SUMMARY
Yeast prions (infectious proteins) were discovered by their outré genetic properties, and have become important models for an array of human prion and amyloid diseases. A single prion protein can become any of many distinct amyloid forms (called prion variants or strains), each of which is self-propagating, but with different biological properties (e.g., lethal vs. mild). The folded in-register parallel β sheet architecture of the yeast prion amyloids naturally suggests a mechanism by which prion variant information can be faithfully transmitted for many generations. The yeast prions rely on cellular chaperones for their propagation, but can be cured by various chaperone imbalances. The Btn2/Cur1 system normally cures most variants of the [URE3] prion that arise. Although most variants of the [PSI+] and [URE3] prions are toxic or lethal, some are mild in their effects. Even the most mild forms of these prions are rare in the wild, indicating that they too are detrimental to yeast. The beneficial [Het-s] prion of Podospora anserina poses an important contrast in its structure, biology and evolution, to the yeast prions characterized thus far.
Keywords: Sup35, Ure2, [PSI+], [URE3], Btn2, anti-prion system, protein gene, chaperone, [Het-s]
INTRODUCTION
A protein altered form that can convert the normal form of the protein into the same altered form can be an infectious protein, if it has a way to move from one individual to another. Amyloids, linear polymers of largely β-sheet structure, are one way that this can happen (but not the only way). The classical mammalian prion diseases are the uniformly fatal transmissible spongiform encephalopathies (TSEs), based on self-propagating amyloid of the PrP protein (reviewed in (Prusiner 2004; Aguzzi and Lakkaraiu 2015)). Humans are also subject to a number of more common non-infectious amyloidoses such as Alzheimer's disease, Parkinson's disease, type II diabetes mellitus and others. The yeast and fungal prions were first detected as non-chromosomal genes, conferring nonsense suppression ([PSI+] in Saccharomyces cerevisiae, (Cox 1965)), derepressed nitrogen regulation ([URE3] in S. cerevisiae, (Lacroute 1971)) and heterokaryon incompatibility ([Het-s] in Podospora anserina, (Rizet 1952)). Only much later were they shown to be prions (Wickner 1994; Coustou et al. 1997), but their status as genes remains true, with vertical as well as horizontal transmission of information. As will be detailed below, the yeast prions can have heritable alleles, some as stable as chromosomal gene alleles, and will (rarely) mutate from one allele to another.
We will see that these genes (prions) are based on proteins templating their conformation for other protein molecules, in analogy to the templating of sequence by DNA and RNA molecules. Self-propagating, self-templating amyloids are the basis of these prions, linear filamentous polymers of the respective prion protein. A mechanism has been proposed for the templating (Wickner et al. 2008; Wickner et al. 2013), based on their known architecture (Shewmaker et al. 2006; Baxa et al. 2007; Wickner et al. 2008) and the sequence-independence of prion-forming ability (Ross et al. 2004; Ross et al. 2005).
Yeast and fungal prions fit the definition of 'epigenetic phenomenon', namely, “mitotically and/or meiotically heritable changes in gene function that cannot be explained by changes in DNA sequence” (Riggs et al. 1996). Like the chromatin-based epigenetic phenomena, the amyloid-based prions have provided an alternative means of transmission of heritable/infectious information.
Yeast and fungal prions have proven to display many phenomena that parallel the mammalian TSEs, and these parallels will be detailed below. However, the many tools available to the yeast geneticist have facilitated the rapid development of the yeast prion field, resulting in rapid progress in this area, in spite of the relatively modest number of research groups involved. In some cases, models proposed in the TSE field (like the existence of prions!) were first proven for yeast prions. In other cases (such as the extensive role of chaperones and the Btn2/Cur1 prion-curing system) the yeast system has led the way.
DISCOVERY OF YEAST PRIONS
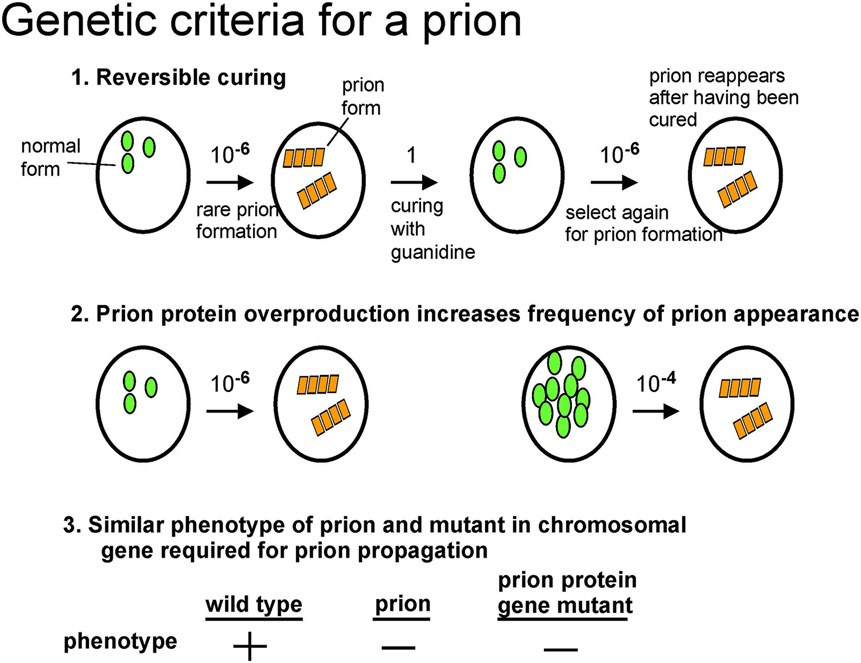
That [URE3] and [PSI+] were prions of Ure2p and Sup35p, respectively was first evident from their unusual genetic properties, each inconsistent with [URE3] or [PSI+] being DNA or RNA replicons, but actually expected for a prion (Wickner 1994)(Figure 1). These genetic criteria were a) infectivity, as shown by non-chromosomal inheritance; b) reversible curability: each prion could be efficiently cured, but would (rarely) re-appear in the same strain; c) overproduction of the corresponding protein dramatically increased the frequency of the prion's appearance; and d) the phenotype of the prion was essentially that of a recessive mutant in the corresponding chromosomal gene for the prion protein, and yet this chromosomal gene was necessary for the propagation of the corresponding prion (Wickner 1994)(reviewed in (Wickner et al. 2015)). That amyloid of the respective prion proteins is the basis of the [PSI+] and [URE3] prions (Fig. 2) was shown by the protease - resistance of Ure2p in [URE3] strains (Masison and Wickner 1995), the aggregated state of Sup35p in [PSI+] cells (Patino et al. 1996; Paushkin et al. 1996), and of Ure2p in [URE3] cells (Edskes et al. 1999), and amyloid formation by Sup35p (King et al. 1997), seeded by extracts of [PSI+] cells (Glover et al. 1997; Paushkin et al. 1997), and of Ure2p (Taylor et al. 1999). Of course, infectivity of amyloid produced in vitro from the corresponding recombinant prion protein, but not by the non-amyloid form, is definitive (Maddelein et al. 2002; King and Diaz-Avalos 2004; Tanaka et al. 2004; Brachmann et al. 2005), but not useful as a screening procedure. Other methods of finding prions will be discussed below.
Fig. 1. Genetic criteria for a yeast or fungal prion.
The [URE3] and [PSI+] prions follow all three criteria (Wickner 1994) because their phenotype is often due to the deficiency of the normal protein form, but [PIN+] and [Het-s] do not (Coustou et al. 1997; Derkatch et al. 2001) because their phenotypes are due to novel effects of the amyloid form. Modified from Wickner et al., 2013.
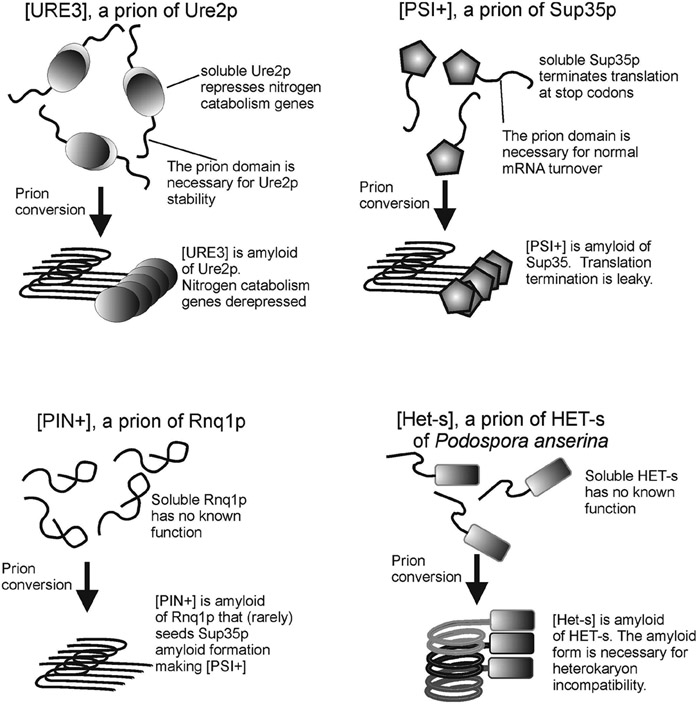
Fig.2. Yeast and fungal prions.
The prions [URE3], [PSI+], and [PIN+] of S. cerevisiae and [Het-s] of Podospora anserina are formed by amyloid formation of Ure2p, Sup35p, Rnq1p and HET-s, respectively. The proteins' normal functions and prion phenotypes are shown. The prion domains of Ure2p and Sup35p have normal functions, explaining their retention in evolution in spite of rare prion formation. The amyloids of Ure2p, Sup35p and Rnq1p are each folded in-register parallel β-sheet structures with one molecule per one β-strand layer along the long axis of the filament, while that of HET-s is a β-helix with one molecule per two β-strand layers. Modified from Wickner et al., 2008.
PRION DOMAINS
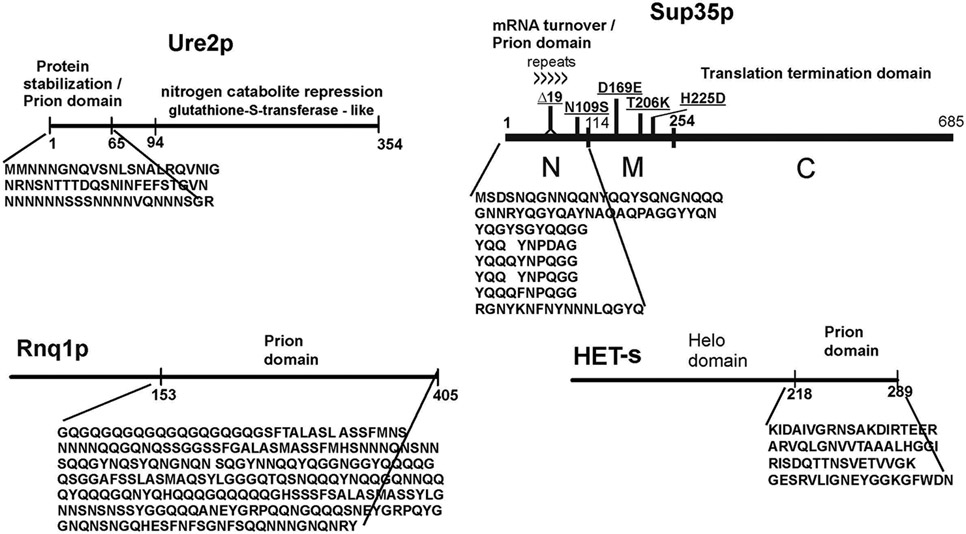
The N-terminal Q/N rich regions of Ure2p (Masison and Wickner 1995; Masison et al. 1997) and Sup35p (TerAvanesyan et al. 1994) are each sufficient for the propagation of the [URE3] and [PSI+] prions, respectively (Figure 3). The Ure2p prion domain can do so in the complete absence of the C-terminal part of the molecule, efficiently receiving the prion from the full length Ure2p, propagating the prion and transmitting again to the full length Ure2p (Masison et al. 1997). Deletions in the C-terminal domain of Ure2p can dramatically elevate the efficiency of conversion to the prion form, in vivo (Masison and Wickner 1995) and in vitro (Taylor et al. 1999). Sup35p and HET-s prion formation is also elevated by deletion of regions outside the prion domain (Kochneva-Pervukhova et al. 1998; Balguerie et al. 2003), presumably by destabilizing the native structure of the prion domain. Modifications of the Sup35M (middle) domain also affect prion propagation by affecting the ability of Hsp104 to access the filaments for cleavage (Helsen and Glover 2012). This region, enriched for charged amino acids, separates the N-terminal prion forming domain from the essential C-terminal domain.
Fig.3. Prion domains.
The portion of the molecule that actually forms amyloid and is essential and sufficient for prion propagation is shown diagramatically. Note that the precise borders of the prion domain depend on the prion variant (e.g. (Bradley and Liebman 2004; Chang et al. 2008)). The prion domains of Ure2p and Sup35p each have non-prion functions as well. The common polymorphisms of Sup35p that give rise to intraspecies barriers (Bateman and Wickner 2012) are shown. Modified from Wickner et al., 2008.
The prion domain of Ure2p is important for the stability of the molecule against protein degradation, so that without the prion domain, nitrogen catabolite regulation is defective (Shewmaker et al. 2007). The Sup35p prion domain also has non-prion functions, being important in regulating general mRNA turnover through its interaction with the polyA binding protein and polyA-degrading enzymes (Hosoda et al. 2003; Funakoshi et al. 2007). This region also affects localization of polysomes, directing some to associate with the tubulin cytoskeleton (Li et al. 2014). Thus, prion domains have non-prion functions.
Ross et al. showed that amino acid composition, not sequence, determines the ability of a segment to form a prion, so that randomly shuffling the amino acids in the Ure2p or Sup35p prion domains did not eliminate their prion-forming ability (Ross et al. 2004; Ross et al. 2005). Ross and coworkers have dissected the nature of this composition requirement. and can rather accurately predict prion forming ability (Toombs et al. 2010; Cascarina and Ross 2014). Indeed, it was possible to design prion domains de novo, synthesize the genes, and show that the 'designer' protein could be a prion in vivo (Toombs et al. 2012).
THE ROLL CALL OF PRIONS (Table 1)
Table 1.
Prions of Yeast, Fungi and Mammals
| Prion | Prion protein |
Prion phenotype | Protein function | Reference |
|---|---|---|---|---|
| TSE | PrP | transmissible spongiform encephalopathy | GPI-anchored cell surface glycoprotein, function not clear | (Prusiner 2004) |
| [URE3] | Ure2p | derepressed nitrogen catabolism genes | repression of nitrogen catabolism gene transcription (e.g. DAL5) | (Wickner 1994) |
| [PSI+] | Sup35p | readthru of translation termination codons | subunit of translation termination factor | (Wickner 1994) |
| [PIN+] | Rnq1p | rare priming of formation of other prions | non-essential; function unknown | (Derkatch et al. 2001) |
| [SWI+] | Swi1p | deficient in utilizing non-fermentable carbon sources | chromatin remodeling | (Du et al. 2008) |
| [OCT+] | Cyc8p | slow growing, with defects in mating and sporulation | subunit (with Tup1p) of transcription repressor | (Patel et al. 2009) |
| [ISP+] | Sfp1p | antisuppressor, overproduces Sup35p | transcription factor for ribosome proteins | (Rogoza et al. 2010) |
| [MOT3+] | Mot3p | filamentous growth on low nitrogen | transcription factor for hypoxic, hyperosmotic stress | (Alberti et al. 2009) |
| [MOD5+] | Mod5p | slow growth, fluconazole-resistance | tRNA isopentenyltransferase | (Suzuki et al. 2012) |
| [BETA] | Prb1p | activation of Prb1p precursor by active Prb1p | Vacuolar protease B | (Roberts and Wickner 2003) |
| [Het-s] | HET-s | heterokaryon incompatibility in Podospora anserina | heterokaryon incompatibility (no non-prion function known) | (Coustou et al. 1997) |
PrP is a non-essential mammalian cell surface protein of uncertain function whose self-propagating conversion to an amyloid form underlies the TSEs. These TSEs include scrapie of sheep, bovine spongiform encephalopathy, human Creutzfeldt-Jakob disease (in its many forms) and chronic wasting disease of elk and deer. The absence of effective treatment for these conditions and the enormous cost in time and money in their study makes the yeast and fungal prion systems particularly attractive for understanding the prion phenomena and how cells deal with them.
Sup35p is a subunit of the translation termination factor of S. cerevisiae, and its conversion to a self-propagating amyloid form results in the prion [PSI+], with deficiency of this factor producing frequent read-through of the termination codons of many yeast genes (see references above)(Fig. 2). The prion is usually assayed by read-through of premature termination codons in ADE1 or ADE2 for adenine biosynthesis. In addition to enabling selection of the [PSI+] prion, the red color of ade1 or ade2 colonies enables differentiation of 'strong' (white) or 'weak' (pink) prion variants (see below) based on the efficiency of the read-through (inversely related to the available free Sup35p) as well as the bright red clones that have lost [PSI+].
The [URE3] prion of S. cerevisiae is an amyloid form of Ure2p (see references above), a negative regulator of transcription of genes encoding the enzymes and transporters for poor nitrogen sources (Cooper 2002; Magasanik and Kaiser 2002). Ure2p is active when a good nitrogen source is available, inactive when only a poor nitrogen source is supplied or when Ure2p is in the prion form (Fig. 2). A DAL5-ADE2 fusion gene is usually used to detect or select [URE3] (Schlumpberger et al. 2001; Brachmann et al. 2005). Dal5p is the allantoate transporter and its expression is closely controlled by Ure2p (Rai et al. 1987).
The prion amyloid of Rnq1 (rich in N and Q residues) is called [PIN+] (for [PSI+] - inducibility) or [RNQ+] (Derkatch et al. 1997; Sondheimer and Lindquist 2000; Derkatch et al. 2001) (Fig. 2). [PIN+] dramatically increases the frequency with which [PSI+] arises by relatively rarely serving as a nidus for Sup35p amyloid formation. Rnq1p has no known function (Sondheimer and Lindquist 2000). In detecting the basis of the [PIN+] prion, it was shown that overproduction of other Q/N-rich proteins could also stimulate [PSI+] generation (Derkatch et al. 2001; Osherovich and Weissman 2001). This led to the discovery of several other prions, including [SWI+], [OCT+] and [MOD5+] (Du et al. 2008; Patel et al. 2009; Suzuki et al. 2012).
[SWI+] is an amyloid-based prion of Swi1p (Du et al. 2008), a subunit of a chromatin remodeling complex. Like swi1 mutants, [SWI+] strains are deficient in utilizing non-fermentable carbon sources such as raffinose, galactose, glycerol or sucrose.
[OCT+] is an amyloid-based prion of Cyc8p, a subunit (with Tup1p) of a transcription repressor (Patel et al. 2009). Like cyc8 mutants, [OCT+] cells are slow growing, with defects in mating and sporulation, and are derepressed for expression of Cyc7p, allowing growth on lactate of cyc1 mutants (Patel et al. 2009).
[ISP+] (opposite of [PSI+]) is a prion of Sfp1p, a transcriptional regulator of ribosomal protein genes and ribosome biogenesis genes (Rogoza et al. 2010). [ISP+] strains show decreased read-through of nonsense codons (the opposite of [PSI+]) in strains with certain mis-sense mutations in sup35 and sup45 as a result of increased expression of Sup35p. Sfp1p forms largely intra-nuclear aggregates in [ISP+] cells and, presumably as a result, is transmitted by cytoplasmic mixing only at a low frequency (Rogoza et al. 2010).
Mot3p is a transcription factor that represses genes for adaptation to hypoxic conditions, and positively regulates some genes involved in resisting osmotic stress (Martinez-Montanes et al. 2013). Mot3p can form an amyloid-based prion, [MOT3+] that enhances filamentous growth on poor nitrogen sources (Holmes et al. 2013).
Mod5p is tRNA isopentenyltransferase, a modifier of anticodon-adjacent residues of several tRNAs affecting their decoding properties (Dihanich et al. 1987). Unlike other yeast amyloid-based prions, Mod5p does not have a Q/N-rich region, but nonetheless forms an amyloid-based prion, [MOD+] and was found by its ability to produce a Pin-like effect (ability to enhance [PSI+] generation) (Suzuki et al. 2012). The core of the infectious amyloid of Mod5p is a 24 residue region with 6 charged residues, unlike the charge-poor prion domains of other yeast prion cores (Suzuki et al. 2012). It will be of interest to know the structure of the amyloid underlying this prion. The presence of the [MOD+] prion enhances growth in the presence of azole anti-fungal drugs, probably by allowing greater ergosterol synthesis. The prion impairs growth in the absence of the drug to a similar degree (Suzuki et al. 2012).
The [Het-s] prion of the filamentous fungus Podospora anserina is based on amyloid of the HET-s protein (Coustou et al. 1997; Maddelein et al. 2002)(Fig. 2). This prion is named for its requirement for heterokaryon incompatibility based on the polymorphic het-s / het-S locus (Rizet 1952). Fungal clones are generally not completely separate cells (unlike cells in a clone of yeast), but rather have connections through perforated septa partially separating cells in a mycelium. These perforations allow passage of organelles and other cell contents from cell to cell, making the clone like one large polynucleated cell. When two clones of P. anserina grow toward each other, there is fusion of hyphae from each clone with those of another clone. The first trial hyphal fusions test whether the two clones are identical at about a dozen polymorphic loci scattered about the fungal genome (het loci). A difference at any one of these loci results in death of the cells that underwent the trial fusion, and a barrier to further fusion. These heterokaryon incompatibility systems are designed to protect against harmful viruses and plasmids. One such polymorphic locus is het-s, with 2/3 of wild isolates having the het-s allele and 1/3 having the het-S allele. When het-S and het-s clones meet, the proper incompatibility is only shown if the HET-s protein (product of het-s) is in the prion form.
Not all prions are based on amyloid. The [BETA] prion is simply the active form of the vacuolar protease B of S. cerevisiae. Normally the inactive protease B precursor protein is activated by cleavage by protease A (Jones 1991). However, in the absence of protease A (pep4 mutant), mature active protease B itself can activate its own precursor (Zubenko et al. 1982). As a result, in a pep4 mutant, active protease B can act as a prion, with all the expected properties (Roberts and Wickner 2003). This system is somewhat artificial, because the prion property is only evident in the pep4 mutant, but in that condition, [BETA] is critical for survival in stationary phase and necessary for sporulation (Roberts and Wickner 2003). Several other non-chromosomal genetic elements have been proposed to be prions, but will require further work to confirm their status.
It is not out of place to briefly discuss what is and is not a prion. When Prusiner defined the word "prion" in 1982 (Prusiner 1982), many were at pains to argue that there was/ could be no such thing as a prion. But once we discovered yeast prions in 1994, this attitude changed to "prion envy", a phenomenon that continues to the present. The defining features of prions are a) infectivity and b) the protein-only requirement. As the preceding example illustrates, amyloid is not part of the definition, but is rather a frequent (but not invariant) property of most prions. An amyloid that is not transmissible from one individual to another is not a prion, even if it is self-propagating. Even less qualified is an amyloid whose formation and destruction is regulated by some cellular condition or environment. To be a prion, a protein must have a non-prion form that is quite stable in the absence of the prion form. If the protein, once formed, automatically falls into the amyloid form, for example, then there is no possibility of infection.
PRION VARIANTS/STRAINS AND TRANSMISSION BARRIERS
Prions are poorly transmitted between species, a phenomenon called the species barrier, and first recognized as an extended incubation period on first transmission of scrapie from sheep to goats (compared to sheep-to-sheep or goat-to-goat transmission) (Cuille and Chelle 1939). In some cases the barrier to transmission of TSEs is absolute. The basis of the species transmission barrier was shown to be the PrP amino acid sequence itself (Prusiner et al. 1990), and was one of the most convincing lines of early evidence for the prion model of scrapie because it suggested that the PrP molecule was the information-carrying molecule.
[PSI+] can be formed by the Sup35 prion domains (N) of Candida albicans, Pichia methanolica and Kluyveromyces lactis, fused to the S. cerevisiae Sup35MC (the non-prion part) and expressed in S. cerevisiae (Chernoff et al. 2000; Kushnirov et al. 2000; Santoso et al. 2000). Full length Sup35p's, also expressed in S. cerevisiae, from Candida maltosa, Debaryomyces hansenii, K. lactis, and Zygosaccharomyces rouxii could also form [PSI+] (Nakayashiki et al. 2001). Transmission barriers among these species were evident, although, of course, these species do not mate in the wild. Haploids of different species of Saccharomyces can mate with each other to produce healthy diploids, although their meiotic products are usually inviable. Transmission barriers between the Ure2p's of Saccharomyces species for transmission of [URE3] were also observed (Edskes and Wickner 2002; Edskes et al. 2009), as were barriers for the transmission of [PSI+] between the Sup35's of several Saccharomyces species (Chen et al. 2007). These barriers were based on interspecies sequence differences in the prion domains (Chen et al. 2007; Edskes et al. 2009).
Within S. cerevisiae, there are essentially three groups of Sup35p prion domain sequences (Resende et al. 2003; Bateman and Wickner 2012). These sequence differences produce substantial transmission barriers among these three polymorphs (Bateman and Wickner 2012). Rnq1p polymorphs are also found in wild S. cerevisiae strains and these too produce transmission barriers, or in some cases, complete inability to have a [PIN+] prion (Kelly et al. 2012).
A single peptide with a single amino acid sequence can form any of a large number of prion 'variants' or 'strains'. This phenomenon, long known in mammalian prions (e.g., (Bruce et al. 1991)), was first recognized in yeast prions by Derkatch and Liebman studying [PSI+] (Derkatch et al. 1996), and has been now documented in [URE3] and [PIN+] as well (Schlumpberger et al. 2001; Bradley et al. 2002; Brachmann et al. 2005). This phenomenon suggests that there must be a mechanism by which a prion can template its own structure, but since the sequence of the protein was the same in different prion variants, this must mean that conformation can be templated. Evidence for conformational differences between different strains/variants of a given prion has been found in several systems (Bessen and Marsh 1992; Toyama et al. 2007; Chang et al. 2008), but only with the elucidation of the architecture of the yeast prions has a possible mechanism for this template emerged (see below). The array of prion variants is surprisingly large (Table 2), a fact which at one time led many to doubt the existence of prions because there was not even a hypothesis about how protein conformation could be transmitted from molecule to molecule. Mammalian prion variants can be distinguished by (at least) incubation period, regions of the brain typically affect, neurological symptoms and signs, degree of protease-resistance of PrP in diseased tissue and ability to infect other species than the species of origin. Yeast prion variants have been classified by strength of the prion phenotype (strong vs. weak), stability of prion propagation, reaction to overproduction or deficiency of various chaperones and cofactors, toxicity to the host (lethal, toxic or mild), sensitivity to antiprion systems (such as Btn2/Cur1), ability to cross intraspecies or interspecies transmission barriers, or ability to propagate in an array of mutant prion proteins (Table 2).
Table 2.
Prion variants/strains
| Prion | Variant discriminant | Ref. |
|---|---|---|
| TSE | Incubation period | |
| Region of brain affected | ||
| Interspecies transmission efficiency | ||
| Clinical signs: dementia, ataxia, glucose intolerance… | ||
| [PSI+] | Strong/Weak: efficiency of terminator read-thru | (Derkatch et al. 1996) |
| Stability: directly related to seed number | (Derkatch et al. 1996) | |
| Lethal/Toxic/Mild | (McGlinchey et al. 2011) | |
| Pattern of sensitivity to intraspecies transmission barriers from polymorphic Sup35 prion domain | (Bateman and Wickner 2012; Bateman and Wickner 2013) | |
| Sensitivity to interspecies barriers | (Sharma et al. 2015) | |
| Ability to propagate in various mutant prion proteins | (King 2001) | |
| Sensitivity to chaperone over/under production | (Borchsenius et al. 2006) (Kushnirov et al. 2000) (Lancaster et al. 2013)(Harris et al. 2014) |
|
| [URE3] | Strong/Weak: level of DAL5 promoter activity | (Brachmann et al. 2005) |
| Stability: directly related to seed number | (Brachmann et al. 2005) | |
| Toxic/Mild | (McGlinchey et al. 2011) | |
| Btn2-Cur1 Sensitive/Hypersensitive | (Wickner et al. 2014) | |
| Species barrier sensitive/resistant | (Edskes et al. 2009) | |
| [PIN+] | Strong/weak: efficiency of generating [PSI+] | (Bradley et al. 2002) |
| Pattern of [PSI+] variants generated | (Sharma and Liebman 2013) |
How stable are prion variants? Some experiments using interspecies transfer of TSEs have suggested remarkable stability as long as the transmission barrier to the other species was not too great (reviewed by (Bruce 2003)). In other cases, crossing the species barrier selected a distinct variant that, when returned to the original species showed a different incubation period (e.g. (Kimberlin et al. 1987). Similar results have been found in yeast. Different [URE3] variants show different barriers to transmission to other Saccharomyces species's Ure2p (Edskes et al. 2009). Having overcome a species barrier, [URE3] variants retain their ability to propagate in their host of origin, in contrast to variants arising de novo in the new host (Edskes et al. 2009). If the species barrier to transmission is too high, as between S. cerevisiae and Pichia methanicola, the accuracy of transmission breaks down and rare mistemplated variants are selected (Vishveshwara and Liebman 2009).
The three polymorphs of Sup35p in wild strains of S. cerevisiae produce [PSI+] transmission barriers between polymorphs, called intraspecies barriers (Bateman and Wickner 2012). The extent of these barriers depends dramatically on the [PSI+] variant being tested (Bateman and Wickner 2012). Remarkably, simple extensive mitotic propagation of one strain carrying one of these variants gives rise to each of the other variants (Bateman and Wickner 2013). Since no selective pressure was applied, this result implies the existence of a mixture of variants that resulted in segregation of relatively pure isolates. However, further extensive propagation (without selection) of each of the segregated variants produced, again, each of the other variants, implying that there must be a prion mutation process that continues to renew the heterogeneity of variants within a single cell (Bateman and Wickner 2013). This "prion cloud" model was first proposed to explain the existence of prion variants in mice (Collinge and Clarke 2007), and evidence consistent with this idea were obtained using prion variants resistant to an anti-prion drug (Li et al. 2010), but in those experiments induction of variant mutation by the drug could not be eliminated. This phenomenon suggests that the treatment of amyloid diseases may be complicated by the selection of drug-resistant variants, similar to the problems in treating microbial infectious diseases. The non-selective segregation and mutation of [PSI+] variants at the prion level (Bateman and Wickner 2013) resembles the neutral genetic drift model of Kimura so prominent in explaining DNA-level evolution (Kimura 1968).
STRUCTURE OF INFECTIOUS AMYLOID OF YEAST AND FUNGAL PRIONS
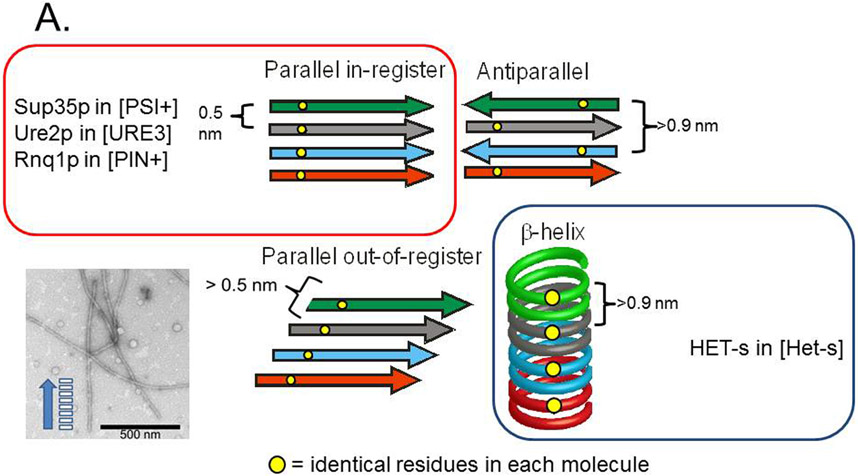
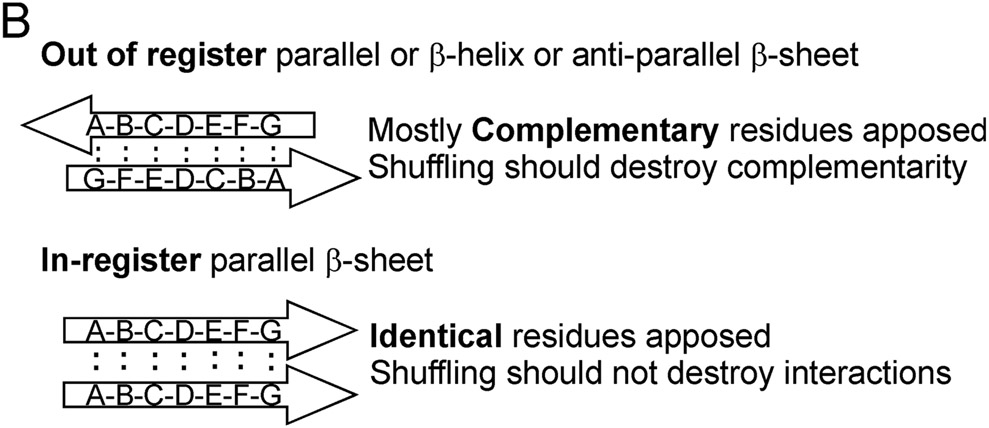
Amyloid is a filamentous unbranched polymer of proteins or peptides containing mostly β sheet structure, with the β strands perpendicular to the long axis of the filament. The major types of β sheet are anti-parallel, parallel in-register, parallel out-of-register and β helix (Fig. 4A) (Tycko 2006). Several human pathologic amyloids have been shown to be parallel in-register (reviewed in (Tycko and Wickner 2013).
Fig. 4. Possible amyloid β-sheet structures.
A. Each color represents a separate molecule, and each small circle represents a particular atom, the same in each molecule. Labeling a single C atom with 13C and using solid-state NMR to measure the distance to the nearest neighbor 13C allows one to distinguish the in-register parallel β-sheet structure (~0.5 nm expected) from the other structures (> 0.5 nm expected). B. The strict sequence specificity for prion propagation implies an interaction between residues in the filament and those of a molecule newly joining the end. If the structure is antiparallel, β-helix, or out of register, that relationship must be one of complementarity. If the structure is in-register parallel, it is identity. Shuffling is expected to destroy the complementarity relationships (as it would with DNA strands), but not identity interactions, which could still all occur, just in a different order along the peptide chain (Ross et al. 2005). Modified from Ross et al., 2005.
[Het-s]Infectious amyloid of the HET-s prion domain forms a single structure as indicated by sharp peaks on multidimensional solid-state NMR (Ritter et al. 2005; Wasmer et al. 2008). The structure is a β helix with each molecule comprising two turns of the helix (Fig. 4A)(Wasmer et al. 2008). In addition to the detailed solid-state NMR data (Ritter et al. 2005; Wasmer et al. 2008), mass per length data showed ~0.5 molecules per 4.7 angstroms, the distance between two β strands (Sen et al. 2006). This means that each molecule comprises two β strands along the long axis of the filament, exactly as expected for the β helix model (Figure 4A). In a β helix structure, at least half of the hydrogen bonds supporting the structure are intramolecular. Intramolecular bonds will be kinetically favored, and so it is likely that they form first, before intermolecular bonds, and fix the conformation of the monomer. Over a longer time frame, monomers then probably stack to form the filaments. This could explain why there is only one conformer, and only one prion variant.
Yeast prion amyloids.
The prion domains of Ure2p or Sup35p could be shuffled (simply re-ordering the amino acids without changing the amino acid content) and yet still generate prions (in all five shuffles of each prion domain) (Ross et al. 2004; Ross et al. 2005). However, prion propagation requires near sequence identity between donor and recipient molecules (e.g. (Prusiner et al. 1990)), and in some cases, even a single amino acid difference can block prion propagation (Priola et al. 1994; Santoso et al. 2000; King 2001). This seeming paradox led to the suggestion that the amyloid underlying these prions has an in-register parallel architecture (Ross et al. 2005)(Fig. 4B). Amyloids of Ure2p and Sup35p are known to have β sheet structures (Glover et al. 1997; King et al. 1997; Taylor et al. 1999). If the arrangement of β strands were antiparallel, or β helix, or parallel but out of register, the sequence specificity for propagation would have to be due to an interaction of complementarity between residues on the new molecule joining the end of the filament, and the largely non-identical residues of the last molecule already on the end of the filament. Shuffling the sequence would almost always destroy such complementarity and thus destroy prion-forming potential. However, if the structure were in-register parallel β sheet, then a residue on the molecule newly joining the end of the filament would interact with the identical residue of the molecule already present on the end of the filament. Shuffling the sequence would leave the same residues interacting with their identical partner, but just in a different order along the peptide chain (Ross et al. 2005).
Using selective labeling schemes of infectious amyloid of the prion domains of Sup35p, Ure2p and Rnq1p, and solid-state NMR, it was shown that the architecture of each is, indeed, an in-register parallel β sheet (Shewmaker et al. 2006; Baxa et al. 2007; Wickner et al. 2008; Gorkovskiy et al. 2014) (Fig. 5). The prion amyloid of the Candida albicans Ure2p prion domain also has this architecture (Engel et al. 2011). As predicted from the shuffling experiments, shuffled prion domains also had the same architecture (Shewmaker et al. 2008). These experiments show that the distance between a particular residue in the prion domain and the same residue on the next molecule in the filament is about the 4.7 angstrom distance between β strands. These results rule out antiparallel β sheets, the β helix, and parallel out-of-register structures (Fig. 4A). Electron microscopy showed that the sheets must be folded several times along the long axis of the filaments (Fig. 4 a). This architecture was supported by mass per length measurements of each of these filaments, which all show a single molecule per 4.7 angstroms length along the filament (Baxa et al. 2003; Diaz-Avalos et al. 2005; Chen et al. 2009). These measurements specifically rule out β-helix models which predict 0.5 or less of a molecule per 4.7 angstroms, as observed in amyloid of HET-s (Sen et al. 2006) (see Fig. 4A). Electron spin resonance measurements of Ure2p amyloid confirm the in-register parallel β sheet model (Ngo et al. 2011).
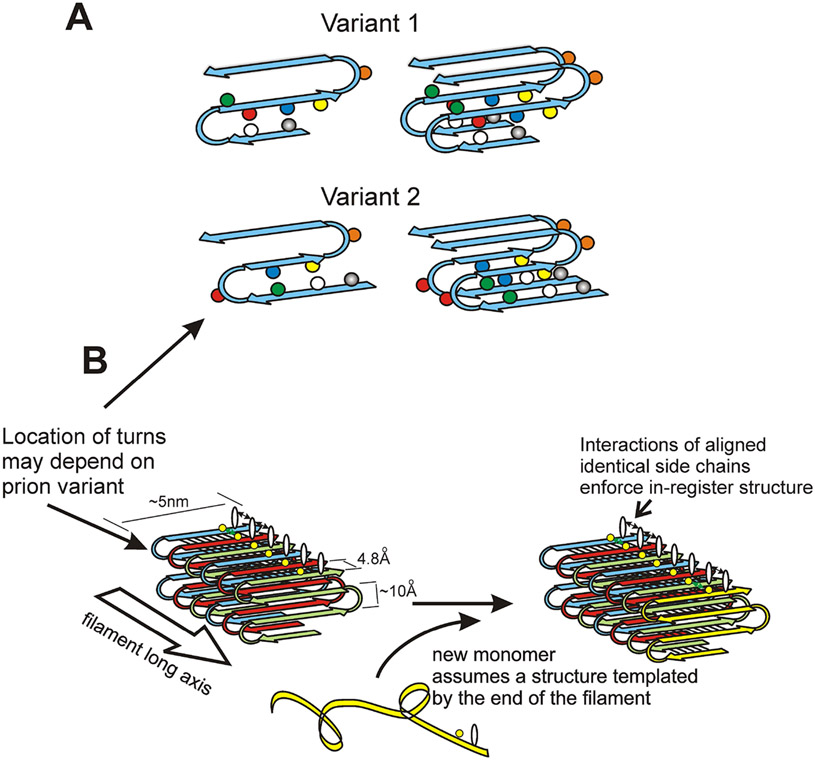
Fig. 5. Structure of the prion amyloids suggests a mechanism of conformation templating.
A. It is proposed that different prion variants have the folds of the β-sheet (turns in the molecules) in different locations (Wickner et al. 2008; Wickner et al. 2013). B. Molecules with the folded in-register parallel β-sheet architecture have rows of identical side chains along the long axis of the filament. Positive interactions (hydrogen bonds or hydrophobic interactions) between the identical side chains maintain the in-register architecture. In order to have those same positive interactions, the new unstructured prion domain joining the end of the filament must adopt a structure in which its turns are in the same location as those in the molecules already in the filament. This explains how prion proteins can template their conformation (Wickner et al. 2008). Modified from Shewmaker et al., 2006.
Recently, site-specific labeling was used to confirm the in-register parallel architecture and to find some potential locations of folds in the sheet of the Sup35p prion domain (the fold locations could differ in different prion variants as discussed below) (Gorkovskiy et al. 2014). Electron spin resonance experiments have similarly suggested the locations of folds in Ure2p amyloid (Ngo et al. 2012). Wong and King have used chemical cross-linking of amyloid filaments made of a modified Sup35p short prion domain seeded by filaments purified from either of two different [PSI+] prion strains to determine the location of folds in the β sheets of the filaments (Wong and King 2015). They detected clear differences between the filaments seeded by different variants. Importantly, the filaments generated in vitro produced only the respective original variants when transfected into yeast cells implying that the templating was faithful (Wong and King 2015). These results support the folded in-register parallel β sheet model and promise to clarify more detail than has been possible so far.
A detailed atomic-level structure of the yeast prion amyloids has so far been impossible because of inherent disorder in the filament preparations and the relatively large size of the minimal infectious peptides. However, some assignments of residues in the Sup35 prion domain have been possible with confirmation of the β sheet secondary structure of this region (Luckgei et al. 2013).
The folded in-register parallel architecture found for the yeast prions studied so far is similar to that found for nearly all of the human pathological amyloids, including Aβ peptide (Alzheimer's disease), amylin (type II diabetes), alpha-synuclein (Parkinson's disease) and others (reviewed in (Tycko 2011)). Yeast prions have thus become important model systems for the very common human amyloidoses.
YEAST PRION AMYLOID STRUCTURE EXPLAINS INHERITANCE OF CONFORMATION
A parallel in-register β sheet amyloid has rows of identical amino acid side chains extending along the long axis of the filaments (Figure 5B). The register is maintained by favorable interactions among the identical side chains. For example, a row of glutamine residues can form a row of hydrogen bonds joining the amide groups of their side chains and extending the length of the filament. The same structure can form with asparagine, serine or threonine residues. A line of any hydrophobic residues will be stabilized by hydrophobic interactions between the side chains. Charged residue side chains would be unfavorable, and there are (probably for this reason) very few charged residues in the prion domains of known prion proteins with this architecture.
The folded in-register parallel β sheet architecture of yeast prion domains leaves relatively little flexibility for variation, although a single peptide sequence can form amyloid with any of many different detailed structures, each self-propagating. The location and size of peptide chain loops at the locations of the folds, and the extent of the parallel in-register structure could well differ among prion variants (Fig. 5A). We have suggested that the same favorable interactions between identical side chains that keep the structure in-register must be the main factor allowing conformational templating by prion amyloids (Ross et al. 2005; Wickner et al. 2008; Wickner et al. 2013). The molecule joining the end of the amyloid filament will adjust its turns to be in the same location as those of the last molecule on the end of the filament in order to have these favorable hydrogen bonds and hydrophobic interactions that are found between molecules already in the filament (Figure 5B). This is the only model that has been proposed so far for the central event in prions, the templating of protein conformation.
PRION BIOLOGY
While most amyloids in humans are pathogenic, functional amyloids in various species have been described. Pmel17 is a melanosome protein that functions in an amyloid form, and is important for melanin biogenesis, (Fowler et al. 2006). Fish embryos (Podrabsky et al. 2001) and fungal cell surfaces (Mackay et al. 2001) use amyloids as a protective layer. Proteins forming a gel in the zona pelludica of mouse eggs are amyloids (Egge et al. 2015), and regulated amyloid formation of Rim4p in S. cerevisiae controls stages of meiosis (Berchowitz et al. 2015). Are some prions functional?
The [Het-s] prion of Podospora anserina
The het-s/het-S system has at least two, and probably three effects on its host, P. anserina. The HET-s protein must be in the amyloid ([Het-s] prion) form in order that a het-s/het-S heterokaryon show the proper incompatibility with programmed death of the heterokaryon cells and formation of a barrier to further hyphal fusions (Rizet 1952; Coustou et al. 1997). As mentioned earlier, the heterokaryon (vegetative) incompatibility reaction is a self/non-self recognition system seen in most or all filamentous fungi whose function is to limit the spread of harmful viruses and plasmids (Caten 1972; Hartl et al. 1975; Debets et al. 1994). In fact the het-s/het-S system can block transmission of a harmful plasmid in Podospora (Debets et al. 2012). It was thus proposed that this was the first case of a functional prion (Coustou et al. 1997; Wickner 1997). However, in contrast to this effect on the mitotic heterokaryon formation, [Het-s] has another, less favorable, effect, this on the meiotic cycle. Mating of female het-s [Het-s] strains with male het-S strains results in a high frequency selective death of het-S meiotic segregants (Bernet 1965; Dalstra et al. 2003). Crossing het-s [Het-s*] (no prion) with het-S strains does not produce this spore lethality. This is an example of meiotic drive, a gene or locus promoting its spread in the population, not by favoring fitness of the individual, but by impairing inheritance of other alleles at that locus. Segregation Distorter of Drosophila melanogaster and the "t locus" of mice are well known examples. The meiotic drive effect of [Het-s] favors the inheritance of the het-s allele and could result in elimination of the het-S allele. However, the heterokaryon incompatibility function of the het-s/het-S locus is most effective if the two alleles are present at roughly equal frequency in the population (Debets et al. 2012). That 2/3 of wild strains have the het-s genotype is explained as a balance between these two effects (Debets et al. 2012). As expected for a beneficial infectious element, [Het-s] is present in 92% of wild het-s strains (Debets et al. 2012).
Adjacent to the het-s/S gene is nwd2, encoding a gene with similarity to a family of pathogen recognition proteins of plants and animals, called STAND proteins, and including an N-terminal domain similar to the prion domain of HET-s. The nwd2 gene is disrupted in all het-s strains, but intact in het-S strains (Debets et al. 2012). It is proposed that an as yet unknown ligand of NWD2 can induce amyloid formation by NWD2 and that, like the cell death induced by the interaction of HET-S with the prion amyloid of HET-s, the amyloid of NWD2 induces HET-S to cause cell death (Paoletti and Saupe 2009). Evidence for this reaction has been obtained, although the nature of the normal signal remains unknown (Daskalov et al. 2015). It is proposed that from this NWD2 : HET-S interaction evolved the [Het-s] prion by successive disruption of nwd2, and mutation of het-S to het-s (Daskalov and Saupe 2015).
These three biological effects of the het-s/S locus have combined to shape its evolution. This system is so far unique in being a demonstrated beneficial prion, but with side-effects whose details and implications are continuing to be elucidated.
Yeast Prions.
Can they be beneficial?
Following the discovery of the beneficial [Het-s] prion, it was reported that the yeast [PSI+] prion made cells resistant to the effects of high temperature or high ethanol concentration and that [PSI+] was thus an advantage to yeast (Eaglestone et al. 1999). A subsequent study compared five [PSI+] and [psi−] isogenic strain pairs and did not find protection against heat or ethanol by [PSI+], nor any consistent protection by [PSI+] under any condition (True and Lindquist 2000). However, in about 1/4 of the conditions where [PSI+] made a difference in a particular strain pair, the [PSI+] strain grew better (True and Lindquist 2000). It was inferred that [PSI+] could help yeast evolve by helping some strains under some conditions. Another group, using the same strains, could not reproduce these results (Namy et al. 2008). A later study, reporting that in the rare [PSI+] wild strains, the prion aided cell growth under certain conditions (Halfmann et al. 2012) was again not confirmed using the same strains (Wickner et al. 2015). Thus, it remains controversial whether [PSI+] is beneficial under any but the sort of artificial conditions used in the assay (read-through of a premature termination codon in a biosynthetic gene).
It may be impossible to resolve the issue of whether [PSI+] or [URE3] are beneficial or detrimental by laboratory tests. Even finding [PSI+] or [URE3] to be reproducibly beneficial under some condition, would not imply that such condition constitutes a significant part of the S. cerevisiae ecological niche. A report that certain stress conditions induce the appearance of [PSI+] also included the finding that under most such conditions, [PSI+] was detrimental (Tyedmers et al. 2008), and the stress-induction could not be repeated (Kelly et al. 2012; Westergard and True 2014).
Can [PSI+] and [URE3] be toxic/lethal?
[PSI+] and [URE3] can be quite detrimental or even lethal: most [URE3] isolates slow growth noticeably (Schwimmer and Masison 2002) and most variants are highly toxic (McGlinchey et al. 2011). Isolating [PSI+] in the presence of minimal expression of Sup35C (lacking the prion domain) allowed detection of lethal and near-lethal variants of [PSI+], which were, again, more common than the mild form of the prion (McGlinchey et al. 2011). Note that the studies reporting benefits of yeast prions only considered the mildest variants (Eaglestone et al. 1999; True and Lindquist 2000; Halfmann et al. 2012). A realistic estimate of the fitness value of a cell's forming a prion must weigh the potential benefits against the real risks of prion formation. [PSI+], [URE3], [PIN+] and [SWI+] in wild strains. Even the uniformly fatal mammalian prion diseases are not rare in the wild. Chronic Wasting disease of deer and elk are epidemic in many parts of the US (Saunders et al. 2012). Like other lethal infectious viral or bacterial diseases, their infectivity outruns their destruction of their hosts. If there were an infectious agent that actually benefited its host, infectivity and host effects would both act to spread the agent in the population, and it would soon be found in most isolates. The [Het-s] prion, found in 92% of wild het-s isolates (Debets et al. 2012), is a clear example of this general idea. Therefore, an infectious agent that is rare in the wild must be detrimental to its host. Examining the incidence of [PSI+] and [URE3] in the wild is a measure of how beneficial/detrimental are the mildest, most stable variants of these prions, and gives only an upper limit on any potential positive effect on the fitness of maintaining prion-forming ability.
Two early studies found no [PSI+] isolates among a few wild strains, but did find two [PIN+] strains (Chernoff et al. 2000; Resende et al. 2003). A further study of 70 wild isolates found none with [PSI+] or [URE3], but again did detect [PIN+] in 11 of the strains (Nakayashiki et al. 2005). The selfish RNA viruses, L-BC, 20S and 23S RNA and the DNA plasmid 2 micron DNA, encode only factors promoting their own replication and segregation, but were found in 8, 14, 1 and 38 of the 70 strains, respectively (Nakayashiki et al. 2005). The 2 micron DNA plasmid was particularly useful because studies by three groups have shown that this parasitic plasmid slows cell growth by 1-3% (Futcher and Cox 1983; Mead et al. 1986; Futcher et al. 1988; Kelly et al. 2012). The 2 micron DNA plasmid is somewhat unstable in mitosis, and can only arise de novo over geologic time, whereas nonlethal [URE3], [PSI+] and [PIN+] arise about once in 106 cells. Nonetheless, these three prions are less widespread, implying that even their mildest variants must have a greater detrimental effect than the 1-3% growth slowing shown for 2 micron DNA (Kelly et al. 2012). Another larger study of 690 wild isolates found 9 [PSI+] strains (Halfmann et al. 2012), statistically indistinguishable from the 0 of 70 in the earlier report (Nakayashiki et al. 2005). The incidence of [PIN+] was slightly lower, at 6%.
Although the low frequency of [PIN+] indicates, by this comparison with the 2 micron DNA plasmid, that [PIN+] is detrimental, the frequency of its occurrence in nature is still greater than the frequency of its arising de novo. The spread of [PIN+] could be a result of selection for an advantageous trait determined by [PIN+] or it could be a result of outcross mating (Kelly et al. 2014). As predicted by the latter explanation wild [PIN+] strains tend to show evidence of having arisen from outcross matings, namely, heterozygosity for common polymorphic alleles (Kelly et al. 2014). The report that 1/3 of wild strains had guanidine - curable (and thus prion-determined) traits could not be reproduced with a sample of these strains (Kryndushkin et al. 2013). The [SWI+] prion was also absent from the 70 strain collection, indicating that it too has a negative effect on fitness (Bateman and Wickner 2012).
Species Distribution of Prion-forming Ability.
The N-terminal domains of Sup35s of several species have been found capable of substituting for the S. cerevisiae Sup35p prion domain in forming a prion in S. cerevisiae cells: Sup35N from Candida albicans, Kluyveromyces lactis, Pichia methanolica and several species of Saccharomyces (Chernoff et al. 2000; Kushnirov et al. 2000; Santoso et al. 2000; Nakayashiki et al. 2001; Chen et al. 2007; Afanasieva et al. 2011). These results were interpreted as implying conservation of a desirable function (prion formation) selected by evolution. However, a wider survey of Sup35s from yeasts and fungi showed that the Sup35s of many species, including Schizosaccharomyces pombe, Ashbya gossypii, Aspergillus nidulans, Aspergillus fumigatus, Magnaporthe grisea, Ustilago maydis and Cryptococcus neoformans could not form prions by the same test (Edskes et al. 2014). Of course, the presence of a trait in many species does not imply that that trait is a function conserved for the organism's benefit. Occasional broken limbs is a trait widely found in vertebrates.
[URE3] formation by full length Ure2p from other Saccharomyces species has likewise been tested in S. cerevisiae, and most can do so (Edskes and Wickner 2002; Baudin-Baillieu et al. 2003; Edskes et al. 2009), but that of Saccharomyces castellii cannot (Edskes et al. 2009). The Ure2p of Kluyveromyces lactis is unable to form [URE3] in either S. cerevisiae or in K. lactis itself (Safadi et al. 2011). The Ure2p of Candida albicans can form [URE3] in either S. cerevisiae or in Candida glabrata, but the Ure2p of C. glabrata is unable to form the prion in either S. cerevisiae or its native host, C. glabrata (Edskes et al. 2011; Engel et al. 2011; Edskes and Wickner 2013). Notably, the Ure2p of C. glabrata is very close in sequence to that of S. cerevisiae, but cannot form a prion, while the more distantly related C. albicans Ure2p can form a prion. This is not what is expected for conservation of prion formation.
All of the studies comparing the putative prion domains (which do not always form prions) of Sup35p and Ure2p have shown that the rate of evolutionary change is far greater for these domains than for the remainder of the molecule (e.g. (Santoso et al. 2000; Edskes and Wickner 2002; Bateman and Wickner 2012)). Because even a single amino acid difference between the prion protein of the infection donor and recipient can, in some cases, block propagation of a prion (e.g. (Priola et al. 1994; Santoso et al. 2000; King 2001), the accumulation of mutations in the prion domains can be viewed as selected to block infection of the organism with a potentially lethal disease. Kuru is an epidemic form of Creutzfeldt-Jakob disease that occurred among the Fore people of New Guinea as a result of their ritual funereal cannibal custom (Collinge and Alpers 2008). This epidemic led to the emergence of the PrP G127V mutation because it confers complete resistance to the disease (Asante et al. 2015). The polymorphs found among S. cerevisiae strains in the Sup35p prion domain likewise produce a barrier to transmission of [PSI+] and are likely to have been selected for this reason (Bateman and Wickner 2012). The prion domains of Ure2p and Sup35p are known to have important functions (see above), and so have not been simply lost. Their composition has likely been selected to minimize the frequency of prion formation given the constraints of these non-prion functions. Paul et al. found that proteins with composition somewhat similar to that of prions could be converted to prions by a very small number of changes, suggesting that their composition has been selected to minimize prion formation (Paul et al. 2015).
It remains possible that there is some special niche of yeast in which one or more of these prions is helpful for the host, but a general role in aiding stress-tolerance (Eaglestone et al. 1999) or evolvability (True and Lindquist 2000), as has been proposed by others, seems unlikely. Some have proposed a 'bet hedging' role for prion generation under stress (Newby and Lindquist 2013), but the prevalence of lethal or highly toxic [URE3] and [PSI+] variants makes prion generation a bad bet for the cell.
Other yeast prions.
Pseudohyphal growth is a highly regulated process which allows starved cells to search for nutrients (Gimeno et al. 1992). The [MOT3+] prion can affect development of pseudohyphal growth by the absence of Mot3p's normal regulatory effect on FLO11 transcription (Holmes et al. 2013). While it is argued that [MOT3+] is thus adaptive, it would seem that, like other prions affecting gene expression, [MOT3+] simply reduces the flexibility of cells to adapt to changing conditions. [MOT3+] was found in six of 96 isolates (Halfmann et al. 2012). Other yeast prions (Table 1) have not yet been surveyed in wild strains or studied in enough detail to know if they are advantageous or detrimental.
It is often argued that prions or retrotransposons are desirable agents because they generate the diversity that natural selection needs in order that organisms may evolve in competition with other organisms or adapt to changing environments. It should be noted that diversity is not in short supply in the biological world. A myriad of assaults on our DNA genomes are opposed by a similarly bewildering array of repair and recombination processes to limit the changes and attempt to pass on a relatively intact version of the genome to the next generation. These repair processes are inevitably imperfect, as the diversity we see testifies, so there is no need for a 'diversifier' function. The immune system's diversifying activity may seem an exception to this rule, but because the diversifications are not inherited, this is rather a case of differentiation of cell function. It would seem that selecting for a mutator in a sexual organism is limited by the fact that most, like the prions, produce mostly detrimental changes.
CELLULAR SYSTEMS AFFECTING YEAST PRIONS.
Chaperones and chaperone co-factors.
The early recognition of the critical role of the disaggregating chaperone, Hsp104 in prion propagation (Chernoff et al. 1995) opened this area as a means of learning features of prions as well as a method of studying chaperones (reviewed by (Masison and Reidy 2015)). Hsp104 is necessary for the propagation of all of the amyloid-based yeast prions (Chernoff et al. 1995; Derkatch et al. 1997; Moriyama et al. 2000; Du et al. 2008; Alberti et al. 2009; Patel et al. 2009; Suzuki et al. 2012) except [ISP+] (Volkov et al. 2002). Overexpression of Hsp104 cures [PSI+] (Chernoff and Ono 1992; Chernoff et al. 1995), but not other prions.
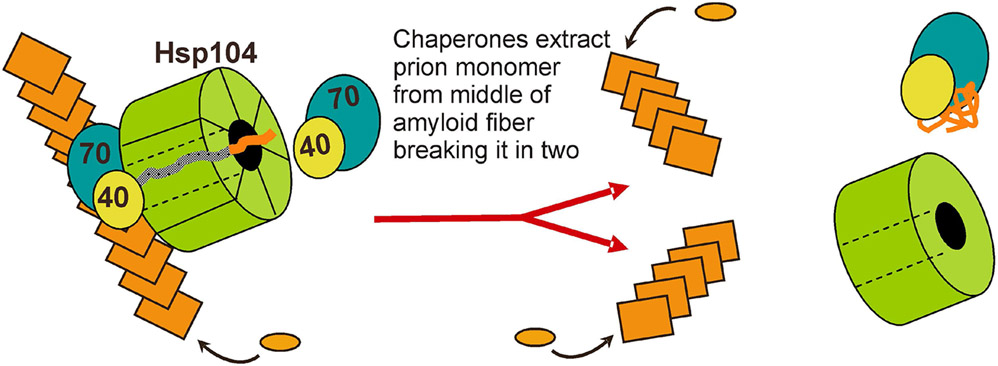
Millimolar guanidine HCl cures [PSI+] (Tuite et al. 1981) and indeed all of the amyloid based yeast prions. This prion curing effect is due to inhibition of Hsp104 (Ferreira et al. 2001; Jung and Masison 2001; Jung et al. 2002; Grimminger et al. 2004). Hsp104's disaggregase activity includes the ability to pull the chain of the aggregated protein through a pore formed by the hexameric Hsp104, thereby denaturing it and giving it a new chance to renature (Lum et al. 2004; Weibezahn et al. 2004). Mutations impairing this activity also impair prion propagation proportionately (Hung and Masison 2006; Tessarz et al. 2008). This suggests that Hsp104 (with collaborating factors) pull a molecule from the middle of an amyloid fiber, thereby breaking the fiber and creating new seeds/propagons (Fig. 6). [Note that 'seed' means prion propagating unit in the cell, presumably a single amyloid filament.] Based on the evidence that guanidine inhibition of Hsp104 blocks the splitting of prion amyloid (Paushkin et al. 1996; Ness et al. 2002; Kryndushkin et al. 2003), and thus the generation of new seeds, seed-counting methods were developed that have been useful in further studies (Cox et al. 2003).
Fig. 6. Chaperone-catalyzed filament cleavage generates new prion seeds.
Hsp104, in collaboration with Hsp70 and Hsp40, extract a prion protein monomer from the middle of a filament, breaking the filament into two filaments, each of which can continue to grow and propagate the prion. Adapted from reference (Masison et al. 2009).
Hsp104 acts with Hsp40s and Hsp70s to disaggregate proteins in vitro (Glover and Lindquist 1998), and it was soon learned that Hsp70s affect both the overproduction curing of [PSI+] by Hsp104 (Newnam et al. 1999), and are necessary for its stable propagation (Jung et al. 2000; Jones and Masison 2003; Hines et al. 2011). Hsp40s are also needed for prion propagation (Sondheimer et al. 2001; Aron et al. 2007; Higurashi et al. 2008; Tipton et al. 2008; Hines et al. 2011; Troisi et al. 2015). This collaboration was confirmed by showing that ClpB, the E. coli homolog of Hsp104, cannot propagate the yeast prions [PSI+], [URE3] or [PIN+] when substituted for Hsp104, but when the E. coli Hsp70 (DnaK) and its nucleotide exchange factor (GrpE) are also expressed, then ClpB can fulfill both the heat shock - resistance and prion propagation roles of Hsp104 (Reidy and Masison 2012). Substitution of the ClpB M domain, known to interact with Hsp70s, with that of Hsp104 also suffices to allow its functioning in yeast (Reidy and Masison 2012). The Hsp104 of S. pombe can substitute for that of S. cerevisiae without other S. pombe components (Reidy et al. 2013).
In addition to being required for propagation of all amyloid-based yeast prions, Hsp104, when overproduced, cures [PSI+] (and not others) (Chernoff et al. 1995). Several lines of evidence indicate that the [PSI+]-curing activity of Hsp104 is quite distinct from its prion-propagating (seed-producing) activity. Mutations in the N-terminal domain of Hsp104, or deletion of the entire N-terminal region to residue 147 does not adversely affect prion propagation or heat shock tolerance, but completely eliminates the ability of Hsp104 overexpression to cure [PSI+] (Hung and Masison 2006). Other cellular components that affect Hsp104 overproduction curing do not affect Hsp104 promoted prion propagation: the ribosome-associated Hsp70s, Ssb1 and Ssb2, assist Hsp104 overproduction curing (Allen et al. 2005); Sti1p, a cochaperone that interacts with Hsp90, Hsp70 and Hsp104, is necessary for Hsp104 overproduction curing (Reidy and Masison 2010); inhibition of Hsp90 with radicicol blocks Hsp104 curing (Reidy and Masison 2010); and increasing or decreasing ubiquitin expression increases or decreases Hsp104 curing (Chernova et al. 2003; Allen et al. 2007). Guanidine, which inhibits the "severing" activity, still inhibits Hsp104 when the chaperone is overexpressed, and yet guanidine has little effect on the efficiency of Hsp104 overproduction curing (Park et al. 2014). This data indicates that curing by Hsp104 overexpression is not a result of inhibition of the activity.
Nystrom's group has described a system that assymetrically segregates damaged (e.g. oxidized) proteins, with preferential retention in the mother cell (reviewed in (Nystrom and Liu 2014)). This system involves Hsp104 (e.g.(Erjavec et al. 2007)) and so it was possible that Hsp104 overproduction was curing [PSI+] by assymetric segregation of the prion aggregates. However, sorting of cells being cured of [PSI+] by Hsp104 overproduction curing gave equal curing of mother and daughter cells (Park et al. 2014). Further, the distribution of prion aggregates between mother and bud during Hsp104 overproduction was in proportion to their relative volumes (Park et al. 2014).
Direct observation of cells undergoing [PSI+] curing by Hsp104 overexpression indicate that the foci are being gradually dissolved (Park et al. 2014). It is suggested that this represents removal of Sup35p molecules from the ends of the filaments ("trimming"). The influence of ubiquitin and other chaperone factors on Hsp104 overproduction curing is explained by proteasome degradation of the amyloid core after trimming (Park et al. 2014).
The chaperone environment also shapes what prion variants arise and the frequency of prion generation. Ssb1 and Ssb2 are nearly identical ribosome-associated members of the Hsp70 family, and are believed to aid in the proper folding of newly synthesized proteins (Nelson et al. 1992; James et al. 1997). The Ssb's are attached to the ribosome by another Hsp70 family member, Ssz1, and an Hsp40, Zuo1p (Gautschi et al. 2001; Huang et al. 2005). Overproduction of Ssb1/2 increases the efficiency with which Hsp104 overproduction cures [PSI+] (Chernoff et al. 1999), showing that the Ssb's have anti prion effects. Deletion of both SSB genes results in an increase in [PSI+] generation, either spontaneously or induced by overexpression of the Sup35 prion domain (Chernoff et al. 1999), and this induction is independent of [PIN+] (Kiktev et al. 2015). Like ssb1 ssb2 strains, mutations of zou1 or ssz1 also increase the rate of [PSI+] generation (Amor et al. 2015; Kiktev et al. 2015), but have the opposite effects of ssb1 ssb2 on overproduction curing by Hsp104 (Kiktev et al. 2015). Disruption of zou1 or ssz1 results in freeing of much of the Ssb1/2 from the ribosomes, where it competes with Ssa's for binding to Sup35p amyloid, thereby, it is proposed, aiding in the Hsp104 overproduction curing (Kiktev et al. 2015).
Overexpression of Hsp104 increases [URE3] prion generation by the Ure2p of Candida albicans by 1000-fold (Kryndushkin et al. 2011). Sis1p is necessary for propagation of all of the yeast prions tested (Higurashi et al. 2008), but not all of the protein is needed (Kirkland et al. 2011). Interestingly, deletion of either the C-terminal domain or the adjacent glycine - methionine rich region as well, resulted in a normally mild variant of [PSI+] being highly toxic or lethal (Kirkland et al. 2011). The mechanism of toxicity in these cells would be of considerable interest.
As mentioned above, inhibition of Hsp90 with radicicol inhibits curing of [PSI+] by overproduction of Hsp104. Recently, evidence has appeared that [URE3] propagation requires interaction of Hsp90 with its co-chaperone Cpr7 (Kumar et al. 2015). Deletion of either the C-terminal motif of Hsp90 that interacts with Cpr7p, or deletion of the CPR7 gene itself results in inability to propagate [URE3] (Kumar et al. 2015). The precise role of Hsp90s and their co-chaperones in [URE3] propagation remain to be determined.
Btn2/Cur1 cure [URE3] (not [PSI+]) by sequestering seeds
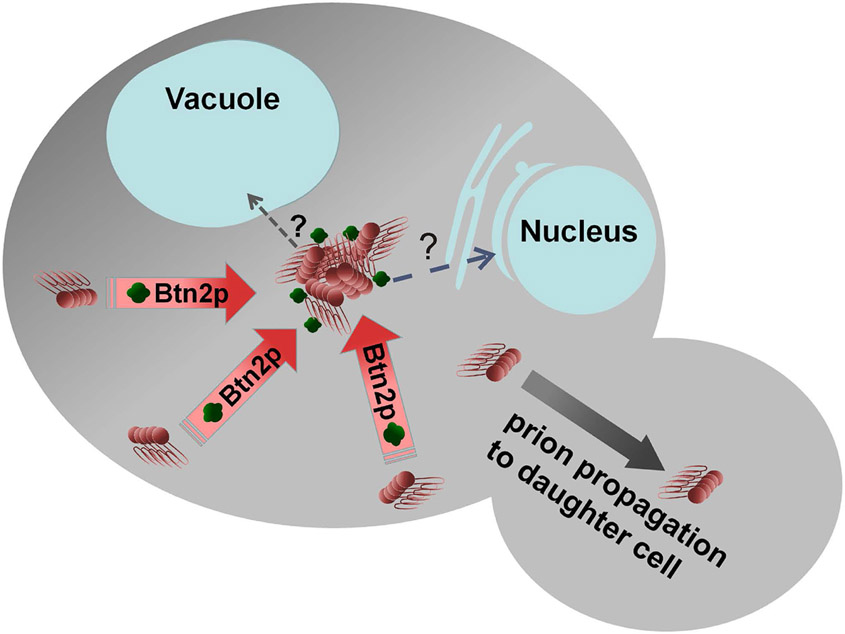
A screen for proteins whose overproduction cures [URE3] produced Btn2p and its paralog, Cur1p (Kryndushkin et al. 2008). Overproduced Btn2p or Cur1p also cure an artificial prion (Malinovska et al. 2012). Btn2p is involved in endosomal protein sorting (Kama et al. 2007) and is distantly related to human HOOK1, a member of a family of proteins involved in transport of large cargos along the microtubules (Maldonado-Baez et al. 2013). In cells in which overproduced Btn2p is curing [URE3], the aggregates of Ure2p appear collected at a single site coincident with Btn2p (Kryndushkin et al. 2008) (Fig. 7). Most [URE3] prion variants isolated in the absence of Btn2p and Cur1p can be cured by replacement of normal levels of either protein (Wickner et al. 2014). As discussed above, there are many methods of curing yeast prions, particularly by over- or under-expressing some chaperones or other cellular components, but Btn2p and Cur1p appear to be acting to cure the [URE3] prion at their normal expression levels. The distinction between which [URE3] variants are cured by normal levels of Btn2p and Cur1p and which are only cured by overexpression appears to be the seed number. Those [URE3] with low seed number (and thus a 'weak' phenotype) are cured by normal levels of Btn2p and Cur1p while the higher seed number [URE3]s are only cured on overproduction of the antiprion components (Wickner et al. 2014). Hsp42 is known to function in collecting aggregated proteins (Specht et al. 2011), and it interacts in vivo with Btn2p (Malinovska et al. 2012). Hsp42 was found to be important for the prion - curing action of overproduced Btn2p, but neither of Btn2p or Cur1p is dependent on the other in their curing (Wickner et al. 2014). Overproduction of Hsp42 also cures [URE3] in a process dependent on Cur1p (Wickner et al. 2014).
Fig. 7. Btn2p collects protein aggregates, thereby curing the [URE3] prion.
Btn2p, acting with Hsp42, gathers prion filaments (and other aggregates) to one place in the cell. Following cell division, one of the daughter cells is prion-free. Low seed number [URE3] prions are cured by normal levels of Btn2p, while higher propagon number [URE3]s are only cured on overproduction of Btn2p. Modified from Wickner et al., 2015.
Btn2p also collects non-prion non-amyloid aggregates (Kryndushkin et al. 2012; Malinovska et al. 2012), however overproduced Btn2p or Cur1p do not cure a [PSI+] prion variant, although there is some degree of colocalization of Btn2p and Sup35p in such cells(Kryndushkin et al. 2008). Cur1p overproduction also increases lethality of [PSI+] in strains heterozygous for a sup45 missense mutation (Kiktev et al. 2011), again suggesting interaction of Cur1p with the Sup35p amyloid. The site to which Btn2p appears to bring Ure2p aggregates in a [URE3] cell is not clear at this time, and there is some disagreement about where non-prion aggregates are collected (Malinovska et al. 2012; Miller et al. 2015). Moreover, the relation of Btn2p's prion-curing activity and its endosome transport role remains to be elucidated.
There is a wide array of aggregate-accumulating sites defined recently in yeast. These include the IPOD (Kaganovich et al. 2008), JUNQ (Kaganovich et al. 2008), aggresome (Wang et al. 2009), P-bodies (Sheth and Parker 2003), Q-bodies (Escusa-Toret et al. 2013), age-related aggregates (Aguilaniu et al. 2003), Btn2p-sites (Kryndushkin et al. 2008), Hsp42-sites (Specht et al. 2011) and stress granules (Hoyle et al. 2007).
Actin cytoskeleton, ubiquitin, heat shock and prions.
Although prolonged growth at elevated temperatures does not cure yeast prions, a short term heat shock (30 - 60 min at 39 C) cures a weak [PSI+] variant from a substantial proportion of cells (Newnam et al. 2011). This curing occurs with an asymmetric preferential loss of [PSI+] from daughter cells (Newnam et al. 2011; Ali et al. 2014). [PSI+] propagation is also impaired by growth in the presence of lactrunculin A, a drug that disrupts the actin cytoskeleton (Bailleul-Winslett et al. 2000), or by mutation of sla1, a gene involved in actin cytoskeleton assembly (Bailleul et al. 1999). Sla1p interacts with the Sup35p prion domain in two-hybrid tests (Bailleul et al. 1999). Las17p is an actin nucleation promoting factor, and Lsb1 and Lsb2 are paralogous binding partners of Las17. An lsb1 or lsb2 deletion (or both) destabilize [PSI+] after heat shock (Chernova et al. 2011; Ali et al. 2014), and the stabilizing activity of Lsb1p requires its proteolytic processing (Ali et al. 2014). Lsb2p also has Pin+ activity when overproduced (Derkatch et al. 2001), an effect that is limited by the ubiquitin-mediated rapid turnover of Lsb2p (Chernova et al. 2011). Finally, ubc4Δ (a ubiquitin conjugating enzyme) or ubp6Δ (a ubiquitin deconjugating enzyme) result in decreased curing of [PSI+] by Hsp104 overproduction and increased generation of [PSI+] (Allen et al. 2007). These observations indicate that the ubiquitin system antagonizes [PSI+] generation, but the authors could not detect ubiquitin conjugation of Sup35p itself (Allen et al. 2007).
Perspectives and Controversies.
While the yeast prion world has made a strong showing in spite of being massively outnumbered by those studying human and animal prions, there are a number of outstanding controversies which will only be resolved by more work and development of better methods.
Structure of prion amyloids.
Although the folded in-register parallel β sheet architecture of yeast prions appears to be settled, this does not constitute an atomic level 'structure'. While HET-s naturally forms uniform filaments, such is apparently not the case for the yeast prions that have been studied thus far. Even seeding with extracts or extracted filaments from a cells with a single apparent prion variant does not appear to produce filaments with the uniformity of the HET-s filaments. However, some progress with this approach has been made (Kryndushkin et al. 2011; Frederick et al. 2014; Wong and King 2015). The approach of seeding from cells may be limited by the inherent heterogeneity of prions in the cells themselves, as shown in the 'prion cloud' phenomenon (Bateman and Wickner 2013).
Biology.
While it seems settled that the Podospora anserina prion [Het-s] is largely beneficial and the yeast prions are usually detrimental to their host, it remains possible that there are some circumstances, not yet defined, when the latter may prove to be a benefit. Some yeast prions not yet discovered or well characterized may be advantageous, or even a well studied prion may be found to be common in yeast isolated from a particular niche.
Prion - handling systems.
Whether Hsp104 can act alone to sever fibers, the location of Btn2p (with and without aggregates), the precise role of ubiquitin in prion propagation, the role of the many sites of aggregate accumulation in prion phenomena, … and many other issues of mechanism remain for further work. The Btn2/Cur1 system works on [URE3], but not on [PSI+] or [PIN+]; are there other systems that cure these prions (the Ssb's block [PSI+] generation)?
New Prions.
The large number of prions found in yeast suggests that there are many more to be found in other organisms. Biochemical methods to search for amyloids (e.g. (Kryndushkin et al. 2013)) are more widely applicable while genetic methods depend on special features of an organisms life cycle. Recent developments in the study of amyloidoses suggest that the TSEs are not the only mammalian prions (reviewed by (Jucker and Walker 2013)). Mice with apolipoprotein AII amyloidosis appear to transmit the disease to animals in the same cage (Xing et al. 2001), and amyloidosis A transmission between animals without innoculation has been shown (Zhang et al. 2008). Infectivity by inoculation for mouse models of Alzheimer's disease (Jucker and Walker 2013), and of mice with human Alzheimer's brain (Watts et al. 2014) has now been observed. Finally, evidence of iatrogenic infection with Alzheimer's disease in patients treated with human pituitary-derived growth hormone has appeared (Naunmuktane et al. 2015).
Box 1: Abbreviations and yeast genetic nomenclature.
Yeast prions are non-chromosomal genes and therefore are shown in brackets, e.g. [URE3]. Yeast prions are dominant and so, like dominant chromosomal mutants, are shown in capitals, often italicized. Chromosomal genes are given in italics, e.g., URE2, and recessive (usually mutant) alleles are lower case, e.g. ure2. Protein products of a S. cerevisiae gene are shown as, for example, Ure2p. For some prions, "+" indicates the presence of the prion and "−" or "−o" its absence. For all, the absence of the prion is indicated by the name in lower case, e.g., [psi−] or [ure-o].
Sup35p = subunit of the translation termination factor and the protein that can form the [PSI+] prion.
Ure2p = negative regulator of transcription of genes encoding enzymes and transporters for poor nitrogen sources; active in the presence of a good nitrogen source. Forms the [URE3] prion.
Rnq1p = basis of the [PIN+] prion (also called [RNQ+].
TSE = transmissible spongiform encephalopathy
Acknowledgement:
This work was supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health.
References:
- Afanasieva EG, Kushnirov VV, Tuite MF & Ter-Avanesyan MD (2011). Molecular basis for transmission barrier and interference between closely related prion proteins in yeast. J. Biol. Chem, 286, 15773 – 15780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilaniu H, Gustafsson L, Gigoulet M & Nystrom T (2003). Assymetric inheritance of oxidatively damaged proteins during cytokinesis. Science, 299, 1751 – 1753. [DOI] [PubMed] [Google Scholar]
- Aguzzi A & Lakkaraiu AK (2015). Cell biology of prions and prionoids: a status report. Trends in Cell Biol., 25. [DOI] [PubMed] [Google Scholar]
- Alberti S, Halfmann R, King O, Kapila A & Lindquist S (2009). A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell, 137, 146 – 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M, Chernova TA, Newnam GP, Yin L, Shanks J, Karpova TS, Lee A, Laur O, Subramanian S, Kim D, McNally JG, Seyfried NT, Chernoff YO & Wilkinson KD (2014). Stress-dependent proteolytic processing of the actin assembly protein Lsb1 modulates a yeast prion. J. Biol. Chem, 289, 27625 – 27639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen KD, Chernova TA, Tennant EP, Wilkinson KD & Chernoff YO (2007). Effects of ubiquitin system alterations on the formation and loss of a yeast prion. J Biol Chem, 282, 3004–13. [DOI] [PubMed] [Google Scholar]
- Allen KD, Wegrzyn RD, Chernova TA, Muller S, Newnam GP, Winslett PA, Wittich KB, Wilkinson KD & Chernoff YO (2005). Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]. Genetics, 169, 1227–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor AJ, Castanzo DT, Delany SP, Selechnick DM, van Ooy A & Cameron DM (2015). The ribosome-associated complex antagonizes prion formation in yeast. Prion, 9, 144 – 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron R, Higurashi T, Sahi C & Craig EA (2007). J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds for prion propagation. EMBO J., 26, 3794 – 3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asante EA, Smidak M, Grimshaw A, Houghton R, Tomlinson A, Jeelani A, Jakubcova T, Hamdan S, Richard-Londt A, Linehan JM, Brandner S, Alpers M, Whitfield J, Mead S, Wadsworth JDF & Collinge J (2015). A naturally occurring variant of the human prion protein completely prevents prion disease. Nature, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailleul-Winslett PA, Newnam GP, Wegrzyn RD & Chernoff YO (2000). An antiprion effect of the anticytoskeletal drug latrunculin A in yeast. Gene Expr, 9, 145–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailleul PA, Newnam GP, Steenbergen JN & Chernoff YO (1999). Genetic study of interactions between the cytoskeletal assembly protein Sla1 and prion - forming domain of the release factor Sup35 (eRF3) in Saccharomyces cerevisiae. Genetics, 153, 81 – 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balguerie A, Dos Reis S, Ritter C, Chaignepain S, Coulary-Salin B, Forge V, Bathany K, Lascu I, Schmitter J-M, Riek R & Saupe S (2003). Domain organization and structure-function relationship of the HET-s prion protein of Podospora anserina. Embo J, 22, 2071 – 2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman D & Wickner RB (2013). The [PSI+] prion exists as a dynamic cloud of variants. Plos Genet., 9, e1003257. doi: 10.1371/journal.pgen.1003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman DA & Wickner RB (2012). [PSI+] prion transmission barriers protect Saccharomyces cerevisiae from infection: intraspecies 'species barriers'. Genetics, 190, 569 – 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin-Baillieu A, Fernandez-Bellot E, Reine F, Coissac E & Cullin C (2003). Conservation of the prion properties of Ure2p through evolution. Mol. Biol. Cell, 14, 3449 – 3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxa U, Taylor KL, Wall JS, Simon MN, Cheng N, Wickner RB & Steven A (2003). Architecture of Ure2p prion filaments: the N-terminal domain forms a central core fiber. J. Biol. Chem, 278, 43717 – 43727. [DOI] [PubMed] [Google Scholar]
- Baxa U, Wickner RB, Steven AC, Anderson D, Marekov L, Yau W-M & Tycko R (2007). Characterization of β-sheet structure in Ure2p1-89 yeast prion fibrils by solid state nuclear magnetic resonance. Biochemistry, 46, 13149 – 13162. [DOI] [PubMed] [Google Scholar]
- Berchowitz LE, Kabachinski G, Walker MR, Carlile TM, Gilbert WV, Schwartz TU & Amon A (2015). Regulated formation of an amyloid-like translational repressor governs gametogenesis. Cell, 163, 406 – 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernet J (1965). Mode d'action des gènes de barrage et relation entre l'incompatibilité cellulaire et l'incompatibilité sexuelle chez le Podospora anserina. Ann. Sci. Natl. Bot, 6, 611–768. [Google Scholar]
- Bessen RA & Marsh RF (1992). Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J. Virol, 66, 2096 – 2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchsenius AS, Muller S, Newnam GP, Inge-Vechtomov SG & Chernoff YO (2006). Prion variant maintained only at high levels of the Hsp104 disaggregase. Curr. Genet, 49, 21 – 29. [DOI] [PubMed] [Google Scholar]
- Brachmann A, Baxa U & Wickner RB (2005). Prion generation in vitro: amyloid of Ure2p is infectious. Embo J, 24, 3082 – 3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley ME, Edskes HK, Hong JY, Wickner RB & Liebman SW (2002). Interactions among prions and prion "strains" in yeast. Proc Natl Acad Sci U S A, 99 (Suppl. 4), 16392 – 16399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley ME & Liebman SW (2004). The Sup35 domains required for maintenance of weak, strong or undifferentiated yeast [PSI+] prions. Mol. Microbiol, 51, 1649 – 1659. [DOI] [PubMed] [Google Scholar]
- Bruce ME (2003). TSE strain variation: an investigation into prion disease diversity. Br Med Bull, 66, 99 – 108. [DOI] [PubMed] [Google Scholar]
- Bruce ME, McConnell I, Fraser H & Dickinson AG (1991). The disease characteristics of different strains of scrapie in Sinc congenic mouse lines: implications for the nature of the agent and host control of pathogenesis. J. Gen. Virol, 72, 595 – 603. [DOI] [PubMed] [Google Scholar]
- Cascarina SM & Ross ED (2014). Yeast prions and human prion-like proteins: sequence features and prediction methods. Cell. Mol. Life Sci, 71, 2047 – 2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caten CE (1972). Vegetative incompatibility and cytoplasmic infection in fungi. J. Gen. Microbiol, 72, 221 – 229. [DOI] [PubMed] [Google Scholar]
- Chang H-Y, Lin J-Y, Lee H-C, Wang H-L & King C-Y (2008). Strain-specific sequences required for yeast prion [PSI+] propagation. Proc Natl Acad Sci U S A, 105, 13345 – 13350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Newnam GP & Chernoff YO (2007). Prion species barrier between the closely related yeast proteins is detected despite coaggregation. Proc Natl Acad Sci U S A, 104, 2791–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Thurber KR, Shewmaker F, Wickner RB & Tycko R (2009). Measurement of amyloid fibril mass-per-length by tilted-beam transmission electron microscopy. Proc. Natl. Acad. Sci. USA, 106, 14339 – 14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Galkin AP, Lewitin E, Chernova TA, Newnam GP & Belenkiy SM (2000). Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Molec. Microbiol, 35, 865 – 876. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B-I, Inge-Vechtomov SG & Liebman SW (1995). Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science, 268, 880 – 884. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Newnam GP, Kumar J, Allen K & Zink AD (1999). Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone Ssb in formation, stability and toxicity of the [PSI+] prion. Mol. Cell. Biol, 19, 8103 – 8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO & Ono B-I (1992). Dosage-dependent modifiers of PSI-dependent omnipotent suppression in yeast. Protein synthesis and targeting in yeast. Brown AJP, Tuite MF and McCarthy JEG. Berlin, Springer-Verlag: 101–107. [Google Scholar]
- Chernova TA, Allen KD, Wesolowski LM, Shanks JR, Chernoff YO & Wilkinson KD (2003). Pleiotropic effects of Ubp6 loss on drug sensitivities and yeast prion are due to depletion of the free ubiquitin pool. J. Biol. Chem, 278, 52102 – 52115. [DOI] [PubMed] [Google Scholar]
- Chernova TA, Romanyuk AV, Karpova TS, Shanks JR, Ali M, Moffatt N, Howie RL, O'Dell AO, McNally JG, Liebman SW, Chernoff YO & Wilkinson KD (2011). Prion induction by the short-lived stresss induced protein Lsb2 is regulated by ubiquitination and association with the actin cytoskeleton. Mol. Cell, 43, 242 – 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge J & Alpers MP (2008). The end of kuru: 50 years of research into an extraordinary disease. Philos. Trans. R. Soc. Lond. B Biol. Sci, 363, 3607 – 3763.18672464 [Google Scholar]
- Collinge J & Clarke AR (2007). A general model of prion strains and their pathogenicity. Science, 318, 930 – 936. [DOI] [PubMed] [Google Scholar]
- Cooper TG (2002). Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to th GATA factors: connecting the dots. FEMS Microbiol. Revs, 26, 223–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustou V, Deleu C, Saupe S & Begueret J (1997). The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc. Natl. Acad. Sci. USA, 94, 9773 – 9778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox BS (1965). PSI, a cytoplasmic suppressor of super-suppressor in yeast. Heredity, 20, 505 – 521. [Google Scholar]
- Cox BS, Ness F & Tuite MF (2003). Analysis of the generation and segregation of propagons: entities that propagate the [PSI+] prion in yeast. Genetics, 165, 23 – 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuille J & Chelle PL (1939). Experimental transmission of trembling to the goat. C. R. Seances Acad. Sci, 208, 1058 – 1060. [Google Scholar]
- Dalstra HJP, Swart K, Debets AJM, Saupe SJ & Hoekstra RF (2003). Sexual transmission of the [Het-s] prion leads to meiotic drive in Podospora anserina. Proc Natl Acad Sci U S A, 100, 6616 – 6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalov A, Habenstein B, Martinez D, Debets AJM, Sabate R, Loquet A & Saupe SJ (2015). Signal transduction by a fungal NOD-like receptor based on propagation of a prion amyloid fold. PLoS Biol., 13, e1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalov A & Saupe SJ (2015). As a toxin dies a prion comes to life: a tentative natural history of the [Het-s] prion. Prion, 9, 184 – 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debets AJ, Dalstra HJ, Slakhorst M, Koopmanschap B, Hoekstra RF & Saupe SJ (2012). High natural prevalence of a fungal prion. Proc. Natl. Acad. Sci. USA, 109, 10432 – 10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debets F, Yang X & Griffiths AJ (1994). Vegetative incompatibility in Neurospora: its effect on horizontal transfer of mitochondrial plasmids and senescence in natural populations. Curr. Genet, 26, 113 – 119. [DOI] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Hong JY & Liebman SW (2001). Prions affect the appearance of other prions: the story of [PIN]. Cell, 106, 171 – 182. [DOI] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Zhou P, Chernoff YO & Liebman SW (1997). Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics, 147, 507 – 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG & Liebman SW (1996). Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics, 144, 1375 – 1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Avalos R, King CY, Wall JS, Simon M & Caspar DLD (2005). Strain-specific morphologies of yeast prion amyloids. Proc Natl Acad Sci U S A, 102, 10165 – 10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dihanich ME, Najarian D, Clark R, Gillman EC, Martin NC & Hopper AK (1987). Isolation and characterization of MOD5, a gene required for isopentenylation of cytoplasmic and mitochondrial tRNAs of Saccharomyces cerevisiae. Mol. Cell. Biol, 7, 177 – 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Park K-W, Yu H, Fan Q & Li L (2008). Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat. Genet, 40, 460 – 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglestone SS, Cox BS & Tuite MF (1999). Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. EMBO J., 18, 1974 – 1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes HE, Khamar HJ, Winchester C-L, Greenler AJ, Zhou A, McGlinchey RP, Gorkovskiy A & Wickner RB (2014). Sporadic distribution of prion-forming ability of Sup35p from yeasts and fungi. Genetics, 198, 605 – 616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes HK, Engel A, McCann LM, Brachmann A, Tsai H-F & Wickner RB (2011). Prion-forming ability of Ure2 of yeasts is not evolutionarily conserved. Genetics, 188, 81 – 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes HK, Gray VT & Wickner RB (1999). The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc. Natl. Acad. Sci. USA, 96, 1498 – 1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes HK, McCann LM, Hebert AM & Wickner RB (2009). Prion variants and species barriers among Saccharomyces Ure2 proteins. Genetics, 181, 1159 – 1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes HK & Wickner RB (2002). Conservation of a portion of the S. cerevisiae Ure2p prion domain that interacts with the full - length protein. Proc. Natl. Acad. Sci. USA, 99 (Suppl. 4), 16384–16391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes HK & Wickner RB (2013). The [URE3] prion in Candida. Euk. Cell, 12, 551 – 558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egge N, Muthusubramanian A & Cornwall GA (2015). Amyloid properties of the mouse egg zona pellucida. PLoS One, 10, e0129907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel A, Shewmaker F, Edskes HK, Dyda F & Wickner RB (2011). Amyloid of the Candida albicans Ure2p prion domain is infectious and has a parallel in-register β-sheet structure. Biochemistry, 50, 5971 – 5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erjavec N, Larsson L, Grantham J & Nystrom T (2007). Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev., 21, 2410 – 2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escusa-Toret S, Vonk WIM & Frydman J (2013). Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat. Cell Biol, 15, 1231 – 1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira PC, Ness F, Edwards SR, Cox BS & Tuite MF (2001). The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol Microbiol, 40, 1357–69. [DOI] [PubMed] [Google Scholar]
- Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE & Kelly JW (2006). Functional amyloid formation within mammalian tissue. Plos Biology, 4, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick KK, Debelouchina GT, Kayatekin C, Dominy T, Jacvone AC, Griffin RG & Lindquist S (2014). Distinct prion strains are defined by amyloid core structure and chaperone binding site dynamics. Chem. Biol, 21, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi Y, Doi Y, Hosoda N, Uchida N, Osawa M, Shimada I, Tsujimoto M, Suzuki T, Katada T & Hoshino S (2007). Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev., 21, 3135 – 3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futcher AB & Cox BS (1983). Maintenance of the 2 μm circle plasmid in populations of Saccharomyces cerevisiae. J Bacteriol, 154, 612 – 622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futcher B, Reid E & Hickey DA (1988). Maintenance of the 2 micron circle plasmid of Saccharomyces cerevisiae by sexual transmission: an example of selfish DNA. Genetics, 118, 411 – 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi M, Lilie H, Funfschilling U, Mun A, Ross S, Lithgow T, Rucknagel P & Rospert S (2001). RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc. Natl. Acad. Sci. USA, 98, 3762 – 3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA & Fink GR (1992). Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth regulation by starvation and RAS. Cell, 68, 1077 – 1090. [DOI] [PubMed] [Google Scholar]
- Glover JR, Kowal AS, Shirmer EC, Patino MM, Liu J-J & Lindquist S (1997). Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell, 89, 811 – 819. [DOI] [PubMed] [Google Scholar]
- Glover JR & Lindquist S (1998). Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell, 94, 73–82. [DOI] [PubMed] [Google Scholar]
- Gorkovskiy A, Thurber KR, Tycko R & Wickner RB (2014). Locating the folds of the in-register parallel β-sheet of the Sup35p prion domain infectious amyloid. Proc. Natl. Acad. Sci. USA, 111, E4615–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimminger V, Richter K, Imhof A, Buchner J & Walter S (2004). The prion curing agent guanidinium chloride specifically inibits ATP hydrolysis by Hsp104. J. Biol. Chem, 279, 7378 – 7383. [DOI] [PubMed] [Google Scholar]
- Halfmann R, Jarosz DF, Jones SK, Chang A, Lancster AK & Lindquist S (2012). Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature, 482, 363 – 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JM, Nguyen PP, Patel MJ, Sporn ZA & Hines JK (2014). Functional diversification of hsp40: distinct J-protein functional requirements for two prions allow for chaperone-dependent prion selection. PloS Genet., 10, e1004510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Dempster ER & Brown SW (1975). Adaptive significance of vegetative incompatibility in Neurospora crassa. Genetics, 81, 553 – 569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsen CW & Glover JR (2012). Insight into molecular basis of curing of [PSI+] prion by overexpression of 104-kDa heat shock protein (Hsp104). J. Biol. Chem, 287, 542 – 556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higurashi T, Hines JK, Sahi C, Aron R & Craig EA (2008). Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc. Natl. Acad. Sci. USA, 105, 16596 –16601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines JK, Li X, Du Z, Higurashi T, Li L & Craig EA (2011). [SWI], the prion formed by the chromatin remodeling factor Swi1, is highly sensistive to alterations in Hsp70 chaperone system activity. PloS Genet., 7, e1001309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes DL, Lancaster AK, Lindquist S & Halfmann R (2013). Hertable remodeling of yeast multicellularity by an environmentally responsive prion. Cell, 153, 153 – 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda N, Kobayashii T, Uchida N, Funakoshi Y, Kikuchi Y, Hoshino S & Katada T (2003). Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J. Biol. Chem, 278, 38287 – 38291. [DOI] [PubMed] [Google Scholar]
- Hoyle NP, Castelli LM, Campbell SG, Holmes LE & Ashe MP (2007). Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spacially distinct from P-bodies. J. Cell Biol, 179, 65 – 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Gautschi M, Walter W, Rospert S & Craig EA (2005). The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nat. Struct. Mol. Biol, 12, 497 – 504. [DOI] [PubMed] [Google Scholar]
- Hung GC & Masison DC (2006). N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics, 173, 611 – 620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Pfund C & Craig EA (1997). Functional specificity among Hsp70 molecular chaperones. Science, 275, 387–9. [DOI] [PubMed] [Google Scholar]
- Jones EW (1991). Three proteolytic systems in the yeast Saccharomyces cerevisiae. J. Biol. Chem, 266, 7963 – 7966. [PubMed] [Google Scholar]
- Jones GW & Masison DC (2003). Saccharomyces cerevisiae Hsp70 Mutations Affect [PSI(+)] Prion Propagation and Cell Growth Differently and Implicate Hsp40 and Tetratricopeptide Repeat Cochaperones in Impairment of [PSI(+)]. Genetics, 163, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jucker M & Walker LC (2013). Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature, 501, 45 – 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Jones G & Masison DC (2002). Amino acid residue 184 of yeast Hsp104 chaperone is critical for prion-curing by guanidine, prion propagation, and thermotolerance. Proc. Natl. Acad. Sci. USA, 99, 9936 – 9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Jones G, Wegrzyn RD & Masison DC (2000). A role for cytosolic Hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics, 156, 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G & Masison DC (2001). Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr. Microbiol, 43, 7 – 10. [DOI] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R & Frydman J (2008). Misfolded proteins partition between two distinct quality control compartments. Nature, 454, 1088 – 1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kama R, Robinson M & Gerst JE (2007). Btn2, a Hook1 ortholog and potential Batten disease-related protein, mediates late endosome-Golgi protein sorting in yeast. Mol. Cell. Biol, 27, 605 – 621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AC, Busby B & Wickner RB (2014). Effect of domestication on the spread of the [PIN+] prion in Saccharomyces cerevisiae. Genetics, May 8, epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AC, Shewmaker FP, Kryndushkin D & Wickner RB (2012). Sex, prions and plasmids in yeast. Proc. Natl. Acad. Sci. USA, 109, E2683 – E2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiktev DA, Chernoff YO, Archipenko AV & Zhouravleva GA (2011). Identification of genes influencing synthetic lethality of genetic and epigenetic alterations in translation termination factors in yeast. Dokl. Biochem. Biophys, 438, 117 – 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiktev DA, Melomed MM, Lu CD, Newnam GP & Chernoff YO (2015). Feedback control of prion formation and propagation by the ribosome-associated chaperone complex. Mol. Microbiol, 96, 621 – 632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberlin RH, Cole S & Walker CA (1987). Temporary and permanent modifications to a single strain of mouse scrapie on transmission to rats and hamsters. J. Gen. Virol, 68, 1875 – 1881. [DOI] [PubMed] [Google Scholar]
- Kimura M (1968). Evolutionary rate at the molecular level. Nature, 217, 624 – 626. [DOI] [PubMed] [Google Scholar]
- King C-Y, Tittmann P, Gross H, Gebert R, Aebi M & Wuthrich K (1997). Prion-inducing domain 2-114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc. Natl. Acad. Sci. USA, 94, 6618 – 6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CY (2001). Supporting the structural basis of prion strains: induction and identification of [PSI] variants. J Mol Biol, 307, 1247–60. [DOI] [PubMed] [Google Scholar]
- King CY & Diaz-Avalos R (2004). Protein-only transmission of three yeast prion strains. Nature, 428, 319 – 323. [DOI] [PubMed] [Google Scholar]
- Kirkland PA, Reidy M & Masison DC (2011). Functions of yeast Hsp40 chaperone Sis1p dispensable for prion propagation but important for prion curing and protection from prion toxicity. Genetics, 188, 565 – 577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochneva-Pervukhova NV, Poznyakovski AI, Smirnov VN & Ter-Avanesyan MD (1998). C-terminal truncation of the Sup35 protein increases the frequency of de novo generation of a prion-based [PSI+] determinant in Saccharmyces cerevisiae. Curr. Genet, 34, 146 – 151. [DOI] [PubMed] [Google Scholar]
- Kryndushkin D, Ihrke G, Piermartiri TC & Shewmaker F (2012). A yeast model of optineurin proteinopathy reveals a unique aggregation pattern associated with cellular toxicity. Mol. Microbiol, Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Kryndushkin D, Pripuzova N, Burnett B & Shewmaker F (2013). Non-targeted identification of prions and amyloid-forming proteins from yeast and mammalian cells. J. Biol. Chem, 288, 27100–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin D, Shewmaker F & Wickner RB (2008). Curing of the [URE3] prion by Btn2p, a Batten disease-related protein. EMBO J., 27, 2725 – 2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD & Kushnirov VV (2003). Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J. Biol. Chem, 278, 49636 – 49643. [DOI] [PubMed] [Google Scholar]
- Kryndushkin DS, Engel A, Edskes HK & Wickner RB (2011). Molecular chaperone Hsp104 can promote yeast prion generation. Genetics, 188, 339 – 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin DS, Wickner RB & Tycko R (2011). The core of Ure2p prion fibrils is formed by the N-terminal segment in a parallel cross-β structure: evidence from solid-state NMR. J. Mol. Biol, 409, 263 – 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Gaur D, Gupta A, Puri A & Sharma D (2015). Hsp90-associated immunophilin homolog Cpr7 is required for the mitotic stability of [URE3] prion in Saccharomyces cerevisiae. PloS Genet., 11, e1005567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov VV, Kochneva-Pervukhova NV, Cechenova MB, Frolova NS & Ter-Avanesyan MD (2000). Prion properties of the Sup35 protein of yeast Pichia methanolica. EMBO J., 19, 324 – 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov VV, Kryndushkin D, Boguta M, Smirnov VN & Ter-Avanesyan MD (2000). Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr. Biol, 10, 1443 – 1446. [DOI] [PubMed] [Google Scholar]
- Lacroute F (1971). Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J. Bacteriol, 106, 519 – 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster DL, Dobson CM & Rachubinski RA (2013). Chaperone proteins select and maintain [PIN+] prion conformations in Saccharomyces cerevisiae. J. Biol. Chem, 288, 1266 – 1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Browning S, Mahal SP, Oelschlegel AM & Weissmann C (2010). Darwinian evolution of prions in cell culture. Science, 327, 869 – 872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Rayman JB, Kandel ER & Derkatch IL (2014). Functional role of Tia1/Pub1 and Sup35 prion domains: directing protein synthesis machinery to the tubulin cytoskeleton. Mol. Cell, 55, 1 – 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckgei N, Schutz AK, Bousset L, Habenstein B, Souigues Y, Gardiennet C, Meier BH, Melki R & Bockmann A (2013). The conformation of the prion domain of Sup35p in isolation and in the full-length protein. Biomol. NMR Assign, 52, 12741 – 4. [DOI] [PubMed] [Google Scholar]
- Lum R, Tkach JM, Vierling E & Glover JR (2004). Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J. Biol. Chem, 279, 29139 – 29146. [DOI] [PubMed] [Google Scholar]
- Mackay JP, Matthews JM, Winefield RD, Mackay LG, Haverkamp RG & Templeton MD (2001). The hydrophobin EAS is largely unstructured in solution and functions by forming amyloid-like structures. Structure (Camb), 9, 83–91. [DOI] [PubMed] [Google Scholar]
- Maddelein ML, Dos Reis S, Duvezin-Caubet S, Coulary-Salin B & Saupe SJ (2002). Amyloid aggregates of the HET-s prion protein are infectious. Proc Natl Acad Sci U S A, 99, 7402–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magasanik B & Kaiser CA (2002). Nitrogen regulation in Saccharomyces cerevisiae. Gene, 290, 1–18. [DOI] [PubMed] [Google Scholar]
- Maldonado-Baez L, Cole NB, Kramer H & Donaldson JG (2013). Microtubule-dependent endosomal sorting of clathrin-independent cargo by Hook1. J. Cell. Biol, 201, 233 – 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovska L, Kroschwald S, Munder MC, Richter D & Alberti S (2012). Molecular chaperones and stress-inducible protein-sorting factors coordinate the spaciotemporal distribution of protein aggregates. Mol. Biol. Cell, 23, 3041 – 3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Montanes F, Rienzo A, Poveda-Huertes D, Pascual-Ahuir A & Proft M (2013). Activator and repressor functions of the Mot3 transcription factor in the osmostress reponse of Saccharomyces cerevisiae. Euk. Cell, 12, 636 – 647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masison DC, Kirkland PA & Sharma D (2009). Influence of Hsp70s and their regulators on yeast prion propagation. Prion, 3, 65 – 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masison DC, Maddelein M-L & Wickner RB (1997). The prion model for [URE3] of yeast: spontaneous generation and requirements for propagation. Proc. Natl. Acad. Sci. USA, 94, 12503–12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masison DC & Reidy M (2015). Yeast prions are useful for studying protein chaperones and protein quality control. Prion, 9, 174 – 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masison DC & Wickner RB (1995). Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science, 270, 93 – 95. [DOI] [PubMed] [Google Scholar]
- McGlinchey R, Kryndushkin D & Wickner RB (2011). Suicidal [PSI+] is a lethal yeast prion. Proc. Natl. Acad. Sci. USA, 108, 5337 – 5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead DJ, Gardner DCJ & Oliver SG (1986). The yeast 2 μ plasmid: strategies for the survival of a selfish DNA. Mol. Gen. Genet, 205, 417 – 421. [DOI] [PubMed] [Google Scholar]
- Miller SBM, Ho C-T, Winkler J, Khokhrina M, Neuner A, Mohamed MYH, Guilbride DL, Richter K, Lisby M, Schiebel E, Mogk A & Bukau B (2015). Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J., 34, 778 – 797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama H, Edskes HK & Wickner RB (2000). [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol. Cell. Biol, 20, 8916 – 8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayashiki T, Ebihara K, Bannai H & Nakamura Y (2001). Yeast [PSI+] "prions" that are crosstransmissible and susceptible beyond a species barrier through a quasi-prion state. Mol Cell, 7, 1121–30. [DOI] [PubMed] [Google Scholar]
- Nakayashiki T, Kurtzman CP, Edskes HK & Wickner RB (2005). Yeast prions [URE3] and [PSI+] are diseases. Proc Natl Acad Sci U S A, 102, 10575–10580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O, Galopier A, Martini C, Matsufuji S, Fabret C & Rousset C (2008). Epigenetic control of polyamines by the prion [PSI+]. Nat. Cell. Biol, 10, 1069 – 1075. [DOI] [PubMed] [Google Scholar]
- Naunmuktane Z, Mead S, Ellis M, Wadsworth JDF, Nicoll AJ, Kenny J, Launchbury F, Linehan JM, Richard-Leondt A, Walker AS, Rudge P, Collinge J & Brandner S (2015). Evidence for human transmission of amyloid-β pathology and cerebral amyloid angiopathy. Nature, 525, 247 – 250. [DOI] [PubMed] [Google Scholar]
- Nelson RJ, Ziegilhoffer T, Nicolet C, Werner-Washburne M & Craig EA (1992). The translation machinery and 70 kDal heat shock protein cooperate in protein synthesis. Cell, 71, 97 – 105. [DOI] [PubMed] [Google Scholar]
- Ness F, Ferreira P, Cox BS & Tuite MF (2002). Guanidine hydrochloride inhibits the generation of prion "seeds" but not prion protein aggregation in yeast. Mol. Cell. Biol, 22, 5593 – 5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby GA & Lindquist S (2013). Blessings in disguise: biological benefits of prion-like mechanisms. Trends in Cell Biol., 23, 251 – 259. [DOI] [PubMed] [Google Scholar]
- Newnam GP, Birchmore JL & Chernoff YO (2011). Destabilization and recovery of a yeast prion after mild heat shock. J. Mol. Biol, 408, 432 – 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newnam GP, Wegrzyn RD, Lindquist SL & Chernoff YO (1999). Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell. Biol, 19, 1325 – 1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo S, Chiang V & Guo Z (2012). Quantitative analysis of spin exchange interactions to identify β strand and turn regions in Ure2 prion domain fibrils with site-directed spin labeling. J. Struct. Biol, 180, 374 – 381. [DOI] [PubMed] [Google Scholar]
- Ngo S, Gu L & Guo Z (2011). Hierarchical organization in the amyloid core of yeast prion protein Ure2. J. Biol. Chem, 286, 29691 – 29699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystrom T & Liu B (2014). Protein quality control in time and space - links to cellular aging. FEMS Yeast Res, 14, 40 – 48. [DOI] [PubMed] [Google Scholar]
- Osherovich LZ & Weissman JS (2001). Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI+] prion. Cell, 106, 183 – 194. [DOI] [PubMed] [Google Scholar]
- Paoletti M & Saupe SJ (2009). Fungal incompatibility: evolutionary origin in pathogen defense? Bioessays, 31, 1201 – 1210. [DOI] [PubMed] [Google Scholar]
- Park Y-N, Zhao X, Yim Y-I, Todor H, Ellerbrook R, Reidy M, Eisenberg E, Masison DC & Greene LE (2014). Hsp104 overexpression cures Saccharomyces cerevisiae [PSI+] by causing dissolution of the prion seeds. Euk. Cell, 13, 635 – 647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel BK, Gavin-Smyth J & Liebman SW (2009). The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat. Cell Biol, 11, 344 – 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino MM, Liu J-J, Glover JR & Lindquist S (1996). Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science, 273, 622 – 626. [DOI] [PubMed] [Google Scholar]
- Paul KR, Hendrich CG, Waechter A, Harman MR & Ross ED (2015). Generating new prions by targeted mutation or segment duplication. Proc. Natl. Acad. Sci. USA, published ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin SV, Kushnirov VV, Smirnov VN & Ter-Avanesyan MD (1996). Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J., 15, 3127 – 3134. [PMC free article] [PubMed] [Google Scholar]
- Paushkin SV, Kushnirov VV, Smirnov VN & Ter-Avanesyan MD (1997). In vitro propagation of the prion-like state of yeast Sup35 protein. Science, 277, 381 – 383. [DOI] [PubMed] [Google Scholar]
- Podrabsky JE, Carpenter JF & Hand SC (2001). Survival of water stress in annual fish embryos: dehydration avoidance and egg amyloid fibers. Am. J. Physiol. Regulatory Integrative Comp. Physiol, 280, R123 – R131. [DOI] [PubMed] [Google Scholar]
- Priola SA, Caughey B, Race RE & Chesebro B (1994). Heterologous PrP molecules interfere with accumulation of protease-resistant PrP in scrapie-infected murine neuroblastoma cells. J Virol, 68, 4873–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB (1982). Novel proteinaceous infectious particles cause scrapie. Science, 216, 136 – 144. [DOI] [PubMed] [Google Scholar]
- Prusiner SB, Ed. (2004). Prion Biology and Diseases. Cold Spring Harbor, NY, Cold Spring Harbor Laboratory Press. [Google Scholar]
- Prusiner SB, Scott M, Foster D, Pan K-M, Groth D, Mirenda C, Torchia M, Yang S-L, Serban D, Carlson GA, Hoppe PC, Westaway D & DeArmond SJ (1990). Transgenic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell, 63, 673 – 686. [DOI] [PubMed] [Google Scholar]
- Rai R, Genbauffe F, Lea HZ & Cooper TG (1987). Transcriptional regulation of the DAL5 gene in Saccharomyces cerevisiae. J. Bacteriol, 169, 3521 – 3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidy M & Masison DC (2010). Sti1 regulation of Hsp70 and Hsp90 is critical for curing of Saccharomyces cerevisiae [PSI+] prions by Hsp104. Mol. Cell. Biol, 30, 3542 – 3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidy M & Masison DC (2012). Prokaryotic chaperones support yeast prions and thermotolerance and define disaggregation machinery interactions. Genetics, 192, 185 – 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidy M, Sharma R & Masison DC (2013). Schizosaccharomyces pombe disaggregation machinery chaperones support Saccharomyces cerevisiae growth and prion propagation. Euk. Cell, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende CG, Outeiro TF, Sands L, Lindquist S & Tuite MF (2003). Prion protein gene polymorphisms in Saccharomyces cerevisiae. Mol. Microbiol, 49, 1005 – 1017. [DOI] [PubMed] [Google Scholar]
- Riggs AD, Martienssen RA & Russo VEA (1996). Introduction. Epigenetic mechanisms of gene regulation. Russo VEA, Martienssen RA and Riggs AD. Cold Spring Harbor, N.Y., Cold Spring Harbor Laboratory Press: 1 – 4. [Google Scholar]
- Ritter C, Maddelein ML, Siemer AB, Luhrs T, Ernst M, Meier BH, Saupe SJ & Riek R (2005). Correlation of structural elements and infectivity of the HET-s prion. Nature, 435, 844–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizet G (1952). Les phenomenes de barrage chez Podospora anserina: analyse genetique des barrages entre les souches s et S. Rev. Cytol. Biol. Veg, 13, 51 – 92. [Google Scholar]
- Roberts BT & Wickner RB (2003). A class of prions that propagate via covalent auto-activation. Genes Dev., 17, 2083 – 2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogoza T, Goginashvili A, Rodionova S, Ivanov M, Viktorovskaya O, Rubel A, Volkov K & Mironova L (2010). Non-mendelian determinant [ISP+] in yeast is a nuclear-residing prion form of the global transcriptional regulator Sfp1. Proc. Natl. Acad. Sci. USA, 107, 10573 – 10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross ED, Baxa U & Wickner RB (2004). Scrambled prion domains form prions and amyloid. Mol Cell Biol, 24, 7206–7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross ED, Edskes HK, Terry MJ & Wickner RB (2005). Primary sequence independence for prion formation. Proc Natl Acad Sci U S A, 102, 12825 – 12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross ED, Minton AP & Wickner RB (2005). Prion domains: sequences, structures and interactions. Nat. Cell Biol, 7, 1039–1044. [DOI] [PubMed] [Google Scholar]
- Safadi RA, Talarek N, Jacques N & Aigle M (2011). Yeast prions: could they be exaptations? The URE2/[URE3] system in Kluyveromyces lactis. FEMS Yeast Res., 11, 151 – 153. [DOI] [PubMed] [Google Scholar]
- Santoso A, Chien P, Osherovich LZ & Weissman JS (2000). Molecular basis of a yeast prion species barrier. Cell, 100, 277 – 288. [DOI] [PubMed] [Google Scholar]
- Saunders SE, Bartelt-Hunt SL & Bartz JC (2012). Occurence, transmission, and zoonotic potential of chronic wasting disease. Emerg. Inf. Dis, 18, 369 – 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlumpberger M, Prusiner SB & Herskowitz I (2001). Induction of distinct [URE3] yeast prion strains. Mol Cell Biol, 21, 7035–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwimmer C & Masison DC (2002). Antagonistic interactions between yeast [PSI+] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol. Cell. Biol, 22, 3590 – 3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A, Baxa U, Simon MN, Wall JS, Sabate R, Saupe SJ & Steven AC (2006). Mass analysis by scanning transmission electron microscopy and electron diffraction validate predictions of stacked beta-solenoid model of HET-s prion fibrils. J. Biol. Chem, 282, 5545 – 5550. [DOI] [PubMed] [Google Scholar]
- Sharma A, Bruce KL, Chen B, Gyoneva S, Behrens SH, Bommarius AS & Chernoff YO (2015). Contributions of the prion protein sequence, strain and environment to the species barrier. J. Biol. Chem, .M115.684100. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma J & Liebman SW (2013). Exploring the basis of [PIN+] variant differences in [PSI+] induction. J. Mol. Biol, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U & Parker R (2003). Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science, 300, 805 – 808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewmaker F, Mull L, Nakayashiki T, Masison DC & Wickner RB (2007). Ure2p function is enhanced by its prion domain in Saccharomyces cerevisiae. Genetics, 176, 1557 – 1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewmaker F, Ross ED, Tycko R & Wickner RB (2008). Amyloids of shuffled prion domains that form prions have a parallel in-register β-sheet structure. Biochemistry, 47, 4000–4007. [DOI] [PubMed] [Google Scholar]
- Shewmaker F, Wickner RB & Tycko R (2006). Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc. Natl. Acad. Sci. USA, 103, 19754 – 19759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondheimer N & Lindquist S (2000). Rnq1: an epigenetic modifier of protein function in yeast. Molec. Cell, 5, 163 – 172. [DOI] [PubMed] [Google Scholar]
- Sondheimer N, Lopez N, Craig EA & Lindquist S (2001). The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J., 20, 2435–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht S, Miller SBM, Mogk A & Bukau B (2011). Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J. Cell. Biol, 195, 617 – 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki G, Shimazu N & Tanaka M (2012). A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science, 336, 355 – 359. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chien P, Naber N, Cooke R & Weissman JS (2004). Conformational variations in an infectious protein determine prion strain differences. Nature, 428, 323 – 328. [DOI] [PubMed] [Google Scholar]
- Taylor KL, Cheng N, Williams RW, Steven AC & Wickner RB (1999). Prion domain initiation of amyloid formation in vitro from native Ure2p. Science, 283, 1339 – 1343. [DOI] [PubMed] [Google Scholar]
- TerAvanesyan A, Dagkesamanskaya AR, Kushnirov VV & Smirnov VN (1994). The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics, 137, 671 – 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessarz P, Mogk A & Bukau B (2008). Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol. Microbiol, 68, 87 – 97. [DOI] [PubMed] [Google Scholar]
- Tipton KA, Verges KJ & Weissman JS (2008). In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol. Cell, 32, 584 – 591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toombs JA, McCarty BR & Ross ED (2010). Compositional determinants of prion formation in yeast. Mol. Cell. Biol, 30, 319 – 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toombs JA, Petri M, Paul KR, Kan GY & Ross ED (2012). De novo design of synthetic prion domains. Proc. Natl. Acad. Sci. USA, 109, 6519 – 6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama BH, Kelly MJ, Gross JD & Weissman JS (2007). The structural basis of yeast prion strain variants. Nature, 449, 233 – 237. [DOI] [PubMed] [Google Scholar]
- Troisi EM, Rockman ME, Nguyen PP, Oliver EE & Hines JK (2015). Swa2, the yeast homolog of mammalian auxilin, is specifically required for the propagation of the prion variant [URE3-1]. Mol. Microbiol, 97, 926 – 941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True HL & Lindquist SL (2000). A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature, 407, 477–483. [DOI] [PubMed] [Google Scholar]
- Tuite MF, Mundy CR & Cox BS (1981). Agents that cause a high frequency of genetic change from [psi+] to [psi−] in Saccharomyces cerevisiae. Genetics, 98, 691 – 711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko R (2006). Molecular structure of amyloid fibrils: insights from solid-state NMR. Quart. Revs. Biophys, 1, 1–55. [DOI] [PubMed] [Google Scholar]
- Tycko R (2011). Solid-state NMR studies of amyloid fibril structure. Annu. Rev. Phys. Chem, 62, 279 – 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko R & Wickner RB (2013). Molecular structures of amyloid and prion fibrils: consensus vs. controversy. Accounts of Chem. Res, 46, 1487 – 1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J, Madariaga ML & Lindquist S (2008). Prion switching in response to environmental stress. PLoS Biol., 6, e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishveshwara N & Liebman SW (2009). Heterologous cross-seeding mimics cross-species prion conversion in a yeast model. BMC Biol., 7, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov KV, Aksenova YA, Soom MJ, Sipov KV, Svitin AV, Kurischko C, Shkundina IS, Ter-Avanesyan MD, Inge-Vechtomov SG & Mironova LN (2002). Novel non-Mendelian determinant involved in the control of translation accuracy in Saccharomyces cerevisiae. Genetics, 160, 25 – 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Meriin AB, Zaarur N, Romanova NV, Chernoff YO, Costello CE & Sherman MY (2009). Abnormal proteins can form aggresome in yeast: aggresome-targeting signals and components of the machinery. FASEB J., 23, 451 – 463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R & Meier BH (2008). Amyloid fibrils of the HET-s(218-279) prion form a beta solenoid with a triangular hydrophobic core. Science, 319, 1523 – 1526. [DOI] [PubMed] [Google Scholar]
- Watts JC, Condello C, Stohr J, Oehler A, Lee J, DeArmond SJ, Lannfelt L, Ingelsson M, Giles K & Prusiner SB (2014). Serial propagation of distinct strains of Aβ prions from Alzheimer's disease patients. Proc. Natl. Acad. Sci. USA, 111, 10323 – 10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibezahn J, Tessarz P, Schlieker C, Zhan R, Maglica Z, Lee S, Zentgraf H, Weber-Ban EU, Dougan DA, Tsai FT, Mogk A & Bukau B (2004). Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell, 119, 653 – 665. [DOI] [PubMed] [Google Scholar]
- Westergard L & True HL (2014). Extracellular environment modulates the formation and propagation of particular amyloid structures. Mol. Microbiol, 92, 698 – 715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB (1994). [URE3] as an altered URE2 protein: evidence for a prion analog in S. cerevisiae. Science, 264, 566 – 569. [DOI] [PubMed] [Google Scholar]
- Wickner RB (1997). A new prion controls fungal cell fusion incompatibility. Proc. Natl. Acad. Sci. USA, 94, 10012 – 10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Beszonov E & Bateman DA (2014). Normal levels of the antiprion proteins Btn2 and Cur1 cure most newly formed [URE3] prion variants. Proc. Natl. Acad. Sci. USA, 111, E2711–E2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Dyda F & Tycko R (2008). Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register β-sheet structure. Proc Natl Acad Sci U S A, 105, 2403 – 2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Edskes HK, Bateman DA, Kelly AC, Gorkovskiy A, Dayani Y & Zhou A (2013). Amyloids and yeast prion biology. Biochemistry, 52, 1514 – 1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Shewmaker F, Kryndushkin D & Edskes HK (2008). Protein inheritance (prions) based on parallel in-register β-sheet amyloid structures. Bioessays, 30, 955 – 964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Shewmaker FP, Bateman DA, Edskes HE, Gorkovskiy A, Dayani Y & Beszonov EE (2015). Yeast prions: structure, biology and prion-handling systems. Microbiol. Mol. Biol. Rev, 79, 1 – 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SH & King CY (2015). Amino acid proximities in two Sup35 prion strains revealed by chemical cross-linking. J. Biol. Chem, 290, 25062 – 25071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Nakamura A, Chiba T, Kogishi K, Matsushita T, Li F, Guo Z, Hosokawa M, Mori M & Higuchi K (2001). Transmission of mouse senile amyloidosis. Lab. Invest, 81, 493 – 499. [DOI] [PubMed] [Google Scholar]
- Zhang B, Une Y, Fu X, Yan J, Ge F, Yao J, Sawashita J, Mori M, Tomozawa H, Kametani F & Higuchi K (2008). Fecal transmission of AA amyloidosis in the cheetah contributes to high incidence of disease. Proc. Natl. Acad. Sci. USA, 105, 7263 – 7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubenko GS, Park FJ & Jones EW (1982). Genetic properties of mutations at the PEP4 locus in Saccharomyces cerevisiae. Genetics, 102, 679 – 690. [DOI] [PMC free article] [PubMed] [Google Scholar]