Abstract
Mitochondrial cristae membranes are the oxidative phosphorylation sites in cells. Crista junctions (CJs) form the highly curved neck regions of cristae and are thought to function as selective entry gates into the cristae space. Little is known about how CJs are generated and maintained. We show that the central coiled-coil (CC) domain of the mitochondrial contact site and cristae organizing system subunit Mic60 forms an elongated, bow tie–shaped tetrameric assembly. Mic19 promotes Mic60 tetramerization via a conserved interface between the Mic60 mitofilin and Mic19 CHCH (CC-helix-CC-helix) domains. Dimerization of mitofilin domains exposes a crescent-shaped membrane-binding site with convex curvature tailored to interact with the curved CJ neck. Our study suggests that the Mic60-Mic19 subcomplex traverses CJs as a molecular strut, thereby controlling CJ architecture and function.
The Mic60-Mic19 complex assembles into a tetrameric molecular strut that controls crista junction architecture.
INTRODUCTION
Mitochondria are highly dynamic double membrane-bound organelles crucial for cellular metabolism, energy conversion, signaling, and apoptosis (1–4). They are characterized by extended and intricately folded inner membrane structures termed cristae that were described in the early days of electron microscopy (EM) and later recognized as the main sites of oxidative phosphorylation. Cristae are highly adaptive and variable in shape and size depending on cell type, metabolic state, and developmental stage (4–6). Key determinants for cristae morphology are oligomeric F1Fo–adenosine 5′-triphosphate (ATP) synthase complexes that shape the tips and rims of cristae (7), whereas filaments of dynamin-like Mgm1/OPA1 are thought to stabilize and deform cristae from the intracristal space in an energy-dependent manner (8, 9). Cristae are connected to the mitochondrial inner boundary membrane via crista junctions (CJs) (fig. S1, A and B) (5, 10–12). These highly curved tubular openings with a circular or slit-like cross section have been suggested to function as selective pores for proteins and metabolites controlling passageways in and out of the intracristal space (10, 11).
The conserved multisubunit mitochondrial contact site and cristae organizing system (MICOS) localizes to CJs (13–18). It is crucial for the formation and stabilization of CJs from yeast to humans and likely plays an important role for the regulation of CJ permeability. MICOS is composed of the Mic60 and Mic10 subcomplexes that both have membrane-shaping activity (fig. S1) (19–25). Mic60 is anchored in the inner mitochondrial membrane (IMM) via an N-terminal transmembrane (TM) segment and exposes a large domain into the intermembrane space (Fig. 1A) that associates with Mic19 (and in metazoa additionally Mic25). The Mic60 module of MICOS links CJs to the mitochondrial outer membrane through the formation of membrane contact sites with different partner protein complexes, such as the sorting and assembly machinery for β barrel proteins (SAM complex) (5, 6, 14–16, 26, 27). Mic12 in yeast or MIC13/QIL1 in higher eukaryotes connects the Mic60 and Mic10 modules of MICOS (21, 28). Loss of MICOS components leads to a massively altered cristae morphology in all organisms examined so far. The strongest phenotype with a nearly complete loss of CJs and accumulation of detached sheets of lamellar cristae membranes are observed upon ablation of Mic60 (5, 12–16, 18). How exactly MICOS controls CJ architecture and function is, however, unclear since no structural information of any MICOS component is available.
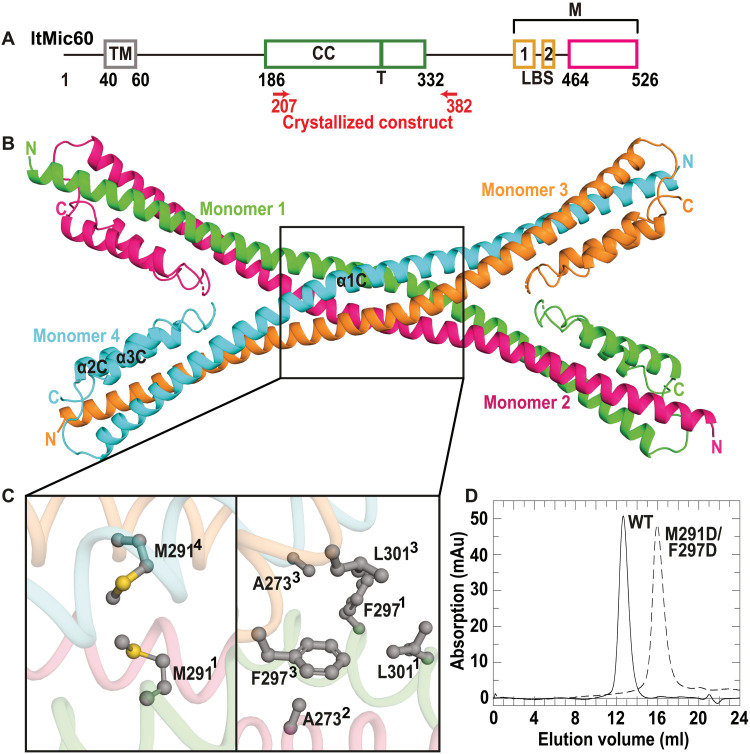
Fig. 1. Mic60 CC domain forms an antiparallel tetramer.
(A) Domain architecture of Mic60 (amino acid numbers refer to Mic60 from L. thermotolerans). LBS 1 and LBS 2, lipid-binding site; M, mitofilin domain. The green bar indicates the tetramer interface (T), and the magenta box denotes the previously reported, sequence-based boundaries of the mitofilin domain. (B) Cartoon representation of the Mic60 CC domain (ltMic60CC). N and C termini of each monomer are labeled. (C) Close-up view of the tetramer interface. (D) SEC profile of ltMic60CC and ltMic60CCM291D/F297D. mAu, milli-absorption units.
Here, we found that the central coiled-coil (CC) domain of Mic60 forms a bow tie–shaped tetrameric assembly. Mic19 promotes Mic60 tetramerization. The C-terminal mitofilin domains of Mic60 dimerize to form two crescent-shaped membrane-binding modules on each side of the CC. Our structural study suggests that the Mic60-Mic19 complex traverses CJs, therefore controlling their diameter and function.
RESULTS
The CC domain of Mic60 forms an antiparallel tetramer
In tomograms of fixed Saccharomyces cerevisiae cells, we frequently observed filamentous densities in the mitochondrial CJs (fig. S1, C and E, and movie S1). Because of its specific localization, we reasoned that the density may constitute part of the MICOS complex and, for its elongated shape, the predicted CC region of Mic60. To obtain high-resolution structural information of the CC domain, we determined its crystal structure from the thermostable yeast Lachancea thermotolerans (amino acids 207 to 382; ltMic60CC) (Fig. 1, A and B, and tables S1 and S2). ltMic60CC forms an elongated α helix (α1C) with two short α helices (α2C and α3C) tightly packed onto the C-terminal ends of α1C (Fig. 1B). In agreement with size exclusion chromatography (SEC) and Blue native polyacrylamide gel electrophoresis (BN-PAGE) analysis, four ltMic60CC molecules assembled into a tetramer via a hydrophobic, highly conserved interface (Fig. 1, B to D; and figs. S2A; S3, A and B; and S4). An antiparallel dimeric CC further dimerized to form a bow tie–shaped tetrameric assembly. In agreement with the structure, a double amino acid substitution in this interface (M291D/F297D) leads to disruption of the ltMic60CC tetramer into monomers (Fig. 1D and fig. S2A).
Mic19 promotes Mic60 tetramerization to stabilize CJs
Since longer constructs of L. thermotolerans Mic60 (ltMic60) could not be expressed in a soluble form, we resorted to Mic60 from Chaetomium thermophilum (ctMic60) for further biochemical analysis. In agreement with previous data (23), an almost full-length construct of ctMic60 excluding the TM region (residues 208 to 691, ctMic60sol) migrated as a dimer in BN-PAGE, with some minor higher-order assemblies (Fig. 2, A and B, and table S2). Notably, addition of purified ctMic19 induced the formation of a heteromeric species, likely containing four molecules of Mic60 and Mic19 each (Fig. 2B).
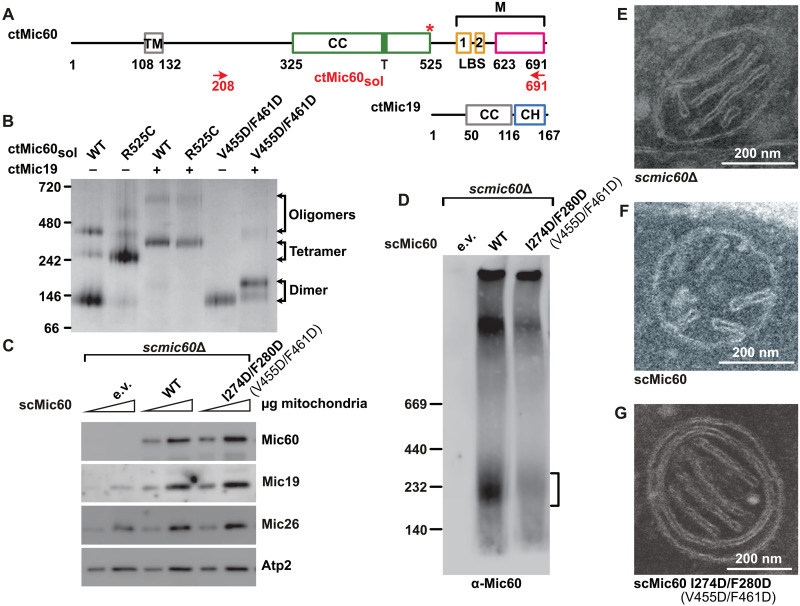
Fig. 2. Mic19 promotes Mic60 tetramerization to stabilize CJs.
(A) Domain architecture of ctMic60 and ctMic19, as in Fig. 1A. CH, CC-helix-CC-helix domain. The red asterisk indicates the position of the artificially introduced disulfide bridge. (B) Representative BN-PAGE analysis showing oligomerization of different ctMic60sol variants and their complexes with ctMic19. (C) Steady-state levels of selected mitochondrial proteins in the indicated S. cerevisiae strains determined by SDS-PAGE and Western blot. e.v., empty vector. Numbering of C. thermophilum proteins shown in parentheses. (D) BN-PAGE and immunoblot analysis of Mic19–Protein A immunoprecipitation elution fractions showing the oligomeric state of isolated Mic60-containing complexes. The deduced position of the tetrameric Mic60 species is indicated by the bracket. Numbering of the corresponding C. thermophilum residues is shown in parentheses. (E to G) Ultrathin sections of S. cerevisiae mitochondria in the indicated strains (see Fig. 4D and fig. S6B for quantification).
To prove the involvement of the tetrameric interface in this assembly, we introduced a structure-based disulfide bridge in ctMic60sol (R525C) that can only form in the tetrameric context (fig. S3C). Under oxidizing conditions, ctMic60solR525C formed a tetramer even in the absence of Mic19 (Fig. 2B and fig. S2B). In the presence of Mic19, the assembly was shifted to a hetero-oligomeric complex of comparable size to ctMic60sol-Mic19 in BN-PAGE. This indicates that the cross-link stabilizes a native form of the Mic60-Mic19 assembly. Furthermore, a double amino acid substitution in the tetrameric interface, V455D/F461D, greatly reduced higher-order assembly of Mic60 in the absence and presence of Mic19 (Fig. 2B).
ctMic60sol protein cosedimented with Folch liposomes derived from bovine brain lipids and dragged ctMic19 into the pellet fraction (fig. S2, C and D) (23). ctMic60solR525C and ctMic60solV455D/F461D proteins cosedimented with liposomes to a similar extent as ctMic60sol. The ctMic60solV455D/F461D variant showed reduced recruitment of Mic19 to Folch liposomes (fig. S2D) and a 30-fold reduced affinity to Mic19 in isothermal titration calorimetry (ITC) experiments compared to ctMic60sol (fig. S5, A, B, and L). This suggests that oligomerization of Mic60 via the tetrameric interface is required for a tight interaction with Mic19.
To analyze the physiological role of Mic60 tetramerization, we used S. cerevisiae as a model. Mic60-deficient S. cerevisiae cells showed massively reduced levels of Mic19 (Fig. 2C), as previously described (14, 16). Expression of a tetramer-disruptive S. cerevisiae Mic60 (scMic60) variant (I274D/F280D; fig. S4) restored mitochondrial accumulation of Mic19 in these cells (Fig. 2C) but interfered with the appearance of Mic60-containing tetrameric complexes, as revealed by BN-PAGE (Fig. 2D). In agreement with the BN-PAGE data obtained with ctMic60sol, the tetramerization-deficient Mic60 variant did not completely abolish self-assembly, as assessed in pull-down experiments, but the homotypic Mic60 interaction was weakened compared to the wild-type (WT) situation (fig. S6A). Mic60-deficient cells showed the expected loss of CJs and fragmentation of the mitochondrial network (16), which was rescued by reexpression of scMic60 (Fig. 2, E and F, and figs. S6B and S7A). Notably, reexpression of tetramerization-defective scMic60 variant in mic60Δ cells did neither restore CJ architecture (Fig. 2G) nor mitochondrial network morphology (fig. S7A). In EM tomograms of these mitochondria, the few remaining CJs did not show filamentous densities (fig. S1, D and E, and movie S2). Furthermore, their mean CJ diameter was reduced from 13.8 to 10.4 nm (fig. S1F). We conclude that tetramerization of Mic60 is required for the formation and/or maintenance of CJs.
The Mic60 mitofilin domain binds to the Mic19 CHCH domain via a conserved interface
We next aimed to characterize the molecular basis of the Mic60-Mic19 interaction, which requires the C-terminal mitofilin domain of Mic60 and the C-terminal CHCH domain of Mic19 (23). Because isolated mitofilin domain constructs tended to precipitate after purification, we determined the crystal structure of a fusion construct (termed Mito1_CHCH), containing the C-terminal region of the ctMic60 mitofilin domain linked to the ctMic19 CHCH domain (table S1 and S2).
Each Mito1_CHCH monomer consists of one α helix from the mitofilin domain (α3M) and α1CH and α2CH of the Mic19 CHCH domain, which together form a three helical bundle (Fig. 3A). The mitofilin-CHCH domain interface has an area of 660 Å2 and is dominated by conserved, hydrophobic interactions (Fig. 3B and figs. S3E, S4, and S8). Mutagenesis of the hydrophobic residues in the context of the longer ctMic60sol and ctMic19 constructs impaired formation of higher-order oligomers in BN-PAGE (fig. S2E) and strongly reduced binding affinity in ITC experiments (see color code in Fig. 3B and fig. S5, A and E to M) and Mic60-mediated recruitment of Mic19 to liposomes (Fig. 3C and fig. S2D). Thus, the interaction in the fusion construct faithfully reflects the interaction of Mic60 and Mic19. In contrast, disruption of the peripheral polar interactions in the interface showed only minor effects on oligomerization and a moderate reduction in binding affinity (Fig. 3B and figs. S2E and S5).
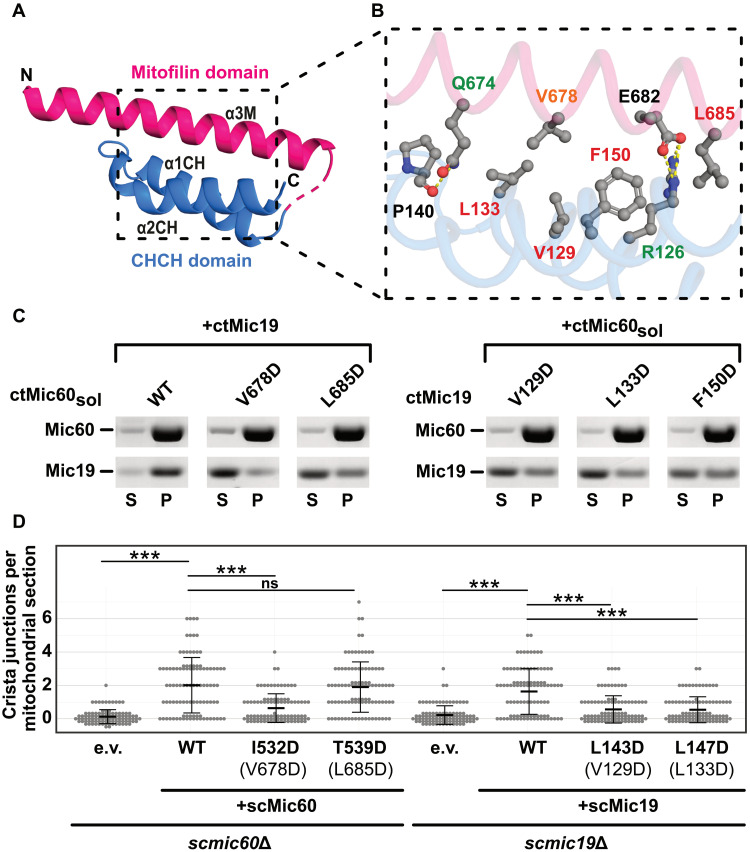
Fig. 3. Structural characterization of the Mic60 and Mic19 interaction.
(A) Structure of Mito1_CHCH fusion construct. Domains are colored as in Fig. 2A. The unresolved linker region is indicated as dashed line. (B) Magnification of the ctMic60-ctMic19 interface. Label colors represent the effect of single amino acid substitutions, as analyzed by ITC experiments: Green, up to 15× reduced; orange, up to 100× reduced; red, no binding, compared to the ctMic60sol-ctMic19 interaction. See fig. S5 for ITC data. (C) Liposome cosedimentation assays of ctMic60sol and ctMic19. S, supernatant; P, pellet. See also fig. S2 (C and D). (D) Quantification of CJs per mitochondrial section in S. cerevisiae mitochondria. ***P ≤ 0.001. ns, not significant. See also fig. S6B. Numbering of the corresponding ctMic60 residues is shown in parentheses.
When reintroduced into Mic60-deficient S. cerevisiae cells, the scMic60 I532D variant with a defective Mic60-Mic19 interface displayed reduced levels of mitochondrial Mic19 in comparison to cells expressing WT scMic60 (figs. S4 and S6C). Isolated Mic60-containing oligomeric complexes were reduced for this variant (fig. S6F). Accordingly, scMic60 I532D–containing mitochondria were almost devoid of CJs and showed a strongly fragmented overall morphology (Fig. 3D and figs. S6B and S7A). By contrast, the scMic60 T539D substitution at the periphery of the interface did not reduce the number of CJs but still affected mitochondrial tubular network formation in cells (Fig. 3D and figs. S6B and S7A). When coexpressed with Protein A–tagged Mic19, both Mic60 variants showed reduced coisolation (fig. S6G) in pull-down experiments, but more pronounced for scMic60 I532D.
Similarly, scMic19 variants with amino acid substitutions in the Mic60-Mic19 interface (scMic19 L143D and L147D; fig. S8) could not be coisolated with Protein A–tagged Mic60 (fig. S6H). When the Mic19 variants were reexpressed in a MIC19 deletion strain, reduced accumulation of Mic19 in mitochondria, a decreased number of CJs, and increased mitochondrial fragmentation were observed (Fig. 3D and figs. S6, B and D, and S7B). These data reveal the critical importance of the hydrophobic Mic60-Mic19 interface for protein stability and MICOS integrity in living cells.
The mitofilin dimer forms a convex membrane-binding site
Besides the N-terminal TM anchor, Mic60 interacts with membranes via two distinct lipid-binding sites (LBS 1 and LBS 2) in the C-terminal region of the protein (Fig. 2A) (23). Constructs including LBS 1 and LBS 2 did not crystallize. Coevolution analysis (29) predicted that LBS 2 is flexible (fig. S3D) and might therefore interfere with crystallization. Sourcing this information, we determined the crystal structure of the mitofilin domain including LBS 1, but without LBS 2, again fused to the CHCH domain of ctMic19 (Mito2_CHCH; tables S1 and S2).
The Mito2_CHCH structure revealed that the mitofilin domain is built of a four-helix bundle: α1M, α2M, and the LBS 1 from one monomer interact with α3M from an opposing monomer to form an interdomain swapped dimer with an interface area of 2000 Å2 (Fig. 4A). The interaction of α3M with the CHCH domain of Mic19 is identical to the previously described structure containing the truncated mitofilin domain construct (Fig. 3A). L676 in the dimer interface points into a hydrophobic pocket of the interacting monomer (Fig. 4A and fig. S3F).
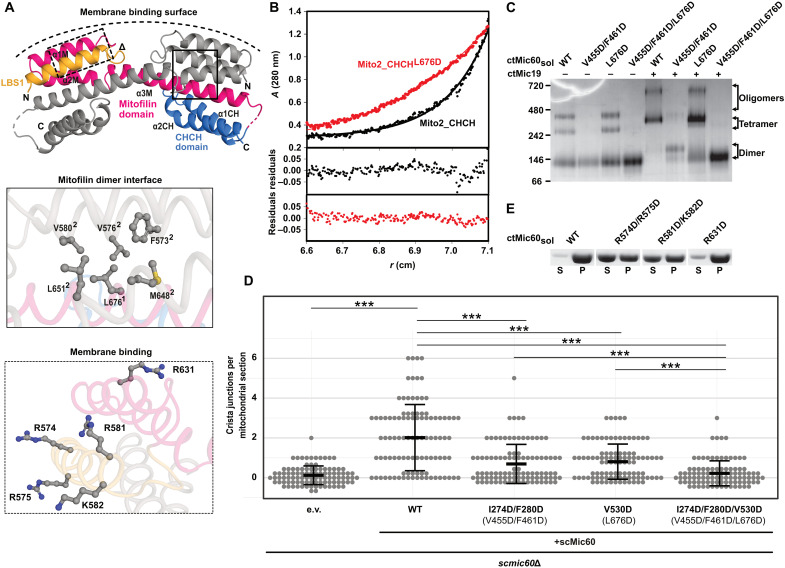
Fig. 4. The mitofilin dimer forms a convex membrane-binding site.
(A) Cartoon representation of the mitofilin dimer in complex with the Mic19 CHCH domain (Mito2_CHCH). One monomer is colored as in Fig. 2A. Dimer interface residues (boxed in straight lines) and potential membrane binding residues (boxed in dashed lines) are shown in the close-up views. The deduced curved membrane binding surface is indicated. (B) Association states of Mito2_CHCH (black) and Mito2_CHCHL676D (red) were analyzed at a concentration of 1 mg/ml by sedimentation equilibrium analysis. Top: The original data (dots) and the fit of the data (line): Mito2_CHCH: molecular mass (Mr) = 34 ± 4 kDa. Mito2_CHCHL676D: Mr = 16 ± 2 kDa (molecular weight of the monomer: 17 kDa). Bottom: The deviation of the fit to the data. (C) BN-PAGE analysis of different ctMic60sol variants and their complexes with ctMic19. (D) Quantification of CJs per mitochondrial section in S. cerevisiae mitochondria. ***P ≤ 0.001. See also see fig. S6B. Numbering of the corresponding C. thermophilum residues is shown in parentheses. (E) Liposome cosedimentation assays of ctMic60sol and membrane binding variants (see also fig. S2C).
In agreement with the structural data, the Mito2_CHCH construct was a dimer in analytical ultracentrifugation experiments, whereas the L676D amino acid substitution rendered the protein monomeric (Fig. 4B). In the longer ctMic60sol construct (amino acids 208 to 691), the L676D variant showed a similar behavior as the WT protein in BN-PAGE, forming mostly a dimer (Fig. 4C). Liposome binding of the variant was also comparable to that of unmodified ctMic60sol (fig. S2C). However, complex formation with ctMic19 in BN-PAGE (Fig. 4C) and ITC (fig. S5, C and L), as well as Mic60-mediated Mic19 recruitment to liposomes, was reduced (fig. S2D), indicating that dimerization of the Mic60 mitofilin domain supports Mic19 recruitment. Simultaneous disruption of the tetrameric and the dimeric interface in the V455D/F461D/L676D variant had an even more drastic effect, completely preventing higher-order oligomer formation of ctMic60sol alone and in complex with ctMic19 (Fig. 4C). In addition, Mic60-dependent Mic19 recruitment to liposomes was severely affected (fig. S2D). Having both interfaces disrupted, the number of CJs per mitochondrial section and the formation of a tubular mitochondrial network in the respective S. cerevisiae variant (scMic60I274D/F280D/V530D) were impaired to a similar extent as in the mic60 knockout strain (Fig. 4D and fig. S6, B and E, and S7A), showing the additive effect of both assembly sites for tetramerization.
The dimeric arrangement positions the two positively charged LBS 1 helices on the outside of the dimer on a convex membrane-binding surface (Fig. 4A). Replacement of the positively charged amino acids on this convex surface led to reduced membrane binding (Fig. 4, A and E, fig. S2C, and table S2), supporting the idea that the convex surface in the mitofilin dimer comprises the membrane-binding site of Mic60.
DISCUSSION
Proper mitochondrial ultrastructure is a prerequisite for mitochondrial functions in cellular energy metabolism and signaling. CJs form a highly curved membrane microdomain at the transition between the inner boundary and cristae membrane. With this study, we provide the first structural insights into CJ architecture leading to an advanced mechanistic understanding of CJ formation, stabilization, and function. The CC domain of Mic60 forms an elongated bow tie–shaped antiparallel CC. Mic19 promotes tetramerization of Mic60 into an active MICOS subcomplex. We reveal the structural basis of the heteromeric interaction of Mic60 and Mic19 and show that the mitofilin domain of Mic60 forms a convex-shaped dimeric membrane-binding module that is tailored to bind to the highly curved CJ membrane. Complementary mutational analysis in yeast demonstrates the critical role of these structural elements for CJ formation and MICOS function in vivo. Together, our findings can be compiled into a structural model of the Mic60-Mic19 complex at CJs (Fig. 5).
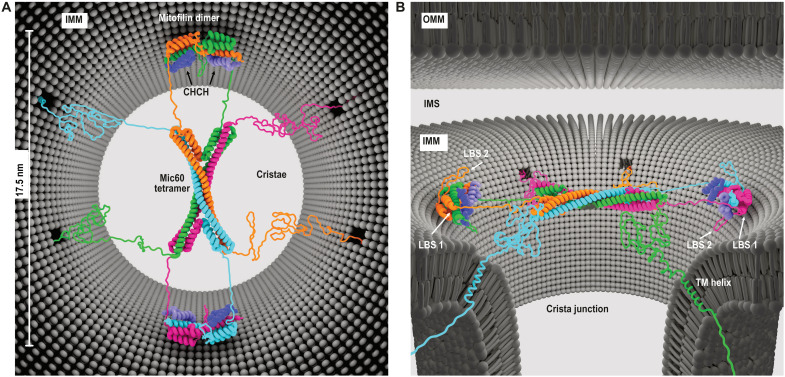
Fig. 5. Model of Mic60-Mic19 function at CJs.
(A) Top view and (B) side view showing the proposed architecture of the Mic60-Mic19 complex at CJs. Each monomer has a different color. Regions not determined by x-ray crystallography are modeled as unstructured elements.
Each of the widely separated endings of the antiparallel Mic60 CC tetramer contains two connections to the mitofilin domain. Since the linker between the two domains is short, the two dimeric membrane-binding mitofilin modules within a tetramer must be localized on opposite sides of the CC. In a cellular context, this implies that the tetrameric CC spans over the CJ, and the two mitofilin domain dimers bind to opposite membrane surfaces in the CJ (Fig. 5, A and B). In this scenario, the convex-shaped membrane-binding sites of the mitofilin dimer are complementary to the membrane curvature of the CJ. The N-terminal TM regions of Mic60 would anchor the complex in the CJ membrane.
The proposed model of the Mic60-Mic19 complex is in agreement with the filamentous density in CJs observed in tomograms of fixed yeast mitochondria (fig. S1). However, the exact molecular identity of the filamentous density and its detailed localization in CJs is not known, and we cannot exclude that it is only indirectly related to the presence of the Mic60 tetramer. Previously proposed ring-like assemblies of Mic60 in CJs are difficult to reconcile with the fourfold symmetry of the tetrameric CC and the twofold symmetry of the mitofilin domain. Super-resolution microscopy of human Mic60 suggests that up to 10 Mic60 molecules are located at one CJ (30), corresponding to two to three tetramers.
Our model has functional implications for the Mic60-Mic19 complex that can be tested in future experiments. For example, the N-terminal region of Mic19, together with parts of Mic60, is available to extend to the outer mitochondrial membrane (OMM). By interacting with SAM and translocase of the outer membrane (TOM) complexes (14, 16, 31), membrane contact sites would be formed (fig. S1). These contact sites have been implicated, among others, in lipid transfer between the IMM and OMM (32). In this context, our structural data would support the idea of an arch dome-like assembly that vaults the entry into cristae.
Our model also explains the uniform diameters of circular or slit-like CJs, which would be governed by the length of the traversing tetrameric CC of the Mic60-Mic19 complex. In line with this idea, vertebrates have longer Mic60 CC sequences and larger CJ diameters. The soluble portion of Mic60 has membrane-remodeling activity that is stimulated by Mic19 binding (23). Thus, one function of the Mic60-Mic19 complex may relate to the generation of membrane curvature at CJs, for example, by membrane scaffolding via the convex-shaped membrane-binding site or by the membrane insertion of hydrophobic residues from LBS 1 and 2. In addition, destabilization of the Mic60 tetramer through site-directed mutagenesis leads to significantly reduced CJ diameters in yeast cells (fig. S1F). This suggests a second function of the Mic60-Mic19 tetramer in preventing the collapse of CJs, for example, by counteracting membrane curvature generation of other membrane-remodeling proteins at cristae. Consequently, the lack of CJs in the absence of Mic60 may be explained by increased CJ scission induced by other membrane-remodeling proteins at CJs. A candidate for membrane curvature generation is Mic10, which forms a small wedge-shaped TM hairpin. Its oligomerization into higher-order assemblies within the membrane was shown to induce membrane curvature (19, 20). Further contributions to cristae remodeling may origin from dynamin-like Mgm1/OPA1 filaments or ATP synthase dimers (9, 33). A recent study shed first light on how these complexes may work together and orchestrate their membrane-remodeling functions, which has paved the ground for further studies on this exciting topic (34).
Last, by spanning across CJs, the Mic60-Mic19 complex is tailored to serve as a physical barrier preventing the free diffusion of proteins and metabolites in and out of the cristae space. Notably, an intrinsically disordered region is predicted between the N-terminal TM and the central CC domain of Mic60 proteins (Fig. 5, A and B) (35). Similar to the FG (phenylalanine-glycine) repeats in nuclear pore complexes, this region may contribute to the formation of a sieve-like diffusion barrier (36) to control selective passage across CJs.
Together, our structure function study provides molecular insights into CJs representing a cellular membrane hub that had remained a terra incognita of structural biology. It reveals fundamental mechanisms of eukaryotic cell biology that have implications for the formation and architecture of membrane contact sites and the selective exchange of molecules between different cellular compartments.
MATERIALS AND METHODS
Cloning and plasmids
To obtain structural information, about 200 Mic60 and Mic19 constructs from different species were cloned and expressed. Codon-optimized constructs of ltMic60 (UniProt ID: C5E325, synthesized by Eurofins Genomics), ctMic60 (UniProt ID: G0SHY5), and Mic19 (ctMic19; UniProt ID: G0S140) were cloned into the pET26b vector (ltMic60) or modified pET28a (ctMic60 and ctMic19), encoding a C-terminal His6-tagged and a human rhinovirus 3C (HRV-3C) protease cleavable N-terminal His6-tagged fusion construct, respectively. Constructs Mito1_CHCH and Mito2_CHCH were generated using overlap extension polymerase chain reaction (PCR) and cloned into pETDuet-1 vector (Merck) encoding a HRV-3C protease cleavable N-terminal His6-tagged fusion protein. Variants of ltMic60, ctMic60, and ctMic19 were generated using site directed mutagenesis (37).
Expression and purification
Expression plasmids were freshly transformed into Escherichia coli BL21 (DE3) cells. Protein expression was carried out in terrific broth supplemented with kanamycin (50 μg/ml) or ampicillin (100 μg/ml). The cultures were grown until OD600 (optical density at 600 nm) reached 0.8 at 37°C and 80 rpm and protein expression subsequently induced by the addition of 300 μM isopropyl β-d-1-thiogalactopyranoside and incubated at 20°C for another 20 hours. The cells were centrifuged at 4000g, collected, and frozen at −20°C until needed.
ltMic60 expression plasmid containing cells were diluted in lysis buffer [50 mM Hepes/NaOH (pH 8.0), 500 mM NaCl, 20 mM imidazole, 10% glycerol, 1 mM 3,3′,3′′-phosphanetriyltripropanoic acid (TCEP), and deoxyribonuclease I (1 mg/ml) (Roche)] before disruption using a microfluidizer (Microfluidics). To remove insoluble parts, the solution was centrifuged at 100,000g for 45 min at 4°C. The cleared supernatant was applied onto a prepacked Ni2+ sepharose High Performance IMAC (immobilized metal affinity chromatography) resin (GE Healthcare Life Science) containing gravity flow column loaded with 100 mM nickel sulfate and equilibrated in lysis buffer. The column was washed using lysis buffer and bound proteins eluted with lysis buffer containing 50 and 500 mM imidazole, respectively. To remove the C-terminal His6-tag, carboxypeptidase A (from bovine pancreas, Sigma-Aldrich) treatment was applied during overnight dialysis at 4°C against dialysis buffer [50 mM Hepes/NaOH (pH 8.0), 500 mM NaCl, and 10% glycerol]. A second Ni2+ sepharose column was used to separate cleaved from uncleaved protein. Last, a SEC run using a S200 column and SEC buffer [50 mM Hepes/NaOH (pH 8.0), 500 mM NaCl, 10% glycerol, and 1 mM TCEP] (GE Healthcare Life Science) was applied to separate pure protein from aggregates and carboxypeptidase A. Pure protein was concentrated to 31 mg/ml, flash-frozen in liquid nitrogen, and stored at −80°C until further use.
All ctMic60 and ctMic19 constructs were purified in a similar manner as described for ltMic60. However, the lysis buffer contained 50 mM Hepes/NaOH (pH 7.5), 500 mM NaCl, and 20 mM imidazole. The N-terminal His6-tag was cleaved using recombinant His6-tagged HRV-3C protease during overnight dialysis using 50 mM Hepes/NaOH (pH 7.5), 150 mM NaCl, and 25 mM imidazole. The final SEC was performed using S200 and S75 SEC columns (GE Healthcare Life Science) equilibrated in 20 mM Hepes/NaOH (pH 7.5) and 150 mM NaCl for ctMic60 and ctMic19, respectively. ctMic60solR525C was purified in a similar manner in the presence of 2 to 5 mM dithiothreitol (DTT).
Mass spectrometry
Mass spectrometry (MS) analysis using liquid chromatography–electrospray ionization–time-of-flight MS (LC-ESI-TOF-MS) indicated that a disulfide bridge was formed in all ctMic19 CHCH domain-containing constructs domain–containing constructs, as the calculated and the measured molecular mass showed a difference of exactly 2 Da. Protein intact mass analyses were conducted on an Agilent 1290 Infinity II UHPLC system coupled to an Agilent 6230B TOF LC/MS instrument equipped with an AJS (Agilent Jet Stream Technology) ion source operated in positive ion mode (denaturing conditions). Protein samples were desalted using a Zorbax 300SB-C3 guard column (2.1 mm by 12.5 mm, 5 μm). Protein solutions were diluted in 0.1% formic acid (in H2O) to 0.06 mg/ml. Approximately 0.3 μg of sample was injected for each analysis. LC/MS parameters were adapted from (38). The ion source was operated with the capillary voltage at 4000 V, nebulizer pressure at 50 psi, drying and sheath gas at 350°C, and drying and sheath gas flow rate at 12 and 11 liters/min, respectively. The instrument ion optic voltages were as follows: fragmentor, 250 V; skimmer, 65 V; and octopole radio frequency voltage, 750 V. MS data were analyzed using the protein deconvolution feature of the MassHunter BioConfirm Version 10.0 software (Agilent) that uses the maximum entropy algorithm for accurate molecular mass calculation. Deconvolution was performed between mass range of 800 to 2500 m/z (mass-to-charge ratio), using peaks with a ratio of signal to noise greater than 30:1. The deconvoluted mass range was set at 5 to 25 kDa, and the step mass was 1 Da.
Analytical SEC
Analytical SEC was performed using 100 μl of a protein solution (3 mg/ml) on a Superdex S200 10/300 (GE Healthcare Life Science) column at a flow rate of 0.5 ml/min at 4°C. The running buffer contained 50 mM Hepes/NaOH (pH 8) and 500 mM NaCl.
Crystallization, data collection, refinement, and other tools
Initial crystallization conditions were identified with the vapor diffusion method in 96-well sitting drop format at 20°C using an automated dispensing robot (Art Robbins Instruments). Optimized and plate-like protein crystals grew within 3 to 5 days by mixing 1 μl of ltMic60CC (31 mg/ml) and 1 μl of reservoir containing 17.5% polyethylene glycol (PEG) 1500, 0.1 M MMT buffer (pH 7.1) (a 1:2:2 molar mixture of dl-malic acid, MES, and tris base), and 0.1 M d-sorbitol. The drop was equilibrated against 500 μl of the same reservoir solution in 24-well hanging drop format at 20°C. To obtain high-quality diffracting protein crystals, dehydration was performed. Therefore, protein crystals were transferred into a new drop containing 35% PEG 1500, 250 mM NaCl, 0.1 M MMT buffer (pH 7.2), and 0.1 d-(-)-fructose and equilibrated against 500 μl of the same solution for 24 hours before flash cooling into liquid nitrogen.
Diffraction quality crystals of Mito1_CHCH and Mito2_CHCH grew within 2 to 10 days and were obtained in 96-well sitting drop format at 20°C by automated mixing 0.2 μl of protein solution (17 to 20 mg/ml) and 0.2 μl of reservoir solution (80 μl total reservoir volume). Ten minutes before crystallization trials, Mito1_CHCH was mixed with 1% trypsin (w/w) and incubated at 4°C. The final reservoir solutions contained 33% (v/v) Jeffamine M-600 and 0.1 M Hepes/NaOH (pH 7.2) (Mito1_CHCH) or 30% (w/v) Jeffamine ED-2001 and 0.1 M Hepes/NaOH (pH 7.0) (Mito2_CHCH). After they stopped growth, crystals were directly flash-cooled in liquid nitrogen. Diffraction data were collected at −173°C and 0.9184 Å on beamline BL14.1 operated by Helmholtz-Zentrum Berlin at the BESSY II electron storage ring (Berlin-Adlershof, Germany) (39) and indexed, integrated, and scaled with XDSAPP (40). The structure of ltMic60CC and Mito1_CHCH was solved by molecular replacement using AMPLE from the CCP4 software packaging (41, 42). For ltMic60CC, four ideal α helices build by 20 alanine residues have been placed, and AutoBuild from the PHENIX suite was used for initial model building (43, 44). For Mito1 _CHCH, three ideal α helices built by 30 alanine residues could be placed. Additional helices have been identified using Phaser-MR (44, 45) from the PHENIX suite. SHELXE (46) and AutoBuild were used to obtain the initial model. The structure of Mito2_CHCH was solved by molecular replacement with Phaser-MR using the final refined structure of Mito1_CHCH as search model. ltMic60CC crystallized in space group P4212 with one monomer, Mito1_CHCH in P1 with six monomers and Mito2_CHCH in P21 with four monomers arranged as two dimers in the asymmetric unit. For ltMic60CC, only amino acids 235 to 382 are visible in the structure. Residues ctMic60661–691 and ctMic19118–158 are visible in all monomers of Mito1_CHCH. In case of Mito2_CHCH, ctMic60565–586, ctMic60622–689, and ctMic19118–160 are visible in all monomers.
Refinement was carried out using iterative steps of manual model building in Coot (47) and maximum likelihood refinement with individual B factors, Translation-Libration-Screw-rotation (TLS), and secondary structure restraints using phenix.refine (48). Final structure validation was carried out with MolProbity (49). All statistics for data collection and refinement as well as the corresponding Protein Data Bank (PDB) codes can be found in table S1.
Surface conservation plot were created using the ConSurf Server (50) with standard settings and multiple sequence alignments using MULTALIN64. Figures were prepared with PyMOL (The PyMOL Molecular Graphics System, Version 1.8.2.3 Schrödinger LLC). PDBePISA web server (51) was used to calculate the interface area between the mitofilin domain and the CHCH domain.
Evolutionary coupling analysis
Structure prediction of the C-terminal region of Mic60 was done using the EVCoupling server (29). Structure predictions of ctMic60557–685 were obtained using the input sequence ctMic60550–693 (monomer pipeline, version 1, bitscore 0.1). The highest scoring folding candidate is shown.
Oxidation of cysteines
To artificially induce disulfide bridge formation, 30 μM of the respective protein has been dialyzed against 20 mM tris-HCl (pH 7.5) and 150 mM NaCl overnight. The next day, oxidation of free cysteines was performed using 500 μM CuSO4 for 15 min at 4°C, followed by the addition of 50 mM EDTA. The residual CuSO4 and EDTA were removed using a PD-10 column (GE Healthcare Life Science). The concentration of the final oxidized protein was set to 1 mg/ml for further analysis.
Blue native polyacrylamide gel electrophoresis
BN-PAGE analysis of recombinant purified proteins was performed using the NativePAGE bis-tris system (Thermo Fisher Scientific). Five micrograms (μg) of single proteins or 5 μg of Mic60 variants incubated for 15 min with 2 μg of Mic19 variants were applied on 4 to 16% acrylamide BN-PAGE gradient gels and run for 2 hours at 150 V on ice. Proteins were visualized by Coomassie Brilliant Blue (CBB) staining. Immunoprecipitated mitochondrial protein complexes eluted in digitonin buffer (see below) were mixed with 10× BN-PAGE loading buffer [5% (w/v) CBB G250, 500 mM 6-aminocaproic acid, and 100 mM bis-tris/HCl (pH 7.0)] and loaded on home-made 4 to 13% BN-PAGE gradient gels that were run for 2 to 3 hours at 150 V in a water-cooled Hoefer gel chamber at 6°C. Proteins were blotted on polyvinylidene difluoride (PVDF) membranes and visualized using specific antibodies together with an enhanced chemiluminescence (ECL) detection system.
Isothermal titration calorimetry
ITC experiments were performed using the PEAQ-ITC system (Malvern) in 20 mM Hepes/NaOH (pH 7.5) and 150 mM NaCl at 10°C. Mic60 concentration in the sample cell varied between the variants in the range of 44 to 81 μM, and Mic19 concentration in the syringe varied between 391 and 809 μM. The Malvern analysis software was used to integrate the binding isotherms and calculate the binding parameters. At least two biological replicates per run were conducted, while one of them is shown.
Liposome cosedimentation assay
Folch lipids (total bovine brain lipids fraction I, Sigma-Aldrich) were dried under an argon stream, dissolved in 20 mM Hepes/NaOH (pH 7.5) and 150 mM NaCl, incubated overnight at 4°C, and sonified for 15 min in a sonification bath. Forty microliters of a reaction mixture containing liposomes (0.6 mg/ml) and 5 μM protein (or complex) was incubated for 30 min at room temperature (RT) and centrifuged at 200,000g for 16 min at 20°C. The respective supernatant and pellet fractions were analyzed by SDS-PAGE, and the protein bands were quantified using ImageJ (version 1.50i) (52).
Analytical ultracentrifugation
The fusion constructs Mito2_CHCH and Mito2_CHCHL676D were analyzed at protein concentrations of 0.05 to 1 mg/ml in 20 mM Hepes/NaOH (pH 7.5) and 150 mM NaCl using a Beckman Optima XL-I centrifuge equipped with an An50Ti rotor and double sector cells. Sedimentation equilibrium measurements were carried out at 20°C and 16,000 rpm. The data were recorded at a wavelength of 230 or 280 nm and analyzed using the software Sedfit (53). No concentration-dependent assembly was observed for either construct in the applied concentration range.
S. cerevisiae strains and plasmids
S. cerevisiae strains are derivates of YPH499 (54). Deletion strains mic60∆ and mic19∆ were described previously (16). For generation of different Mic60 and Mic19 variants containing individual amino acid substitutions, PCR fragments containing either the MIC60 or the MIC19 open reading frames together with their natural promoter and terminator regions were cloned into plasmid pRS416, and the respective mutations were generated via site-directed mutagenesis (see table S3 for a list of plasmids and table S4 for a list of S. cerevisiae strains used in this study). S. cerevisiae strains expressing Mic19 fused to a C-terminal Protein A tag for immunoprecipitation were generated by homologous recombination using a transformation cassette that consists of a tobacco etch virus protease cleavage site, a ZZ domain of S. aureus Protein A for immunoglobulin G (IgG) binding, and a HIS3 marker gene for selection (55). For isolation of mitochondria, S. cerevisiae cells were grown in liquid minimal glycerol medium [0.67% (w/v) yeast nitrogen base, 0.07% (w/v) complete supplement mixture (CSM) amino acid mix minus uracil, and 3% (v/v) glycerol] at 30°C.
Isolation of S. cerevisiae mitochondria
Cells were grown in minimal glycerol medium to midlog phase and harvested by centrifugation at 1200g for 5 min at RT. Pellets were resuspended in wet weight DTT softening buffer (2 ml/g) [0.1 M tris/H2SO4 (pH 9.4) and 10 mM DTT] and incubated for 20 min at 30°C. After centrifugation (2000g for 5 min at RT), cell pellets were washed with Zymolyase buffer [1.2 M sorbitol and 20 mM KPi (pH 7.4)]. Cells were resuspended in 6.5 ml of Zymolyase buffer containing 4 mg of Zymolyase per gram of cells (wet weight) and incubated for 30 min at 30°C for enzymatic digestion of the cell wall. The resulting spheroplasts were harvested by centrifugation (2000g for 5 min at RT), washed again with Zymolyase buffer, and resuspended in 6.5 ml of homogenization buffer [0.6 M sorbitol, 10 mM tris/HCl (pH 7.4), 1 mM EDTA, 0.2% (w/v) bovine serum albumin, and 1 mM phenylmethylsulfonyl fluoride (PMSF)] per gram of cells. Spheroplasts were then homogenized using a glass-teflon Dounce homogenizer (15 strokes). The suspension was centrifuged (1500g for 5 min at 4°C) to remove cell debris. The supernatant was transferred into a new tube and again centrifuged (15,000g for 10 min at 4°C). The mitochondria-containing pellet was resuspended in SEM buffer [250 mM sucrose, 10 mM MOPS (pH 7.2), and 1 mM EDTA]. Protein concentration was measured via a Bradford assay and adjusted to 10 mg of total protein/ml of suspension. Isolated mitochondria were flash-frozen in liquid nitrogen and stored at −80°C.
Steady-state levels of mitochondrial proteins
For comparing the steady-state levels of individual proteins in WT and mutant mitochondria, frozen samples were thawed slowly on ice and centrifuged (15,000g for 10 min at 4°C). The mitochondrial pellet was resuspended in Laemmli buffer [60 mM tris/HCl (pH 6.8), 2% (w/v) SDS, 10% (v/v) glycerol, 0.01% bromophenol blue, and 1% β-mercaptoethanol). The samples were incubated for 10 min at 65°C and loaded on SDS-PAGE gels for protein separation. Subsequently, proteins were blotted on PVDF membranes and visualized using specific antibodies together with an ECL detection system (see table S5 for a list of the used antibodies).
Immunoprecipitation
For IgG affinity chromatography, 0.9 mg of isolated S. cerevisiae mitochondria (total protein content) containing Protein A–tagged Mic19 or Mic60 was resuspended in solubilization buffer [20 mM tris/HCl (pH 7.4), 50 mM NaCl, 0.1 mM EDTA, 10% (v/v) glycerol, 2 mM PMSF, 1× Roche protein inhibitor cocktail, and 1% (w/v) digitonin] and incubated for 30 min at 4°C. Mitochondrial detergent extracts were centrifuged (20,000g for 10 min at 4°C), and the supernatant was incubated with 50 μl of human IgG–coupled Sepharose beads [pre-equilibrated with 0.5 M acetate (pH 3.4)] for 90 min at 4°C. Beads were washed 10× with digitonin buffer [20 mM tris/HCl (pH 7.4), 0.5 mM EDTA, 60 mM NaCl, 10% (v/v) glycerol, 2 mM PMSF, and 0.3% digitonin], followed by centrifugation (700g for 30 s at 4°C). Bound proteins were eluted either by tobacco etch virus protease cleavage over night at 4°C and centrifugation (1200g for 30 s at 4°C) or by incubation with 50 mM glycine (pH 2.5) and subsequent neutralization with 1 M tris (pH 8.0), followed by centrifugation (1200g for 1 min at 4°C).
Quantification of Western blots
Western blots analyses of immunoprecipitation experiments were quantified using ImageJ2 Version 2.3.0/1.53q. Background-corrected signals obtained with the relevant antibodies were quantified in total and eluate samples. The signal intensities of the total samples were set to 100% to determine the relative amount of signal in eluate samples. Relative coisolation efficiencies were calculated by determining the ratio of WT/WT and WT/variant setting the WT/WT ratio to 100% for each protein analyzed.
EM of S. cerevisiae mitochondria
S. cerevisiae cells grown in minimal medium supplemented with 2% glucose for 24 hours at 30°C were diluted in minimal glycerol medium and grown until early log phase. Cells were handled as previously described (56). Cells were fixed for 3 hours with freshly prepared 4% (w/v) paraformaldehyde and 0.5% (v/v) glutaraldehyde (GA) in 0.1 M citrate buffer adjusted to growth conditions for pH and temperature (57). After washing with citrate buffer, cells were permeabilized for 1 hour with 1% (w/v) sodium metaperiodate at 4°C. Cells were washed with 0.1 M phosphate buffer and then embedded in 12% gelatine by cooling the 37°C warm gelatine in ice after 10 min of incubation. Cubes (1 mm3) were cut, infiltrated overnight with 2.3 M sucrose, mounted onto specimen chucks, and frozen in liquid nitrogen. Ultrathin sections were cut using an UC7 ultramicrotome (Leica) at −110°C and collected on formvar/carbon-coated copper grids (Plano). The gelatine was removed by washing with PBS at 37°C and water before staining with 3% (w/v) silicotungstic acid hydrate (Fluka) in 2.8% (w/v) polyvinyl alcohol (Sigma-Aldrich) in water for 5 min. Grids were imaged after drying with the transmission electron microscope EM910 (Zeiss) operating at 80 kV and equipped with a Quemesa charge-coupled device (CCD) camera and the imaging software iTEM (Emsis) at ×10,000 magnification. All data are presented as means ± SD, and value differences were compared statistically. Data analysis and plotting were done with the statistic program R. The normal distribution was tested using the Kolmogorov-Smirnov test and the Q-Q plot. Since data were not normally distributed, the two-sided Wilcoxon rank sum test for independent samples with continuity correction was used. For all groups, n = 100 mitochondrial cross sections were analyzed. Differences of P ≤ 0.05 were considered significant (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001).
S. cerevisiae cells used for tomography analysis were treated in a Zymolase-containing buffer [50 mM tris (pH 7.5), 5 mM MgCl2, 1.4 M sorbitol, 0.5% mercaptoethanol, and Zymolase 20T (0.15 mg/ml)] to remove the cell wall. Upon centrifugation at 2000 rpm in a Stat Spin Microprep 2 table top centrifuge, the pellet was fixed for 60 min at RT by immersion in 2% glutaraldehyde in 0.1 M cacodylate buffer at pH 7.4. The fixed pellet was immobilized with 2% agarose in sodium cacodylate buffer at pH 7.4. Small pieces of the immobilized pellet were fixed using buffered 1% osmium tetroxide, followed by aqueous 1% uranyl acetate. Samples were then dehydrated and embedded in Agar 100. Alternatively, pelleted cells were vitrified using a BAL-TEC HPM-010 high-pressure freezer. The samples were substituted over 72 hours at −90°C in a solution of 2% OsO4, 0.1% uranyl acetate, and 5% H2O in anhydrous acetone. After a further incubation over 20 hours at −20°C, samples were warmed up to +4°C and washed with anhydrous acetone subsequently. The samples were embedded at RT in Agar 100 (Epon 812 equivalent) at 60°C over 24 hours. After ultrathin sectioning (230 nm), section were counterstained with lead citrate. Images were taken in a Talos L120C transmission electron microscope (Thermo Fisher Eindhoven, The Netherlands). Tilt series from 230-nm-thick sections were recorded in 4K mode using the dose symmetric scheme from −65° to 65° at 2° intervals. Tomograms were calculated using Etomo (http://bio3d.colorado.edu/) (58). Size determination of CJs was calculated using 3dmod.
Analysis of mitochondrial networks
S. cerevisiae cells were grown in liquid minimal medium [0.67% (w/v) yeast nitrogen base, 0.07% (w/v) CSM amino acid mix minus uracil, and 3% (w/v) glucose] at 24°C until an OD600 of 1.0. Cells were pelleted by centrifugation for 3 min at 400g. Mitochondrial networks of the S. cerevisiae cells were stained using 175 nM 3,3′-dihexyloxacarbocyanine iodide in buffer A [10 mM Hepes (pH 7.2) and 5% glucose] for 10 min at RT. Cells were washed once with buffer A. For imaging, cells were transferred on a microscopy slide, and the sample was covered with a cover slip. Images were taken immediately at a Leica DMi8 fluorescent microscope with a 63×/1.40 objective and a Leica DFC3000 G CCD camera. Z-stacks, covering the whole cell, were recorded for each image. For each cell, sections of 8 μm by 8 μm were cropped and deconvoluted with Huygens Essential (Scientific Volume Imaging, Netherlands, http://svi.nl). Maximum intensity projections were created in Fiji. For quantification of mitochondrial morphology, tubular versus nontubular mitochondrial networks were assessed by the experimenter in cells from three independent and blinded experiments (for each individual culture, approximately 300 cells were counted). Percentages were calculated and the P values were derived by an unpaired t test using GraphPad Prism.
Preparation of the Mic60-Mic19 model within CJs
The model of Mic60 and Mic19 in CJs was created and prepared by E. Werner, RNS Berlin (www.rns.berlin), who granted copyrights. The model comprises the crystal structures of the CC region of Mic60 from L. thermotolerans (ltMic60CC, residues 235 to 382), the LBS 1 and mitofilin domain of ctMic60, and the CHCH domain of ctMic19 (Mito2_CHCH). In this, the sequence of ltMic60 was used as template. All other parts of Mic60 were modeled as unstructured regions. A CJ diameter of 17.5 nm was used (13). The model was generated using the Maya software from Autodesk Inc. (www.autodesk.com/products/maya/) and Modeling kit and Rigging kit of the plugin Molecular Maya (mMaya) from Digizyme Inc. (https://clarafi.com/tools/mmaya/).
Acknowledgments
We thank Y. Roske for help with crystallographic data collection, E. Werner (Research Network Services Ltd., Berlin, Germany) for the creation of the CJ model figure, and the entire BESSY team for support during data collection at beamline MX14.1, MX14.2, or MX14.3. We are grateful to C. Schiel (Electron Microscopy, MDC) for preparing ultrathin sections of S. cerevisiae cells, A. Schütz (Protein Production and Characterization Platform, MDC) for support with the MS analysis, K. Noll (Saarland University Medical School) for assistance with the biochemical characterization of mitochondrial protein complexes, and S. Stoldt (University Medical Center Göttingen) for the preparation of S. cerevisiae for electron tomography.
Funding: This project was supported by grants from the Deutsche Forschungsgemeinschaft (FOR 2848/P06 to O.D., FOR 2848/Z1 to S.Ja. and D.R., and SFB 894/P20 and IRTG 1830 to M.v.d.L.), the ERC grant MitoShape (ERC-2013-CoG-616024 to O.D.), the ERC grant MitoCristae (ERCAdG no. 835102 to S.Ja.), a Humboldt fellowship to J.K.N., a Boehringer Ingelheim Fonds fellowship to F.W., and a DOC Fellowship of the Austrian Academy of Sciences to M.H.
Author contributions: T.B.-B. and K.F. designed MICOS constructs, grew crystals, solved their structures, and performed biochemical experiments, with support from C.B., E.L., F.W., J.L., and S.Ju. A.v.d.M. and K.v.d.M. performed structure-based functional experiments in S cerevisiae. J.K.N. conducted and analyzed evolutionary coupling predictions. E.L., S.K., and D.R. provided EM analyses of S. cerevisiae mitochondria. M.H. designed the Mic60-Mic19 fusion construct and grew initial crystals. H.L. performed analytical ultracentrifugation analyses. T.B.-B., K.F., F.W., S.Ja., M.v.d.L., and O.D. designed research and interpreted structural, biochemical, and EM data. T.B.-B., K.F., M.v.d.L., and O.D. wrote the manuscript with inputs from all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The atomic coordinates of ltMic60CC, Mito1_CHCH, and Mito2_CHCH have been deposited in the PDB with accession numbers 7PUZ, 7PV0, and 7PV1.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Tables S1 to S5
References
Other Supplementary Material for this manuscript includes the following:
Movies S1 and S2
REFERENCES AND NOTES
- 1.Nunnari J., Suomalainen A., Mitochondria: In sickness and in health. Cell 148, 1145–1159 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Youle R. J., Mitochondria-striking a balance between host and endosymbiont. Science 365, eaaw9855 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Chan D. C., Mitochondrial dynamics and its involvement in disease. Annu. Rev. Pathol. 15, 235–259 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Giacomello M., Pyakurel A., Glytsou C., Scorrano L., The cell biology of mitochondrial membrane dynamics. Nat. Rev. Mol. Cell Biol. 21, 204–224 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Schorr S., van der Laan M., Integrative functions of the mitochondrial contact site and cristae organizing system. Semin. Cell Dev. Biol. 76, 191–200 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Colina-Tenorio L., Horten P., Pfanner N., Rampelt H., Shaping the mitochondrial inner membrane in health and disease. J. Intern. Med. 287, 645–664 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Davies K. M., Anselmi C., Wittig I., Faraldo-Gomez J. D., Kuhlbrandt W., Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc. Natl. Acad. Sci. U.S.A. 109, 13602–13607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glytsou C., Calvo E., Cogliati S., Mehrotra A., Anastasia I., Rigoni G., Raimondi A., Shintani N., Loureiro M., Vazquez J., Pellegrini L., Enriquez J. A., Scorrano L., Soriano M. E., Optic atrophy 1 is epistatic to the core MICOS component MIC60 in mitochondrial cristae shape control. Cell Rep. 17, 3024–3034 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faelber K., Dietrich L., Noel J. K., Wollweber F., Pfitzner A. K., Mühleip A., Sánchez R., Kudryashev M., Chiaruttini N., Lilie H., Schlegel J., Rosenbaum E., Hessenberger M., Matthaeus C., Kunz S., von der Malsburg A., Noé F., Roux A., van der Laan M., Kühlbrandt W., Daumke O., Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1. Nature 571, 429–433 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frey T. G., Renken C. W., Perkins G. A., Insight into mitochondrial structure and function from electron tomography. Biochim. Biophys. Acta 1555, 196–203 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Mannella C. A., Structure and dynamics of the mitochondrial inner membrane cristae. Biochim. Biophys. Acta 1763, 542–548 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Kondadi A. K., Anand R., Reichert A. S., Cristae membrane dynamics - A paradigm change. Trends Cell Biol. 30, 923–936 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Rabl R., Soubannier V., Scholz R., Vogel F., Mendl N., Vasiljev-Neumeyer A., Körner C., Jagasia R., Keil T., Baumeister W., Cyrklaff M., Neupert W., Reichert A. S., Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J. Cell Biol. 185, 1047–1063 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harner M., Körner C., Walther D., Mokranjac D., Kaesmacher J., Welsch U., Griffith J., Mann M., Reggiori F., Neupert W., The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 30, 4356–4370 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoppins S., Collins S. R., Cassidy-Stone A., Hummel E., DeVay R. M., Lackner L. L., Westermann B., Schuldiner M., Weissman J. S., Nunnari J., A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J. Cell Biol. 195, 323–340 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von der Malsburg K., Müller J. M., Bohnert M., Oeljeklaus S., Kwiatkowska P., Becker T., Loniewska-Lwowska A., Wiese S., Rao S., Milenkovic D., Hutu D. P., Zerbes R. M., Schulze-Specking A., Meyer H. E., Martinou J. C., Rospert S., Rehling P., Meisinger C., Veenhuis M., Warscheid B., van der Klei I., Pfanner N., Chacinska A., van der Laan M., Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev. Cell 21, 694–707 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Jans D. C., Wurm C. A., Riedel D., Wenzel D., Stagge F., Deckers M., Rehling P., Jakobs S., STED super-resolution microscopy reveals an array of MINOS clusters along human mitochondria. Proc. Natl. Acad. Sci. U.S.A. 110, 8936–8941 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stephan T., Brüser C., Deckers M., Steyer A. M., Balzarotti F., Barbot M., Behr T. S., Heim G., Hübner W., Ilgen P., Lange F., Pacheu-Grau D., Pape J. K., Stoldt S., Huser T., Hell S. W., Möbius W., Rehling P., Riedel D., Jakobs S., MICOS assembly controls mitochondrial inner membrane remodeling and crista junction redistribution to mediate cristae formation. EMBO J. 39, e104105 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bohnert M., Zerbes R. M., Davies K. M., Mühleip A. W., Rampelt H., Horvath S. E., Boenke T., Kram A., Perschil I., Veenhuis M., Kühlbrandt W., van der Klei I. J., Pfanner N., van der Laan M., Central role of Mic10 in the mitochondrial contact site and cristae organizing system. Cell Metab. 21, 747–755 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Barbot M., Jans D. C., Schulz C., Denkert N., Kroppen B., Hoppert M., Jakobs S., Meinecke M., Mic10 oligomerizes to bend mitochondrial inner membranes at cristae junctions. Cell Metab. 21, 756–763 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Guarani V., McNeill E. M., Paulo J. A., Huttlin E. L., Fröhlich F., Gygi S. P., van Vactor D., Harper J. W., QIL1 is a novel mitochondrial protein required for MICOS complex stability and cristae morphology. eLife 4, e06265 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedman J. R., Mourier A., Yamada J., McCaffery J. M., Nunnari J., MICOS coordinates with respiratory complexes and lipids to establish mitochondrial inner membrane architecture. eLife 4, e07739 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hessenberger M., Zerbes R. M., Rampelt H., Kunz S., Xavier A. H., Purfürst B., Lilie H., Pfanner N., van der Laan M., Daumke O., Regulated membrane remodeling by Mic60 controls formation of mitochondrial crista junctions. Nat. Commun. 8, 15258 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tarasenko D., Barbot M., Jans D. C., Kroppen B., Sadowski B., Heim G., Möbius W., Jakobs S., Meinecke M., The MICOS component Mic60 displays a conserved membrane-bending activity that is necessary for normal cristae morphology. J. Cell Biol. 216, 889–899 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tirrell P. S., Nguyen K. N., Luby-Phelps K., Friedman J. R., MICOS subcomplexes assemble independently on the mitochondrial inner membrane in proximity to ER contact sites. J. Cell Biol. 219, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bohnert M., Wenz L. S., Zerbes R. M., Horvath S. E., Stroud D. A., von der Malsburg K., Müller J. M., Oeljeklaus S., Perschil I., Warscheid B., Chacinska A., Veenhuis M., van der Klei I. J., Daum G., Wiedemann N., Becker T., Pfanner N., van der Laan M., Role of mitochondrial inner membrane organizing system in protein biogenesis of the mitochondrial outer membrane. Mol. Biol. Cell 23, 3948–3956 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ott C., Dorsch E., Fraunholz M., Straub S., Kozjak-Pavlovic V., Detailed analysis of the human mitochondrial contact site complex indicate a hierarchy of subunits. PLOS ONE 10, e0120213 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zerbes R. M., Hoss P., Pfanner N., van der Laan M., Bohnert M., Distinct roles of Mic12 and Mic27 in the mitochondrial contact site and cristae organizing system. J. Mol. Biol. 428, 1485–1492 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Hopf T. A., Green A. G., Schubert B., Mersmann S., Schärfe C. P. I., Ingraham J. B., Toth-Petroczy A., Brock K., Riesselman A. J., Palmedo P., Kang C., Sheridan R., Draizen E. J., Dallago C., Sander C., Marks D. S., The EVcouplings Python framework for coevolutionary sequence analysis. Bioinformatics 35, 1582–1584 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pape J. K., Stephan T., Balzarotti F., Büchner R., Lange F., Riedel D., Jakobs S., Hell S. W., Multicolor 3D MINFLUX nanoscopy of mitochondrial MICOS proteins. Proc. Natl. Acad. Sci. U.S.A. 117, 20607–20614 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ott C., Ross K., Straub S., Thiede B., Götz M., Goosmann C., Krischke M., Mueller M. J., Krohne G., Rudel T., Kozjak-Pavlovic V., Sam50 functions in mitochondrial intermembrane space bridging and biogenesis of respiratory complexes. Mol. Cell. Biol. 32, 1173–1188 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.F. Sadeqi, K. Stroh, M. Vache, D. Riedel, A. Janshoff, H. J. Risselada, M. Meinecke, Membrane interactions of mitochondrial lipid transfer proteins. bioRxiv 2022.04.05.487160 [Preprint]. 7 April 2022. 10.1101/2022.04.05.487160. [DOI]
- 33.Blum T. B., Hahn A., Meier T., Davies K. M., Kuhlbrandt W., Dimers of mitochondrial ATP synthase induce membrane curvature and self-assemble into rows. Proc. Natl. Acad. Sci. U.S.A. 116, 4250–4255 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rampelt H., Wollweber F., Licheva M., de Boer R., Perschil I., Steidle L., Becker T., Bohnert M., van der Klei I., Kraft C., van der Laan M., Pfanner N., Dual role of Mic10 in mitochondrial cristae organization and ATP synthase-linked metabolic adaptation and respiratory growth. Cell Rep. 38, 110290 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu C., Wu J., Sun H., Briganti F., Meder B., Wei W., Steinmetz L. M., Single-molecule, full-length transcript isoform sequencing reveals disease-associated RNA isoforms in cardiomyocytes. Nat. Commun. 12, 4203 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hulsmann B. B., Labokha A. A., Gorlich D., The permeability of reconstituted nuclear pores provides direct evidence for the selective phase model. Cell 150, 738–751 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Liu H., Naismith J. H., An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8, 91 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chalk R., Mass spectrometric analysis of proteins. Methods Mol. Biol. 1586, 373–395 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Mueller U., Förster R., Hellmig M., Huschmann F. U., Kastner A., Malecki P., Pühringer S., Röwer M., Sparta K., Steffien M., Ühlein M., Wilk P., Weiss M. S., The macromolecular crystallography beamlines at BESSY II of the Helmholtz-Zentrum Berlin: Current status and perspectives. Eur. Phys. J. Plus 130, 141 (2015). [Google Scholar]
- 40.Sparta K. M., Krug M., Heinemann U., Mueller U., Weiss M. S., XDSAPP2.0. J. Appl. Crystallogr. 49, 1085–1092 (2016). [Google Scholar]
- 41.Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G. W., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S., Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bibby J., Keegan R. M., Mayans O., Winn M. D., Rigden D. J., AMPLE: A cluster-and-truncate approach to solve the crystal structures of small proteins using rapidly computed ab initio models. Acta Crystallogr. D Biol. Crystallogr. 68, 1622–1631 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Liebschner D., Afonine P. V., Baker M. L., Bunkóczi G., Chen V. B., Croll T. I., Hintze B., Hung L. W., Jain S., McCoy A. J., Moriarty N. W., Oeffner R. D., Poon B. K., Prisant M. G., Read R. J., Richardson J. S., Richardson D. C., Sammito M. D., Sobolev O. V., Stockwell D. H., Terwilliger T. C., Urzhumtsev A. G., Videau L. L., Williams C. J., Adams P. D., Macromolecular structure determination using x-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Zwart P. H., Hung L. W., Read R. J., Adams P. D., Iterative model building, structure refinement and density modification with the PHENIX auto buildwizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J., Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheldrick G. M., A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., Adams P. D., Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams C. J., Headd J. J., Moriarty N. W., Prisant M. G., Videau L. L., Deis L. N., Verma V., Keedy D. A., Hintze B. J., Chen V. B., Jain S., Lewis S. M., Arendall W. B. III, Snoeyink J., Adams P. D., Lovell S. C., Richardson J. S., Richardson D. C., MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., Ben-Tal N., ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH image to imageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schuck P., Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sikorski R. S., Hieter P., A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., Schiebel E., Epitope tagging of yeast genes using a PCR-based strategy: More tags and improved practical routines. Yeast 15, 963–972 (1999). [DOI] [PubMed] [Google Scholar]
- 56.Slot J. W., Geuze H. J., Cryosectioning and immunolabeling. Nat. Protoc. 2, 2480–2491 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Tokuyasu K. T., A technique for ultracryotomy of cell suspensions and tissues. J. Cell Biol. 57, 551–565 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mastronarde D. N., Held S. R., Automated tilt series alignment and tomographic reconstruction in IMOD. J. Struct. Biol. 197, 102–113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sutton R. B., Fasshauer D., Jahn R., Brunger A. T., Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature 395, 347–353 (1998). [DOI] [PubMed] [Google Scholar]
- 60.Gao S., von der Malsburg A., Paeschke S., Behlke J., Haller O., Kochs G., Daumke O., Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature 465, 502–506 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Tables S1 to S5
References
Movies S1 and S2