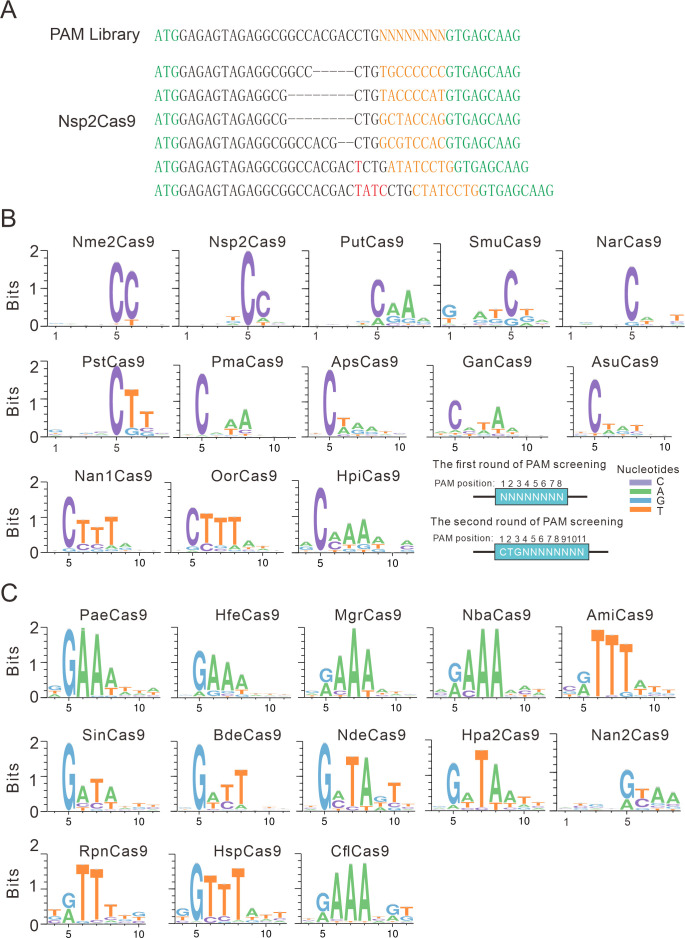
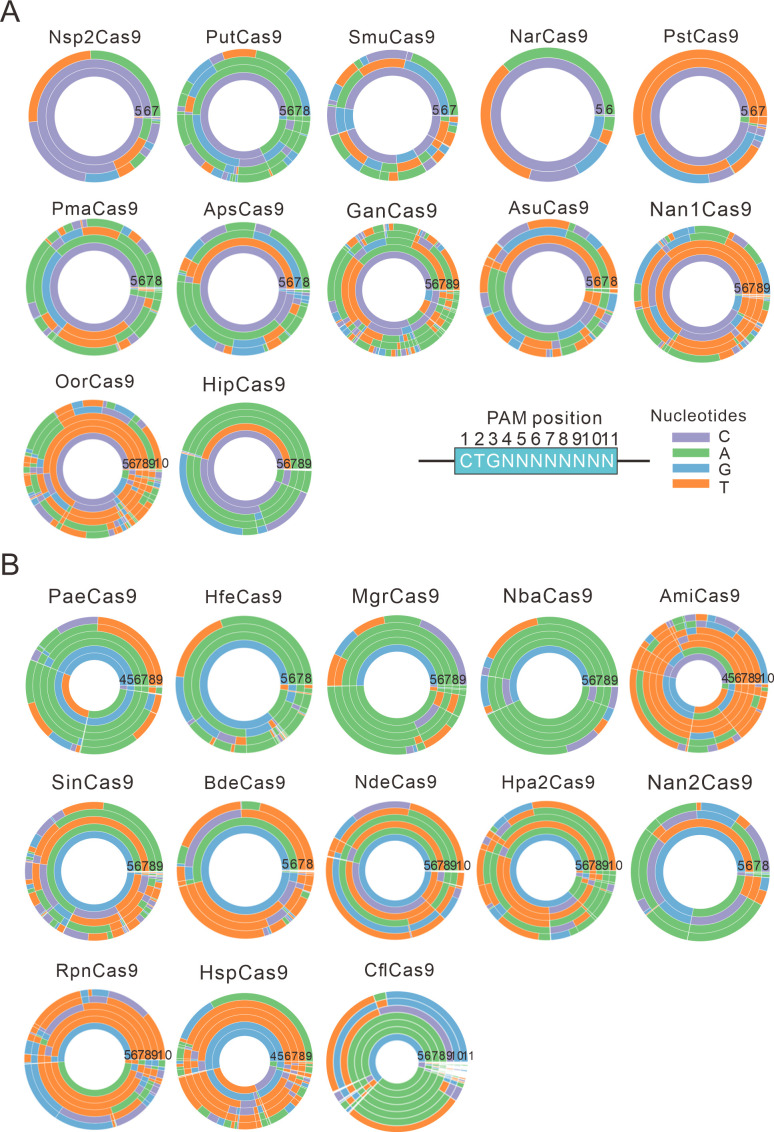
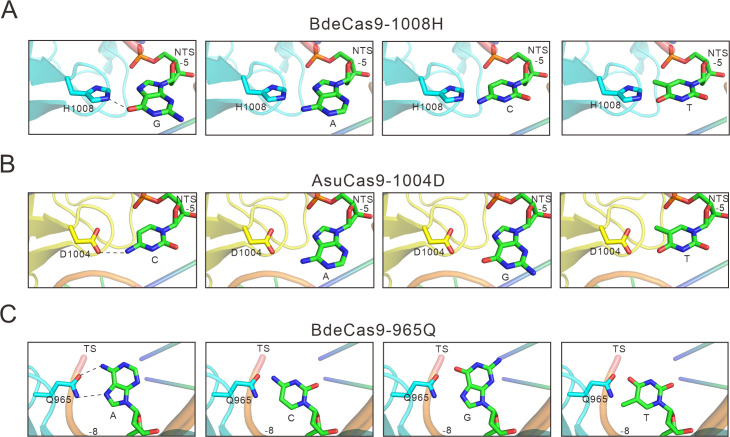
Figure 2. PAM analysis for each Cas9 nuclease.
(A) Example of indel sequences measured by deep sequencing for Nsp2Cas9. The GFP coding sequences are shown in green; an 8 bp random sequence is shown in orange; black dashes indicate deleted bases; red bases indicate insertion mutations. (B) The PAM WebLogos for Nme1Cas9 orthologs containing an aspartate residue corresponding to the Nme1Cas9 H1024. PAM positions for each WebLogo are shown below. The PAM WebLogos for Nme2Cas9, Nsp2Cas9, PutCas9, SmuCas9, NarCas9, PstCas9 are generated from the first round of PAM screening and the PAM WebLogos for others are generated from the second round of PAM screening. PAM positions in the screening assay are shown on the bottom right. (C) The PAM WebLogos for Nme1Cas9 orthologs containing histidine, or asparagine residues corresponding to the Nme1Cas9 H1024. PAM positions for each WebLogo are shown below. The PAM WebLogo for Nan2Cas9 is generated from the first round of PAM screening and the PAM WebLogos for others are generated from the second round of PAM screening.