Abstract
T4 gene 2 mutants have a pleiotropic phenotype: degradation of injected phage DNA by exonuclease V (ExoV) in the recBCD+ host cell cytoplasm and a low burst size due, at least in part, to a decreased ability for head-to-tail (H-T) joining. The more N terminal the mutation, the more pronounced is the H-T joining defect. We have overexpressed and purified the recombinant gene 2 product (rgp2) to homogeneity in order to test its role in H-T joining, during in vitro reconstitution. When we mix extracts of heads from a gp2+ phage infection (H+) with tails from a gp2+ or gp2− phage infection (T+ or T−), the H-T joining is fast and all of the reconstituted phage grow equally well on cells with or without ExoV activity. When heads from gene 2 amber mutants (H−) are used, addition of rgp2 is required for H-T joining. In this case, H-T joining is slow and only about 10% of the reconstituted phage can form plaques on ExoV+ cells. When extracts of heads with different gene 2 amber mutations are mixed with extracts of tails (with a gene 2 amber mutation) in the presence of rgp2, we find that the size of the gp2 amber peptide of the head extract is inversely related to the fraction of reconstituted phage with a 2+ phenotype. We conclude that free rgp2 is biologically active and has a direct role in H-T joining but that the process is different from H-T joining promoted by natural gp2 that is incorporated into the head in vivo. Furthermore, it seems that gp2 has a domain which binds it to the head. Thus, the presence of the longer gp2am mutants (with this domain) inhibits their replacement by full-length rgp2.
Epstein et al. (11) found that when a phage T4 gene 2am mutant (2am) is grown on a restrictive (Su0) host, the heads are not joined to the tails. Some of the heads have DNA, and some are empty. They concluded that the product of gene 2 (gp2) is involved in the joining of heads to tails (H-T joining). In addition to the separated heads and tails found in T4 2− lysates by Epstein et al., King (15) found many complete phage particles of normal appearance, in which tails are attached to heads full of DNA. However, since these complete phage particles were noninfectious, he concluded that gene 2 is also involved in functions other than H-T joining.
Granboulan et al. (13) showed that an extract of T4 2am phage was complemented by T4 2+ extract only when the 2+ heads in the extract were unattached to the tails and the tails in the 2− extract were unattached to the heads. This suggested that gp2 normally enters the phage head during morphogenesis and can no longer do so after H-T joining. Granboulan et al. (13) also quantitated the T4 particles in extracts of phage T4 2−-infected nonpermissive hosts. They found about 40 full-headed and 20 empty-headed phage particles per infected cell and about 10 full and 10 empty, detached heads (i.e., without attached tails) per infected cell. Though only about 0.07 infective particles were produced per infected cell (ca. 0.1% of the wild-type burst), there were 40 killer particles/cell. They concluded that most heads can join normally and firmly to tails during morphogenesis of T4 2am in nonpermissive cells, but because of the fragility of the heads, the DNA is lost from many of these particles. At this time, it was not yet clear why the full-headed particles were not infective. The source of the killer particles, whether from empty- or full-headed phage (or both), was also not clear, especially since there were more killers than empty-headed phage. (They did not report on the tail fiber content of phage particles or of the tails). They also found that heads of gene 64 mutants, now recognized as an N-terminal class of gene 2 mutants (19), lose their DNA more easily than wild-type heads. In addition, they showed that the H-T joint of the T4 gene 64 mutant is more fragile than the H-T joint of wild-type or gene 2 mutant particles. Finally, they showed that neither 2− lysates nor free 2− heads (from a 2− 10− lysate) nor 64− lysates can be complemented in vitro by gp2 or gp64 headless (23am) lysates. Only detached 64− heads (from a 64− 10− lysate) are complemented in vitro by a headless, 23− lysate. They concluded from this that only free virgin heads, and not decapitated 64− heads (broken off from the fragile H-T junction), can be joined to tails to produce infective phage and then only at low efficiency (about 8%; calculated from Table 7 or reference 13). The conclusion from their paper is that gp64 (now called gp2 [see below]) from a 23−, headless lysate can complement 64− heads in vitro, before the heads join to tails from the same headless lysate. (Gene 64 mutations are only a more N-terminal class of gene 2 mutations, which will be discussed below.)
The high yield of seemingly intact T4 2− phage particles which are not infective led to the rumor that they are defective in DNA injection. When we found that these particles attach to and kill the host cells with a multiplicity of infection (MOI) of 1 (23), we thought that their inviability might result from restriction. We showed that the DNA in the head is injected and then degraded and that the DNA hydrolysis was caused exclusively by exonuclease V (ExoV) in the cytoplasm (24). We then showed more specifically (21) that degradation of the entering T4 chromosome is exonucleolytic. This phenomenon, called exonucleolytic restriction (9), is more usually seen in vivo, only after specific exonucleolytic restriction cleavage which creates unprotected DNA termini that become the substrate for ExoV. Subsequent ExoV digestion of unprotected DNA termini prevents chromosome repair and also expression of genes in or dependent upon the region contiguous to the endonuclease site which is hydrolyzed by ExoV (10). When Lipinska et al. (19) showed that gene 64 mutants are only an N-terminal class of gene 2 mutants, it became clear that neither 2− nor 64− phage particles (now called 2−) are complemented by a headless (23−) extract containing gp2. The reason for this seemed to be that the H-T junction prevents entry of gp2 into the head to join to the DNA (13).
A model explaining these phenomena is that gp2 is attached to the DNA terminus in the mature phage head and thereby protects it from ExoV action during transfer into the host cell cytoplasm. Nevertheless, it does not rule out the possibility that gp2 mediates a modification of DNA termini which prevents degradation by RecBCD.
We have now cloned, expressed, and purified gp2 to homogeneity. In order to test directly the idea of Granboulan et al. (13) that gp2 can be added to the free 2− head, we developed a three-component, in vitro reconstitution system using (i) extracts of T4 23−, lacking heads (T), with or without gp2 (T+ or T−, respectively); (ii) extracts lacking tails, with or without gp2 (H+ or H−, respectively); (iii) gp2, either natural, in phage-infected extracts, or as rgp2. Our criterion for functional gp2 entry into the head of the reconstituted T4 particle is protection of phage DNA (during subsequent infection) from exonucleolytic restriction by ExoV in Escherichia coli B40 (SuI ExoV+) (26). We show that gp2 is required for H-T joining during morphogenesis and show that rgp2 incorporation into gp2am heads (before H-T joining) can protect the DNA from restriction during subsequent injection. Finally, we show by complementation with rgp2 that it is easier to replace small amber fragments of gp2 in the unattached 2− head than it is to replace larger amber fragments. These results corroborate the work of Granboulan et al. (13), who showed that an extract containing gp2 can complement a free amC86(64−) head but not a free amN51(2−) head which has a longer gp2 fragment.
MATERIALS AND METHODS
Bacteria, bacteriophages, and plasmids.
All strains are described in Table 1.
TABLE 1.
Bacterial strains, phages, and plasmids
| Strain, phage, or plasmid | Genotype | Source or reference |
|---|---|---|
| E. coli strains | ||
| B | sup0 (Su−) (ExoV+) | Lab strain |
| Bb | sup0 (Su−) (ExoV+) | Lab strain |
| B40SuI | argF40(Am) supD60 (SuI ExoV+) | Lab strain |
| BL21(DE3) | F−ompT hsdS (rB− mB−) gal (int::PlacUV5-T7 gene 1 imm 21 min5) | W. F. Studier |
| JC7735 | F−supD60 (SuI) recB21 (ExoV−), leu(Am) trpE9829 strR321 B1− | A. J. Clark |
| MC1000 | F−araD139 Δ(ara-leu)7679 galU galK Δ(lac)X74 rpsL thi | 8 |
| MC1061 | F−araD139 Δ(ara-leu)7679 galU rpsL galK Δ(lac)X74 hsdR2 (rK− mK+)mcr thi | 8 |
| Phage strains | ||
| M13mp7 | M13 with inverted repeat polylinker | 20 |
| T4D+ | Wild type | Lab strain |
| T4 2− amN51 | Mutation in gene 2 | Lab strain |
| T4 2− amC86 | Mutation in gene 2 (formerly in gene 64) | Lab strain |
| T4 2− amE1102 | Mutation in gene 2 | Lab strain |
| T4 10− amB255 | Mutation in gene 10 (defective in baseplate formation; it is used as a source of heads) | Lab strain |
| T4 23− amB17 | Mutation in gene 23 (defective in head formation; it is used as a source of tails) | Lab strain |
| Plasmids | ||
| pACYC184 | Cmr Tetr | 27 |
| pACYC2+3am · 9 | pACYC184 with the T4 insert from pBB5; Cmr | This work |
| pgp2 (pAV3 · 2+3am · 1) | pET-3 with the T4 insert from pBB5 oriented for expression; Ampr | This work |
| pAV3 · 2+3am · 2 | Same as pAV3 · 2+3am · 1 but with opposite orientation; Ampr | This work |
| pBB5 | pUC8 with the same insert 2+3am as pKA2+3am; Ampr | This lab |
| pET-3 | pBR322 derivative used for gene expression; Ampr | 26 |
| pLysS | pACYC184 derivative carrying T7 gene 3.5 (T7 lysozyme); Cmr | 27 |
(i) Bacteria.
E. coli B and Bb (Su0 ExoV+) were used as nonpermissive hosts for T4 amber mutants to produce extracts. E. coli B40 (SuI ExoV+) was used as the amber permissive host but is restrictive for gp2− phage particles lacking gp2 due to exonucleolytic digestion of the infecting DNA. It is used to determine the titer of phenotypically 2+ particles, i.e., reconstituted particles containing gp2 which can protect injecting DNA from ExoV restriction. E. coli JC7735 (SuI ExoV−) is used as an amber permissive host which, in contrast to B40, is also nonrestrictive to infecting gp2− DNA. It is used to determine the titer of total phage (2+ plus 2−). E. coli BL21(DE3)/pLysS, which contains a defective λ prophage carrying the gene for T7 RNA polymerase under the control of the lacUV5 promoter (4), and a plasmid expressing T7 lysozyme (27) were provided by F. W. Studier. This strain was used to express gp2 from the plasmid pgp2.
(ii) Phage.
Bacteriophage T4D+ is the wild-type strain. The double mutants 2− 10− and 2− 23− can have any of the three gene 2 mutations listed above, as specified in the text. However, unless otherwise noted, amE1102 (the shortest gp2 peptide) was used. All amber mutants were grown in B40 to make phenotypically normal phage stocks and in the nonpermissive strain Bb to make extracts.
(iii) Plasmids.
pACYC184 was a gift of M. Malamy; pET-3 was a gift of F. W. Studier.
Media and buffers.
(Most media and buffers are described in reference 22.) L broth is 1% Bacto Tryptone, 0.5% yeast extract, and 0.5% NaCl brought to pH 7.0 with 1 N NaOH. M9 is 1 g of NH4Cl, 3 g of KH2PO4, 6 g of Na2HPO4, 4 g of glucose, and 1 ml of 1 M MgSO4 in 1 liter of H2O. M9-ZB contains all components of M9 and ZB (see below). Plain phage broth is 1% Bacto Peptone, 0.5% NaCl, 0.3% beef extract, and 0.1% dextrose. T2 plates contain 1% Bacto Tryptone, 0.8% NaCl, 0.2% Na-citrate, 0.1% glucose, and 1.2% agar neutralized with 1 N NaOH. T2 top agar is like T2 plates but with 0.6% agar. ZB is 10 g of N-Z-amine A and 5 g of NaCl in 1 liter of water (26). Antibiotics were incorporated into growth media at the following concentrations: 100 μg of ampicillin per ml, 25 μg of chloramphenicol (CAM) per ml, and 25 μg of tetracycline per ml.
Buffer A is 50 mM Tris-HCl (pH 8.0) and 25 mM NaCl. Buffer B is 10 mM Tris-HCl (pH 7.9), 0.1 mM EDTA, 0.1 mM dithiothreitol (DTT), and 5% glycerol. Buffer C is 20 mM Tris-HCl (pH 7.9), 10 mM MgCl2, 1 mM EDTA, 0.1 mM DTT, 0.1 mM phenylmethylsulfonyl fluoride, and 30% glycerol. Buffer D is 50 mM Tris-HCl (pH 8.0), 2 mM EDTA, and 0.1 mM DTT. Dilution fluid is 0.1% Bacto Peptone, 0.3% NaCl, and 0.025% MgSO4. Disruption buffer is 2% sodium dodecyl sulfate (SDS) in 60 mM Tris-HCl (pH 6.8). Lysis buffer is 50 mM Tris-HCl (pH 8.0), 2 mM EDTA, 0.1 mM DTT, 0.1 M NaCl, 5% glycerol, and 200 mg of lysozyme/ml. Phosphate-buffered saline is 10 mM NaPO4 (pH 7.2) and 0.9% NaCl. Washing fluid is 10 mM Tris-HCl (pH 7.4), 1 mM MgCl2, 0.1% NaCl, and 0.01% gelatin. All reagents are of analytical grade, and enzymes and proteins were obtained commercially.
Plasmid constructs.
All plasmids are described in Table 1. Unless otherwise noted, all methods were essentially as previously described in reference 22. The constructs pAV3 · 2+3am · 1 and pAV3 · 2+3am · 2 were made as follows. We isolated a 1.6-kb fragment of T4 DNA, containing gene 2+ and gene 3am, from pBB5 by digestion with EcoRI and PstI. We prepared EcoRI-BamHI and PstI-BamHI linkers from replicative-form DNA of phage M13mp7. We ligated these linkers to the respective EcoRI and PstI termini of the 1.6-kb fragment and then inserted the BamHI-terminated 1.6-kb fragment into the BamHI site (in the tet gene) of a dephosphorylated pACYC184 plasmid which we then transformed into E. coli MC1061. We selected Camr transformants on l-CAM plates and screened for Tets. We chose a Camr Tets transformant which tested positive for marker rescue of a gene 2amN51 mutation and called the plasmid pACYC2+3am · 9. When the plasmid was digested with BamHI, we obtained a 1.6-kb DNA fragment (with EcoRI and PstI sites just internal to the terminal BamHI sites) which we purified from a low-melting-temperature agarose gel. We ligated this fragment (containing genes 2 and 3am) into the dephosphorylated BamHI site of pET-3, and the ligation reaction mixture was transformed into E. coli MC1000. Ampr transformants were selected and rechecked for gene 2 by marker rescue of phage T4 amN51. We obtained two types of plasmid: pAV3 · 2+3am · 1 (henceforth called pgp2 for simplicity and clarity [Table 1]), with gene 2 properly oriented to the tet promoter, and pAV3 · 2+3am · 2, with gene 2 oriented opposite the transcription direction.
Marker rescue test for gene 2.
Marker rescue plates for screening were prepared by overlaying a T2 plate with 3 ml of T2 soft agar containing 4 drops of a fresh, mid-log E. coli B culture. Three-milliliter aliquots of T4 amN51 dilutions were spotted on the plates first. Then 3 ml of mid-log cultures from transformant colonies was overlaid on the spots. If the plasmid carried the corresponding gene 2+ marker, a zone of lysis formed after overnight incubation at 37°C.
Preparation of crude cell extracts with rgp2.
All operations were at 4°C unless specifically noted otherwise. We grew E. coli BL21(DE3)/pLysS/pgp2 to an optical density at 600 nm (OD600) of 0.5 at 37°C in M9-ZB plus 0.2% glucose with ampicillin and CAM and then induced the cells with isopropyl-β-d-thiogalactopyranoside (IPTG). We took aliquots at 30-min intervals, harvested the cells and resuspended them in lysis buffer, and sonicated the extracts to reduce the viscosity. The crude, concentrated extracts were run on SDS-polyacrylamide gels. To quantitate gp2, we stained extracts with Coomassie brilliant blue R-250 (18) and scanned the gels optically on an LKB Ultroscan XL scanner with Gelscan XL software.
Preparation of antibody to rgp2.
The proteins in the crude extract containing rgp2 were separated by SDS–12% polyacrylamide gel electrophoresis (PAGE). We washed the gel with deionized water, stained it with 0.05% Coomassie blue, prepared it in water, washed it again with water, and excised a weakly stained band of gp2 which we emulsified in complete Freund's adjuvant and injected intradermally into a rabbit. This was followed by four booster injections at 7- to 14-day intervals, and we collected serum after 40 days.
Western blot analysis.
Western blots were used to positively identify the gp2 band. We transferred proteins from parallel gels onto nitrocellulose or polyvinylidene difluoride membranes (Mini Trans-Blot; Bio-Rad), essentially as described by Towbin et al. (31). The membranes were probed with the rabbit anti-gp2 serum. After washing, we incubated the blots with anti-rabbit immunoglobulin G-alkaline phosphatase conjugate and developed them colorimetrically with nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate (NBT-BCIP).
Purification of rgp2.
Crude, concentrated extracts containing gp2 were prepared as described above except that we incubated them after induction for up to 3 h before lysis by sonication. We sedimented the sonicated extract at 17,000 × g for 20 min at 4°C and found an aggregate of rgp2 (basic) and DNA (acidic) in the sediment. After washing the pellet with buffer A, we solubilized rgp2 and DNA from the pellet by stirring in a small amount of buffer A containing 2 M NaCl. After sedimentation, we diluted the supernatant to 1 M NaCl and supplemented it with 0.1% polyethyleneimine, to precipitate DNA. We sedimented the mixture, diluted the supernatant 2.5-fold with buffer A, and added ammonium sulfate to 60% saturation. We resuspended the precipitate in a small volume of buffer B, dialyzed it, and loaded it onto a DEAE–Sephadex A-50 column (9 by 130 mm) equilibrated with the same buffer. The column was eluted with buffer B, and rgp2 was eluted in a single peak (monitored at 280 nm) at the front of the column. We pooled fractions containing gp2, precipitated them in 60% ammonium sulfate, resuspended the protein in buffer B, and rechromatographed it on the column. The rgp2 fractions (monitored at 280 nm) were concentrated, dialyzed against buffer C, and stored at −20°C. We used the 280-nm/260-nm ratio to estimate the protein/DNA ratio and assayed protein concentration directly (6), using a bovine serum albumin standard.
N-terminal protein sequencing.
Purified rgp2 was concentrated with a Centricon microconcentrator 10 (Amicon) and washed with water (to remove Tris and glycerol which might interfere with sequencing). We determined the gp2 sequence by automated Edman degradation.
Kinetics of gp2 expression in phage-infected cells.
E. coli cells, grown to an OD600 of 0.5, were infected with phage T4 at an MOI of 8. We took 10-ml aliquots at 0, 5, 7, 9, 11, 13, 15, and 20 min; added crushed frozen L broth; and stored the aliquots on ice. We sedimented these cultures, resuspended the pellets in disruption buffer, heated them for 5 min at 95°C, sonicated them for 3 s to reduce the viscosity, electrophoresed samples on SDS-polyacrylamide gels, and transferred from the gels onto nitrocellulose membranes for Western blot analysis.
Identification of gp2 in T4 phage particles.
To make concentrated phage preparations, E. coli cells were grown to an OD600 of 0.5 in L broth (plus 0.2% glucose) at 37°C with aeration and infected with T4 phage at an MOI of 8. After an hour, we lysed the infected cells with chloroform and sedimented the lysate at 3,000 × g for 10 min to remove debris. We incubated the supernatant for 1 h at 30°C with DNase and RNase and sedimented it at 50,000 × g for 30 min. To concentrate the phage, we washed the pellet once and resuspended the phage in the desired volume of buffer.
We disrupted the concentrated phage preparation by heating it for 8 min at 95°C in an equal volume of 2× disruption buffer and sonicated it for 10 s. We dialyzed the extract against 50 mM Tris-HCl (pH 8.0) at room temperature overnight in order to remove SDS from the sample, sedimented the debris, and supplemented the supernatant with ammonium sulfate to 50% saturation. The resulting protein precipitate was resuspended in 100 μl of buffer D (1/20 volume of the concentrated T4 phage sample) and used for SDS-PAGE and Western blots.
Preparation of extracts for reconstitution.
We made extracts containing wild-type heads (H+) and 2− heads (H−) (i.e., with mutant gp2), by infecting E. coli Bb at 30°C with T4 10− or T4 2− 10−, respectively, at an MOI of 5, and superinfected extracts with the same MOI 10 min later. After 50 min, we gently sedimented the infected cells at 1,100 × g for 6 min. The pellet was resuspended in 2 mM MgSO4 at 1/100 of the original volume and lysed with CHCl3. We removed the debris at low speed, discarded the pellet, and stored the extracts at 4°C for not more than a few days before use. In order to minimize the risk of disruption, we did not freeze and thaw the heads.
We made extracts containing wild-type tails with free gp2 (T+) or without free gp2 (T−) in essentially the same way, except that T4 23− or T4 2− 23− phage were used. To lyse the infected cells, we resuspended them in 0.01 M Tris-HCl (pH 7.0), containing 0.02 M MgSO4 and 5 μg of DNase I per ml, and freeze-thawed them three times in dry ice-ethanol. We kept the extract in small aliquots at −70°C and freeze-thawed it just before use.
Preparation of partially purified and washed heads.
H+ or H− extracts were prepared as described above except that the low-speed pellet (after CHCl3 lysis) was resuspended in an equal volume of 2 mM MgSO4 and resedimented, and the two supernatants (containing the heads) were combined. We pelleted the heads from the combined supernatants by sedimentation at 36,000 × g for 35 min (in a Beckman Ti50 rotor), resuspended the pellets in 1/100 of the original volume of 2 mM MgSO4, and stored them for not more than 2 days at 4°C.
To wash the heads, we sedimented H+ extracts in a Beckman Airfuge at 30 lb/in2 for 5 min. We resuspended the pellets in an equal volume of 2 mM MgSO4 and used them on the same day.
Reconstitution of phage particles.
We incubated a 60-μl volume, containing 20 μl of each component (or buffer), at 30°C for the indicated time. We determined titers of reconstituted phage on B40SuI (ExoV+) to assay 2+ phage and on JC7735SuI (ExoV−) for total phage (i.e., 2+ plus 2−).
RESULTS
Gene 2 expression and purification.
Lipinska et al. (19), using the pT7-13 expression vector (27), demonstrated a 30-kDa radioactive gp2 band on SDS-polyacrylamide gels, but protein expression was too low for detection by Coomassie blue staining. (This was due to background expression of gp2 which is very toxic to the cell [3a].) Therefore, we subcloned gene 2 into a pET-3 vector to obtain pgp2 and introduced it into E. coli BL21(DE3)/pLysS to express gp2 under the tight control of a phage T7 promoter. The equal growth rates of uninduced BL21(DE3)/pLysS and BL21(DE3)/pLysS/pgp2 strains suggested that there was little or no leakage of gene 2 expression (19). Western blots of induced cell extracts revealed immunoreactive rgp2 bands (data not shown). By 150 min after induction with IPTG, rgp2 accounted for up to 16% of the total cell protein (data not shown). When we induced BL21(DE3)/pLysS/pAV3 · 2+3am · 2 (with gene 2 in the opposite direction, as a control), it showed no rgp2 expression.
We purified gp2 to homogeneity as determined by SDS-PAGE. The purity of the gp2 preparation was also evident by the lack of significant secondary peaks in the Edman degradation. Since gp2 is a very basic protein (pI = 10.94), it is difficult to remove all traces of nucleic acid which may bind to it specifically, or adventitiously, in the aggregates. The polyethyleneimine-mediated precipitation of DNA, after disaggregation by 2 M NaCl and repeated ion-exchange chromatography, yielded a final gp2 preparation with <1% nucleic acid (as determined by the 260-nm/280-nm ratio).
Sequence of gp2.
We found an apparent molecular mass for gp2 of about 30 kDa on SDS-polyacrylamide gels, though we had expected a molecular mass of 27.5 kDa based on the apparent open reading frame (ORF) (19). We thought that the mobility was aberrant due to the basic character of the protein. Subsequently, Koch et al. (16) resequenced gene 2 and found 40 additional N-terminal residues due to an extra T inserted (in the Lipinska sequence) between C13 and A14 of the Koch sequence. To resolve this issue, we sequenced the 10 N-terminal residues of purified rgp2 and found the sequence MAIFQIINES, which is identical to the first 10 amino acids of the extended Koch et al. ORF and consistent with the 31.5-kDa mass calculated from the revised ORF of Koch et al. and found by SDS-PAGE. Therefore, we have proven that the Koch et al. N-terminal extension of the ORF is correct. There are a few other unresolved discrepancies between the two sequences including a 6-codon segment from bp 112 to 128 of the Koch sequence in which C112 is deleted in the Lipinska sequence and a C is added after A128 to restore the proper frame. Two other changes are GA53–54 to AG and T972 to A.
Kinetics of gp2 expression in T4 phage-infected cells.
Based on DNA-RNA hybridization, Broida and Abelson (7) reported that T4 gene 2 transcription occurs late in the phage cycle, commencing at about 12 min at 30°C. In order to verify this finding and to establish the time of initial gp2 synthesis, we looked for the first appearance of gp2 in T4-infected cells by Western blotting. We found that gp2 is not detectable in T4-infected E. coli until after 13 min at 37°C (unpublished data). Translation of gp2 at 13 min is consistent with late transcription, as found by Broida and Abelson (7).
Demonstration of gp2 in the T4 phage particle.
In order to demonstrate the presence of gp2 in the phage particle, we disrupted a preparation of concentrated phage with SDS, sonicated it to reduce viscosity (due to DNA), and concentrated gp2 and some other proteins with ammonium sulfate. We then separated these proteins on SDS-polyacrylamide gels and identified gp2 by Western blot analysis to show that gp2 is present in T4 wild-type particles (data not shown).
Biological activity of purified rgp2.
In order to evaluate the role of gp2 in the morphogenesis of T4 phage, we developed a three-component reconstitution system containing heads, tails, and gp2. The role of gp2 in the reconstitution of infective particles with 2+ and 2− phenotypes is illustrated in Table 2. Line 1 shows that reconstitution by a mixture of H+ (an extract of T4 10am-infected E. coli Bb containing 2+ heads, no tails, and possibly free gp2) and T+ (an extract of T4 23am-infected E. coli Bb containing tails, no heads, and probably free gp2) yields a 1,000-fold increase of total phage over the background level. The last column of line 1 shows that all of the reconstituted phage are phenotypically 2+ (i.e., they can grow on ExoV+ as well as on ExoV− hosts), reflecting the 2+ phenotype of heads in the head extract. Line 2 shows that addition of rgp2 does not increase the yield of phage (and may even interfere a bit with H-T joining). Thus, the amount of gp2 supplied by the 2+ extracts (whether incorporated into heads or free) was not limiting for reconstitution in line 1.
TABLE 2.
Complementation of T4 phage by gp2 in vitroa
| Line | Mixture | Total phage | 2+ phage | 2+/total ratiob |
|---|---|---|---|---|
| 1 | H+ + T+ | 6.2 × 109 (6.0 × 106)c | 6.3 × 109 (6.6 × 106) | 1.0 (1.1) |
| 2 | H+ + T+ + rgp2 | 3.2 × 109 | 2.8 × 109 | 0.9 |
| 3 | H− + T− | 7.2 × 107 (4.8 × 107) | 1.0 × 107 (2.5 × 106) | 0.31 (0.05) |
| 4 | H− + T− + rgp2 | 9.2 × 109 | 1.4 × 109 | 0.15 |
| 5 | H− + T+ | 6.7 × 108 (4.0 × 106) | 4.4 × 107 (4.5 × 106) | 0.06 (1.1) |
| 6 | H− + T+ + rgp2 | 2.9 × 108 | 2.1 × 107 | 0.06 |
| 7 | H+ + T− | 2.3 × 109 (2.5 × 106) | 2.5 × 109 (2.5 × 106) | 1.1 (1.0) |
| 8 | H+ + T− + rgp2 | 1.8 × 109 | 2.0 × 109 | 1.1 |
| 9 | H+(w)d + T− | 3.2 × 109 (2.5 × 106) | 2.8 × 109 (2.5 × 106) | 0.88 (1.0) |
| 10 | H+(w) + T− + rgp2 | 2.2 × 109 | 2.1 × 109 | 0.95 |
Incubation was at 30°C for 1 h.
Ratio of newly reconstituted phage after subtracting background. (The ratio of the background 2+ to total phage is in parentheses.)
Background values of total phage (phenotypically 2+ plus 2−) and of 2+ phage are calculated from titers of the individual components on JC7735 and on B40SuI, respectively. The background values of the individual components for lines 1, 2, and 5 to 10 are as follows: H+, 4.3 × 106 and 4.3 × 106; T+, 7.8 × 106 and 8.9 × 106; H−, 2.5 × 105 and 2.0 × 105; T−, 7.5 × 105 and 6.5 × 105. For lines 3 and 4, the values are as follows: H−, 7.5 × 105 and 2.0 × 106; T−, 9.6 × 107 and 3.0 × 106. There was no detectable background of phage in the gp2 preparation.
Heads were washed (w) by sedimentation to remove only cytoplasmic gp2 (external to the head).
Line 3 shows that a reconstitution mixture of H− (amE1102) and T− produces very few infective phage over the background of total phage. (The background in the preparation of T− used in lines 3 and 4 is unusually high). Line 1 shows about 250-fold-more total phage produced (after subtracting background) than line 3. This suggests that gp2 is required for H-T joining. Line 4 shows that when rgp2 is present in the mixture of H− and T−, the titer of total phages rises almost 200-fold over background to reach approximately the titer achieved in lines 1 and 2. This strongly reinforces the notion that gp2 is required for H-T joining. The titer of phenotypically 2+ phage in line 4 is increased about 500-fold over background. This implies that some rgp2 was incorporated into the head. Thus we have shown that rgp2, purified from aggregates, has both biological activities: (i) it promotes H-T joining during reconstitution, and (ii) it protects previously packaged DNA from degradation by ExoV upon injection into the host cytoplasm (during the next round of infection). Only 15% of the phage particles reconstituted in the presence of rgp2 (line 4, last column), can grow on B40SuI ExoV+. This may reflect a lower efficiency of in vitro incorporation of rgp2 and/or reduced ability of rgp2 to protect DNA termini from ExoV. The reduced efficiency of rgp2 in vitro (compared to natural gp2) for production and morphogenesis in vivo may be due to (i) alteration of a large fraction of the cloned rgp2 during purification, thereby reducing its ability to protect DNA from exonuclease digestion; (ii) inhibition of rgp2 entry into most of the heads by some natural morphogenetic component (e.g., the mutant gp2am fragment) in the head; or (iii) the lack of some component (normally present in phage-infected cells) to increase gp2 incorporation.
Since the reason for the lower frequency of phenotypically 2+ phage to total phage in line 4 (compared to lines 1 and 2) was not apparent, we compared natural gp2 in the T− extract with rgp2. Line 5 shows that when we mixed extracts of H− (with no wild-type gp2 in the heads or in the cytoplasm) and T+ (from infected cells, which should have natural gp2 in a “free” state), we found approximately a 150-fold increase (over background) in total infective phage, of which only 6% had a 2+ phenotype. Line 6 shows that addition of purified gp2 does not increase the extent of reconstitution of total phage or of 2+ phage (and in fact decreases both in equal proportion, similar to the case in lines 1 and 2). Comparison of lines 5 and 6 with lines 3 and 4 shows that natural gp2 in a T+ extract, like rgp2, can mediate H-T joining in vitro but is no more effective than rgp2 in conferring a 2+ phenotype on a 2− head in an H− extract.
It would appear, from the ratio of 2+ to total phage reconstituted in lines 6 and 4 compared to line 2, that the role of free gp2 during in vitro H-T joining is decoupled from its role in protecting reconstituted phage from ExoV during subsequent infection. It is possible that the rate of incorporation of gp2 into 2− heads (presumably a necessary prerequisite for subsequent ExoV protection) may be lower than that of H-T joining. This raises the question whether incorporation of natural gp2 during in vivo morphogenesis induces H-T joining as well.
To test this idea, we mixed H+ with T− to see if H-T joining would be normal in the absence of added free gp2. Line 7 shows that, when H+ and T− extracts are mixed, we reconstitute the normal complement of total phage and all of them are of 2+ phenotype. The addition of rgp2 (line 8) produces no significant change in the titers. This suggests either that there is sufficient free gp2 in H+ extracts to promote maximal H-T joining or that gp2-containing heads do not need free gp2 for H-T joining.
In order to test more directly whether only gp2 built into heads during morphogenesis in vivo is sufficient to promote attachment to tails, we mixed T− with washed 2+ heads (to remove any free gp2 from the extract). The washed 2+ heads were prepared by sedimenting an H+ extract and resuspending the pellet in buffer to minimize the amount of free cytoplasmic gp2 in the head preparation. Line 9 shows that washed 2+ heads, H+(w), produced phage as well as the unwashed H+ extract did (line 7). Line 10 shows that addition of rgp2 did not change the overall result. Therefore, we conclude that although free gp2 can function to join 2− heads to tails, no free gp2 is required to join 2+ heads to tails. The fact that about 90% of the reconstituted phage have a 2+ phenotype strongly suggests that gp2 that is incorporated into the heads during in vivo morphogenesis is much more efficient in promoting H-T joining and DNA protection.
Is the mechanism of H-T joining different for 2+ heads and 2− heads?
Our results show that 2− heads need free gp2 for H-T joining during reconstitution, whereas 2+ heads do not. The requirement for a third component, gp2, for H-T joining with H− heads suggests that the kinetics of infective particle production during reconstitution of 2− heads might be different from that of 2+ heads. If the free gp2 interaction with heads is rate limiting in the three-component system, reconstitution of 2+ heads should be faster than that of 2− heads.
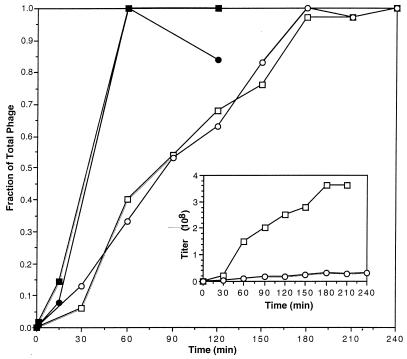
The inset in Fig. 1 shows the kinetics of total phage reconstitution from a mixture of H− and T− extracts with rgp2. The maximum yield, reached by 180 min, is 3.8 × 108 total phage/ml and 0.3 × 108 2+ phage/ml. Relative rates of these reconstitutions, normalized to the maximum, are shown with the same symbols in Fig. 1. The calculated rates of phage reconstitution are 2.1 × 106 total phage/min and 1.7 × 105 2+ phage/min. The ratio of these rates, 0.08, is constant over the 180-min period.
FIG. 1.
The role of gp2 in the kinetics of reconstitution. The inset illustrates the kinetics of reconstitution of a mixture of H−, T−, and gp2. The ordinate quantitates the titer (108) of phage (the maximum titer after subtracting the background at 0 min is 3.7 × 108). The upper curve shows the kinetics of total phage reconstitution, and the lower shows the kinetics for phenotypically 2+ particles (maximum titer, 0.3 × 108). In the main graph, the rightmost pair of curves shows the same data as in the inset but as a relative titer, normalized to the highest titer of each. The leftmost pair of curves represents the data found when an H+ extract was used. These curves are normalized to 2.4 × 109 and 2.5 × 109 for the total and 2+ phage titers, respectively. —— , total phage produced using H− heads; ○——○, 2+ phage produced using H− heads; ▪——▪, total phage produced using H+ heads; ●——●, 2+ phage produced using H+ heads.
The two leftmost curves in Fig. 1 show the kinetics of reconstitution of phage for the mixture of H+ and T− extracts with rgp2. The maximum yield, reached after 1 h (or less) is 2.5 × 109 phage/ml. In this case, total phage and 2+ phage are always about equal and the rate of reconstitution (calculated in the 15- to 60-min interval) is at least 5 × 107 phage/min. The 24-fold reduction in rate of total phage production (i.e., H-T joining) might be expected when comparing the three-component system (H−-T−-rgp2) to the two-component system (H+-T−), in which 2+ is preincorporated into the head. However, the further 12-fold reduction in the rate of 2+ phage production seems to show that rgp2 facilitates H-T joining more rapidly than DNA protection, since H-T joining would preclude subsequent DNA protection (13). Thus, in vivo, it is probable that gp2 is incorporated into the head with the DNA before H-T joining, in order to protect it. Whether this discrepancy between in vitro and in vivo mechanisms is a result of defects in the composition or components of the in vitro system remains to be seen.
Pleiotropy of the gene 2 phenotype is region dependent.
All gene 2 amber mutants investigated so far exhibit an inability to grow on cells with ExoV activity (19). Thus, even our most C-terminal amber mutation in gene 2 is ExoV sensitive. Nevertheless, amN51 mutants have a burst size 50% of normal when titers are determined on ExoV− cells (23). Mutations closer to the N terminus show very little H-T joining, and of those that are joined, a relatively large fraction are askew and probably inviable (11, 13). These observations indicate that, in vivo, some gp2 N-terminal mutations can drastically reduce DNA protection with a relatively small effect on H-T joining.
To test the effect of the size of different gp2am truncations on reconstitution, we used three different gene 2 amber mutant head extracts. The semiquantitative order of length of the genes, based on marker rescue experiments (19), is as follows: gp2+ (1.0) > gp2amN51 [H(N)] (≥0.67) > gp2amC86 [H(C)] (≤0.39 but ≥0.27) > gp2amE1102 [H(E)] (≤0.27). (The relative size of a gp2 peptide is in parentheses. The size is derived from the map data in Fig. 1 of reference 19, adjusted for the sequence as extended in reference 16. The symbol of the head with the specific fragment is in brackets.) Each of the head donor strains also had the gene 10 mutation, amN255, to prevent tail assembly. In all cases, we used T− (E1102, the shortest gp2am fragment) extract, T(E), for the tails. In Table 3, we give the averaged results of these experiments (each repeated five times).
TABLE 3.
Effect of phenotype of head on H-T joining and ExoV protectiona
| gp2 | H+
|
H(N)
|
H(C)
|
H(E)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | 2+ | 2+/total ratio | Total | 2+ | 2+/total ratio | Total | 2+ | 2+/total ratio | Total | 2+ | 2+/total ratio | |
| − | 19 | 19 | 1.0 | 9.5 | 0.63 | 0.066 | 0.43 | 0.12 | 0.27 | 3.3 | 1.4 | 0.44 |
| + | 36 | 40 | 1.1 | 90 | 1.34 | 0.015 | 94 | 5.3 | 0.057 | 65 | 8.7 | 0.13 |
| Δ | 17 | 21 | 1.25 | 80 | 0.71 | 0.009 | 94 | 5.2 | 0.055 | 62 | 7.3 | 0.12 |
| H(X) | 0.034 | 0.029 | 0.046 | 0.060 | 0.01 | 0.019 | 0.39 | 0.096 | ||||
| T(E) | 0.29 | 0.043 | 0.29 | 0.043 | 0.29 | 0.043 | 0.29 | 0.043 | ||||
Values in rows 1 to 3 are titers (108) of reconstitution mixtures after incubation at 30°C for 60 min. Total phage titers were determined on E. coli JC7735, and 2+ phage titers were determined on E. coli B40. Values in rows 4 and 5 are titers (108) of unmixed extracts.
In the H+ columns, row 1 shows that when a control H+ extract is mixed with a T− extract, all of the reconstituted phage are phenotypically 2+. Prior addition of rgp2 to the mix (row 2) doubles the phage production, and all of the reconstituted phage are still phenotypically 2+. In the H(N) columns, total phage production is lower than that for H+ (by half), but only 7% of the total phage are phenotypically 2+. If the amN51 heads are reconstituted in the presence of rgp2, the titer of total phage rises 10-fold, but only 1.5% of these newly made phage are 2+ (row 2). In row 3, (Δ = row 2 − row 1), we calculate the efficiency of rgp2 in H-T joining and in DNA protection in vitro. The 2+/total ratio is 0.009, calculated from the difference between the titers (± gp2) when using the H(N) extract. This is more than 100-fold lower than the result found using H+ extract. One interpretation of the results using amN51 mutant heads is that the gp2amN51 fragment inside of the head can serve to mediate H-T joining from within but not quite as well as gp2+ does. This is in agreement with previous findings (13, 23). (In a similar experiment, Granboulan et al. [reference 13, Table 7, row 1] found a similar ratio of 0.004, if total phage is 90 as implied in the control value below the table.) In the presence of gp2+, the H-T joining is more efficient due to the additional action of external gp2 in H-T joining of previously incompetent heads, possibly because they had too few associated amN51 fragments to join. Nevertheless, wild-type gp2 cannot displace gp2amN51, sufficiently or effectively, inside the head in vitro and therefore cannot render the DNA in the phage head resistant to ExoV degradation during infection.
When reconstitution of an H(C) extract is compared with that of an H(N) extract, the total phage produced in the absence of rgp2 is 20-fold lower. This shows that a shorter gp2am fragment in the head is less efficient in H-T joining. When rgp2 is added to the reconstitution mixture (row 2), total phage rises more than 200-fold, to the level of the corresponding amN51 sample, demonstrating the many competent heads potentially available for H-T joining. The results of reconstitution in the presence of rgp2 (row 2) also show that fourfold-more 2+ phage are produced by reconstitution with added rgp2 in a mixture with H(C) than with H(N) extract. When we estimated the precise rgp2 effect, we found (row 3) that the ratio of 2+ to total, 0.055, is sixfold higher for H(C) than for H(N). (In a similar experiment, Granboulan et al. [reference 13, Table 7, row 2] found a ratio of 0.08.) These results suggest that rgp2 displaces gp2amC86 [called gp2(C)] more easily than it can displace gp2(N). Since the gp2(C) peptide is not longer than the first 40% of gp2, whereas gp2(N) comprises at least 67% of the peptide, we postulate that a region near the center of the gp2 sequence is needed for binding gp2 to the heads. The reduced total phage produced without rgp2 suggests that the shorter gp2(C) is even less effective than gp2(N) for H-T joining.
When an H(E) extract is mixed with a T(E) extract in the absence of added rgp2, total phage rises to 10 times the calculated background level (average of lines 4 and 5). Surprisingly, almost half of the phage produced have a 2+ phenotype. We cannot explain this high value (and it is not the case in Table 2). In any case, it shows that heads with very short 2am fragments are very poor at H-T joining, and the only ones producing infective phage are the background wild-type heads, containing full-length gp2. In the presence of rgp2, the level of reconstitution of total phage rises another 20-fold to the approximate levels achieved by the other mixtures with gp2. The fraction of phenotypically 2+ phage produced by this mixture, 0.13, is higher than that for reconstitutions with the other two mutants. The corrected value for DNA protection by rpg2 is 0.12. This is 2-fold higher than that for H(C) and 13-fold higher than that for H(N). This may reflect an even weaker binding of gp2amE1102 due to the loss of the rest of a binding region located in the central portion of gp2. On the basis of these experiments, we suggest that (i) as the length of gp2 in the heads is shortened, its ability to promote effective H-T joining decreases (11, 13) (Table 3, row 1); and (ii) the ability to acquire a 2+ phenotype by adding rgp2 during in vitro reconstitution rises as the gp2am fragment gets shorter because it is more easily replaced by rgp2 during reconstitution.
Table 2 (line 3) shows that the amount of H-T joining using H(E) heads in the absence of rgp2 was not significant. In Fig. 1, we see that H-T joining using wild-type heads is faster than when using H(E) heads, even in the presence of rgp2. Table 3 shows that the amount of total phage produced in the absence of rgp2 is proportionate to the size of the gp2 fragment. Therefore, we think it likely that the rate of H-T joining may depend upon the size of the natural gp2 fragment in the head, even in the presence of free rgp2. If so, it is possible that the increased ratio of 2+ to total phage as the natural gp2 fragment in the head is decreased may reflect a longer period, before H-T joining, when rgp2 can enter the head to protect DNA. Another factor might be that rgp2 in vitro is much less efficient for DNA protection than is gp2 in vivo. To distinguish between the state of the protein and the morphogenetic process to explain this difference in affinity, an experiment using rgp2 in vivo during morphogenesis would be desirable.
DISCUSSION
We have detected gp2 in the virion and found that purified washed T4+ heads, H+(w), can serve as the only source of gp2 for H-T joining and DNA protection (Table 2, line 9). This finding corroborates the pleiotropic roles of gp2 and the presence of gp2 in T4 phage heads. We have also shown that rgp2 can serve as the sole source of gp2 for H-T joining and DNA protection; however, it is slower and less efficient than H+(w). This shows that gp2 is required for both H-T joining and DNA protection (9, 13, 21, 24). We have shown, by gel shift assay, that rgp2 binds to DNA (G. R. Wang, unpublished data). rgp2 showed no specificity; single- and double-stranded DNA, closed and open, with and without overhang, were all retarded by rgp2 under our conditions. Since cloned and purified rgp2 activity in vitro is quite inefficient (roughly calculated at 104 molecules/reconstituted phage), specific binding of active molecules may be obscured by the nonspecific binding of inactive protein. There is no clear proof that gp2 protection of T4 DNA termini is a direct result of steric hindrance. Indirect binding (for example, as a result of chemical [enzymatic] modification resulting in ligation of 3′-OH to 5′-PO4 termini of adjacent strands, which occurs in N15 phage [30] and linear plasmids of Borrelia burgdorferi [5]) might also be invoked. Whether the activity of gp2 is direct or not, we think that its functional effect is directly on the DNA and not as an ExoV inhibitor, as is the case in Mu gam (1, 2, 29, 33), λ gam (12, 14, 32), and T4 anti-ExoV (9, 21a, 24, 28a). The late transcription of gp2 (7), the extremely basic character of the protein, and the small amount of gp2 in the phage particle are all compatible with gp2 acting on the DNA. However, the best evidence so far for the action of gp2 at DNA termini in vivo is from the work of Lipinska et al. (19), who show that inducible expression of cloned gp2 in an ExoV+ host will prevent degradation of DNA from infecting T4 2− phage. If phage are added even 1 min after induction, there is no breakdown of the DNA. There is a similar phenomenon for T4 phage whose DNA is not glucosylated. Such phage are restricted endonucleolytically at a number of sites by the E. coli rgl system (9) and then further degraded by ExoV at the newly created DNA termini. But the DNA can be protected from degradation by rgp2 in vivo (19). In this case, the level of protection from DNA hydrolysis, by ExoV, increases with the period between gp2 induction and DNA injection. We think that this reflects the lack of sufficient gp2 shortly after induction to prevent the hydrolysis of the many restriction-induced termini, shortly after DNA injection. As more gp2 is built up before injection, more of the DNA termini can be rapidly protected by gp2 before ExoV hydrolysis ensues.
Snustad predicted that gp4 plays a catalytic role in T4 morphogenesis (25). Granboulan et al. (13) also found that genes 4 and 50 (now known to be the N- and C-terminal parts, respectively, of gene 4) are fragile in their H-T attachment. We have found (D. Oliver, unpublished results) that the rate of reconstitution of 4− heads and 4− tails is proportional to the concentration of added gp4 in the mixture, consistent with Snustad's findings and his prediction. It is possible that the level of gp4 in the extract is too low for the maximum rate of H-T joining and that the addition of more gp4 (or rgp4) might close the gap between the curves for H+ and H− head extracts illustrated in Fig. 1. However, we have not done such an experiment.
Aksiote-Benbasat and Bloomfield (3) showed that H-T joining is diffusion controlled in a second-order reaction and depends upon an ionizable imidazole-like group (pK = 6.8), where a decrease in pH, to approximately 5.8, increases the rate constant. This implies that the reaction brings together groups of opposite charges. Since the head and tail proteins of known or calculable charge, at that time, were all of acidic pI, it was expected that increasing ionic strength would shield the repulsion and thereby raise the rate of joining. However, the opposite was the case. The prescient explanation proffered by Aksiote and Bloomfield was that “some positively charged proteins are attached to the head vertex or tail tip and locally outweighed the net negative charge.” More recent work has shown that all of the major structural proteins of the head, the neck, and the tail, except for two, have low isoelectric points [protein (pI)]: head, 13 (4.92), 14 (4.47), 20 (5.34), 23 (5.29), 24 (4.62); tail, 3 (4.22), 15 (4.77), 18 (4.80), 19 (4.55), 29 (4.93) [17]). The exceptions are gp2 (10.94) (16) and gp4 (9.79) (17). We suggest that the positively charged protein predicted by Aksiote and Bloomfield is gp2, and/or possibly gp4. Because of its localization at the head vertex (bound to a DNA terminus from inside and/or bound on the outside of the head, e.g., to gp20, -23, or -24), its presence could not only outweigh the local negative charge but might also specifically interact with a protein(s) of the tail tube (e.g., gp15 or gp3) to promote H-T attachment. (Of course, the pI of a protein does not necessarily reflect the overall surface charge or the charge at the site of interaction.) Thus, heads already containing gp2 would join more rapidly than those mixed with “free gp2” (which would increase the degrees of freedom), while those without gp2 would be the slowest, if they joined at all. This hypothesis could be tested by varying the pH and ionic strength, as well as determining the order of the reaction, analogous to the experiments done in Bloomfield's lab.
The selective evolutionary changes which go hand in hand with environmental changes, species development, and the rise of organelle formation and parasitism might be traced in phages. Phages would seem to arise from the bacteria as a sexual organelle for transduction. T4 might be considered an evolutionarily aberrant parasitic organelle which feeds on renewable E. coli sources (in situ or in sewage). It would be of interest to trace the genetic origins of various mechanisms of phage infection, morphogenesis, restriction, and suppression, to help unravel the evolutionary stages of phages.
ACKNOWLEDGMENTS
We thank Douglas Hamilton for his early and successful efforts in our lab, in developing the initial three-part in vitro reconstitution system. We thank S. Tabor, C. Richardson, W. Studier, A. J. Clark, T. Mattson, A. Wright, and M. Malamy for strains and F. Eiserling, D. Raychaudry, and P. Hyman for their comments on the manuscript.
This grant was supported in part by NIGMS grant 13511, NSF grant 9834603, ONR grant N00014-98-1-0784, and by a special grant from the dean's office of the Tufts University School of Medicine.
REFERENCES
- 1.Abraham Z H L, Symonds N. Purification of overexpressed gamgene protein from bacteriophage Mu by denaturation-renaturation techniques and a study of its DNA-binding properties. Biochem J. 1990;269:679–684. doi: 10.1042/bj2690679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akroyd J, Clayson E, Higgins N P. Purification of the gam gene-product of bacteriophage Mu and determination of the nucleotide sequence of the gamgene. Nucleic Acids Res. 1986;14:6901–6914. doi: 10.1093/nar/14.17.6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aksiote-Benbasat J, Bloomfield V. Kinetics of head-tail joining in bacteriophage T4D studied by quasi-elastic light scattering: effects of temperature, pH, and ionic strength. Biochemistry. 1981;20:5018–5025. doi: 10.1021/bi00520a031. [DOI] [PubMed] [Google Scholar]
- 3a.Appasani K, Thaler D S, Goldberg E B. Bacteriophage T4 gp2 interferes with cell viability and with bacteriophage lambda red recombination. J Bacteriol. 1999;181:1352–1355. doi: 10.1128/jb.181.4.1352-1355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arditti R R, Scaife J, Beckwith J. The nature of mutants in the lacpromoter region. J Mol Biol. 1968;38:421–426. doi: 10.1016/0022-2836(68)90396-3. [DOI] [PubMed] [Google Scholar]
- 5.Barbour A, Garon C F. Linear plasmids of the bacterium Borrelia burgdorferihave covalently closed ends. Science. 1987;237:409–411. doi: 10.1126/science.3603026. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Broida J, Abelson J. Sequence organization and control of transcription in the bacteriophage T4 tRNA region. J Mol Biol. 1985;185:545–563. doi: 10.1016/0022-2836(85)90071-3. [DOI] [PubMed] [Google Scholar]
- 8.Casadaban M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 9.Dharmalingam K, Goldberg E B. Mechanism, localization and control of restriction cleavage of phage T4 and λ chromosomes in vivo. Nature. 1976;260:406–410. doi: 10.1038/260406a0. [DOI] [PubMed] [Google Scholar]
- 9a.Dharmalingam K, Goldberg E B. Phage coded protein prevents restriction of unmodified progeny T4 DNA. Nature. 1976;260:454–456. doi: 10.1038/260454a0. [DOI] [PubMed] [Google Scholar]
- 10.Dharmalingam K, Revel H R, Goldberg E B. Physical mapping and cloning of bacteriophage T4 anti-restriction-endonuclease gene. J Bacteriol. 1982;149:354–362. doi: 10.1128/jb.149.2.694-699.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Epstein R H, Bolle A, Steinberg C M, Kellenberger E, Boy de la Tour E, Chevalley R, Edgar R S, Susman M, Denhardt G H, Lielausis A. Physiological studies of conditional lethal mutants of bacteriophage T4D. Cold Spring Harbor Symp Quant Biol. 1963;28:375–392. [Google Scholar]
- 12.Friedman S A, Hays J B. Protective inhibition of Escherichia coliRecBC activities by plasmid-encoded GamS function of phage lambda. Gene. 1986;43:255–263. doi: 10.1016/0378-1119(86)90214-3. [DOI] [PubMed] [Google Scholar]
- 13.Granboulan P, Sechaud J, Kellenberger E. On the fragility of phage T4-related particles. Virology. 1971;46:407–425. doi: 10.1016/0042-6822(71)90042-0. [DOI] [PubMed] [Google Scholar]
- 14.Karu A E, Sakaki Y, Echols H, Linn S. The protein specified by bacteriophage λ: structure and inhibitory activity for the recBC enzyme of Escherichia coli. J Biol Chem. 1975;250:7377–7387. [PubMed] [Google Scholar]
- 15.King J. Assembly of the tail of bacteriophage T4. J Mol Biol. 1968;32:231–262. doi: 10.1016/0022-2836(68)90007-7. [DOI] [PubMed] [Google Scholar]
- 16.Koch T, Lamm N, Ruger W. Sequencing, cloning and overexpression of genes of bacteriophage T4 between map positions 74.325 and 77.184. Nucleic Acids Res. 1989;17:4392. doi: 10.1093/nar/17.11.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kutter E, Stidham T, Guttman B, Kutter E, Batts D, Peterson S, Djavakhishvili T, Arisaka F, Mesyanzhinov V, Rüger W, Mosig G. Genomic map of bacteriophage T4. In: Karam J D, Drake J W, Kreuzer K N, Mosig G, Hall D H, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, Miller E S, editors. Molecular biology of bacteriophage T4. Washington, D.C.: American Society for Microbiology; 1994. pp. 491–519. [Google Scholar]
- 18.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Lipinska B, Krishnarao A S M, Bolten B M, Balakrishnan R, Goldberg E B. Cloning and identification of bacteriophage T4 gene 2 product gp2 and action of gp2 on infecting DNA in vivo. J Bacteriol. 1989;171:488–497. doi: 10.1128/jb.171.1.488-497.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Messing J, Crea R, Seeburg P H. A system for DNA shotgun sequencing. Nucleic Acids Res. 1981;9:309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oliver D B, Goldberg E B. Protection of parental T4 DNA from a restriction exonuclease by the product of gene 2. J Mol Biol. 1977;116:877–881. doi: 10.1016/0022-2836(77)90276-5. [DOI] [PubMed] [Google Scholar]
- 21a.Sakaki Y. Inactivation of the ATP-dependent DNase of Escherichia coliafter infection with double-stranded DNA phages. Virology. 1974;13:1611–1612. doi: 10.1128/jvi.14.6.1611-1612.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 23.Silverstein J L, Goldberg E B. T4 DNA injection. I. Growth cycle of a gene 2 mutant. Virology. 1976;72:195–211. doi: 10.1016/0042-6822(76)90323-8. [DOI] [PubMed] [Google Scholar]
- 24.Silverstein J L, Goldberg E B. T4 DNA injection. II. Protection of entering DNA from host exonuclease V. Virology. 1976;72:212–223. doi: 10.1016/0042-6822(76)90324-x. [DOI] [PubMed] [Google Scholar]
- 25.Snustad D P. Dominance interactions in Escherichia coli cells mixedly infected with T4D wild-type and amber mutants and their possible implications as to type of gene product function: catalytic vs. stoichiometric. Virology. 1968;35:550–563. doi: 10.1016/0042-6822(68)90285-7. [DOI] [PubMed] [Google Scholar]
- 26.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 27.Studier F W. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J Mol Biol. 1991;219:37–44. doi: 10.1016/0022-2836(91)90855-z. [DOI] [PubMed] [Google Scholar]
- 28.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Tanner D, Oishi M. The effect of bacteriophage T4 infection on an ATP dependent DNase in Escherichia coli. Biochim Biophys Acta. 1971;228:764–769. doi: 10.1016/0005-2787(71)90747-7. [DOI] [PubMed] [Google Scholar]
- 29.Thaler D S, Stahl M M, Stahl F W. Evidence that the normal route of replication-allowed Red-mediated recombination involves double-chain ends. EMBO J. 1987;6:3171–3176. doi: 10.1002/j.1460-2075.1987.tb02628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tilly K. Independence of bacteriophage N15 lytic and linear plasmid replication from the heat shock proteins DnaJ, DnaK, and GrpE. J Bacteriol. 1991;173:6639–6642. doi: 10.1128/jb.173.20.6639-6642.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unger R C, Clark A J. Interaction of the recombination pathway of bacteriophage λ and its host Escherichia coliK-12: effects on exonuclease V activity. J Mol Biol. 1972;70:539–548. doi: 10.1016/0022-2836(72)90558-x. [DOI] [PubMed] [Google Scholar]
- 33.Williams J G K, Radding C M. Partial purification of an exonuclease inhibitor induced by bacteriophage Mu-1. J Virol. 1981;39:548–558. doi: 10.1128/jvi.39.2.548-558.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]