Abstract
The open reading frames sll1625 and sll0823, which have significant sequence similarity to genes coding for the FeS subunits of succinate dehydrogenase and fumarate reductase, were deleted singly and in combination in the cyanobacterium Synechocystis sp. strain PCC 6803. When the organic acid content in the Δsll1625 and Δsll0823 strains was analyzed, a 100-fold decrease in succinate and fumarate concentrations was observed relative to the wild type. A similar analysis for the Δsll1625 Δsll0823 strain revealed that 17% of the wild-type succinate levels remained, while only 1 to 2% of the wild-type fumarate levels were present. Addition of 2-oxoglutarate to the growth media of the double mutant strain prior to analysis of organic acids in cells caused succinate to accumulate. This indicates that succinate dehydrogenase activity had been blocked by the deletions and that 2-oxoglutarate can be converted to succinate in vivo in this organism, even though a traditional 2-oxoglutarate dehydrogenase is lacking. In addition, reduction of the thylakoid plastoquinone pool in darkness in the presence of KCN was up to fivefold slower in the mutants than in the wild type. Moreover, in vitro succinate dehydrogenase activity observed in wild-type membranes is absent from those isolated from the double mutant and reduced in those from the single mutants, further indicating that the sll1625 and sll0823 open reading frames encode subunits of succinate dehydrogenase complexes that are active in the thylakoid membrane of the cyanobacterium.
Synechocystis sp. strain PCC 6803, a unicellular cyanobacterium, contains a respiratory electron transport chain on both the cytoplasmic and thylakoid membranes (11, 22). The cytoplasmic membrane forms the inner boundary of the periplasmic space and is known to contain proteins typically associated with respiratory electron transport, such as NAD(P)H dehydrogenase (NDH-1), cytochrome b6f, and terminal oxidases (presumably predominantly a quinol oxidase) (10, 11, 22). The thylakoid membrane contains both a photosynthetic electron transport chain that includes photosystem (PS) I and II and a respiratory electron transport chain containing NDH-1 and a cytochrome aa3-type terminal oxidase (16, 23). The respiratory and photosynthetic electron transport chains in this membrane share common electron carriers, including the cytochrome b6f complex, the plastoquinone (PQ) pool, and soluble redox-active proteins (21, 26).
Other than linear photosynthetic and respiratory electron transfer, auxiliary electron transport pathways appear to exist in thylakoids of chloroplasts and cyanobacteria. Examples include cyclic electron transfer around PS I (2, 8) and the possible presence of succinate dehydrogenase (SDH) (4, 18), for which genes may exist in the cyanobacterial genome (13). Cyclic electron flow around PS I has been investigated with inhibitors and mutants (3, 5, 24, 27). NDH-1 has been found to play a central role in this cyclic electron flow, but alternate pathways clearly occur (12, 24). The functional presence of a SDH activity in the chloroplast has been implied based on the observation of oxygen uptake when PS II and NDH-1 have been inhibited (27). However, concrete evidence on the presence or activity of succinate:quinol oxidoreductase(s) (the term used for the family of proteins that includes both SDH and fumarate reductase [FRD] complexes) in thylakoids is still lacking. Succinate:quinol oxidoreductases donate electrons to quinone via the oxidation of succinate in the case of SDH, or transfer two electrons from quinol to fumarate in the case of FRD (9).
Synechocystis sp. strain PCC 6803 is often used as a model organism with which to study photosynthesis and related processes. For molecular-genetic studies, this cyanobacterium has several advantages over other prokaryotic and eukaryotic photosynthetic organisms. Synechocystis sp. strain PCC 6803 is spontaneously transformable, meaning that it takes up DNA from the surrounding media without pretreatment or electroporation, and once the DNA is inside, it can be incorporated into the organism's genome by double homologous recombination (25). Synechocystis sp. strain PCC 6803 grows photoautotrophically and photoheterotrophically, allowing for the deletion of essential genes for photosynthesis or respiration without generally leading to a lethal phenotype. Since Synechocystis sp. strain PCC 6803 is the only cyanobacterium for which a complete genomic sequence has been determined (13), comprehensive molecular biological approaches are feasible that can be used to understand processes related to photosynthesis in vivo.
This work set out to determine the presence of succinate:quinol oxidoreductase activity in thylakoid membranes of Synechocystis sp. strain PCC 6803, because this information is important to understand electron transfer pathways into and out of the PQ pool in thylakoid membranes. Open reading frames expected to code for polypeptides with significant amino acid sequence identity to the FeS-containing B-subunit of SDH or FRD (SdhB or FrdB), sll1625 and sll0823, were identified from the genome sequence (13). It is not possible from the sequence alone to discern whether a gene codes for a subunit of an SDH type or of an FRD type of succinate:quinol oxidoreductase (1). In this study, the expression of sll1625 and sll0823 and the function of their gene products in thylakoid electron transport and in the central organic acid metabolism of the cyanobacterium were investigated.
MATERIALS AND METHODS
Growth conditions.
The wild-type and mutant strains of Synechocystis sp. strain PCC 6803 were grown in liquid BG-11 medium (20) at 30°C. Cultures were grown at a light intensity of 45 to 50 μmol of photons m−2 s−1, except where indicated that cultures were grown at low light intensity (<5 μmol of photons m−2 s−1). Cells grown for organic acid analysis were harvested at an optical density at 730 nm (OD730) of 0.5 (as determined with a Shimadzu UV 160 spectrophotometer), which corresponds to the mid-exponential phase.
Deletion mutant construction and segregation.
Regions of the Synechocystis sp. strain PCC 6803 genome containing either the sll1625 or sll0823 open reading frames with flanking regions on either end (nucleotides 1320979 to 1319117 and 2862235 to 2860717 in the genome sequence according to the numbering of CyanoBase [http://www.kazusa.or.jp/cyano]) were amplified via PCR with primers with unique restriction sites engineered into them (Table 1). The PCR products were cloned into pUC19 plasmids; the resulting plasmids were named psll1625 and psll0823, respectively. A 0.3-kb HincII-MscI fragment (between nucleotides 1320279 and 1319992 in the genome sequence) was deleted from the psll1625 plasmid, and a kanamycin resistance cassette from the plasmid pUC4K was inserted in its place, creating the pΔsll1625 plasmid. To generate a construct to inactivate sll0823, a 0.5-kb HindIII-BamHI fragment (2861623 to 2861113 in the genome sequence) in the psll0823 plasmid was replaced with a 1.3-kb chloramphenicol cassette originating from the pACYC184 plasmid. The resulting plasmid was named pΔsll0823. Wild-type Synechocystis sp. strain PCC 6803 was then transformed with either plasmid, and transformants were subcultured in the presence of increasing concentrations of the appropriate antibiotic to aid in segregation of wild-type and mutant genome copies. Segregation analysis was performed by PCR with primers specific for the flanking regions of sll1625 or sll0823. After segregation of the deletion mutant had been confirmed by PCR, subsequent subcultures on plates included a modest concentration of the appropriate antibiotic (25 μg ml−1). The Δsll1625 Δsll0823 double mutant strain was created by transforming the already segregated Δsll1625 strain with the pΔsll0823 plasmid. Segregation was confirmed as indicated above.
TABLE 1.
Sequence of primers used in this study and their relative positions within the genome of Synechocystis sp. strain PCC 6803
| Primer | Sequencea | Position |
|---|---|---|
| 5′ sll0823 | 5′ AAC GCC TTA ATG cAT GCC CAA GGC 3′ | 2862235–2862212 |
| 3′ sll0823 | 5′ TGG CCA AGG gAA TTC CCT GTG ACC 3′ | 2860717–2860740 |
| 5′ sll1625 | 5′ GGA AGC GGT GcA TgC GGC CCA GGC 3′ | 1320979–1320956 |
| 3′ sll1625 | 5′ TTT GGT GGA GAA TTC TCA GGG TGC 3′ | 1319117–1319140 |
| 5′ sll1625b | 5′ GTG GAG CCC GGT GCC ACC 3′ | 1320469–1320452 |
| 3′ sll1625b | 5′ GGC TCC ATC CCG GGA GTC GG 3′ | 1319963–1319982 |
| 5′ RT-PCR sll0823 | 5′ CAC CAT CAG TCC CCT GGG GAA 3′ | 2861557–2861537 |
| 3′ RT-PCR sll0823 | 5′ GGG CTT CGA GGC GGG CCT TAG TGG 3′ | 2861238–2861261 |
| 5′ RT-PCR sll1625 | 5′ CCG ACT CCC GGG ATG GAG CCA CAG 3′ | 1319982–1319959 |
| 3′ RT-PCR sll1625 | 5′ CCG GCG GAA GGT TCA AAG CCC AGG 3′ | 1319610–1319633 |
| 3′ RT sll0823 | 5′ GGG CTT CGA GGC GGG CCT 3′ | 2861238–2861255 |
| 3′ RT sll1625 | 5′ CCG GCG GAA GGT TCA AAG 3′ | 1319610–1319627 |
Lowercase letters denote base changes used for initial cloning.
RNA isolation and RT-PCR.
Fifty milliliters of cells in mid-exponential phase were harvested by centrifugation, resuspended in 800 μl of 0.3 M sucrose (pH 8.0), and transferred to a microcentrifuge tube. The cell suspension was spun down, frozen at 77 K, and thawed on a water-ice mixture. The supernatant was removed, and 60 μl of EDTA (0.5 M [pH 8.0]) was added along with 0.3 M sucrose (pH 8.0) to a total volume of 600 μl. The cells were resuspended by vortexing, and 60 μl of 50 mM sodium acetate (pH 4.5) and 60 μl of 20% (wt/vol) sodium dodecyl sulfate were added. After mixing, the suspension was incubated at 65°C for 5 min. Then, 600 μl of phenol (65°C) that had been equilibrated with Tris-HCl (pH 8.0) was added. The suspension then was shaken three to five times to mix and left at 65°C for 5 min. Tubes were cooled quickly at −80°C for 1 min and centrifuged for 5 min at 14,000 × g, and the top phase (pink) was removed to a new RNase-free tube. Three hundred microliters of 65°C phenol and 300 μl of chloroform were added, and the tubes were shaken and centrifuged for 5 min at 14,000 × g. The supernatant was washed again with chloroform, spun as before, and the supernatant was removed to a new RNase-free microcentrifuge tube. An aliquot of LiCl (10 M) (2/9 of the volume of the supernatant) was added along with 100% ethanol (3 supernatant volumes). Precipitation was carried out for 45 min at −20°C. RNA was spun down (14,000 × g) for 30 min at room temperature, resuspended in 30 μl, and treated with RNase-free DNase (1 U/10 μg of RNA) (15).
For reverse transcription-PCR (RT-PCR), 2 μg of the RNA sample was treated again with DNase (1 U) and incubated at 70°C for 20 min, and to this reaction mixture (10-μl total volume), 2 μl of SuperScript first-strand buffer (250 mM Tris-HCl (pH 8.3), 375 mM KCl, 15 mM MgCl2), 1 μl of SuperScript reverse transcriptase (200 U) (Gibco-BRL), 1 μl of deoxynucleoside triphosphates (dNTPs [10 mM [each] dATP, dCTP, dTTP, and dGTP), and 1 μl of RT primers (20 pmol) (Table 1) were added. The reaction was carried out at 45°C for 1 h. The solution was diluted with 29 μl of double-distilled water, 5 μl of PCR buffer, 0.5 μl of dNTPs (10 mM each), and 1 μl of RT-PCR primers (Table 1) to a total reaction volume of 51 μl and used for PCR.
Organic acid extraction.
Extraction, purification, and derivatization of organic acids from Synechocystis sp. strain PCC 6803 cells were essentially done by a modification of the procedures outlined in reference 7. One liter of an OD730 = 0.5 culture was harvested by centrifugation and sucked onto a filter (0.45-μm pore size). Cells were rinsed with chilled double-distilled water (3 ml) three times; five rinses were used if organic acids had been added to the medium. Within 30 s after the last rinse, the filter with the cells was immersed in 5 ml of 5% (wt/vol) perchloric acid. After 10 min, the filter was removed, and 2 M potassium bicarbonate was added dropwise to bring the pH to 3.2 to 3.5. The extract was centrifuged for 30 min at 48,000 × g, and the supernatant was collected.
Organic acid purification and derivatization.
Sep-Pak C18 cartridges (Waters Assoc., Deerfield, Mich.) were preconditioned with 10 ml of methanol and 10 ml of H2O. Directly following preconditioning, 0.5 ml of extract mixed with 0.5 ml of 1 mM HCl was loaded onto the column. Organic acids were subsequently eluted with 1 ml of 1 M HCl and 2 ml of 30% acetonitrile in 1 M HCl. A 0.5-ml sample of the eluent and 20 μl of 1 mM adipate (internal dicarboxylate standard) were then evaporated until dry with a Speed Vac rotary evaporator and resuspended in 25 μl of pyridine. An equal volume of N-methyl-N-tert-butyldimethylsilyltrifluoroacetamide (TBDMSFA) (derivatization grade) (Aldrich, Milwaukee, Wis.) was added, and the sample was placed in a sonicating water bath at 60°C for 3 h (7).
GC/MS analysis.
Gas chromatography-mass spectral (GC/MS) analysis was performed with a Shimadzu 17-A gas chromatograph and Shimadzu QP5000 mass spectrometer linked to a data processor (Class-5K GC/MS software; Shimadzu). One microliter of the extract-adipate mixture in pyridine-TBDMSFA was injected. Injection was performed in the splitless mode at 260°C with a hold time of 1.2 min followed by continuous venting at 30 ml/min. The DB-5 column (30 mm by 0.25 μm inside diameter, 0.25-μm film thickness) was set at an initial temperature of 60°C for 2 min and then was increased to 150°C at a rate of 20°C min−1. A heating rate of 6°C min−1 was then employed until 300°C was reached. This temperature was then held for 17 min. The detector interface temperature was set at 300°C. The mass spectrometer (1.8 keV, electron impact mode) was set up to record the spectrum between 60 and 500 m/z units beginning at 60 min after injection to avoid detector saturation (7).
Chlorophyll fluorescence measurements.
Fluorescence measurements were taken with a Walz fluorometer (PAM 101, 102), with the intensity of the measuring light at its minimum (<0.01 μmol of photons m−2 s−1) and the damping setting at its maximum (time constant of 960 ms). Wild-type and mutant cells were harvested in the mid-exponential growth phase (OD730 = 0.5), spun down, and resuspended in 10 mM HEPES-NaOH (pH 7.0) buffer at a chlorophyll concentration of 10 μg ml−1. The fluorescence level (F0) was measured for 15 s with the measuring light on, the measuring light was turned off, and KCN was injected to a final concentration of 1 mM. The measuring light (<0.01 μmol of photons m−2 s−1) was turned on again at 15-s intervals (10 s off, 5 s on) for 60 s and then at 35-s intervals (30 s off, 5 s on) for the duration of the 6-min time course. The measuring light was kept on long enough to observe a steady-state fluorescence yield. The measuring light itself did not have a noticeable actinic effect. The fluorescence yield values that were measured were then plotted as a function of the incubation time with KCN.
Succinate-dependent reduction in isolated membranes.
The isolation of membranes from cells of the wild-type and mutant strains that were in the mid-exponential growth phase was carried out as previously described (29). The resulting membrane preparation consists predominantly of thylakoids, but also contains cytoplasmic membranes. Membranes (300 μg of chlorophyll per ml) treated with β-dodecyl maltoside (2 mg/5 mg of chlorophyll) for 30 min at 4°C were incubated for 5 min at 37°C in 80 mM potassium phosphate buffer (pH 7.0) containing 50 mM succinate and 5 mM KCN. Assays were conducted by monitoring the reduction of dichlorophenolindophenol (DCPIP) (50 μM per assay) at 598 nm with an HP8452 diode array spectrophotometer. The DCPIP reduction rate was calculated by using an extinction coefficient of 21 mM−1 cm−1 (28).
RESULTS
Insertional deletion.
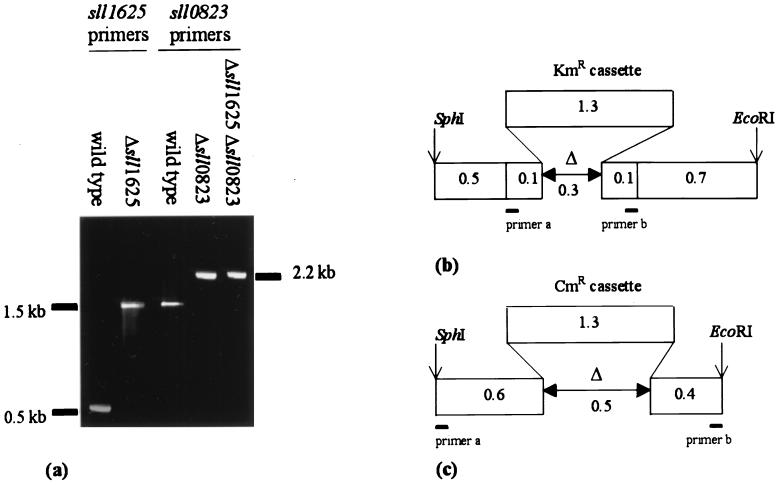
Two open reading frames, sll1625 and sll0823, that were expected to code for polypeptide with significant sequence similarity to the FeS subunit of SDH of E. coli (31% identity) were found in the Synechocystis sp. strain PCC 6803 genome (13). Alignment of the translated sequence of these open reading frames to B subunits of FRD and SDH from several organisms revealed that the cysteine residues that bind the three FeS centers and that are characteristic of this type of protein are conserved in Sll1625 and Sll0823 (see reference 9 for a sequence alignment). The sll1625 and sll0823 open reading frames were cloned, and deletion constructs (pΔsll1625 and pΔsll0823) were created in which regions coding for the FeS binding domains were replaced by a cassette conferring resistance to kanamycin and chloramphenicol, respectively. These plasmids were used for transformations of the Synechocystis sp. strain PCC 6803 wild type, and pure Δsll1625 and Δsll0823 strains as well as a Δsll1625 Δsll0823 double mutant were obtained (Fig. 1).
FIG. 1.
Segregation analysis of the deletion mutants. Genomic DNA from the wild-type, Δsll1625, Δsll0823, and Δsll1625 Δsll0823 strains was used as a template for amplification of the sll1625 and sll0823 genes to verify segregation in the mutants (a). The segregated Δsll1625 strain was used as the background strain for creating the Δsll1625 Δsll0823 strain; therefore, only the sll0823 segregation analysis is shown (a, far right lane). Schematic maps of the deletion constructs used to create Δsll1625 (b) and Δsll0823 (c) are shown with the PCR primers used for segregation analysis represented by bars (Table 1). KmR, kanamycin resistance; CmR, chloramphenicol resistance. In panel b, primer a is 5′ sll1625b, and primer b is 3′ sll1625b. In panel c, primer a is 5′ sll0823, and primer b is 3′ sll0823.
Growth rate analysis of the individual mutant strains.
Wild-type Synechocystis sp. strain PCC 6803 had a doubling time of 12.5 ± 1.5 h for photoautotrophic growth. The strains carrying single deletions (Δsll1625 and Δsll0823) and the double mutant (Δsll1625 Δsll0823) had essentially the same growth rate, with doubling times of 11.7 ± 2.1, 13.2 ± 0.9, and 12.2 ± 1.1 h, respectively. The growth rate of the mutants was also similar to that of the wild type when cells were grown photomixotrophically at normal or low light intensity (<5 μE m−2 s−1) with 5 mM glucose or fumarate as a fixed carbon source.
Transcript analysis.
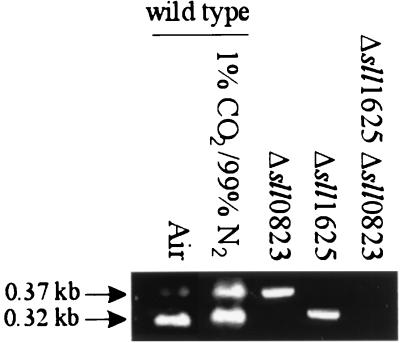
Transcripts of sll1625 and sll0823 were not detectable by Northern blot analysis in the wild type or the deletion strains created (data not shown). Therefore, the more qualitative RT-PCR assay was used to monitor the presence of sll1625 and sll0823 transcripts under different growth conditions. Both sll1625 and sll0823 transcripts could be amplified from RNA samples from cells that had been propagated photoautotrophically while being bubbled with air or a 1% CO2–99% N2 mix (Fig. 2). Primers used for RT were 3′ RT sll1625 and 3′ RT sll0823. Subsequent PCR was performed with primer sets 5′ RT-PCR sll1625 and 3′ RT-PCR sll1625 and 5′ RT-PCR sll0823 and 3′ RT-PCR sll0823, respectively (Table 1). The control reactions without reverse transcriptase gave no bands after PCR amplification (data not shown) indicating that no significant DNA contamination existed in the RNA samples. If reactions were carried out with RNA from the single-deletion strains, transcripts corresponding to the remaining open reading frames were detected (Fig. 2). As expected, in the double-deletion mutant, no sll1625 or sll0823 transcripts were amplified.
FIG. 2.
Transcript analysis. Ethidium bromide-stained gels of RT-PCR products with transcripts isolated from the wild type grown under photoautotrophic conditions when bubbled with air or with a 1% CO2–99% N2 mix (left two lanes) and from the three deletion mutant strains grown photoautotrophically in ambient air (right three lanes). The 0.37-kb band represents amplification of the sll1625 transcript, and the 0.32-kb band represents amplification of the sll0823 transcript.
GC/MS analysis of organic acid content.
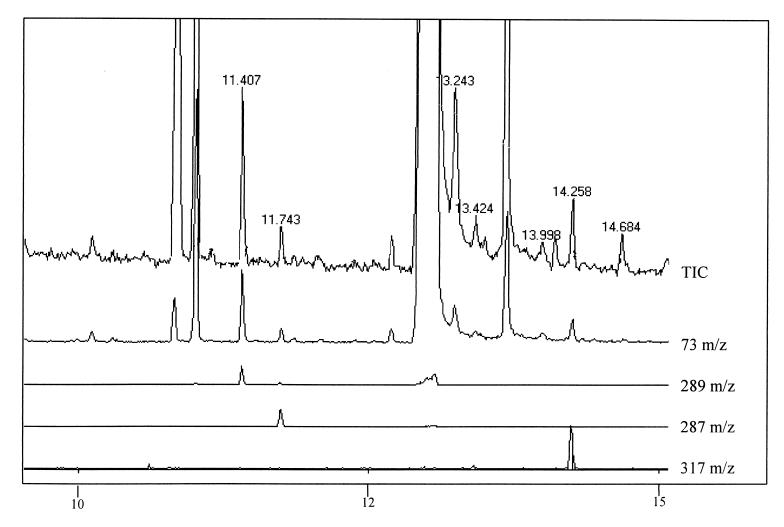
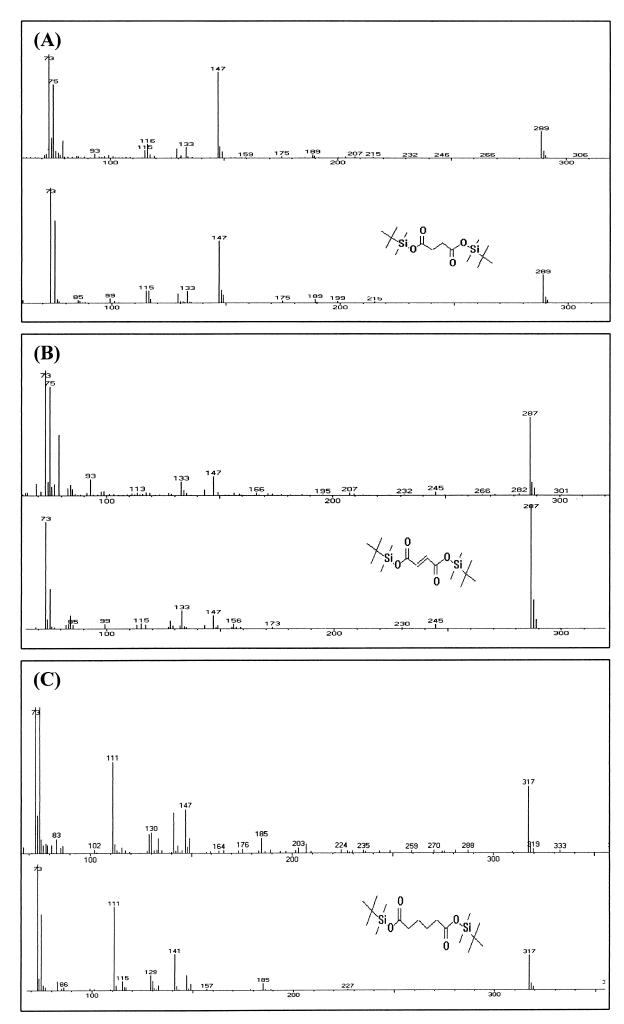
To determine whether sll1625 and sll0823 encode components of a complex with predominantly SDH or FRD activity, the relative fumarate and succinate content of the wild-type and mutant cells was measured by GC/MS. Cells were grown to an OD730 of 0.5 (mid-exponential phase), and organic acids were extracted and derivatized from at least three different cultures of the same strain as described in Materials and Methods. The primary advantage of using GC/MS in our study for organic acid analysis is that derivatized succinate and fumarate give rise to clear, well-separated peaks. Furthermore, all tert-butyldimethylsilyl (TBDMS) based derivatives have a “base peak” of 73 m/z units due to fragmenting of the TBDMS moiety itself, along with a peak at total derivative mass minus 57 (m−57) m/z units, representing loss of the butyl group (7). The GC/MS spectrum of the m/z signal compatible with succinate (base peak at 73 m/z units together with an m−57 peak at 289 m/z units), fumarate (73 and 287 m/z units), and adipate (73 and 317 m/z units) is presented in Fig. 3. Indeed, the mass spectrum of these compounds (Fig. 4A to C, upper traces) is similar to spectra of these compounds found in the National Institute of Standards and Technology library (Fig. 4A to C, lower traces), thus confirming the peak assignments.
FIG. 3.
GC/MS analysis of organic acid derivatives prepared from cell extracts. (Top to bottom) Chromatograms generated from the total impact spectrum (TIC; 60 to 500 m/z), the TBDMS-specific 73-m/z fragment, the bis-TBDMS succinate-specific 289-m/z fragment, the bis-TBDMS fumarate 287-m/z fragment, and the bis-TBDMS adipate 317-m/z fragment. Comparison of the top two traces shows the improvement in the signal/noise ratio by using single-ion-fragment (73 m/z)-generated chromatograms.
FIG. 4.
MS fractionation identification of derivatized compounds. Mass fragment spectra from the peaks at 11.407, 11.743, and 14.258 min (A, B, and C, respectively [upper traces]) were identified as the derivatives of succinate, fumarate, and adipate, respectively, based on similarity of fractionation to standards found within the National Institute of Standards and Technology mass fractionation spectral library (A, B, and C, respectively [lower traces]).
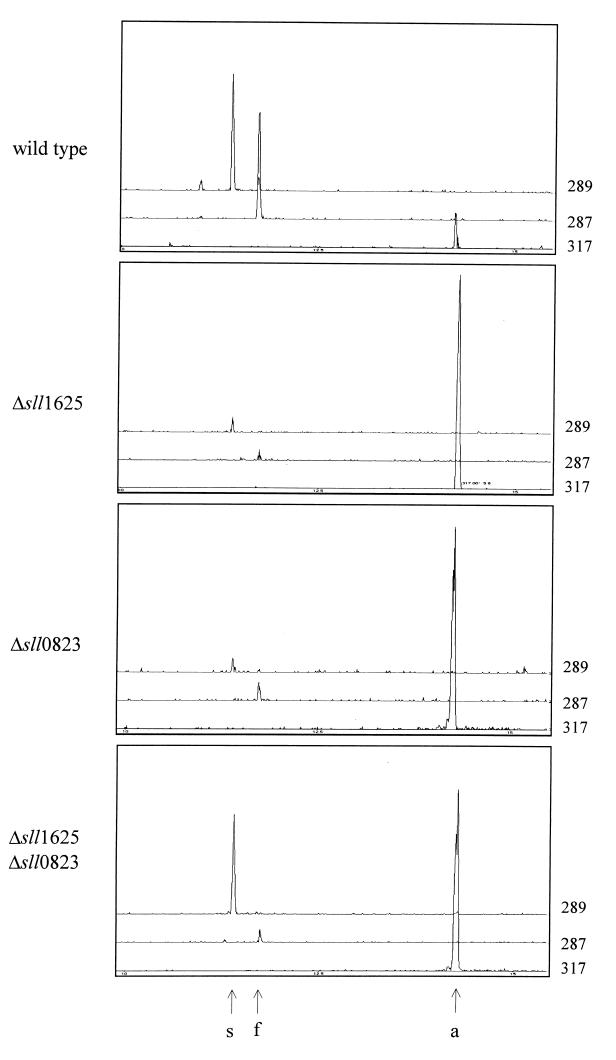
Quantitative information on succinate and fumarate levels can be obtained by comparison of the area of the signal with that of the internal standard, bis-(butyl-t-dimethylsilyl) adipate. Extracts from the single mutants (Δsll1625 and Δsll0823) exhibited an 80- to 100-fold-reduced amount of succinate and fumarate compared to wild-type extracts (Fig. 5). The ratio of succinate to fumarate in these two mutants was similar to that in the wild type. In contrast, the succinate level of the Δsll1625 Δsll0823 double mutant strain was 16 to 23% of that of the wild-type control, whereas the fumarate level was similar to that in the single mutants (Fig. 5).
FIG. 5.
Relative levels of succinate and fumarate in extracts from the wild type and deletion mutants. Single-ion-fragment GC/MS-generated chromatograms for bis-TBDMS succinate (289 m/z), bis-TBDMS fumarate (287 m/z), and the bis-TBDMS adipate (317 m/z) internal standard are shown. Organic acids were extracted from strains as indicated. Relative levels of succinate and fumarate in the cells can be determined by comparison of the peak areas of the derivatives of these organic acids with the area of the internal standard peak (adipate). Succinate (s), fumarate (f), and adipate (a) peak designations are indicated at the bottom with arrows.
Addition of organic acids.
Even though the results obtained with the single mutants were surprising, the results obtained with the Δsll1625 Δsll0823 double mutant (Fig. 5, bottom panel) suggest that both the sll1625 and sll0823 genes code for SDH components: the succinate levels are increased compared to the situation in the single mutants. To test this further, fumarate or 2-oxoglutarate was added to the growth medium of the double mutant and the wild type. Upon addition of fumarate (5 mM final concentration in the medium) 1 h prior to extraction, the fumarate level in cells of the double mutant increased 50-fold, whereas the succinate level remained constant (Table 2). Upon addition of 2-oxoglutarate (5 mM final concentration in the medium) 1 h prior to derivatization, fumarate levels in the double mutant remained low, whereas succinate levels increased two- to threefold compared to the untreated samples (Table 2). When the same fumarate or 2-oxoglutarate incubation and extraction was performed with wild-type cells, both fumarate and succinate levels increased about three- to fourfold, while the relative ratio of succinate to fumarate did not change (Table 2). These results corroborate the working hypothesis of both sll1625 and sll0823 being components of a succinate dehydrogenase.
TABLE 2.
Succinate and fumarate levels after treatment with organic acidsa
| Strain | Treatment | Peak areab
|
|
|---|---|---|---|
| Succinate | Fumarate | ||
| Wild type | None | 1.61 ± 0.09 | 1.54 ± 0.10 |
| + Fumarate | 6.20 ± 0.12 | 6.41 ± 0.15 | |
| + 2-Oxoglutarate | 4.80 ± 0.10 | 5.0 ± 0.12 | |
| Δsll1625 Δsll0823 | None | 0.28 ± 0.04 | 0.01 ± 0.04 |
| + Fumarate | 0.35 ± 0.05 | 0.48 ± 0.09 | |
| + 2-Oxoglutarate | 0.76 ± 0.12 | 0.02 ± 0.05 | |
Fumarate or 2-oxoglutarate (5 mM each) was added to the growth medium 1 h prior to organic acid extraction.
Peak areas are normalized to the peak area of the internal standard.
Succinate:DCPIP oxidoreductase activity in isolated membranes.
To further substantiate this notion, succinate-dependent reduction of DCPIP was measured in isolated membranes. This assay constitutes an in vitro determination of SDH activity. Wild-type membranes yielded rates of succinate-dependent DCPIP reduction of 34 ± 4 μmol mg of chlorophyll−1 h−1. When membranes from the Δsll1625 Δsll0823 double-deletion mutant were used for the assay, no succinate-dependent DCPIP reduction could be observed. In membrane preparations from the single mutants succinate-dependent DCPIP reduction could be observed, but at rates significantly lower than those of the wild type (18 ± 3 μmol mg of chlorophyll−1 h−1 for Δsll1625 membranes and 14 ± 5 μmol mg of chlorophyll−1 h−1 for Δsll0823 membranes).
Chlorophyll fluorescence as a probe for the redox state of the PQ pool.
Now that an in vitro activity of SDH has been established, it is important to determine whether the SDH enzyme complex associated with Sll1625 or Sll0823 is functionally relevant in vivo and may donate electrons to the PQ pool. To this aim, the redox state of the PQ pool in thylakoids was monitored indirectly by determining the chlorophyll fluorescence yield in darkness (no PS II or PS I activity) upon addition of KCN (inhibiting oxidase activity). The fluorescence yield depends on the redox state of QA−, the first electron-accepting PQ in PS II: the chlorophyll fluorescence yield is high when QA is reduced and low when QA is oxidized. QA is in redox equilibrium with the PQ pool, and the Em,7 (midpoint redox potential at neutral pH) of QA/QA− is about 80 mV more negative than that of PQ/PQH2 in the thylakoid (14). Therefore, there is redox equilibrium between QA and PQ (Keq = 23). If oxidation of the PQ pool is blocked by the presence of KCN and absence of light, the rate of reduction of the PQ pool therefore can be monitored qualitatively by measuring the chlorophyll fluorescence yield elicited by weak pulses of measuring light.
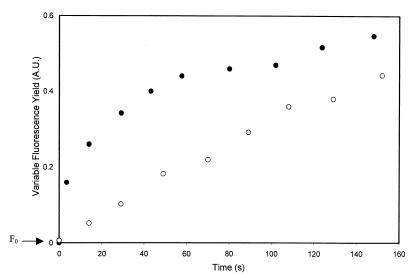
In the wild type, cells exhibited a rapid increase (half-life [t1/2] = 19 s) in the fluorescence yield upon CN− addition, reflecting an increase in the QA− level due to PQ pool reduction (Fig. 6). In cells of the Δsll1625 Δsll0823 double-deletion mutant, the initial fast rise in fluorescence yield was absent, and only a slow rise was observed (t1/2 = 92 s) (Fig. 6). The kinetics of the Δsll1625 and Δsll0823 single mutant strains were intermediate between those of the wild-type and double-deletion strains, with t1/2s of the initial rise of 54 and 49 s, respectively.
FIG. 6.
Changes in chlorophyll fluorescence yield in darkness after addition of KCN. Variable fluorescence (arbitrary units [A.U.]) of the wild type (●) and the Δsll1625 Δsll0823 strain (○) was measured by very weak illumination that did not have any actinic effect. Otherwise, cells were kept in darkness. KCN (1 mM final concentration) was added at t = 0 s. Curves were normalized so that a value of 1 on the y axis corresponds to maximal variable fluorescence yield. Constant fluorescence (F0) has been subtracted.
DISCUSSION
Two open reading frames were identified in the genomic sequence of Synechocystis sp. strain PCC 6803 that appeared to encode the FeS-containing B subunits of a succinate:quinol oxidoreductase (13). The function of these open reading frames has been investigated in this study.
Expression.
The amplification of sll1625 and sll0823 transcripts by RT-PCR indicates that both of the open reading frames are expressed under the growth conditions tested (Fig. 2). Because the transcripts could not be detected by Northern blot analysis, transcript levels are low, and we preferred not to attempt to quantify changes in the amounts of transcripts under different growth conditions, because such quantification on the basis of accumulation of RT-PCR products is not very reliable.
Function of Sll1625 and Sll0823.
The phenotype of the Δsll1625 Δsll0823 double mutant is most instructive regarding the role of Sll1625 and Sll0823 in Synechocystis sp. strain PCC 6803. This double mutant accumulates significant amounts of succinate, but essentially lacks fumarate. Membranes from this strain lack the ability to catalyze the succinate-dependent reduction of DCPIP. Moreover, the rapid reduction of the quinone pool in thylakoids upon KCN addition is eliminated in this mutant; a slower reduction is present in the single mutants. On the basis of these observations, we conclude that both Sll1625 and Sll0823 are components of SDH complexes. Because the two single mutants are intermediate between the double mutant and the control strain in the in vivo assay of KCN-induced QA reduction and the in vitro DCPIP reduction assay, we suggest that Sll1625 and Sll0823 are functionally similar and that both are present in association with thylakoids. Because only a single gene for a flavin-binding SDH-FRD subunit (SdhA and FrdA) appears to be present in the genome sequence of Synechocystis sp. strain PCC 6803, Sll1625 and Sll0823 may be functionally interchangeable, and both may be accommodated in functional form in a native SDH complex on the membrane. In view of the observation that in both single mutants the rise in chlorophyll fluorescence upon KCN addition has been slowed down in comparison with that of the wild type, it appears that both SdhB subunits can be associated with the thylakoid membrane, and neither would be expected to be associated solely with the cytoplasmic membrane.
The presence of two genes that have moderate sequence similarity to one another (<40% identity at the amino acid level) and that both encode a component of an SDH is not generally found in other prokaryotes. For example, Escherichia coli contains a gene set encoding a SDH and a set of genes coding for FRD but not two of either one. If two genes coding for one subunit of the same complex exist in one organism, they have been found to be very similar in terms of amino acid sequence (>80% identity) (6).
An important question is what is the cause of the large decrease in succinate and fumarate levels in the two single mutants. It is apparent that SDH activity is reduced in these mutants compared to that in the wild type (Fig. 5), which would explain the decreased fumarate level. However, the succinate levels also decrease. A possibility is that the cells have a mechanism by which they down-regulate the levels of succinate when conversion of fumarate has been slowed down and the succinate accumulation that may result from decreased SDH activity may trigger induction of other succinate utilization pathways or decrease succinate formation.
Succinate and fumarate levels in the cells.
We can estimate the amount of succinate and fumarate in cells based upon the GC/MS data presented in Fig. 5. The peak areas of the derivatized adipic acid internal standard, of which 400 pmol was injected, are directly proportional to the areas of the succinate and fumarate derivative peaks. For each GC/MS injection, the extract from about 108 cells (0.02% of the total sample) was used, mixed with a derivatized adipate (see Materials and Methods). Assuming a cellular volume of 1 μm3, the succinate and fumarate concentrations in the cell in vivo in the single-deletion strains appear to be in the 0.02 to 0.05 mM range versus 2.5 to 5.0 mM for the wild-type strain. The relatively high levels of succinate and fumarate in the wild-type cells serve to further emphasize that the SDH complex may play a significant role in PQ pool reduction in the thylakoids. Furthermore, the high levels of succinate and fumarate in the cell may indicate a possible reason for their apparent down-regulation of the concentration of these organic acids in the absence of a subunit of one or more SDH complexes.
2-Oxoglutarate conversion.
Cyanobacteria have long been realized to lack a classical 2-oxoglutarate dehydrogenase and thereby were considered to lack a complete tricarboxylic acid cycle (17). Indeed, genes resembling those coding for a traditional 2-oxoglutarate dehydrogenase complex are not found in the genome. However, upon addition of 2-oxoglutarate to either the wild type or the Δsll1625 Δsll0823 mutant, an increase in the succinate level was observed (Table 2), suggesting a conversion of 2-oxoglutarate to succinate. The enzymes involved in this conversion have not yet been elucidated, but reactions catalyzed by Fe-containing dioxygenases (such as aspartyl beta hydroxylases) (19) as well as several multistep reactions are feasible, and potential genes for the corresponding enzymes may be present in the Synechocystis sp. strain PCC 6803 genome sequence.
Relative activity of NDH-1 and SDH.
Our work indicates that in darkness in the presence of cyanide, the rate of PQ pool reduction is very much decreased in the absence of Sll1625 and Sll0823. This indicates that under these conditions, SDH activity is the major pathway of electrons into the PQ pool and is much larger than NDH-1 activity associated with the thylakoid membranes. The fluorescence yield data indicate that the quinone reduction due to SDH activity levels off after part of the QA population has been reduced. This partial reduction can be attributed to a redox equilibrium that is reached between succinate and PQ. Our data also indicate the presence of a slower PQ reduction, probably due to NDH-1 activity, that continues in our assays for a much longer period of time. Based upon the slope of the initial rate of QA reduction, it appears that if thermodynamically possible, in the wild type the PQ pool would be fully reduced within 20 s after KCN addition. The double mutant under the same conditions would take nearly 270 s to fully reduce the PQ pool in darkness. Assuming 20 PQ per PS II and a Keq of 23 for reduction of QA via PQH2, the initial rate of SDH activity was calculated to be about 150 μmol of PQ reduced mg of chlorophyll−1 h−1 for the wild type and 10 μmol of PQ reduced mg of chlorophyll−1 h−1 for the double mutant strain. The initial rate of electron transfer by SDH to the PQ pool in the wild type therefore is quite significant compared to rates of linear photosynthetic electron flow.
The lower yield of total QA− accumulation due to SDH activity versus that due to NDH-1 activity may be due to the expected difference in equilibrium constants between succinate, PQ, and QA (Em,7 values of succinate/fumarate, PQ/PQH2, and QA/QA− are 0, +10, and −80 mV, respectively) versus that between NAD(P)/NAD(P)H (Em,7 = −320 mV) and PQ and QA. The driving force for uphill electron transfer from PQ to QA provided by succinate oxidation is not strong enough to fully reduce QA. The small redox gradient between succinate and PQ may imply a role for succinate in the reduction of the PQ pool when this pool is predominantly oxidized.
Now that the presence and activity of an SDH complex in thylakoid membranes have been established, it is clear that SDH contribution to overall electron transport in cyanobacterial photosynthesis and respiration is considerable and needs to be taken into account. This makes an overall understanding of electron flow through this membrane and its regulation more complex, but at the same time more challenging.
ACKNOWLEDGMENTS
We are grateful to Satoshi Tabata (Kazusa Research Institute) for access to the genome sequence prior to publication. We also thank Karl Booksh (Department of Chemistry and Biochemistry, Arizona State University) for the use of his Shimadzu GC/MS apparatus.
Support for this research was provided by a grant from the Human Frontiers Science Program (RG 0051/1997M). Jason Cooley was supported by a Graduate Research Training grant from the National Science Foundation (DGE-9553456).
REFERENCES
- 1.Ackrell B A C, Armstrong F A, Cochran B, Sucheta A, Yu T. Classification of fumarate reductases and succinate dehydrogenases based upon their contrasting behavior in the reduced benzylviologen fumarate assay. FEBS Lett. 1993;326:92–94. doi: 10.1016/0014-5793(93)81768-u. [DOI] [PubMed] [Google Scholar]
- 2.Bendall D S, Manasse R S. Cyclic photophosphorylation and electron transport. Biochim Biophys Acta. 1995;1229:23–38. [Google Scholar]
- 3.Berger S, Ellersiek U, Steinmüller K. Cyanobacteria contain a mitochondrial complex-I homologous NADH-dehydrogenase. FEBS Lett. 1991;286:129–132. doi: 10.1016/0014-5793(91)80957-5. [DOI] [PubMed] [Google Scholar]
- 4.Biggins J. Respiration in blue-green algae. J Bacteriol. 1969;99:570–575. doi: 10.1128/jb.99.2.570-575.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binder A, Hauser R, Krogmann D. Respiration in energy-transducing membranes of the thermophilic cyanobacterium Mastigocladus laminosus. Biochim Biophys Acta. 1984;765:241–246. [Google Scholar]
- 6.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 7.Chen Z L, Landman P, Colmer T D, Adams M A. Simultaneous analysis of amino and organic acids in extracts of plant leaves as tert-butyldimethylsilyl derivatives by capillary gas chromatography. Anal Biochem. 1998;259:203–211. doi: 10.1006/abio.1998.2659. [DOI] [PubMed] [Google Scholar]
- 8.Fork D C, Herbert S K. Electron transport and photophosphorylation by Photosystem I in vivo in plants and cyanobacteria. Photosynth Res. 1993;36:149–168. doi: 10.1007/BF00033035. [DOI] [PubMed] [Google Scholar]
- 9.Hägerhäll C. Succinate:quinone oxidoreductases—variations on a conserved theme. Biochim Biophys Acta. 1997;1320:107–141. doi: 10.1016/s0005-2728(97)00019-4. [DOI] [PubMed] [Google Scholar]
- 10.Howitt C A, Smith G D, Day D A. Cyanide-insensitive oxygen uptake and pyridine nucleotide dehydrogenases in the cyanobacterium Anabaena PCC 7120. Biochim Biophys Acta. 1993;1141:313–320. [Google Scholar]
- 11.Howitt C A, Vermaas W F J. Quinol and cytochrome oxidases in the cyanobacterium Synechocystis sp. PCC 6803. Biochemistry. 1998;37:17944–17951. doi: 10.1021/bi981486n. [DOI] [PubMed] [Google Scholar]
- 12.Jeanjean R, Bedu S, Havaux M, Matthijs H C P, Joset F. Salt-induced photosystem I cyclic electron transfer restores growth on low inorganic carbon in a type 1 NAD(P)H dehydrogenase deficient mutant of Synechocystis PCC6803. FEMS Microbiol Lett. 1998;167:131–137. [Google Scholar]
- 13.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. , 185–209. [DOI] [PubMed] [Google Scholar]
- 14.Krieger A, Rutherford A W, Johnson G N. On the determination of redox midpoint potential of the primary quinone electron acceptor, QA, in photosystem II. Biochim Biophys Acta. 1995;1229:193–201. [Google Scholar]
- 15.Mohamed A, Jansson C. Influence of light on photosynthesis-specific transcripts in the cyanobacterium Synechocystis 6803. Plant Mol Biol. 1989;13:693–700. doi: 10.1007/BF00016024. [DOI] [PubMed] [Google Scholar]
- 16.Molitor V, Trnka M, Peschek G A. Isolated and purified plasma and thylakoid membranes of the cyanobacterium Anacystis nidulans contain immunologically cross-reactive aa3-type cytochrome oxidase. Curr Microbiol. 1987;14:263–268. [Google Scholar]
- 17.Pearce J, Leach C K, Carr N G. The incomplete citric acid cycle in the blue-green alga Anabaena variabilis. J Gen Microbiol. 1969;49:301–313. doi: 10.1099/00221287-55-3-371. [DOI] [PubMed] [Google Scholar]
- 18.Peschek G A. Electron transport reactions in respiratory particles of hydrogenase-induced Anacystis nidulans. Arch Microbiol. 1980;125:123–131. [Google Scholar]
- 19.Prescott A G, John P. Dioxygenases: molecular structure and role in plant metabolism. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:245–271. doi: 10.1146/annurev.arplant.47.1.245. [DOI] [PubMed] [Google Scholar]
- 20.Rippka R, Deruelles J, Waterbury J B, Herdman M, Stanier R Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;181:1–60. [Google Scholar]
- 21.Scherer S. Do photosynthetic and respiratory electron transport chains share redox proteins? Trends Biochem Sci. 1990;15:458–462. doi: 10.1016/0968-0004(90)90296-n. [DOI] [PubMed] [Google Scholar]
- 22.Schmetterer G. Cyanobacterial respiration. In: Bryant D A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 409–435. [Google Scholar]
- 23.Sturzl E, Scherer S, Böger P. Interaction of respiratory and photosynthetic electron transport and evidence for membrane-bound pyridine-nucleotide dehydrogenase in Anabaena variabilis. Physiol Plant. 1984;60:479–483. [Google Scholar]
- 24.Tanaka Y, Katada S, Ogawa I H T, Takabe T. Electron flow from NAD(P)H dehydrogenase to photosystem I is required for adaptation to salt shock in the cyanobacterium Synechocystis sp. PCC6803. Plant Cell Physiol. 1997;38:1311–1318. [Google Scholar]
- 25.Vermaas W. Molecular genetics of the cyanobacterium Synechocystis sp. PCC 6803: principles and possible biotechnology applications. J Appl Phycol. 1996;8:263–273. [Google Scholar]
- 26.Vermaas W F J, Shen G, Styring S. Electrons generated by photosystem II are utilized by an oxidase in the absence of photosystem I in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 1994;337:103–108. doi: 10.1016/0014-5793(94)80638-1. [DOI] [PubMed] [Google Scholar]
- 27.Willeford K O, Gombos Z, Gibbs M. Evidence for chloroplastic succinate-dehydrogenase participating in the chloroplastic respiratory and photosynthetic electron-transport chains of Chlamydomonas reinhardtii. Plant Physiol. 1989;90:1084–1087. doi: 10.1104/pp.90.3.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X, Yang L, He D, Yu C-A. The quinone binding site in succinate-ubiquinone reductase from Escherichia coli. J Biol Chem. 1998;273:31916–31923. doi: 10.1074/jbc.273.48.31916. [DOI] [PubMed] [Google Scholar]
- 29.Yu J, Vermaas W F J. Synthesis and turnover of photosystem II reaction center polypeptides in cyanobacterial D2 mutants. J Biol Chem. 1993;268:7407–7413. [PubMed] [Google Scholar]