Abstract
The protein arginine methyltransferase 5 (PRMT5), which is highly expressed in tumour tissues, plays a crucial role in cancer development. However, the mechanism by which PRMT5 promotes cancer growth is poorly understood. Here, we report that PRMT5 contributes to lipid metabolism reprogramming, tumour growth and metastasis depending on the SIRT7-mediated desuccinylation of PRMT5 K387 in tumours. Mass spectrometric analysis identified PRMT5 lysine 387 as its succinylation site. Moreover, the desuccinylation of PRMT5 K387 enhances the methyltransferase activity of PRMT5. SIRT7 catalyses the desuccinylation of PRMT5 in cells. The SIRT7-mediated dessuccinylation of PRMT5 lysine 387 fails to bind to STUB1, decreasing PRMT5 ubiquitination and increasing the interaction between PRMT5 and Mep50, which promotes the formation of the PRMT5-Mep50 octamer. The PRMT5-Mep50 octamer increases PRMT5 methyltransferase activity, leading to arginine methylation of SREBP1a. The symmetric dimethylation of SREBP1a increases the levels of cholesterol, fatty acid, and triglyceride biogenesis in the cells, escaping degradation through the ubiquitin-proteasome pathway. Functionally, the desuccinylation of PRMT5 K387 promotes lipid metabolism reprogramming, tumour growth and metastasis in vitro and in vivo in tumours.
Keywords: PRMT5, methyltransferase activity, desuccinylation, SIRT7, lipid metabolism reprogramming
Introduction
Arginine methylation is a key modification in the progression of cancer. It has been reported that protein arginine methyltransferase 5 (PRMT5) plays a vital role in a wide range of cellular processes, such as genomic organisation, gene transcription, differentiation and cell cycle control [1]. The epigenetic factor PRMT5 complexed with methylosome protein 50 (Mep50, also known as WDR77) catalyses arginine methylation on histones and other proteins [2–5]. However, the significance of PRMT5 in tumours is elusive.
Depending on its methyltransferase activity, PRMT5 mediates the symmetrical dimethylation of arginine residues within histones H2A, H3, and H4 [6–8] and other proteins, such as p53, SPT5, SREBP1a and MBD2 [9–12]. Specifically, PRMT5 catalyses H3R8me2s and H4R3me2s, which in turn alter chromatin structure to regulate gene expressions, such as cyclin E1 (CCNE1) and tumour suppressor genes (RB1, RBL1 and RBL2) [13, 14]. In addition, certain tumour suppressors, such as the metastasis inhibition factor Nm23, can be epigenetically silenced by PRMT5 [8]. Moreover, components of the transcription machinery are substrates for PRMT5-catalysed methylation modifications, such as SPT5 and E2F1 [10, 15]. Thus, PRMT5 primarily functions as a tumour-promoting factor. PRMT5 methyltransferase activity is elevated in cancer cells. Its expression is highly correlated with poor prognosis in many human tumours [16–21], indicating that PRMT5 has therapeutic potential for treating human cancer. However, the mechanism by which PRMT5 displays its methyltransferase activity in cancer is poorly understood.
Succinylation modification is a posttranslational modification (PTM) of lysine residues [22]. Succinylation of lysine residues will change the charge status more than other acetyl groups (from +1 to −1), and it is comparable to that caused by protein phosphorylation (from 0 to −2 charges). In addition, succinylation adds a larger structural moiety than acetylation or methylation, which are two PTMs known to have important cellular roles. Since succinylation causes a greater change in charge and a more dramatic structural alteration, succinylation leads to more significant changes in protein structure and function [23]. New types of nonacetyl acylation usually share the same enzymes mediating the writing and erasing of acetylation [24]. The erasers of succinylation include SIRT3, SIRT4, SIRT5 and SIRT7 [25, 26]. It has been reported that PRMT5 activity can be regulated by phosphorylation [27]. However, whether succinylation modification is involved in the methyltransferase activity of PRMT5 is unclear.
In this study, we attempted to identify the mechanism by which PRMT5 enhances tumour growth. Strikingly, we observed the succinylation of PRMT5 in the cells. Then, the succinylation site of PRMT5 at lysine residue 387 (K387) was identified. Moreover, the SIRT7-mediated desuccinylation of PRMT5 K387 contributed to its methyltransferase activity by promoting the formation of the PRMT5-Mep50 octamer, leading to lipid metabolism reprogramming, tumour growth and metastasis of cancer cells. Our findings provide new insights into the mechanism by which PRMT5 enhances tumorigenesis.
Materials and methods
Cell lines and cell culture
HEK293T cells, the human breast carcinoma cell line MCF7, and the human hepatocellular carcinoma cell line HepG2 were cultured in Dulbecco’s Modified Eagle’s Medium (Invitrogen, Carlsbad, California, USA) supplemented with 10% (vol/vol) foetal bovine serum (FBS), 2 mM glutamine, penicillin (100 U/mL) and streptomycin (100 mg/mL) in 5% CO2 at 37 °C. Human pancreatic carcinoma PANC-1 cells were cultured in RPMI-1640 medium supplemented with 10% FBS and 1% streptomycin/penicillin. PRMT5 knockdown HepG2 cell lines (HepG2-shPT5) were established by using the shRNA system. The shRNA sequences of PRMT5 were as follows: shRNA#1, 5′-GCGTTTCAAGAGGGAGTTCA-3′, shRNA#2, 5′-CCCATCCTCTTCCCTATTAA-3′, and shRNA#3 (target PRMT5 3′UTR sequence), 5′-GGCTCAAGCCACCAATCTAT-3′. Lentiviral stocks were infected in HEK293T cells to generate the PRMT5-stable cell line. HepG2 and HepG2-shPT5 cells were transduced with a pCDH-Puro lentivirus carrying the PRMT5 or PRMT5K387R expression sequence in the presence of 5 mg/mL polybrene (Sigma-Aldrich). After 24 h of culturing, transduced cells were selected with puromycin (2 mg/mL, Amresco, USA). All the cell lines were mycoplasma-free by PCR.
Plasmid or siRNA generation and transfection
Genscript Biotechnology Co., Ltd constructed all the plasmids used in this study. The plasmids used in this study for construction are listed in Table S1. All siRNAs were synthesised by Sangon Biotech (Shanghai, Chain). The siRNA sequences used in this study are listed in Table S2. According to the manufacturer’s protocol, the transfections were performed using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA).
RNA extraction and quantitative real-time PCR (RT-qPCR)
Total RNA was extracted from cells (or nude tumour tissues) using TRIzol reagent (Solarbio, Beijing, China). First-strand cDNA was synthesised using the Hifair III 1st strand cDNA synthesis supermix kit (Yeasen Biotech, Shanghai, China). Quantitative real-time PCR was performed by a StepOnePlus real-time PCR machine (Bio-Rad) using qPCR SYBR Green Master Mix (Yeasen Biotech, Shanghai, China). Relative transcriptional folds were calculated as 2-ΔΔCt. The primers used were listed in Table S3.
Western blot analysis
Total protein lysates were extracted from hepatoma cells or liver tissues with RIPA buffer according to the manufacturer’s protocol. Histones were extracted using the Histone Extraction Kit (Abcam, Cambridge, UK). The assays were performed as described previously [28]. The antibodies used for Western blot analysis are listed in Table S4.
Immunofluorescence analysis
The assays were performed as described previously [29]. Cells grown on acid-treated glass coverslips in 6-well plates were washed three times with precooled PBS and fixed with 4% paraformaldehyde for 10 min, followed by permeabilization for 10 min at room temperature with 0.5% Triton X-100. After incubation for 1 h with 5% BSA to block nonspecific binding, primary antibodies were added and incubated overnight at 4 °C. The bound antibodies were visualised by incubation with secondary antibodies. Images were acquired using a confocal microscope or fluorescence microscopy. The antibodies used were listed in Table S4.
Analysis of tumorigenicity in nude mice
All animal experiments were in accordance with the Guide for the Care and Use of Laboratory Animals (NIH publications 86-23 revised 1985) and were performed according to institutional ethical guidelines. The Institute Research Ethics Committee at Nankai University approved the study protocol. The tumour-bearing male nude mice model was established to evaluate the tumour proliferation ability. HepG2 cells were treated and grouped, including shCtrl, shPT5, shPT5 + siSREBP1, shPT5 + PT5, shPT5 + PT5K387R, and shPT5 + PT5K387R + siSREBP1. The mice were randomly grouped and subcutaneously injected with treated tumour cells (5 × 106), with six mice per group. Tumour growth was measured 9 days after injection and then every 3 days. At 27 days after injection, mice were sacrificed, and tumours were removed for analysis.
An orthotopic liver tumour mouse model was generated to evaluate the tumour metastasis ability. HepG2 cells were treated and grouped, including shCtrl, shPT5, shPT5 + siSREBP1, shPT5 + PT5, shPT5 + PT5K387R, and shPT5 + PT5K387R + siSREBP1. The cells were injected into the livers of nude mice (1 × 107 cells/mouse, n = 6 mice/group). The mice were sacrificed, and intrahepatic/lung metastasis burden was assessed in 30 days. Lung metastatic lesions were directly counted on tissue sections using H&E staining.
Immunohistochemistry (IHC)
The tumour tissue from nude mice was fixed and embedded in paraffin after those mice were sacrificed. The IHC assays were performed as described previously [28]. First, the samples were dewaxed. Then, antigen retrieval was applied at 95 °C with citrate buffer (pH 6.0) for 15 min. The slides were treated with 3% H2O2 for 10 min and blocked with goat serum for 1 h. Then, slides were incubated with the monoclonal antibody at 4 °C overnight. After washing three times with PBS, samples were incubated with goat anti-rabbit or anti-mouse IgG coupled to horseradish peroxidase (ORIGENE, Beijing, China) for 30 min at 37 °C. Immunostaining was performed using chromogen 3,3′-diaminobenzidine and counterstained with Mayer’s hematoxylin (ZSBG-BIO, China). The slides were then dehydrated and covered with a coverslip. The antibodies used for IHC staining are listed in Table S4.
Coimmunoprecipitation (CoIP) assays
The assays were performed as described previously [27]. Cells were washed with cold PBS and lysed with cold cell lysis buffer for 30 min at 4 °C. Then, 500 μg of cellular extract was incubated with appropriate specific antibodies or normal rabbit/mouse immunoglobin G (IgG) at 4 °C overnight with constant rotation, followed by the addition of Protein A/G magnetic beads (Merck-Millipore, Darmstadt, Germany) and incubation for 2 h at 4 °C. Beads were then washed five times with cell lysis buffer (20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 20 mM KCl, 1.5 mM MgCl2, 15% glycerol, 1 mM EDTA, 0.5% NP-40, and 1% protease inhibitor). The immune complex was subjected to SDS-PAGE followed by immunoblotting with the indicated antibodies. Immunodetection was performed using Super ECL Detection Reagent (Yeasen Biotech, Shanghai, China) according to the manufacturer’s instructions. The antibodies used were listed in Table S4.
Immunoprecipitation and protein purification
Total cells were lysed with cell lysis buffer for Western &IP according to the manufacturer’s instructions (GenStar, China). Immunoprecipitation was carried out by incubating with anti-DDDK-tag mAb-magnetic agarose (M2 beads) or anti-HA-tag mAb-magnetic agarose (MBL, Japan) according to the manufacturer’s instructions. Then, the bounded proteins were eluted with 0.1 mol/L glycine (pH 2.5) and neutralised with 1 M Tris buffer. The elution was analysed by Western blot analysis.
Identification of PRMT5 PTM sites
HepG2 cells were transfected with Flag-PRMT5 plasmids and treated with 10 mM NAM and 1 μM TSA for 6 h before harvest. Cells were lysed by lysis buffer and immunoprecipitated with M2 beads overnight. Immunoprecipitated PRMT5 proteins were subjected to 10% SDS-PAGE gel, and PRMT5 bands were excised from the gel and analysed by mass spectrometry. Distinct modification sites were chosen for further potential succinylation site analysis.
In vitro methylation assays
Flag-PRMT5 WT and mutants were purified from transfected 293 T cells by anti-Flag immunoprecipitation. The immobilised proteins were then incubated with 20 μL of HMTase buffer (20 mM Tris, pH 8.8, 25 mM NaCl, 4 mM EDTA, 1 mM PMSF, 0.5 mM DTT) supplemented with 10 μg of purified histone H4 or histone and 2 mCi 3H-SAM (Amersham) at 30 °C for 2 h. The reaction was stopped by the addition of sample loading buffer for immunoblotting.
Analysis of cell proliferation
Cells were seeded into 96-well plates (2000 cells/well) for 8 h before transfection. The number of cells was counted according to the protocol of the CCK8 assay kit according to the manufacturer’s instructions (Abcam). The EdU assay was carried out by using the Cell-LightTM EdU imaging detection kit according to the manufacturer’s instructions (RiboBio, Guangzhou, China).
Analysis of colony formation
For colony formation analysis, 48 h after transfection, 1000 viable transfected cells were placed in 6-well plates and maintained in complete medium for 2 weeks. Colonies were fixed with methanol and stained with methylene blue.
Wound healing assays
Cells in different states were seeded into 6-well cell culture plates, and cells overgrew the dish for ~24 h. Then, the cells were wounded with a sterile yellow plastic tip. Cell migration was observed and photographed by microscopy (Eclipse TS100, Nicon) several hours later.
Oil Red O staining
Cells were washed twice with phosphate saline and fixed with 10% formalin. Oil Red O staining was performed according to the manufacturer’s instructions.
Total triglyceride, fatty acids and cholesterol assays
Cellular triglycerides were assayed using a tissue triglyceride assay kit (Applygen Technologies Inc., Beijing, China). Cellular fatty acids were assayed using a tissue fatty acid assay kit (Tongwei Technologies Inc., Shanghai, China). Cellular cholesterols were assayed using a tissue total cholesterol assay kit (Applygen Technologies Inc., Beijing, China). The experiments were performed according to the manufacturer’s recommended protocol.
Patient samples
HCC tissue samples and their corresponding peritumour tissues utilised in this study were obtained from Tianjin Medical University Cancer Institute and Hospital (Tianjin, P.R. China) after surgical resection. Written consent, approving the use of their tissues for research purposes after the operation, was obtained from each patient. All study procedures were in compliance with the regulations of the Institute of Research Ethics Committee at Nankai University (Tianjin, China). The medical records of the patients are listed in Table S5.
Mass spectrometry (MS)
To identify the interacting proteins of PRMT5 and PRMT5K387R in Flag-PRMT5-expressing HepG2 cells, MS analysis was carried out by Cary Biomedical Technology Co., Ltd. Briefly, 100 μg of protein per condition was transferred into a new Eppendorf tube for digestion. Then, the sample was analysed by online nanospray LC-MS/MS on a Q Exactive™ Plus mass spectrometer (Thermo Fisher Scientific, MA, USA) coupled to an EASY-nanoLC 1000 system (Thermo Fisher Scientific, MA, USA). Tandem mass spectra were processed by PEAKS Studio version X + (Bioinformatics Solutions Inc., Waterloo, Canada). PEAKS DB was set up to search the UniPort Homo sapiens database (Ver201907, 20414 Entries) assuming trypsin as the digestion enzyme. The raw data are listed in Supplementary Material 2.
RNA sequencing data
Total RNA was extracted from HepG2-shCtrl and HepG2-shPT5 cell lines and subjected to RNA-seq analysis performed by Shanghai Majorbio Biopharm Technology Co. Ltd. The data were analysed on the free online platform of the Majorbio Cloud Platform (www.majorbio.com). The RNA sequencing (RNA-seq) data are listed in Supplementary Material 3.
Untargeted metabolomics analysis (untargeted LC–MS)
Untargeted metabolomics analysis of HepG2-shCtrl and HepG2-shPT5 cell lines was performed by Shanghai Majorbio Biopharm Technology Co., Ltd. The data reported in this paper were analysed on the free online platform of the Majorbio Cloud Platform (www.majorbio.com). The raw data are listed in Supplementary Material 4.
TCGA database
Public TCGA (https://portal.gdc.cancer.gov/) data repositories for liver hepatocellular carcinoma (LIHC) (Cancer Genome Atlas Network, 2014) were used to analyse the expression of SIRT7.
Statistical analysis
Each experiment was repeated at least three times. All analyses and graphs were generated with GraphPad Prism 5. Statistical significance was assessed by comparing mean values (± SD) using Student’s t test or ANOVA and was assumed for *P < 0.05; **P < 0.01; ***P < 0.001 and NS (not significant).
Results
Identification of succinylation of PRMT5 K387 and its role in methyltransferase activity of PRMT5
In this study, we attempted to identify the mechanism by which PTMs modulate PRMT5 methyltransferase activity. Protein succinylation is likely to lead to more significant changes than acetylation in protein structure and function due to its dramatic structural alteration and great changes in charge [23]. Given that PRMT5 activity could be regulated by phosphorylation [27], we hypothesised that succinylation might be involved in the regulation of PRMT5 methyltransferase activity. As expected, we observed the succinylation of exogenously (or endogenously) expressed PRMT5 protein (abbreviated as PT5) in HepG2 cells by CoIP analysis using a specific pan anti-succinyllysine (Pan-Ksuc) antibody. Meanwhile, the levels of succinylation were increased by nicotinamide (NAM, an inhibitor of SIRT family deacetylases) and trichostatin A (TSA, an inhibitor of HDAC class I and class II) (Fig. 1a, Supplementary Fig. S1a), suggesting that PRMT5 is succinylated in the cells. Interestingly, the levels of PRMT5 succinylation were higher in the cells treated with NAM than in the cells treated with TSA (Supplementary Fig. S1b), suggesting that the SIRT family is involved in the desuccinylation of PRMT5.
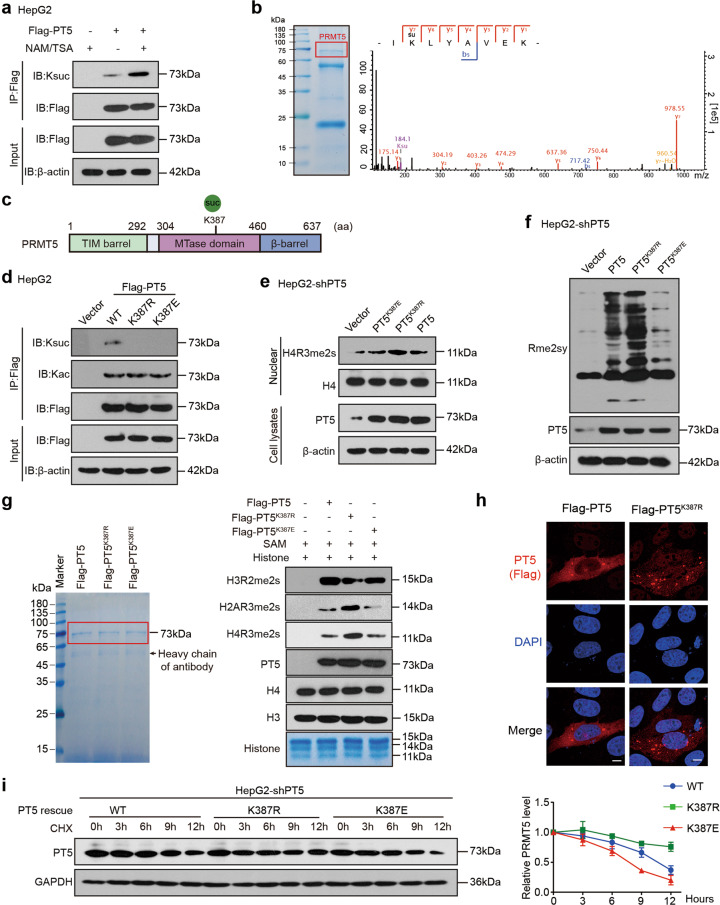
Fig. 1. Identification of succinylation of PRMT5 K387 and its role in methyltransferase activity of PRMT5.
a Succinylation levels of exogenous PRMT5 were tested by immunoprecipitation (IP) analysis in HepG2 cells treated with NAM and TSA as indicated. b Succinylation sites of PRMT5 were examined by liquid chromatography-mass spectrometry (LC-MS). PRMT5 was immunoprecipitated from HepG2 cells, and the loading proteins were stained with Coomassie blue (left panel). K represents potential succinylation sites in PRMT5 analysed by LC-MS (right panel). c A schematic model of the structure and succinylation sites of PRMT5. d The succinylation and acetylation levels of Flag-tagged PRMT5 or PRMT5 mutants were examined by IP analysis in HepG2 cells transfected with the indicated plasmids. e, f The methylation of H4R3 or global proteins was detected by Western blot analysis using H4R3me2s or Rme2sy antibodies in HepG2-shPT5 cells transfected with the indicated plasmids. g In vitro methylation assays were performed using recombinant histone proteins and purified Flag-PRMT5. Flag-tagged PRMT5 or PRMT5 mutants purified from HEK293T cells were incubated with SAM and recombinant histones. The loading of the proteins was visualised using Coomassie blue staining (left). The methylation of H4 R3, H3 R2 and H2A R3 was evaluated by Western blot analysis using H4R3me2s, H3R2me2s and H2AR3me2s antibodies (right). h The localisation of exogenous Flag-tagged PRMT5 (red) was determined by immunofluorescence staining in HepG2-shPRMT5 cells. Scale bars, 5 μm. i HepG2-shPT5 cells were transfected with WT or mutant PRMT5 plasmids, followed by treatment with cycloheximide (CHX, 100 μg/mL) for the indicated times. The levels of PRMT5 were detected by Western blot analysis (left). The quantitative analysis of PRMT5 expression is shown (right).
To identify the succinylation sites of PRMT5, we purified the PRMT5 protein from HepG2 cells and examined its succinylation sites by performing liquid chromatography–mass spectrometry (LC–MS), in which only one lysine residue 387 (K387) was identified as a candidate site (Fig. 1b, c). Then, we generated two mutants, namely, K387R (lysine to arginine, mimicking the desuccinylated state) and K387E (lysine to glutamic acids, mimicking the negatively charged succinyllysine modification). We failed to observe the succinylation (but not acetylation) of Flag-tagged PRMT5 mutants (K387R and K387E) in HepG2, PANC1 and MCF-7 cells (Fig. 1d, Supplementary Fig. S1c), suggesting that K387 is a succinylation site of PRMT5.
PRMT5 catalyses the formation of symmetric dimethyl arginine (sDMA) on histone and nonhistone proteins, and PRMT5 has been shown to methylate histones H2A, H3, and H4 in vitro and in vivo [2, 6, 9]. Symmetric methylation of histone H4 arginine 3 (H4R3me2s) is an established substrate of PRMT5 [30]. To test the effect of the PRMT5 mutants (K387R and K387E) on its methyltransferase activity, we conducted enzyme activity assays in vivo and in vitro. Because PRMT5 knockout led to obvious apoptosis and cell death, we generated a stable PRMT5 knockdown HepG2 cell line (HepG2-shPT5). The knockdown efficiency of PRMT5 was assessed by Western blot analysis (Supplementary Fig. S1d), and shPRMT5#3 targeting the 3′UTR region of PRMT5 was used for subsequent experiments. The reintroduction of PRMT5 rescued the shPRMT5#3 (termed shPRMT5)-mediated decrease in PRMT5 and H4R3me2s in HepG2 cells (Supplementary Fig. S1e). Then, we generated HepG2-shPT5 cell lines with stable PRMT5, PRMT5K387R, and PRMT5K387E expression (HepG2-shPT5 + PT5, HepG2-shPT5 + PT5K387R, HepG2-shPT5 + PT5K387E). Compared to wild-type PRMT5, overexpression of PRMT5K387R greatly increased the levels of H4R3me2s and global levels of symmetrical methylation of arginine residues (Rme2sy), while PRMT5K387E reduced the levels of H4R3me2s and Rme2sy in HepG2-shPT5 cell lines (Fig. 1e, f), suggesting that the succinylation of PRMT5 K387 reduces its methyltransferase activity in vivo. We obtained similar results in PANC1 and MCF-7 cell lines (Supplementary Fig. S1f). The in vitro methylation assays showed that PRMT5K387R greatly enhanced the methylation of H4R3 and H2AR3, but H3R2 and PRMT5K387E displayed the opposite effects (Fig. 1g), suggesting that the succinylation of PRMT5 K387 greatly impairs its methyltransferase activity in vitro. Using the PRMT5-rescued HepG2-shPT5 cell lines, we found that the PRMT5K387R mutant failed to influence protein localisation, possibly enhancing protein stability compared with wild-type PRMT5 (Fig. 1h, i). Taken together, we conclude that PRMT5 is succinylated at K387, which affects its methyltransferase activity.
SIRT7 mediates the desuccinylation of PRMT5 to enhance its methyltransferase activity
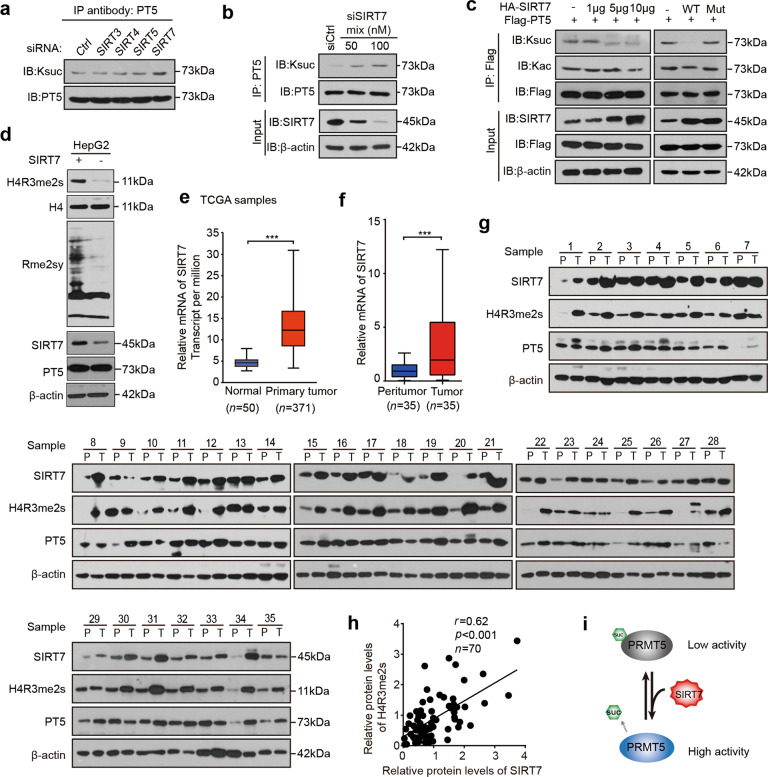
Because SIRT family members were preferentially involved in PRMT5 succinylation (Supplementary Fig. S1b), we next screened the major deacetylase members of the SIRT family. The siRNA efficiency of the deacetylases was verified by RT-qPCR and Western blot analysis in the HepG2 cell line (Supplementary Fig. S2a, b). Our results showed that siSIRT7 remarkably increased the succinylation levels of PRMT5 in HepG2 cells (Fig. 2a). Notably, SIRT7 knockdown (or overexpression) increased (or decreased) the succinylation levels of endogenous PRMT5 in a dose-dependent manner, and the mutant of SIRT7 without catalytic activity failed to affect the succinylation levels of PRMT5 in the cells (Fig. 2b, c, Supplementary Fig. S2c), indicating that SIRT7 is an eraser of PRMT5 succinylation. Importantly, the overexpression of SIRT7 increased the levels of Rme2sy and H4R3me2s in HepG2 cells (Fig. 2d), suggesting that SIRT7-mediated desuccinylation of PRMT5 increases its methyltransferase activity. Clinically, TCGA data analysis revealed that the expression levels of SIRT7 were elevated in HCC samples compared with normal samples (Fig. 2e). Moreover, high levels of SIRT7 correlated with poor patient survival (Supplementary Fig. S2d). As expected, our data validated that the mRNA levels of SIRT7 were significantly increased in 35 clinical HCC tissues relative to their corresponding peritumour tissues (Fig. 2f). The succinylation levels of PRMT5 proteins were examined in clinical liver cancer and peritumour samples (Supplementary Fig. S2e). Western blot analysis revealed that SIRT7 and H4R3me2s were overexpressed in the above HCC tissues relative to their corresponding peritumour tissues (Fig. 2g). Then, the relationship between SIRT7 expression levels and H4R3me2s levels was analysed by ImageJ software in the above samples (Fig. 2h), supporting that SIRT7 is associated with the methyltransferase activity of PRMT5 in HCC. Thus, we conclude that SIRT7 mediates the desuccinylation of PRMT5 to enhance its methyltransferase activity in a model (Fig. 2i).
Fig. 2. SIRT7 mediates the desuccinylation of PRMT5 to enhance its methyltransferase activity.
a HepG2 cells were treated with different siRNAs targeting SIRT family members. PRMT5 was immunoprecipitated, and the succinylation of PRMT5 was analysed by Western blot analysis. b HepG2 cells were treated with different concentrations of siSIRT7 Mix (siSIRT7#1, siSIRT7#2, siSIRT7#3). PRMT5 was immunoprecipitated, and the succinylation of PRMT5 was analysed by Western blot analysis. c HepG2 cells were cotransfected with plasmids containing Flag-tagged PRMT5 and HA-tagged SIRT7 or SIRT7 mutant. Flag-tagged PRMT5 was immunoprecipitated with M2 beads, and the acetylation and succinylation levels of PRMT5 were analysed by Western blot analysis. d Methylation of global proteins and H4R3 was detected by Western blot analysis in HepG2 cells transfected with or without SIRT7 plasmids using Rme2sy and H4R3me2s antibodies. e Expression of SIRT7 in normal and LIHC (liver hepatocellular carcinoma) tumour samples in TCGA database. f The mRNA levels of SIRT7 were evaluated by RT-qPCR in 35 pairs of clinical liver cancer tissues. g The protein levels of SIRT7, H4R3me2s and PRMT5 were determined by Western blot analysis in 35 pairs of clinical liver cancer tissues. h The relationship between the levels of SIRT7 expression and the levels of H4R3me2s in the above samples was analysed by ImageJ software. i A model showed that SIRT7 desuccinylates PRMT5 to enhance the methyltransferase activity of PRMT5. Statistically significant differences are indicated: ***P < 0.001; Student’s t test.
SIRT7-mediated desuccinylation of PRMT5 K387 enhances the formation of the PRMT5-Mep50 octamer
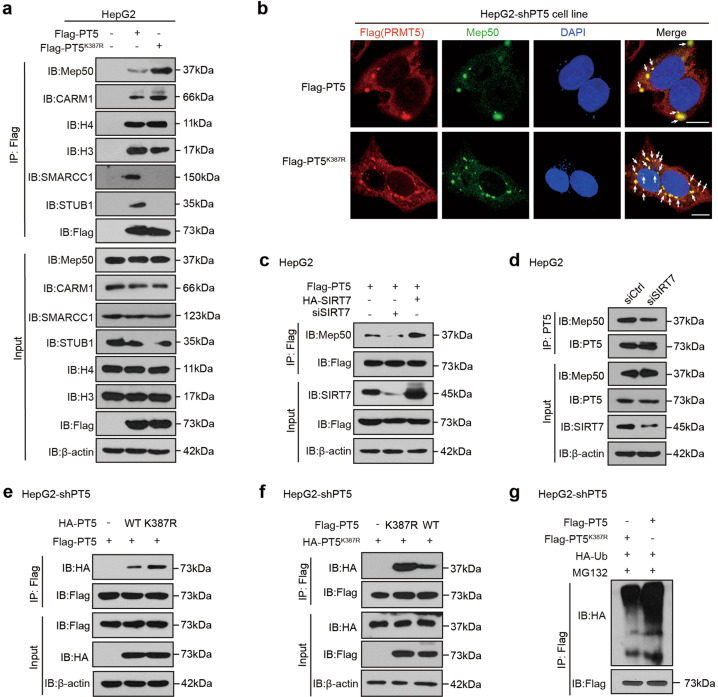
In mammalian cells, PRMT5 is tightly bound by Mep50, and this interaction is required for PRMT5 activity [31]. Meanwhile, other binding proteins can function as modulators or adaptors of the PRMT5-Mep50 complex [32]. In our system, we validated that Mep50 markedly enhanced the methyltransferase activity of PRMT5 in HepG2-shPT5 cells (Supplementary Fig. S3a). Considering that lysine succinylation induced substantial changes to a protein’s chemical properties, we examined whether the succinylation of PRMT5 affected the interaction of PRMT5 with its binding proteins. LC-MS analysis of the immunoprecipitated proteins of PRMT5 or PRMT5K387R mutants in HepG2 cells showed that the mutant of PRMT5 K387R remarkably influenced the interaction of PRMT5 with its binding proteins (Supplementary Fig. S3b, c). Then, CoIP assays confirmed that PRMT5K387R increased (reduced) its binding protein levels of Mep50 and CARM1 (H3, SMARCC1 and STUB1) relative to wild-type PRMT5 (Fig. 3a). Immunofluorescence staining showed that PRMT5K387R increased the colocalization of PRMT5 with Mep50 in HepG2-shPT5 cells (Fig. 3b), suggesting that the desuccinylation of PRMT5 promotes the binding of Mep50 to PRMT5. As expected, the overexpression (or knockdown) of SIRT7 enhanced (or reduced) the interaction between PRMT5 and Mep50 in HepG2 cells (Fig. 3c, d), suggesting that SIRT7-mediated desuccinylation of PRMT5 enhances the interaction of PRMT5 and Mep50. Our data showed that PRMT5K387R increased the interaction between PRMT5 and PRMT5 in the PRMT5-Mep50 octamer (Fig. 3e, f). Interestingly, Western blot analysis showed that treatment with CHX decreased the protein levels of PRMT5, while MG132 (a proteasome inhibitor) reversed the effect induced by CHX in HepG2 cells. Considering that STUB1 regulates PRMT5 via ubiquitin-dependent proteasomal degradation [33], we validated the interaction of PRMT5 with STUB1 in our system (Fig. 3a, Supplementary Fig. S3d). Moreover, ubiquitination assays showed that the mutant of PRMT5 K387R resulted in a substantial decrease in PRMT5 ubiquitination in HepG2 cells (Fig. 3g), suggesting that the desuccinylation of PRMT5 inhibits the ubiquitination of PRMT5. Taken together, we conclude that the SIRT7-mediated desuccinylation of PRMT5 K387 enhances the formation of the PRMT5-Mep50 octamer in the cells.
Fig. 3. SIRT7-mediated desuccinylation of PRMT5 K387 enhances the formation of the PRMT5-Mep50 octamer.
a Immunoprecipitation analysis of the interaction between Mep50 (or CARM1, H4, H3, SMARCC1, and STUB1) and WT or mutant PRMT5 in HepG2 cells transfected with Flag-tagged PT5 or Flag-tagged PT5K387R. b The colocalization between endogenous Mep50 and Flag-tagged PRMT5 was examined by immunofluorescence staining in HepG2-shPT5 cells. Scale bars, 10 μm. c Immunoprecipitation analysis of the interaction between endogenous Mep50 and Flag-PRMT5 in HepG2 cells treated with HA-tagged SIRT7 or siSIRT7. d Immunoprecipitation analysis of the interaction between Mep50 and PRMT5 in HepG2 cells treated with or without siSIRT7. e, f The effect of PRMT5K387R on the interaction of PRMT5 and PRMT5 was measured by CoIP analysis in HepG2-shPT5 cells. g The ubiquitination levels of PRMT5 and PRMT5K387R were detected by CoIP analysis in HepG2 cells.
Desuccinylation of PRMT5 K387 confers lipid metabolism reprogramming by inducing arginine methylation of SREBP1a
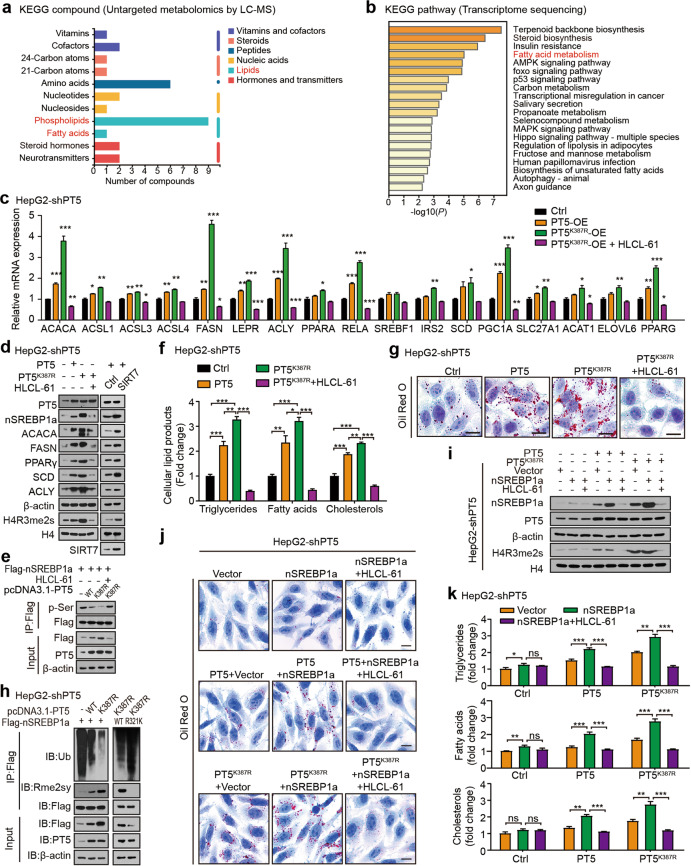
PRMT5 is a type II arginine methyltransferase with a wide range of biological and physiological effects [34]. Next, we explored the effect of desuccinylation of PRMT5 K387 on tumour cells. Untargeted metabolomics analysis (untargeted LC-MS) showed that numerous metabolites, including phospholipids, fatty acids, steroid hormones, neurotransmitters, nucleosides, nucleotides and amino acids were influenced by PRMT5 in hepatoma cells (Fig. 4a, Supplementary Fig. S4a, b), suggesting that PRMT5 plays a crucial role in the metabolic reprogramming of tumour cells. Moreover, RNA-seq revealed that PRMT5 conferred lipid metabolism, steroid biosynthesis, insulin resistance, the AMPK, the p53, the MAPK, the Hippo signalling pathways and autophagy in hepatoma cells (Fig. 4b, Supplementary Fig. S4c), suggesting that PRMT5 globally functions in the development of cancer. Then, we verified the effect of PRMT5 and PRMT5K387R on the expression of lipid metabolism-related genes by RT-qPCR and Western blot. RT-qPCR showed that PRMT5 upregulated the mRNA levels of ACACA, ACSL1, ACSL3, ACSL4, FASN, LEPR, ACLY, RELA, PGC1A, SLC27A1 and PPARG in HepG2-shPT5 cells. Western blot analysis demonstrated that PRMT5 increased the protein levels of nSREBP1a, FASN, ACACA, PPARγ, SCD, and ACLY in the cells. Interestingly, PRMT5K387R displayed a stronger effect than wild-type PRMT5 in the system, which could be inhibited by treatment with HLCL-61, an inhibitor of PRMT5 (Fig. 4c–e), suggesting that desuccinylation of PRMT5 K387 upregulates lipid metabolism-associated factors by increasing its methyltransferase activity in tumour cells. The overexpression of SIRT7 increased the protein levels of nSREBP1a, FASN, ACACA, PPARγ, and SCD in the cells (Fig. 4d). IP assays showed that PRMT5K387R reduced the phosphorylation of SREBP1a (Fig. 4e). Moreover, PRMT5K387R elevated the levels of triglycerides, fatty acids, cholesterols and lipid droplets, in which HLCL-61 blocked the effect of PRMT5K387R on the factors in hepatoma cells (Fig. 4f, g), supporting that the desuccinylation of PRMT5 K387 promotes lipid metabolism reprogramming by increasing its methyltransferase activity in tumour cells.
Fig. 4. Desuccinylation of PRMT5 K387 confers lipid metabolism reprogramming by inducing arginine methylation of SREBP1a.
a The KEGG pathway database was used to analyse the metabolites affected by PRMT5 in HepG2 cells. b The KEGG pathway enrichment analysis of PRMT5-regulated target genes in HepG2 cells. c The effect of PRMT5 or PRMT5K387R on the expression of lipid metabolism-associated genes was measured by RT-qPCR analysis in HepG2-shPT5 cells. d The effect of PRMT5, PRMT5K387R and SIRT7 on the protein levels of the indicated proteins was determined by Western blot analysis in HepG2-shPT5 cells. e The effect of PRMT5 and PRMT5K387R on the phosphorylation levels of SREBP1a proteins was tested by IP analysis in HepG2-shPT5 cells. f The levels of intracellular triglycerides, fatty acids and cholesterols were evaluated after the indicated treatments, such as pcDNA3.1-PT5, pcDNA3.1-PT5K387R, or pcDNA3.1-PT5K387R + HLCL-61, in HepG2-shPT5 cells. g The effect of PRMT5 or PRMT5K387R on lipogenesis was determined by Oil Red O staining in HepG2-shPT5 cells. Scale bars, 10 μm. h The ubiquitination and methylation levels of wild-type (WT) SREBP1a and mutant (R321K) SREBP1a were detected by CoIP analysis in HepG2-shPT5 cells transfected with vector, PRMT5 and PRMT5K387R plasmids. i The protein levels of SREBP1a, PRMT5 and H4R3me2s were examined by Western blot analysis in HepG2-shPT5 cells with the indicated treatment. j The effect of PRMT5 or PRMT5K387R with SREBP1a on lipogenesis was determined by Oil Red O staining in HepG2-shPT5 cells. Scale bars, 10 μm. k The levels of intracellular triglycerides, fatty acids and cholesterols were evaluated after the indicated treatment in HepG2-shPT5 cells. Statistically significant differences are indicated: ns, no significance; *P < 0.05; **P < 0.01; ***P < 0.001; one-way ANOVA.
SREBP1a is a vital transcription factor involved in cholesterol, fatty acid, and triglyceride biogenesis [35]. It has been reported that PRMT5 symmetrically dimethylates SREBP1a on R321, leading to its evasion from degradation through the ubiquitin-proteasome pathway [12]. Therefore, we were interested in the effect of desuccinylation of PRMT5 K387 on SREBP1a. Based on the data that PRMT5K387R remarkably increased the protein levels of SREBP1a but not the mRNA levels in HepG2-shPT5 cells as above, we examined the effect of PRMT5K387R on the ubiquitination of SREBP1a in the cells. CoIP assays revealed that PRMT5K387R increased the levels of arginine methylation of nSREBP1a and reduced its ubiquitination using Rme2sy (pan-symmetric dimethylation) and Ub (pan-ubiquitin) antibodies in the cells, and the mutant of SREBP1a (R321K) without SREBP1 methylation failed to affect the event (Fig. 4h), suggesting that PRMT5K387R increases the levels of SREBP1a protein by reducing its proteasomal-dependent ubiquitination degradation in the cells. Moreover, PRMT5K387R significantly increased the protein levels of nSREBP1a by enhancing the enzyme activity of PRMT5 in the cells (Fig. 4i). PRMT5K387R enhanced the levels of lipid droplets, triglycerides, fatty acids and cholesterols, in which the overexpression of nSREBP1a elevated the role of PRMT5K387R and HLCL-61 blocked the effect of PRMT5K387R and SREBP1a on the factors in the system (Fig. 4j, k), suggesting that the desuccinylation of PRMT5 K387 leads to lipid metabolism reprogramming through targeting SREBP1a in tumour cells. Overall, we summarise that the desuccinylation of PRMT5 K387-enhanced methyltransferase activity contributes to the lipid metabolism reprogramming of tumour cells by inducing the arginine methylation of SREBP1a.
Desuccinylation of PRMT5 K387 promotes the proliferation, migration and invasion of cancer cells in vitro
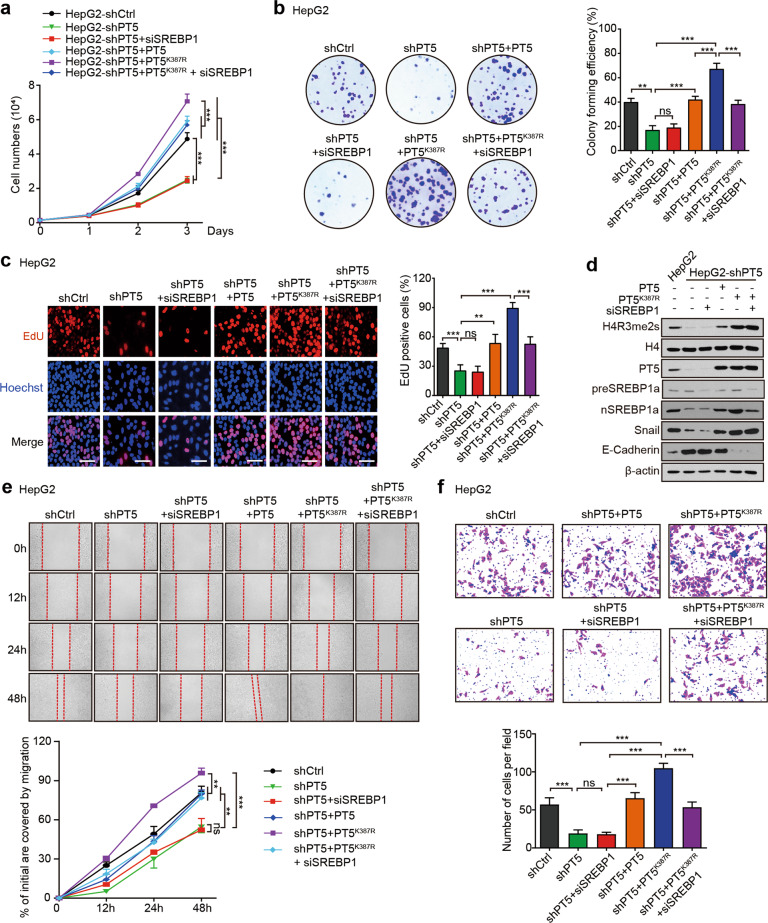
Given that PRMT5 is highly expressed in cancers and associated with the development of cancer [32], we next investigated the effect of PRMT5 succinylation modification on tumorigenesis. CCK-8, colony formation, EdU assays, Western blot analysis, wound healing and Transwell invasion assays revealed that knockdown of PRMT5 (shPT5) dramatically decreased cell proliferation, migration and invasion, which could be strongly rescued by PRMT5K387R relative to wild-type PRMT5 in HepG2 cells, indicating that the desuccinylation of PRMT5 promotes the proliferation, migration, and invasion of cancer cells. Moreover, SREBP1a knockdown (siSREBP1a) blocked the enhancing effect of PRMT5K387R on the proliferation, migration and invasion of tumour cells (Fig. 5a–f), suggesting that SREBP1a is involved in the events in which desuccinylation of PRMT5 promotes the proliferation, migration and invasion of cancer cells. Moreover, shPRMT5 apparently decreased the protein level of nSREBP1a, whereas the preSREBP1a level had no change in HepG2 cells, suggesting that PRMT5 modulates the protein SREBP1a at the posttranscriptional level (Fig. 5d). Meanwhile, CCK-8, colony formation, and Transwell invasion assays were used to repeat the data in MCF-7 and PANC1 cells (Supplementary Fig. S5a–c). Thus, we conclude that the desuccinylation of PRMT5 K387 promotes the proliferation, migration and invasion of cancer cells in vitro.
Fig. 5. Desuccinylation of PRMT5 K387 promotes the proliferation, migration and invasion of cancer cells in vitro.
The proliferation ability of cancer cells was evaluated by CCK-8 (a), colony formation assays (b) and EdU assays (c) in HepG2 cells with the indicated treatments, such as shCtrl, shPT5, shPT5 + siSREBP1, shPT5 + PT5, shPT5 + PT5K387R, and shPT5 + PT5K387R + siSREBP1. d The protein levels of H4R3me2s, PRMT5, SREBP1a, Snail, and E-cadherin were detected by Western blot analysis in HepG2 cells with the indicated treatment. e The migration ability of cancer cells was detected by wound-healing assays in HepG2 cells with the indicated treatment. f The invasion ability of cancer cells was determined by Transwell cell invasion assays in HepG2 cells with the indicated treatment. Statistically significant differences are indicated: ns, no significance; **P < 0.01; ***P < 0.001; one-way ANOVA or two-way ANOVA.
Desuccinylation of PRMT5 K387 accelerates tumour growth and metastasis in vivo
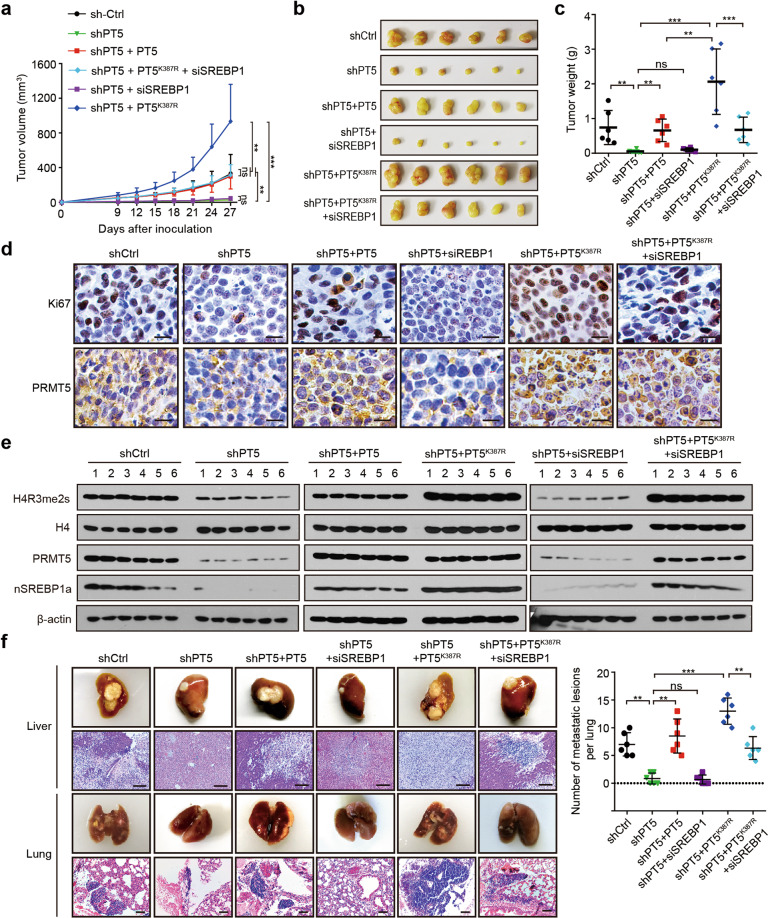
Next, we further investigated the effect of desuccinylation of PRMT5 K387 on tumorigenesis in vivo. Interestingly, we observed that the shPT5 + PT5K387R group had enhanced tumour growth relative to the shPT5 + PT5 group in the HepG2 tumour-bearing nude mice model, in which SREBP1a knockdown (siSREBP1) blocked the effect (Fig. 6a–c). IHC staining assays indicated that the expression of Ki67 was higher in the tumour tissues from the shPT5 + PT5K387R group than that of Ki67 in the tumour tissues from the shPT5 + PT5 group, in which SREBP1 knockdown blocked the effect (Fig. 6d). Western blot analysis validated that the levels of H4R3me2s and SREBP1a were elevated in the tumour tissues from shPT5 + PT5K387R group mice (Fig. 6e), suggesting that the desuccinylation of PRMT5 K387 accelerates tumour growth by increasing the protein levels of SREBP1a. Moreover, the shPT5 + PT5K387R group had increased metastatic lesions per lung relative to the shPT5 + PT5 group in the HepG2 orthotopic mouse model, in which SREBP1a knockdown blocked the effect (Fig. 6f), suggesting that the desuccinylation of PRMT5 K387 promotes tumour metastasis through SREBP1a. Taken together, we conclude that the desuccinylation of PRMT5 K387 accelerates tumour growth and metastasis in vivo.
Fig. 6. Desuccinylation of PRMT5 K387 accelerates tumour growth and metastasis in vivo.
a Growth curves of tumours from tumour-bearing nude mice. HepG2 cells were treated as indicated, such as shCtrl, shPT5, shPT5 + siSREBP1, shPT5 + PT5, shPT5 + PT5K387R, and shPT5 + PT5K387R + siSREBP1, and then injected subcutaneously into the flanks of nude mice (n = 6 mice). Tumour diameters were measured at the indicated time points, and tumour volumes were calculated. b Photographs of dissected tumours from nude mice. c Average tumour weight of each group. d The expression of Ki67 and PRMT5 was detected by IHC in tumour tissues from nude mice. Scale bars, 25 μm. e The protein levels of H4R3me2s, PRMT5, and SREBP1a were examined by Western blot analysis in tumour tissues from tumour-bearing nude mice. f Photographs and H&E staining of the liver and lung tissues from the HepG2 orthotopic liver tumour mice model. HepG2 cells were treated as indicated. The cells were injected into the liver of nude mice (n = 6 mice). Scale bars, 200 μm. Statistically significant differences are indicated: ns, no significance; **P < 0.01; ***P < 0.001; one-way ANOVA.
Discussion
PRMT5 plays important roles in the development of cancer, in which the methyltransferase activity of PRMT5 is crucial for its function [36]. However, the role of PRMT5 in the modulation of tumour growth is poorly understood. In recent years, our understanding of protein PTMs has rapidly expanded. Succinylation leads to significant changes in protein structure and function in tumours [28]. In this study, we investigated the role of succinylation of PRMT5 in the development of cancer.
Arginine methylation, a PTM, is widely found in mammalian cells. The nine members of the human PRMT family are grouped into three types according to their mode of methylation: type I (PRMT1, PRMT2, PRMT3, PRMT4, PRMT6 and PRMT8), which catalyses the formation of ω-NG-monomethylarginines (MMA), ω-NG, NG-asymmetric dimethylarginines; type II (PRMT5 and PRMT9), which catalyses the formation of ω-NGMMA and ω-NG; N′G-sDMA; and type III (PRMT7), which is only known to catalyse the formation of ω-NG-MMA [37]. As the major sDMA methyltransferase, PRMT5 plays indispensable roles in cell development. Indeed, numerous studies have solidified PRMT5 as a major regulator of epigenetic-mediated gene expression, the DNA damage response, mRNA splicing, stem cell function, and the immune response processes or cellular states that are hijacked by the tumour to ensure its survival [9, 34, 38–44]. PRMT5 can methylate H2A and H4 efficiently and H3 weakly, and H4R3me2s is a key histone methylation mark deposited by PRMT5 [2, 30]. Considering that PRMT5 activity can be regulated by phosphorylation [27], we attempted to identify other PTMs involved in the modulation of the methyltransferase activity of PRMT5. Interestingly, we found that PRMT5 was succinylated at the K387 site in vitro and in vivo, and the desuccinylation of PRMT5 K387 greatly enhanced its methyltransferase activity. New types of nonacetyl acylations usually share the same enzymes mediating the erasing of acetylation [24]. Emerging evidence suggests that sirtuins can remove acyl groups in addition to acetyl groups from lysine residues [45, 46]. Then, we screened the PRMT5 desuccinylase from the major deacetylase members of the SIRT family. Our data showed that SIRT7 was an eraser of PRMT5 succinylation. Our results agree with reports that SIRT7 is a desuccinylase [26, 47]. We found that the desuccinylation of PRMT5 K387 was required for its methyltransferase activity in tumours.
We next examined the underlying mechanism by which the desuccinylation of PRMT5 affected the methyltransferase activity of PRMT5. Human PRMT5 binds Mep50 to form a stable hetero-octameric complex [31]. The PRMT5-Mep50 complex had a significantly higher level of methyltransferase activity compared with PRMT5 alone, owing to higher affinities for both the binding partners, substrates and SAM [31]. As an obligate binding partner of PRMT5 that directs substrates to the active site, the upregulation of Mep50 results in an increase in the methyltransferase activity of PRMT5 [48]. Our studies showed that the desuccinylation of PRMT5 K387 enhanced its methyltransferase activity by promoting the binding of Mep50 to PRMT5. PRMT5 directly associates with a range of other protein factors that may alter its subcellular localisation and protein substrate selection, such as CARM1, SWI/SNF and CHIP (STUB1) [8, 33, 49]. Interestingly, our data revealed that the desuccinylation of PRMT5 K387 altered the interaction of PRMT5 with its binding proteins, such as CARM1, SMARCC1 and STUB1. However, the desuccinylation of PRMT5 K387 failed to bind to STUB1, resulting in a substantial decrease in PRMT5 ubiquitination. This suggests that the desuccinylation of PRMT5 K387 enhances its methyltransferase activity by increasing the formation of the hetero-octameric complex.
PRMT5 plays important roles in cancer cell function and survival [38, 39]. Based on RNA-seq and a nontargeted metabolomics approach, we found that PRMT5 significantly modulated lipid metabolism in tumour cells. Our data agree with recently published studies showing that PRMT5 regulates fatty acid metabolism and lipid droplet biogenesis in white adipose tissues [29]. Functionally, we validated that the desuccinylation of PRMT5 K387 contributed to lipid metabolism reprogramming in tumours by inducing arginine methylation of SREBP1a. This suggests that the desuccinylation of PRMT5 K387 accelerates tumour growth and metastasis by targeting SREBP1a. Therapeutically, PRMT5 may serve as a precise target for cancers.
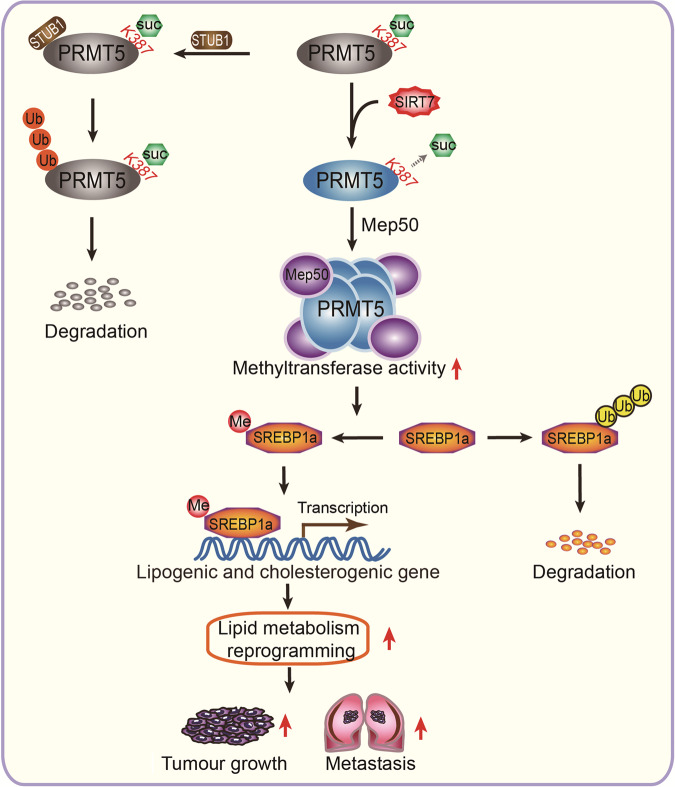
In conclusion, we summarise a model of SIRT7-mediated desuccinylation of PRMT5 K387 in the modulation of tumour growth in Fig. 7. In this model, the desuccinylation of PRMT5 at K387 enhances its methyltransferase activity in the cells. SIRT7, as an eraser of PRMT5 K387, mediates the desuccinylation of PRMT5 K387 in cells. SIRT7-mediated desuccinylation of PRMT5 K387 promotes the formation of the PRMT5-Mep50 complex. Moreover, the desuccinylation of PRMT5 significantly promotes lipid metabolism reprogramming, tumour growth and metastasis of cancer cells in vivo and in vitro by inducing the arginine methylation of SREBP1a. Our findings provide new insights into the mechanism of PRMT5 methyltransferase activity in tumorigenesis.
Fig. 7. A model of desuccinylation of PRMT5 K387 modulating its methyltransferase activity in tumours.
In this model, the desuccinylation of PRMT5 at K387 enhances its methyltransferase activity in the cells. SIRT7, as an eraser of PRMT5 K387, mediates the desuccinylation of PRMT5 K387 in cells. SIRT7-mediated desuccinylation of PRMT5 K387 promotes the formation of the PRMT5-Mep50 complex. Moreover, the desuccinylation of PRMT5 significantly promotes lipid metabolism reprogramming, tumour growth and metastasis of cancer cells in vivo and in vitro by inducing the arginine methylation of SREBP1a.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31670769), National Natural Science Foundation of China (No. 81970519), National Natural Science Foundation of China (No. 82173317), Programme for JLU Science and Technology Innovative Research Team (2017TD-08), Fundamental Research Funds for the Central Universities, TianQing Liver Disease Research Fund Subject (No. TQGB20200118), and National Science and Technology Major Project of China (No. 2018ZX10732101).
Author contributions
HFY designed, performed experiments, analysed data, and wrote the manuscript. MZ designed, performed experiments, and analysed data. LNZ and HLY performed experiments and analysed data. GY, YY, YG, YFW and WZ performed experiments. JQN and TQS designed, oversaw experiments, technical and material support, and obtained funding. XDZ designed, oversaw experiments, interpreted data, technical and material support, obtained funding and wrote the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Hong-feng Yuan, Man Zhao.
Contributor Information
Tian-qiang Song, Email: tjmuch@hotmail.com.
Jun-qi Niu, Email: junqiniu@aliyun.com.
Xiao-dong Zhang, Email: zhangxiaodong@tjmuch.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41401-021-00841-y.
References
- 1.Li Y, Chitnis N, Nakagawa H, Kita Y, Natsugoe S, Yang Y, et al. PRMT5 is required for lymphomagenesis triggered by multiple oncogenic drivers. Cancer Discov. 2015;5:288–303. doi: 10.1158/2159-8290.CD-14-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burgos ES, Wilczek C, Onikubo T, Bonanno JB, Jansong J, Reimer U, et al. Histone H2A and H4 N-terminal tails are positioned by the MEP50 WD repeat protein for efficient methylation by the PRMT5 arginine methyltransferase. J Biol Chem. 2015;290:9674–89. doi: 10.1074/jbc.M115.636894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho MC, Wilczek C, Bonanno JB, Xing L, Seznec J, Matsui T, et al. Structure of the arginine methyltransferase PRMT5-MEP50 reveals a mechanism for substrate specificity. PLoS One. 2013;8:e57008. doi: 10.1371/journal.pone.0057008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang M, Xu RM, Thompson PR. Substrate specificity, processivity, and kinetic mechanism of protein arginine methyltransferase 5. Biochemistry. 2013;52:5430–40. doi: 10.1021/bi4005123. [DOI] [PubMed] [Google Scholar]
- 5.Liu J, Zhang E, Ma Z, Wu W, Kosinska A, Zhang X, et al. Enhancing virus-specific immunity in vivo by combining therapeutic vaccination and PD-L1 blockade in chronic hepadnaviral infection. PLoS Pathog. 2014;10:e1003856. doi: 10.1371/journal.ppat.1003856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, et al. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006;8:623–30. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- 7.Branscombe TL, Frankel A, Lee JH, Cook JR, Yang Z, Pestka S, et al. PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J Biol Chem. 2001;276:32971–6. doi: 10.1074/jbc.M105412200. [DOI] [PubMed] [Google Scholar]
- 8.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24:9630–45. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B, et al. Arginine methylation regulates the p53 response. Nat Cell Biol. 2008;10:1431–9. doi: 10.1038/ncb1802. [DOI] [PubMed] [Google Scholar]
- 10.Kwak YT, Guo J, Prajapati S, Park KJ, Surabhi RM, Miller B, et al. Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol Cell. 2003;11:1055–66. doi: 10.1016/S1097-2765(03)00101-1. [DOI] [PubMed] [Google Scholar]
- 11.Tan CP, Nakielny S. Control of the DNA methylation system component MBD2 by protein arginine methylation. Mol Cell Biol. 2006;26:7224–35. doi: 10.1128/MCB.00473-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L, Zhao X, Zhao L, Li J, Yang H, Zhu Z, et al. Arginine methylation of SREBP1a via PRMT5 promotes de novo lipogenesis and tumor growth. Cancer Res. 2016;76:1260–72. doi: 10.1158/0008-5472.CAN-15-1766. [DOI] [PubMed] [Google Scholar]
- 13.Fabbrizio E, El Messaoudi S, Polanowska J, Paul C, Cook JR, Lee JH, et al. Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep. 2002;3:641–5. doi: 10.1093/embo-reports/kvf136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Pal S, Sif S. Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Mol Cell Biol. 2008;28:6262–77. doi: 10.1128/MCB.00923-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pastore F, Bhagwat N, Pastore A, Radzisheuskaya A, Karzai A, Krishnan A, et al. PRMT5 inhibition modulates E2F1 methylation and gene-regulatory networks leading to therapeutic efficacy in JAK2(V617F)-mutant MPN. Cancer Discov. 2020;10:1742–57. doi: 10.1158/2159-8290.CD-20-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bao X, Zhao S, Liu T, Liu Y, Liu Y, Yang X. Overexpression of PRMT5 promotes tumor cell growth and is associated with poor disease prognosis in epithelial ovarian cancer. J Histochem Cytochem. 2013;61:206–17. doi: 10.1369/0022155413475452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung J, Karkhanis V, Tae S, Yan F, Smith P, Ayers LW, et al. Protein arginine methyltransferase 5 (PRMT5) inhibition induces lymphoma cell death through reactivation of the retinoblastoma tumor suppressor pathway and polycomb repressor complex 2 (PRC2) silencing. J Biol Chem. 2013;288:35534–47. doi: 10.1074/jbc.M113.510669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ibrahim R, Matsubara D, Osman W, Morikawa T, Goto A, Morita S, et al. Expression of PRMT5 in lung adenocarcinoma and its significance in epithelial-mesenchymal transition. Hum Pathol. 2014;45:1397–405. doi: 10.1016/j.humpath.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 19.Scoumanne A, Zhang J, Chen X. PRMT5 is required for cell-cycle progression and p53 tumor suppressor function. Nucleic Acids Res. 2009;37:4965–76. doi: 10.1093/nar/gkp516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang S, Ma Y, Hu X, Zheng Y, Chen X. Targeting PRMT5/Akt signalling axis prevents human lung cancer cell growth. J Cell Mol Med. 2019;23:1333–42. doi: 10.1111/jcmm.14036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheng X, Bowen N, Wang Z. GLI pathogenesis-related 1 functions as a tumor-suppressor in lung cancer. Mol Cancer. 2016;15:25. doi: 10.1186/s12943-016-0508-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z, Tan M, Xie Z, Dai L, Chen Y, Zhao Y. Identification of lysine succinylation as a new post-translational modification. Nat Chem Biol. 2011;7:58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabari BR, Zhang D, Allis CD, Zhao Y. Metabolic regulation of gene expression through histone acylations. Nat Rev Mol Cell Biol. 2017;18:90–101. doi: 10.1038/nrm.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao S, Zhang X, Li H. Beyond histone acetylation-writing and erasing histone acylations. Curr Opin Struct Biol. 2018;53:169–77. doi: 10.1016/j.sbi.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell. 2013;50:919–30. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Shi L, Yang S, Yan R, Zhang D, Yang J, et al. SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat Commun. 2016;7:12235. doi: 10.1038/ncomms12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu F, Zhao X, Perna F, Wang L, Koppikar P, Abdel-Wahab O, et al. JAK2V617F-mediated phosphorylation of PRMT5 downregulates its methyltransferase activity and promotes myeloproliferation. Cancer Cell. 2011;19:283–94. doi: 10.1016/j.ccr.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang G, Yuan Y, Yuan H, Wang J, Yun H, Geng Y, et al. Histone acetyltransferase 1 is a succinyltransferase for histones and non-histones and promotes tumorigenesis. EMBO Rep. 2021;22:e50967. doi: 10.15252/embr.202050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia Z, Yue F, Chen X, Narayanan N, Qiu J, Syed SA, et al. Protein arginine methyltransferase PRMT5 regulates fatty acid metabolism and lipid droplet biogenesis in white adipose tissues. Adv Sci. 2020;7:2002602. doi: 10.1002/advs.202002602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol. 2009;16:304–11. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antonysamy S, Bonday Z, Campbell RM, Doyle B, Druzina Z, Gheyi T, et al. Crystal structure of the human PRMT5:MEP50 complex. Proc Natl Acad Sci USA. 2012;109:17960–5. doi: 10.1073/pnas.1209814109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stopa N, Krebs JE, Shechter D. The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cell Mol Life Sci. 2015;72:2041–59. doi: 10.1007/s00018-015-1847-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang HT, Zeng LF, He QY, Tao WA, Zha ZG, Hu CD. The E3 ubiquitin ligase CHIP mediates ubiquitination and proteasomal degradation of PRMT5. Biochim Biophys Acta. 2016;1863:335–46. doi: 10.1016/j.bbamcr.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richters A. Targeting protein arginine methyltransferase 5 in disease. Future Med Chem. 2017;9:2081–98. doi: 10.4155/fmc-2017-0089. [DOI] [PubMed] [Google Scholar]
- 35.Lee JH, Phelan P, Shin M, Oh BC, Han X, Im SS, et al. SREBP-1a-stimulated lipid synthesis is required for macrophage phagocytosis downstream of TLR4-directed mTORC1. Proc Natl Acad Sci USA. 2018;115:E12228–E34. doi: 10.1073/pnas.1813458115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kryukov GV, Wilson FH, Ruth JR, Paulk J, Tsherniak A, Marlow SE, et al. MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science. 2016;351:1214–8. doi: 10.1126/science.aad5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zurita-Lopez CI, Sandberg T, Kelly R, Clarke SG. Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming omega-NG-monomethylated arginine residues. J Biol Chem. 2012;287:7859–70. doi: 10.1074/jbc.M111.336271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiang K, Zielinska AE, Shaaban AM, Sanchez-Bailon MP, Jarrold J, Clarke TL, et al. PRMT5 is a critical regulator of breast cancer stem cell function via histone methylation and FOXP1 expression. Cell Rep. 2017;21:3498–513. doi: 10.1016/j.celrep.2017.11.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin Y, Zhou J, Xu F, Jin B, Cui L, Wang Y, et al. Targeting methyltransferase PRMT5 eliminates leukemia stem cells in chronic myelogenous leukemia. J Clin Invest. 2016;126:3961–80. doi: 10.1172/JCI85239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andreu-Perez P, Esteve-Puig R, de Torre-Minguela C, Lopez-Fauqued M, Bech-Serra JJ, Tenbaum S, et al. Protein arginine methyltransferase 5 regulates ERK1/2 signal transduction amplitude and cell fate through CRAF. Sci Signal. 2011;4:ra58. doi: 10.1126/scisignal.2001936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Auclair Y, Richard S. The role of arginine methylation in the DNA damage response. DNA Repair. 2013;12:459–65. doi: 10.1016/j.dnarep.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 42.Mounir Z, Korn JM, Westerling T, Lin F, Kirby CA, Schirle M, et al. ERG signaling in prostate cancer is driven through PRMT5-dependent methylation of the androgen receptor. Elife. 2016;5:e13964. doi: 10.7554/eLife.13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Yang Y, Liu X, Long Y, Zheng Y. PRMT5 promotes human lung cancer cell apoptosis via Akt/Gsk3beta signaling induced by resveratrol. Cell Transpl. 2019;28:1664–73. doi: 10.1177/0963689719885083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin Y, Hu Q, Xu J, Ji S, Dai W, Liu W, et al. PRMT5 enhances tumorigenicity and glycolysis in pancreatic cancer via the FBW7/cMyc axis. Cell Commun Signal. 2019;17:30. doi: 10.1186/s12964-019-0344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feldman JL, Baeza J, Denu JM. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem. 2013;288:31350–6. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bao X, Wang Y, Li X, Li XM, Liu Z, Yang T, et al. Identification of ‘erasers’ for lysine crotonylated histone marks using a chemical proteomics approach. Elife. 2014;3:e02999. doi: 10.7554/eLife.02999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu S, Dong Z, Ke X, Hou J, Zhao E, Zhang K, et al. The roles of sirtuins family in cell metabolism during tumor development. Semin Cancer Biol. 2019;57:59–71. doi: 10.1016/j.semcancer.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 48.Qi H, Shi X, Yu M, Liu B, Liu M, Song S, et al. Sirtuin 7-mediated deacetylation of WD repeat domain 77 (WDR77) suppresses cancer cell growth by reducing WDR77/PRMT5 transmethylase complex activity. J Biol Chem. 2018;293:17769–79. doi: 10.1074/jbc.RA118.003629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nie M, Wang Y, Guo C, Li X, Wang Y, Deng Y, et al. CARM1-mediated methylation of protein arginine methyltransferase 5 represses human gamma-globin gene expression in erythroleukemia cells. J Biol Chem. 2018;293:17454–63. doi: 10.1074/jbc.RA118.004028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.