Abstract
MYC as a transcriptional factor plays a crucial role in breast cancer progression. However, the mechanisms underlying MYC deubiquitination in breast cancer are not well defined. Here, we report that OTUB1 is responsible for MYC deubiquitination. OTUB1 could directly deubiquitinate MYC at K323 site, which blocks MYC protein degradation. Moreover, OTUB1 mediated MYC protein stability is also confirmed in OTUB1-knockout mice. Stabilized MYC by OTUB1 promotes its transcriptional activity and induces HK2 expression, which leads to enhance aerobic glycolysis. Therefore, OTUB1 promotes breast tumorigenesis in vivo and in vitro via blocking MYC protein degradation. Taken together, our data identify OTUB1 as a new deubiquitination enzyme for MYC protein degradation, which provides a potential target for breast cancer treatment.
Subject terms: Cancer metabolism, Cancer metabolism
Introduction
Ubiquitination plays a supportive role in regulating protein degradation, which is involved in many physiological and pathological processes [1]. OTUB1 belongs to the ovarian tumor domain protease (OTU) subfamily of deubiquitinases, which could block ubiquitination leading to protein stability [2]. OTUB1 as an oncogene could deubiquitinate multiple substrates including p53 [3], TRAF3 [4], c-Maf [5], PD-L1 [6], Cyclin E1 [7], FOXM1 [8], p100 [9], SLC7A11 [10], Snail [11], RAS [12], SMAD2/3 [2], ATF6 [13], c-IAP1 [14], MSH2 [15] and Tau [16], which regulates many cancer progression. Furthermore, high expression of OTUB1 in prostate cancer enhances cell invasion and tumorigenesis [7, 17]. In addition, OTUB1 as a marker of colorectal cancer promotes metastasis, which leads to poor survival [18]. In hepatocellular carcinoma, OTUB1 is overexpressed to enhance cell migration and invasion [19]. Moreover, overexpression of OTUB1 promotes migration of human glioma cells, which is correlated with a poor prognosis [20]. However, the functions of OTUB1 on breast tumorigenesis are still largely unknown.
MYC also named c-MYC as a transcription factor belongs to the basichelix-loop-helix-leucine zipper family located in the cell nucleus, which contributes to cell growth, death, differentiation, and metabolism [21]. MYC is frequently mutated and overexpressed in multiple of cancers [22]. More energy needs to be produced in rapidly proliferating tumor cells, and aerobic glycolysis could satisfy this process [23]. MYC is ubiquitous and highly expressed in proliferating cancer cells, which is the master regulator of aerobic glycolysis [24]. MYC promotes expressions of glycolysis key enzymes, such as HK2, PKM2 and LDHA, which could enhance aerobic glycolysis to enable cancer cells to proliferate faster [25]. MYC is highly expressed in breast cancer, especially in the triple negative breast cancer [26]. Post translational modification of MYC protein plays a key role in tumorigenesis [27]. However, the upstream deubiquitination enzymes that regulate MYC ubiquitination are still poorly understood. Here, we discover a new molecular mechanism responsible for breast tumorigenesis between OTUB1 and MYC. OTUB1 deubiquitinates MYC, and promotes its protein stability. Interestingly, OTUB1 increases aerobic glycolysis via MYC mediated HK2 expression, which promotes breast tumor growth. Taken together, we find a molecular mechanism based therapeutic strategy of targeting breast tumorigenesis via OTUB1-MYC-HK2 axis.
Materials and methods
Cell culture and transfection
The HEK293T, MCF-7 and MB231 cells were from Chinese Academy of Sciences, which were maintained at 37 °C supplied with 5%CO2. All cells were cultured in DMEM medium supplemented with 10% FBS, 50 μg/ml streptomycin, and 50 U/mL penicillin. Cell transfection was performed by using Lipofectamine 3000. The information of indicated plasmid was provided in supplementary materials.
Quantitative real-time PCR
Total RNA was extracted from indicated cells using TRIzolTM (Takara) reagent according to the protocol, and the RNA was reversely transcribed as previously described [28]. The expression levels of mRNA were quantified using the TB Green® Fast qPCR Mix (Takara). The information of primer sequence was listed in Supplementary Materials.
Western blot
The cells were lysed with lysis buffer (150 mM NaCl, 50 mM Tris-HCl [pH 7.5], and 0.5% NP40) with multiple protease inhibitors (Sigma–Aldrich), and the protein concentrations were quantified [29, 30]. The extracted proteins were determined by SDS-PAGE and transferred onto PVDF membranes. The membranes were incubated with indicated antibody at 4 °C overnight. The membranes were visualized by odyssey instrument. The information of indicated antibodies was provided in supplementary materials.
Immunoprecipitation (IP) and GST pull-down assays
Cell lysates were prepared by lysis buffer with multiple protease inhibitors (Sigma–Aldrich) [28–30]. The protein A/G agarose beads were added into the cell lysates with indicated antibodies at 4 °C overnight. Then, the beads were washed with IP buffer before performed by immunoblotting. GST-pull down assay was performed as described previously [31, 32].
In vitro deubiquitination assays
HEK293T cell were co-transfected with HA-tagged ubiquitin and Flag-tagged MYC, and treated with MG132 to increase the ubiquitination level of MYC. Then, the IP assay was performed to get ubiquitinated Flag-tagged MYC. The MYC proteins were incubated with His-OTUB using deubiquitinating buffer (60 mM HEPES, 5 mM MgCl2, 4% glycerol, pH 7.5) at 30 °C for 4 hr [6]. The deubiquitinated proteins were tested by western blotting.
Construction of stable cell pools
pLVX-shRNA1 was constructed as short-hairpin RNA vectors. Indicated cells were infected with viruses and the single stable cells were screened. The knockdown efficiency was tested after selection with puromycin for 2 weeks. Indicated genes were constructed into the pLVX-IRES-Neo vectors and transfected with psPAX2 and pMD2.G package vectors in HEK293T cells. The indicated breast cancer cells were then infected indicated lentiviruses, following a selection with G418 for 2 weeks. Then, the single stable cells were generated. The information of shRNA sequence was provided in supplementary materials.
Xenograft mouse model
The procedures related to animal studies were approved by the animal care committee of Weifang medical University (Weifang, China). The female 4 week old BALB/c nude mice were injected subcutaneously with 5 × 106 / 100 μL PBS MCF-7 cells with OTUB1 depletion reconstituted stable overexpression of MYC. Tumor volume was recorded by formula: Tumor volume = (length × width2)/2. Tumors were dissected and weighed after 3 weeks.
Statistical analysis
Statistical analyses were determined using Graphpad Prism 7.0 software, and presented as mean ± s.e.m. Differences between variables were considered to be statistically significant at P < 0.05. *P < 0.05 and n.s. was not significant.
Results
OTUB1 interacts with MYC
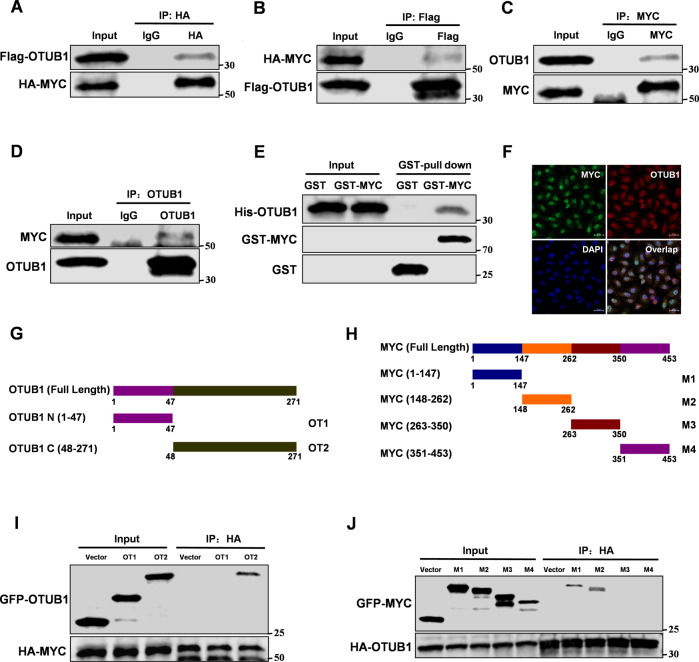
To study the functions of OTUB1 in breast cancer, we used OTUB1 as bait to identify MYC as a new binding partner (Supplementary Fig. S1A). To further confirm the interactions between OTUB1 and MYC, we performed Co-IP assays. The results showed that exogenously overexpressed Flag-tagged OTUB1 interacted with exogenously overexpressed HA-tagged MYC (Fig. 1A, B). Furthermore, endogenous interactions between OTUB1 and MYC were confirmed in the MCF-7 cells (Fig. 1C, D). GST-pull down assays showed that OTUB1 directly bound to MYC in vitro (Fig. 1E). As we expected, OTUB1 could co-localize with MYC both in the cytoplasm and nucleus of MCF-7 cells (Fig. 1F and Supplementary Fig. S1B). Based on their interaction, we next identified the binding regions between OTUB1 and MYC. We generated two fragments of OTUB1, amino acids 1-47 (OT1), and 48-271 (OT2) (Fig. 1G). Based to its structure, MYC protein was also divided into four fragments, amino acids 1–147 (M1), 148–262 (M2), 263–350 (M3), and 351–453 (M4) (Fig. 1H). The data showed that the C-terminal OTU domain of OTUB1 and N-terminal domain of MYC (1–262) were necessary for their interaction (Fig. 1I, J). Both amino acids 1–141 (M1) and 148–262 (M2) of MYC interacted with OTUB1, suggesting that two domains might format a complex structure to promote its interaction. Taken together, our data show that OTUB1 directly interacts with MYC.
Fig. 1. OTUB1 interacts with MYC.
Immunoprecipitation and immunoblotting analyses were performed with the indicated antibodies. A, B Immunoprecipitation (IP) and western blot analysis of the exogenous OTUB1/MYC proteins interaction in the 293 T cells co-transfected with Flag-tagged OTUB1 and HA-tagged MYC. C, D IP and western blot analysis of the endogenous OTUB1/MYC proteins interaction in the MCF-7 cells. E GST-pull down assay analysis of OTUB1/MYC proteins interaction using purified GST-tagged MYC and His-OTUB1. F Confocal immunofluorescence microscopy analysis of the OTUB1/MYC proteins interaction in the MCF-7 cells. G, I IP and western blot analysis of the HA-tagged MYC/GFP-tagged OTUB1 fragments proteins interaction in the 293 T cells. H, J IP and western blot analysis of the HA-tagged OTUB1/GFP-tagged MYC fragments proteins interaction in the 293 T cells.
OTUB1 enhances MYC protein stability
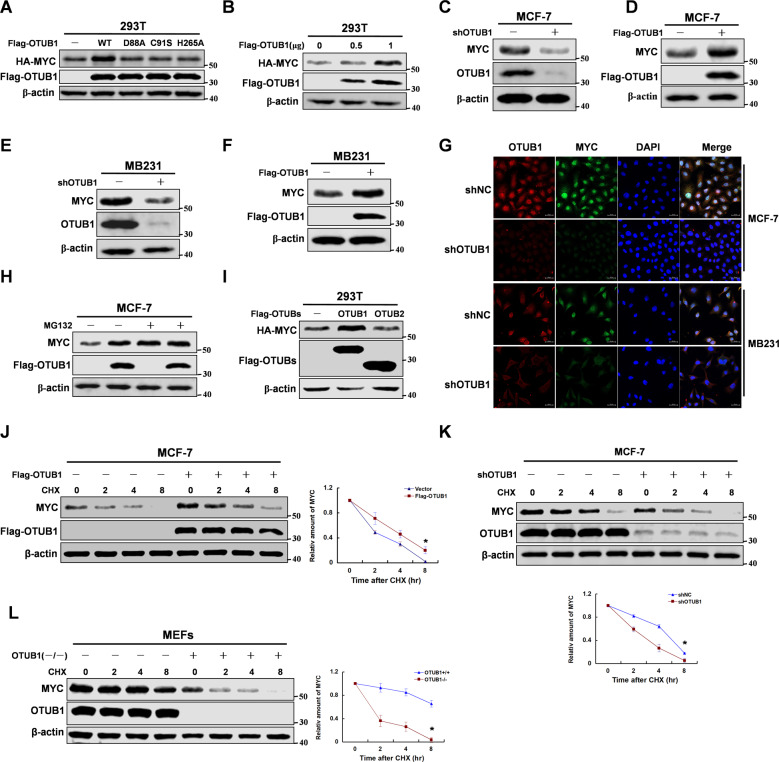
OTUB1 as a deubiquitinating enzyme could deubiquitinate multiple substrates, which could regulate protein stability [33]. So we speculated that OTUB1 might promote MYC protein stability. As we expected, overexpression of OTUB1 increased MYC protein stability, but the enzyme inactivated mutant did not (Fig. 2A, B). We used USP13 as a negative control which was a tumor suppressor in breast cancer [34], and used USP22 [35], USP28 [36], USP36 [37] or USP37 [38] as positive control which promoted stability of MYC. The data showed that OTUB1 was a new positive regulator for MYC (Supplementary Fig. S1C, D). To test whether deubiquitination and stabilization of MYC by OTUB1 relies on its ability to inhibit E2 enzymes, we constructed OTUB1 mutant (△N). Interestingly, deletion N-terminal of OTUB1 could still promote MYC protein stability (Supplementary Fig. S1E). The efficient of OTUB1 knockdown was determined in MCF-7 cells, and its expressions in breast cancer cells were also detected (Supplementary Fig. S2A, B). Furthermore, we manipulated the expression of OTUB1 in breast cancer cells, and demonstrated that OTUB1 enhanced MYC protein stability (Fig. 2C–G). To test whether OTUB1 regulates MYC protein stability via the proteasome pathway, we treated the cells with MG132. The data showed that MG132 abrogated the OTUB1-mediated stability of MYC protein, suggesting that OTUB1 regulated MYC protein stability via the ubiquitin proteasome pathway (Fig. 2H). Interestingly, OTUB2 as another member of OUT family could not regulate MYC protein stability (Fig. 2I). Moreover, to explore the protein half-life of MYC, the cells were treated with cycloheximide (CHX). As we expected, OTUB1 could increase the half-life of MYC protein (Fig. 2J–L), but the mutants of OTUB1 could not (Supplementary Fig. S2C). Collectively, the data show that OTUB1 promotes MYC protein stability via the ubiquitin proteasome pathway.
Fig. 2. OTUB1 enhances MYC protein stability.
Immunoblotting analyses were performed with the indicated antibodies. A 293 T cells were overexpressed HA-tagged MYC and Flag-tagged OTUB1 (WT or mutants) proteins. Immunoblotting analyse was performed. B 293 T cells were overexpressed HA-tagged MYC and Flag-tagged OTUB1 (0,0.5 or 1 μg) proteins. Immunoblotting analyse was performed. C, E MCF-7 or MB-231 cells were knocked down OTUB1 with shRNA. Immunoblotting analyse was performed. D, F MCF-7 or MB-231 cells were overexpressed Flag-tagged OTUB1 proteins. Immunoblotting analyse was performed. G MCF-7 or MB-231 cells were knocked down OTUB1 with shRNA. Immunofluorescence was performed. H MCF-7 cells with overexpression Flag-tagged OTUB1 protein were treated with MG132 for 8 hr. Immunoblotting analyse was performed. I 293 T cells were overexpressed the indicated HA-tagged MYC and Flag-tagged OTUB1 or OTUB2 proteins. Immunoblotting analyse was performed. J MCF-7 cells with overexpression of Flag-tagged OTUB1 were treated with CHX for indicated time. Immunoblotting analyse was performed. K MCF-7 cells with knockdown of OTUB1 were treated with CHX for indicated time. Immunoblotting analyse was performed. L MEFs cells with knockout of OTUB1 were treated with CHX for indicated time. Immunoblotting analyse was performed.
OTUB1 deubiquitinates MYC at K323
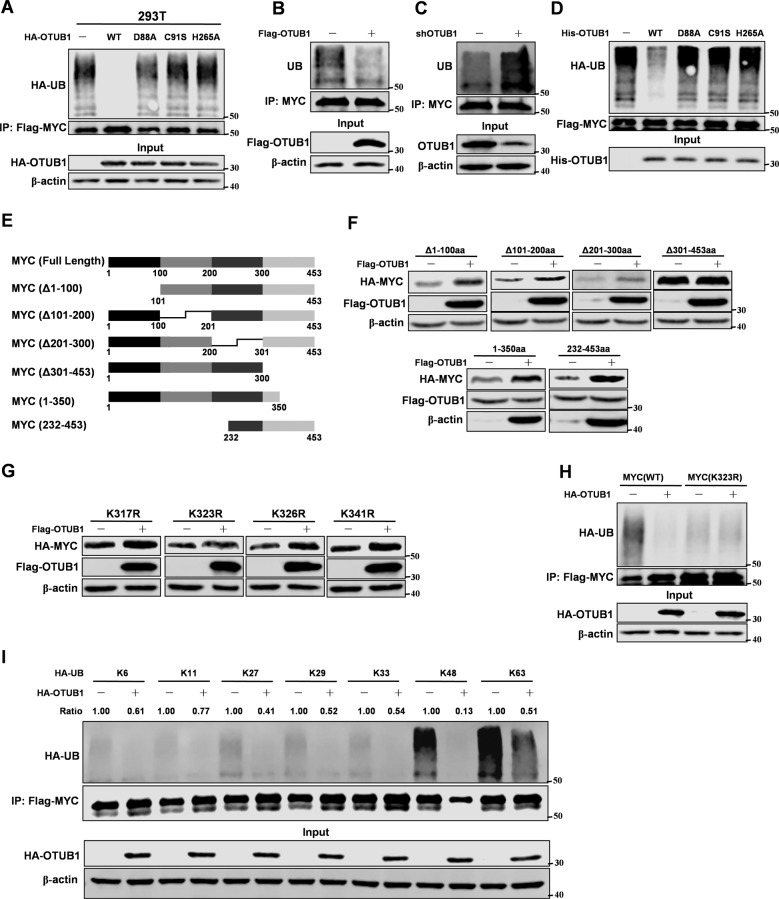
In previous experiments, we found that OTUB1 maintained MYC protein stability through the ubiquitin proteasome pathway. So we next examined whether OTUB1 could directly regulate the ubiquitination level of MYC. Compared to OTUB1 mutants, overexpression wild type OTUB1 could decrease the ubiquitination level of MYC in 293 T cells (Fig. 3A). Moreover, we found that manipulated expression of OTUB1 could change the ubiquitination level of MYC (Fig. 3B, C). Consistently, in vitro experiment showed that OTUB1 directly deubiquitinated MYC (Fig. 3D). Furthermore, we isolated nucleus and cytoplasm proteins from OTUB1 overexpression MCF-7 cells. The data showed that OTUB1 decreased ubiquitination of MYC both in the cytoplasm and nucleus, which was consistent with the PLA results (Supplementary Fig. S3A). To identify the lysine residues on MYC protein deubiquitinated by OTUB1, we constructed several mutants of MYC protein (Fig. 3E). Interestingly, OTUB1 did not affect the protein stability of MYC mutant (▵301–350aa) (Fig. 3F). To further confirm which lysine residue of MYC was deubiquitinated by OTUB1, we mutated all lysine residues on MYC (301–350aa). Interestingly, OTUB1 could not increase protein stability of MYC-K323R mutant, which suggested that OTUB1 deubiquitinated MYC at K323 (Fig. 3G). Moreover, OTUB1 also failed to deubiquitinate MYC-K323R mutant (Fig. 3H and Supplementary Fig. S3B). To determine which lysine (K)-linked polyubiquitin chain conjugated with MYC was deubiquitinated by OTUB1, we overexpressed several ubiquitin mutants. We found that MYC preferentially conjugated with lysine (K) 48-linked polyubiquitin chain, which was deubiquitinated by OTUB1 (Fig. 3I). Collectively, we conclude that OTUB1 deubiquitinates MYC at K323.
Fig. 3. OTUB1 deubiquitinates MYC at K323.
Mmunoblotting analyses were performed with the indicated antibodies. A 293 T cells with overexpression of both HA-tagged OTUB1 (WT, D88A, C91S or H265A) and Flag-tagged MYC proteins. After 48 hr transfection, cells were treated with MG132 for 8 hr followed by IP and western blot. B MCF-7 cells were overexpressed Flag-tagged OTUB1. After 48 hr transfection, cells were treated with MG132 for 8 hr followed by IP and western blot. C MCF-7 cells were stably expressed shRNA-OTUB1. Cells were treated with MG132 for 8 hr followed by IP and western blot. D MYC proteins were purified from 293 T cells transfected with Flag-tagged MYC by IP with Flag beads and elution with Flag peptide. His-tagged OTUB1 proteins were prified from E.coli. His-OTUB1 and Flag-MYC were added into a reaction mixture at 30 °C for 4 hr. Immunoblotting analyse was performed. E The MYC truncation mutants used in this study. F 293 T cells were co-overexpressed both Flag-tagged OTUB1 and different mutants of HA-tagged MYC. Immunoblotting analyse was performed. G 293 T cells were co-overexpressed both HA-tagged MYC (K323R) and Flag-tagged OTUB1. After 48 hr transfection, cells were treated with MG132 for 8 hr followed by IP and western blot. H 293 T cells with overexpression of both HA-tagged OTUB1 and Flag-tagged MYC (WT or K323R) proteins. After 48 hr transfection, cells were treated with MG132 for 8 hr followed by IP and western blot. I 293 T cells transfected with different ubiquitin mutants were co-overexpressed both HA-tagged OTUB1 and Flag-tagged MYC. After 48 hr transfection, cells were treated with MG132 for 8 hr followed by IP and western blot.
OTUB1 promotes MYC transcriptional activity
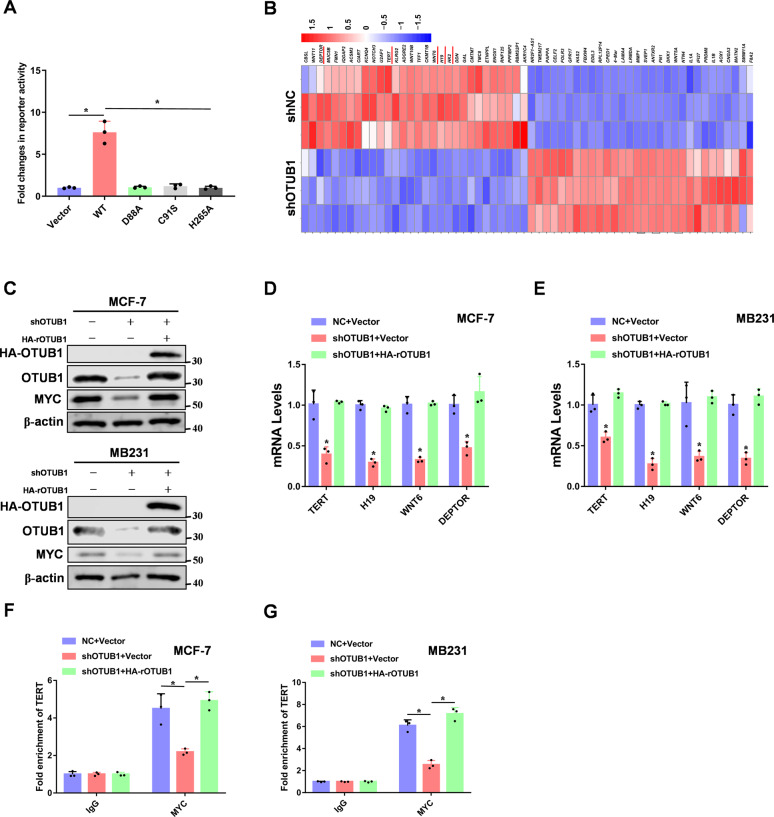
Our study showed that OTUB1 bound to MYC and increased its protein stability. So we speculated that OTUB1 would promote MYC transcriptional activity. To test this hypothesis, luciferase reporter assays demonstrated that overexpression of OTUB1 significantly increased MYC transcriptional activity (Fig. 4A). Furthermore, we knocked down OTUB1 in breast cancer cells, and performed RNA-sequencing analysis. From the heat map, OTUB1-depleted cells showed decreased expression of previously reported MYC transcriptional target genes such as TERT [39], H19 [40], WNT6 [41], and DEPTOR [42] (Fig. 4B). Moreover, we rescued OTUB1 expression in OTUB1-knockdown breast cancer cells (Fig. 4C). Consistently, qRT-PCR data further confirmed that depletion of OTUB1 decreased RNA expression of TERT, H19, WNT6, and DEPTOR (Fig. 4D, E). To study whether OTUB1 enhances more MYC binding to its target gene promoters via promoting its protein stability, we performed CHIP assays. We found that OTUB1 increased MYC binding to the TERT and DEPTOR promoters, which were previously reported to be MYC target genes (Fig. 4F–G and Supplementary Fig. S4A, B). Thus, OTUB1 promotes the transcriptional activity of MYC.
Fig. 4. OTUB1 promotes MYC transcriptional activity.
A 293 T cells were transfected Vector or Flag-tagged OTUB1 (WT, D88A, C91S or H265A) with Myc-luc reporter plasmids, and detected by luciferase reporter assay. B Heat map representing the most significantly regulated genes detected in RNA-seq analysis with knockdown of OTUB1 in MCF-7 cells. The red underline genes were candidates for next study. C MCF-7 or MB231 cells expressing OTUB1 shRNA were reconstituted expression of HA-rOTUB1. Protein levels were quantitated by WB. D, E Examination of the mRNA levels of TERT, H19, WNT6 and DEPTOR by RT-PCR analyses in breast cancer cells. F, G CHIP assays were performed using OTUB1 knockdown breast cancer cells. The results were normalized against the values of IgG controls. (All data represent mean ± SEM n = 3), *p < 0.05.
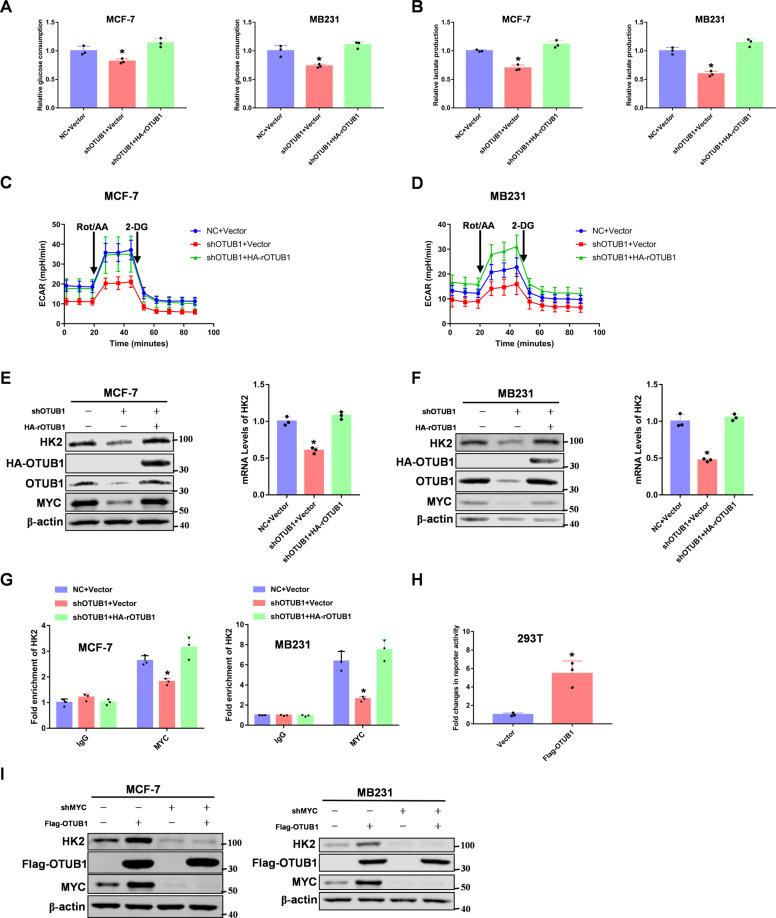
OTUB1 promotes aerobic glycolysis via MYC induced HK2 expression
MYC acts as a master in regulating aerobic glycolysis and mediates multiple of cancer progressions [43]. Our RNA-sequencing results showed that OTUB1 knockdown downregulated expression of HK2, a key enzyme in aerobic glycolysis. So we made a hypothesis that OTUB1 promoted aerobic glycolysis via MYC mediated HK2 expression. To this end, we examined the glucose consumption and lactate production in OTUB1 rescued breast cancer cells. As we expected, OTUB1 promoted glucose consumption and lactate production (Fig. 5A, B). We next measured glycolytic rate in response to OTUB1 knockdown by using Seahorse Bioscience Flux Analyzer. We found that knockdown of OTUB1 reduced glycolytic rate in breast cancer cells (Fig. 5C, D), but had no effect on mitochondrial oxidative phosphorylation (Supplementary Fig. S5A, B). Furthermore, we found that OTUB1 could enhance HK2 expression in breast cancer cells (Fig. 5E, F). In addition, OTUB1 increased MYC binding to the HK2 promoter (Fig. 5G), which was previously reported to be a MYC target gene [44]. Furthermore, luciferase reporter assay showed that overexpression of OTUB1 significantly promoted HK2 transcriptional activity (Fig. 5H). Interestingly, we found OTUB1 mediated HK2 expression depended on MYC in breast cancer cells (Fig. 5I). Taken together, OTUB1 promotes aerobic glycolysis via MYC mediated HK2 expression.
Fig. 5. OTUB1 promotes aerobic glycolysis via MYC induced HK2 expression.
A Glucose consumptions were determined in OTUB1-depleted breast cancer cells rescued with HA-OTUB1. B Lactate productions were determined in OTUB1-depleted breast cancer cells rescued with HA-OTUB1. C, D OTUB1-depleted MCF-7 or MB231 cells were reconstituted expression of HA-OTUB1. ECAR levels were determined by seahorse. E, F OTUB1-depleted MCF-7 or MB231 cells were reconstituted expression of HA-OTUB1. HK2 mRNA and protein levels were determined. G CHIP assays were performed using OTUB1 knockdown breast cancer cells. The results were normalized against the values of IgG controls. H 293 T cells were transfected Vector or Flag-tagged OTUB1 with HK2 reporter plasmids, and detected by luciferase reporter assay. I MCF-7 or MB231 cells were both overexpressed Flag-tagged OTUB1 and knocked down MYC. Immunoblotting analyse was performed. (All data represent mean ± SEM n = 3), *p < 0.05.
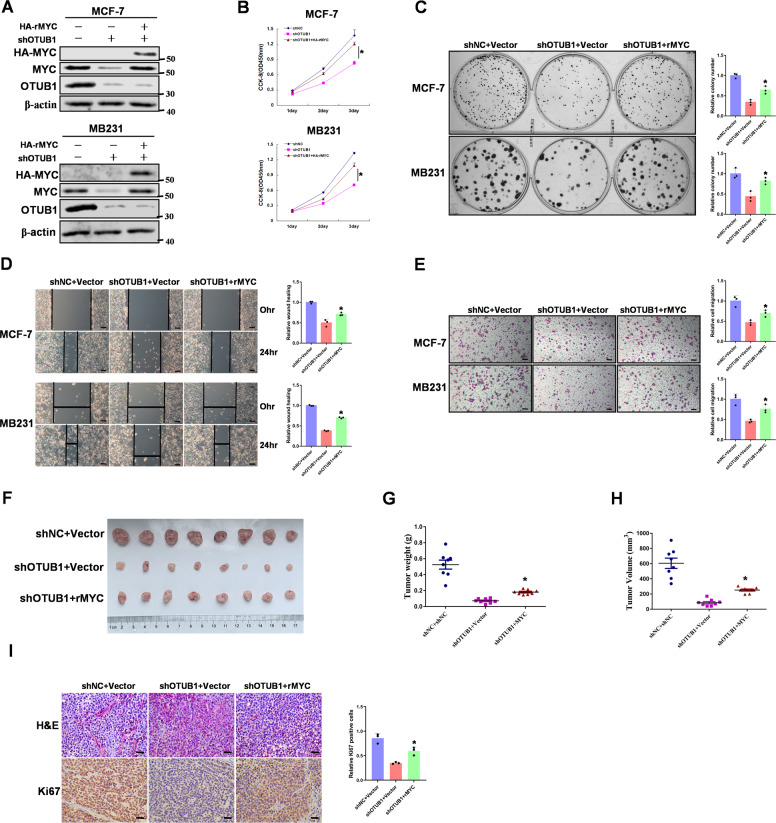
MYC contributes to OTUB1 mediated breast tumorigenesis in vitro and in vivo
Previous studies have reported that OTUB1 is an oncogene in multiple cancers [45]. To further study the functions of the OTUB1/MYC signaling pathway in breast tumorigenesis, we transfected OTUB1-knockdown breast cancer cells with stable overexpression MYC vectors (Fig. 6A). Knocking down OTUB1 significantly decreased breast cell proliferation but overexpression of MYC counteracted this effect (Fig. 6B). Consistently, colony formation assay demonstrated that OTUB1 promoted cell proliferation through its regulation of MYC (Fig. 6C). Moreover, cell-scratch test and transwell migration assay showed that MYC was required for OTUB1 induced cell migration (Fig. 6D, E). To determine whether MYC is involved in OTUB1 induced breast tumor growth in vivo, we performed a xenograft tumor model in nude mice. Compare to the control group, knockdown of OTUB1 resulted in a decrease of tumor growth, whereas rescued overexpression of MYC significantly reversed this suppression (Fig. 6F–H). To test the expression of Ki67 in tumor specimens, we performed IHC staining in collected tumor samples. Consistently, knockdown of OTUB1 led to a great reduction of cell proliferation marker Ki67. In contrast, rescued overexpression of MYC showed a rapid elevation in Ki67 positive staining cells (Fig. 6I). Moreover, we determined the OTUB1, MYC and HK2 protein expressions in theses tissues (Supplementary Fig. S6). These results support the conclusion that MYC is involved in OTUB1 induced breast tumorigenesis in vitro and in vivo.
Fig. 6. MYC contributes to OTUB1 mediated breast tumorigenesis in vitro and in vivo.
A MCF-7 or MB231 cells with OTUB1 depletion reconstituted stable overexpression of MYC. Total cell lysates were prepared, and immunoblotting analyse was performed. B MCF-7 or MB231 cells with OTUB1 depletion reconstituted stable overexpression of MYC. CCK-8 assays were performed. C MCF-7 or MB231 cells with OTUB1 depletion reconstituted stable overexpression of MYC. Clone formation assays were performed. D MCF-7 or MB231 cells with OTUB1 depletion reconstituted stable overexpression of MYC. Wound healing assays were performed (Scale bar, 50 μm). E MCF-7 or MB231 cells with OTUB1 depletion reconstituted stable overexpression of MYC. Cell invasion assays were performed (Scale bar, 50 μm). F–H MCF-7 cells with OTUB1 depletion reconstituted stable overexpression of MYC were subcutaneously injected into nude mice. After 3 weeks, the mice were sacrificed and dissected at the endpoint. Tumor growth and weight were examined. I Representative images of H/E staining and Ki67 staining of tumor samples (Scale bar, 20 μm). (All data represent mean ± SEM n ≥ 3), *p < 0.05.
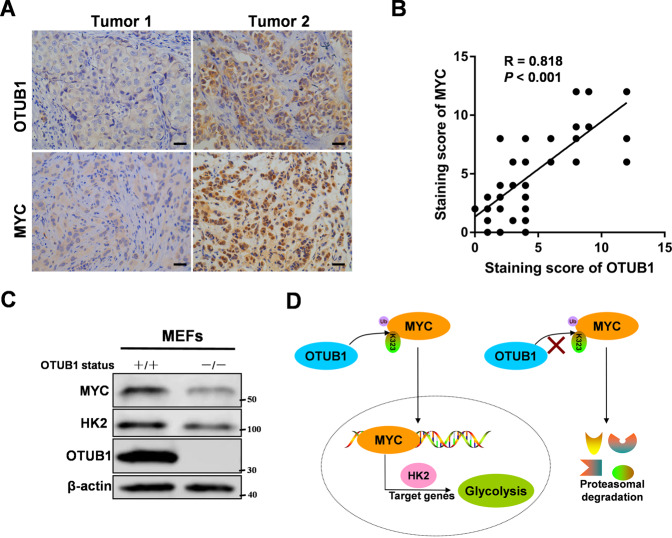
OTUB1 expression is positively correlated with MYC in breast cancer
To further assess the expression relevance of OTUB1 and MYC in breast cancer, we used IHC to analyze expression in 80 cases of breast cancer tissues. Obviously, consistent with our previous observations, OTUB1 and MYC were both highly expressed in breast cancer tissues (Fig. 7A). Furthermore, the expression levels of OTUB1 and MYC positively correlated with each other (Fig. 7B), which was consistent in the triple negative breast cancer (Supplementary Fig. S7A). Moreover, we used an OTUB1 knockout mouse model to derive mouse embryonic fibroblasts (MEFs) from the 13.5 day embryos. Consistently, OTUB1 knockout caused a decreased expression of MYC protein (Fig. 7C), and high expression of OTUB1 in MB231 cell line also had high expressions of MYC and HK2 compared to MCF-7 cell line (Supplementary Fig. S7B). Furthermore, we used the Kaplan–Meier survival analysis to study whether the expression of OTUB1 and MYC correlates with patient survival. It showed that low OTUB1 and MYC expression predicted better patient survival (Supplementary Fig. S7C, D). So the data support the clinical potential of targeting OTUB1-MYC axis as breast cancer therapeutics.
Fig. 7. OTUB1 expression is positively correlated with MYC in breast cancer.
A Representative images of IHC staining of tumor tissues from two breast cancer patients were shown (scale bar, 20 µm). B Pearson correlation analysis was performed to determine correlation between OTUB1 and MYC expression in breast cancer tissues. C OTUB1 depletion causes decrease of MYC protein level in MEFs cells. Immunoblotting analyse was performed. D Schematic model of mechanism that OTUB1 functions as an oncogene to regulate aerobic glycolysis to promote breast tumorigenesis via up-regulating HK2 expression. (All data represent mean ± SEM n ≥ 3), *p < 0.05.
Discussion
MYC as a transcriptional factor is expressed abnormally in 70% human cancers, which controls the cellular metabolism to maintain the tumor growth [46]. Deubiquitination plays a crucial role in the regulation of MYC protein functions. Deubiquitination of MYC by USP37 promotes lung cancer cell proliferation and the Warburg effect in cancer cells [38]. Furthermore, USP22 and USP28 bind to MYC, and enhance its protein stability, which is essential for breast cancer cell proliferation [35, 36]. Moreover, USP16 is found to be a new deubiquitinase of MYC, which promotes castration-resistant prostate cancer cell proliferation [47]. However, there is still no report that the OTU subfamily of deubiquitinases could regulate MYC protein stability. In this study, we identified the OTUB1 as a novel regulator of MYC stability. Our study demonstrated that OTUB1 stablized MYC by direct deubiquitination at K323. Importantly, OTUB1 regulated aerobic glycolysis and breast cell proliferation via MYC mediated HK2 expression. So the OTUB1/MYC axis provides signals to promote glycolysis and thus contributes to cancer cell proliferation.
HK2 as a key enzyme promotes aerobic glycolysis, and reprograms the metabolic flux in cancer cells. HK2 expression is regulated by several transcriptional factors, such as androgen receptor (AR) [48], STAT3 [49], NF-κB [50], MYC [44], and HIF-1 [44]. At the transcriptional level, we identified OTUB1 was a new regulator for HK2 expression via promoting MYC protein stability, and illuminated that the new oncogene function of OTUB1 could promote aerobic glycolysis. The upstream of factors regulating OTUB1 pathway may need further study.
Taken together, we find a new substrate of deubiquitinase OTUB1, the transcriptional factor MYC, and build the relationship between OTUB1 and aerobic glycolysis. We illuminate the molecular mechanism of OTUB1 mediated MYC protein stability through the ubiquitin proteasome pathway, and demonstrate the functions of OTUB1 in promoting breast tumorigenesis in vitro and in vivo (Fig. 7D). Thus, OTUB1 serves as a potential therapeutic target in breast cancer.
Supplementary information
Author contributions
XH, CR and CL performed experiments and analyzed data. PQ and TY provided access to material and facilities and contributed reagents. ZY designed, supervised the project, and wrote the manuscript.
Funding
The study was supported by research grants from National Natural Science Foundation of China (Grant no. 81972489 and 82003201), National Natural Science Foundation of Shandong Province (Grant no. ZR2020YQ58 and ZR2020QH255), Taishan Scholar Program of Shandong Province (Grant no. tsqn202103113), Shandong Province College Science and Technology Plan Project (Grant no. J17KA254), Projects of medical and health technology development program in Shandong province (Grant no. 2018WS057).
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The procedures related to human subjects were approved by the Ethics Committee of Weifang Medical University. Animal experiments were performed according to the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals, and were approved by the ethics committee of Weifang Medical University (Weifang, China).
Footnotes
Edited by S Kumar
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xue Han, Chune Ren, Chao Lu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-022-00971-8.
References
- 1.Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24:3353–9. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herhaus L, Al-Salihi M, Macartney T, Weidlich S, Sapkota GP. OTUB1 enhances TGFbeta signalling by inhibiting the ubiquitylation and degradation of active SMAD2/3. Nat Commun. 2013;4:2519. doi: 10.1038/ncomms3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun XX, Challagundla KB, Dai MS. Positive regulation of p53 stability and activity by the deubiquitinating enzyme Otubain 1. EMBO J. 2012;31:576–92. doi: 10.1038/emboj.2011.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li S, Zheng H, Mao AP, Zhong B, Li Y, Liu Y, et al. Regulation of virus-triggered signaling by OTUB1- and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. J Biol Chem. 2010;285:4291–7. doi: 10.1074/jbc.M109.074971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Y, Xu M, Tong J, Tang X, Chen J, Chen X, et al. Targeting the Otub1/c-Maf axis for the treatment of multiple myeloma. Blood. 2021;137:1478–90. doi: 10.1182/blood.2020005199. [DOI] [PubMed] [Google Scholar]
- 6.Zhu D, Xu R, Huang X, Tang Z, Tian Y, Zhang J, et al. Deubiquitinating enzyme OTUB1 promotes cancer cell immunosuppression via preventing ER-associated degradation of immune checkpoint protein PD-L1. Cell Death Differ. 2021;28:1773–89. doi: 10.1038/s41418-020-00700-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liao Y, Wu N, Wang K, Wang M, Wang Y, Gao J, et al. OTUB1 promotes progression and proliferation of prostate cancer via deubiquitinating and stabling cyclin E1. Front Cell Dev Biol. 2020;8:617758. doi: 10.3389/fcell.2020.617758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karunarathna U, Kongsema M, Zona S, Gong C, Cabrera E, Gomes AR, et al. OTUB1 inhibits the ubiquitination and degradation of FOXM1 in breast cancer and epirubicin resistance. Oncogene. 2016;35:1433–44. doi: 10.1038/onc.2015.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Yang JY, Xie X, Jie Z, Zhang L, Shi J, et al. Preventing abnormal NF-kappaB activation and autoimmunity by Otub1-mediated p100 stabilization. Cell Res. 2019;29:474–85. doi: 10.1038/s41422-019-0174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu T, Jiang L, Tavana O, Gu W. The deubiquitylase OTUB1 mediates ferroptosis via stabilization of SLC7A11. Cancer Res. 2019;79:1913–24. doi: 10.1158/0008-5472.CAN-18-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou H, Liu Y, Zhu R, Ding F, Cao X, Lin D, et al. OTUB1 promotes esophageal squamous cell carcinoma metastasis through modulating Snail stability. Oncogene. 2018;37:3356–68. doi: 10.1038/s41388-018-0224-1. [DOI] [PubMed] [Google Scholar]
- 12.Baietti MF, Simicek M, Abbasi Asbagh L, Radaelli E, Lievens S, Crowther J, et al. OTUB1 triggers lung cancer development by inhibiting RAS monoubiquitination. EMBO Mol Med. 2016;8:288–303. doi: 10.15252/emmm.201505972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang HH, Li C, Ren JW, Liu L, Du XH, Gao J, et al. OTUB1 facilitates bladder cancer progression by stabilizing ATF6 in response to endoplasmic reticulum stress. Cancer Sci. 2021;112:2199–209. doi: 10.1111/cas.14876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koschel J, Nishanth G, Just S, Harit K, Kroger A, Deckert M, et al. OTUB1 prevents lethal hepatocyte necroptosis through stabilization of c-IAP1 during murine liver inflammation. Cell Death Differ. 2021;28:2257–75. doi: 10.1038/s41418-021-00752-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Q, Huang Y, Gu L, Chang Z, Li GM. OTUB1 stabilizes mismatch repair protein MSH2 by blocking ubiquitination. J Biol Chem. 2021;296:100466. doi: 10.1016/j.jbc.2021.100466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang P, Joberty G, Buist A, Vanoosthuyse A, Stancu IC, Vasconcelos B, et al. Tau interactome mapping based identification of Otub1 as Tau deubiquitinase involved in accumulation of pathological Tau forms in vitro and in vivo. Acta Neuropathol. 2017;133:731–49. doi: 10.1007/s00401-016-1663-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iglesias-Gato D, Chuan YC, Jiang N, Svensson C, Bao J, Paul I, et al. OTUB1 de-ubiquitinating enzyme promotes prostate cancer cell invasion in vitro and tumorigenesis in vivo. Mol Cancer. 2015;14:8. doi: 10.1186/s12943-014-0280-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Y, Wu J, Fu X, Du W, Zhou L, Meng X, et al. OTUB1 promotes metastasis and serves as a marker of poor prognosis in colorectal cancer. Mol Cancer. 2014;13:258. doi: 10.1186/1476-4598-13-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ni Q, Chen J, Li X, Xu X, Zhang N, Zhou A, et al. Expression of OTUB1 in hepatocellular carcinoma and its effects on HCC cell migration and invasion. Acta Biochim Biophys Sin (Shanghai) 2017;49:680–8. doi: 10.1093/abbs/gmx056. [DOI] [PubMed] [Google Scholar]
- 20.Xu L, Li J, Bao Z, Xu P, Chang H, Wu J, et al. Silencing of OTUB1 inhibits migration of human glioma cells in vitro. Neuropathology. 2017;37:217–26. doi: 10.1111/neup.12366. [DOI] [PubMed] [Google Scholar]
- 21.Dejure FR, Eilers M. MYC and tumor metabolism: chicken and egg. EMBO J. 2017;36:3409–20. doi: 10.15252/embj.201796438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baluapuri A, Wolf E, Eilers M. Target gene-independent functions of MYC oncoproteins. Nat Rev Mol Cell Biol. 2020;21:255–67. doi: 10.1038/s41580-020-0215-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsieh AL, Walton ZE, Altman BJ, Stine ZE, Dang CV. MYC and metabolism on the path to cancer. Semin Cell Dev Biol. 2015;43:11–21. doi: 10.1016/j.semcdb.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng X, Boyer L, Jin M, Mertens J, Kim Y, Ma L. Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. Elife. 2016;5:e13374. doi: 10.7554/eLife.13374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horiuchi D, Camarda R, Zhou AY, Yau C, Momcilovic O, Balakrishnan S, et al. PIM1 kinase inhibition as a targeted therapy against triple-negative breast tumors with elevated MYC expression. Nat Med. 2016;22:1321–9. doi: 10.1038/nm.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hann SR. Role of post-translational modifications in regulating c-Myc proteolysis, transcriptional activity and biological function. Semin Cancer Biol. 2006;16:288–302. doi: 10.1016/j.semcancer.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 28.Ren C, Han X, Lu C, Yang T, Qiao P, Sun Y. Ubiquitination of NF-kappaB p65 by FBXW2 suppresses breast cancer stemness, tumorigenesis, and paclitaxel resistance. Cell Death Differ. 2022;29:381–392. doi: 10.1038/s41418-021-00862-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu C, Qiao P, Sun Y, Ren C, Yu Z. Positive regulation of PFKFB3 by PIM2 promotes glycolysis and paclitaxel resistance in breast cancer. Clin Transl Med. 2021;11:e400. doi: 10.1002/ctm2.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu C, Ren C, Yang T, Sun Y, Qiao P, Han X, et al. Fructose-1, 6-bisphosphatase 1 interacts with NF-kappaB p65 to regulate breast tumorigenesis via PIM2 induced phosphorylation. Theranostics. 2020;10:8606–18. doi: 10.7150/thno.46861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang T, Ren C, Lu C, Qiao P, Han X, Wang L, et al. Phosphorylation of HSF1 by PIM2 Induces PD-L1 expression and promotes tumor growth in breast cancer. Cancer Res. 2019;79:5233–44. doi: 10.1158/0008-5472.CAN-19-0063. [DOI] [PubMed] [Google Scholar]
- 32.Yang T, Ren C, Qiao P, Han X, Wang L, Lv S, et al. PIM2-mediated phosphorylation of hexokinase 2 is critical for tumor growth and paclitaxel resistance in breast cancer. Oncogene. 2018;37:5997–6009. doi: 10.1038/s41388-018-0386-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Q, Fu Y, Li L, Liu CH, Zhang L. The functions and regulation of Otubains in protein homeostasis and diseases. Ageing Res Rev. 2021;67:101303. doi: 10.1016/j.arr.2021.101303. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J, Zhang P, Wei Y, Piao HL, Wang W, Maddika S, et al. Deubiquitylation and stabilization of PTEN by USP13. Nat Cell Biol. 2013;15:1486–94. doi: 10.1038/ncb2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim D, Hong A, Park HI, Shin WH, Yoo L, Jeon SJ, et al. Deubiquitinating enzyme USP22 positively regulates c-Myc stability and tumorigenic activity in mammalian and breast cancer cells. J Cell Physiol. 2017;232:3664–76. doi: 10.1002/jcp.25841. [DOI] [PubMed] [Google Scholar]
- 36.Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R, et al. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol. 2007;9:765–74. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- 37.Sun XX, He X, Yin L, Komada M, Sears RC, Dai MS. The nucleolar ubiquitin-specific protease USP36 deubiquitinates and stabilizes c-Myc. Proc Natl Acad Sci USA. 2015;112:3734–9. doi: 10.1073/pnas.1411713112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan J, Deng Q, Jiang C, Wang X, Niu T, Li H, et al. USP37 directly deubiquitinates and stabilizes c-Myc in lung cancer. Oncogene. 2015;34:3957–67. doi: 10.1038/onc.2014.327. [DOI] [PubMed] [Google Scholar]
- 39.Wu KJ, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, et al. Direct activation of TERT transcription by c-MYC. Nat Genet. 1999;21:220–4. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- 40.Barsyte-Lovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, Andrulis IL, et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66:5330–7. doi: 10.1158/0008-5472.CAN-06-0037. [DOI] [PubMed] [Google Scholar]
- 41.Schaale K, Brandenburg J, Kispert A, Leitges M, Ehlers S, Reiling N. Wnt6 is expressed in granulomatous lesions of Mycobacterium tuberculosis-infected mice and is involved in macrophage differentiation and proliferation. J Immunol. 2013;191:5182–95. doi: 10.4049/jimmunol.1201819. [DOI] [PubMed] [Google Scholar]
- 42.Wang Q, Zhou Y, Rychahou P, Harris JW, Zaytseva YY, Liu J, et al. Deptor Is a Novel Target of Wnt/beta-Catenin/c-Myc and contributes to colorectal cancer cell growth. Cancer Res. 2018;78:3163–75. doi: 10.1158/0008-5472.CAN-17-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong Y, Tu R, Liu H, Qing G. Regulation of cancer cell metabolism: oncogenic MYC in the driver’s seat. Signal Transduct Target Ther. 2020;5:124. doi: 10.1038/s41392-020-00235-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim JW, Gao P, Liu YC, Semenza GL, Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol. 2007;27:7381–93. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saldana M, VanderVorst K, Berg AL, Lee H, Carraway KL. Otubain 1: a non-canonical deubiquitinase with an emerging role in cancer. Endocr Relat Cancer. 2019;26:R1–14. doi: 10.1530/ERC-18-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lourenco C, Resetca D, Redel C, Lin P, MacDonald AS, Ciaccio R, et al. MYC protein interactors in gene transcription and cancer. Nat Rev Cancer. 2021;21:579–91. doi: 10.1038/s41568-021-00367-9. [DOI] [PubMed] [Google Scholar]
- 47.Ge J, Yu W, Li J, Ma H, Wang P, Zhou Y, et al. USP16 regulates castration-resistant prostate cancer cell proliferation by deubiquitinating and stablizing c-Myc. J Exp Clin Cancer Res. 2021;40:59. doi: 10.1186/s13046-021-01843-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moon JS, Jin WJ, Kwak JH, Kim HJ, Yun MJ, Kim JW, et al. Androgen stimulates glycolysis for de novo lipid synthesis by increasing the activities of hexokinase 2 and 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2 in prostate cancer cells. Biochem J. 2011;433:225–33. doi: 10.1042/BJ20101104. [DOI] [PubMed] [Google Scholar]
- 49.Li M, Jin R, Wang W, Zhang T, Sang J, Li N, et al. STAT3 regulates glycolysis via targeting hexokinase 2 in hepatocellular carcinoma cells. Oncotarget. 2017;8:24777–84. doi: 10.18632/oncotarget.15801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Londhe P, Yu PY, Ijiri Y, Ladner KJ, Fenger JM, London C, et al. Classical NF-kappaB metabolically reprograms sarcoma cells through regulation of hexokinase 2. Front Oncol. 2018;8:104. doi: 10.3389/fonc.2018.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.