in Table 1, with an extended version in Supplemental Table S1, Supplemental digital content 1, http://links.lww.com/EJEM/A334. The patient cohort had a substantial number of older patients with severe diseases [median age 73 years, 76 patients (24.4%) with qSOFA ≥2]; 86 patients (27.6%) had active malignancies and 65 patients (20.8%) had immunosuppression as defined in Supplemental Table S2, Supplemental digital content 1, http://links.lww.com/EJEM/A334. In total 58 patients (18.6%) experienced 72-h multiorgan failure and 22 patients (7.1%) died in hospital.
Table 1.
Patient characteristics segmented by in-hospital mortality
| Characteristics | All | Survival and/or discharge | In-hospital mortality | P values |
|---|---|---|---|---|
| (n = 312) | n = 290 (92.9%) | n = 22 (7.1%) | ||
| Age (years) | 72.5 (57.0, 80.0) | 72.0 (57.0, 80.0) | 77.5 (69.5, 84.8) | 0.02 |
| Sex (female) | 132 (42.3%) | 119 (41.0%) | 13 (59.1%) | 0.12 |
| Vital signs at presentation | ||||
| Respiratory rate (/min) | 303; 21 (18, 26) | 281; 21 (18, 25) | 23 (19, 32) | 0.15 |
| SBP (mmHg) | 311; 123 (104, 138) | 289; 124 (107, 139) | 107 (98, 133) | 0.05 |
| Altered mentation | 59 (18.9%) | 48 (16.6%) | 11 (50.0%) | <0.01 |
| qSOFA ≥2 | 76 (24.4%) | 65 (22.4%) | 11 (50.0%) | <0.01 |
| NEWS2 | 5 (3, 8) | 5 (3, 8) | 9 (6, 12) | <0.01 |
| Comorbidity | ||||
| Malignancy | 86 (27.6%) | 77 (26.6%) | 9 (40.9%) | 0.21 |
| T2D | 63 (20.2%) | 58 (20.0%) | 5 (22.7%) | 0.78 |
| COPD | 40 (12.8%) | 38 (13.1%) | 2 (9.1%) | 0.75 |
| Immunocompromised | 65 (20.8%) | 61 (21.0%) | 4 (18.2%) | 0.35 |
| Biomarkers | ||||
| WBC (109cells/L) | 11.1 (8.0, 15.3) | 11.0 (7.9, 14.9) | 15.0 (9.1, 22.0) | 0.01 |
| CRP (mg/L) | 70.8 (21.7, 178.1) | 68.3 (20.5, 157.8) | 189.1 (61.7, 260.4) | <0.01 |
| Procalcitonin (µg/L) | 305; 0.3 (0.1, 1.1) | 283; 0.3 (0.1, 0.9) | 1.7 (0.3, 26.6) | <0.01 |
| Lactate (mmol/L) | 301; 1.9 (1.4, 2.5) | 279; 1.8 (1.4, 2.3) | 3.1 (2.2, 4.3) | <0.01 |
| IMX-SEV-2 | 0.04 (0.03, 0.07) | 0.04 (0.03, 0.06) | 0.09 (0.07, 0.19) | <0.01 |
| Outcomes | ||||
| Mechanical ventilation | 21 (6.7%) | 13 (4.5%) | 8 (36.4%) | <0.01 |
| ICU admission | 71 (22.8%) | 56 (19.3%) | 15 (68.2%) | <0.01 |
| 72 h multiorgan failure | 58 (18.6%) | 39 (13.4%) | 19 (86.4%) | <0.01 |
Continuous variables are presented with median and interquartile range and compared using Mann–Whitney U Test. Nominal variables are presented with frequency and column percentage and compared using Fisher’s exact test. The number of cases with valid data is shown for variables with missing data (e.g. 301 patients had lactate concentration measurements).
COPD, chronic obstructive pulmonary disease; CRP, C-reactive protein; NEWS2, National Early Warning Score 2; qSOFA, Quick Sequential Organ Failure Assessment; T2D, Type two diabetes; WBC, White blood cell count.
Performance of IMX-SEV-2
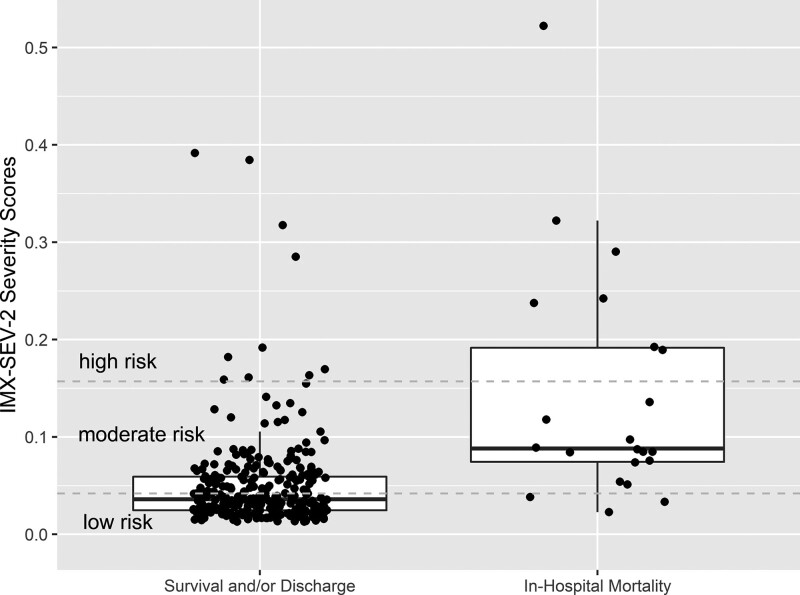
Figure 2 shows the distribution of IMX-SEV-2 severity scores segmented by in-hospital mortality vs. survival and/or discharge; scores were higher in patients who died in the hospital, and most patients who died were in the moderate and high-risk score ranges whereas most survivors were in the low-risk scores. To compare the performance of IMX-SEV-2 as a continuous metric with other standard prognostic biomarkers, we calculated AUROCs for in-hospital mortality and 72-h multiorgan failure (Fig. 3a–c). For in-hospital mortality, IMX-SEV-2 had an AUROC of 0.84 [95% confidence intervals (CI), 0.76–0.93]. In comparison, lactate, qSOFA and NEWS2 had significantly lower AUROCs (P = 0.015, 0.001 and 0.013 compared to IMX-SEV-2, respectively).
Fig. 2.
IMX-SEV-2 shows ability to separate patients with in-hospital mortality vs. survival or discharge. Distribution of IMX-SEV-2 severity scores segmented by in-hospital mortality vs. survival and/or discharge. Horizontal lines indicate the preset threshold cutoffs which divide the score into three interpretation bands: high (≥0.157), moderate (0.042–0.157) and low severity (<0.042).
Fig. 3.
Performance of the IMX-SEV-2 severity score compared with other prognostic markers. Receiver operating characteristic (ROC) for distinguishing (a) in-hospital mortality, and (b) 72-h multiorgan failure. (c) Area under the receiver operating characteristics (AUROCs) with 95% confidence intervals for in-hospital mortality, and 72-h multiorgan for IMX-SEV-2 compared to other clinical markers.
For identifying and predicting 72-h multiorgan failure, IMX-SEV-2 had an AUROC of 0.76 (95% CI, 0.68–0.83). Lactate, qSOFA and NEWS2 had similar AUROCs, none of which differed significantly from IMX-SEV-2 (P = 0.341, 0.846 and 0.215, respectively).
The area under the precision-recall curve (AUPRC) is a useful accuracy metric for data with unbalanced outcomes. Unlike AUROCs, where the baseline value is 0.5, the baseline AUPRC is equal to the outcome prevalence (0.071 for in-hospital mortality and 0.186 for multiorgan failure). For in-hospital mortality, IMX-SEV-2 had an AUPRC of 0.36 (95% CI, 0.19–0.58), significantly higher than NEWS2 and qSOFA, but not lactate (P = 0.045, 0.016 and 0.322, respectively). For multiorgan failure, IMX-SEV-2 had an AUPRC of 0.51, significantly higher than lactate, but not NEWS or qSOFA (P = 0.022, 0.940 and 0.081 respectively). The PRC plots and AUPRCs are shown in Supplemental Figure S1, Supplemental Digital Content 1, http://links.lww.com/EJEM/A334.
IMX-SEV-2 clinical interpretation bands
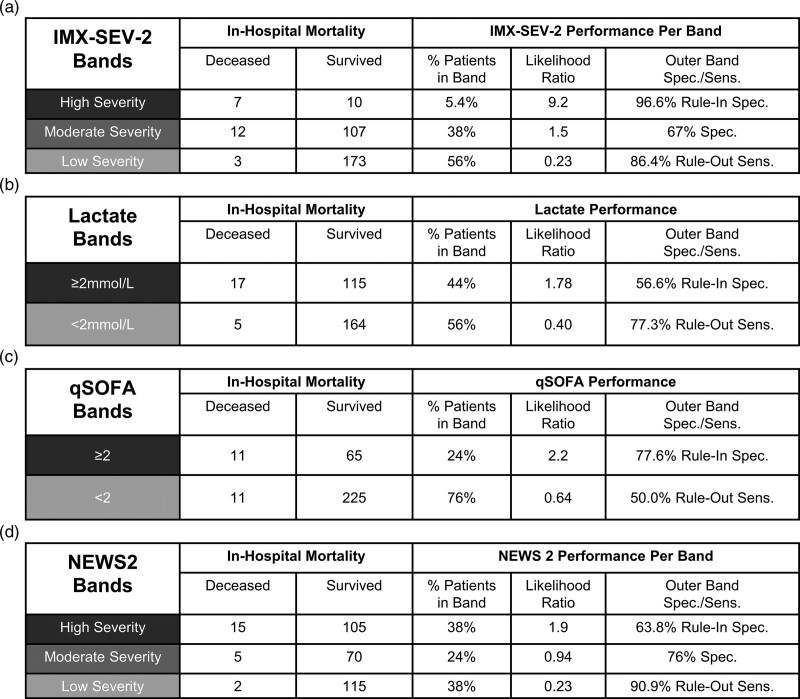
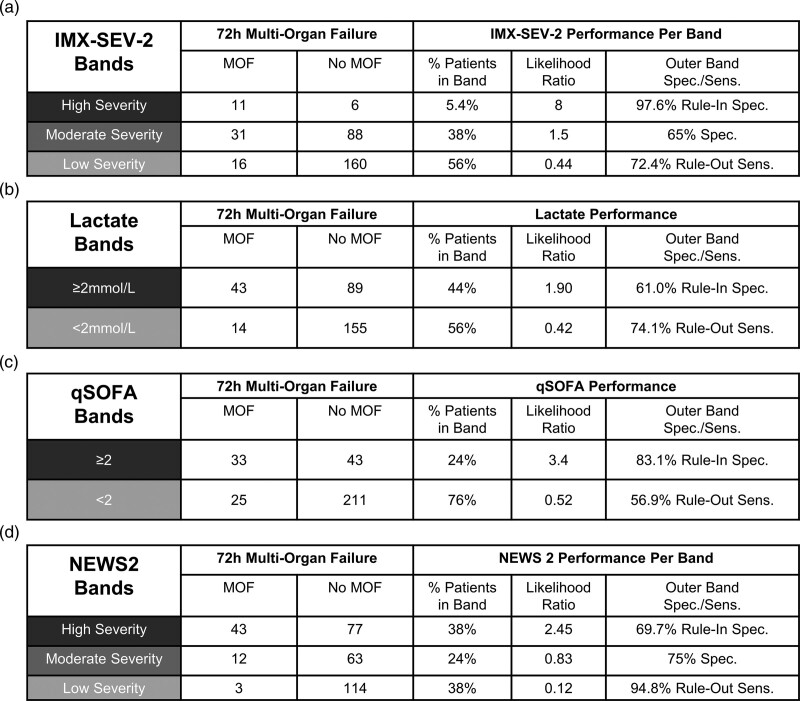
IMX-SEV-2 incorporates three bands to improve clinical utility. The performance of the bands is shown in Fig. 4 for in-hospital mortality and Fig. 5 for 72-h multiorgan failure, each compared to lactate, qSOFA and NEWS2. IMX-SEV-2 demonstrated a specificity of 96.6% for the high severity rule-in band and a sensitivity of 86.4% for the low severity rule-out band compared to a specificity of 63.8% and a sensitivity of 90.9% for the rule-in and rule-out bands in NEWS2. This is also reflected in the likelihood ratios: NEWS2 demonstrated a lower likelihood ratio of 1.9 for the high severity band compared to 9.2 for the IMX-SEV-2 classifier; for the middle bands indicating moderate severity, only IMX-SEV-2 generated a likelihood ratio of >1. For the rule-out low severity bands both NEWS2 and IMX-SEV-2 showed likelihood ratios of 0.23. Importantly, despite having the same likelihood ratio for the low severity interpretation band, IMX-SEV-2 placed a higher percentage of patients into the low severity band compared to NEWS2 (56% vs. 38%).
Fig. 4.
Performance of interpretation bands for in-hospital mortality. Performance of IMX-SEV-2, qSOFA, lactate and NEWS2 bands for predicting in-hospital mortality (a–d). IMX-SEV-2 bands are generated using the predefined thresholds. NEWS2 bands are defined as low risk (0–4), medium risk (5–6, and/or ≥3 in any single category) and high risk (≥7). Lactate results are presented for the 301 patients with concentrations measured at presentation.
Fig. 5.
Performance of interpretation bands for multiorgan failure. Performance of IMX-SEV-2, qSOFA, lactate and NEWS2 bands for identifying and predicting 72-h multiorgan failure (a–d). IMX-SEV-2 bands are generated using the predefined thresholds. NEWS2 bands are defined as low risk (0–4), medium risk (5–6, and/or ≥3 in any single category) and high risk (≥7). Lactate results are presented for the 301 patients with concentrations measured at presentation.
Overall NEWS2 placed more patients in the outer bands (76% for NEWS2 vs. 62% for IMX-SEV-2), albeit with a lower high-risk likelihood ratio (1.9, compared to 9.2).
In comparison, when applying two interpretation bands with a cutoff of 2 mmol/L, lactate showed likelihood ratios of 1.78 (rule-in high severity band) and 0.40 (rule-out low severity band) compared to 2.2 and 0.64 for qSOFA ≥2. Therefore, among the four predictors, only IMX-SEV-2 and NEWS2 demonstrated a result with high sensitivity for ruling out mortality (low severity band). Among those two predictors, only IMX-SEV-2 demonstrated a high specificity rule-in (high severity) band.
To allow for a better comparison with lactate and qSOFA, which are clinically used with a single threshold, single-threshold versions of IMX-SEV-2 (using its two thresholds separately) are shown in Supplementary Table S3 alongside the comparators (Supplementary Table S3, Supplemental digital content 1, http://links.lww.com/EJEM/A334).
Subgroup analysis and confounding factors
Of the 312 adult patients with suspected acute infections, 76 (24.4%) had a qSOFA score ≥2 at admission, representing a cohort of patients with sepsis or a high likelihood of developing sepsis [9]. Among these patients, IMX-SEV-2 and lactate showed increased AUROCs compared to the overall cohort, while the accuracy of NEWS2 decreased [IMX-SEV-2 for in-hospital mortality of 0.92 (0.85–0.98), compared to 0.83 (0.69–0.96) for lactate, P = 0.08 and 0.70 (0.55–0.86) for NEWS2, P = 0.011]. The decision tree in Supplemental Figure S2, Supplemental digital content 1, http://links.lww.com/EJEM/A334 shows how IMX-SEV-2 could help reassess subjects initially stratified by qSOFA or NEWS2. For example, 33 patients with qSOFA ≥2 were correctly placed into the low severity band by IMX-SEV-2 and did not develop fatal outcomes.
A linear regression model was used to assess whether IMX-SEV-2 was impacted by patient characteristics including age, immunosuppression, bacterial infection, viral infection and sex after controlling for severity (in-hospital mortality and multiorgan failure) (Supplemental Table S4, Supplemental digital content 1, http://links.lww.com/EJEM/A334). Immunosuppression, age, bacterial infection, viral infection, and sex showed no statistically significant impact on IMX-SEV-2 scores.
Discussion
This study describes the validation of IMX-SEV-2, a 29-host response mRNA severity classifier in a prospective cohort of 312 patients presenting with suspected acute infection to a tertiary care center ED in Germany.
This study builds upon previous work in developing a prognostic biomarker for risk-stratifying patients with acute infection and sepsis [22–24]. We show that IMX-SEV-2 is accurate in predicting in-hospital mortality with an AUROC of 0.84 and does so with significantly higher accuracy than lactate, qSOFA or NEWS2. This finding is important, as IMX-SEV-2 could assist emergency physicians more reliably in identifying ED patients at risk of deterioration. In combination with the diagnostic classifier IMX-BVN-2 [19], this assay has the potential to both simultaneously prove an infection and identify imminent organ dysfunction, thereby providing a single, objective test for sepsis.
In line with previous research, our cohort showed low sensitivities and specificities for qSOFA in regard to in-hospital mortality [11]. In critically ill patients with a qSOFA≥2, IMX-SEV-2 predicted in-hospital mortality with an AUROC of 0.92. This is of interest, as the more severely ill subgroup better reflects the training cohorts used to generate the classifier [21], as such an improved performance was expected. Additionally, we would suggest that the ease of determining qSOFA scores combined with the relatively low specificity presents a possible triage for IMX-SEV-2 once developed as a rapid point-of-care test to accurately assess severity (Supplementary Figure S2, Supplemental digital content 1, http://links.lww.com/EJEM/A334). In our cohort, of the patients classified by qSOFA as at the highest risk for clinical deterioration, IMX-SEV-2 was able to place 33 (43.4%) in the lowest band, with no fatalities.
The IMX-SEV-2 classifier uses two pre-set cutoffs which segment the severity score into three interpretation bands (low severity, moderate severity and high severity). The banding showed that, compared to lactate and qSOFA, IMX-SEV-2 and NEWS2 are more accurate for ruling out mortality, and that among these two, IMX-SEV-2 had the better rule-in band performance. This banding approach allows for an easy yet nuanced stratification of a diverse patient cohort, ranging from uncomplicated infections to those with imminent risk of deteriorating and organ dysfunction (Figs. 4 and 5).
For 72-h multiorgan failure, the AUROC of IMX-SEV-2 was 0.76, not significantly different from qSOFA (0.77), lactate (0.73) or NEWS2 (0.81). This is an important early endpoint. However, lower performance was expected, as IMX-SEV-2 was originally trained to predict 30-day mortality [23]. The results show that the clinical signs of organ failure, as shown by qSOFA, as well as the hypoperfusion associated with septic shock, as implicated by higher lactate, are similarly represented by the transcriptomic test. Importantly, new machine learning classifiers based on the same assay but trained to predict multiorgan failure could refine this result and add to the clinical utility of the transcriptome assay. While our results show that this endpoint is still best served by clinical scores, such as NEWS2 and qSOFA, the proof of principle in this area offers insight into future use and development of host-response scores.
A strength of our trial is the study design, which incorporates as few exclusion criteria as possible to best reflect a real-world ED setting. While patients with immunosuppression, and/or malignancies are often excluded from studies evaluating host response biomarkers [25–27], we purposely chose to include all of these patient subtypes as their clinical presentation can be similar to that of patients with acute infection and suspected sepsis. Importantly, regression analysis did not reveal that immunosuppression or age impacted the performance of the severity score. Finally, the demographic and clinical differences between the classifier training data and the patients from this cohort demonstrate the high generalizability of the assay.
Limitations
The primary limitation of this study was the fact that among our final sample of 312 ED patients, only 22 patients died in the hospital, reducing the statistical power. While mortality was indeed a prospective endpoint, included in the trial registration, the low number of deaths is a result of the cohort design and power analysis having been based on analyses of diagnostic performance [19], instead of severity. Specifically, predicting the deterioration of patients presenting initially as stable would be of significant clinical interest. However, the case number required to power such an analysis would be well beyond that of this cohort. Furthermore, we chose endpoints that reflect the clinical utility of a severity test as accurately as possible by purposely reducing disruptive factors, such as unrelated death or palliative treatment plans. Importantly, this refined selection results in lower numbers of patients with the respective outcomes, which limits the study’s power.
Conclusion
In a much-needed field of research, biomarkers for infection severity, IMX-SEV-2 is part of a new generation of transcriptomic tests. This is the first large-scale prospective validation of a transcriptomic sepsis severity classifier in a real-world ED setting. When translated into a point-of-care assay, it has the potential to quantify and predict the severity of acute infections, saving hospital resources and allowing for appropriate triage to higher-level care thereby addressing an unmet need for improved patient management. An interventional trial will be necessary to assess the true clinical utility of the IMX-SEV-2 and IMX-BVN-2 classifiers.
Acknowledgements
Conflicts of interest
The institution of N.G., E.D.W., D.L., N.M., W.H.B., R.T., K.K., R.S. and W.B received funding from Inflammatix. J.W., O.L., M.M., L.B., R.L. and T.E.S are employees of and option shareholders in Inflammatix. N.G. received a travel grant from Inflammatix, Inc. to attend a conference.
Supplementary Material
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website (www.euro-emergencymed.com).
References
- 1.Kaukonen KM, Bailey M, Pilcher D, Cooper DJ, Bellomo R. Systemic inflammatory response syndrome criteria in defining severe sepsis. N Engl J Med 2015; 372:1629–1638. [DOI] [PubMed] [Google Scholar]
- 2.Yealy DM, Mohr NM, Shapiro NI, Venkatesh A, Jones AE, Self WH. Early care of adults with suspected sepsis in the emergency department and out-of-hospital environment: a consensus-based task force report. Ann Emerg Med 2021; 78:1–19. [DOI] [PubMed] [Google Scholar]
- 3.McCreery RJ, Florescu DF, Kalil AC. Sepsis in immunocompromised patients without human immunodeficiency virus. J Infect Dis 2020; 222:S156–S165. [DOI] [PubMed] [Google Scholar]
- 4.Wang HE, Shah MN, Allman RM, Kilgore M. Emergency department visits by nursing home residents in the United States. J Am Geriatr Soc 2011; 59:1864–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saeed K, Wilson DC, Bloos F, Schuetz P, van der Does Y, Melander O, et al. The early identification of disease progression in patients with suspected infection presenting to the emergency department: a multi-centre derivation and validation study. Crit Care 2019; 23:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xantus G, Allen P, Norman S, Kanizsai P. Antibiotics administered within 1 hour to adult emergency department patients screened positive for sepsis: a systematic review. Eur J Emerg Med 2020; 27:260–267. [DOI] [PubMed] [Google Scholar]
- 7.González-Del Castillo J, Nuñez-Orantos MJ, Llopis F, Martín-Sánchez FJ. Risk stratification of infected patients in emergency department. Crit Care Med 2016; 44:e455. [DOI] [PubMed] [Google Scholar]
- 8.de Groot B, Stolwijk F, Warmerdam M, Lucke JA, Singh GK, Abbas M, et al. The most commonly used disease severity scores are inappropriate for risk stratification of older emergency department sepsis patients: an observational multi-centre study. Scand J Trauma Resusc Emerg Med 2017; 25:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The third international consensus definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016; 315:801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen FC, Kung CT, Cheng HH, Cheng CY, Tsai TC, Hsiao SY, Su CM. Quick sepsis-related organ failure assessment predicts 72-h mortality in patients with suspected infection. Eur J Emerg Med 2019; 26:323–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans L, Rhodes A, Alhazzani W, Antonelli M, Coopersmith CM, French C, et al. Surviving sepsis campaign: international guidelines for the management of sepsis and septic shock 2021. Crit Care Med 2021; 49:1974–1982. [DOI] [PubMed] [Google Scholar]
- 12.Royal College of Physicians. National Early Warning Score (NEWS) 2: Standardising the assessment of acute-illness severity in the NHS. Updated report of a working party. London: RCP, 2017. [Google Scholar]
- 13.Silcock DJ, Corfield AR, Staines H, Rooney KD. Superior performance of National Early Warning Score compared with quick Sepsis-related Organ Failure Assessment Score in predicting adverse outcomes: a retrospective observational study of patients in the prehospital setting. Eur J Emerg Med 2019; 26:433–439. [DOI] [PubMed] [Google Scholar]
- 14.Brink A, Alsma J, Verdonschot RJCG, Rood PPM, Zietse R, Lingsma HF, Schuit SCE. Predicting mortality in patients with suspected sepsis at the Emergency Department; a retrospective cohort study comparing qSOFA, SIRS and National Early Warning Score. PLoS One 2019; 14:e0211133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puskarich MA, Shapiro NI, Massey MJ, Kline JA, Jones AE. Lactate clearance in septic shock is not a surrogate for improved microcirculatory flow. Acad Emerg Med 2016; 23:690–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Opal SM, Wittebole X. Biomarkers of infection and sepsis. Crit Care Clin 2020; 36:11–22. [DOI] [PubMed] [Google Scholar]
- 17.Burnham KL, Davenport EE, Radhakrishnan J, Humburg P, Gordon AC, Hutton P, et al. Shared and distinct aspects of the sepsis transcriptomic response to fecal peritonitis and pneumonia. Am J Respir Crit Care Med 2017; 196:328–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsalik EL, Langley RJ, Dinwiddie DL, Miller NA, Yoo B, van Velkinburgh JC, et al. An integrated transcriptome and expressed variant analysis of sepsis survival and death. Genome Med 2014; 6:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bauer W, Kappert K, Galtung N, Lehmann D, Wacker J, Cheng HK, et al. A novel 29-messenger RNA host-response assay from whole blood accurately identifies bacterial and viral infections in patients presenting to the Emergency Department with suspected infections: a prospective observational study. Crit Care Med 2021; 49:1664–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freund Y, Lemachatti N, Krastinova E, Van Laer M, Claessens YE, Avondo A, et al.; French Society of Emergency Medicine Collaborators Group. Prognostic accuracy of sepsis-3 criteria for in-hospital mortality among patients with suspected infection presenting to the emergency department. JAMA 2017; 317:301–308. [DOI] [PubMed] [Google Scholar]
- 21.Mayhew MB, Buturovic L, Luethy R, Midic U, Moore AR, Roque JA, et al. A generalizable 29-mRNA neural-network classifier for acute bacterial and viral infections. Nat Commun 2020; 11:1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ducharme J, Self WH, Osborn TM, Ledeboer NA, Romanowsky J, Sweeney TE, et al. A multi-mRNA host-response molecular blood test for the diagnosis and prognosis of acute infections and sepsis: proceedings from a clinical advisory panel. J Pers Med 2020; 10:E266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sweeney TE, Perumal TM, Henao R, Nichols M, Howrylak JA, Choi AM, et al. A community approach to mortality prediction in sepsis via gene expression analysis. Nat Commun 2018; 9:694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore AR, Roque J, Shaller BT, Asuni T, Remmel M, Rawling D, et al. Prospective validation of an 11-gene mRNA host response score for mortality risk stratification in the intensive care unit. Sci Rep 2021; 11:13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Houten CB, de Groot JAH, Klein A, Srugo I, Chistyakov I, de Waal W, et al. A host-protein based assay to differentiate between bacterial and viral infections in preschool children (OPPORTUNITY): a double-blind, multicentre, validation study. Lancet Infect Dis 2017; 17:431–440. [DOI] [PubMed] [Google Scholar]
- 26.Self WH, Rosen J, Sharp SC, Filbin MR, Hou PC, Parekh AD, et al. Diagnostic accuracy of FebriDx: a rapid test to detect immune responses to viral and bacterial upper respiratory infections. J Clin Med 2017; 6:E94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller RR, 3rd, Lopansri BK, Burke JP, Levy M, Opal S, Rothman RE, et al. Validation of a host response assay, SeptiCyte LAB, for discriminating sepsis from systemic inflammatory response syndrome in the ICU. Am J Respir Crit Care Med 2018; 198:903–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.