Abstract
Wastewater-based surveillance (WBS) has been an effective tool for monitoring and understanding potential SARS-CoV-2 transmission across small and large-scale communities. In this study at the University of Saskatchewan, the assessment of SARS-CoV-2 was done over eight months during the 2021–2022 academic year. Wastewater samples were collected using passive samplers that were deployed in domestic sewer lines near adjacent campus residences and extracted for viral RNA, followed by Reverse Transcription quantitative Polymerase Chain Reaction (RT-qPCR). The results showed similar trends for SARS-CoV-2 detection frequencies and viral loads across university residences, the whole campus, and from related WBS at Saskatoon Wastewater Treatment Plant. The maximum daily detection frequency for seven dormitories considered was about 75 %, while maximum daily case numbers for the residences and campus-wide were about 11 and 75 people, respectively. In addition, self-reported rates of infection on campus peaked during similar time frames as increases in viral load were detected at the Saskatoon wastewater treatment plant. These similarities indicate the usefulness and cost-effectiveness of monitoring the spread of COVID-19 in small-scale communities using WBS.
Keywords: SARS-CoV-2, COVID-19, Wastewater-based surveillance (WBS), Passive sampling, University residences
Graphical abstract
1. Introduction
Wastewater-based surveillance (WBS) has been used worldwide for surveillance of municipal wastewaters to determine the spatiotemporal spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) responsible for the COVID-19 pandemic (Daughton, 2020; Xie et al., 2022). The monitoring of municipal wastewaters has been of increasing value to stakeholders as WBS has been shown to be a leading indicator (2 to 7 days; Peccia et al., 2020; D'Aoust et al., 2021) of COVID-19 outbreaks and viral caseloads given its ability to monitor asymptomatic and pre-symptomatic populations shedding virus in feces (Ahmed et al., 2020; Graham et al., 2021; Habtewold et al., 2022). WBS for COVID-19 has been accomplished at several scales (i.e., from individual buildings to large cities) using a variety of sampling methods (i.e., grab to composite sampling) with analysis of SARS-CoV-2 RNA concentrations being done with a diverse suite of instruments (i.e., traditional ‘gold-standard’ to proprietary systems such as the currently assessed LuminUltra platform). The initial focus of COVID-19 WBS globally was on larger, city-size sewersheds typically served by composite autosamplers using costly, limited availability, and time-intensive custom real-time quantitative PCR workflows (Kitajima et al., 2020). However, more recent attention has been on the development of passive sampling strategies with analysis using ‘off the shelf’ low cost and effort instrumentation that can be made more readily available to stakeholders at a smaller scale.
Recently WBS has included monitoring of apartment buildings (Wong et al., 2021), student hostels (Chelvan, 2021), worker dormitories (Mohan, 2020), and university residences (Corchis-Scott et al., 2021; Scott et al., 2021; Vo et al., 2022; Wang et al., 2022), the latter of which are the focus of the current study. Continuous sampling using autosamplers has been considered to be the optimal method for wastewaters given they allow for attenuation of flow fluctuations over time with resultant samples being a more accurate composition of daily wastewater flows (Habtewold et al., 2022). However, Habtewold et al. (2022) also clearly express the limitations of autosamplers including potential issues with sites such as limited security, access to power, location of sampling, etc. A potential option to overcome these obstacles is the use of grab sampling (e.g., Scott et al., 2021 for university campus sampling), however, these are sub-optimal given these are only a ‘snapshot’ of the wastewater at the time of the sampling. To resolve this apparent dilemma, passive sampling methods have been developed and have become of great interest recently. These techniques normally encompass placing sorbent materials in the wastewater stream for a duration of about 24 to 72 h. This passive sampling has been assessed including a variety of materials (e.g., Moore swab electronegative membrane filters) (Hayes et al., 2021) and sampling durations (Habtewold et al., 2022). Interestingly, the Moore swab methodology has been used for two recent university campus-based wastewater surveillance studies (Corchis-Scott et al., 2021; Wang et al., 2022). Clearly, for smaller-scale WBS, passive sampling is a cost-effective and efficient methodology for sample collection.
Initially, the wastewater surveillance of COVID-19 only used costly time and labour-intensive RNA extraction and quantification methodologies. In addition, sample transportation and shipping logistics to centralized laboratories capable of performing the analysis typically led to delays of hours to days for determination of SARS-CoV-2 viral loads, thus negating the ‘early warning’ potential of this type of surveillance (Larsen and Wigginton, 2020; Daigle et al., 2022). Over time, there has been a development of fast and relatively inexpensive instruments such as the GeneXpert (Daigle et al., 2022) and the LuminUltra GeneCount system (Hayes et al., 2021), among others. While these methods are still based on RT-qPCR chemistries, kits and instruments have been optimized to be easily employed in point-of-care settings or by wastewater treatment plant (WWTP) operators, respectively. Many such technologies have shown promise in their ease of use, low costs, and accurate detection of SARS-CoV-2 virus when compared to ‘gold-standard’ methods. Thus, these types of instruments are quickly becoming viable options for WBS during the COVID-19 pandemic with potential future benefits for WBS of other viruses.
Clearly the combination of passive sampling and off-the-shelf, simple instrumentation has become a promising method for WBS of SARS-CoV-2, especially for smaller-scale applications. However, further study is necessary to determine the robustness of passive sampling and the effectiveness of simple instruments for determination of SARS-CoV-2 in wastewater. Thus, the current study presents the assessment of SARS-CoV-2 in wastewater of student residences at the University of Saskatchewan, Saskatoon, Canada, using the LuminUltra GeneCount instrument. This study includes a 5-day a week assessment of the virus for seven residence locations using the passive sampling technique over an 8-month duration. In addition, results of the surveillance are compared with both the residence and campus-wide daily self-reported cases. Further, a comparison of the campus results is made against the City of Saskatoon wastewater treatment plant viral loads starting in August 2021, which have been determined as part of a research collaboration involving co-authors of this current study (Xie et al., 2022; Oloye et al., 2022). This combination of results provides a unique perspective comparing the small-scale and large-scale analysis, while also comparing wastewater surveillance and clinical case data at the small-scale.
2. Methods
2.1. Passive sampler processing and deployment sites
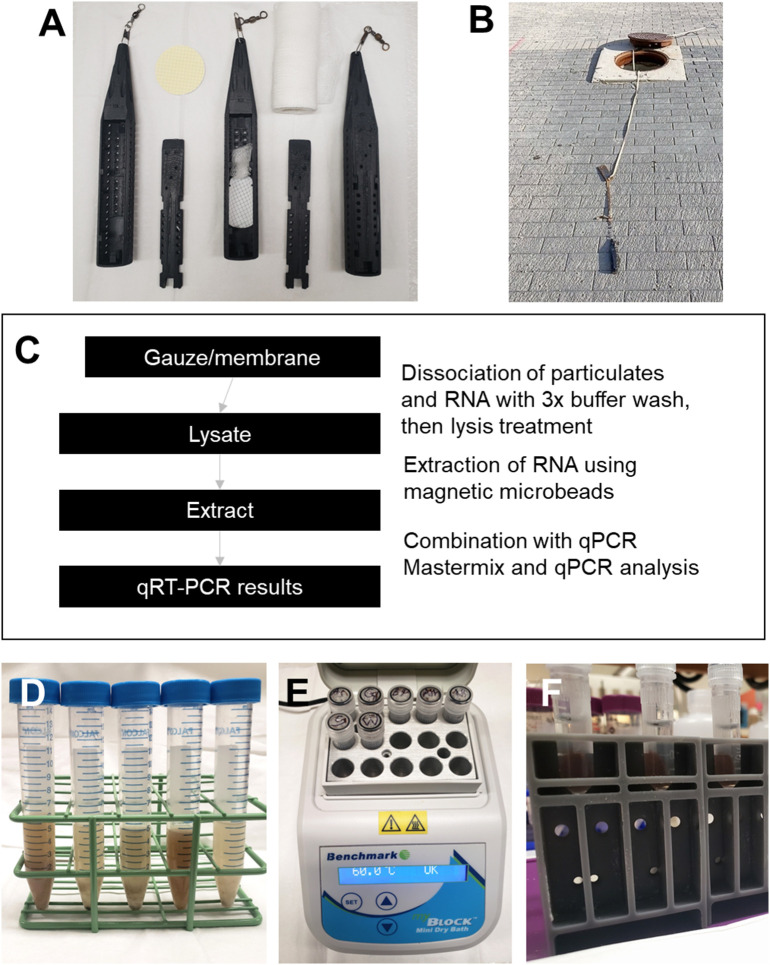
The passive samplers used in this study were based on a novel design meant to resemble a torpedo as developed and tested by Schang et al. (2021) (Fig. 1A). These samplers were designed to allow for easy shedding of debris in the wastewater line while also being weighted enough to allow the sampling of the wastewater stream. Each passive torpedo sampler contained a laboratory-grade electronegative cellulose nitrate membrane filter (Sartorius, Germany) and medical-grade cotton gauze to ensure contact between wastewater and filter. Seven sampling sites were selected representing wastewater effluents from seven University of Saskatchewan residences having capacities of 125 to 399 people. To ensure protection of privacy of students in these residences, we will refer to these residences as locations 1–7. All passive samplers in this study were deployed and collected between 7:00 and 8:00 AM from Monday to Friday (24 hour sampling during the week, 72 h over the weekend) from seven manholes located at wastewater lines adjacent to residence halls between July 5th, 2021 and April 28th, 2022. Samplers were tethered to the manhole cover via a high tensile wire (Fig. 1B).
Fig. 1.
Collection and extraction of SARS-CoV-2 virus via passive sampling. (A) Torpedo sampler pictured empty (left side), with electronegative filter membrane and cotton gauze (middle), and assembled (right side); (B) demonstration of manhole torpedo sampler deployment setup showing sampler connected to manhole cover using high tensile wire; (C) flowchart describing the procedural steps for measuring SARS-CoV-2; (D) raw wastewater samples mixed with elution buffer in 15 mL centrifuge tubes; (E) Benchmark Block™ Mini Dry Bath incubator used for RNA concentration and extraction; and (F) magnetic racks with 2 mL samples tubes containing magnetic beads and eluted RNA sample.
Once collected, the samplers were immediately brought to the University of Saskatchewan Environmental Laboratory for extraction and analysis following the LuminUltra GeneCount™ SARS-CoV-2 Advanced Wastewater kit manufacturer's protocol and wastewater testing materials (LuminUltra Technologies Ltd., Canada) (Fig. 1C). The membrane filters and cotton gauze were extracted from torpedo samplers and added to 15 mL centrifuge tubes filled with an elution buffer 0.075 % (v/v) Tween 20 in 25 mM TRIS HCl. Tubes were manually shaken vigorously for 1 min, contents were transferred to new 15 mL centrifuge tubes filled with elution buffer, and these tubes were shaken vigorously for 1 min to prepare the wastewater sample for molecular extraction and concentration of RNA (Fig. 1D).
2.2. RNA concentration and extraction
RNA extraction was performed using a magnetic bead-based separation procedure based on the manufacturer's protocol, which will be briefly described herein. First, 1 mL of extracted wastewater in elution buffer was transferred to a centrifuge tube containing Lysis Buffer Concentrate and Lysis Supplement 1A. Tubes were thoroughly mixed and incubated for 10 min at room temperature. After incubation, ethanol and magnetic beads were added, samples were manually mixed, incubated for 10 min at room temperature, and then placed in magnetic racks.
The supernatant was discarded, and samples were washed with the wash buffer five times. Each time the samples were washed, the supernatant was discarded. Next, ethanol was added to well-mixed samples, and contents were transferred to 2 mL tubes. The supernatant was discarded with magnetic separation, and an RNA elution buffer was added before incubating at 60 °C for 5 min (Fig. 1E). After incubation, sample tubes were placed in magnetic racks (Fig. 1F) and eluted RNA was collected for analysis.
2.3. Quantitative RT-qPCR method
All samples were analyzed by RT-qPCR using the manufacturer's protocol. Briefly, 5 μL RNA samples were transferred to PCR strip tubes that contained 15 μL of RT-qPCR Master Mix. Eluted RNA samples were diluted with bovine serum albumin (BSA) (2.5 μL of 1 mg/mL BSA with 2.5 μL eluted RNA) to avoid inhibition and enhance PCR amplification yields. A negative control (15 μL Master Mix and 5 μL Nuclease-free water) and positive control (15 μL Master Mix and 5 μL Positive Control DNA) were also prepared as standards. All RNA sample tubes were gently mixed and processed by RT-qPCR on a GeneCount Q-16 device (LuminUltra Technologies Ltd., Canada) for analysis.
2.4. Statistical analysis and ethics
Statistical analysis included linear regression of matching timepoints for the data including detection frequency vs. (Total daily campus cases, people; Total daily residence cases, people; and Saskatoon WWTP viral load, gc/100 mL). Statistically significant Spearman correlation coefficients (p < 0.0001) are presented with regression lines and 95 % confidence intervals (Supporting information Fig. S1).
Note that total campus and residence cases were all self-reported to the university administration with the explicit consent to publish the numbers on a publicly-accessible online website. All data were provided to us in an anonymized spreadsheet. Thus, no ethics review for this study specifically was necessary.
3. Results and discussion
3.1. Dormitory residence COVID-19
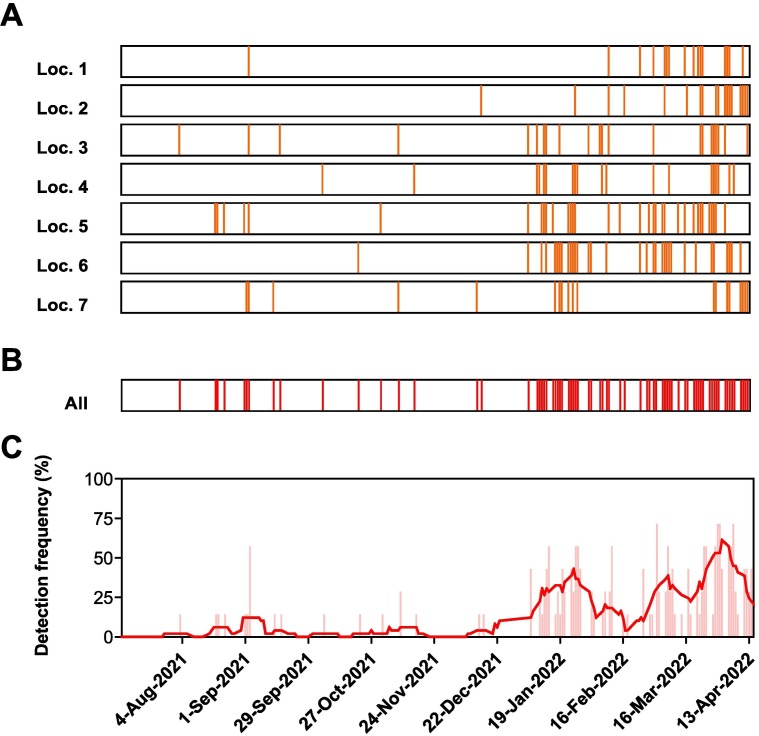
The barcode data indicating virus ‘hits’ for the seven residences is shown in Fig. 2A, while the combination of all ‘hits’ for the residences is presented in Fig. 2B. The overall detection frequency is presented in Fig. 2C including a 7-day moving average of this data which is useful for comparisons presented in Fig. 3 .
Fig. 2.
SARS-CoV-2 detection ‘hits’ for residence wastewaters during the 2021–2022 academic year. (A) Individual SARS-CoV-2 hits from seven different locations across six residences; (B) combined SARS-CoV-2 hits across all six residences; and (C) distribution frequency plot of SARS-CoV-2 detection across the residences including a 7-day moving average of hits (solid red line).
Fig. 3.
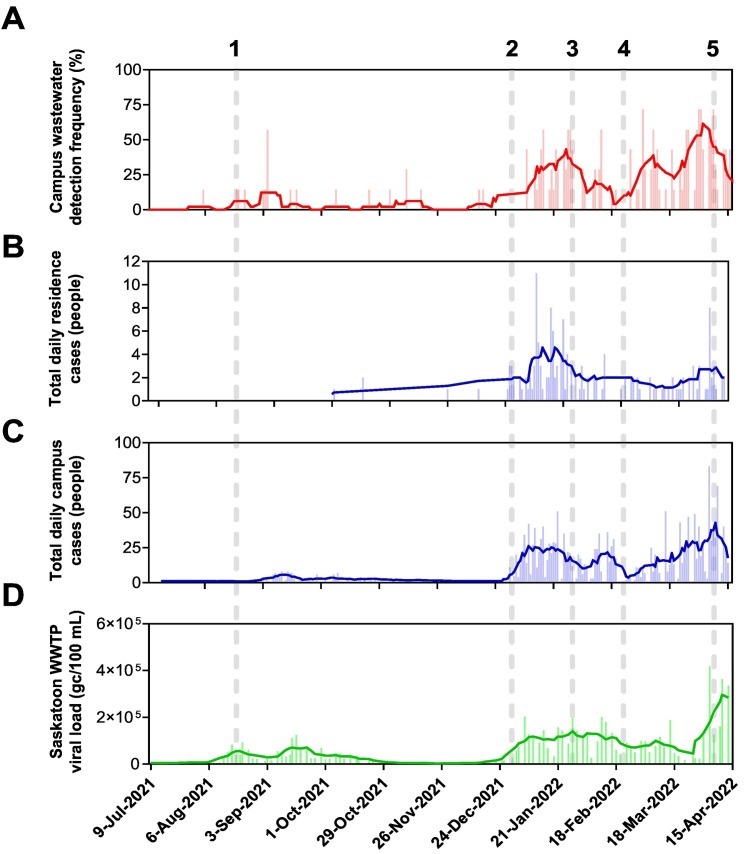
Comparison of daily SARS-CoV-2 metrics between university residences, campus, and Saskatoon Wastewater Treatment Plant data. (A) Distribution frequency plot of SARS-CoV-2 detection across the residences including a 7-day moving average of hits (solid red line); (B) distribution frequency plot of total daily cases (people) reported in residences including 7-day moving average of cases (solid blue line); (C) total reported daily cases for the entire university campus including 7-day moving average of cases (solid blue line); and (D) daily viral load as measured and reported by Saskatoon WWTP including 7-day moving average of load (solid green line). The numbered dashed lines (1–6) represent specific events that will be discussed in the main manuscript.
Interestingly, two of the seven residences had positive results prior to the students moving into the dormitories at the end of August and early September. These hits were found in Locations 3 and 5 and were attributed to contractors working on site in both locations (pers. comm.). As expected, there was a peak of detections in early September with four of the seven residences having positive hits as student numbers increased. However, Locations 2, 4, and 6 did not have positive hits until much later in the year during November, October, and October, respectively (Fig. 2A). The overall hits indicate low numbers of detections until mid-December 2021, which coincides with increases discussed in Fig. 3. The overall hits increased for the remainder of the academic year ending in April 2022. Moving 7-day averages during this peak period fluctuated between about 25 to 50 % detection frequencies for the seven locations (Fig. 2C).
Trends in this study correlate well with studies by Gibas et al. (2021) and Lu et al. (2022), which found that in summer months the low student populations in university residences led to inconsistent positive results. Both studies found that as classes started in September the inundation of students onto campus led to increased SARS-Cov-2 infections on campus and coincidental increases in wastewater virus detections. Betancourt et al. (2021) supported this observation and noted that spikes of infections also occurred due to student behaviour after holiday periods. Small-scale WBS of SARS-CoV-2 can be a valuable tool in monitoring the spread of infection in university settings, or other similar locations. Comparing municipal wastewater testing can illustrate the similarities and differences between small-scale and large-scale monitoring and can help illuminate the actual value of WBS testing.
3.2. Dormitory vs. municipal WBS
The overall residence detection frequency and 7-day moving average data are presented again in Fig. 3A for easier comparisons to other data. Fig. 3B presents the daily self-reported residence case numbers including the 7-day moving averages, while Fig. 3C presents the same information for the campus-wide self-reported daily cases. The final Fig. 3D shows the Saskatoon WWTP viral loads as determined using methods presented in Xie et al. (2022) and available at https://water.usask.ca/covid-19/. The linear regressions for comparison of the dormitory detection frequencies versus daily residence cases, campus cases, and Saskatoon WWTP viral loads are presented in Fig. S1.
Although wastewater surveillance of residences started with preliminary assessment in August 2021 and initial student-filled residences in September 2021, the case numbers specific to dormitory residences were unavailable until October 2021. Prior to the start of the mid-December detection frequency increase (Fig. 3A), the residence case numbers were only one or two cases. Coincident with the increase in detection, the case numbers also increased with a maximum of 11 self-reported new cases in early January 2022 and having 7-day averages around four new cases. Following this surge of cases, the numbers reduced similarly to the detection frequencies until end of March and early April when there was another peak of seven new self-reported cases with 7-day averages of about three new cases.
The campus-wide data (Fig. 3C) was available over the same time period as the residence wastewater data and shows a similar peak in self-reported case numbers at the beginning of September 2021 of about 5–10 new cases. Following this peak, this data showed a similar pattern as the residences with limited cases until mid-December 2021 with an increase at that time to a 7-day average around 25 new cases. Interestingly, this increase did not have a high peak with a maximum number of cases reaching 50 in early December. Unlike the residence cases, the campus-wide 7-day moving average declined until mid-February to a low of about 10 new cases, followed by an increase to their peak of 75 new cases with a coincident 7-day moving average of 50 new cases. Interestingly, the total campus cases were correlated to the detection frequency (R = 0.67, p < 0.0001) while the residence cases were not correlated (R = 0.14, p = 0.0340) (Fig. S1). This may be attributed to the significantly smaller populations within the dormitories as well as variability in self-reporting over time.
The Saskatoon WWTP data (Fig. 3D) collected using a 24-h composite sampler indicates a small peak during August which would not be expected for either the residence or campus data given the limited number of students on campus during this time. However, the remaining trends for the Saskatoon data are similar to the detection (Fig. 3A) and case number data for both the residences (Fig. 3B) and campus (Fig. 3C). The Saskatoon viral loads began increasing in late December reaching a maximum of about 2 × 10−5 gene copies (gc)/100 mL which was maintained until mid-March. The viral load increased at this time to about 2 × 10−5 gc/100 mL, matching the increases in all three metrics found during this time period. Similar to the campus-wide case data, the dormitory detection frequency was correlated with the Saskatoon WWTP data (Fig. S1).
The numbers at the top of Fig. 3 indicate five time points of interest for us in comparisons between the various datasets. Point 1 shown in Fig. 3 represents the move-in date to residence for University of Saskatchewan students and effectively the start of the fall semester. However, this particular semester included remote teaching of classes for the majority of students and disciplines. There was a slight increase in positive hits found in the campus wastewater, in the campus cases, and in the City of Saskatoon wastewater samples. This increase may be due to the increased number of students on campus despite the remote teaching, as well as the increased number of students in elementary and secondary schools in the city. However, all metrics generally stayed relatively low throughout the fall term, likely due to a combination of remote learning, low population density on campus, and the prevalence of the slower spreading Delta variant during this period (Tian et al., 2021). January 2022 was the start of the winter term (Fig. 3, Point 2) where there was a sharp rise in wastewater residence hits/cases, and campus cases that could be attributed to students travelling over the holiday break to visit family and potentially attending large-scale, close-proximity events. In response to this, classes were taught remotely to start the winter term until February 8th (Fig. 3, Point 3), showing wastewater and campus cases trending downward after the initial winter term surge.
The mid-semester break occurred during the week of February 21st, 2022 and coincided with the lowest amount of positive hits at the University of Saskatchewan during the winter term (Fig. 3, Point 4). However, cases trended upwards within two weeks following this break, surpassing previous peaks in the 2021–2022 academic year. This can be explained by the rapid spread of the Omicron variant in late 2021 and throughout 2022 (He et al., 2021; Oloye et al., 2022) in combination with students' probable regional, national, and international travel over the break. This was also reflected by the Saskatoon WWTP viral load trending upwards during the same period. This peak continued until the end of lectures (Fig. 3, Point 5) and the start of the final examination period. Theoretically, a correlation between an increase in cases and in-person learning is shown in the data, with downward trends coinciding with times when students were learning remotely or no longer had in-person lectures.
4. Conclusions
-
•
The spread of SARS-CoV-2 in the University of Saskatchewan residences and general population closely followed the trends of the local and surrounding communities.
-
•
The maximum daily detection frequency for SARS-CoV-2 virus in seven dormitories considered was about 75 %.
-
•
The maximum daily reported case numbers for the residences and campus-wide were about 11 and 75 people, respectively.
-
•
Passive sampling in wastewater-based surveillance can be useful in monitoring the spread of SARS-CoV-2 in a small population.
-
•
RT-qPCR analysis of university residence wastewater can be a cost-effective alternative to invasive individual testing.
The following is the supplementary data related to this article.
Linear regression for matching timepoints with no timeshift or latency considered. Spearman correlation coefficients and p-value for slopes are shown. Values that are significant are shown including 95 % confidence intervals.
Funding/support
This study was funded by the Pandemic Response Team through the University of Saskatchewan.
CRediT authorship contribution statement
Niteesh Jain: Formal analysis, Methodology, Writing – review and editing.
Daniel Hamilton: Formal analysis, Methodology, Writing – review and editing.
Kerry McPhedran: Funding acquisition, Supervision, Writing – review and editing.
Markus Brinkmann: Funding acquisition, Supervision, Writing – review and editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to acknowledge Charles Lytle and other facilities staff for their support during the deployment and retrieval of samplers. LuminUltra Technologies Inc. has been instrumental in adapting their laboratory protocols to passive sampling strategies. Additionally, we would like to acknowledge Dr. Dave McCarthy for developing and sharing of the 3D files for printing the torpedo samplers used in this study.
Editor: Kerry McPhedran
Data availability
The authors do not have permission to share data.
References
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J.F., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt W.Q., Schmitz B.W., Innes G.K., Prasek S.M., Pogreba Brown K.M., Stark E.R., Foster A.R., Sprissler R.S., Harris D.T., Sherchan S.P., Gerba C.P., Pepper I.L. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021;779 doi: 10.1016/j.scitotenv.2021.146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelvan V.P. Channel News Asia. 2021. 437 NUS UTown hostel residents swabbed for COVID-19 after viral material found in wastewater sample.https://www.channelnewsasia.com/news/singapore/nus-utown-hostel-residents-swabbed-covid-19-moh-14473672 Retrieved 10 June 2022 from. [Google Scholar]
- Corchis-Scott R., Geng Q., Seth R., Ray R., Beg M., Biswas N., Charron L. Averting an outbreak of SARS-CoV-2 in a university residence hall through wastewater surveillance. Microbiol. Spec. 2021;9 doi: 10.1128/Spectrum.00792-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle J., Racher K., Hazenberg J., Yeoman A., Hannah H., Duong D., Mohammed U., Spreitzer D., Gregorchuk B.S.J., Head B.M., Meyers A.F.A., Sandstrom P.A., Nichani A., Brooks J.I., Mulvey M.R., Mangat C.S., Becker M.G. A sensitive and rapid wastewater test for SARS-COV-2 and its use for the early detection of a cluster of cases in a remote community. Appl. Environ. Microbiol. 2022;88(5) doi: 10.1128/AEM.01740-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust P.M., Graber T.E., Mercier E., Montpetit D., Alexandrov I., Neault N., Baig A.T., Mayne J., Zhang X., Alain T., Servos M.R., Srikanthan N., MacKenzie M., Figeys D., Manuel D., Jüni P., MacKenzie A.E., Delatolla R. Catching a resurgence: increase in SARS-CoV-2 viral RNA identified in wastewater 48 h before COVID-19 clinical tests and 96 h before hospitalizations. Sci. Total Environ. 2021;770 doi: 10.1016/j.scitotenv.2021.145319l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughton C.G. Wastewater surveillance for population-wide Covid-19: the present and future. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibas C., Lambirth K.C., Mittal N., Juel M., Barua V.B., Roppolo Brazell L., Hinton K., Lontai J., Stark N., Young I., Quach C., Russ M., Kauer J., Nicolosi B., Chen D., Akella S., Tang W., Schlueter J., Munir M. Implementing building-level SARS-CoV-2 wastewater surveillance on a university campus. Sci. Total Environ. 2021;782 doi: 10.1016/j.scitotenv.2021.146749. 146749-146749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham K.E., Loeb S.K., Wolfe M.K., Catoe D., Sinnott-Armstrong N., Kim S., Yamahara K.M., Sassoubre L.M., Mendoza Grijalva L.M., Roldan-Hernandez L., Langenfeld K., Wigginton K.R., Boehm A.B. SARS-CoV-2 RNA in wastewater settled solids is associated with COVID-19 cases in a large urban sewershed. Environ. Sci. Technol. 2021;55(1):488–498. doi: 10.1021/acs.est.0c06191. [DOI] [PubMed] [Google Scholar]
- Habtewold J., McCarthy D., McBean E., Law I., Goodridge L., Habash M., Murphy H.M. Passive sampling, a practical method for wastewater-based surveillance of SARS-CoV-2. Environ. Res. 2022;204 doi: 10.1016/j.envres.2021.112058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes E.K., Sweeney C.L., Anderson L.E., Li B., Erjavec G.B., Gouthro M.T., Krkosek W.H., Stoddart A.K., Gagnon G.A. A novel passive sampling approach for SARS-CoV-2 in wastewater in a Canadian province with low prevalence of COVID-19. Environ. Sci Water Res. Technol. 2021;7:1576–1586. [Google Scholar]
- He X., Hong W., Pan X., Lu G., Wei X. SARS-CoV-2 omicron variant: characteristics and prevention. Med. Comment. 2021;2(4):838–845. doi: 10.1002/mco2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen D.A., Wigginton K.R. Tracking COVID-19 with wastewater. Nat. Biotechnol. 2020;38:1151–1153. doi: 10.1038/s41587-020-0690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu E., Ai Y., Davis A., Straathof J., Halloran K., Hull N., Winston R., Weir M.H., Soller J., Bohrerova Z., Oglesbee M., Lee J. Wastewater surveillance of SARS-CoV-2 in dormitories as a part of comprehensive university campus COVID-19 monitoring. Environ. Res. 2022;212(Pt E) doi: 10.1016/j.envres.2022.113580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M. Channel News Asia. 2020. From manhole to sampling bottle: How wastewater helps indicate presence of COVID-19 in Foreign Worker Dormitories.https://www.channelnewsasia.com/news/singapore/foreignworker-dormitories-sampling-testing-covid19-wastewater-12953408 retrieved 10 June 2022 from. [Google Scholar]
- Oloye F., Xie Y., Asadi M., Cantin J., Challis J., Brinkmann M., McPhedran K., Kristian K., Keller M., Sadowski M., Jones P., Landgraff C., Mangat C., Fuzzen M., Servos M., Giesy J. Rapid transition between SARS-CoV-2 variants of concern Delta and omicron detected by monitoring municipal wastewater from three Canadian cities. Sci. Total Environ. 2022;841 doi: 10.1016/j.scitotenv.2022.156741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schang C., Crosbie N.D., Nolan M., Poon R., Wang M., Jex A., John N., Baker L., Scales P., Schmidt J., Thorley B.R., Hill K., Zamyadi A., Tseng C.W., Henry R., Kolotelo P., Langeveld J., Schilperoort R., Shi B., Einsiedel S., Thomas M., Black J., Wilson S., McCarthy D.T. Passive sampling of SARS-CoV-2 for wastewater surveillance. Environ. Sci. Technol. 2021;55(15):10432–10441. doi: 10.1021/acs.est.1c01530. [DOI] [PubMed] [Google Scholar]
- Scott L.C., Aubee A., Babahaji L., Vigil K., Tims S., Aw T.G. Targeted wastewater surveillance of SARS-CoV-2 on university campus for COVID-19 outbreak detection and mitigation. Environ. Res. 2021;200 doi: 10.1016/j.envres.2021.111374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D., Sun Y., Zhou J., Ye Q. The global epidemic of the SARS-CoV-2 delta variant, key spike mutations and immune escape. Frontiers Immunology. 2021;12 doi: 10.3389/fimmu.2021.751778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo V., Tillett R.L., Chang C.-L., Gerrity D., Betancourt W.Q., Oh E.C. SARS-CoV-2 variant detection at a university dormitory using wastewater genomic tools. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.149930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu P., Zhang H., Ibaraki M., Van Tassell J., Geith K., Cavallo M., Kann R., Saber L., Kraft C.S., Lane M., Shartar S., Moe C. Early warning of a COVID-19 surge on university campus based on wastewater surveillance for SARS-CoV-2 at residence halls. Sci. Total Environ. 2022;821 doi: 10.1016/j.scitotenv.2022.153291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J.C.C., Tan J., Lim Y.X., Arivalan S., Hapuarachchi H.C., Mailepessov D., Griffiths J., Jayarajah P., Setoh Y.X., Tien W.P., Low S.L., Koo C., Yenamandra S.P., Kong M., Lee V.J.M., Ng L.C. Non-intrusive wastewater surveillance for monitoring of residential building for COVID-19 cases. Sci. Total Environ. 2021;786 doi: 10.1016/j.scitotenv.2021.147419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Challis J.K., Oloye F., Asadi M., Cantin J., Brinkmann M., McPhedran K.N., Hogan N., Sadowski M., Jones P.D., Landgraff C., Mangat C., Servos M.R., Giesy J.P. RNA in municipal wastewater reveals magnitudes of COVID-19 outbreaks across four waves driven by SARS-CoV-2 variants of concern. Environ. Sci. Technol. Water. 2022 doi: 10.1021/acsestwater.1c00349&ref=pdf. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Linear regression for matching timepoints with no timeshift or latency considered. Spearman correlation coefficients and p-value for slopes are shown. Values that are significant are shown including 95 % confidence intervals.
Data Availability Statement
The authors do not have permission to share data.