Abstract
Although chemotherapy and recently approved immunotherapies have improved treatment of triple-negative breast cancer (TNBC), the clinical outcome for this deadly disease remains unsatisfactory. We found that both cluster of differentiation 73 (CD73) and transforming growth factor (TGF)β were elevated in TNBC and correlated with the epithelial–mesenchymal transition (EMT), fibrotic stroma, an immune-tolerant tumor environment, and poor prognosis. To explore the efficacy of CD73-TGFβ dual-blockade, we generated a bifunctional anti-CD73-TGFβ construct consisting of the CD73 antibody MEDI9447 fused with the TGFβRII extracellular-domain (termed MEDI-TGFβR). MEDI-TGFβR retained full and simultaneous blocking efficiency for CD73 and TGFβ. Compared with MEDI9447 activity alone, MEDI-TGFβR demonstrated superior inhibitory activity against CD73-dependent cell migration and the EMT in CD73-high TNBC cells and effectively reduced lung metastasis in a syngeneic mouse model of TNBC. Mechanistically, the CD73-TGFβ dual-blockade reverted the EMT and stromal fibrosis and induced tumor cell death, which was accompanied by the accumulation of M1-macrophages and production of tumor necrosis factor α (TNFα). The CD73-TGFβ dual-blockade promoted a multifaceted inflammatory tumor microenvironment, as shown by the diminished levels of myeloid-derived suppressor cells (MDSCs) and M2-macrophages, and substantially increased levels of activated dendritic cells, cytotoxic T cells, and B cells. Collectively, our results have highlighted a novel strategy for TNBC treatment.
Keywords: CD73, NT5E, TGFβ, TNBC, antitumor immunity
Introduction
In the Global Cancer Statistics 2020 report, breast cancer surpassed lung cancer as the most commonly diagnosed cancer worldwide and accounts for 11.7% of all new cancer cases [1]. Patients who lack the estrogen receptor, progesterone receptor, and ERBB2/HER2 are diagnosed with triple-negative breast cancer (TNBC), which constitutes ~15% of all breast cancer cases. TNBC comprises a genetically heterogeneous population with the majority of cases classified as a basal-like phenotype, which manifests a clinically high incidence of metastasis and aggressive clinical course [2]. Although chemotherapy and recently approved immunotherapies are established treatment paradigms for TNBC, the overall survival for metastatic TNBC remains ~18 months [3, 4]. Systemic chemotherapy inevitably incurs rapid drug resistance, and immunotherapy generally results in low response rates [5]. Therefore, there is an urgent need to develop new treatment strategies to improve outcome.
Cluster of differentiation 73 (CD73), also known as 5ʹ-nucleotidase exo (NT5E), is a membrane-bound protein encoded by the NT5E gene. CD73 is the enzyme responsible for the generation of extracellular adenosine (ADO) via enzymatic dephosphorylation of adenosine monophosphate. Several reports have demonstrated that elevated CD73 expression is associated with poor clinical outcomes across diverse cancer cohorts [6–9]. Prolonged periods of inflammation, hypoxia, and anti-cancer drugs can induce aberrant CD73 expression, which is tightly correlated with tumor plasticity, immune evasion, and drug resistance [10]. In recent analyses of clinical data from TNBC patients, a pathological complete response to neoadjuvant chemotherapy was obtained in 53% of cases with low CD73 versus 21% of cases with high CD73 tumor expression [11], and a clear association existed between both low CD73 expression and robust tumor infiltrating lymphocytes and a better treatment outcome [12]. It has been reported that high CD73 expression promotes anthracycline resistance, and CD73 gene expression itself can be induced by chemotherapy [13, 14]. Recently, Weinberg et al. demonstrated that abrogation of CD73 in quasi-mesenchymal carcinoma cells sensitized these tumor cells to anti-CTLA4 immunotherapy [15]. These studies suggest that CD73 plays a role in drug resistance and tumor progression in TNBC and represents a potential target for therapy development.
Transforming growth factor β (TGFβ) is a well-established immunosuppressive cytokine and potent regulator of the epithelial–mesenchymal transition (EMT). TGFβ is abundantly expressed in the TNBC microenvironment and positively correlates with higher histologic tumor grade and reduced disease-free survival (DFS) [16]. TGFβ forms a regenerative feedback loop with CD73 in both tumor cells [17, 18] and various immune cells [19–23]. Cooperation between CD73 and TGFβ can further aggravate the EMT process, cell adhesion and dissemination, and immune evasion. Chalmin et al. [19] and Li et al. [20] reported that TGFβ stimulated CD73 expression in Th17 cell and MDSC, respectively, through distinct signaling pathways. Furthermore, elevated levels of ADO inhibited T cell and NK cell activity, thereby protecting tumor cells from the cytotoxic effects of chemotherapy. In addition, a TGFβ-rich tumor milieu confers resistance to anti-4-1BB immunotherapy by sustaining CD73 expression primarily on the surface of infiltrating CD8+ T cells in several tumor models [23]. Targeting TGFβ alone has not been highly successful in the clinic [24] and may be associated with cardiac safety concerns [25]. However, given the co-presence of aberrantly high levels of CD73 and TGFβ in the tumor milieu, specifically targeting both of these proteins in CD73-high tumor cells and/or in the tumor environment may offer a more effective therapeutic strategy.
Herein, we have generated a bifunctional anti-CD73-TGFβ agent (termed MEDI-TGFβR) to simultaneously inhibit CD73 and TGFβ pathways. Bioinformatics analysis, in vitro experiments, and preclinical mouse models validated high CD73 expression and rich TGFβ accumulation in TNBC tissue, and we explored the potential utility of CD73-TGFβ dual-blockade as a novel cancer therapy.
Materials and methods
CD73 transcriptome analysis and tumor tissue microarrays
Genome expression profile data of breast cancer patient samples were obtained from The Cancer Genome Atlas (TCGA, n = 990, https://www.cancer.gov). The classification standard for basal-like/TNBC was described previously [26]. Survival analysis was conducted in GraphPad Prism 8.0, and the log-rank test was used to compare DFS between groups. The transcriptome data sets of 28 luminal and 30 basal breast cancer cell lines [27] were obtained from the Cancer Cell Line Encyclopedia (CCLE, https://portals.broadinstitute.org/ccle). All of the heat maps, box diagrams, and correlation plots were generated using GraphPad Prism 8.0. CIBERSORT, an analytical tool developed by Newman et al. [28], was used to analyze the immune cell fractions of all samples from TCGA. CD73 or CD8 protein was assayed using breast cancer tissue microarrays (TMA) (Outdo Biotech, Cat. #HBreD050Bc01) with anti-CD73 (Cell Signaling Technology, Cat. #13160) or anti-CD8 (Abcarta, Cat. #PA067).
Cell culture, gene knockdown, and surface CD73 levels in cancer cell lines
MDA-MB-231 (MDA-231), MDA-MB-453 (MDA-453), and HEK293T cell lines were obtained from the American Type Culture Collection (ATCC). The cell line 4T1 was a gift from Professor Qi-zhi Zhang (Fudan University). Cells were cultured using standard methods and reagents (Invitrogen). The cell lines were periodically tested for mycoplasma contamination. MDA-231 cells were stably infected with the pTRIPZ-based short hairpin (sh)RNA lentiviral vectors for human CD73/NT5E or vectors containing a nontargeting shRNA sequence (Open Biosystems, Cat. #V3LH_359876, Cat. #RHS_4346). CD73 gene depletion was induced by treating the cells with 1 μg/mL doxycycline for 5–7 days. Cell surface expression of CD73 in various mouse and human tumor lines was quantified by flow cytometry using the anti-CD73 antibody MEDI9447 (see below).
Construction, production, and quantification of MEDI-TGFβR
MEDI9447 (MEDI) is a clinical-stage anti-human/mouse CD73 monoclonal antibody [29]. MEDI-TGFβR is an anti-CD73 antibody-TGFβ receptor II fusion protein. Its light chain (LC) is identical to the LC of MEDI. The heavy chain (HC) of MEDI-TGFβR was constructed by double digestion of MEDI’s HC and ligation with the N-terminal extracellular domain of TGFβRII via a flexible (Gly4Ser)4Gly linker [30]. The HC and LC expression plasmids were co-transfected into HEK293T cells using Lipofectamine™ 2000 (Thermo Fisher, Cat. #11-668-019), and the cells were cultured in serum-free medium for 5–6 days before harvest. The antibody and fusion proteins were purified with protein G (GE healthcare, Cat. #17061801) and quantified with standard protocols. The purified proteins were examined by SDS-PAGE under reducing and non-reducing conditions.
MEDI-TGFβR target binding and inhibition of CD73 enzymatic activity
The ability of MEDI-TGFβR to bind TGFβ was measured using a standard enzyme-linked immunosorbent assay (ELISA) protocol. Briefly, 96-well ELISA plates were coated with recombinant human TGFβ1 (0.5 μg/mL), and serial dilutions (1:3) of human IgG1 (hIgG1), anti-TGFβ (Invitrogen, Cat. #16-9243-85), MEDI, or MEDI-TGFβR were added to the wells and incubated for 2 h at room temperature. Binding was detected with a horseradish peroxidase-labeled secondary antibody (Jackson ImmunoResearch, Cat. #109-035-003). The binding of MEDI-TGFβR to CD73 on the cell surface was assayed using flow cytometry. Serial dilutions (1:3) of hIgG1, MEDI, or MEDI-TGFβR were added to 1 × 105 tumor cells in 96-well U-bottom plate and incubated for 1 h at 4 °C. The cells were stained with phycoerythrin-conjugated anti-human IgG-Fc (Abcam, Cat. #ab98956) and analyzed by flow cytometry. An enzyme inhibition assay was used to determine CD73 enzymatic activity in various tumor cell lines as previously described [31].
Cell migration and lung metastasis
Tumor cells were suspended in 200 μL of serum-free medium containing 66.7 nmol/L hIgG1, anti-TGFβ, MEDI, or MEDI-TGFβR and added to the upper chambers of a Trans-well system with 8-μm pore size (Corning, Cat. #3422). The lower chambers contained medium with 10% serum and the identical concentrations of drugs used in the upper chamber. After an 8–10 h incubation, the migrated tumor cells were stained with 0.2% crystal violet and counted under a microscope. Cells from each treatment were counted in five representative viewing fields at ×200 magnification.
In vivo efficacy studies were performed under protocols approved by the Institutional Animal Care and Use Committee of Fudan University. In the first set of experiments, 5 × 105 4T1 cells were pre-incubated with 50 μg hIgG1 or MEDI for 30 min at 4 °C before tail vein injection into female Balb/c mice (5–6 weeks old). In the second set of experiments, a therapeutic treatment protocol was used. Balb/c mice were injected intravenously with 10 mg/kg hIgG1, MEDI, or 12.5 mg/kg MEDI-TGFβR. The next day (Day 0), 5 × 105 4T1 cells were injected into the tail vein. The mice were treated intravenously on days 1 and 8 after 4T1 cell injection with the identical doses of drugs as described above. Mice were euthanized and dissected on day 15, and lung tissues were fixed and subjected to histological analysis.
Vimentin or surface CD73 immunofluorescence in cultured cells
Tumor cells were seeded on culture chamber slides and treated with 33.3 nmol/L hIgG1, anti-TGFβ, MEDI, or MEDI-TGFβR for 24 h. The slides were processed and incubated with anti-CD73 (MEDI9447) or anti-vimentin (Abcam, Cat. #ab92547) antibodies at 4 °C overnight. Immunofluorescence was detected by subsequent staining with phycoerythrin-conjugated anti-human IgG-Fc (Abcam, Cat. #ab98956) or Alexa Fluor 488-conjugated anti-rabbit IgG (Jackson ImmunoResearch, Cat. #711-545-152). Images were acquired using a Leica immunofluorescent microscope (model DMI4000D) with ×200 magnification.
Lung tissue immunohistochemistry and immunofluorescence
Mouse lung tissues were fixed in 4% formalin, embedded in paraffin, and sectioned. Tissue sections were stained with hematoxylin-eosin or Masson’s trichrome stains or by immunohistochemistry (IHC) using standard protocols. In some assays, multiplexed IHC was performed with tyramide signal amplification [32] and dyed with Tyr-488 and Tyr-CY3 (Record Biological Technology). The IHC antibodies are listed as follows: anti-α-SMA (Servicebio, Cat. #GB13044), anti-CD73 (Cell Signaling Technology, Cat. #13160), anti-TGFB1 (Abcam, Cat. #ab215715), anti-vimentin (Abcam, Cat. #ab92547), anti-F4/80 (CST, Cat. #70076), anti-CD206 (Abcam, Cat. #ab64693), anti-TNFα (Arigo, Cat. #ARG10158), anti-CD11c (Servicebio, Cat. #GB11059), anti-CD103 (Abcam, Cat. #ab224202), anti-CD8A (Sino Biological, Cat. #50389-T26), anti-CD19 (Abcam, Cat. #ab245235), and anti-GZMB (Abcam, Cat. #ab255598). Images were acquired using the Leica microscope with ×200 magnification.
Statistical analysis
All numerical data are presented as mean ± standard deviation (SD). Numerical data processing and statistical analyses were performed with Microsoft Excel and GraphPad Prism 8 software. For IHC of lung sections, results from 3 to 5 independent images per lung (n = 3) were pooled, analyzed, and quantified using ImageJ software. P values were calculated using unpaired, two-tailed Student t-tests. In all tests, P values < 0.05 were considered to be statistically significant.
Results
High CD73 expression in basal-like/TNBC correlates with elevated EMT, fibrotic stroma, an immune-tolerant tumor microenvironment, and poor prognosis
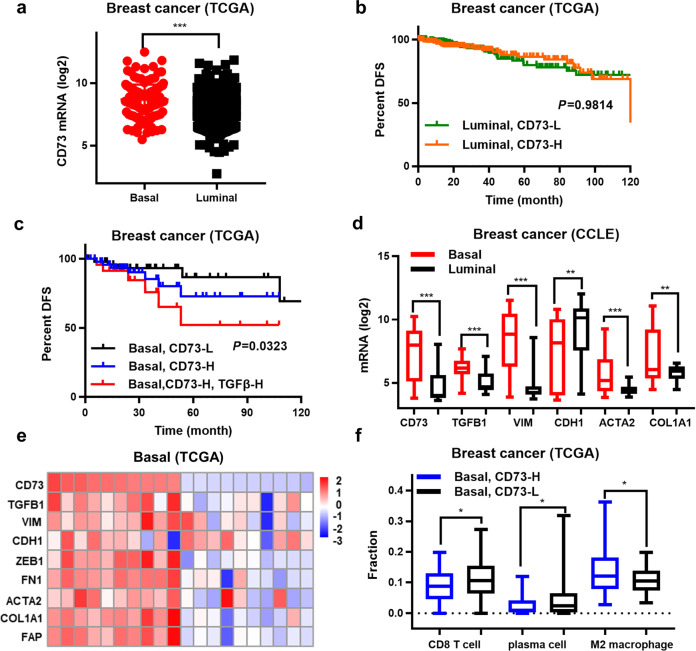
Approximately 70% of TNBC tumors present with a “basal-like” phenotype based on gene expression analysis [2, 5]. We clustered breast cancer patient samples from TCGA database into “basal-like” or “luminal-like” groups according to their transcriptome profiles [26]. The transcriptome analysis revealed higher CD73/NT5E mRNA expression in the basal-like/TNBC group compared with that of the luminal-like group (P < 0.001) (Fig. 1a), and high CD73 expression was associated with poor DFS (Fig. 1b, c). Additionally, high TGFB1 mRNA expression in the basal-like/TNBC and CD73-high groups further reduced DFS (P < 0.05) (Fig. 1c). High CD73 mRNA expression was co-enriched for the gene signatures of EMT (TGFB1, VIM, FN1, ZEB1) and high-density fibrotic stroma (ACTA2, COL1A1, FAP) when both TCGA and CCLE datasets were analyzed (Fig. 1d, e). To explore the relationship between CD73 expression and immune cell infiltration, the CIBERSORT algorithm was applied to the TCGA cohort. CD8+ T lymphocyte and plasma cell populations were lower in the CD73-high group than the CD73-low group. However, the levels of M2-like tumor-associated macrophages were higher in the CD73-high group than in the CD73-low group (P < 0.05) (Fig. 1f). These results indicate that the aberrant CD73 transcriptome in TNBC may contribute to a high EMT state, fibrotic stroma, and an immune-tolerant tumor environment, which promotes disease progression.
Fig. 1. High CD73 expression in basal-like/triple-negative breast cancer (TNBC) correlates with elevated EMT, fibrotic stroma, an immune-tolerant tumor microenvironment, and poor prognosis.
a CD73 mRNA expression in basal-like/TNBC (n = 57) vs. luminal-like (n = 57) patients (The Cancer Genome Atlas [TCGA] datasets were accessed in March 2021). Kaplan–Meier analysis of disease-free survival (DFS) was stratified according to CD73 or TGFB1 mRNA expression in the b luminal-like group and c basal-like/TNBC group. d The transcriptome signature of the EMT and extracellular matrix markers in basal-like/TNBC vs. luminal cell lines. e EMT and extracellular matrix markers in 20 basal-like/TNBC patients with the highest and lowest CD73 expression. f Fractions of CD8+ T cells, plasma cells, and M2 macrophages in the TCGA cohort were analyzed by CIBERSORT. *P < 0.05, **P < 0.01, ***P < 0.001.
Anti-CD73 antibody MEDI9447 reduces cell migration and lung metastasis in a mouse model of TNBC
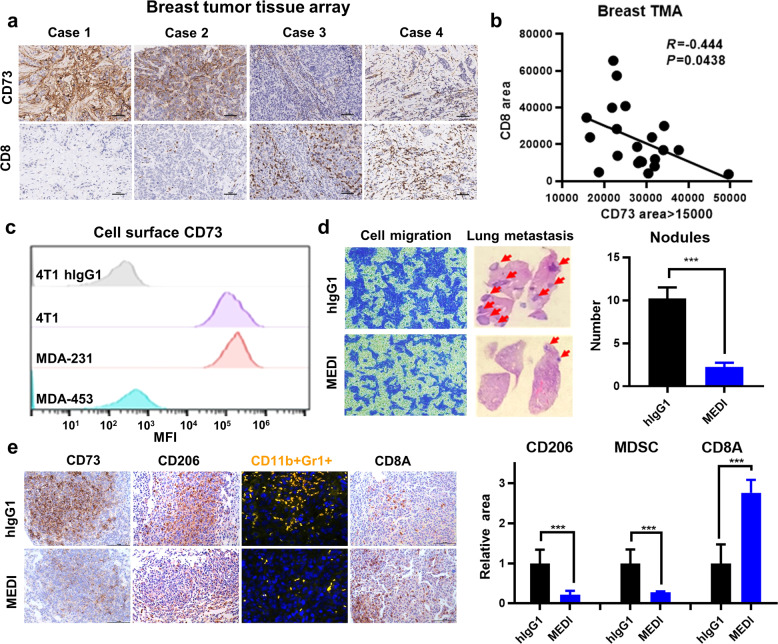
Based on the transcriptome analysis above, we performed IHC using breast cancer TMA. The level of CD73 protein was inversely correlated with the tissue infiltration of CD8+ T cells (P < 0.05) (Fig. 2a, b). To investigate the effect of CD73 inhibition on TNBC lung metastasis, we treated 4T1 cells with hIgG1 or MEDI [29]. The 4T1 cell line is a well-established mouse TNBC line with elevated CD73 surface expression (Fig. 2c). Cell migration of 4T1 cells was markedly inhibited by the anti-CD73 MEDI antibody compared with that in the hIgG1 control group (Fig. 2d). When 4T1 cells were pre-incubated for 30 min with MEDI or hIgG1 and then injected into Balb/c mice, the hematogenous metastasis to the lung was substantially reduced in the MEDI-treated group compared with that in the hIgG1 group (P < 0.001) (Fig. 2d). We examined immune infiltration patterns in the metastatic lung nodules. In the MEDI-treated group, CD73 expression in the tumor area was decreased and accompanied by substantial decreases in CD206+ tumor-associated macrophages and CD11b + Gr1+ MDSCs, while CD8A+ T cell infiltration was markedly increased compared with that of the hIgG1 group (P < 0.001) (Fig. 2e). These results suggest that CD73 plays an important role in metastasis, which may be associated with a tumor-promoting immune-tolerant microenvironment.
Fig. 2. The anti-CD73 antibody MEDI9447 reduces cell migration and lung metastasis in a mouse model of TNBC.
a Representative immunohistochemistry (IHC) images of breast cancer tissue microarray (TMA) cases showing CD73 and CD8 protein expression. b The TMA IHC staining scores for CD73 and CD8 expression were correlated. c The CD73 cell surface levels for the indicated mouse and human cell lines were measured by flow cytometry using the anti-CD73 antibody MEDI9447. d Cell migration (left) and lung metastatic nodules (right) in the TNBC 4T1 cell culture and mouse models, respectively. e Lung tissues were analyzed by IHC staining with anti-CD73, anti-CD206, and anti-CD8A antibodies, and dual immunofluorescence was performed with anti-CD11b and anti-Gr1 antibodies. The IHC shows tumor infiltration by various immune cells within the metastatic nodules. ***P < 0.001.
Design and characterization of MEDI-TGFβR for dual-blockade of CD73 and TGFβ
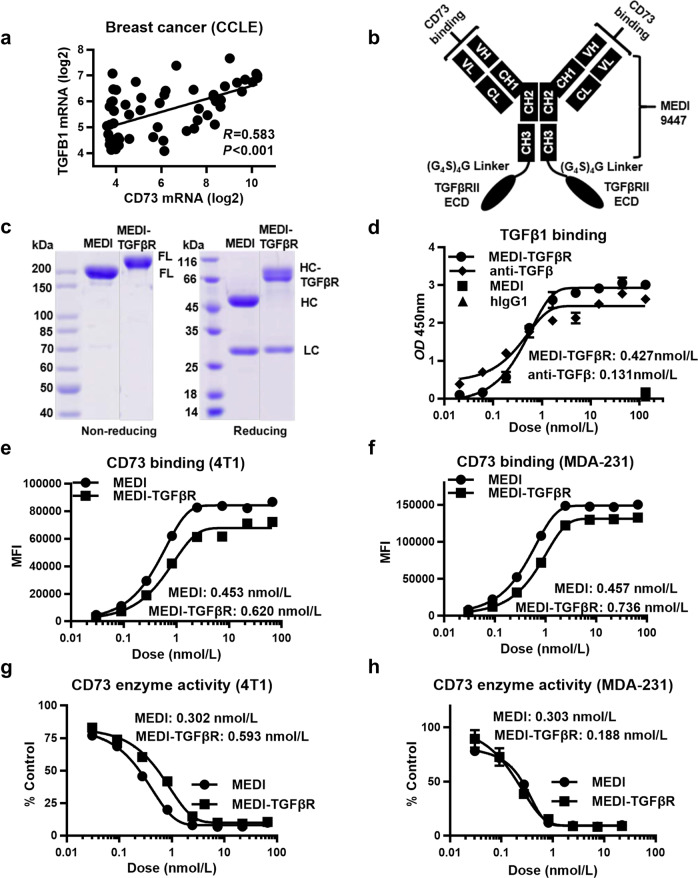
TGFβ may play an important role in TNBC by regulating the EMT and drug resistance [33]. We found that TGFβ1 expression was positively associated with CD73 expression (Fig. 3a). The coexistence and interplay of CD73 and TGFβ presents a dynamic and complicated problem for treating TNBC, and a systemic anti-TGFβ treatment may not produce a favorable therapeutic window [24, 25]. Therefore, we created a bifunctional MEDI-TGFβR fusion protein to specifically target both CD73 and TGFβ in tumors with high CD73 expression. The fusion construct was designed (Fig. 3b) and prepared as described in the Methods section. In this scenario, the MEDI-TGFβR was expected to accumulate in CD73-high tumors and downregulate CD73 activity, while also binding to TGFβ in the tumor environment. SDS-PAGE under reducing and non-reducing conditions verified the molecular identity and integrity of the fusion protein (Fig. 3c). In the ELISA assays, both anti-TGFβ and MEDI-TGFβR efficiently bound to free TGFβ1 with EC50 values of 0.131 ± 0.004 nmol/L and 0.427 ± 0.029 nmol/L, respectively (Fig. 3d). MEDI-TGFβR retained binding abilities similar to MEDI for cell surface CD73. EC50 values for MEDI-TGFβR and MEDI were 0.620 ± 0.341 and 0.453 ± 0.099 nmol/L in mouse 4T1 cells and 0.736 ± 0.179 and 0.457 ± 0.124 nmol/L in human MDA-231 cells, respectively (Fig. 3e, f). Inhibition of CD73 enzymatic activity was also similar between MEDI-TGFβR and MEDI with IC50 values of 0.593 ± 0.147 and 0.302 ± 0.039 nmol/L in 4T1 cells and 0.188 ± 0.053 and 0.303 ± 0.025 nmol/L in MDA-231 cells, respectively (Fig. 3g, h). These results demonstrate that MEDI-TGFβR elicits favorable target binding and potent inhibition of CD73 enzyme activity.
Fig. 3. Design and characterization of the MEDI-TGFβRII fusion protein for dual-blockade of CD73 and TGFβ.
a The associations between CD73 and TGFB1 transcriptome expression in cancer cell lines from the Cancer Cell Line Encyclopedia. b Schematic representation of the MEDI-TGFβR construct. c SDS-PAGE showing the purified MEDI and MEDI-TGFβR proteins. The molecular weights of full length (FL), heavy chain (HC), and light chain (LC) proteins are indicated. d TGFβ binding affinities of anti-TGFβ and MEDI-TGFβR as measured by ELISA. Comparisons of CD73 binding affinities of MEDI versus MEDI-TGFβR in e 4T1 cells or f MDA-231 cells. Comparisons of CD73 enzyme inhibitory potency for MEDI versus MEDI-TGFβR in g 4T1 cells and h MDA-231 cells.
Bispecific MEDI-TGFβR is efficacious in downregulating the EMT and lung metastasis and promoting a favorable tumor stroma
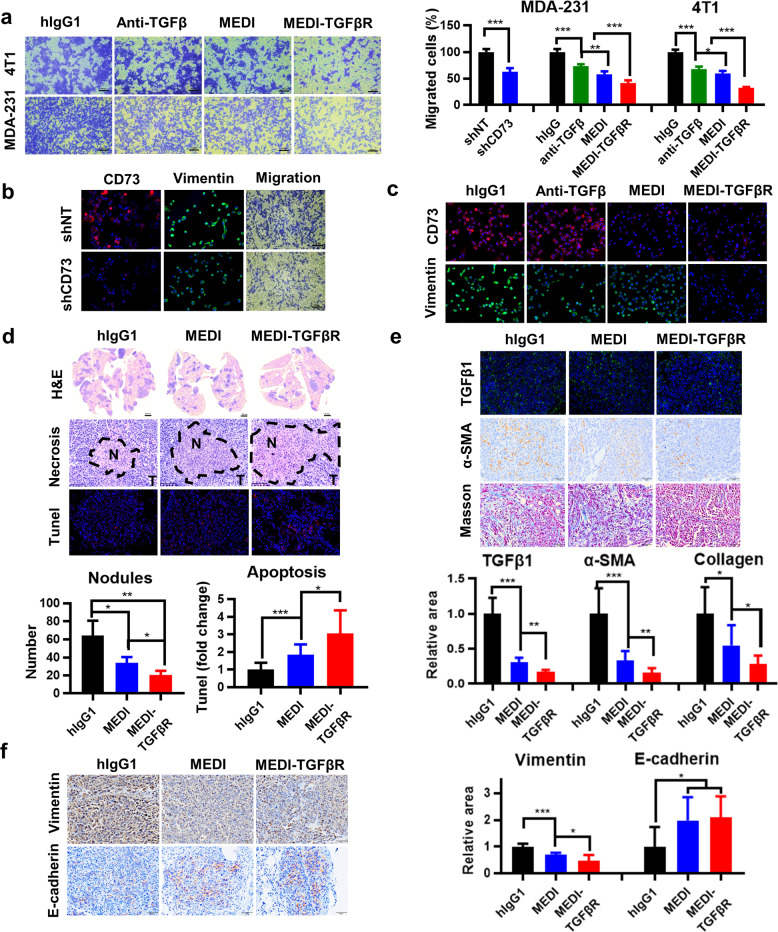
We examined the pharmacological effects of MEDI-TGFβR by employing both in vitro and in vivo TNBC models. Treatment of MDA-231 or 4T1 cells with MEDI-TGFβR each resulted in profound suppression of cell migration in vitro compared with migration after MEDI or anti-TGFβ treatment (Fig. 4a, b). The reduced cell migration was accompanied by a substantial downregulation of the EMT biomarker vimentin, which was diminished considerably in the MEDI-TGFβR-treated cells compared with that in MEDI-treated, anti-TGFβ-treated, or CD73-depleted cells (Fig. 4b, c). In the syngeneic mouse model, tail vein injection of 4T1 cells led to rapid metastasis to the lungs in Balb/c mice. Intravenous weekly treatments with MEDI-TGFβR demonstrated a substantial overall reduction in tumor burden compared with that of the MEDI-treated group (P < 0.05) (Fig. 4d). The strong reduction in tumor burden was associated with increases in necrosis and apoptosis that occurred within the metastasized tumor nodules, which was more prominent in the lungs from the MEDI-TGFβR-treated than MEDI-treated mice (P < 0.05) (Fig. 4d). While we observed an abundant TGFβ1 signal in the metastatic nodules of hIgG1-treated mice, treatment with MEDI-TGFβR led to a greater suppression of TGFβ levels compared with that after MEDI treatment (P < 0.05) (Fig. 4e). TGFβ promotes growth of cancer-associated fibroblasts [34], and treatment with MEDI-TGFβR potentiated the suppression of α-SMA expression (P < 0.01) and collagen deposition (P < 0.05) (Fig. 4e). Both MEDI and MEDI-TGFβR efficiently downmodulated the EMT signature with reduced vimentin and enhanced E-cadherin (Fig. 4f). These results suggest that blocking CD73 and TGFβ pathways concurrently with MEDI-TGFβR can achieve greater efficacy in suppressing metastasis and tumor growth in part by regulating tumor plasticity and promoting a favorable tumor stroma.
Fig. 4. The bispecific MEDI-TGFβR fusion protein is more efficacious in downregulating the EMT and lung metastasis and promoting a favorable tumor stroma.
a Trans-well assays were performed using 4T1 or MDA-231 cells treated with 66.7 nmol/L hIgG1, anti-TGFβ, MEDI, or MEDI-TGFβR, and results were analyzed by ImageJ software. b MDA-231 cells carrying the doxycycline-inducible short hairpin RNA (sh) control or shCD73 lentivirus constructs were treated to deplete CD73 before analysis of cell migration and determination of CD73 and vimentin expression by immunofluorescence staining. c CD73 and vimentin expression was assayed in MDA-231 cells treated with 66.7 nmol/L hIgG1, anti-TGFβ, MEDI, or MEDI-TGFβR. d Lung metastatic nodules after injection of 4T1 cells in a syngeneic mouse model and the areas of necrosis or apoptosis (n = 3). e Immunohistochemistry (IHC) staining for TGFβ1 and α-SMA and Masson’s trichrome staining for collagen. f IHC staining for vimentin and E-cadherin. The representative views and quantification of positive areas are shown. *P < 0.05, **P < 0.01, ***P < 0.001.
CD73-TGFβ dual-blockade improves innate and adaptive immune profiles in the tumor microenvironment
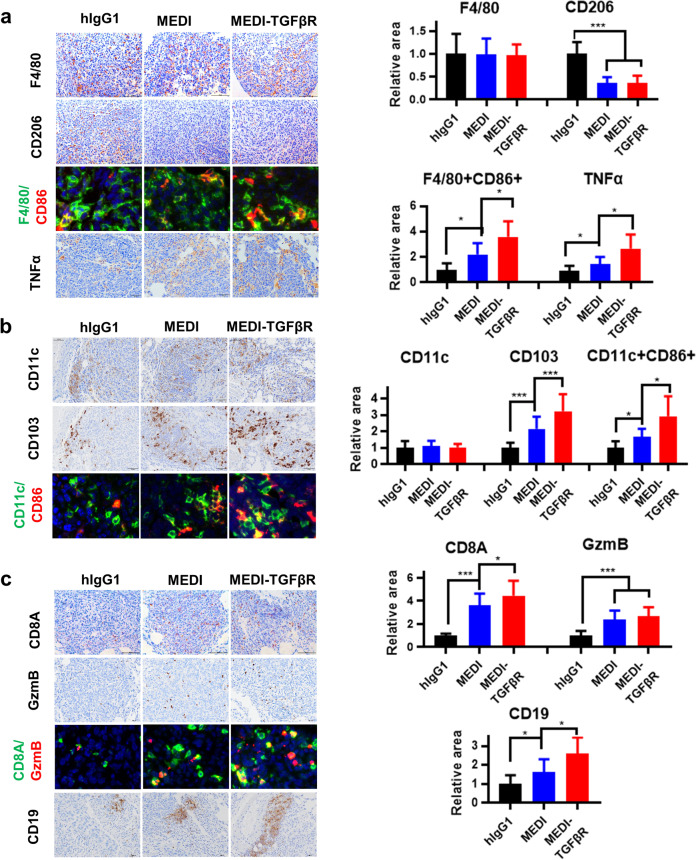
A complex interaction between CD73 with TGFβ resulting in immune-tolerance of TNBC was revealed (Figs. 1 and 3a). Therefore, we investigated both innate and adaptive immune cell infiltration in the metastatic lung tumor microenvironment. First, we performed IHC or multiplexed IHC staining using macrophage biomarkers. While the overall levels of F4/80+ macrophage pan-marker did not fluctuate between the mouse groups, CD206+ M2-like macrophages were substantially reduced in both MEDI and MEDI-TGFβR groups (P < 0.001) (Fig. 5a). In contrast, M1-like macrophages (F4/80+ CD86+ ) and TNFα, which is a major tumor-killing cytokine secreted by M1-like macrophages, were markedly increased in the MEDI-TGFβR group relative to that of the hIgG1 or MEDI groups (P < 0.05) (Fig. 5a). We then examined dendritic cells and found that CD11c expression was similar among the groups. However, MEDI-TGFβR-treated mice showed the greatest increase in CD103+ cells in the tumor nodules compared with that in the other groups (P < 0.001) (Fig. 5b). Consistently, multiplexed IHC of CD11c and CD86 revealed a more robust activation of dendritic cells in the MEDI-TGFβR-treated tumor nodules (P < 0.05) (Fig. 5b). Next, we investigated adaptive immunity, including T and B cells. IHC analysis of lung tissues showed improved tumor infiltration of CD8+ T cells and CD19+ B cells in MEDI-TGFβR-treated mice relative to that of MEDI-treated animals (P < 0.05) (Fig. 5c). Furthermore, colocalization analysis of CD8A and granzyme B (GZMB) confirmed that the MEDI-TGFβR-treated group had the greatest enhancement of CD8+ GZMB+ cytotoxic T cell infiltration compared with that in the other groups (Fig. 5c). Collectively, these results indicate that MEDI-TGFβR induces a more potent proinflammatory and antitumor microenvironment by dual-blockade of CD73 and TGFβ pathways.
Fig. 5. CD73-TGFβ dual-blockade improves innate and adaptive immune profiles in the tumor microenvironment.
Lung sections as described in Fig. 4d were subjected to immunohistochemistry (IHC) or multiplexed IHC (tyramide signal amplification method) analysis. a IHC staining for the macrophage markers F4/80, CD206 (M2-like), and dual-staining for F4/80 and CD86 (M1-like) or tumor necrosis factor (TNF)α. b IHC staining for the dendritic cell markers CD11c, CD103, and dual-staining for CD11c and CD86. c IHC staining for the T and B cell markers CD8A and CD19, and dual-staining for CD8A and granzyme B (GZMB). Representative images (1/8 of the size captured from the original images, Supplementary Fig. S1) and quantification of positive areas are shown. *P < 0.05, ***P < 0.001.
Discussion
In this study, we investigated the importance of CD73 and TGFβ interactions in basal-like/TNBC. Analyses of patient tumor transcriptome database sets and tumor tissue arrays demonstrated that elevated CD73 expression occurs frequently and is associated with poor prognosis, EMT, and an immunosuppressive tumor environment. Importantly, high TGFβ expression further worsened DFS in patients with CD73-high basal-like/TNBC. The regenerative feedback loop between CD73 and TGFβ aggravated tumor plasticity, drug resistance, and immune evasion [19, 20, 23]. Our strategy focused on dual-blockade of CD73 and TGFβ pathways by creating a bifunctional MEDI-TGFβR fusion protein, which potently inhibited both CD73 and TGFβ when evaluated at the biochemical, cellular, and animal levels. In a mouse model of TNBC, MEDI-TGFβR achieved a superior efficacy compared with CD73 blockade alone. Mechanistically, CD73-TGFβ dual-blockade elicited a substantially heightened level of tumor cell death that was mediated through both apoptosis and necrosis and accompanied by increased infiltration of inflammatory M1-like macrophages. The dual-blockade also reduced tumor stromal fibrosis and extracellular matrix deposition, which are expected to retard tumor growth and facilitate drug penetration and response. Predictably, inhibition of CD73 and TGFβ in mouse tumors resulted in a robust transformation from an immunosuppressive tumor environment (CD206+, CD11b+Gr1+) to a potent proinflammatory microenvironment as demonstrated by the coordinated enhancements in cytotoxic T cells (CD8+, CD8+GZMB+), B cells (CD19+), and dendritic cells (CD103+, CD11c+CD86+). Overall, these findings provide a rationale for investigating a CD73-TGFβ dual-targeting strategy in the clinic.
The EMT is one of the most widely studied mechanisms of tumor plasticity and metastasis and is also increasingly recognized as an underlying cause of a wide spectrum of therapy resistance [15, 35, 36]. TGFβ is a well-known inducer of the EMT and immune evasion in cancer, and CD73 plays a role in cytoskeleton remodeling in the β-catenin-dependent EMT process, resulting in metastasis [37]. In our study, elevated CD73 expression was positively associated with the EMT process, and inhibition of CD73 alone substantially decreased the vimentin/E-cadherin mesenchymal index, which was further reduced by treatment with MEDI-TGFβR. Targeting CD73 was shown to enhance the antitumor activity of anti-PD-1 and anti-CTLA4 monoclonal antibodies in preclinical models [38, 39]. Furthermore, melanoma patients with high CD73 tumor levels presented with progressive disease after anti-PD-1 therapy [40], which argues for an adaptive resistance mechanism. These observations support the concept that the combined targeting of CD73 and TGFβ may be more effective in re-sensitizing therapeutic responsiveness in patients with TNBC, including those undergoing chemotherapy, targeted therapy, or anti-PD-1/PD-L1/CTLA4 immunotherapy.
At present, several anti-CD73 monoclonal antibodies and selective small molecule inhibitors are being tested in early phase clinical trials [41]. The systemic application of TGFβ inhibitors presents challenges because of the complex role of TGFβ in normal tissue homeostasis [24, 25]. However, several therapies targeting the TGFβ pathway are now undergoing clinical trials, including fusion proteins that simultaneously targets TGFβ and PD-L1 [30] or CTLA4 [42], and have demonstrated manageable safety profiles in patients [43]. We propose that neutralization of both TGFβ and CD73 will enhance the therapeutic efficacy window for treating subsets of patients with CD73/TGFβ-high TNBC.
In conclusion, we demonstrated the importance of CD73 and its relationship with TGFβ in TNBC. The improved therapeutic activity of MEDI-TGFβR included reversion of the EMT state and generation of a favorable tumor stroma and tumor-suppressive immune-microenvironment. We propose CD73-TGFβ dual-blockade as a novel treatment option for TNBC.
Supplementary information
Acknowledgements
This work was funded by Fudan University (EZF301002), NSF of China (81373442), NST Major Project of China (2018ZX09711002-008), and NBR 973 Program of China (2013CB932500). The authors thank the Fudan School of Pharmacy Animal Facility and Instrument Center for technical support. We thank Susan Zunino, PhD, from Liwen Bianji (Edanz) (www.liwenbianji.cn/), for editing the English text of a draft of this manuscript.
Author contributions
Conception and design: KY, ZQR. Development of methodology: YX, ZQR, RJ. Acquisition of data: YX, ZQR, RJ, LL, JPP. Analysis and interpretation of data: YX, ZQR, KY. Writing, review, and/or revision of the manuscript: YX, ZQR, LL, KY. Administrative, technical, or material support: YX, ZQR, KY. Study supervision: YX, ZQR, KY.
Competing interests
The authors declare no competing interests.
Supplementary information
The online version contains supplementary material available at 10.1038/s41401-021-00840-z.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Dent R, Hanna WM, Trudeau M, Rawlinson E, Sun P, Narod SA. Pattern of metastatic spread in triple-negative breast cancer. Breast Cancer Res Treat. 2009;115:423–8. doi: 10.1007/s10549-008-0086-2. [DOI] [PubMed] [Google Scholar]
- 3.Gucalp A, Traina TA. Triple-negative breast cancer: adjuvant therapeutic options. Chemother Res Pract. 2011;2011:1–13. doi: 10.1155/2011/696208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narayan P, Wahby S, Gao JJ, Amiri-Kordestani L, Ibrahim A, Bloomquist E, et al. FDA Approval Summary: atezolizumab plus paclitaxel protein-bound for the treatment of patients with advanced or metastatic TNBC whose tumors express PD-L1. Clin Cancer Res. 2020;26:2284–9. doi: 10.1158/1078-0432.CCR-19-3545. [DOI] [PubMed] [Google Scholar]
- 5.Vagia E, Mahalingam D, Cristofanilli M. The landscape of targeted therapies in TNBC. Cancers. 2020;12:916. doi: 10.3390/cancers12040916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu XX, Chen YT, Feng B, Mao XB, Yu B, Chu XY. Expression and clinical significance of CD73 and hypoxia-inducible factor-1α in gastric carcinoma. World J Gastroenterol. 2013;19:1912–8. doi: 10.3748/wjg.v19.i12.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turcotte M, Spring K, Pommey S, Chouinard G, Cousineau I, George J, et al. CD73 is associated with poor prognosis in high-grade serous ovarian cancer. Cancer Res. 2015;75:4494–503. doi: 10.1158/0008-5472.CAN-14-3569. [DOI] [PubMed] [Google Scholar]
- 8.Wu XR, He XS, Chen YF, Yuan RX, Zeng Y, Lian L, et al. High expression of CD73 as a poor prognostic biomarker in human colorectal cancer. J Surg Oncol. 2012;106:130–7. doi: 10.1002/jso.23056. [DOI] [PubMed] [Google Scholar]
- 9.Buisseret L, Pommey S, Allard B, Garaud S, Bergeron M, Cousineau I, et al. Clinical significance of CD73 in triple-negative breast cancer: Multiplex analysis of a phase III clinical trial. Ann Oncol. 2018;29:1056–62. doi: 10.1093/annonc/mdx730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang H, Yao F, Davis PF, Tan ST, Hall SRR. CD73, tumor plasticity and immune evasion in solid cancers. Cancers. 2021;13:1–27. doi: 10.3390/cancers13020177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerbelli B, Botticelli A, Pisano A, Pernazza A, Campagna D, De Luca A, et al. CD73 expression and pathologic response to neoadjuvant chemotherapy in triple negative breast cancer. Virchows Arch. 2020;476:569–76. doi: 10.1007/s00428-019-02722-6. [DOI] [PubMed] [Google Scholar]
- 12.Cerbelli B, Scagnoli S, Mezi S, De Luca A, Pisegna S, Amabile MI, et al. Tissue immune profile: A tool to predict response to neoadjuvant therapy in triple negative breast cancer. Cancers. 2020;12:1–12. doi: 10.3390/cancers12092648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loi S, Pommey S, Haibe-Kains B, Beavis PA, Darcy PK, Smyth MJ, et al. CD73 promotes anthracycline resistance and poor prognosis in triple negative breast cancer. Proc Natl Acad Sci USA. 2013;110:11091–6. doi: 10.1073/pnas.1222251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samanta D, Park Y, Ni X, Li H, Zahnow CA, Gabrielson E, et al. Chemotherapy induces enrichment of CD47+/CD73+/PDL1+ immune evasive triple-negative breast cancer cells. Proc Natl Acad Sci USA. 2018;115:1239–48. doi: 10.1073/pnas.1718197115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dongre A, Rashidian M, Eaton EN, Reinhardt F, Thiru P, Zagorulya M, et al. Direct and indirect regulators of epithelial–mesenchymal transition– mediated immunosuppression in breast carcinomas. Cancer Discov. 2021;11:1286–305. doi: 10.1158/2159-8290.CD-20-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang M, Wu J, Mao K, Deng H, Yang Y, Zhou E, et al. Role of transforming growth factor-β1 in triple negative breast cancer patients. Int J Surg. 2017;45:72–6. doi: 10.1016/j.ijsu.2017.07.080. [DOI] [PubMed] [Google Scholar]
- 17.García-Rocha R, Monroy-García A, Hernández-Montes J, Weiss-Steider B, Gutiérrez-Serrano V, del Carmen Fuentes-Castañeda M, et al. Cervical cancer cells produce TGF-β1 through the CD73-adenosine pathway and maintain CD73 expression through the autocrine activity of TGF-β1. Cytokine. 2019;118:71–9. doi: 10.1016/j.cyto.2018.09.018. [DOI] [PubMed] [Google Scholar]
- 18.Ávila-Ibarra LR, Mora-Garcia MDL, García-Rocha R, Hernández-Montes J, Weiss-Steider B, Montesinos JJ, et al. Mesenchymal stromal cells derived from normal cervix and cervical cancer tumors increase CD73 expression in cervical cancer cells through TGF-β1 production. Stem Cells Dev. 2019;28:477–88. doi: 10.1089/scd.2018.0183. [DOI] [PubMed] [Google Scholar]
- 19.Chalmin F, Mignot G, Bruchard M, Chevriaux A, Végran F, Hichami A, et al. Stat3 and Gfi-1 transcription factors control Th17 cell immunosuppressive activity via the regulation of ectonucleotidase expression. Immunity. 2012;36:362–73. doi: 10.1016/j.immuni.2011.12.019. [DOI] [PubMed] [Google Scholar]
- 20.Li J, Wang L, Chen X, Li L, Li Y, Ping Y, et al. CD39/CD73 upregulation on myeloid-derived suppressor cells via TGF-β-mTOR-HIF-1 signaling in patients with non-small cell lung cancer. Oncoimmunology. 2017;6:1–13. doi: 10.1080/2162402X.2017.1320011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryzhov SV, Pickup MW, Chytil A, Gorska AE, Zhang Q, Owens P, et al. Role of TGF-β signaling in generation of CD39+CD73+myeloid cells in tumors. J Immunol. 2014;193:3155–64. doi: 10.4049/jimmunol.1400578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ni C, Fang QQ, Chen WZ, Jiang JX, Jiang Z, Ye J, et al. Breast cancer-derived exosomes transmit lncRNA SNHG16 to induce CD73+γδ1 Treg cells. Signal Transduct Target Ther. 2020;5:41. doi: 10.1038/s41392-020-0129-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen S, Fan J, Zhang M, Qin L, Dominguez D, Long A, et al. CD73 expression on effector T cells sustained by TGF-β facilitates tumor resistance to anti-4-1BB/CD137 therapy. Nat Commun. 2019;10:150. doi: 10.1038/s41467-018-08123-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohn A, Lahn MM, Williams KE, Cleverly AL, Pitou C, Kadam SK, et al. A phase I dose-escalation study to a predefined dose of a transforming growth factor-β1 monoclonal antibody (TβM1) in patients with metastatic cancer. Int J Oncol. 2014;45:2221–31. doi: 10.3892/ijo.2014.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitra MS, Lancaster K, Adedeji AO, Palanisamy GS, Dave RA, Zhong F, et al. A potent pan-TGFβ neutralizing monoclonal antibody elicits cardiovascular toxicity in mice and cynomolgus monkeys. Toxicol Sci. 2020;175:24–34. doi: 10.1093/toxsci/kfaa024. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Li Q, Zhao H, Ma L, Meng T, Qian J, et al. Pathological expression of tissue factor confers promising antitumor response to a novel therapeutic antibody SC1 in triple negative breast cancer and pancreatic adenocarcinoma. Oncotarget. 2017;8:59086–102. doi: 10.18632/oncotarget.19175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neve RM, Chin K, Fridlyand J, Yeh J, Frederick L, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2009;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geoghegan JC, Diedrich G, Lu X, Rosenthal K, Sachsenmeier KF, Wu H, et al. Inhibition of CD73 AMP hydrolysis by a therapeutic antibody with a dual, non-competitive mechanism of action. MAbs. 2016;8:454–67. doi: 10.1080/19420862.2016.1143182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lan Y, Zhang D, Xu C, Hance KW, Marelli B, Qi J, et al. Enhanced preclinical antitumor activity of M7824, a bifunctional fusion protein simultaneously targeting PD-L1 and TGF-β. Sci Transl Med. 2018;10:5488. doi: 10.1126/scitranslmed.aan5488. [DOI] [PubMed] [Google Scholar]
- 31.Jin R, Liu L, Xing Y, Meng T, Ma L, Pei J, et al. Dual mechanisms of novel CD73-targeted antibody and antibody–drug conjugate in inhibiting lung tumor growth and promoting antitumor immune-effector function. Mol Cancer Ther. 2020;19:2340–52. doi: 10.1158/1535-7163.MCT-20-0076. [DOI] [PubMed] [Google Scholar]
- 32.Tóth ZE, Mezey É. Simultaneous visualization of multiple antigens with tyramide signal amplification using antibodies from the same species. J Histochem Cytochem. 2007;55:545–54. doi: 10.1369/jhc.6A7134.2007. [DOI] [PubMed] [Google Scholar]
- 33.Xu X, Zhang L, He X, Zhang P, Sun C, Xu X, et al. TGF-β plays a vital role in triple-negative breast cancer (TNBC) drug-resistance through regulating stemness, EMT and apoptosis. Biochem Biophys Res Commun. 2018;502:160–5. doi: 10.1016/j.bbrc.2018.05.139. [DOI] [PubMed] [Google Scholar]
- 34.Derynck R, Turley SJ, Akhurst RJ. TGFβ biology in cancer progression and immunotherapy. Nat Rev Clin Oncol. 2021;18:9–34. doi: 10.1038/s41571-020-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.David JM, Dominguez C, Palena C. Pharmacological and immunological targeting of tumor mesenchymalization. Pharmacol Ther. 2016;170:212–25. doi: 10.1016/j.pharmthera.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Z, Gu C, Yao X, Guo W, Wang H, Lin T, et al. CD73 promotes tumor metastasis by modulating RICS/RhoA signaling and EMT in gastric cancer. Cell Death Dis. 2020;11:202. doi: 10.1038/s41419-020-2403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allard B, Pommey S, Smyth MJ, Stagg J. MEDI9447 enhances the antitumor activity of anti-PD-1 and anti-CTLA-4 mAbs. Clin Cancer Res. 2013;19:5626–35. doi: 10.1158/1078-0432.CCR-13-0545. [DOI] [PubMed] [Google Scholar]
- 39.Hay CM, Sult E, Huang Q, Mulgrew K, Fuhrmann SR, McGlinchey KA, et al. Targeting CD73 in the tumor microenvironment with MEDI9447. Oncoimmunology. 2016;5:1–10. doi: 10.1080/2162402X.2016.1208875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reinhardt J, Landsberg J, Schmid-Burgk JL, Ramis BB, Bald T, Glodde N, et al. MAPK signaling and inflammation link melanoma phenotype switching to induction of CD73 during immunotherapy. Cancer Res. 2017;77:4697–709. doi: 10.1158/0008-5472.CAN-17-0395. [DOI] [PubMed] [Google Scholar]
- 41.Roh M, Wainwright DA, Wu JD, Wan Y, Zhang B. Targeting CD73 to augment cancer immunotherapy. Curr Opin Pharmacol. 2020;53:66–76. doi: 10.1016/j.coph.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ravi R, Noonan KA, Pham V, Bedi R, Zhavoronkov A, Ozerov IV, et al. Bifunctional immune checkpoint-targeted antibody-ligand traps that simultaneously disable TGFβ enhance the efficacy of cancer immunotherapy. Nat Commun. 2018;9:741. doi: 10.1038/s41467-017-02696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strauss J, Heery CR, Schlom J, Madan RA, Cao L, Kang Z, et al. Phase I trial of M7824 (MSB0011359C), a bifunctional fusion protein targeting PD-L1 and TGFbeta, in advanced solid tumors. Clin Cancer Res. 2018;24:1287–95. doi: 10.1158/1078-0432.CCR-17-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.