Abstract
Metabolic reprogramming is a distinctive characteristic of SARS-CoV-2 infection, which refers to metabolic changes in hosts triggered by viruses for their survival and spread. It is current urgent to understand the metabolic health status of COVID-19 survivors and its association with long-term health consequences of infection, especially for the predominant non-severe patients. Herein, we show systemic metabolic signatures of survivors of non-severe COVID-19 from Wuhan, China at six months after discharge using metabolomics approaches. The serum amino acids, organic acids, purine, fatty acids and lipid metabolism were still abnormal in the survivors, but the kynurenine pathway and the level of itaconic acid have returned to normal. These metabolic abnormalities are associated with liver injury, mental health, energy production, and inflammatory responses. Our findings identify and highlight the metabolic abnormalities in survivors of non-severe COVID-19, which provide information on biomarkers and therapeutic targets of infection and cues for post-hospital care and intervention strategies centered on metabolism reprogramming.
Keywords: COVID-19 survivors, Metabolic abnormalities, Metabolomics, Health consequences
COVID-19 survivors; Metabolic abnormalities; Metabolomics; Health consequences.
1. Introduction
Coronavirus disease 2019 (COVID-19), a global pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has resulted in 523,786,368 confirmed cases with 6,279,667 deaths, as of 24 May 2022 (https://covid19.who.int/). About 80% of confirmed COVID-19 patients presented with non-severe symptoms, and the proportion is increasing as vaccination and treatment protocols mature (Huang et al., 2020; Costanzo et al., 2022a). Patients were generally discharged within one month, but they may not have “truly recovered” due to the long-term sequelae and debilitation. The epidemiological cohort studies have reported that COVID-19 survivors mainly suffered from fatigue or muscle weakness, sleep difficulties, anxiety or depression and dyspnoea at six months after infection, and these symptoms retained even one year (Huang et al., 2021a, 2021b). Besides, confirmed persistent symptoms in survivors of SARS and Middle East respiratory syndrome (MERS) also call attention on “long COVID-19”.
Metabolic reprogramming is initiated by virus for their replication and survival and also modulated by the immune response after SARS-CoV-2 infection (Shen and Wang, 2021; Xiao et al., 2021). Several metabolic pathways, mainly including amino acids, energy, lipids and immune-related metabolism, were reprogrammed in hospitalized COVID-19 patients (Su et al., 2020; Shen et al., 2020; Costanzo et al., 2022b). These metabolic changes had not completely returned to normal even the discharge criteria of the official care protocol for COVID-19 were met (Wu et al., 2020). Metabolites are more sensitive than clinical indicators to reflect the condition of the body. Considering the fact that metabolic changes usually progress to a chronic state, we propose that the effects of SARS-CoV-2 infection on metabolism may be long-lasting, especially in the absence of intervention and treatment after discharge.
Metabolic health plays a crucial role in regulating the long-term health consequences and convalescent duration for COVID-19 patients, given the close relationship between metabolism and pathophysiological states. However, metabolic health status of COVID-19 survivors after discharge has not been completely elucidated. In the present study, we characterized the serum metabolic signatures of survivors of non-severe COVID-19 at six months after discharge from the first pandemic wave in Wuhan, China, and further explored the potential adverse outcomes associated with these metabolic abnormalities. The findings are expected to provide more information on understanding of coronavirus pathology and biomarkers and the health cares and intervention targets of COVID-19 survivors after discharge.
2. Results
2.1. Survivors exhibit differences in demographics, clinical characteristics and metabolic profile from controls
Twenty-one past-infected volunteers (survivors) with non-severe COVID-19 symptoms and 22 control volunteers without SARS-CoV-2 infection history were recruited at Renmin Hospital of Wuhan University, Wuhan, China. Their clinical data are presented in Table S1. Survivors of non-severe COVID-19 had a median age of 50.2 years (young: 31.0 ± 4.4; elder: 56.2 ± 6.7) and 28.5% were male, and control volunteers had a median age of 54.5 years (young: 29.6 ± 2.9; elder: 61.9 ± 8.3) in which 36.4% were male. Compared to controls, red blood cell counts, hemoglobin, albumin, total protein and aspartate aminotransferase (AST) were significantly decreased in COVID-19 survivors, but high-density lipoprotein cholesterol (HDL-c) decreased only in elder survivors. Changes in the first two indices suggested the hematopoiesis in the bone marrow and the oxygen transport in blood was inhibited to a certain extent, and that in others might be associated with hepatic dysfunction.
We integrated non-targeted and targeted metabolomic approaches to characterize the difference of serum metabolic profile of survivors of non-severe COVID-19 and controls. Total of 116 polar metabolites and 455 lipids were detected in serum samples (Table S2). Orthogonal partial least squares discriminant analysis (OPLS-DA) found a dramatic difference between the serum metabolic profiles of survivors and controls (Figure 1a). Our results suggest that the metabolic-health effects of SARS-CoV-2 infection still existed six months after discharge. According to the screening criteria for significantly differential metabolites (in the Methods section), 83 of 165 metabolites with |log2(fold change)| ≥ 0.25, and p < 0.05 in volcano plot were significantly varied by meeting variable importance in the projection (VIP) > 1.0 and area under curve (AUC) > 0.75, mainly including amino acids, organic acids, fatty acids and lipids (Figure 1b and Table S3). As shown in Figure 1c and 1d, the numbers of differential metabolites in elder group were higher than that of young group (159 vs 30, Table S3), suggesting the metabolic abnormality in elder survivors might be more profound although the sample size of young survivors is small. Change trends of the differential metabolites from each class in both age groups were basically consistent. The top differential metabolites (VIP ≥2.0) in all, young and elder samples were integrated and analyzed, and the change trends showed that most of them were reduced in serum from survivors of non-severe COVID-19 (Figure 1e). The significantly differential metabolites in all survivors were further performed pathway analysis, and the result showed that the primary abnormal metabolic pathways of survivors of non-severe COVID-19 were amino acid metabolism, TCA cycle, lipid metabolism and fatty acid metabolism (Figure 1f and Table S4). Moreover, similar metabolic abnormalities were also found in blood of in-hospital COVID-19 patients (Mahmud and Garrett, 2020; Aggarwal et al., 2021), demonstrating that metabolic reprogramming induced by SARS-CoV-2 infection had not returned to normal at six months after discharge. The persistent metabolomic abnormalities in COVID-19 survivors may require a longer recovery period (Li et al., 2022).
Figure 1.
Abnormal serum metabolites associated with the history of SARS-CoV-2 infection. (a) OPLS-DA score plot (R2Y = 0.96, p = 0.003; Q2 = 0.60, p < 0.001 for permutation test) obtained from modelling metabolic profile in the survivors of non-severe COVID-19 (red) and controls (blue). Volcano plots for differential metabolites in all (b), young (c), and elder (d) samples. (e) Heat maps of top differential metabolites in all, young, and elder samples. The color of each column represents the logarithm of fold change of metabolites with base 2, and p values were calculated using a student's t-test, ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. (f) The main KEGG pathways disturbed in survivors of non-severe COVID-19.
2.2. Abnormal amino acid and purine metabolism indicate hepatic injury and mental disorders
Amino acids are mainly synthesized in the liver, and play important roles in the immune system and maintaining redox status. Hepatic injury is frequently present in hospitalized COVID-19 patients (Zhang et al., 2020; Caterino et al., 2021). The main manifestations of hepatic injury, elevated alanine aminotransferase and aspartate aminotransferase levels, were not observed in discharged COVID-19 patients in our cohort, but the decreased albumin, total protein and AST might be associated with hepatic dysfunction. As shown in Figure 2, serum ornithine, aspartate, asparagine, alanine, glycine, serine, proline, histidine and threonine were significantly down-regulated (p < 0.05) in COVID-19 survivors compared with controls, while arginine, glutamate, valine, taurine, phenylalanine, lysine, glutamine and methionine were significantly up-regulated. Moreover, the change trends were similar in young and elder survivors (Figure S1). Further pathway enrichment analysis results show these significantly altered amino acids in COVID-19 survivors are involved in the urea cycle, ammonia recycling, glycine and serine metabolism, arginine and proline metabolism, aspartate metabolism and glutamate metabolism (Figure S2). Urea cycle converts toxic ammonia into urea in the liver, and its disorder also is the common feature of hepatic injury, such as acute and chronic liver dysfunction and liver carcinogenesis (Bigot et al., 2017). In COVID-19 patients, most of urea cycle-related metabolites were decreased (Shen et al., 2020; Song et al., 2020). We found that the urea cycle was still disordered with the decrease of ornithine and increase of arginine in survivors of non-severe COVID-19. Therefore, hepatic injury caused by SARS-CoV-2 infection has not been recovered six months after discharge, and might mainly result from the urea cycle blocked in the arginine node.
Figure 2.
Disorders in targeted metabolites in survivors of non-severe COVID-19. Heatmap representation of Z-score transformed concentrations of the amino acids and amino acid derivatives, organic acids and neurotransmitters. Each column represents a single sample, and the dark red in color scale is for the highest value, white is for the midpoint value and dark blue is for the lowest value.
Anxiety, depression, disturbed sleep and post-traumatic stress disorder (PTSD) are common adverse mental health symptoms to the COVID-19 pandemic (Rajkumar, 2020; Taquet et al., 2021). Glutamate is the main excitatory neurotransmitter in the central nervous system, and its disorder affects mental health (Nasir et al., 2020). The serum level of glutamate was significantly increased in survivors of non-severe COVID-19, and can contribute to the abovementioned mental symptoms through neuron damage (Zeredo et al., 2019). PTSD was the most common condition among COVID-19 survivors, and the high occurrence rate (42.9%) of denial or avoidance of infection history was found in our cohort (Table S1), and the higher glutamate levels was found in elder survivors with PTSD (Figure S3). Elevated glutamate in survivors also suggested that the energy metabolism of their central nervous system was weakened in survivors, which could explain their remaining symptoms of fatigue and sleep disturbance (Tarazona et al., 2021; Drews et al., 2020). Furthermore, purine metabolism was inhibited in survivors of non-severe COVID-19 with the reduction of inosine, deoxyinosine and hypoxanthine. As end-product of purine metabolism, low serum levels of uric acid in elder survivors (Table S3) might increase the risks of multiple sclerosis, neurodegenerative diseases and optic neuritis (Kutzing and Firestein, 2008). These results suggested that the raising awareness of long-term mental health care in COVID-19 survivors is urgent.
2.3. Altered organic acid and carbohydrate metabolites indicate inhibition of cellular energy production
SARS-CoV-2 infection can decrease serum levels of organic acids, and further severely inhibit energy metabolism (Song et al., 2020). We found that organic acids involved in the TCA cycle, succinic acid and 2-ketoglutaric acid remained significantly down-regulated in serum samples from survivors of non-severe COVID-19 (Figure 2 and Figure S1), suggesting that the damaged energy metabolism has not returned to normal. Notably, the levels of citric acid, malic acid, fumaric acid and aconitic acid showed no significant difference in survivors compared to controls, demonstrating that the TCA cycle inhibited by SARS-CoV-2 infection was recovering. Glycolysis is adopted to facilitate virus replication and cytokine production after SARS-CoV-2 infection, and the elevated glucose was observed in COVID-19 patients (Wu et al., 2020). Glycolysis remained inhibited in young survivors of non-severe COVID-19, with significant increase in glucose-6-phosphate (G6P), and which might be a major risk factor for new-onset diabetes and kidney injury in COVID-19 survivors (Rubino et al., 2020; Sundaram et al., 2021). The TCA cycle and glycolysis are metabolic pathways for the generation of ATP, but the former is more highly efficient pathway. The significant down-regulations of lactic acid and AST were also evidence for the disrupted energy metabolism (Figure S1 and Figure 2e). Therefore, ATP production of COVID-19 survivors was mainly attenuated by weakened TCA cycle, further resulted in fatigue or muscle weakness of the COVID-19 survivors (41.7%, in Table S1).
2.4. Disturbed glycerophospholipid and glycerolipid metabolism are more significant in elder survivors
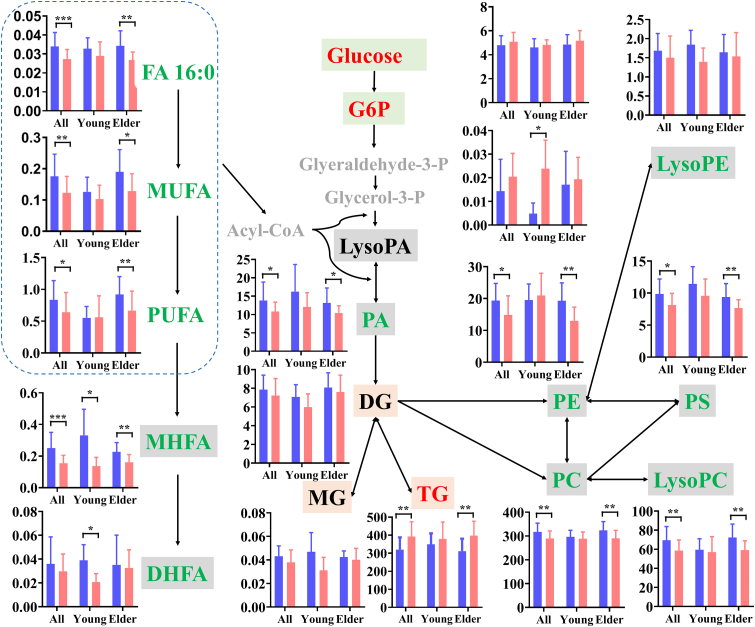
Lipids are envelope components of SARS-CoV-2, and play important roles in host-pathogen interactions and pathogenesis (Abu-Farha et al., 2020). Dysregulated lipid metabolism in COVID-19 patients was ubiquitous and closely related to the levels of disease severity (Su et al., 2020). Even when patients has met clinical discharge criteria, their lipid metabolism still remains abnormal (Wu et al., 2020). In our study, lipid metabolic dysregulations sustained in survivors of non-severe COVID-19 six months after discharge (Figure S4), and it may be more obvious in elder cases (Table S3). As shown in Figure 3, the total levels of lysophosphatidylcholines (LysoPC), phosphatidic acids (PA), phosphatidylcholines (PC), phosphatidylethanolamines (PE), phosphatidylserines (PS), and ceramides (Cer) in elder survivors were significantly down-regulated, except for triglycerides (TG). In young cases, no lipid classes presented significant change (p > 0.05). These initial observations suggest that glycerophospholipid and sphingolipid metabolism could be inhibited in elder survivors, but glycerolipid metabolism could be activated. The serum lipid metabolic disruptions in survivors of non-severe COVID-19 differed from that of SARS survivors twelve years after infection, with increase of phosphatidylinositols (PI), lysophosphatidylinositols (LysoPI) and LysoPC (Wu et al., 2017), and a possible explanation for this is the high-dose steroid treatment for SARS patients.
Figure 3.
Metabolic network of significantly changed lipid classes in all, young and elder survivors of non-severe COVID-19. Bars in all graphs represent mean abundance of controls (light blue column) and survivors (pink column) in different subgroups, error bars represent standard deviation. The normalized total peak abundance of individual lipid class is shown on the Y-axis. Red, green, black and gray colors are significantly up-regulated, down-regulated, unchanged and undetected lipids or metabolites, and the arrows indicate the direction of metabolism. The dotted-line box includes all free fatty acids (FFA), which convert to Acyl-CoA for participate the synthesis of LPA and PA. p values were calculated using a student's t-test, ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. MUFA: monounsaturated fatty acids, PUFA: polyunsaturated fatty acids, MHFA: mono-hydroxyl fatty acids; DHFA: di-hydroxyl fatty acids, G6P: glucose-6-phosphate, Glyeraldehyde-3-P: glyeraldehyde-3-phosphate, Glycerol-3-P: glycerol-3-phosphate, PC: phosphatidylcholines, LysoPC: lysophosphatidylcholines, PA: phosphatidic acids, LysoPA: lyso phosphatidic acids, PE: phosphatidylethanolamines, LysoPE: lisophosphatidylethanolamines, PS: phosphatidylserines, MG: monoglycerides, DG: diglycerides, and TG: triglyceride.
As important components of bio-membranes, glycerophospholipids are essential for intracellular metabolism and transmembrane signaling. Total abundance of PC, PE, and PS were significantly down-regulated in elder survivors, indicating that their transformation was markedly inhibited in the whole glycerophospholipid metabolism, and cell membrane structure could be destructed. As major precursors of PS and PC, the significant decrease of serine and choline in the serum of elder survivors also proved this inhibition (Figure S5). Cytosolic phospholipase A2α (PLA2α), which hydrolyzes glycerophospholipids to free fatty acids (FFAs) and lysophospholipids, is activated after SARS-CoV-2 infection (Shen et al., 2020). The down-regulation of total lysoPC and FFA in elder survivors demonstrated the abnormal activation of PLA2α had returned to normal. Most of PC and PE species with at least one unsaturated fatty acyl chain were significantly reduced in elder COVID-19 survivors six months after discharge, and several PA, PI and PS species were also reduced (Table S3). As important energy storage units, total TG levels were significantly up-regulated in elder survivors, demonstrating the biosynthesis of TG might be accelerated along with the reduction of MG, to maintain the balance of the energy metabolism in the body. It was closely associated with fatigue or muscle weakness syndrome of the COVID-19 survivors in our cohort. The accumulation of TG was also found in the plasma of COVID-19 survivors after discharge (Xu et al., 2021; Li et al., 2022). Besides, elevated TG and reduced HDL-c might increase the risk of coronary heart diseases in elder survivors (Krauss and Siri, 2004).
2.5. Alterations in inflammation mediators are associated with age
As bioactive signaling mediators, FFAs and their oxygenated products are involved in numerous functions, including inflammation, coagulation and apoptosis (Dennis and Norris, 2015). Fatty acid metabolism was significantly inhibited in survivors of non-severe COVID-19, and varied with the ages of survivors (Figure 3). The total levels of mono-hydroxyl fatty acids (MHFAs) were significantly down-regulated in both young and elder survivors, while FFAs reduced only in elder cases, and di-hydroxyl fatty acids (DHFAs) reduced only in young cases. MHFAs are active anti-inflammatory and pro-resolution mediators (Serhan et al., 2008), two and five of MHFA species were respectively reduced in young and elder discharged patients, whose reduction demonstrates a potential deficit in the resolve of inflammatory in survivors of non-severe COVID-19. Li et al. (2022) also found that two MHFA species were downregulated in plasma of severe and non-severe COVID-19 survivors 6 months after discharge from the hospital.
In elder survivors, 38.8% of FFA species, including FA 16:0, FA 18:0, FA 18:1, FA 18:2, FA 20:1, FA 20:4 and FA 22:6, were significantly down-regulated (Table S3), demonstrating that the elder survivors might have a poor resistance to infection and/or disease. It was different from the elevation of serum FFAs in recovered SARS patients twelve years after infection (Wu et al., 2017). Unsaturated fatty acids can inactivate enveloped viruses and inhibit their proliferation, but their changes were inconsistent in human blood after SARS-CoV-2 infection (Su et al., 2020; Shen et al., 2020). Further, DHFAs with eighteen carbon atoms were significantly reduced in young survivors (Table S3). Among them, DHFA from FA 18:2 can inhibit inflammatory and immune responses, regulate the vascular development and repair process, negatively regulate the pain, enhance increase exercise and skeletal muscle regulation (Hildreth et al., 2020). Thus, a high fatty acid diet is suggested for survivors of non-severe COVID-19 for the elevation of the levels of FFAs and their oxygenated products both in blood and tissues to boost their immune system.
2.6. Recovered immune-related metabolites indicate the macrophages in normal state
Immune dysregulation is common in COVID-19 patients. The kynurenine pathway participates in the regulation of the body's immune system, and is significantly activated in SARS-CoV-2 patients (Thomas et al., 2020). Activation of this pathway can help to enhance the phagocytic ability of macrophages and turn on the switch for macrophage effector responses by autonomic synthesis of NAD+ (Minhas et al., 2019). We found the serum levels of metabolites in the kynurenine pathway, including tryptophan, kynurenine, kynurenic acid, xanthurenic acid and 5-hydroxy-tryptamine were not significantly altered from those of the controls, suggesting that kynurenine pathway in survivors of non-severe COVID-19 has been returned to normal. As an anti-inflammatory metabolite secreted by macrophages, itaconic acid is significantly up-regulated in the early stages of SARS-CoV-2 infection (Song et al., 2020). However, itaconic acid in survivors of non-severe COVID-19 was also not significantly different from that in the control cases. In short, recovered kynurenine pathway metabolites and itaconic acid indicated macrophages activated by SARS-CoV-2 infection might have returned to normal.
3. Discussion
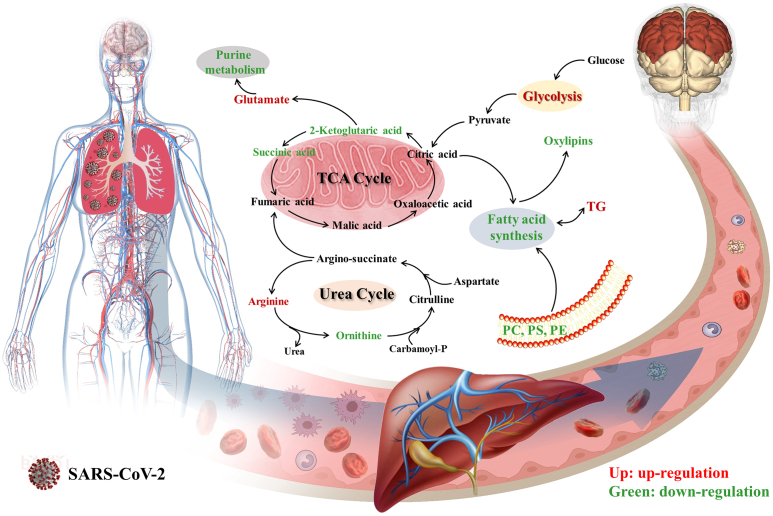
SARS-CoV-2 infection triggers immune response and inflammatory storm accompanied by metabolic reprogramming. Up to now, most studies on metabolic changes in COVID-19 patients focused on acute infection and recovery periods for exploring the pathogenesis, prognosis, and potential biomarkers of infection (Su et al., 2020; Shen et al., 2020; Wu et al., 2020; Hasan et al., 2021). Reprogramming of amino acids, glucose, organic acids, lipids and fatty acids is common in hospitalized COVID-19 patients. These reprogrammed metabolisms not only offer energy and essential substrates for SARS-CoV-2 replication, but also regulate immune and inflammation response. Notably, the long-lasting effects of infection on metabolic health in post-convalescence may be very considerable, not just during acute infection and recovery periods (Ayres, 2020). Although the metabolic activities were increased in COVID-19 patients after clinical treatment relative to acute infection period, the partial metabolic changes continued until discharge, maybe even months after discharge (Wu et al., 2020; Zheng et al., 2021; Shi et al., 2021; Li et al., 2022). This study aim to illustrate the systemic metabolic abnormalities of survivors of non-severe COVID-19 at six months after discharge by serum metabolomics analyses. We found that amino acids, organic acids, purine, fatty acids and lipid metabolism of the survivors still are abnormal compared to controls (Figure 4), but the kynurenine pathway metabolites and itaconic acid had returned to normal levels. It was indicated that the effects of SARS-CoV-2 infection on health are profound. As reported, plasma glycerophospholipid, purine, and phenylalanine metabolism, and TCA cycle in the moderate/severe/critical patients without previous underlying diseases at three months after discharge were different from those of controls (Zhang et al., 2021). Moreover, metabolic profiles of survivors with organ dysfunctions, such as abnormal pulmonary function, decreased glomerular filtration rate or cerebral white matter alterations also have been changed (Xu et al., 2021; Zhou et al., 2021; Yang et al., 2021).
Figure 4.
Abnormalities of body metabolism in survivors of non-severe COVID-19.
Metabolic reprogramming is not only a prominent feature of SARS-CoV-2 infection, but also an important contributor to the progression and prognosis. Sequelae are very common among COVID-19 survivors, fatigue or muscle weakness and PTSD were dominant in our study cohort. The metabolic features of hepatic injury, mental disorders, fatigue or muscle weakness, and inflammation in survivors have been found. Abnormal energy metabolism for ATP production is a typical characteristic of fatigue or muscle weakness, and also include amino acids, nitrogen, oxidative stress and hormone metabolic imbalance (Armstrong et al., 2014). Coronaviruses are neurotropic, neurovirulent, and neuroinvasive, and the point prevalence of PTSD was 32.2% in the post-illness stage of SARS and MERS (Rogers et al., 2020). Glutamate is involved in the pathophysiology of PTSD via its actions on the initiation and maintenance of the hypothalamic-pituitary-adrenal response and learning and memory formation (Averill et al., 2017). Energy metabolism and inflammatory pathways associated with stress responses, such as urea cycle, TCA cycle, purine metabolism and glycolysis, also occurred in PTSD mice (Miller et al., 2020). Furthermore, lipid and fatty acid metabolism are closely associated with PTSD-related psychopathology (Karabatsiakis et al., 2015). In conclusion, the systemic metabolic changes induced by infection are still present and might further affect the long-term health outcomes of COVID-19 survivors. The long-term physiological and mental health care to promote metabolic health are needed for COVID-19 survivors. To this end, targeting these metabolic pathways may be an important approach to prevent and treat complications and further improve health-related quality of life.
4. Limitations of the study
The chemical signatures of COVID-19 survivors were identified using metabolomics in the present study, but there were a number of limitations. First, we cannot rule out the possibility that the survivors had taken medication, especially for elders with previous underlying diseases. Second, we recruited as many recovered patients as possible, but the sample size was small. Third, serum samples reflect the systemic metabolic abnormalities of survivors, being the sum of the metabolic contributes of many organs and tissues. Fourth, these findings are from patients infected by the original strain in China, and different virus variant, vaccination, racial, ethnic, and geographical cohorts were not considered in our study. Finally, confirmatory studies is needed for extending the current understanding of metabolic health in COVID-19 survivors.
Declarations
Author contribution statement
Fang Li and Lei Fu: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Xiaoxiong Liu, Yueguang Lv, Zhiyu Chen and Wenbo Li: Performed the experiments.
Xin-an Liu: Conceived and designed the experiments; Performed the experiments.
Yong Liang, Qian Luo and Fan Pan: Conceived and designed the experiments.
Zhiyi Yang and Ang Guo: Analyzed and interpreted the data.
Funding statement
Fang Li was supported by National Natural Science Foundation of China [22076197]. Prof. Qian Luo was supported by National Natural Science Foundation of China [82127801], Scientific Instrument Developing Project of the Chinese Academy of Sciences [YJKYYQ20200034], Shenzhen Science and Technology Innovation Commission [JCYJ20200109115405930, JCYJ20210324115811031].
Data availability statement
Data will be made available on request.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We thank Drs. Bin Qiao, Wanli Jiang and Zhi Zeng at Renmin Hospital of Wuhan University, China, for their contributions and assistance in the study.
Appendix.
STAR methods
Resource availability
Lead contact
Further information and requests for reagents may be directed to and will be fulfilled by the Lead contact, Qian Luo (qian.luo@siat.ac.cn).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All data and codes to understand and assess the conclusion of this research are available from the corresponding author after publication upon reasonable request. The proposal with detailed aims/plan and other materials may be required to guarantee the rationality of requirement and the security of the data. The patient data will be shared after review and approval of the submitted proposal and any related requested materials.
Experimental model and subject details
Study participants and serum samples
Between 15 August and 30 September 2020, total of 21 survivors (5 under-44-year-old individuals as young survivors and 16 over-45-year-old individuals as elder survivors) with non-severe COVID-19 symptoms and 22 controls (5 young and 17 elder individuals) without SARS-CoV-2 infection history were recruited at Renmin Hospital of Wuhan University (Wuhan, China). Enrolled survivors met the following criteria: non-severe COVID-19 adult patients, and the severity was judged according to the guidelines for the diagnosis and management of COVID-19 patients (7th edition) by National Health Commission of China; met the uniform discharge criteria according to the Chinese clinical guidance for SARS-CoV-2 pneumonia diagnosis and treatment issued by the National Health Commission before discharge hospital; at least six months and not re-infected after discharge. We excluded patients who were pregnant. All subjects underwent at SARS-CoV-2 nucleic acid test before serum sampling, and were negative. Serum samples were collected and stored at −80 °C until to metabolomics analysis. Demographic and clinical data were extracted from electronic medical record. The history of SARS-CoV-2 infection was firstly obtained by inquiry diagnosis and medical records, and infection history and symptom were further confirmed by later telephone follow-up. The study protocol was approved by the Research Ethics Commission of Renmin Hospital of Wuhan University (approval No. WDRY2020-K044) and Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences (approval No. SIAT-IRB-210415-H0561).
Metabolomics analysis
Targeted metabolomics analysis
Forty polar metabolites, including amino acids and amino acid derivatives, organic acids and neurotransmitters, were quantitatively analyzed by using an ultra-high performance liquid chromatography coupled with triple quadrupole mass spectrometry. A 50 μL aliquot of serum was mixed with 200 μL of methanol, and the mixture was vigorously vortex-mixed for 10 min to extract metabolites, precipitate proteins and inactivate any potential viruses, followed by centrifugation at 12,000 ×g for 10 min at 4 °C. The supernatant was separated and transferred to vials, then 1.0 μL was injected into LC-MS/MS for analysis.
A Shimadzu UPLC system was interfaced with A Shimadzu 8060 QQQ-MS/MS equipped with an electrospray ionization (ESI) source. Amino acids and organic acids were separated on an ACQUITY UPLC® HSS C18 Column (Waters, 2.1 mm × 100 mm, 1.8 μm) with the column temperature at 40 °C. For amino acids, the mobile phase included 0.1% formic acid in water (A) and 0.1% formic acid in methanol (B) at a flow rate of 0.2 mL min−1, and the gradient procedure was listed as follows: 0 min, 5% B, 2.5 min, 60% B, 3.5 min, 60% B, 3.51 min, 5% B, then followed by 2 min for equilibration. For organic acids, the mobile phase included 10 mM ammonium acetate in water (A) and 10 mM ammonium acetate in methanol (B), and was eluted at 0.2 mL min-1 according to the gradient procedure as follows: 0 min, 5% B, 3 min, 100% B, 5 min, 100% B, 5.01 min, 5% B, then followed by 2.0 min for equilibration. Neurotransmitters were separated on an ACQUITY UPLC® HSS PFP Column (Waters, 2.1 mm × 100 mm, 1.8 μm) with the column temperature at 40 °C. The mobile phase included 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B). The mobile phase was eluted at 0.2 mL/min according to the gradient procedure as follow: 0 min, 5% B, 3 min, 20% B, 5 min, 100% B, 6.5 min, 100% B, 6.51 min, 5% B, and additional 2 min post-run at initial conditions.
The main working parameters were set as follows: the capillary voltage was 4.0 kV at positive mode and 3.0 kV at negative mode. Nebulization gas, curtain gas and heating gas were all nitrogen, and the flow rate respectively was 3.0 L min-1, 10 L min-1 and 10 L min-1. Collision induced dissociation gas was argon with pressure of 17 kPa. The interface temperature, DL temperature and heating block temperature was 300 °C, 250 °C and 400 °C, respectively. The MRM transitions, retention times and other conditions of amino acids, organic acids and neurotransmitters were shown in Table S5. Instrument control, data acquisition, and analysis were performed using Shimadzu LabSolutions software. The concentrations of these metabolites were calculated by external standard method.
Non-targeted metabolomics analysis
An aliquot of 50 μL deactivated serum samples were added to 300 μL methanol (containing 10 μg/mL PE 17:0/17:0, PC 15:0/15:0, SM d18:1/17:0, PS 17:0/17:0, Cer d18:1/17:0, d5-TG 17:0/17:1/17:0, LPC 15:0, PA 17:0/17:0 and PG 17:0/17:0) and homogenized 10 min to precipitate proteins and inactivate any potential viruses, then 1.0 mL methyl tert-butyl ether was added and vortex-mixed, followed by the addition of 200 μL H2O. The mixture was orderly vortex-mixed and centrifuged (at 10,000 × g at 4 °C for 15 min). Finally, the non-polar (upper) and polar (lower) phase was separately collected and dried under a gentle stream of high-purity nitrogen in a biosafety hood. The dried non-polar and polar extracts were re-dissolved and analyzed by a UPLC system coupled with Q-Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific). The mass spectrometer was operated in the heated electrospray ionization source with a mass range of m/z 50–1000. The conditions were as follows: spray voltage, 3.5 kV (positive ion mode, PIM) and 3.0 kV (negative ion mode, NIM); stopped collision energy, 25 eV, 35 eV and 45 eV for PIM and 20 eV, 25 eV and 30 eV for NIM; capillary temperature, 320 °C; heater temperature, 350 °C. The full scan and data-dependent LC-MS/MS spectra were collected with the resolution of 70,000 and 17,500, respectively. The separation of non-polar metabolites was performed on a UPLC BEH C8 column (Waters, 2.1 × 100 mm, 1.8 μm) and the column oven temperature was 40 °C. An injection volume of 5.0 μL was used for analysis. For non-polar metabolites, the mobile phase A and B were 10 mM ammonium formate in acetonitrile/water (v/v, 6:4) and 10 mM ammonium formate in acetonitrile/isopropanol (v/v, 1:9) at a flow rate of 0.4 mL min−1, respectively. The gradient condition was as follows: 0 min, 58% A; 1.5 min, 58% A; 15.5 min, 15% A; 15.6 min, 14% A, hold for 2.4 min; 18.1 min, returned to the starting gradient and kept for 22.0 min. For polar metabolites, the mobile phases A and B were 0.1% formic acid in acetonitrile and 0.1% formic acid in water at a flow rate of 0.4 mL min−1, respectively. The gradient condition was as follows: 0 min, 100% B; 4.0 min, 100%B, 13.0 min, 100% A, hold for 4 min; 17.1 min, returned to 100% B and kept for 5.0 min.
Raw data files were processed and analyzed using Progenesis QI v2.3 software (Nonlinear Dynamics) for both qualitative and quantitative analysis. A three-dimensional matrix including retention time (RT), mass to charge ratio and peak abundance was obtained after retention time alignment, data normalization and peak picking. The interested m/z were chosen with the relative standard deviation of the peak volume ≤30% in quality control samples and the detectable rate ≥80% in samples, and identified by comparing ions to the Lipid Maps Database, Human Metabolome Database, and MetaScope theoretical fragment search. Final identification results were obtained by comparing results of Progenesis QI and Compound Discoverer software (Thermo Fisher Scientific). A total of 76 polar metabolites (excluding metabolites in targeted metabolomics analysis) were identified by both progenesis QI and Compound Discoverer, and 455 lipids (including 18 FFAs, 12 MHFA, 9 DHFA, 2 di-hydroxyl fatty acids, 10 cholesteryl esters, 16 ceramides, 4 glycosylceramides, 20 sphingomyelins, 8 MG, 31 DG, 96 TG, 3 LysoPA, 25 LysoPC, 13 LysoPE, 1 lysophosphatidylinositol, 2 lysophosphatidylserines, 3 lysophosphatidylglycerols, 20 PA, 81 PC, 37 PE, 9 phosphatidylglycerols, 5 phosphatidylinositols, 30 PS, in Table S2) were identified by combining the results from soft basing on the precursor ion and fragment ions tolerance with a <5 ppm and rule of RT. Statistical analysis and visualization of pathway were performed in the MetaboAnalyst 5.0 web portal.
Data analysis
Statistical analysis was performed by IBM SPSS 22.0 and MetaboAnalyst 5.0, and the Student's t-test was used to investigate the differences between controls and survivors from different groups. The graphs in this study were drawn by GraphPad Prism 8 and Origin 8.0. In the MetaboAnalyst 5.0 web portal, concentration/peak abundance of metabolites were uploaded for statistical analysis, and then the remaining missing values were replaced by a small value (1/5 of the minimum positive value of each variable), followed by no data filtering, data normalization by sum and auto scaling. The screening criteria for significantly differential metabolites were as follows: |log2(fold change)| ≥ 0.25, p < 0.05, VIP > 1.0 and AUC >0.75. To compare changes in different lipid classes, the total peak abundance of individual lipid class was obtained by adding normalized abundance (normalization to sum) of all detected species in the same class.
Key Resources Table
| REAGENT OR RESOURCES | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals | ||
| Acetonitrile | J&K | Cat#932537 |
| Methanol | J&K | Cat#952707 |
| Formic acid | J&K | Cat#942988 |
| Ammonium acetate | J&K | Cat#992378 |
| Ammonium formate | Aladdin | Cat#A100187 |
| Isopropyl alcohol | J&K | Cat#201015 |
| Methyl tert-butyl ether | J&K | Cat#471256 |
| PE17:0/17:0 | Avanti Polar Lipids | Cat#830756P |
| PC 15:0/15:0 | Avanti Polar Lipids | Cat#850350P |
| SM d18:1/17:0 | Avanti Polar Lipids | Cat#860585P |
| PS 17:0/17:0 | Avanti Polar Lipids | Cat#840028P |
| Cer d18:1/17:0 | Avanti Polar Lipids | Cat#860517P |
| d5-TG 17:0/17:1/17:0 | Avanti Polar Lipids | Cat#860903P |
| LPC 15:0 | Avanti Polar Lipids | Cat#855576P |
| PA 17:0/17:0 | Avanti Polar Lipids | Cat#830856P |
| PG 17:0/17:0 | Avanti Polar Lipids | Cat#830456P |
| Serotonin/5-HT (≥98%) | Sigma-Aldrich | Cat#14927 |
| Choline (≥99%) | Sigma-Aldrich | Cat#C7017 |
| Creatine (≥98%) | Sigma-Aldrich | Cat#C3630 |
| Creatinine (≥98%) | Sigma-Aldrich | Cat#C4255 |
| L-glutamate (≥99%) | Macklin | Cat#L810368 |
| L-glutamine (≥99%) | Macklin | Cat#L810391 |
| Kynurenic acid (≥98%) | Sigma-Aldrich | Cat#K3375 |
| L-kynurenine (≥98%) | Macklin | Cat#L864410 |
| L-phenylalanine (≥99%) | Macklin | Cat#L816180 |
| Taurine (≥99%) | Macklin | Cat#T6017 |
| L-tryptophan (≥98%) | Sigma-Aldrich | Cat#T8941 |
| L-tyrosine (≥99%) | Macklin | Cat#L818844 |
| Xanthurenic acid (≥96%) | Aladdin | Cat#X113958 |
| L-alanine (≥99%) | Macklin | Cat#L800640 |
| L-arginine (≥98%) | Sigma-Aldrich | Cat#A5131 |
| L-asparagine (≥99%) | Macklin | Cat#L800456 |
| L-aspartate (≥99%) | Macklin | Cat#L800690 |
| L-carnitine (≥98%) | TCI | Cat#C0049 |
| L-citrulline (≥98%) | Sigma-Aldrich | Cat#C7629 |
| L-cysteine (≥99%) | Macklin | Cat#L804954 |
| L-cystine (≥98%) | TCI | Cat#C0518 |
| L-glycine (≥99%) | Macklin | Cat#G800880 |
| L-histidine (≥99%) | Macklin | Cat#L811073 |
| L-isoleucine (≥99%) | Macklin | Cat#L811667 |
| L-leucine (≥99%) | Macklin | Cat#L812333 |
| L-lysine (≥99%) | Macklin | Cat#L812314 |
| L-methionine (≥98%) | Macklin | Cat#L812760 |
| Niacinamide (≥99.5%) | Sigma-Aldrich | Cat#72340 |
| L-ornithine (≥99%) | Sigma-Aldrich | Cat#57197 |
| L-proline (≥99%) | Macklin | Cat#L816039 |
| L-serine (≥99%) | Macklin | Cat#L817495 |
| L-threonine (≥99%) | Macklin | Cat#L819290 |
| L-valine (≥99%) | Macklin | Cat#L820396 |
| 2-ketoglutaric acid (≥99%) | Sigma-Aldrich | Cat#75890 |
| Trans-aconitic acid (≥98%) | Sigma-Aldrich | Cat#A0127 |
| Citric acid (≥99.5%) | Sigma-Aldrich | Cat#46933 |
| Fumaric acid (≥99%) | Sigma-Aldrich | Cat#47910 |
| Itaconic acid (≥99%) | Sigma-Aldrich | Cat#I29204 |
| Lactic acid (≥98%) | Sigma-Aldrich | Cat#L6402 |
| Malic acid (≥99%) | Sigma-Aldrich | Cat#02288 |
| Succinic acid (≥99%) | Sigma-Aldrich | Cat#S3674 |
| Software and Algorithms | ||
| Progenesis QI | Waters | https://www.nonlinear.com/ |
| Compound Discoverer | Thermo Fisher | https://www.thermofisher.cn/ |
| LabSolutions | Shimadzu | https://www.ssi.shimadzu.com/ |
| GraphPad Prism | GraphPad Software | https://www.graphpad.com/scientificsoftware/ |
| MetaboAnalyst 4.0 | Xia Lab @ McGill | https://www.metaboanalyst.ca/ |
Appendix ASupplementary data
The following is the supplementary data related to this article:
References
- Abu-Farha M., Thanaraj T.A., Qaddoumi M.G., Hashem A., Abubaker J., Al-Mulla F. The role of lipid metabolism in COVID-19 virus infection and as a drug target. Int. J. Mol. Sci. 2020;21(10):3544. doi: 10.3390/ijms21103544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal S., Acharjee A., Mukherjee A., Baker M.S., Srivastava S. Role of multiomics data to understand host-pathogen interactions in COVID-19 pathogenesis. J. Proteome Res. 2021;20(2):1107–1132. doi: 10.1021/acs.jproteome.0c00771. [DOI] [PubMed] [Google Scholar]
- Armstrong C.W., McGregor N.R., Butt H.L., Gooley P.R. Metabolism in chronic fatigue syndrome. Adv. Clin. Chem. 2014;66:121–172. doi: 10.1016/b978-0-12-801401-1.00005-0. [DOI] [PubMed] [Google Scholar]
- Averill L.A., Purohit P., Averill C.L., Bosel M.A., Krystal J.H., Abdallah C.G. Glutamate dysregulation and glutamatergic therapeutics for PTSD: evidence from human studies. Neurosci. Lett. 2017;649:147–155. doi: 10.1016/j.neulet.2016.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres J.S. A metabolic handbook for the COVID-19 pandemic. Nat. Metab. 2020;2(7):572–585. doi: 10.1038/s42255-020-0237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigot A., Tchan M.C., Thoreau B., Blasco H., Maillot F. Liver involvement in urea cycle disorders: a review of the literature. J. Inherit. Metab. Dis. 2017;40(6):757–769. doi: 10.1007/s10545-017-0088-5. [DOI] [PubMed] [Google Scholar]
- Caterino M., Costanzo M., Fedele R., Cevenini A., Gelzo M., Di Minno A., Andolfo I., Capasso M., Russo R., Annunziata A., Calabrese C., Fiorentino G., D’Abbraccio M., Dell’Isola C., Maria Fusco F., Parrella R., Fabbrocini G., Gentile I., Castaldo G., Ruoppolo M. The serum metabolome of moderate and severe COVID-19 patients reflects possible liver alterations involving carbon and nitrogen metabolism. Int. J. Mol. Sci. 2021;22(8):9548. doi: 10.3390/ijms22179548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., De Giglio M.A.R., Roviello G.N. CAnti-Coronavirus vaccines: past investigations on SARS-CoV-1 and MERS-CoV, the approved vaccines from BioNTech/Pfizer, Moderna, Oxford/AstraZeneca and others under development against SARSCoV-2 infection. Curr. Med. Chem. 2022;29(1):4–18. doi: 10.2174/0929867328666210521164809. [DOI] [PubMed] [Google Scholar]
- Costanzo M., Caterino M., Fedele R., Armando C., Pontillo M., Barra L., Ruoppolo M. COVIDomics: the proteomic and metabolomic signatures of COVID-19. Int. J. Mol. Sci. 2022;23(5):2414. doi: 10.3390/ijms23052414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis E.A., Norris P.C. Eicosanoid storm in infection and inflammation. Nat. Rev. Immunol. 2015;15(8):511–523. doi: 10.1038/nri3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews L., Zimmermann M., Westhoff P., Brilhaus D., Poss R.E., Bergmann L., Wiek C., Brenneisen P., Piekorz R.P., Mettler-Altmann T., Weber A.P.M., Reichert A.S. Ammonia inhibits energy metabolism in astrocytes in a rapid and glutamate dehydrogenase 2-dependent manner. Dis. Model Mech. 2020;13(10) doi: 10.1242/dmm.047134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan M.R., Suleiman M., Pérez-López A. Metabolomics in the diagnosis and prognosis of COVID-19. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.721556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildreth K., Kodani S.D., Hammock B.D., Zhao L. Cytochrome P450-derived linoleic acid metabolites EpOMEs and DiHOMEs: a review of recent studies. J. Nutr. Biochem. 2020;86 doi: 10.1016/j.jnutbio.2020.108484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.L., Wang Y.M., Li X.W., Ren L.L., Zhao J.P., Hu Y., Zhang L., Fan G.H., Xu J.Y., Gu X.Y., Cheng Z.S., Yu T., Xia J.A., Wei Y., Wu W.J., Xie X.L., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J.G., Wang G.F., Jiang R.M., Gao Z.C., Jin Q., Wang J.W., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.L., Huang L.X., Wang Y.M., Li X., Ren L.L., Gu X.Y., Kang L., Guo L., Liu M., Zhou X., Luo J.F., Huang Z.H., Tu S.J., Zhao Y., Chen Li., Xu D.C., Li Y.P., Li C.H., Peng L., Li Y., Xie W.X., Cui D., Shang L.H., Fan G.H., Xu J.Y., Wang Y., Zhong J.C., Wang C., Wang J.W., Zhang D.Y., Cao B. 6-month consequences of COVID-19 in patients discharged from hospital: a cohort study. Lancet. 2021;397:220–232. doi: 10.1016/S0140-6736(20)32656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L.X., Yao Q., Gu X.Y., Wang Q.Y., Ren L.L., Wang Y.M., Hu P., Guo L., Liu M., Xu J.Y., Zhang X.Y., Qu Y.L., Fan Y.Q., Li X., Li C.H., Yu T., Xia J.A., Wei M., Chen L., Li Y.P., Xiao F., Liu D., Wang J.W., Wang X.G., Cao B. 1-year outcomes in hospital survivors with COVID-19: a longitudinal cohort study. Lancet. 2021;398:747–758. doi: 10.1016/S0140-6736(21)01755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabatsiakis A., Hamuni G., Wilker S., Kolasssa S., Renu D., Kaderait S., Schauer M., Hennessy T., Kolassa I.-T. Metabolite profiling in posttraumatic stress disorder. J Mol. Psychiatr. 2015;3(1):2. doi: 10.1186/s40303-015-0007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss R.M., Siri P.W. Metabolic abnormalities: triglyceride and low-density lipoprotein. Endocrinol. Metabol. Clin. 2004;33(2):405–415. doi: 10.1016/j.ecl.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Kutzing M.K., Firestein B.L. Altered uric acid levels and disease states. J. Pharmacol. Exp. Therapeut. 2008;324(1):1–7. doi: 10.1124/jpet.107.129031. [DOI] [PubMed] [Google Scholar]
- Li H.W., Li X., Wu Q., Wang X., Qin Z.H., Wang Y.G., He Y.B., Wu Q., Li L., Chen H.Y. Plasma proteomic and metabolomic characterization of COVID-19 survivors 6 months after discharge. Cell Death Dis. 2022;13(3):235. doi: 10.1038/s41419-022-04674-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmud I., Garrett T.J. Mass spectrometry techniques in emerging pathogens studies: COVID-19 Perspectives. J. Am. Soc. Mass Spectrom. 2020;31:2013–2024. doi: 10.1021/jasms.0c00238. [DOI] [PubMed] [Google Scholar]
- Miller J.S., Rodriguez-Saona L., Hackshaw K.V. Metabolomics in central sensitivity syndromes. Metabolites. 2020;10(4):164. doi: 10.3390/metabo10040164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minhas P.S., Liu L., Moon P.K., Joshi A.U., Dove C., Mhatre S., Contrpois K., Wang Q., Lee B.A., Coronado M., Bernstein D., Snyder M.P., Migaud M., Mochly-Rosen D., Rabinowitz J.D., Andreasson K.I. Macrophage de novo NAD(+) synthesis specifies immune function in aging and inflammation. Nat. Immunol. 2019;20:50–63. doi: 10.1038/s41590-018-0255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir M., Trujillo D., Levine J., Dwyer J.B., Rupp Z.W., Bloch M.H. Glutamate systems in DSM-5 anxiety disorders: their role and a review of glutamate and GABA psychopharmacology. Front. Psychiatr. 2020;11 doi: 10.3389/fpsyt.2020.548505. ARTN 548505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkumar R.P. COVID-19 and mental health: a review of the existing literature. Asian J Psychiatr. 2020;52 doi: 10.1016/j.ajp.2020.102066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J.P., Chesney E., Oliver D., Pollak T.A., McGuire P., Fusar-Poli P., Zandi M.S., Lewis G., David A.S. Psychiatric and neuropsychiatric presentations associated with severe coronavirus infections: a systematic review and meta-analysis with comparison to the COVID-19 pandemic. Lancet Psychiatr. 2020;7(7):611–627. doi: 10.1016/S2215-0366(20)30203-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubino F., Amiel S.A., Zimmet P., Alberti G., Bornstein S., Eckel R.H., Mingrone G., Boehm B., Cooper M.E., Chai Z.L., Prato S.D., Ji L.N., Hopkins D., Herman W.H., Khunti K., Mbanya J.-C., Renard E. New-onset diabetes in covid-19. N. Engl. J. Med. 2020;383(8):PMC7304415. doi: 10.1056/NEJMc2018688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan C.N., Yacoubian S., Yang R. Anti-inflammatory and proresolving lipid mediators. Annu. Rev. Pathol. 2008;3:279–312. doi: 10.1146/annurev.pathmechdis.3.121806.151409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen T., Wang T.T. Metabolic reprogramming in COVID-19. Int. J. Mol. Sci. 2021;22(21) doi: 10.3390/ijms222111475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Yi X., Sun Y.T., Bi X.J., Du J.P., Zhang C., Quan S., Zhang F.F., Sun R., Qian L.J., Ge W.G., Liu W., Liang S., Chen H., Zhang Y., Li J., Xu J.Q., He Z.,B., Chen B.F., Wang J., Yan H.X., Zheng Y.F., Wang D.L., Zhu J.S., Komh Z.Q., Kang Z.Y., Liang X., Ding X., Ruan G., Xiang N., Cai X., Gao H.H., Li L., Li S.N., Xiao Q., Lu T., Zhu Y., Liu H.F., Chen H.X., Guo T.N. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182(1):59–72. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D., Yan R., Lv L.X., Jiang H.Y., Lu Y.F., Sheng J.F., Xie J.J., Wu W.R., Xia J.F., Xu K.J., Gu S.L., Chen Y.F., Huang C.J., Guo J., Du Y.L., Li L.J. The serum metabolome of COVID-19 patients is distinctive and predictive. Metabolism. 2021;118 doi: 10.1016/j.metabol.2021.154739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.W., Lam S.M., Fan X., Cao W.J., Wang S.Y., Tian H., Chua G.H., Zhang C., Meng F.P., Xu Z., Fu J.L., Huang L., Xia P., Yang T., Zhang S.H., Li B.W., Jiang T.J., Wang R.X., Wang Z.H., Shi M., Zhang J.Y., Wang F.S., Shui G.H. Omics-driven systems interrogation of metabolic dysregulation in COVID-19 pathogenesis. Cell Metabol. 2020;32(2):188–202. doi: 10.1016/j.cmet.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y.P., Chen D., Yuan D., Lausted C., Choi J.C., Dai C.Z.L., Voillet V., Duvvuri V.R., Scherler K., Troisch P., Baloni P., Qin G.R., Smith B., Kornilov S.A., Rostomily C., Xu A., Li J., Dong S., Rothchild A., Zhou J., Murray K., Edmark R., Hong S.G., Heath J.E., Earls J., Zhang R.Y., Xie J.Y., Li S., Roper R., Jones L., Zhou Y., Rowen L., Liu R., Mackay S., O’Mahony D.S., Dale C.R., Wallick J.A., Algren H.A., Zager M.A., ISB-Swedish COVID19 Biobanking Unit. Wei W., Price N.D., Huang S., Subramanian N., Wang K., Magis A.T., Hadlock J.J., Hood L., Aderem A., Bluestone J.A., Lanier L.L., Greenberg P.D., Gottardo R., Davis M.M., Goldman J.D., Heath J.R. Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell. 2020;183(6):1479–1495. doi: 10.1016/j.cell.2020.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram S., Soni M., Annigeri R. Urine abnormalities predict acute kidney injury in COVID-19 patients: an analysis of 110 cases in Chennai, South India. Diabetes Metabol. Syndr. 2021;15(1):187–191. doi: 10.1016/j.dsx.2020.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taquet M., Geddes J.R., Husain M., Luciano S., Harrison 6-month neurological and psychiatric outcomes in 236379 survivors of COVID-19: a retrospective cohort study using electronic health records. Lancet Psychiatr. 2021;8(5):416–427. doi: 10.1016/S2215-0366(21)00084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona S., Carmona H., Conesa A., Llansola M., Felipo V. A multi-omic study for uncovering molecular mechanisms associated with hyperammonemia-induced cerebellar function impairment in rats. Cell Biol. Toxicol. 2021;37:129–149. doi: 10.1007/s10565-020-09572-y. [DOI] [PubMed] [Google Scholar]
- Thomas T., Stefanoni D., Reisz J.A., Nemkov T., Bertolone L., Francis R.O., Hudson K.E., Zimring J.C., Hansen K.C., Hod E.A., Spitalnik S.L., D’Alessandro A. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI. Insight. 2020;5 doi: 10.1172/jci.insight.140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Zhou L.N., Sun X., Yan Z.F., Hu C.X., Wu J.P., Xu L., Li X., Liu H.L., Yin P.Y., Li K., zhao J.Y., Li Y.L., Wang X.L., Li Y., Zhang Q.Y., Xu G.W., Chen H.Y. Altered lipid metabolism in recovered SARS patients twelve years after infection. Sci. Rep. 2017;7(1):9110. doi: 10.1038/s41598-017-09536-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Shu T., Yang X.B., Song J.X., Zhang M.L., Yao C.Y., Liu W., Huang M.H., Yu Y., Yang Q.Y., Zhu T.J., Xu J.Q., Mu J.F., Wang Y.X., Wang H., Tang T., Ren Y.J., Wu Y.R., Lin S.H., Qiu Y., Zhang D.Y., Shang Y., Zhou X. Plasma metabolomic and lipidomic alterations associated with COVID-19. Natl. Sci. Rev. 2020;7(7):1157–1168. doi: 10.1093/nsr/nwaa086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao N., Nie M., Pang H.H., Wang B.H., Hu J.L., Meng X.J., Li K., Ran X.R., Long Q.X., Deng H.J., Chen N., Li S., Tang N., Huang A.L., Hu Z.P. Integrated cytokine and metabolite analysis reveals immunometabolic reprogramming in COVID-19 patients with therapeutic implications. Nat. Commun. 2021;22:68. doi: 10.1038/s41467-021-21907-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.J., Zhou M., Luo P., Yin Z.R., Wang S.F., Liao T.T., Yang F., Wang Z., Yang D., Peng Y., Geng W., Li Y.Y., Zhang H., Jin Y. Plasma metabolomic profiling of patients recovered from COVID-19 with pulmonary sequelae 3 months after discharge. Clin. Infect. Dis. 2021;73(12):2228–2238. doi: 10.1093/cid/ciab147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Zhou M., Li L.L., Luo P., Fan W.L., Xu J.J., Chen Q., Pan F., Lei P., Zheng C.S., Jin Y. Characteristics of mental health implications and plasma metabolomics in patients recently recovered from COVID-19. Transl. Psychiatry. 2021;11:307. doi: 10.1038/s41398-021-01426-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeredo J.L., Quah S.K.L., Wallis C.U., Alexander L., Cockcroft G.J., Santangelo A.M., Xia J., Shiba Y., Dalley J.W., Cardinal R.N., Roberts A.C., Clarke H.F. Glutamate within the marmoset anterior hippocampus interacts with area 25 to regulate the behavioral and cardiovascular correlates of high-trait anxiety. J. Neurosci. 2019;39(16):3094–3107. doi: 10.1523/JNEUROSCI.2451-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Shi L., Wang F.S. Liver injury in COVID-19: management and challenges. Lancet Gastroenterol. 2020;5(5):428–430. doi: 10.1016/S2468-1253(20)30057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.J., Luo P., Xu J.J., Yang L., Ma P., Tan X.Y., Chen Q., Zhou M., Song S.W., Xia H., Wang S.F., Ma Y.L., Yang F., Liu Y., Li Y.M., Ma G.Z., Wang Z.H., Duan Y.R., Jin Y. Plasma metabolomic profiles in recovered COVID-19 patients without previous underlying diseases 3 months after discharge. J. Inflamm. Res. 2021;14:4485–4501. doi: 10.2147/JIR.S325853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Jin S.W., Li T., Ying W.Y., Ying B.Y., Chen D., Ning J., Zheng C.F., Li Y.P., Li C., Chen C.S., Li X.K., Gao H.C. Metabolomics reveals sex-specific metabolic shifts and predicts the duration from positive to negative in non-severe COVID-19 patients during recovery process. Comput. Struct. Biotechnol. J. 2021;19:1863–1873. doi: 10.1016/j.csbj.2021.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Tan X.Y., Luo P., Xu J.J., Yin Z.R., Liao T.T., Wang S.F., Wang Z.H., Jin Y. Changes in glomerular filtration rate and metabolomic differences in severely ill coronavirus disease survivors 3 months after discharge. BBA-Mol. Basis Dis. 2021;1868(1) doi: 10.1016/j.bbadis.2021.166289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.
All data and codes to understand and assess the conclusion of this research are available from the corresponding author after publication upon reasonable request. The proposal with detailed aims/plan and other materials may be required to guarantee the rationality of requirement and the security of the data. The patient data will be shared after review and approval of the submitted proposal and any related requested materials.