Abstract
Since 1892, it has been widely assumed that somatic mutations are evolutionarily irrelevant in animals because they cannot be inherited by offspring. However, some nonbilaterians segregate the soma and germline late in development or never, leaving the evolutionary fate of their somatic mutations unknown. By investigating uni- and biparental reproduction in the coral Acropora palmata (Cnidaria, Anthozoa), we found that uniparental, meiotic offspring harbored 50% of the 268 somatic mutations present in their parent. Thus, somatic mutations accumulated in adult coral animals, entered the germline, and were passed on to swimming larvae that grew into healthy juvenile corals. In this way, somatic mutations can increase allelic diversity and facilitate adaptation across habitats and generations in animals.
Somatic mutations can be transferred from animal parents to their offspring and thus contribute to evolution and adaptation.
INTRODUCTION
Over an organism’s lifetime, somatic genetic mutations accumulate due to environmental damage and errors during cellular processes such as DNA replication and repair (1, 2). Somatic mutations generally occur at higher rates than mutations in the germline (3–5). To limit damage from potentially deleterious somatic mutations, animals typically avoid passing them on to their offspring (1, 2, 6). Most bilaterians achieve this by segregating the germline from the soma early in development.
In 1892, Weismann first proposed the theory that somatic mutations acquired during an animal’s lifetime are evolutionarily irrelevant because they cannot cross the assumed barrier between the soma and germline (known as Weismann’s barrier) and thus cannot contribute to genetic variation of the next generation (7). Important nuances of Weismann’s original proposition have been lost in the long history of discourse about this theory (8) and its relevance to clonal animals has been questioned (2, 9). In the modern interpretation of Weismann’s barrier, its existence is regarded as universal (7). The timing of germline segregation, however, is variable across animals (10, 11). Several animals including sponges, hydrozoans, flatworms, polychaetes, sea urchins, and some chordates segregate their germline at later stages of development, long after embryogenesis is complete (11–18). Some of these taxa maintain multipotent progenitor cells throughout their lives; thus, somatic mutations could potentially cross Weismann’s barrier and be transmitted to the next generation. In this way, somatic mutations could generate adaptive alleles that pass through a zygotic unicellular stage, a role that was thought to be reserved for germline mutations.
While parental somatic mutations were previously observed in coral gametes (19, 20), the technical difficulty of tracking somatic mutations from parents, into gametes, through sexual reproduction, and into offspring has prevented researchers from determining whether somatic mutations can be passed to viable offspring and thus play a role in coral evolution (21, 22). Here, we discover a class of uniparental, coral offspring and we leverage this phenomenon to show that parental somatic mutations are inherited by healthy coral offspring. We also investigate whether somatic mutations can be inherited by biparental offspring and find preliminary evidence that this occurs as well. Our findings provide strong evidence of somatic mutation inheritance. Thus, we solidify the potential role for heritable somatic genetic variation in animal adaptation and evolution.
RESULTS
Spontaneous development of eggs from a large coral genet in Curaçao
Scleractinian corals are characterized by modular growth (23), long life spans (24), and simultaneous asexual and sexual reproduction. The hermaphroditic Caribbean coral Acropora palmata (Lamarck, 1816) reproduces sexually by releasing large amounts of gametes during annual broadcast spawning events. Eggs are typically fertilized externally by nonself-sperm. After development, pelagic larvae disperse up to hundreds of kilometers away and eventually settle onto the benthos (fig. S1A). Through asexual reproduction (polyp budding and colony fragmentation), an individual A. palmata juvenile can grow into multiple, large coral colonies. Each of these physically separate colonies (or “ramets”) belongs to the same original genotype (or “genet”). While individual polyps and entire ramets may die for a variety of reasons, the genet itself can survive for centuries and grow to cover hundreds of square meters (24). As a coral colony grows, newly emerged somatic mutations can be passed on to new groups of polyps (“modules”) and accumulate through time, leading to genetic mosaicism of the colony and allelic diversity in the genet overall (24, 25).
In 2018, we observed the spontaneous development of A. palmata eggs that were collected from a single parent colony in Curaçao; this occurred after the removal of self-sperm and before the addition of nonself-sperm, or immediately after the addition of nonself-sperm from Florida (see details in Materials and Methods). Although low levels (1 to 2%) of spontaneous development, indicated by cleaving eggs, are often seen in no-sperm controls, we observed nearly 100% development, indicating the occurrence of either outcrossing (due to sperm contamination or colony-level chimerism), self-fertilization (due to the breakdown of self-incompatibility), meiotic parthenogenesis, or mitotic (clonal) parthenogenesis. To determine which of these mechanisms was responsible for the spontaneous development, we investigated whether any sources of nonself-sperm may have contaminated the cultures of eggs. Tissue samples of two to three polyps were collected from multiple locations across the parent colony (n = 10), and one sample was collected from each of five nearest neighbor colonies (all within 2.25 m of the parent colony; Fig. 1A), which were considered putative parents, i.e., potential sources of contaminating sperm.
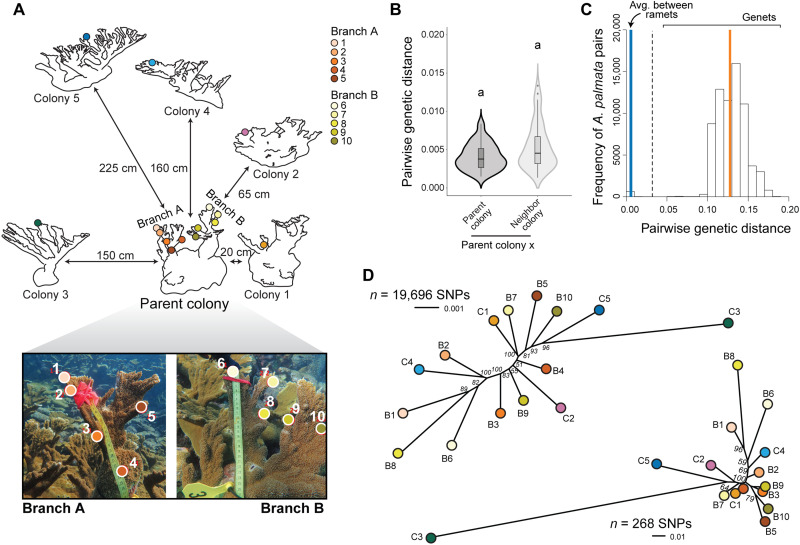
Fig. 1. Detection and mapping of somatic mutations across a parent coral colony and five nearest neighbor colonies (ramets) from the same coral genet.
(A) Sampling map of the parent colony and its five nearest neighbor colonies. Colonies are arranged according to their location on the reef with linear distances between colonies indicated next to the arrows. Colored circles indicate the locations of the tissue samples that were genotyped. Individual colonies in the line drawing are enlarged to show detail and not drawn to scale. Photo credit: Kate L. Vasquez Kuntz, The Pennsylvania State University. (B) Plot of Prevosti’s pairwise genetic distance within the parent colony and between the parent colony and the five neighboring colonies. All genetic distances were smaller than the distance threshold for genet assignment; therefore, all six colonies belonged to the same coral genet. (C) Histogram showing the frequency of pairwise genetic distances for 263 A. palmata samples collected from across the Caribbean in a previous study. The genet threshold is 0.032 (dashed black line). Average genetic distance was 0.0057 ± 0.0054 among ramets of the same genet (blue line, n = 148 ramets from 34 genets). Average genetic distance was 0.128 ± 0.025 among genets that fall above the genet threshold (orange line) [modified with permission from Kitchen et al. (26)]. (D) Neighbor-joining trees for all samples taken from the parent colony (B1 to B10) and nearest neighbor colonies (C1 to C5) showing all loci analyzed (upper tree, n = 19,696) and only those loci identified as mutations (lower tree, n = 268). Nodal values represent support from 100 bootstrap replicates.
We genotyped samples at 19,696 single-nucleotide polymorphism (SNP) loci via a microarray (26) to determine whether the neighboring colonies represented different genets. However, the parent colony and all five of the neighboring colonies belonged to the same A. palmata genet. Average pairwise genetic distances between samples were extremely low, both among the 10 parent colony samples (0.0041 ± 0.0017 SD, n = 10) and among all six of the sampled colonies (0.0054 ± 0.0030 SD; Fig. 1B and data S1). In contrast, the average Caribbean-wide genetic distance among A. palmata genets is 0.128 ± 0.025 SD (n = 263 samples; Fig. 1C) (26). These data therefore eliminated both types of outcrossing (nonself-sperm contamination from nearby colonies and chimerism of the parent colony itself) as likely explanations for the spontaneous development of the eggs (see note S1). Furthermore, the low pairwise genetic distances measured within and between colonies pointed to the existence of somatic mutations in the parent coral and neighboring ramets of the same genet, which could potentially be identified in the resulting offspring.
Identification and mapping of somatic mutations in the parent coral and neighboring ramets
To identify somatic mutations, we compared allele calls for all individual tissue samples using all loci that had complete data from all six colonies (n = 18,110; data S2). For variable loci, minority SNP calls (present in ≤7 samples) were classified as somatic mutations (n = 268). The number of somatic mutations varied by ramet, ranging from 2 to 149 (table S1). Two hundred four of the mutations were unique to one ramet, while six were shared by seven ramets (table S2). All somatic mutations were fixed or nearly fixed in each DNA sample (because low-frequency mutations are unlikely to be detected with the genotyping array; fig. S2 and note S2). A subset of SNP calls was additionally validated via restriction fragment length polymorphism (RFLP; see below). A neighbor-joining phylogeny based on the genetic distances did not support the existence of a correlation between genetic and physical distance in the distribution of variants, either within or among colonies (Fig. 1D).
Of the 268 somatic mutations identified, most mutations (72.01%) were transitions and fewer (27.61%) were transversions (data S3). Mutations occurred in protein-coding (13.43%) and nonprotein-coding regions (86.57%, data S4). After detecting somatic mutations in the parent genet, we investigated whether any of these mutations were inherited by its uniparental offspring, and therefore crossed Weismann’s barrier.
Uniparental offspring inherit parent’s somatic mutations
Eggs collected from the parent colony developed either spontaneously without addition of nonself-sperm or immediately after the addition of sperm from a colony from Florida. These eggs progressed through embryogenesis and developed into swimming larvae that successfully settled, acquired symbionts, and matured into multipolyp juveniles. After 4 months of propagation, we extracted DNA from 81 juvenile colonies and randomly chose 41 of these samples to submit for SNP analysis (26). Because the juveniles from the no-sperm and Florida sperm treatments were reared together in a common pool, we used the genetic distance between each offspring and the parent genet for cohort assignment. We identified 11 offspring of biparental origin, i.e., offspring from the parent colony and the nonself-sperm from Florida. These offspring had genetic distances comparable to an outcross of two parent colonies from Curaçao (27). In contrast, the remaining 30 offspring had genetic distances 2.6× lower than those of biparental origin (0.0361 ± 0.0064, n = 30; fig. S3A and data S1), indicating that they were derived from the one parent colony as a result of either self-fertilization or parthenogenesis. The fact that these larvae were uniparental in origin, as well as the high fidelity of microarray-based genotyping, eliminated many of the technical barriers, such as deciphering both parental mutations and non-Mendelian inheritance patterns, that would otherwise prevent the successful tracking of somatic mutations from parent to gamete to offspring. Thus, we next investigated whether these uniparental offspring had inherited the somatic mutations present in the parent colony.
Of the 268 parental somatic mutations, 50% (n = 134) were inherited by at least one offspring and each offspring harbored between 11 and 50 of these mutations (table S1). We also detected 2331 mutations unique to the offspring only (data S5). Loci of inherited somatic mutations most often changed from a homozygous to a heterozygous state [n = 86, gain of heterozygosity (GOH)]. More rarely, loci changed from a heterozygous to a homozygous state [n = 47, loss of heterozygosity (LOH)], and one locus changed from homozygous allele A/A to homozygous allele C/C (Fig. 2A and data S3). While 94% of parental LOH mutations were inherited by at least one offspring, only 39.6% of the GOH mutations were inherited (Fig. 2A).
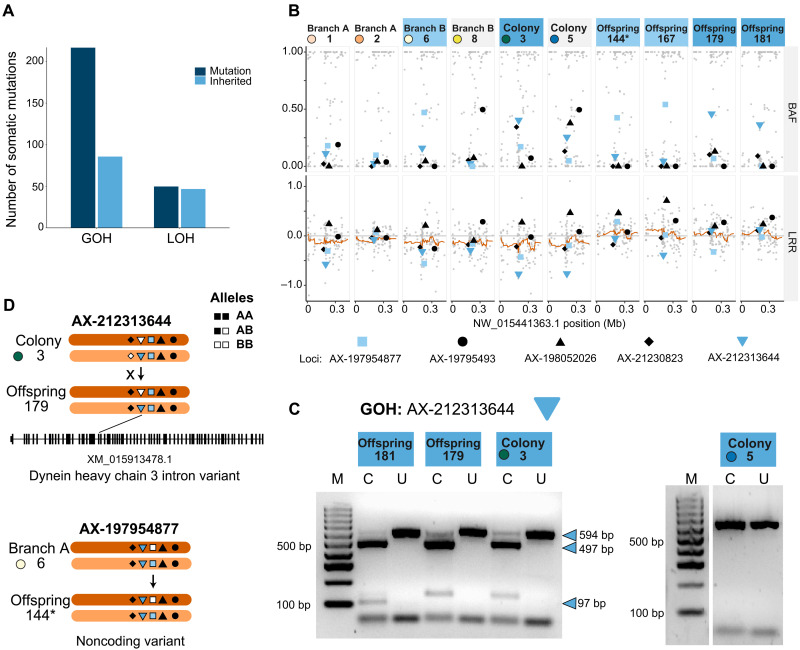
Fig. 2. Characterization and validation of parental somatic mutations inherited by uniparental coral offspring.
(A) Bar plot showing the number of total mutations (dark blue bars) categorized as a GOH or LOH, and the number of mutations in each category that were passed on to offspring (light blue bars). GOH mutations outnumbered LOH mutations, but a greater proportion of LOH mutations were inherited by offspring. (B) Plots of two metrics used to investigate somatic mutations: BAF (B allele frequency; top row) and LRR (log R ratios; bottom row). The orange line represents the 20-bp sliding average of LRR along the scaffold (position in megabases). Blue and black symbols represent specific mutations. Black shapes: not inherited; blue shapes: inherited. Above each plot, adult coral colonies are labeled as in Fig. 1A and juveniles are denoted as “Offspring”. Sample labels are filled according to specific, shared mutations (light blue, locus AX-197954877; medium blue, locus AX-212313644). Samples without those mutations have a white label fill. (C) RFLP validation of inherited GOH mutation AX-212313644. For each sample, the uncut (U) and cut (C) polymerase chain reaction products are shown next to a size standard (lane M). The two offspring (179 and 181) both share the heterozygous mutation also detected in colony 3, resulting in three bands, while colony 5, predicted to have the nonmutant homozygous state for this site, produced only one uncut band. (D) Schematic of inheritance of somatic mutations. RFLP-validated somatic mutation AX-212313644 (down triangle) is found within the intron of the dynein heavy chain 3. GOH mutation AX-197954877 (square) is found on the same scaffold. Symbols represent the five mutations detected along this scaffold in at least one sample in (B). Filled shapes, A allele; open shapes, B allele. Arrows denote inheritance and crosses denote lack of inheritance. Offspring 144 was parthenogenetic (denoted by an asterisk).
Because the tissue samples contained a mixture of cell types, we determined whether the detected somatic mutations were a product of underlying cell mosaicism or copy number variations (CNVs) at the genome level. Two common metrics for detecting ploidy differences and cell mosaicism were calculated: (i) B allele frequency (total allelic intensities) and (ii) log R ratio (relative allelic intensities) (28). B allele frequencies revealed that five GOH mutations were found in at least one parent sample with values near 0.5 (heterozygous) (Fig. 2B). Two of those mutations were also found in the offspring (mutations AX-212313644 and AX-197954877). Combining these findings, we estimated that 61% (n = 82) of inherited somatic mutations in these diploid offspring had two copies, while 39% (n = 52) of inherited somatic mutations had some level of mosaicism (figs. S4 and S5, data S6 and S7, and note S3).
We validated the somatic mutations that were first detected via the genotyping array by designing RFLP markers for 10 of the loci (data S8 and note S4). Of these loci, we verified that two GOH mutations in the offspring were identical to a heterozygous mutation in colony 3 at loci AX-212313644 and AX-212294854 (Fig. 2C and fig. S6). Both loci occur in noncoding regions. Locus AX-212313644 is found within the intron of the dynein heavy chain 3 (LOC107347526) (Fig. 2D), and locus AX-212294854 is found upstream of the uncharacterized gene LOC107346921.
The remaining 8 loci could not be validated using RFLP (8 of 10), suggesting that a subset of the predicted somatic mutations was incorrectly genotyped. These false positives could be attributed to biological variation (i.e., null alleles and copy number variants), sample preservation or preparation artifacts (26), or instrument error (29). However, this false-positive rate should not be extrapolated to be the overall genotyping error rate; loci were not chosen randomly for validation because there were limited restriction enzyme cut sites available to test SNP combinations using RFLP. Previous work identified an array mistyping rate of ≤0.5% (26), a rate that falls within the range of other array-based studies (30, 31) and that is equivalent to less than half the number of somatic mutations predicted in this study (268 of 19,696, or approximately 1.4%). In sum, using an independent method, we confirmed that a subset of somatic mutations in the parent were shared in uniparental offspring, and this signal remained clear against the known error rates of the genotyping array.
Alternative reproductive pathways in corals: Uniparental, meiotic offspring
We next used genetic distances, recombination rates, and heterozygosity estimates to determine whether the 30 uniparental coral offspring identified above were the result of colony-level selfing (i.e., the union of eggs and sperm from the same genet) or whether they were formed through parthenogenesis (i.e., the production of diploid eggs via either meiosis or mitosis; fig. S1). The evolutionary dynamics of inherited somatic mutations will change depending on whether the mutations are transmitted through mitotic or meiotic processes. Mitotic transmission involves no chromosomal recombination and thus preserves the parental genetic arrangement, producing clonal offspring, whereas meiotic transmission (via selfing or meiotic parthenogenesis) involves chromosomal recombination and thus moves parental somatic mutations into new genetic backgrounds (fig. S1B) (32). Such recombination uncouples beneficial from deleterious mutations faster than reproduction without recombination, slowing Muller’s ratchet (33, 34). Parthenogenetic individuals can share between 50 and 100% of the heterozygous loci with their parent (32), but the total number of recombination events present in an offspring depends on whether they were produced through mitotic parthenogenesis, meiotic parthenogenesis, or another uniparental process such as self-fertilization (fig. S1B).
We concluded that all 30 of the uniparental offspring were of meiotic origin because (i) the genetic distances between parent and offspring samples were larger than the genetic distances among samples of the parent genet [one-way analysis of variance (ANOVA) Tukey post hoc P < 0.001; fig. S3A and data S2] and (ii) numerous recombination events had occurred between the parent and each offspring (fig. S3C). However, because the heterozygosity of the offspring was significantly lower (7.1 to 11.5%) than in the parent genet and in A. palmata genets from across the Caribbean (12.8 to 17.3%; one-way ANOVA Tukey post hoc P < 0.0001; fig. S3B and table S1), it was apparent that they originated via self-fertilization or meiotic parthenogenesis and not via outcrossing. For 28 offspring (93%), the mean number of inferred recombination events [mean = 50.67 ± 12.20 (SD)] was consistent with the union of two recombinant parental haplotypes through self-fertilization (fig. S1B). The remaining two uniparental offspring differed from this pattern. Smaller genetic distances to the parent genet than the self-fertilization cohort (fig. S3A) and a small number of inferred recombination events (n = 19 and 22; fig. S3C) suggest that these two individuals were derived from meiotic parthenogenesis, through either restitutional meiosis where anaphase II is skipped or terminal fusion of chromosomes after meiosis (fig. S1B) (31).
Inheritance of somatic mutations is not limited to uniparental offspring
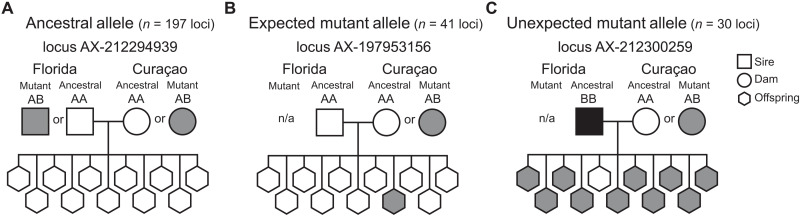
To determine whether the somatic mutations in the parent coral colony from Curaçao could only be inherited by uniparental offspring, we investigated the allele combinations of 11 biparental offspring stemming from the cross between the parent colony in Curaçao and a Florida colony whose sperm had been cryopreserved (27). Given the ancestral alleles of both the egg donor (Curaçao colony) and sperm donor (Florida colony) (data S9), we identified that all offspring had allele combinations that deviated from the expected proportion of alleles based on Mendelian inheritance. Of the 268 somatic mutations identified in the Curaçao genet, we found 41 (15.3%) that were potentially inherited by the biparental offspring (Fig. 3). Most of the remaining loci followed the predictions of Mendelian inheritance (73.5%), while a subset of 30 loci (11.1%) had allele proportions that did not match a pattern of Mendelian inheritance or the inheritance of somatic mutations (Fig. 3). These alleles could have arisen from other sources such as germline mutations, postembryonic mutations, or technical artifacts. While the exact source of these mutations is unknown, germline mutations found exclusively in coral sperm from Acropora hyacinthus made up 8.5% of all identified mutations (19), in line with our findings here. Future work is required to confirm somatic mutation inheritance by biparental offspring as estimated in our analysis.
Fig. 3. Evidence for the inheritance of parental somatic mutations by biparental coral offspring.
Biparental offspring resulted from a cross between the focal A. palmata parent colony in Curaçao and a Florida colony whose sperm had been cryopreserved (27). Previously identified somatic mutations at 268 loci in the Curaçao parent were tracked into 11 outcrossed biparental offspring that were identified during genotyping (n = 11). Most loci matched the expectations of Mendelian inheritance for the Curaçao ancestral allele [(A), 73.5%]. At the remaining loci, offspring either putatively inherited the Curaçao mutant allele [(B), 15.3%] or exhibited unexpected inheritance patterns inconsistent with the Curaçao ancestral or mutant allele [(C), 11.1%]. For example, at locus AX-212300259, one offspring had an unexpected mutant AA genotype, while the parent from Florida did not have an A allele to contribute. These unexpected alleles in the offspring may have resulted from technical errors (26) or represent de novo (germline) mutations (19, 35). n/a, no mutant allele detected.
Supporting these observations from Curaçao, using samples collected in 2017 in Florida (at the other end of the species’ range), we also detected the transmission of parental somatic mutations into both uniparental and biparental offspring (see fig. S7, data S10, and note S5). In sum, we show that the coral A. palmata can produce uniparental, healthy offspring containing parental somatic mutations. These mutations emerged during the parent colony’s adulthood and were passed through meiosis into the germ cells and subsequently into the offspring.
DISCUSSION
By investigating uniparental and biparental, meiotic offspring from the coral A. palmata, we show that somatic mutations at multiple loci, which were acquired over the lifetime of a parent animal, can be inherited by its offspring (Figs. 1 and 2 and fig. S7). These findings were reproducible across two spawning years and in two locations at opposite ends of the species’ range (note S5). Because coral genets can persist for hundreds to thousands of years, somatic mutations can rise to high frequency in modules (polyps) of a genet due to stochasticity or selection (35, 36). Strongly deleterious or lethal mutations might lead to module or colony death, but not genet death, and can thus be removed from the genet’s gene pool while preserving the genet itself. Meanwhile, neutral and beneficial somatic mutations can accumulate in tissues, spread to new modules via polyp budding, and be dispersed over small spatial scales through colony fragmentation [ca. 70 m, (37)]. After these mutations are inherited by offspring, fitness variance is redistributed from the realm of within-colony to between-organism selection. Furthermore, these somatic mutations have the potential to disperse over much longer distances [hundreds of kilometers, (38)] by pelagic coral larvae that have inherited the mutations. Thus, the discovery of heritable somatic mutations in coral offspring represents a previously unconfirmed source for coral adaptation and evolution.
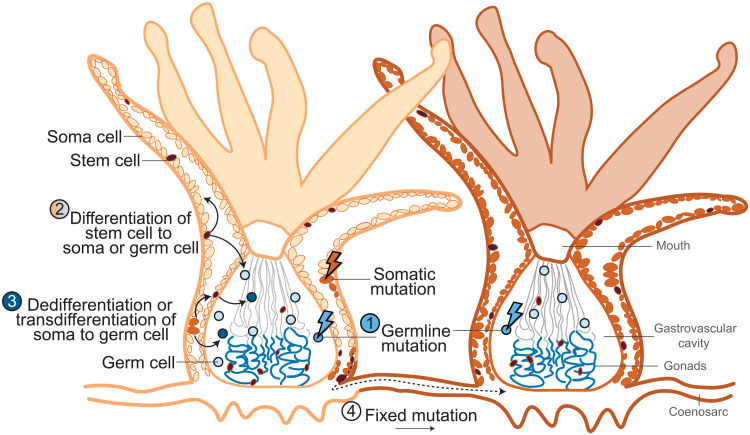
The mechanism by which adult A. palmata transmit somatic mutations to offspring remains to be found (Fig. 4). Mutations may have originated in the soma, dedifferentiated into stem cells, and then redifferentiated into germ cells, or somatic cells may have transdifferentiated directly into germ cells (39). Stem cells have not yet been identified in corals; however, the regenerative properties of anthozoans (40, 41) and the identification of progenitor/undifferentiated cells with stem cell characteristics in a sea anemone (42) and in coral cell lines (43) both point to their existence (19, 35). In any case, the mutations identified and tracked in this study must have occurred after embryogenesis of the primary polyp that founded the genet was complete because the mutations were not shared among all polyps of the adult parent genet or all ramets of the genet. This implies that multipotent progenitor or stem-like cells are not moving freely throughout a colony, setting up competition among cells of different stem cell lineages.
Fig. 4. Modes of inheritance of genetic mutations in animals.
(1) If animals differentiate and segregate germline cells (light blue) from somatic cells (light orange) early in development, then only germline mutations (medium blue) can be inherited by offspring (1). (2) Planarians, sponges, and some cnidarians continuously segregate a germline and somatic tissue from a population of stem cells (brown) as they grow, allowing for an accumulation of mutations that are heritable (11). (3) Cnidarian somatic cells may de- or transdifferentiate into germline cells, passing on mutations that are somatic in origin (dark blue) (14). The cellular source (soma, germ, or stem) for polyp growth is an active area of research. (4) Somatic mutations (dark orange) may rise to fixation in new modules (polyps) through budding from a limited number of soma cells. Lightning bolts represent mutation-causing events. Figure modified with permission from Reusch et al. (21).
Immediately after a somatic cell mutates, it undergoes competition with nonmutated cells in a process called developmental selection (36, 44–46). This “struggle of the parts,” as described by Wilhelm Roux in 1881 and later recognized by Weismann as “intraorganismal selection,” is distinct from germline selection (47, 48) and can occur at the molecular, chromosome, or cellular level. Propagation of the somatic mutation then depends on either successfully outcompeting or coexisting with other somatic lineages during cell growth and proliferation (21, 49). Beneficial (or neutral) mutations that survive developmental selection can therefore be disproportionately represented in the cells of a genet (50), an advantage that germline mutations do not have. Somatic mutations with beneficial fitness effects in clonal organisms may be more common than previously thought (51, 52) and may allow ramets to withstand environmental fluctuations. Here, we show that gametes carrying somatic mutations survive to form healthy juvenile corals.
A high percentage of the coral offspring analyzed here were uniparental (73%) compared to previous studies [~1 to 10% uniparental offspring; (53, 54)]. While this is not common in animals nor is it typical for most broadcast spawning corals, uniparental reproduction is common in plants. Plant species frequently switch from biparental to uniparental reproduction when sexual partners are scarce, e.g., at the edges of the species range, when introduced to new habitats, or after large-scale disturbances (55, 56). Hence, like plants, corals may rely on the generation of uniparental larvae to persist during times when sexual partners are rare. Within-genet selection before gametogenesis may effectively purge lethal recessive mutations that would otherwise be exposed to selection only during mating between genets and so reduce the cost of selfing/uniparental inheritance (36, 57). Thus, the production of uniparental offspring that harbor parental somatic mutations might help buffer against the losses of genetic diversity and consequences of inbreeding (56) that would otherwise occur in uniparental mating, i.e., as a result of self-fertilization (32).
Modular species are found in multiple groups (e.g., multicellular algae, fungi, and animals) across the eukaryotic tree of life (21), and a small number of researchers have suggested that somatically generated variation should be considered to understand evolution in these taxa (36, 58). However, the common assumption that Weismann’s barrier is universal in animals (7) has led biologists to disregard somatic mutations as a potentially important source of new genetic variation to shape animal evolution. Our demonstration of transgenerational inheritance of acquired genetic variation challenges this long-held assumption. Like many other modular, long-lived marine invertebrates, terrestrial plants, and even seagrasses, coral genets experience substantial environmental pressures over their long life spans, and somatic mutations may play a major role in their adaptation to these changes (21, 59). Our findings further illustrate the narrowing differences known to exist in the evolutionary dynamics characterizing plants and nonbilaterian animal groups such as corals.
MATERIALS AND METHODS
Curaçao spawning collection and DNA extraction
Gametes were collected from a subset of A. palmata colonies from Spanish Water reef (latitude, 12.0636°N; longitude, 68.8532°W) in Curaçao on 2 September 2018, seven nights after the full moon. Eggs and sperm were separated in preparation for fertilization by pooled, cryopreserved sperm from Curaçao or Florida. However, for one egg donor colony, eggs in the no-sperm controls underwent cell division despite only being exposed to self-sperm during handling. These offspring were putatively designated as uniparental in origin. In addition, in separate experimental crosses with the same egg donor, cleavage began within minutes of exposure to nonself-sperm from Curaçao or Florida. Because primary cleavage typically begins 60 to 90 minutes after fertilization in A. palmata, we suspected that most of the eggs displaying cleavage within minutes of the addition of nonself-sperm were also likely to be uniparental in origin, while a smaller fraction may have been ultimately fertilized by the added nonself-sperm from Curaçao or Florida. Despite efforts to rinse sperm more quickly from eggs after spawning on two subsequent nights, eggs from this donor colony continued to display apparent self-fertilization. This was followed by normal larval development and normal larval swimming behaviors. Thus, cohorts from all three spawning nights were presumed to include uniparental offspring. Larvae were reared in containers of filtered seawater (FSW) (spun polypropylene filters, 0.5 μm) and then shipped to Mote Marine Laboratory in Florida for settlement, grow out, and further study. At 4 months after fertilization, tissue samples from 81 offspring were preserved in 96% ethanol and shipped to Pennsylvania State University for genetic marker analysis. The coral colony that produced the putative uniparental offspring was sampled in five locations along two of its branches (n = 10 samples total). In addition, one sample was taken from each of the five nearest neighboring colonies (n = 5; Fig. 1A). These 15 samples were also preserved in 96% ethanol for genetic marker analysis. To collect tissue from the adult colonies, whole polyps including skeletal material were sampled. These adult colonies had spawned gametes the previous night, and thus, tissue samples contained mostly somatic tissue and few, if any, gametes (fig. S3A). We extracted genomic DNA from 41 offspring and 15 parent samples using the DNeasy kit (Qiagen, USA) following the manufacturer’s protocol with slight modifications optimized for corals (doi: 10.17504/protocols.io.bgjqjumw).
Genotyping genomic DNA mixtures of two Curaçao genets as mock chimeras
We tested whether the SNP microarray could detect the presence of DNA from two closely related genets in one sample, simulating a chimera from colony fusion (60). Two tissue samples from two related Curaçao genets, genet A and genet B, were selected (fig. S3). The pairwise genetic distance between genet A and genet B is 0.0709 based on SNP analysis; this value is near the upper end of the SD for full siblings (average 0.0642 ± 0.0068 SD) (26). Neither genet by itself was known to be a chimera (based on genotyping of five microsatellite loci; for details, see the “Florida larval DNA extraction and microsatellite analysis” section below). Two genomic DNA mixtures were created: one with a proportion of 85:15 of genet A:genet B and one with a proportion of 58:42 of genet A:genet B. These proportions were calculated on the basis of measurements of the DNA concentration of each sample using Picogreen (Molecular Probes, OR).
SNP analysis and detection of somatic mutations
Offspring and parent samples were genotyped along with biparental offspring from a Curaçao outcross (27) and genomic DNA mixtures using an Affymetrix genotyping array (26) and analyzed using a standardized workflow [https://coralsnp.science.psu.edu/galaxy/, (26)]. A total of 19,696 genotyping loci were exported for downstream analyses using the vcfR package in R (61). Genotype calls were converted into “0/0,” “0/1,” or “1/1.” A call of 0/0 was homozygous for allele A at that locus, a call of 1/1 was homozygous for allele B at that locus, and a call of 0/1 meant that the sample was A/B heterozygous at that locus. Missing data were denoted as “NA,” and heterozygosity for each sample was calculated as the number of probes with NA or 0/1 calls out of all probes divided by the total number of probes (62). Differences in percent heterozygosity were tested using a one-way ANOVA followed by a Tukey post hoc test (see table S1). Prevosti’s genetic distance was calculated using the bitwise function in poppr (62, 63). Differences in the mean pairwise genetic distances were tested with a one-way ANOVA followed by a Tukey post hoc test. Genetic distances were used to construct one neighbor-joining tree of all loci (n = 19,696) and a second tree including only the subset of loci with identified somatic mutations (n = 268). Phylogenetic inference was performed with the aboot function in R package poppr (62) with 100 bootstrap replicates. To infer parental haplotype blocks and recombination events, the genotype data from the uniparental offsprings were phased using the bmh and recombination functions in the hsphase R package (64) and plotted with ComplexHeatmap (65).
Somatic mutations in the parent colony (n = 10 samples) and neighboring colonies (n = 5 samples) were tallied for each locus across samples. Based on our prediction that the ancestral allele should be more frequent than the mutated or alternate allele, alleles were designated as ancestral if eight or more samples shared the allele (table S2). Parental mutant alleles were similarly tallied in the uniparental and biparental offspring following Mendelian inheritance in the latter group. The B allele frequency and log R ratio were calculated using the “affy2vcf” bcftools plugin (https://github.com/freeseek/gtc2vcf) as part of the standard genotyping workflow. CNVs were called separately for the parents and offspring using the CGHcall R package (66). The log R ratios were median-normalized, segmented, and fit to a five-class model (double deletion, hemizygous deletion, normal, gain three to four copies, and amplification). The probability of a CNV gain or loss that exceeded 0.5 for each sample and each locus was counted as a CNV from the in silico predictions. For predicted somatic mutations, each log R plot was also visually inspected for CNVs (data S6 and S7). Chi-square tests were used to assess the differences in the proportion of CNVs for parent and offspring and the mutant loci. Genomic location and predicted effect of the mutations were found with snpEff v4.3 (67) using the Acropora digitifera genome assembly (68).
To test the detection limits of the genotyping array platform when two genotypes are combined, either through contamination or chimerism, we mixed DNA of two donor genets (genet A and genet B from Curaçao) and compared genotyping results of the mixtures to results from pure DNA extracts of each genet. The DNA mixtures were composed of 85:15 genet A:genet B (M1) and 58:42 genet A:genet B (M2). The relationship between the DNA mixtures and the two pure “donor” DNA extracts was analyzed first by calculating Prevosti’s genetic distance (62, 63). Missing data and heterozygosity were also calculated for all samples as described above. First, the dataset was filtered to homozygous loci that differed in the two donor genets (e.g., genet A = AA and genet B = BB). Then, the percentage of genotype calls that matched either of the donor genets, the percentage that was heterozygous, and the percentage that was missing were calculated for each mixture.
Within Axiom Analysis Suite (Thermo Fisher Scientific, USA), mean nucleotide signal intensities were calculated separately for each group (DNA mixtures, donor genets, parent and neighbor ramets, and offspring) to determine whether signal varied for the DNA mixtures compared to the single DNA samples. The signal intensities for each nucleotide were subjected to a one-way ANOVA with a Tukey post hoc test between the DNA mixtures, donor genets, parent and neighbor ramets, and offspring.
RFLP validation of somatic mutations
Of the 134 mutations shared by the parental genet and uniparental offspring, 28 were screened for restriction enzyme cut sites. Primers were designed using Primer 3 (69) on the basis of 500 base pairs (bp) of flanking sequence around the SNP extracted with the bedtools getfasta utility v2.27.1 (70) from the A. digitifera genome (68). Restriction enzyme cut sites were identified in 19 of the 28 variant mutations, but only 10 were tested for validation (data S8). SNP-containing regions were amplified by polymerase chain reaction (PCR) in a 10-μl reaction volume containing water, 1× NH4 buffer (Bioline, Boston, MA), 3 mM MgCl (Bioline, Boston, MA), 1 mM deoxynucleotide triphosphate (dNTP) (Bioline, Boston, MA), 250 nmol of forward and reverse primers (IDT, Coralville, Iowa), 1 U of Biolase Taq (Bioline, Boston, MA), and 1 μl of template DNA. PCR products were then denatured at 94°C for 5 minutes (min), 35 cycles of PCR were performed [94°C for 20 seconds (s), 55.2°C for 20 s, and 72°C for 30 s], and PCR products were held for a final extension at 72°C for 30 min. PCR products were then directly digested by the respective restriction enzyme (listed in data S8) in a 10-μl reaction volume containing 5 μl of PCR product, 1× CutSmart buffer, 5 U of enzyme, and water to volume. Digests were incubated at their respective temperatures following the manufacturer’s protocols. PCR and digest products were visualized using electrophoresis on a 2% tris-EDTA (TE) agarose gel.
Florida spawning collection and crosses from known mutants
A. palmata colonies with known mutations at microsatellite loci were analyzed as described by Baums et al. (37). Gamete bundles were collected on 11 August 2017, from two reefs in the Florida Keys: Sand Island Reef (SIR; 25.0179°N, 80.368617°W) and Elbow Reef (ELR; 25.15185°N, 80.2497°W). At SIR, we collected gametes from two colonies known as SIR “Blue Mutant” and SIR “Orange.” At ELR, gametes were collected from one colony known as ELR “Green Mutant.” The first cross was between SIR Blue Mutant and SIR Orange, and the second cross was between SIR Blue Mutant and ELR Green Mutant. For each cross, we combined 1 ml of eggs (ca. 3000 to 5000 eggs) and 1 ml of sperm (ca. concentration 106 cells/ml) from each parent and waited 1.5 hours to allow fertilization to take place. Following fertilization, zygotes were rinsed two times with 0.2 μm FSW and placed in a 1-liter container with FSW to develop overnight. For each parent, eggs and sperm from the same colony were also combined in selfing controls. Three samples of 20 to 100 eggs/embryos from each cross were collected at 6 to 8 hours after fertilization and preserved in a 4% formaldehyde/4% glutaraldehyde solution in 1× phosphate-buffered saline to score fertilization. Fertilization rate was calculated as the ratio of coral embryos in the prawn-chip stage (irregularly shaped cellular bilayer) over the total number of eggs and embryos in the tube. After 1.5 hours, no dividing embryos were observed in the selfing controls for either SIR Blue Mutant or SIR Orange, while ELR Green Mutant did self, with a self-fertilization rate of 0.64% ± 0.34% on the first night and 32.06% ± 6.99% on the second night. Water was changed twice per day until larvae began to swim at the end of the third day. At 96 hours after fertilization, swimming larvae from each cross (SIR Blue Mutant × SIR Orange n = 300; SIR Blue Mutant × ELR Green Mutant n = 200) were preserved individually in 96% nondenatured ethanol and stored at −20°C until shipment.
Florida larval DNA extraction and microsatellite analysis
DNA was extracted from 95 larvae from each of the two crosses. The larvae were rinsed once with fresh 96% ethanol to remove debris. Then, the ethanol was replaced with 20 μl of 5% Chelex solution and 2 μl of proteinase K (20 mg/ml). Samples were vortexed for 2 s, digested overnight at 55°C, heated to 95°C for 15 min, and then cooled to 4°C.
PCR-amplified microsatellite markers were multiplexed as previously described (37). Microsatellite markers were amplified by PCR in a 10-μl reaction volume containing 1× Promega Reaction Buffer, 25 mM MgCl, 10 mM dNTP, 5 μM primer, 1 U of GoTaq Flexi DNA Polymerase (Promega, WI, USA), and 1 μl of DNA template. Each PCR mixture was denatured at 94°C for 5 min followed by 35 cycles of 94°C for 20 s, 54°C for 20 s, and 72°C for 30 s. Samples were held at 72°C for 30 min for a final extension. Samples were checked for successful sequence amplification by running 4 μl of PCR product on a 2% TE agarose gel. Once amplification was verified, samples were sent to the Genomics Core Facility at Pennsylvania State University for fragment analysis on the Applied Biosystems 3730XL DNA analyzer. Allele sizes were called independently by two researchers for each microsatellite locus using Genemapper 5.0 software (Thermo Fisher Scientific). In A. palmata, somatic mutations often manifest as additional alleles in microsatellite chromatographs. The mutant allele is most often one repeat size larger or smaller than the ancestral allele of that genet (24). The ancestral allele is established by comparing at least five samples from a genet and determining the majority allele. Through this analysis, SIR Blue Mutant had a third allele at microsatellite locus 166 that the ancestral SIR Blue genet did not have (24) while sharing all other alleles across loci with the ancestral genet. The ELR Green Mutant had a third allele at microsatellite locus 192 that the ancestral ELR Green genet did not have. Somatic mutations were not detected in SIR Orange at any of the five microsatellite loci and inheritance of somatic mutations was not detected in the SIR Blue Mutant × SIR Orange cross.
Acknowledgments
We would like to thank K. Stankiewicz and A. Nekrutenko for their bioinformatics assistance and D. Williams and M. Miller for field support of the 2017 experiment. We also thank M. Devlin-Durante for assistance with the microsatellite analysis. M. Hagedorn helped secure funding, led the cryopreservation work in Curaçao during coral spawning, and helped enable the larval shipment to Mote Marine Laboratory, as well as provided the crosses containing a high percentage of spontaneously developing eggs. K. O’Neil helped secure funding, reared coral larvae, and propagated microfragments to preserve access to the unique crosses and genotypes used in this study. We also thank A. Shantz, D. Flores, L. Tichy, C. Lager, K. Lohr, K. Latijnhouwers, and V. Chamberland for dive and laboratory support, as well as C. Osborne for manuscript edits.
Funding: This work was supported by NOAA Office for Coastal Management grant NA17NOS4820083 (to I.B.B. and S.A.K.), National Science Foundation grant OCE-1537959 (to I.B.B.), the Human Frontier Science Program grant RGP0042/2020 (to I.B.B.), National Science Foundation grant IOS-1848671 (to K.L.M.), and the Paul G. Allen Family Foundation (to K.L.M.).
Author contributions: Conceptualization: K.L.V.K., S.A.K., and I.B.B. Methodology: K.L.V.K., S.A.K., T.L.C., and I.B.B. Field work: K.L.V.K., S.A.K., A.N.C., C.P., M.J.A.V., K.L.M., and I.B.B. Investigation: K.L.V.K., S.A.K., T.L.C., S.A.V., and I.B.B. Funding acquisition: S.A.K., K.L.M., M.J.A.V., and I.B.B. Project administration and supervision: I.B.B. Writing—original draft: K.L.V.K., S.A.K., and I.B.B. Writing—review and editing: K.L.V.K., S.A.K., T.L.C., A.N.C., S.A.V., M.J.A.V., K.L.M., and I.B.B.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: SNP data for the samples in this study are available at https://scholarsphere.psu.edu/resources/76e0ccdd-fbe0-4190-92cf-2a91e02bd73b. Code is available on GitHub: https://github.com/skitchen19/Acropora_Mutation_Vasquez_Kitchen_et_al.
Supplementary Materials
This PDF file includes:
Notes S1 to S5
Figs. S1 to S7
Tables S1 and S2
References
Other Supplementary Material for this manuscript includes the following:
Data S1 to S10
REFERENCES AND NOTES
- 1.A. Weismann, Das Keimplasma. Eine Theorie der Vererbung (Fisher, 1892).
- 2.Buss L. W., Evolution, development, and the units of selection. Proc. Natl. Acad. Sci. U.S.A. 80, 1387–1391 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynch M., Evolution of the mutation rate. Trends Genet. 26, 345–352 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L., Vijg J., Somatic mutagenesis in mammals and its implications for human disease and aging. Annu. Rev. Genet. 52, 397–419 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimura M., On the evolutionary adjustment of spontaneous mutation rates. Genet. Res. 9, 23–34 (1967). [Google Scholar]
- 6.L. W. Buss, The Evolution of Individuality (Princeton Univ. Press, 2014). [Google Scholar]
- 7.Bline A. P., Le Goff A., Allard P., What is lost in the Weismann barrier? J. Dev. Biol. 8, 35 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayr E., Weismann and evolution. J. Hist. Biol. 18, 295–329 (1985). [DOI] [PubMed] [Google Scholar]
- 9.Berrill N. J., Liu C. K., Germplasm, Weismann, and Hydrozoa. Q. Rev. Biol. 23, 124–132 (1948). [DOI] [PubMed] [Google Scholar]
- 10.Buss L. W., Diversification and germ-line determination. Paleobiology 14, 313–321 (1988). [Google Scholar]
- 11.Extavour C. G., Akam M., Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development 130, 5869–5884 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Whittle C. A., Extavour C. G., Causes and evolutionary consequences of primordial germ-cell specification mode in metazoans. Proc. Natl. Acad. Sci. U.S.A. 114, 5784–5791 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Littlefield C. L., Germ cells in Hydra oligactis males. I. Isolation of a subpopulation of interstitial cells that is developmentally restricted to sperm production. Dev. Biol. 112, 185–193 (1985). [DOI] [PubMed] [Google Scholar]
- 14.Gold D. A., Jacobs D. K., Stem cell dynamics in Cnidaria: Are there unifying principles? Dev. Genes Evol. 223, 53–66 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishimiya-Fujisawa C., Sugiyama T., Genetic analysis of developmental mechanisms in Hydra: XX. Cloning of interstitial stem cells restricted to the sperm differentiation pathway in Hydra magnipapillata. Dev. Biol. 157, 1–9 (1993). [DOI] [PubMed] [Google Scholar]
- 16.Borisenko I. E., Adamska M., Tokina D. B., Ereskovsky A. V., Transdifferentiation is a driving force of regeneration in Halisarca dujardini (Demospongiae, Porifera). PeerJ 3, e1211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagner D. E., Wang I. E., Reddien P. W., Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332, 811–816 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juliano C., Wessel G., Versatile Germline Genes. Science 329, 640–641 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.E. H. López-Nandam, R. Albright, E. A. Hanson, E. A. Sheets, S. R. Palumbi, Mutations in coral soma and sperm imply lifelong stem cell renewal and cell lineage selection. bioRxiv 2021.07.20.453148 [Preprint]. 4 February 2022. 10.1101/2021.07.20.453148. [DOI] [PMC free article] [PubMed]
- 20.Schweinsberg M., Pech R. A. G., Tollrian R., Lampert K. P., Transfer of intracolonial genetic variability through gametes in Acropora hyacinthus corals. Coral Reefs 33, 77–87 (2014). [Google Scholar]
- 21.Reusch T. B., Baums I. B., Werner B., Evolution via somatic genetic variation in modular species. Trends Ecol. Evol. 36, 1083–1092 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Van Oppen M. J. H., Souter P., Howells E. J., Heyward A., Berkelmans R., Novel genetic diversity through somatic mutations: Fuel for adaptation of reef corals? Diversity 3, 405–423 (2011). [Google Scholar]
- 23.Goffredo S., Lasker H. R., Modular growth of a gorgonian coral can generate predictable patterns of colony growth. J. Exp. Mar. Biol. Ecol. 336, 221–229 (2006). [Google Scholar]
- 24.Devlin-Durante M. K., Miller M. W.; Caribbean Acropora Research Group, Precht W. F., Baums I. B., How old are you? Genet age estimates in a clonal animal. Mol. Ecol. 25, 5628–5646 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Lopez E. H., Palumbi S. R., Somatic mutations and genome stability maintenance in clonal coral colonies. Mol. Biol. Evol. 37, 828–838 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Kitchen S. A., Von Kuster G., Kuntz K. L. V., Reich H. G., Miller W., Griffin S., Fogarty N. D., Baums I. B., STAGdb: A 30K SNP genotyping array and Science Gateway for Acropora corals and their dinoflagellate symbionts. Sci. Rep. 10, 12488 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagedorn M., Page C. A., O’Neil K. L., Flores D. M., Tichy L., Conn T., Chamberland V. F., Lager C., Zuchowicz N., Lohr K., Blackburn H., Vardi T., Moore J., Moore T., Baums I. B., Vermeij M. J. A., Marhaver K. L., Assisted gene flow using cryopreserved sperm in critically endangered coral. Proc. Natl. Acad. Sci. U.S.A. 118, e2110559118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Attiyeh E. F., Diskin S. J., Attiyeh M. A., Mossé Y. P., Hou C., Jackson E. M., Kim C., Glessner J., Hakonarson H., Biegel J. A., Genomic copy number determination in cancer cells from single nucleotide polymorphism microarrays based on quantitative genotyping corrected for aneuploidy. Genome Res. 19, 276–283 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saunders I. W., Brohede J., Hannan G. N., Estimating genotyping error rates from Mendelian errors in SNP array genotypes and their impact on inference. Genomics 90, 291–296 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Hong H., Xu L., Liu J., Jones W. D., Su Z., Ning B., Perkins R., Ge W., Miclaus K., Zhang L., Park K., Green B., Han T., Fang H., Lambert C. G., Vega S. C., Lin S. M., Jafari N., Czika W., Wolfinger R. D., Goodsaid F., Tong W., Shi L., Technical reproducibility of genotyping SNP arrays used in genome-wide association studies. PLOS ONE 7, e44483 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee Y. G., Jeong N., Kim J. H., Lee K., Kim K. H., Pirani A., Ha B. K., Kang S. T., Park B. S., Moon J. K., Kim N., Jeong S.-C., Development, validation and genetic analysis of a large soybean SNP genotyping array. Plant J. 81, 625–636 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Engelstädter J., Asexual but not clonal: evolutionary processes in automictic populations. Genetics 206, 993–1009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charlesworth B., Directional selection and the evolution of sex and recombination. Genet. Res. 61, 205–224 (1993). [DOI] [PubMed] [Google Scholar]
- 34.Muller H. J., Some genetic aspects of sex. Am. Nat. 66, 118–138 (1932). [Google Scholar]
- 35.Barfield S., Aglyamova G. V., Matz M. V., Evolutionary origins of germline segregation in Metazoa: evidence for a germ stem cell lineage in the coral Orbicella faveolata (Cnidaria, Anthozoa). Proc. R. Soc. B 283, 20152128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orive M. E., Somatic mutations in organisms with complex life histories. Theor. Popul. Biol. 59, 235–249 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Baums I. B., Miller M. W., Hellberg M. E., Geographic variation in clonal structure in a reef building Caribbean coral, Acropora palmata. Ecol. Monogr. 76, 503–519 (2006). [Google Scholar]
- 38.Baums I. B., Hughes C. R., Hellberg M. H., Mendelian microsatellite loci for the Caribbean coral Acropora palmata. Mar. Ecol. Prog. Ser. 288, 115–127 (2005). [Google Scholar]
- 39.Wessel G. M., Morita S., Oulhen N., Somatic cell conversion to a germ cell lineage: A violation or a revelation? J. Exp. Zool. B Mol. Dev. Evol. 336, 666–679 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bak R. P. M., Steward-Van Es Y., Regeneration of superficial damage in the scleractinian corals Agaricia agaricites F. purpurea and Porites asteroides. Bull. Mar. Sci. 30, 883–887 (1980). [Google Scholar]
- 41.Burton P. M., Finnerty J. R., Conserved and novel gene expression between regeneration and asexual fission in Nematostella vectensis. Dev. Genes Evol. 219, 79–87 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Sebé-Pedrós A., Saudemont B., Chomsky E., Plessier F., Mailhé M.-P., Renno J., Loe-Mie Y., Lifshitz A., Mukamel Z., Schmutz S., Novault S., Steinmetz P. R. H., Spitz F., Tanay A., Marlow H., Cnidarian cell type diversity and regulation revealed by whole-organism single-cell RNA-Seq. Cell 173, 1520–1534.e20 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Kawamura K., Nishitsuji K., Shoguchi E., Fujiwara S., Satoh N., Establishing sustainable cell lines of a coral, Acropora tenuis. Marine Biotechnol. 23, 1–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pineda-Krch M., Fagerstrom T., On the potential for evolutionary change in meristematic cell lineages through intraorganismal selection. J. Evol. Biol. 12, 681–688 (1999). [Google Scholar]
- 45.Buchholz J. T., Developmental selection in vascular plants. Bot. Gaz. 73, 249–286 (1922). [Google Scholar]
- 46.Whyte L. L., Internal factors in evolution. Acta Biotheor. 17, 33–48 (1964). [DOI] [PubMed] [Google Scholar]
- 47.A. Weismann, On Germinal Selection as a Source of Definite Variation (Open Court Publishing Company, 1896). [Google Scholar]
- 48.W. Roux, Der Kampf der Theile im Organismus (W. Engelmann, 1881).
- 49.Shakiba N., Fahmy A., Jayakumaran G., McGibbon S., David L., Trcka D., Elbaz J., Puri M. C., Nagy A., van der Kooy D., Goyal S., Wrana J. L., Zandstra P. W., Cell competition during reprogramming gives rise to dominant clones. Science 364, eaan0925 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Bódi Z., Farkas Z., Nevozhay D., Kalapis D., Lázár V., Csörgő B., Nyerges Á., Szamecz B., Fekete G., Papp B., Araújo H., Oliveira J. L., Moura G., Santos M. A. S., Székely T. Jr., Balázsi G., Pál C., Phenotypic heterogeneity promotes adaptive evolution. PLOS Biol. 15, e2000644 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Venkataram S., Dunn B., Li Y., Agarwala A., Chang J., Ebel E. R., Geiler-Samerotte K., Hérissant L., Blundell J. R., Levy S. F., Development of a comprehensive genotype-to-fitness map of adaptation-driving mutations in yeast. Cell 166, 1585–1596.e22 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blundell J. R., Schwartz K., Francois D., Fisher D. S., Sherlock G., Levy S. F., The dynamics of adaptive genetic diversity during the early stages of clonal evolution. Nat. Ecol. Evol. 3, 293–301 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fogarty N. D., Vollmer S. V., Levitan D. R., Weak prezygotic isolating mechanisms in threatened Caribbean Acropora corals. PLOS ONE 7, e30486 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baums I. B., Devlin-Durante M. K., Polato N. R., Xu D., Giri S., Altman N. S., Ruiz D., Parkinson J. E., Boulay J. N., Genotypic variation influences reproductive success and thermal stress tolerance in the reef building coral, Acropora palmata. Coral Reefs 32, 703–717 (2013). [Google Scholar]
- 55.Eckert C. G., The loss of sex in clonal plants. Evol. Ecol. 15, 501–520 (2002). [Google Scholar]
- 56.Barrett S. C. H., Influences of clonality on plant sexual reproduction. Proc. Natl. Acad. Sci. U.S.A. 112, 8859–8866 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bobiwash K., Schultz S. T., Schoen D. J., Somatic deleterious mutation rate in a woody plant: estimation from phenotypic data. Heredity 111, 338–344 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gill D. E., Chao L., Perkins S. L., Wolf J. B., Genetic mosaicism in plants and clonal animals. Annu. Rev. Ecol. Syst. 26, 423–444 (1995). [Google Scholar]
- 59.Yu L., Boström C., Franzenburg S., Bayer T., Dagan T., Reusch T. B. H., Somatic genetic drift and multilevel selection in a clonal seagrass. Nat. Ecol. Evol. 4, 952–962 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Puill-Stephan E., van Oppen M. J. H., Pichavant-Rafini K., Willis B. L., High potential for formation and persistence of chimeras following aggregated larval settlement in the broadcast spawning coral, Acropora millepora. Proc. R. Soc. B 279, 699–708 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Knaus B. J., Grunwald N. J., VCFR: A package to manipulate and visualize variant call format data in R. Mol. Ecol. Resour. 17, 44–53 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Kamvar Z. N., Tabima J. F., Grünwald N. J., Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2, e281 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prevosti A., Ocana J., Alonso G., Distances between populations of Drosophila subobscura, based on chromosome arrangement frequencies. Theor. Appl. Genet. 45, 231–241 (1975). [DOI] [PubMed] [Google Scholar]
- 64.Ferdosi M. H., Kinghorn B. P., van der Werf J. H. J., Lee S. H., Gondro C., hsphase: An R package for pedigree reconstruction, detection of recombination events, phasing and imputation of half-sib family groups. BMC Bioinformatics 15, 172 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gu Z. G., Eils R., Schlesner M., Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 66.van de Wiel M. A., Kim K. I., Vosse S. J., van Wieringen W. N., Wilting S. M., Ylstra B., CGHcall: Calling aberrations for array CGH tumor profiles. Bioinformatics 23, 892–894 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Cingolani P., Platts A., Wang L. L., Coon M., Nguyen T., Wang L., Land S. J., Lu X. Y., Ruden D. M., A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w(1118); iso-2; iso-3. Fly 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shinzato C., Shoguchi E., Kawashima T., Hamada M., Hisata K., Tanaka M., Fujie M., Fujiwara M., Koyanagi R., Ikuta T., Fujiyama A., Miller D. J., Satoh N., Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320–323 (2011). [DOI] [PubMed] [Google Scholar]
- 69.Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B. C., Remm M., Rozen S. G., Primer3—New capabilities and interfaces. Nucleic Acids Res. 40, e115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.dela Cruz D. W., Harrison P. L., Enhanced larval supply and recruitment can replenish reef corals on degraded reefs. Sci. Rep. 7, 13985 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Puill-Stephan E., Willis B. L., van Herwerden L., van Oppen M. J. H., Chimerism in wild adult populations of the broadcast spawning coral Acropora millepora on the Great Barrier Reef. PLOS ONE 4, e7751 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rinkevich B., Shaish L., Douek J., Ben-Shlomo R., Venturing in coral larval chimerism: A compact functional domain with fostered genotypic diversity. Sci. Rep. 6, 19493 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Amar K. O., Chadwick N. E., Rinkevich B., Coral kin aggregations exhibit mixed allogeneic reactions and enhanced fitness during early ontogeny. BMC Evol. Biol. 8, 126 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bottega R., Cappellani S., Fabretto A., Spinelli A. M., Severini G. M., Aloisio M., Faleschini M., Athanasakis E., Bruno I., Faletra F., Pecile V., Could a chimeric condition be responsible for unexpected genetic syndromes? The role of the single nucleotide polymorphism-array analysis. Mol. Genet. Genomics 7, e546 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salomon-Torres R., Gonzalez-Vizcarra V. M., Medina-Basulto G. E., Montano-Gomez M. F., Mahadevan P., Yaurima-Basaldua V. H., Villa-Angulo C., Villa-Angulo R., Genome-wide identification of copy number variations in Holstein cattle from Baja California, Mexico, using high-density SNP genotyping arrays. Genet. Mol. Res. 14, 11848–11859 (2015). [DOI] [PubMed] [Google Scholar]
- 77.Peiffer D. A., Le J. M., Steemers F. J., Chang W. H., Jenniges T., Garcia F., Haden K., Li J. Z., Shaw C. A., Belmont J., Cheung S. W., Shen R. M., Barker D. L., Gunderson K. L., High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 16, 1136–1148 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wallace C. C., Reproduction, recruitment and fragmentation in nine sympatric species of the coral genus Acropora. Mar. Biol. 88, 217–233 (1985). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Notes S1 to S5
Figs. S1 to S7
Tables S1 and S2
References
Data S1 to S10