Abstract
It would be apt to say that one of the greatest accomplishments in modern medicine has been the development of vaccines against COVID-19, which had paralyzed the entire world for more than a year. Pfizer and BioNTech codeveloped the first COVID-19 vaccine that was granted emergency-use authorization or conditional approval in several regions globally. This article is an attempt to go ‘behind-the-scenes’ of this development process and highlight key factors that allowed us to move with this unprecedented speed, while adhering to normal vaccine-development requirements to generate the information the regulatory authorities needed to assess the safety and effectiveness of a vaccine to prevent an infectious disease, including quality and manufacturing standards. This is also a story of how Pfizer and BioNTech leveraged our combined skill sets and experience to respond to the global health crisis to progress this program swiftly while ensuring the compliance with our high-quality standards and keeping patient safety at the forefront. We will also highlight multiple other factors that were instrumental in our success.
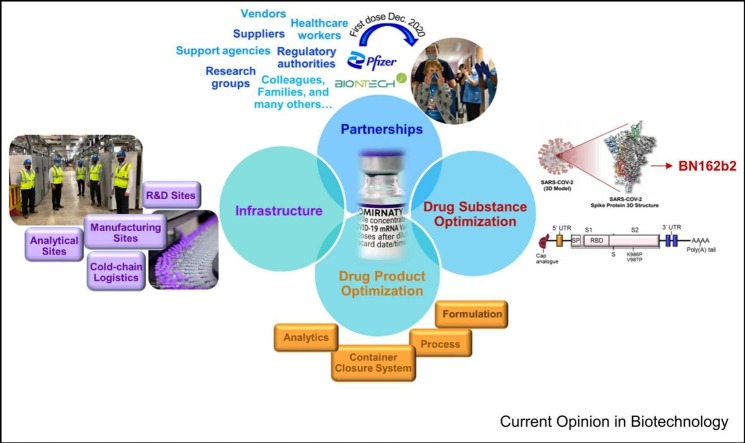
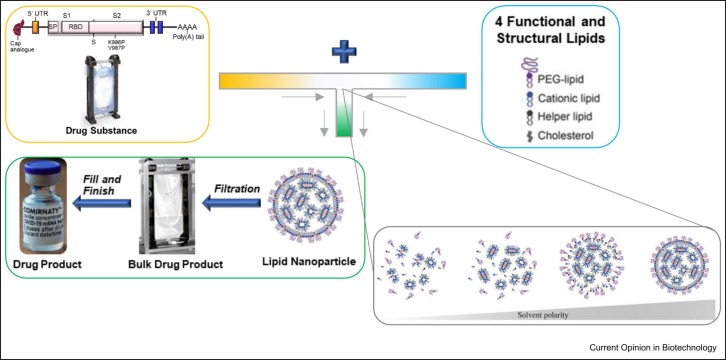
Graphical Abstract
Current Opinion in Biotechnology 2022, 78:102803
This review comes from a themed issue on Pharmaceutical Biotechnology
Edited by Sarah Harcum and Robert Kiss
For complete overview of the section, please refer to the article collection, “Pharmaceutical Biotechnology”
Available online 1st September 2022
https://doi.org/10.1016/j.copbio.2022.102803
0958-1669/© 2022 Elsevier Ltd. All rights reserved.
Introduction
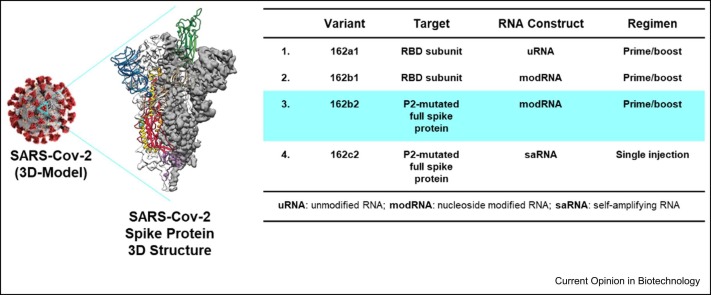
In early 2020, it became evident that SARS-CoV-2, the virus that causes COVID-19, was quickly becoming a global health crisis. With the ongoing Pfizer–BioNTech collaboration for influenza mRNA vaccine, the two companies saw the potential of redirecting their efforts to tackle this historic challenge. To that end, on March 17, 2020, Pfizer and BioNTech began collaborating to accelerate BioNTech’s mRNA-based vaccine program BNT162 that aimed to develop a vaccine to help prevent COVID-19 disease by leveraging expertize and resources of both companies. mRNA vaccines work by delivering mRNA molecules by lipid nanoparticles (LNP) to cells within the body [1]. Once inside the cell, the mRNA is transcribed into protein antigen [2]. For the Pfizer–BioNTech COVID-19 vaccine-development program, our clinical testing included vaccine candidates that generated the SARS-CoV-2 receptor- binding domain subunit protein and the [wild-type] spike protein as the target protein ( Figure 1). The body then recognizes the protein antigen as foreign and mounts an immune response, enabling the body to recognize and help fight future COVID-19 disease 3, 4, 5. The rapid construct design-to-production timelines along with the use of platform processes utilizing cell-free mRNA drug-substance (DS) synthesis provides mRNA vaccines distinct speed advantages over traditional vaccines [1]. The LNPs enabled efficient delivery of the mRNA directly into cells where protein could be produced [7]. The LNPs are composed of four different lipids [8]. These advantages of delivering mRNA using novel LNPs required the use of advanced drug-product (DP) manufacturing technologies that were carefully but rapidly upscaled and used to generate multiple initial constructs. If needed, altogether, the combined use of strategic manufacturing of DS and DP will enable us to quickly pivot to new constructs to address variant strains as they arise once a regulatory path is agreed with the regulatory authorities [9].
Figure 1.
Selection of Pfizer/BioNTech COVID-19 vaccine BN162b2, SARS-Cov-2 spike-protein 3D- structure image produced from Wrapp et al. [10].
Usually, a traditional vaccine-development process takes 10 years or more, but the pandemic required a much faster response 11, 12. With the support from leadership and leveraging Pfizer’s many years of vaccine-development expertize, we took the educated risks to strategically perform activities in parallel that would normally occur sequentially in the traditional setting [13]. For example, and as described in subsequent sections, before any clinical data information has been received, we had triggered discussions with raw material vendors to supply for global pandemic needs as well as preparing scale-up for commercial DS and DP assuming success.
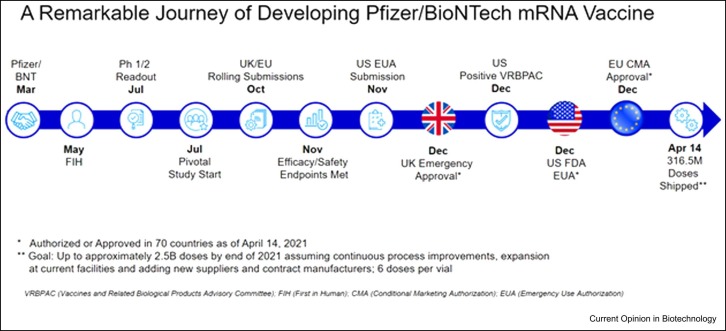
For the first educated risk, we quickly initiated development of four separate vaccine candidates with three different RNA platforms (uRNA, modRNA, and saRNA) (see Figure 1) in parallel, until the clinical data could allow us to focus on lead candidates. As seen in Figure 2, in a record-setting nine months, Pfizer advanced BNT162b2 from research-initiation stage to emergency-use authorization.
Figure 2.
Timeline for the development of the COVID-19 mRNA vaccine.
In the following sections, we identify many of the notable keys to success that made this journey possible.
Partnerships with vendors to establish future supply of critical raw materials
With mRNA being a new modality for vaccine manufacturing, along with the potential need to manufacture billions of doses necessary to respond to a global pandemic, a cross-functional team dedicated to developing raw material sourcing strategies was established at the onset of the program [14]. This team was responsible for forging numerous supply engagements with key vendors to supply the critical raw materials such as enzymes, nucleotides, capping reagents, buffer components, process materials, and lipids. This required rapidly creating and establishing partnerships with vendors who could develop, scale-up, and qualify manufacturing processes as well as assure requisite quality and supply-chain needs not only for the initial supply but also potentially for long-term future supply needs. It involved ordering unprecedented large amounts of the previously mentioned raw materials at risk before clinical outcomes and continuing to work closely with the vendors to forecast potential future supply needs.
Leveraging drug-substance mRNA manufacturing platforms (plasmid DNA template)
One critical starting material for mRNA manufacturing is the DNA template encoding the antigen [15]. At Pfizer, we utilized prior plasmid DNA (pDNA) manufacturing technology expertize from Pfizer’s Gene Therapy Program. This involved quickly screening all four plasmids for each vaccine candidate in parallel in five separate E. coli cell lines that were available. These transfected cell lines were quickly screened in small-scale shake flasks for favorable DNA topology (i.e. high supercoiled DNA, low open circle, and linear DNA) and titer to narrow down to the top two lead cell lines. These top two pDNA cell-line leads were then scaled-up in lab-scale bioreactors and the pDNA was purified, linearized, and analyzed to select the top cell line based on best DNA-topology attributes, cell growth, and titer. A master cell bank (MCB) and working cell bank were then prepared at risk for each of the top lead cell lines for each of the four potential vaccine candidates. Pfizer’s existing large-scale pDNA manufacturing facility in Chesterfield, MO, was utilized to initially manufacture DNA template before the eventual tech transfer to a new DNA template manufacturing facility built at Pfizer’s facility in Andover, MA.
Leveraging drug-substance mRNA manufacturing platforms (mRNA drug-substance)
Starting from the very early preMCB lab-scale supply of linear DNA template, the mRNA process-development team was tasked with first transferring the knowledge for mRNA production to Pfizer bioprocess-development labs in Chesterfield, MO, with the eventual goal of process scale-up to commercial scale in Andover, MA. The lab-scale team supply was used to a) confirm the process transfer, b) supply material for analytical method development and early formulation/stability development, and c) confirm that the process had the necessary prerequisites for scale-up. This required a close three-way collaboration among the Chesterfield team with the Vaccine Research Development (VRD) team located at Pfizer Pearl River, NY and the BioNTech development team in Mainz, Germany.
A section of the existing Chesterfield lab space was converted to supply the RNase-free environment for mRNA production. From the beginning, the decision was made to use disposable (single-use) bioreactor systems for both lab-scale and commercial large-scale manufacturing. This was decided for several reasons: RNase control, quick turnaround of equipment at all scales, and availability of equipment. For RNase control, autoclaving is not sufficient and diethylpyrocarbonate treatment is the typical route for inactivating RNases, but would require extra time and effort 16, 17. Additionally, by using the single-use equipment, no cleaning was required, so both development and manufacturing equipment could be turned around quickly. Further, with speed, we needed to produce material at all scalesworking with the equipment we had available. Trying to purchase additional equipment would have taken too long to receive and install. Leveraging our experience with small-scale disposable bioreactor systems such as the AMBR15 and AMBR250 Modular and their historical fidelity to manufacturing-scale systems 18, 19, the in vitro transcription (IVT) process-development team used these systems to develop the process for scale-up [20]. As we qualified the scale-up process with satellite studies using these systems, these lab systems were then used for process-characterization studies to support the process- performance qualification (PPQ) campaign.
When selecting the potential large-scale bioreactors for the IVT step, it was critical to utilize existing bioreactor systems within the manufacturing network as there was no time to order, install, and qualify new bioreactor systems. We initially used a single-use mixing system and a single-use bioreactor that could be operated at volumes up to 50 L and further scaled up to volumes greater than 100 L for the IVT-unit operation. Owing to the difference in impeller configuration and shape of these bioreactors selected for the IVT-unit operation, we utilized computational fluid dynamics modeling to develop a harmonized mixing control strategy for the different bioreactors to ensure quality and consistency [20]. A clinical manufacturing facility in Andover, MA, was repurposed for the initial scale-up and PPQ. Leveraging the expertize of the Pfizer Global Supply (PGS) quality and validation team was essential for this successful validation/PPQ of the mRNA DS process. This DS process was then also validated at the BioNTech Mainz and Marburg, Germany, manufacturing facilities and in an additional Pfizer facility in Andover, MA.
The strategy of using single-use containers was also useful when selecting and leveraging existing final purified DS bags that required freezing before LNP DP formulation.
Pfizer’s governance system for project management ensured a comprehensive and harmonized control strategy across all the DS manufacturing facilities to ensure continuity of critical quality attributes and process performance. In several instances, the use of Pfizer’s Corporate aviation team was necessary to ensure timelines were met to guarantee shipment of critical supplies due to the limited availability of domestic and international flights during the global pandemic.
The lipid nanoparticles
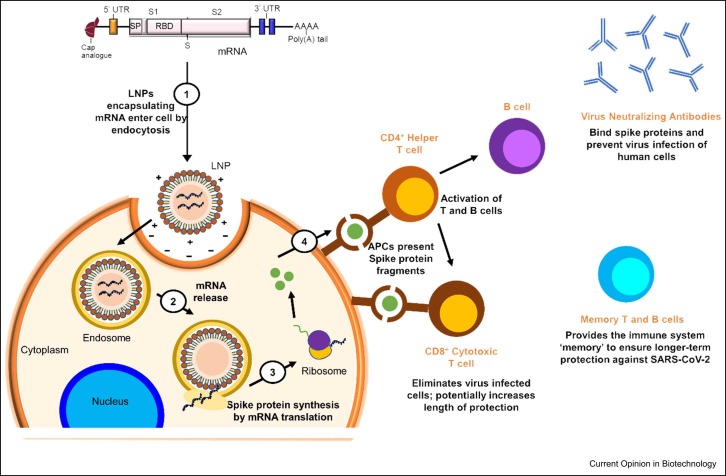
LNPs are a cornerstone of the overall success of the COVID-19 vaccine. mRNA is a large and highly sensitive compound that is readily degraded upon administration into the body [21]. To produce its effect, mRNA is required to transverse cell membranes and enter the cell’s cytosol to be translated into the desired protein. However, due to the large size and negative charges of the compound, it can rarely do so. This is where the role of the LNPs comes in: they encapsulate mRNA and aid the delivery into the cells to enable the downstream effect of encoding for the spike protein to generate an immune response 9, 22, 23. Altogether, the nanosize, lipophilic, and biomimetic features of LNPs facilitate its favorable effect of delivering mRNA into the cells' cytoplasm to begin translation, as demonstrated in Figure 3.
Figure 3.
Proposed mechanism depicting cellular uptake of mRNA-loaded LNPs, spike-protein synthesis, and activation of the immune system to provide long-term protection against SARS-CoV-2 virus.
LNPs are made from four lipids that provide (1) structural integrity, (2) drive mRNA loading, then release inside the cell, and (3) stabilization in the body, as well as for the product’s shelf-life 24, 25, 26. The first LNP-based product was approved by the US Food and Drug Administration (FDA) in 2018, ONPATTRO® (patisiran), which delivers short-interfering RNA to treat polyneuropathies induced by hereditary transthyretin amyloidosis [27]. The four lipids in BNT162b2 comprise an ionizable cationic lipid (i.e. ALC-0315), cholesterol, distearoylphosphatidylcholine, and a polyethylene glycol-conjugated (PEGylated) lipid (i.e. ALC-0159) [28]. The development of LNPs hails from research into nanoparticles that biologically mimic the bilayer of the cell wall and are stealth-like, avoiding rapid clearance from the body 29, 30, 31, 32, 33. The ionizable cationic lipid enables mRNA complexation at low pH, as the lipid possesses a charge to electrostatically bind mRNA, while then becoming neutral at physiological pH to reduce toxicity concerns 34, 35. The ionizable cationic lipid used in the COVID-19 vaccine, ALC-0315, was designed to provide a shorter circulation half-life in the human body (i.e. more biodegradable) and be more efficient to deliver the mRNA into the cell [8]. ALC-315 and the PEGylated lipid, ALC-0519, are proprietary lipids licensed from Acuitas who worked with BioNtech and Pfizer during the effort to produce the first COVID-19 mRNA vaccine.
mRNA drug-product ‘Platform’ process: a herculean effort
The Pfizer–BioNtech COVID-19 vaccine drug product was rigorously developed into a platform process. The program followed as swiftly as possibly while ensuring the normal parameters of vaccine development, including our standard compliance, safety, and quality standards. This effort was possible by leveraging Pfizer’s existing diverse biologics portfolio integrated across four R&D sites, spanning across modalities ranging from molecules in the size order of< 20 kDa to> 500 kDa toward gene therapies and nanomedicine portfolios. Establishing a platform-able approach for critical quality attributes of different modalities while understanding their unique differences is key to such development. Pfizer’s deep understanding and interspersed knowledge of monoclonal antibodies as part of a diverse portfolio was a momentous blessing for Pfizer to apply, including formulation (higher-order structure, route of administration, and indication), process (compounding, filtering, and filling), container closure system (vials, stoppers, overseals, and syringes), and analytics (instrumentation, method development, validation, and data analysis) knowledge toward efficiently developing BNT162b2. High-throughput analytics and highly organized teams were crucial to enable scientifically backed decision-making [13]. Rigorous development focused on utilizing the correct manufacturing and processing parameters to form the LNPs. The correct ratio of all lipidic components is an example of a potentially critical manufacturing parameter. Strategic mixing of the lipids with the mRNA aqueous phase at defined ratios and flow rates through a set flow path results in the self-assembled LNPs loaded with mRNA 34, 36, as demonstrated in Figure 4.
Figure 4.
The mRNA drug-product platform process: mixing of the aqueous stream with the organic stream via a proprietary T-mixing process at fixed ratios and flow rates through a set flow path, provides self-assembly of the LNP formulation with the mRNA into nm-sized particles [36].
The scale-up process of the mRNA–LNP DP was a herculean effort that was strategically focused with strong leadership, thoughtful decision-making, multifaceted parallel development, and dedicated teamwork. Scaling up the process from benchtop-size batches to manufactured batches at scale was rapidly performed. The manufacturing sites at Kalamazoo, MI and Puurs, Belgium, worked in tandem to produce a formulation lab, design an industrial process, and produce the first batch of BNT162b2 within an astounding 100 days. Many efforts were performed at risk, waiting on decisions for what would be the final vaccine candidate and in hope for success in the clinical trials. As COVID-19 was rapidly spreading across the globe causing significant illnesses and deaths, product-development timing was immensely critical. To manufacture at scale, the supply of all ingredients was required in excess. Pfizer developed and produced large-scale quantities of the raw materials required to produce the vaccine. For example, the cationic lipid, ALC-0315, was not readily available at such large quantities to produce millions of vaccine doses. The process of making the cationic lipid, ALC-0315, was developed at the Pfizer site in Groton, CT. It was then scaled up at the Kalamazoo, MI site, where by July 2022, more than 20 kg of lipid was being produced per week, which was enough for millions of vaccine doses.
Distribution and cold-chain logistics
The remarkable vaccine development was followed by an unprecedented challenge of worldwide vaccine distribution amidst a global pandemic [37]. Pfizer tackled the need for the DP to be stored and shipped at ultracold temperatures (−80 °C to −60 °C) by developing rechargeable containers that could maintain the temperature of the vaccines during shipping as well as short-term storage at the destination. These efforts were coherent with justifying stability of the final DP to the FDA at − 20 °C for two weeks in February 2021, and 2–8 °C for 1 month in May 2021 [38].
Conclusion
The development of the first vaccine to receive emergency-use authorization to help prevent COVID-19 was an astounding feat of Pfizer and BioNTech. With the utmost importance, we adhered to normal vaccine development requirements to generate the information required by regulatory authorities and to generate the information needed to assess the safety and effectiveness of a vaccine to help prevent COVID-19, including important quality and manufacturing standards. In response to the global health crisis, we leveraged our vast experience and progressed our program swiftly while ensuring rigorous compliance and quality standards and focusing on safety. In addition, multiple other factors were required for our success, comprising clear target expectations and support from leadership, thoughtful decision-making, 24/7 commitment, everybody ‘All In’, agency engagement, collaboration, rapid review, multifaceted parallel development, moving as fast as the science allows, direct straight-line interactions end to end, communication, clear project planning, roles and responsibilities, and celebrating successes along the way. This feat was not achieved in isolation, where our strong partnerships with experts and specialists in the field worked to the advantage of producing the final product.
Conflict of interest statement
The authors declare the following financial interests/personal relationships that may be considered as potential competing interests: all the authors listed are employees of Pfizer Inc. and may have financial interest in the company in the form of stock, stock options, and other forms of employment-related long-term incentives.
Acknowledgements
The authors would like to acknowledge that the work that is the basis of this article is a result of significant contribution from many colleagues in Pfizer and BioNTech, many of whom did not contribute to the actual authoring of this article, but without whose contributions, there would be no story to tell. The authors here are representing all those colleagues who are responsible for the development and commercialization of this vaccine. We would also like to express our extreme gratitude to healthcare workers, first responders, clinical trial volunteers, the physicians, nurses, and other staff who enabled these trials, governments and regulatory authorities worldwide, essential workers and teachers, our vendors, suppliers, and other support agencies, and all BioNTech and Pfizer colleagues and their families.
Data Availability
The data that have been used are confidential.
References
- 1.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat Rev Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Z., Mateus J., Coelho C.H., Dan J.M., Moderbacher C.R., Gálvez R.I., Cortes F.H., Grifoni A., Tarke A., Chang J., et al. Humoral and cellular immune memory to four COVID-19 vaccines. Cell. 2022;185:2434–2451.e2417. doi: 10.1016/j.cell.2022.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schultze J.L., Aschenbrenner A.C. COVID-19 and the human innate immune system. Cell. 2021;184:1671–1692. doi: 10.1016/j.cell.2021.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pandey S.C., Pande V., Sati D., Upreti S., Samant M. Vaccination strategies to combat novel corona virus SARS-CoV-2. Life Sci. 2020;256 doi: 10.1016/j.lfs.2020.117956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pollard C., De Koker S., Saelens X., Vanham G., Grooten J. Challenges and advances towards the rational design of mRNA vaccines. Trends Mol Med. 2013;19:705–713. doi: 10.1016/j.molmed.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Naderi Sohi A., Kiani J., Arefian E., Khosrojerdi A., Fekrirad Z., Ghaemi S., Zim M.K., Jalili A., Bostanshirin N., Soleimani M. Development of an mRNA-LNP vaccine against SARS-CoV-2: evaluation of immune response in mouse and Rhesus Macaque. Vaccines. 2021;9:1–15. doi: 10.3390/vaccines9091007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kon E., Elia U., Peer D. Principles for designing an optimal mRNA lipid nanoparticle vaccine. Curr Opin Biotechnol. 2022;73:329–336. doi: 10.1016/j.copbio.2021.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reichmuth A.M., Oberli M.A., Jaklenec A., Langer R., Blankschtein D. mRNA vaccine delivery using lipid nanoparticles. Ther Deliv. 2016;7:319–334. doi: 10.4155/tde-2016-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oyston P., Robinson K. The current challenges for vaccine development. J Med Microbiol. 2012;61:889–894. doi: 10.1099/jmm.0.039180-0. [DOI] [PubMed] [Google Scholar]
- 12.Wagner R., Hildt E., Grabski E., Sun Y., Meyer H., Lommel A., Keller-Stanislawski B., Müller-Berghaus J., Cichutek K. Accelerated development of COVID-19 vaccines: technology platforms, benefits, and associated risks. Vaccines. 2021;9:1–11. doi: 10.3390/vaccines9070747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bourla D.A. Moonshot: Inside Pfizer's Nine-Month Race to Make the Impossible Possible. HarperCollins Publishers; 2022. [Google Scholar]
- 14.Rosa S.S., Prazeres D.M.F., Azevedo A.M., Marques M.P.C. mRNA vaccines manufacturing: challenges and bottlenecks. Vaccine. 2021;39:2190–2200. doi: 10.1016/j.vaccine.2021.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohlson J. Plasmid manufacture is the bottleneck of the genetic medicine revolution. Drug Discov Today. 2020;25:1891–1893. doi: 10.1016/j.drudis.2020.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitley J., Zwolinski C., Denis C., Maughan M., Hayles L., Clarke D., Snare M., Liao H., Chiou S., Marmura T., et al. Development of mRNA manufacturing for vaccines and therapeutics: mRNA platform requirements and development of a scalable production process to support early phase clinical trials. Transl Res: J Lab Clin Med. 2022;242:38–55. doi: 10.1016/j.trsl.2021.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green M.R., Sambrook J. How to win the battle with RNase. Cold Spring Harb Protoc. 2019;2019:95–98. doi: 10.1101/pdb.top101857. [DOI] [PubMed] [Google Scholar]
- 18.Janakiraman V., Kwiatkowski C., Kshirsagar R., Ryll T., Huang Y.M. Application of high-throughput mini-bioreactor system for systematic scale-down modeling, process characterization, and control strategy development. Biotechnol Prog. 2015;31:1623–1632. doi: 10.1002/btpr.2162. [DOI] [PubMed] [Google Scholar]
- 19.Velez-Suberbie M.L., Betts J.P.J., Walker K.L., Robinson C., Zoro B., Keshavarz-Moore E. High throughput automated microbial bioreactor system used for clone selection and rapid scale-down process optimization. Biotechnol Prog. 2018;34:58–68. doi: 10.1002/btpr.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manahan M., Nelson M., Cacciatore J.J., Weng J., Xu S., Pollard J. Scale-down model qualification of ambr® 250 high-throughput mini-bioreactor system for two commercial-scale mAb processes. Biotechnol Prog. 2019;35 doi: 10.1002/btpr.2870. [DOI] [PubMed] [Google Scholar]
- 21.Granot Y., Peer D. Delivering the right message: challenges and opportunities in lipid nanoparticles-mediated modified mRNA therapeutics—an innate immune system standpoint. Semin Immunol. 2017;34:68–77. doi: 10.1016/j.smim.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 22.Schoenmaker L., Witzigmann D., Kulkarni J.A., Verbeke R., Kersten G., Jiskoot W., Crommelin D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: structure and stability. Int J Pharm. 2021;601 doi: 10.1016/j.ijpharm.2021.120586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tenchov R., Bird R., Curtze A.E., Zhou Q. Lipid nanoparticles─from liposomes to mRNA vaccine delivery, a landscape of research diversity and advancement. ACS Nano. 2021;15:16982–17015. doi: 10.1021/acsnano.1c04996. [DOI] [PubMed] [Google Scholar]
- 24.Carrasco M.J., Alishetty S., Alameh M.-G., Said H., Wright L., Paige M., Soliman O., Weissman D., Cleveland T.E., Grishaev A., et al. Ionization and structural properties of mRNA lipid nanoparticles influence expression in intramuscular and intravascular administration. Commun Biol. 2021;4 doi: 10.1038/s42003-021-02441-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mui B.L., Tam Y.K., Jayaraman M., Ansell S.M., Du X., Tam Y.Y.C., Lin P.J.C., Chen S., Narayanannair J.K., Rajeev K.G., et al. Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles. Mol Ther Nucleic Acids. 2013;2 doi: 10.1038/mtna.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabnis S., Kumarasinghe E.S., Salerno T., Mihai C., Ketova T., Senn J.J., Lynn A., Bulychev A., McFadyen I., Chan J., et al. A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol Ther. 2018;26:1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akinc A., Maier M.A., Manoharan M., Fitzgerald K., Jayaraman M., Barros S., Ansell S., Du X., Hope M.J., Madden T.D., et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat Nanotechnol. 2019;14:1084–1087. doi: 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- 28.Yanez Arteta M., Kjellman T., Bartesaghi S., Wallin S., Wu X., Kvist A.J., Dabkowska A., Székely N., Radulescu A., Bergenholtz J., et al. Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles. Proc Natl Acad Sci USA. 2018;115:e3351–e3360. doi: 10.1073/pnas.1720542115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eygeris Y., Gupta M., Kim J., Sahay G. Chemistry of lipid nanoparticles for RNA delivery. Acc Chem Res. 2022;55:2–12. doi: 10.1021/acs.accounts.1c00544. [DOI] [PubMed] [Google Scholar]
- 30.Buschmann M.D., Carrasco M.J., Alishetty S., Paige M., Alameh M.G., Weissman D. Nanomaterial delivery systems for mRNA vaccines. Vaccines. 2021;9:1–30. doi: 10.3390/vaccines9010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kulkarni J.A., Darjuan M.M., Mercer J.E., Chen S., van der Meel R., Thewalt J.L., Tam Y.Y.C., Cullis P.R. On the formation and morphology of lipid nanoparticles containing ionizable cationic lipids and siRNA. ACS Nano. 2018;12:4787–4795. doi: 10.1021/acsnano.8b01516. [DOI] [PubMed] [Google Scholar]
- 32.Terada T., Kulkarni J.A., Huynh A., Chen S., van der Meel R., Tam Y.Y.C., Cullis P.R. Characterization of lipid nanoparticles containing ionizable cationic lipids using design-of-experiments approach. Langmuir. 2021;37:1120–1128. doi: 10.1021/acs.langmuir.0c03039. [DOI] [PubMed] [Google Scholar]
- 33.Witzigmann D., Kulkarni J.A., Leung J., Chen S., Cullis P.R., van der Meel R. Lipid nanoparticle technology for therapeutic gene regulation in the liver. Adv Drug Deliv Rev. 2020;159:344–363. doi: 10.1016/j.addr.2020.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kulkarni J.A., Witzigmann D., Leung J., van der Meel R., Zaifman J., Darjuan M.M., Grisch-Chan H.M., Thöny B., Tam Y.Y.C., Cullis P.R. Fusion-dependent formation of lipid nanoparticles containing macromolecular payloads. Nanoscale. 2019;11:9023–9031. doi: 10.1039/c9nr02004g. [DOI] [PubMed] [Google Scholar]
- 35.Lou G., Anderluzzi G., Schmidt S.T., Woods S., Gallorini S., Brazzoli M., Giusti F., Ferlenghi I., Johnson R.N., Roberts C.W., et al. Delivery of self-amplifying mRNA vaccines by cationic lipid nanoparticles: the impact of cationic lipid selection. J Control Release. 2020;325:370–379. doi: 10.1016/j.jconrel.2020.06.027. [DOI] [PubMed] [Google Scholar]
- 36.Evers M.J.W., Kulkarni J.A., van der Meel R., Cullis P.R., Vader P., Schiffelers R.M. State-of-the-art design and rapid-mixing production techniques of lipid nanoparticles for nucleic acid delivery. Small Methods. 2018;2 [Google Scholar]
- 37.Crommelin D.J.A., Anchordoquy T.J., Volkin D.B., Jiskoot W., Mastrobattista E. Addressing the cold reality of mRNA vaccine stability. J Pharm Sci. 2021;110:997–1001. doi: 10.1016/j.xphs.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coronavirus (COVID-19) Update . Update Provides Alternative Temperature for Transportation and Temporary Storage for Frozen Vials Before Dilution. Administration FaD; 2021. FDA allows more flexible storage, transportation conditions for Pfizer-BioNTech COVID-19 vaccine. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that have been used are confidential.