Abstract
The generation and differentiation of cortical projection neurons are extensively regulated by interactive programs of transcriptional factors. Here, we report the cooperative functions of transcription factors Bcl11a and Bcl11b in regulating the development of cortical projection neurons. Among the cells derived from the cortical neural stem cells, Bcl11a is expressed in the progenitors and the projection neurons, while Bcl11b expression is restricted to the projection neurons. Using conditional knockout mice, we show that deficiency of Bcl11a leads to reduced proliferation and precocious differentiation of cortical progenitor cells, which is exacerbated when Bcl11b is simultaneously deleted. Besides defective neuronal production, the differentiation of cortical projection neurons is blocked in the absence of both Bcl11a and Bcl11b: Expression of both pan-cortical and subtype-specific genes is reduced or absent; axonal projections to the thalamus, hindbrain, spinal cord, and contralateral cortical hemisphere are reduced or absent. Furthermore, neurogenesis-to-gliogenesis switch is accelerated in the Bcl11a-CKO and Bcl11a/b-DCKO mice. Bcl11a likely regulates neurogenesis through repressing the Nr2f1 expression. These results demonstrate that Bcl11a and Bcl11b jointly play critical roles in the generation and differentiation of cortical projection neurons and in controlling the timing of neurogenesis-to-gliogenesis switch.
Keywords: Bcl11a, Bcl11b, cortical projection neurons, neurogenesis, Nr2f1
Introduction
Cortical projection neurons, or pyramidal neurons, constitute approximately 70–80% of the neuronal population in the mammalian neocortex, the precise development of which is quintessential for sensory information integration, motor coordination, and cognition (McConnell 1989; Molyneaux et al. 2007; Greig et al. 2013). Cortical neurogenesis begins with the proliferation of neuroepithelial cells which produce cortical radial glial cells (RGCs) located in the ventricular zone (VZ). RGCs divide symmetrically to expand the progenitor pool, or asymmetrically to produce cortical projection neurons (direct neurogenesis) or intermediate progenitor cells (IPCs) that migrate to the subventricular zone (SVZ), where they divide to generate cortical projection neurons (indirect neurogenesis). The projection neurons sequentially populate the cortical layers in an inside-first, outside-last sequences (McConnell 1989, 1995; Leone et al. 2008; Juric-Sekhar and Hevner 2019). As the projection neurons are generated, their subtype identities are established by transcriptional repressors that act in the postmitotic neurons to repress alternate subtype identities (Alcamo et al. 2008; Chen et al. 2008; McKenna et al. 2011, 2015; Tsyporin et al. 2021). At the end of cortical neurogenesis, cortical RGCs switch from generating excitatory projection neurons to producing inhibitory olfactory bulb interneurons and cortical glia, in a process controlled by Shh signaling (Zhang, Liu, et al. 2020; Li et al. 2021).
Recent single-cell RNA-seq analyses (scRNA-seq) have segregated cortical projection neurons into many clusters based on gene expression of individual cells (Tasic et al. 2018). However, cortical projection neurons can also be classified into three broad classes based on axonal projections. The subcerebral neurons that extend their axons into the midbrain, the pons, the medulla oblongata and the spinal cord are located in the layer V. The corticothalamic neurons project their axons into the thalamus. They are located in layer VI. Corticocortical projection neurons extend their axons to other cortical areas either in the ipsilateral (associative projection neurons) or contralateral (commissural projection neurons) hemispheres, and they are distributed across layers II–VI (Molyneaux et al. 2007; Leone et al. 2008; Greig et al. 2013).
A few transcription factors have been identified that are essential for subtype specification and differentiation of cortical projection neurons. For example, Sox5 regulates the migration and identities of corticofugal projection neurons residing in deep cortical layers, and it does so by directly repressing Fezf2 expression (Kwan et al. 2008; Lai et al. 2008; Shim et al. 2012). Tbr1 promotes layer VI corticothalamic neuron subtype identity and represses layer V subcerebral neuronal fate by directly repressing Fezf2 and restricting corticospinal tract outgrowth (Hevner et al. 2001; Bedogni et al. 2010; Han et al. 2011; McKenna et al. 2011). Fezf2 is expressed in both layer V subcerebral neurons and in layer VI corticothalamic neurons and regulates the identities of both subtypes. It promotes the subtype specification of layer V subcerebral neurons and regulates their axon targeting in part by inhibiting Tbr1 and Satb2 expression, the high-level expression of which leads to corticothalamic and corticocortical neuron identities, respectively (Chen et al. 2005, 2008; Molyneaux et al. 2005; Eckler et al. 2014). Recently, it has been shown that in layer VI corticothalamic neurons, Fezf2 recruits a transcriptional co-repressor, Tle4, to repress the expression of genes associated with layer V subcerebral neuron features (Tsyporin et al. 2021). The DNA-binding protein that is involved in chromatin remodeling, Satb2, is required for the subtype specification and differentiation of multiple cortical projection neuron subtypes. In the callosal neurons, Satb2 represses the high-level expression of genes associated with subcerebral neuron identity such as Bcl11b (Alcamo et al. 2008; Britanova et al. 2008). In the layer V subcerebral neurons, Satb2 is required for Fezf2 expression in these neurons, and the mutual regulations between Satb2 and Fezf2 ensure that the subcerebral neuron identity is established, as evidenced by the facts layer V subcerebral neurons and axons are missing in mice deficient in either gene (Chen et al. 2008; McKenna et al. 2015).
Zinc-finger transcription factor family Bcl11 proteins (Bcl11a and Bcl11b) share 55% identity at amino acid level. They were first identified by their functions in the immune and hematopoietic systems (Lessel et al. 2018; Liu et al. 2018; Simon et al. 2020). In the central nervous system, they are mainly expressed in the cerebral cortex, olfactory bulb, striatum, and dorsal spinal cord (Leid et al. 2004). BCL11A (also known as CTIP1) has been implicated in neocortical dysplasia, autism spectrum disorders, neuropsychiatric disorders linked with intellectual deficiency, epileptic encephalopathy, dyspraxia, and hypotonia in human patients (Sanders et al. 2015; Soblet et al. 2018; Yoshida et al. 2018). BCL11B (also known as CTIP2) has been linked to a number of neurodegenerative disorders including Alzheimer’s disease, Huntington’s disease, schizophrenia, and amyotrophic lateral sclerosis (Lennon et al. 2016, 2017).
Recent studies have reported that Bcl11a and Bcl11b are expressed in nonoverlapping populations of neurons in the murine cerebral cortex and have distinct functions (Arlotta et al. 2005; Wiegreffe et al. 2015; Greig et al. 2016; Woodworth et al. 2016). Bcl11b is expressed in layer V subcerebral neurons and is essential for the fasciculation, refinement, and elaboration of the axonal projections to the spinal cord (Arlotta et al. 2005). Bcl11a is expressed in projection neurons in all cortical layers. Within the deep cortical layers, it is not expressed in the Bcl11b+ layer V subcerebral neurons (Woodworth et al. 2016). Bcl11a is critical for the migration of upper layer projection neurons by repressing the expression of Sema3c and regulates the morphology of these neurons in a mechanism independent of Sema3c (Wiegreffe et al. 2015). In the deep cortical layers, Bcl11a promotes the acquisition of corticothalamic and callosal subtype identities and represses the layer V subcerebral identity (Wiegreffe et al. 2015; Woodworth et al. 2016). In addition, Bcl11a regulates cortical arealization by driving sensory-specific differentiation, including gene expression, output connectivity, and formation of topographic maps (Greig et al. 2016). Taken together, these studies indicate that these two related transcriptional factors control distinct development processes of cortical projection neurons, but whether they have joint functions in cortical neurogenesis and projection neuron differentiation remains to be addressed.
Here, we show that Bcl11a is expressed in RGCs and intermediate progenitors, and it regulates the proliferation and differentiation of cortical progenitor cells, as well as the timing of neurogenesis and gliogenesis switch. We show that Bcl11a binds to and likely directly represses the transcription of Nr2f1, a transcription factor previously shown to inhibit the proliferation and promote the differentiation of cortical progenitor cells (Faedo et al. 2008). We discover that beyond their own unique and seemingly antagonizing functions in specifying corticothalamic and deep-layer callosal versus layer V subcerebral neuronal subtype identities, Bcl11a and Bcl11b redundantly promote cortical projection neuron subtype specification and differentiation: In the Bcl11a and Bcl11b double mutant mice, expression of cortical projection neuronal genes, especially of the corticothalamic and layer V subcerebral neuronal genes, is severely reduced or almost absent; and consistent with these cellular and molecular defects, the corticothalamic axons are completely missing in the thalamus and the subcerebral axons fail to reach the pyramidal decussation.
Materials and Methods
Mice
Bcl11aFlox/Flox (Liu et al. 2003; Yu et al. 2012), Bcl11bFlox/Flox (Li et al. 2010), Emx1-cre+/− (Gorski et al. 2002), and ISFlox/Flox (He et al. 2016; Li et al. 2021) mice were previously described. These mice were maintained in a mixed genetic background of C57BL/6J and CD1. The date of vaginal plug detection was designated as embryonic day 0.5 (E0.5) and the day of birth was designated as postnatal day 0 (P0). All animal experiments described in this study were approved in accordance with institutional guidelines at Fudan University Shanghai Medical College.
5-Bromo-2′-deoxyuridine/5-Ethynyl-2′-deoxyuridine Labeling
For cell cycle length analysis, timed pregnant females were intraperitoneally injected with 5-bromo-2′-deoxyuridine (BrdU) (50 mg/kg in 0.9% NaCl) at E13.5 or E15.5; 5-ethynyl-2′-deoxyuridine (EdU) (10 mg/kg in DMSO) was intraperitoneally injected 1.5 h after BrdU injection. Embryonic brains were collected 30 min after EdU injection, followed by immersion fixed for 8 h in 4% PFA, cryoprotected in 30% sucrose for at least 24 h, frozen in the embedding medium, and sectioned at 12 μm using a cryostat. EdU was detected using the Click-iT EdU Cell Proliferation Kit (Thermofisher, USA), following the manufacturer’s instruction (Zhang et al. 2020). Briefly, the sections on slides were fixed in 4% PFA for 15 min, followed by washing in 1× phosphate buffered saline (PBS) twice and permeabilization with 0.5% Triton X-100 for 20 min at room temperature. Reaction cocktail was prepared freshly according to manufacturer’s instruction. The permeabilization buffer was replaced with reaction cocktail and the slides were incubated for 30 min at room temperature, protected from light. We removed the reaction cocktail, washed with 1× PBS, performed the BrdU immunocytochemistry, and proceeded to imaging and analysis.
For analysis of production of deep-layer projection neurons, female mice were given single intraperitoneal injections of BrdU at E13.5. Brains were collected at P0 and processed for BrdU immunocytochemistry.
Tissue Preparation
Postnatal mice were perfused transcardially with 1× PBS (pH 7.4), followed by 4% paraformaldehyde (PFA) in 1× PBS. Postnatal brains were dissected and postfixed overnight in 4% PFA; embryonic brains were immersion fixed for at least 12 h in 4% PFA, cryoprotected in 30% sucrose for at least 24 h, frozen in the embedding medium, and cryosectioned at 8, 12, or 20 μm using a cryostat (CM1950, Leica, USA).
Histology
The sections on slides were fixed in 4% PFA for at least 10 min, washed in double distilled water twice, followed by staining in a cresyl violet solution (Beyotime, C0117) for 1–5 min at room temperature. After cleared in distilled water, the slides were serially dehydrated in 70%, 80%, and 95% and two passes in 100% ethanol (3 min for each step). All slides were then given two 5-min passes in 100% dimethylbenzene and coverslips were applied with neutral balsam (Wang et al. 2018).
Immunohistochemistry
For BrdU detection, 8- or 12-μm-thick coronal sections were treated with 2 N HCl at room temperature for an hour, neutralized in 0.1 M borate buffer (pH 8.6) for 10 min twice, and washed in 1× PBS twice. The sections were incubated overnight at 4°C with a rat anti-BrdU antibody (1:200, Accurate Chemical, OBT0030s) (Li et al. 2018).
For Bcl11a, Bcl11b, Pax6, Eomes, Tuj1, Aldh1l1 immunohistochemistry, sections on slides were boiled in 10 mM sodium citrate briefly for antigen retrieval. The sections were permeabilized with 0.05% Triton X-100 for 30 min and incubated in a blocking buffer (5% donkey serum and 0.05% Triton X-100 in 1× PBS) for 2 h. The blocking buffer was removed, and the sections were incubated overnight at 4°C with the following primary antibodies diluted in the blocking buffer: rabbit anti-Pax6 (1:2000, MBL, PD022), mouse anti-Pax6 (1:400, BD Pharmingen, 561 664), mouse anti-Bcl11a (1:1200, Abcam, ab19487), rat anti-Bcl11b (1:1500, Abcam, ab18465), rabbit anti-Eomes (Tbr2, 1:300, Abcam, ab23345), rat anti-Eomes (Tbr2, 1:500, Thermo Fisher, 12-4875-82), rabbit anti-Dcx (1:2000, Abcam, ab18723), mouse anti-Tuj1 (1:500, Covance, MMS-435P-256), rabbit anti-Tbr1 (1:500, Abcam, ab31940), rabbit anti-PH3 (1:500, Sigma-Aldrich, 06-570), rabbit anti-KI67 (1:500, Vector Labs, VP-K451), mouse anti-NeuN (1:1000, Millipore, MAB-377), rabbit anti-Cleave Caspase-3 (1:500, Cell Signaling, 9616), goat anti-Foxp2 (1:500, Santa Cruz, sc-21 069), rabbit anti-Aldh1l1 (1:1500, Abcam, ab87117), rabbit anti-Gfap (1:2000, Dako, Z0334), goat anti-Egfr (1:500, R&D, AF1280), rabbit anti-Olig2 (1:1000, Millipore, MAB9610), and goat anti-Sox10 (1:300, R&D, AF2864).
The sections were incubated with secondary antibodies against the appropriate species for 90 min at room temperature (all from Jackson, 1:500–1:600). Fluorescently stained sections were then washed, counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma, 200 ng/mL) for 2–5 min, and coverslipped with Gel/Mount (Biomeda, Foster City, CA).
In Situ RNA Hybridization
All in situ RNA hybridization experiments were performed using digoxigenin riboprobes on 20-μm cryostat sections as previously described (Guo et al. 2019; Liu et al. 2019). Riboprobes were made from cDNAs amplified by PCR using the following primers:
Cnr1 Forward: AGAGCTATGGCATACAGGAGTGGTG
Cnr1 Reverse: GACTTGATTATTAGTAGGCGCCCAC
Plk2 Forward: GAGTAGGGAGAGAGACTGGTGCTCG
Plk2 Reverse: ATCTGGAACTGTGTGGCAACAGCT
Rorb Forward: GCCAATGTTCAGAATGTGCTAGGA
Rorb Forward: CTGGGATGTAACCTGCTTGATTGA
Satb2 Forward: AGTTCTTGAACCACCCACCCATTC
Satb2 Reverse: GAGCCTTCCTCACTGTCATTCTCCT
Cux1 Forward: CAAGATGAGAGCGAACAGTCCAGA
Cux1 Reverse: CTACTTCTAGGCTGGATCGGGTCA
Tle4 Forward: CTGTGGCAAATGGTTTGTAAGCA
Tle4 Reverse: AGTTGCCCAAATAGACTCAAAGGAA
Ppp1r1b Forward: ATGAGCCCCAGAGAGATGGAAACT
Ppp1r1b Reverse: ATTACAGCGTAGGAGGGGTTCAGG
Sox5 Forward: ACATCAAGGAAGAGATCCAGGCTG
Sox5 Reverse: CAAACACAGGCTCTTGTTTTCCTCT
Foxp1 Forward: ACACTCTAGGGACATGGCAGATTCA
Foxp1 Reverse: TGGTTCCATTGCCTACATAGTCTGC
Nr2f1 Forward: GAAAACAGCAGGAACCACAACAA
Nr2f1 Reverse: TGAAGAACAGCCTCGACAACATATA
Shtn1 Forward: GAAAACAGGAGAGACGGACAGTTCC
Shtn1 Reverse: GCAGTGAAGGGAGAAAAACAATGTG
Dync1i1 Forward: CACTGTCTCTACCGATGGCAAAATG
Dync1i1 Reverse: AAGATGCCACGGGGATACTGATACA
Otx1 Forward: TCAATTTCAACTCTCCCGACTGTC
Otx1 Reverse: TGGACGCTAAAACAAGAGAGATGC
Nfe2l3 Forward: TGATTTAGGTGCTGTAGGAGGCTG
Nfe2l3 Reverse: GGAACATGTGGATTCTCCCAACTT
Image Acquisition and Analysis
Bright field images (in situ hybridization) and some fluorescent images were imaged with Olympus VS120 slice scanning system using a 4×, 10×, or a 20× objective. Other fluorescent images were taken with Olympus FV1000 confocal microscope system using a 10×, 20×, or a 40× objective. Z-stack confocal images were reconstructed using the FV10-ASW software. All images were merged, cropped, and optimized in Photoshop CC without distortion of the original information.
Analyses were done using GraphPad Prism 7.0 and SPSS Statistics 19.0. Bcl11a expression and its colocalization with Pax6 in the E10.5 (31 250 μm2 area from the VZ) were quantified in three chosen 8-μm sections. PH3+, Eomes+, Pax6+, KI67+, and BrdU+/EdU− cells in the VZ of E15.5 cortices were quantified in three chosen 12 μm sections for each mouse (n = 3 mice). BrdU, NeuN, and Caspase-3 expression in the cortical plates were quantified in three chosen 20-μm sections for each mouse (P0 control, Bcl11a-CKO, Bcl11b-CKO, and Bcl11a/b-DCKO mice were used, n = 3 mice for each genotype). We counted the cell numbers within a 47 610 μm2 area in the cortical plate for each section.
Statistical significance was assessed using unpaired Student’s t-test or one-way ANOVA, followed by a Tukey HSD post hoc test. All quantification results were summarized as the mean ± standard error of mean (SEM). P-values < 0.05 were considered significant.
RNA-Seq Analysis
RNA-Seq analyses were performed as previously described (Guo et al. 2019). The E13.5, E15.5 cortical tissue from Bcl11a-CKO, Bcl11a/b-DCKO mice and littermate controls (Bcl11aFlox/Flox; Bcl11bFlox/Flox), and P0 cortices from Bcl11a-CKO, Bcl11b-CKO, Bcl11a/b-DCKO mice, and littermate controls were dissected (n = 3 mice/genotype/age).
Total RNA was isolated using an RNeasy Mini Kit (QIAGEN) according to the manufacturer’s protocol, quantified using NanoDrop ND-2000 and checked for RNA integrity using a bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). RNA-seq libraries were prepared according to the Illumina TruSeq protocol. An average of 15 million reads per sample was obtained. The gene expression level was reported with fragments per kilobase of exon model per million mapped reads (FPKM) (Trapnell et al. 2012). A gene was considered to be expressed if it had an FPKM > 1. Differentially expressed genes were identified using edgeR. Genes were considered as differentially expressed with a P-value < 0.05. Data from this experiment have been deposited in the GEO database (GSE182990).
CUT&Tag-Seq Analysis
CUT&Tag assay was performed using dissected E14.5 neocortices of wild-type mice, and a Bcl11a mouse polyclonal antibody as described above, using a previously published protocol (Skene and Henikoff 2017; Skene et al. 2018; Kaya-Okur et al. 2019). Briefly, the single-cell suspensions from brain tissue were prepared by mechanically disruption, followed by digestion for 15 min at 37 °C with 1.4 mg/mL papain, 3.3 × 10−3 mg/mL DNase I in DMEM cell culture medium. Digested tissue were filtered and pelleted at 300 g (RCF) for 5 min at room temperature. Then, 1 × 105 cells were washed with 500 μL of wash buffer and centrifuged at 600 g for 3 min at room temperature. Cell pellets were resuspended with 100 μL of wash buffer. Concanavalin A-coated magnetic beads were washed twice with 100 μL binding buffer. Next,10 μL of activated beads were added and incubated with the cells at room temperature for 10 min. Bead-bound cells were resuspended in 50 μL of antibody buffer. Then, 1 μg of primary antibody (mouse polyclonal anti-Bcl11a antibody) or no antibody (negative control) was added and incubated at room temperature for 2 h. The primary antibody was removed using a magnet stand. A secondary antibody (0.5 μg, goat anti-mouse IgG, Vazyme, Ab206-01) was diluted in 50 μL of Dig-wash buffer and cells were incubated at room temperature for 1 h. Cells were washed three times with Dig-wash buffer to remove unbound antibodies. The Hyperactive pG-Tn5 Transposase (Transposase mix, 4 μM, Vazyme) was diluted 1:100 in 100 μL of Dig-300 buffer. Cells were incubated with 0.04 μM Transposase mix at room temperature for 1 h. Cells were washed three times with Dig-300 buffer to remove unbound Transposase mix. Cells were then resuspended in 300 μL of tagmentation buffer and incubated at 37 °C for 1 h. To terminate tagmentation, 10 μL of 0.5 M EDTA, 3 μL of 10% SDS, and 2.5 μL of 20 mg/mL Proteinase K were added to 300 μL of sample and incubated overnight at 37 °C. DNA was purified using phenol–chloroform–isoamyl alcohol extraction, followed by an ethanol precipitation and RNase A treatment. For library amplification, 24 μL of DNA was mixed with 1 μL of TruePrep Amplify Enzyme (TAE, Vazyme), 10 μL of 5× TruePrep Amplify Enzyme buffer, 5 μL of ddH2O, and 5 μL of uniquely barcoded i5 and i7 primers from TruePrep Index Kit V2 for Illumina (Vazyme). A total volume of 50 μL of sample was placed in a Thermocycler using the following program: 72°C for 3 min; 98°C for 30 s; 17 cycles of 98°C for 15 s, 60°C for 30 s, and 72°C for 30 s; 72°C for 5 min and hold at 4°C. To purify the PCR products, 1.2× volumes of VAHTS DNA Clean Beads (Vazyme) were added and incubated at room temperature for 10 min. Libraries were washed twice with 80% ethanol and eluted in 22 μL of ddH2O.
Libraries were sequenced on an Illumina NovaSeq platform and 150-bp paired-end reads were generated. All raw sequence data were quality trimmed to a minimum phred score of 20 using trimmomatic. PCR duplicates were removed using Picard MarkDuplicates vl.107. The reads were aligned to the mm10 mouse genome using Bowtie2 version 2.3.4 and subsequently analyzed using MACS2 software version 2.1.4 to detect genomic regions enriched for multiple overlapping DNA fragments (peaks) that we considered to be putative binding sites. Peaks with a false discovery rate lower than 5% were used for further analyses. Visualization of peak distribution along genomic regions of interested genes was performed with IGV.
scRNA-Seq Dataset Used in this Study
Published scRNA-seq data from 33 976 cells from the mid-gestation human neocortex (3 female donors at GW17, GW17, and GW18, and 1 male donor at GW18) (Polioudakis et al. 2019) (the accession number is dbGaP: phs001836) were downloaded from the website https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001836.v1.p1. Supplementary Figure S1 in this study was generated using an online interface at http://geschwindlab.dgsom.ucla.edu/pages/codexviewer.
The raw gene expression matrix data obtained with Cell Ranger software were processed with standard Seurat workflow. Cells with fewer than 500 detectable genes or with >10% mitochondrial genes were filtered out. A global-scaling normalization method “Log Normalize” was applied. Cell cycle genes were regressed out to mitigate the effects of cell cycle stage. The first 50 PCs were used for clustering. Either TSNE or UMAP was used for dimension-reduction and visualization.
Results
Bcl11a Is Expressed in Cortical RGCs and Intermediate Progenitors
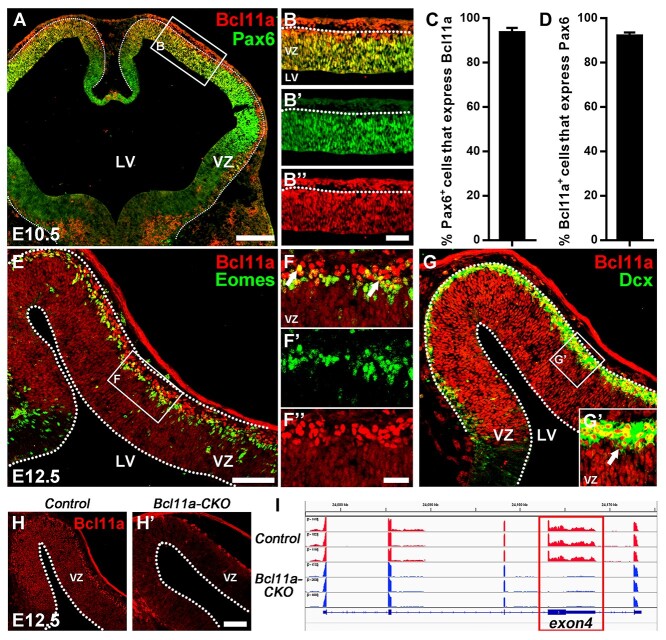
Previous studies reported that Bcl11a protein is expressed in the postmitotic projection neurons (Wiegreffe et al. 2015; Greig et al. 2016; Woodworth et al. 2016). We carefully evaluated whether Bcl11a protein is also expressed in cortical RGCs and intermediate progenitors using immunocytochemistry. At E10.5, we found that 93.5% Pax6-expressing (Pax6+) cells express Bcl11A, while 92% Bcl11a+ cells express Pax6 (Fig. 1A–D). At E12.5, Bcl11a protein is expressed in Eomes+ intermediate progenitors and Dcx+ immature neurons (Fig. 1E–G). To validate the expression of Bcl11a in cortical RGCs and intermediate progenitors, we performed immunohistochemistry on brain sections from cortex-specific Bcl11a conditional knockout mice (Emx1-Cre; Bcl11aFlox/Flox, or Bcl11a-CKO). While Bcl11a protein is detected in the VZ and SVZ in the control mice, the expression is not detected in the VZ/SVZ of the Bcl11a-CKO mice (Fig. 1H,I). Thus, in addition to the cortical projection neurons, Bcl11a protein is expressed in the RGCs, intermediate progenitors, and immature projection neurons during cortical development in mice.
Figure 1.
Bcl11a is expressed in cortical RGCs at early stages of neurogenesis. (A–B″) Bcl11a expression in the mouse cortical VZ at E10.5. Pax6 immunostaining demarcates the VZ. (C, D) Nearly all Pax6+ cells express Bcl11a and vice versa (n = 3, mean ± SEM). (E–F″) Bcl11a expression is detected in Eomes+ cortical projection neuron intermediate progenitors (arrows). (G,G′) Bcl11a is expressed in the Dcx+ immature neurons (arrows). (H,H′) Bcl11a expression is not detected in the neocortex of Bcl11a-CKO mice at E12.5. (I) RNA-Seq data show that the exon 4 (red rectangle) of Bcl11a is deleted in the Bcl11a-CKO mice. LV, lateral ventricle. White dashed lines indicate the LV and VZ boundaries. Scale bars: 200 μm in A for A; 60 μm in B″ for B–B″; 100 μm in E for E, G; 30 μm in F″ for F–F″ and G’; 180 μm in H′ for H–H′.
BCL11A gene has been implicated in human autism spectrum disorder with impaired neocortical development, and it is a candidate for the 2p15-16.1 microdeletion syndrome, which is associated with intellectual deficiency and neocortical dysplasia (Balci et al. 2015; Sanders et al. 2015; Dias et al. 2016). We examined BCL11A expression in fetal brains by analyzing published scRNA-seq data from the mid-gestation human neocortex (GW17-GW18), which provided a high-resolution transcriptome map of 33 976 cortical cells (Supplementary Fig. S1A) (Polioudakis et al. 2019). Similar to its expression in mice, BCL11A expression is present in human cortical RGCs where the cluster is defined by PAX6, SOX2, and SOX9 expression (Supplementary Fig. S1B–F) and in the EOMES+ and NEUROG2+ projection neuron intermediate progenitor cluster (Supplementary Fig. S1B,G,H). In addition, BCL11A is widely expressed in human cortical newly generated and mature projection neurons (Supplementary Fig. S1B,I). BCL11B is mainly expressed in immature and mature projection neurons (Supplementary Fig. S1C,I). We noted that BCL11B is also expressed in some cells in the EOMES+ intermediate progenitor cluster in the human cortex (Supplementary Fig. S1C,G).
Taken together, our results show that besides its wide expression in cortical projection neurons, Bcl11a is expressed in RGCs and projection neuron intermediate progenitors in the developing mouse and human cortices.
Premature Neuronal Differentiation of Cortical RGCs in the Bcl11a-CKO Mice
To determine the function of Bcl11a in cortical neurogenesis, we generated cortex-specific Bcl11a knockout mice using Emx1-Cre (Gorski et al. 2002) and a Bcl11aFlox allele in which the exon 4 of Bcl11a was flanked by two loxP sites (Liu et al. 2003; Yu et al. 2012). RNA-seq analyses of the Bcl11a-CKO cortices revealed an efficient deletion of exon 4, and immunohistochemistry showed the absence of the Bcl11a protein in the mutant cortices (Fig. 1H,H′,I).
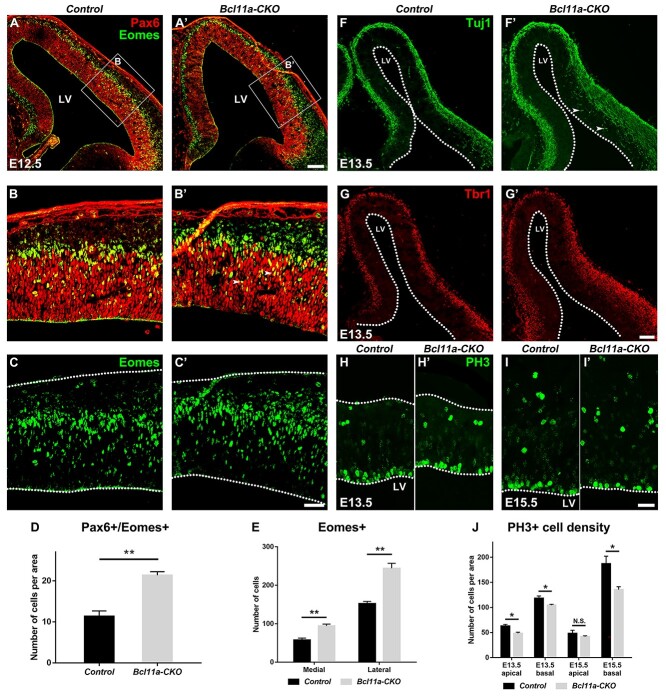
We examined the effect of Bcl11a mutations on cortical neurogenesis. Compared to the littermate control mice, the density of Pax6+/Eomes+ was increased at E12.5 after Bcl11a depletion (1.9-fold, P < 0.01) (Fig. 2A–B′,D); while the numbers of Pax6+ cells did not change at E12.5/E13.5, the numbers of Eomes+ cells were increased among lateral and medial cortex (medial cortex, 1.6-fold of control mice, P < 0.01; lateral cortex, 1.6-fold of control mice, P < 0.01) (Fig. 2C–C′,E). After Bcl11a depletion, RGCs ectopically expressed Eomes, indicating premature neuronal differentiation of cortical RGCs. We examined the expression of the pan-neuronal marker Tuj1 and early-born neuron marker Tbr1 at E13.5. Increased numbers of Tuj1+ and Tbr1+ cells were observed in the mutant cortices than in the control mice (Fig. 2F–G′).
Figure 2.
Bcl11a promotes cortical RGC proliferation and represses their differentiation. (A–C′) More Pax6+/Eomes+ progenitors are present in the Bcl11a-CKO mice than in the control mice at E12.5. (D) Quantification of Pax6+/Eomes+ progenitors at E12.5. (Student’s t-test, **P < 0.01, n = 3, mean ± SEM). (E) Quantification of Eomes+ progenitors at E12.5 in medial and lateral cortical area respectively (Student’s t-test, **P < 0.01, n = 3, mean ± SEM). (F–G′) Increased Tuj1+ (F and F′) and Tbr1+ (G and G′) neurons in the E13.5 Bcl11a-CKO cortices. Arrows indicate precious Tuj1 expression in the progenitors adjacent to the LV. (H–I′) PH3+ cells in the VZ are reduced in the Bcl11a-CKO mice compared to the control mice at E13.5 (H–H′) and E15.5 (I–I′). (J) Quantification of neurons per area expressing PH3 in the apical and basal VZ at E13.5 and E15.5 (Student’s t-test, N.S., not significant, *P < 0.05, n = 3, mean ± SEM). LV, lateral ventricle. White dashed lines indicate the LV and cortex boundaries. Scale bars: 100 μm in A′ for A–A′; 40 μm in C′ for B–C′; 100 μm in G′ for F–G′; 40 μm in I′ for H–I′.
Next, we examined whether Bcl11a regulates progenitor cell proliferation. Using phosphorylated histone 3 (PH3) antibody staining to label M-phase progenitors, we observed significantly decreased numbers of PH3+ cells in the VZ of the Bcl11a-CKO cortices at E13.5 and E15.5 (Fig. 2H–I′). The density of PH3+ cells was reduced in the apical VZ at E13.5 and was reduced in the basal VZ at E13.5/15.5, indicating that the proliferation of both RGCs and IPCs was reduced in the Bcl11a-CKO cortices (Fig. 2J).
In the Bcl11a-CKO mice, the decreased PH3+ cortical RGCs suggested that the RGC proliferation was reduced. Meanwhile increased Eomes+ and Tuj1+ cells suggested an accumulation of intermediate progenitors, which might result from a delay in extinguishment of Eomes expression or from a premature differentiation of RGCs into IPCs.
Bcl11a and Bcl11b Are Transiently Co-expressed in Migrating Cortical Projection Neurons, and Expression of One Is Increased in Compensation when the Other Gene Is Mutated
Bcl11a and Bcl11b are closely related to each other and show approximately 55% similarity of amino acid sequences. Previous studies showed that Bcl11b expression was increased in layer 6 neurons when Bcl11a expression was perturbed either by RNAi or by conditional gene knockout (Canovas et al. 2015; Woodworth et al. 2016). It is possible that Bcl11a and Bcl11b compensate for each other’s function in the single knockout mice.
To test this possibility, we performed immunohistochemistry and confirmed that indeed Bcl11b is mainly expressed in postmitotic neurons, but not in progenitor cells in wild-type cortices (Supplementary Fig. S2A–D). We carefully evaluated whether Bcl11a and Bcl11b are co-expressed in the cortical projection neurons and found that they are co-labeled in nearly all of the migrating neurons in the intermediate zone (IZ) and cortical plate (CP) at the mid-stages of cortical neurogenesis (Supplementary Fig. S2E–F′′′).
We next investigated how mutations in Bcl11a and Bcl11b affected the expression of the other. In the control brains, Bcl11b was expressed at high levels in layer 5 neurons and at low levels in layer 6 neurons. Its expression was moderately increased in layer 6 neurons of the Bcl11a-CKO brains (Supplementary Fig. S2G,G′). It is possible that Bcl11b compensates for the loss of Bcl11a in layer 6 neurons. We also carefully examined Bcl11b expression in the cortical progenitors in the Bcl11a-CKO mice but did not observe increased Bcl11b expression in these cells, indicating Bcl11b is unlikely to compensate for Bcl11a function in the progenitors.
We generated cortex-specific Bcl11b knockout mice (Emx1-Cre; Bcl11bFlox/Flox, or Bcl11b-CKO) by breeding Emx1-cre mice with mice carrying Bcl11bFlox alleles, in which the exon 4 of Bcl11b was flanked by two loxP sites. Some Bcl11b+ cells remained in the cortices of Bcl11b-CKO mice (data not shown), and they were cortical interneurons that originated and migrated from the ventral forebrain. Compared to the control mice, the expression of Bcl11a was increased across all cortical layers in the Bcl11b-CKO mice (Supplementary Fig. S2H,H′). These results indicate that the expression of Bcl11a or Bcl11b is increased in compensation in the post-migration cortical projection neurons when the other gene is mutated.
Reduced Progenitor Proliferation and Precocious Neuronal Differentiation in the Bcl11a/b-DCKO Cortices
Previous studies showed that Bcl11b expression was restricted to the postmitotic neurons and ablation of Bcl11b resulted in reduced proliferation and depletion of neural stem cells in the hippocampus (Simon et al. 2012). They suggested that dentate gyrus postmitotic neurons could provide feedback signals to regulate the proliferation and maintenance of neural stem cells, which were disrupted after Bcl11b depletion. During cortical development, both Bcl11a and Bcl11b were expressed in the postmitotic neurons and could regulate pathfinding or subtype specification, migration, and area identity of cortical projection neurons (Arlotta et al. 2005; Wiegreffe et al. 2015; Greig et al. 2016; Woodworth et al. 2016). It is possible that they function redundantly to control the development of cortical projection neurons, which provide feedback signals to regulate progenitor behaviors. Thus, we performed Bcl11a/b double conditional knockout experiments (Emx1-cre; Bcl11aFlox/Flox; Bcl11bFlox/Flox, or Bcl11a/b-DCKO) to investigate whether Bcl11a and Bcl11b together regulate progenitor proliferation, maintenance, and differentiation in the developing cortex.
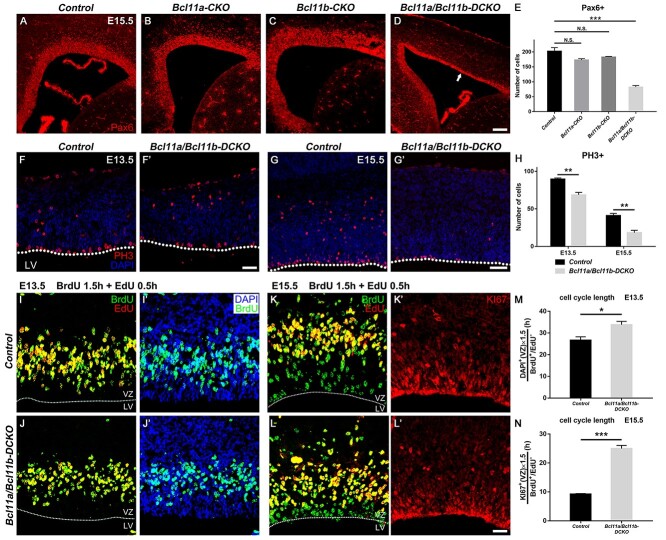
While the density of Pax6+ cells and the VZ thickness were not significantly affected in the Bcl11a-CKO and Bcl11b-CKO mice, the decreased number of the Pax6+ cells in E15.5 double mutant cortices (40.7% of control mice, P < 0.001) led to significantly reduced thickness of the VZ (Fig. 3A–E). Quantification of the numbers of the PH3+ cells in the VZ and SVZ revealed much fewer PH3+ cells in the Bcl11a/b-DCKO mice (E13.5, 76.6% of control mice, P < 0.01; E15.5, 44.5% of control mice, P < 0.01), indicating reduced progenitor cell proliferation in the absence of both Bcl11a and Bcl11b (Fig. 3F–H).
Figure 3.
Depletion of cortical RGCs in Bcl11a/b-DCKO. (A–D) Pax6 expression in VZ in control (A), Bcl11a-CKO (B), Bcl11b-CKO (C), and Bcl11a/b-DCKO mice (D, arrow) at E15.5. (E) The numbers of Pax6+ cells in the cortical VZ of Bcl11a/b-DCKO are significantly fewer than those in the controls at E15.5 (one-way ANOVA, followed by a Tukey HSD post hoc test; N.S., not significant, ***P < 0.001, n = 3, mean ± SEM). (F–G′) Representative images of PH3+ cells in the VZ/SVZ of controls and Bcl11a/b-DCKO at E13.5 (F and F′) and E15.5 (G and G′), respectively. (H) The numbers of PH3+ cells in the cortical VZ are significantly fewer in the Bcl11a/b-DCKO mice than those in the control mice (Student’s t-test, **P < 0.01, n = 3, mean ± SEM). (I–L′) Representative images of BrdU, EdU, and DAPI stainings in the controls and Bcl11a/b-DCKO at E13.5 (I–J′) and BrdU, EdU, and KI67 stainings in the controls and Bcl11a/b-DCKO at E15.5 (K–L′). (M) Quantification of cell cycle length index DAPI+(VZ)/BrdU+ EdU− in the progenitors as indicated in (I–J′) (Student’s t-test, *P < 0.05, n = 3, mean ± SEM). (N) Quantification of cell cycle length index KI67+(VZ)/BrdU+ EdU− in the progenitors as indicated in (K–L′) (Student’s t-test, ***P < 0.001, n = 3, mean ± SEM) LV, lateral ventricle; VZ, ventricular zone. White dashed lines indicate the LV boundaries. Scale bars: 200 μm in D for A–D; 100 μm in F′ for F–F′; 160 μm in G′ for G–G′; 30 μm in L′ for I–L′.
The reduced progenitor cell proliferation in the Bcl11a/b-DCKO mice could be due to either accelerated mitotic cell cycle exit or prolonged cell cycle length. We performed cell cycle quit fraction analyses and found no significant differences in the proportion of progenitors quitting the cell cycle at E13.5 or E15.5 (data not shown). To examine the cell cycle length, we employed pulse-chase of BrdU to label S-phase precursors, followed by an injection of EdU incorporated into proliferative cells 1.5 h later. We collected the brains 0.5 h after the EdU injection and utilized the single labeling cell (BrdU+ EdU−) numbers during the internal period to estimate the cell cycle length. The cell cycle length was calculated using a published method (Martynoga et al. 2005; Seuntjens et al. 2009; Wang et al. 2016). It was significantly prolonged in the Bcl11a/b-DCKO precursors at E13.5 and E15.5 (E13.5, 1.27-fold of control, P < 0.05; E15.5, 2.7-fold of control, P < 0.001), which might partially contribute to the defective proliferation of the mutant mice (Fig. 3I–N).
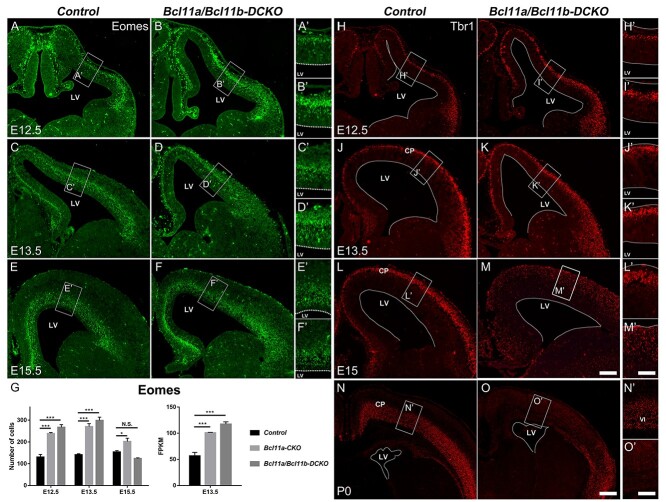
Precocious differentiation of cortical progenitors was observed in the Bcl11a-CKO mice (Fig. 2). We investigated whether lack of both Bcl11a and Bcl11b would exacerbate the phenotype. The numbers of Eomes+ cells were increased in the Bcl11a/b-DCKO mice at E12.5 and E13.5 compared to their respective control mice (E12.5, 2.0-fold of control, P < 0.001; E13.5, 2.1-fold of control, P < 0.001) (Fig. 4A–F′). Compared to the Bcl11a-CKO cortices, the number of Eomes+ cells was slightly increased in the Bcl11a/b-DCKO cortices but did not reach a statistical significance (Fig. 4G, left). At E15.5, the numbers of the Eomes+ intermediate progenitors in the Bcl11a/b-DCKO mice were not significantly different from those in the control mice (Fig. 4E–G).
Figure 4.
The premature differentiation of cortical projection neuron intermediate progenitors in Bcl11a/b-DCKO. (A–F′) In the absence of Bcl11a and Bcl11b, Eomes expression in SVZ is remarkably increased at E12.5 (A–B′) and E13.5 (C–D′) in Bcl11a/b-DCKO compared to the control brains. (G) Quantification of the numbers of Eomes+ cells in the cortices (left) and the FPKM values of Eomes gene expression based on RNA-seq data at E13.5 (right) (Student’s t-test, N.S., not significant, *P < 0.05, ***P < 0.001, n = 3, mean ± SEM). (H–O′) Expression of early-born neuron marker Tbr1 in the control and Bcl11a/b-DCKO cortices at E12.5 (H–I′), E13.5 (J–K′), E15 (L–M′), and at P0 (N–O′). (H–K′) Compared to the control cortices, more neurons in Bcl11a/b-DCKO express Tbr1 at E12.5 and E13.5. (L–M′) Numbers of Tbr1+ cells appear increased at E15, but the expression in each cell is lower than the control. (N–O′) Compared to the control brains at P0, fewer cortical neurons express Tbr1 in the dorsal cortex of Bcl11a/b-DCKO. LV, lateral ventricle; CP, cortical plate. White dashed lines indicate the LV boundaries. Scale bars: 200 μm in M for A–F and H–M; 100 μm in M′ for A′-F′ and H′-M′; 300 μm in O for N,O; 150 μm in O′ for N′–O′.
Consistent with increased numbers of Eomes+ cells at E12.5 and E13.5, we observed more Tbr1+ neurons in the Bcl11a/b-DCKO mice at the same stages (Fig. 4H–K′). However, the number of neurons with strong Tbr1 expression was reduced in the Bcl11a/b-DCKO cortices at E15.5 (Fig. 4L–M′). At P0, Tbr1 expression was virtually invisible in the motor and somatosensory cortices of the Bcl11a/b-DCKO mice (Fig. 4N–O′). These results indicate that Bcl11a and Bcl11b together regulate RGC proliferation and progenitor differentiation in the developing cerebral cortex.
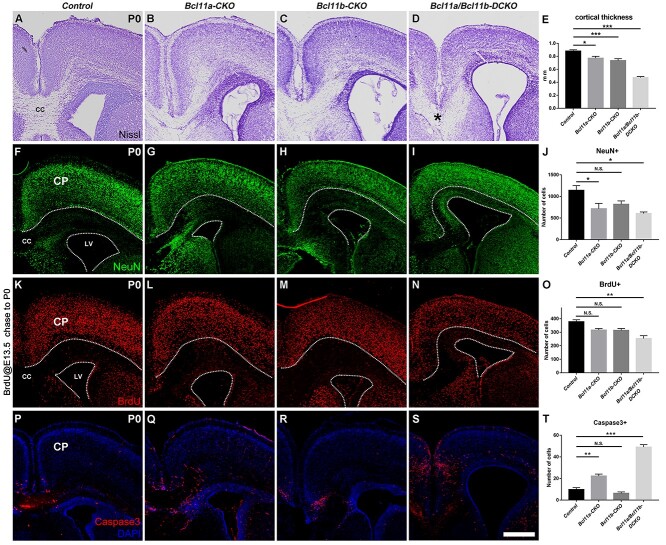
Lack of Bcl11a and Bcl11b Leads to Decreased Cortical Thickness and Increased Cell Death of Cortical Neurons
The Bcl11a/b-DCKO mice die at birth. To determine the long-term effect of mutating both genes on cortical development, we examined the P0 brains from the control, the Bcl11a-CKO, the Bcl11b-CKO, and the Bcl11a/b-DCKO mice. Nissl staining showed the decreases in cortical thickness and enlarged lateral ventricles in the Bcl11a/b-DCKO mice (Fig. 5A–E). The corpus callosum was normal size in the control and Bcl11b-CKO mice, it was reduced in the Bcl11a-CKO mice, and absent in the Bcl11a/b-DCKO mice (Fig. 5A,B,D).
Figure 5.
Defects of cortical structure, production, differentiation, and cell death in the cortices of Bcl11a/b-DCKO mice. (A–D) Nissl staining of cortices in the control mice (A) and conditional knockout mice (B–D). Note the disorganization of laminar structure and loss of corpus callosum in Bcl11a/b-DCKO (asterisk in D). (E) Measurement of the cortical thickness at P0. (one-way ANOVA, followed by a Tukey HSD post hoc test; *P < 0.05, ***P < 0.001, n = 6, mean ± SEM). (F–I) Immunostaining of pan-neuronal marker NeuN at P0. (J) Quantification of NeuN+ neurons in the cortical plate at P0 (one-way ANOVA, followed by a Tukey HSD post hoc test; N.S., not significant, *P < 0.05, n = 3, mean ± SEM). (K–N) BrdU is administered at E13.5; BrdU+ cells in the cortices are analyzed at P0. (O) Quantification of the BrdU+ neurons in the cortical plates at P0 (one-way ANOVA, followed by a Tukey HSD post hoc test; N.S., not significant, **P < 0.01, n = 3, mean ± SEM). (P–S) Immunostaining of cell death marker Cleaved Caspase-3 (red) at P0. (T) Numbers of Caspase-3+ cells are significantly higher in the Bcl11a/b-DCKO than in the controls (one-way ANOVA, followed by a Tukey HSD post hoc test; N.S., not significant, **P < 0.01, ***P < 0.001, n = 3, mean ± SEM). CP, cortical plate; CC, corpus callosum; LV, lateral ventricle. White dashed lines indicate the CP and LV boundaries. Scale bars: 400 μm in S for A–D, F–I, K–N, and P–S.
We examined the numbers of cortical neurons by performing immunohistochemistry for NeuN, a pan-neuronal marker. Compared to the control mice, the numbers of NeuN+ cells in the cortical plates of the Bcl11a-CKO and Bcl11a/b-DCKO mice were significantly reduced (Fig. 5F–J).
To investigate whether loss of Bcl11a/b affects the production and the laminar position of cortical neurons, we injected BrdU into pregnant females at E13.5 and collected the brains at P0. The total numbers of BrdU+ cells were not significantly affected in the Bcl11a-CKO or Bcl11b-CKO mice, but only two-thirds of BrdU+ cells (67.12% of control brains, P < 0.01) were present in the cortical plates of the Bcl11a/b-DCKO mice at P0 (Fig. 5K–O). We divided the IZ and CP into 10 bins and quantified the density of BrdU+ cells in each bin (Supplementary Fig. S3A–D′). While BrdU+ cells labeled at E13.5 preferred to distribute among bins 3–7 in the control or single CKO brains, they were evenly distributed in the Bcl11a/b-DCKO brains. Compared to the BrdU+ cortical cells in the control and single CKO brains, more BrdU+ cells were ectopically distributed in bins 8–10 just above the SVZ (Supplementary Fig. S3E), indicating abnormal neuronal migration and laminar position after Bcl11a/b depletion. Taken together, these results indicated that the production, migration, and laminar position of cortical projection neurons were affected after Bcl11a/b depletion.
We next determined whether mutations in Bcl11a and Bcl11b led to changes in cell viability. Staining of activated Caspase-3 showed a significant increase in cell death in the Bcl11a-CKO at P0, and there was a 5-fold (P < 0.001) increase of apoptosis in the Bcl11a/b-DCKO cortices (Fig. 5P–T). The increased Caspase-3+ cells were present both in the VZ/SVZ and the cortical plate. Thus, Bcl11a and Bcl11b are required for the survival of the cortical progenitors and projection neurons.
Bcl11a and Bcl11b Jointly Regulate Subtype Specification and Differentiation of Multiple Subtypes of Cortical Projection Neurons
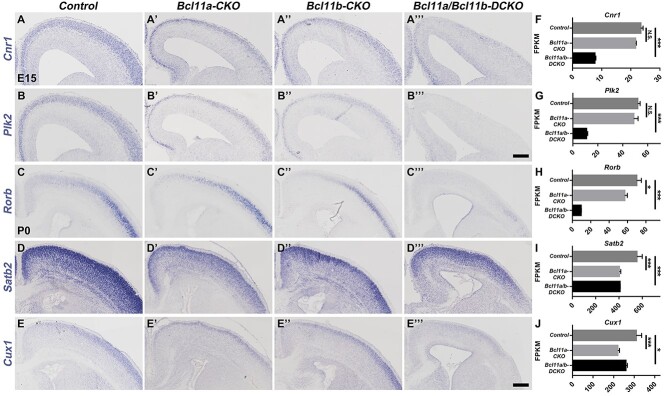
It was reported that Bcl11a regulated the balance between distinct projection neuron subtypes in deep cortical layers (Woodworth et al. 2016). Loss of Bcl11a function resulted in increased numbers of layer V subcerebral neuron development in the sensory cortex at the expense of corticothalamic neurons and deep-layer callosal neurons (Woodworth et al. 2016). Given the co-expression of Bcl11a and Bcl11b in migration neurons when both deep-layer and upper-layer cortical projection neurons are generated, the increased expression of Bcl11a in the layer VI neurons of the Bcl11b-CKO mice, and the increased expression of Bcl11b in cortical neurons in the Bcl11a-CKO mice, it is possible that these two proteins function redundantly in regulating projection neuron subtype specification and differentiation. To test this possibility, we performed in situ hybridization on E15 brain sections. Compared to the control mice, the expression of early pan-neuronal genes including Cnr1, Plk2, Shtn1, and Dync1i1 were reduced in the cortices of both Bcl11a-CKO and Bcl11b-CKO mice, but their expression was absent in the Bcl11a/b-DCKO cortices (Fig. 6A–B′′′, Supplementary Fig. S4A–B′′′), which is consistent with the RNA-seq data (Fig. 6F,G, Supplementary Fig. S4F,G), suggesting that Bcl11a and Bcl11b redundantly regulate the differentiation of cortical projection neurons.
Figure 6.
Expression of some upper-layer neuronal genes is reduced after Bcl11a and Bcl11b depletion. (A–B′′′) In situ hybridization results show severely reduced expression of the pan-neuronal genes Cnr1 (A–A′′′) and Plk2 (B–B′′′) in the cortices of Bcl11a/b-DCKO mice at E15. (C–E′′′) Expression of layer IV projection neurons gene Rorb is missing, whereas Satb2 and Cux1 show reduced expression in the Bcl11a/b-DCKO mice at P0. (F–J) The FPKM values of the genes based on RNA-seq data at the ages corresponding to the in situ experiments (Student's t-test, N.S., not significant, *P < 0.05, ***P < 0.001, n = 3, mean ± SEM). Scale bars: 200 μm in B′′′ for A–B′′′; 300 μm in E′′′ for C–E′′′.
We performed RNA-seq analyses of the control, Bcl11a-CKO, Bcl11b-CKO, and Bcl11a/b-DCKO cortices at P0 and found that the expression of many subtype-specific genes was affected in the single or double mutant mice. We validated the RNA-seq results by performing in situ hybridization or immunohistochemistry. RNA-seq analysis indicated the misregulated expression of genes that mark callosal neurons, including Satb2 and Cux1 (Fig. 6I,J). In situ hybridization confirmed that their expression was reduced in the cortices of the Bcl11a-CKO, Bcl11b-CKO, and the Bcl11a/b-DCKO mice (Fig. 6D–E′′′). The decreased Cux1 expression was more severe in the Bcl11a/b-DCKO mice than in the Bcl11a-CKO and the Bcl11b-CKO mice. RNA-seq analysis similarly revealed reduced expression for layer IV granular neuron gene Rorb in the P0 Bcl11a-CKO, Bcl11b-CKO mice (Fig. 6H), which was exacerbated in the Bcl11a/b-DCKO cortices. In situ hybridization data confirmed the RNA-seq analysis results (Fig. 6C–C′′′).
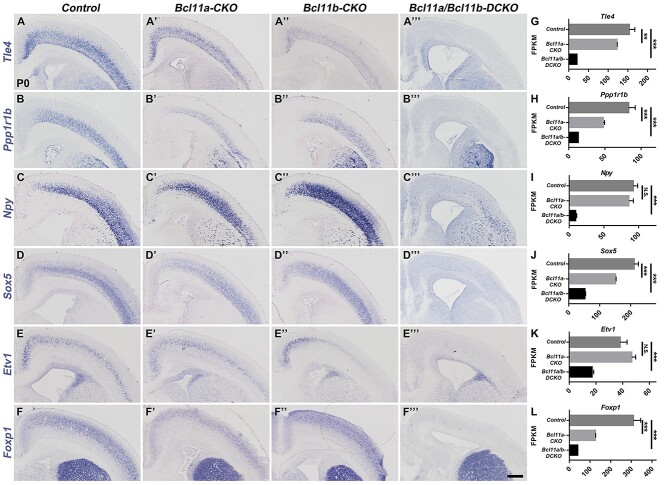
We examined whether and how the genes associated with deep-layer neuronal subtypes were affected in the mutant mice. A previous study (Woodworth et al. 2016) reported that layer V subcerebral neurons expanded at the expense of layer VI corticothalamic and deep-layer callosal neurons in the Bcl11a-CKO mice. Our RNA-seq analysis, in situ hybridization, and immunohistochemistry confirmed the increased expression of some layer V subcerebral neuron genes such as Otx1 (Supplementary Fig. S4C–C′,H) and the decreased expression of layer VI corticothalamic neuronal genes such as Tle4 (Fig. 7A,A′,G), Ppp1r1b (Fig. 7B,B′,H), and Foxp2 (Supplementary Fig. S4E,E′,J), in the Bcl11a-CKO mice. However, our analyses revealed that the expression of these genes and of many other genes associated with layer V subcerebral, layer VI corticothalamic, and subplate neurons was severely reduced or almost absent in the Bcl11a/b-DCKO cortices (Fig. 7 and Supplementary Fig. S4). The dramatic reduction or loss of molecular characteristics associated with deep-layer corticofugal neurons in the Bcl11a/b-DCKO indicates that these two genes redundantly promote the subtype identities of corticofugal neurons.
Figure 7.
Most of the deep-layer projection neuron marker genes are not expressed in the Bcl11a/b-DCKO cortices. (A–D′′′) In situ hybridization shows that expression of the corticothalamic neuron and subplate genes Tle4, Ppp1r1b, Npy, and Sox5 is almost absent in Bcl11a/b-DCKO cortices at P0. (E–F′′′) Some of the specific projection neuron genes confined in layer V, such as Etv1 and Foxp1, show reduced expression in the Bcl11a/b-DCKO cortices. (G–L) The FPKM values of the genes based on RNA-seq data at P0 (Student's t-test, N.S., not significant, **P < 0.01, ***P < 0.001, n = 3, mean ± SEM). Scale bars: 300 μm in F′′′ for A–F′′′.
In summary, both Bcl11a and Bcl11b functions are required for the differentiation of cortical projection neurons, and they redundantly promote neuronal subtype identities, especially for the layer V subcerebral and layer VI corticothalamic neurons.
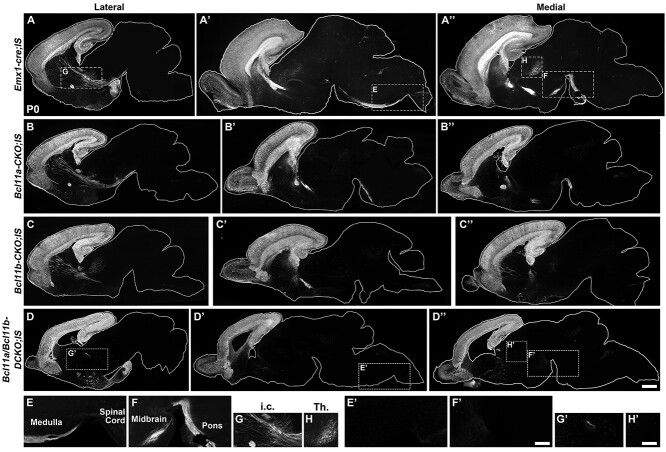
Major Cortical Axon Tracts Are Defective in the Bcl11a/b-DCKO Mice
Next, we examined whether Bcl11a and Bcl11b jointly regulated the axonal projections of the cortical excitatory neurons. To this end, we bred IS reporter mice (Rosa-CAG-LSLFrt-tdTomato-Frt-EGFP) (He et al. 2016; Li et al. 2021) with Emx1-cre mice to directly visualize the axons of cortical neurons. In the control (Emx1-cre; IS) mice, tdTomato+ corticothalamic axons from layer VI neurons, subcerebral axons including the corticospinal tract axons from layer V neurons, and callosal axons were easily identified Fig. 8A–A′′. In the Bcl11a/b-DCKO; IS mice, the corticothalamic axons were absent (Fig. 8A–D′′,H–H′, Supplementary Fig. S5B–B′′′). The cerebral peduncle axons were reduced in the Bcl11b-CKO; IS mice and no tdTomato+ axons were observed extending to the cerebral peduncle or reaching the spinal cord in the Bcl11a/b-DCKO; IS mice (Fig. 8A–D′′,E–G′, Supplementary Fig. S5C–D′′′). Similarly, the callosal axons failed to enter the corpus callosum and to cross the midline in the Bcl11a/b-DCKO; IS mice (Supplementary Fig. S5A–A′′′). The much more severe defects in the Bcl11a/b-DCKO; IS mice than the single mutant mice indicate that Bcl11a and Bcl11b redundantly regulate the development of all major cortical axon tracts, including the corticothalamic, layer V subcerebral, and callosal axons.
Figure 8.
Defects of deep-layer projection neuron axonal projections in Bcl11a/b-DCKO at P0. (A–D′′) Sagittal sections show tdTomato staining in control (Emx1-cre; IS) brains (A–A′′), Bcl11a-CKO; IS brains (B–B″), Bcl11b-CKO; IS brains (C–C″), and Bcl11a/b-DCKO; IS brains (D–D″). (E, F, E′, F′) tdTomato+ axons extend through midbrain, pons, and medulla towards spinal cord in the control brains (E and F), whereas these axons are absent in both Bcl11a/b-DCKO (E′ and F′) and Bcl11b-CKO mice (C–C″). (G, G′) tdTomato+ axons are present in the internal capsule of control mice (G) but rare in Bcl11a/b-DCKO brains (G′). (H, H′) tdTomato+ corticothalamic axons are present in the control brains but absent in the thalamus of Bcl11a/b-DCKO brains. i.c., internal capsule; Th., thalamus. Scale bars: 600 μm in D″ for A–D″; 300 μm in F′ for E–F and E′–F′; 360 μm in H′ for G–H and G′–H′.
Bcl11a Promotes RGC Proliferation and Inhibits Differentiation through Repressing Nr2f1 Expression
We sought to uncover the possible mechanism for Bcl11a and Bcl11b to regulate cortical neurogenesis. Bcl11 proteins were initially isolated through their interactions with Coup-Tf family nuclear receptors (also known as Nr2f nuclear receptors), and Bcl11a can also function as a transcriptional repressor independent of Coup-Tf family members (Avram et al. 2002; Simon et al. 2020). Recently, the protein motifs in the Bcl11a protein that mediate the interaction with members of the Nr2e/f family transcription factors have been identified (Chan et al. 2013).
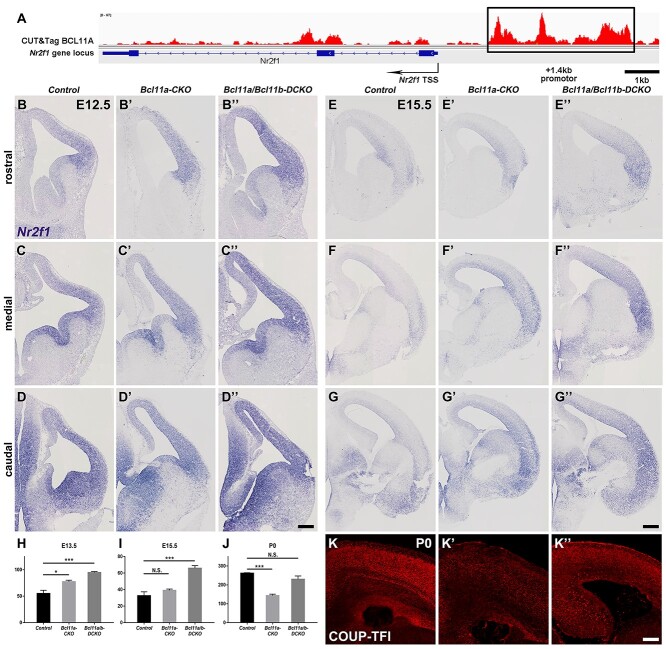
Nr2f1 plays essential roles in promoting differentiation and repressing proliferation of RGCs in the cortex (Faedo et al. 2008; Borello et al. 2014). The increased production of Eomes+ progenitors, and Tuj1+ and Tbr1+ neurons in the Bcl11a-CKO and Bcl11a/b-DCKO cortices was similar to the cortices when Nr2f1 was overexpressed (Faedo et al. 2008). We tested whether Nr2f1 expression was affected in the Bcl11a-CKO and Bcl11a/b-DCKO cortices using in situ hybridization. In the control cortices, the expression of Nr2f1 in VZ at E12.5 exhibits caudal ventral high–rostral dorsal low gradient, consistent with a previous report (Borello et al. 2014). At E12.5 and E15.5, the expression of Nr2f1 in the VZ/SVZ was elevated and shifted dorsally and rostrally in both the Bcl11a-CKO and Bcl11a/b-DCKO cortices, with the increased expression level and dorsal shift being more significant in the Bcl11a/b-DCKO cortices (Fig. 9B–G′′). At P0, Nr2f1 expression in the cortical plate was reduced in the Bcl11a-CKO cortices (Fig. 9K–K′), which is consistent with a previous study that showed decreased Nr2f1 expression in the Bcl11a-CKO cortices as area identity shifts (Greig et al. 2016). Our RNA-seq analysis confirmed the increased Nr2f1 expression in the Bcl11a-CKO and Bcl11a/b-DCKO cortices at early stage and the decreased Nr2f1 expression at later stage (Fig. 9H–J).
Figure 9.
Increased expression of Nr2f1 in the cortices of Bcl11a/b-DCKO mice. (A) CUT&Tag experiments show Bcl11a binds to a conserved region of the Nr2f1 gene. Schematic of the mouse Nr2f1 gene locus (bottom) and CUT&Tag using a Bcl11a antibody (top) are shown. The Bcl11a binding region with a consensus binding site is shown (black rectangle). (B–D″) In situ hybridization shows Nr2f1 expression in the control (B–D), Bcl11a-CKO (B′–D′), and Bcl11a/b-DCKO (B″–D″) mice. (E–G′′) Increased Nr2f1 expression in the ventricular zone and cortical plate of the Bcl11a-CKO (E′–G′) and Bcl11a/b-DCKO (E′′–G′′) mice compared to the controls at E15.5 (E–G). (H–J) The FPKM value of Nr2f1 gene based on RNA-seq data at E13.5, E15.5, and P0 (Student's t-test, N.S., not significant, *P < 0.05, ***P < 0.001, n = 3, mean ± SEM). (K–K″) Immunohistochemistry shows Nr2f1 expression in control (K), Bcl11a-CKO (K′), and Bcl11a/b-DCKO (K″) mice at P0. Scale bars: 200 μm in D″ for B–D″; 300 μm in G′′ for E–G′′, 200 μm in K″ for K–K″.
We performed a CUT&Tag experiment using a Bcl11a antibody and found that Bcl11a binds to the Nr2f1 gene locus (Fig. 9A), suggesting that it may directly inhibit Nr2f1 expression in the developing cortex.
Compared to the Bcl11a-CKO cortices, Nr2f1 expression was further derepressed in the Bcl11a/b-DCKO (Fig. 9B–G′′). Since Bcl11b expression is restricted to postmitotic neurons in both control and Bcl11a-CKO cortices, it could not compensate Bcl11a function in RGCs and progenitors to repress Nr2f1 expression. We showed above that Bcl11a/b were required to specify the deep layer neurons which might provide feedback signaling to RGCs. Loss of deep layer neurons in the Bcl11a/b-DCKO cortices would disrupt the feedback signaling and result in further increase of Nr2f1 expression.
Precocious Neurogenesis to Gliogenesis Switch in the Absence of Bcl11 Genes
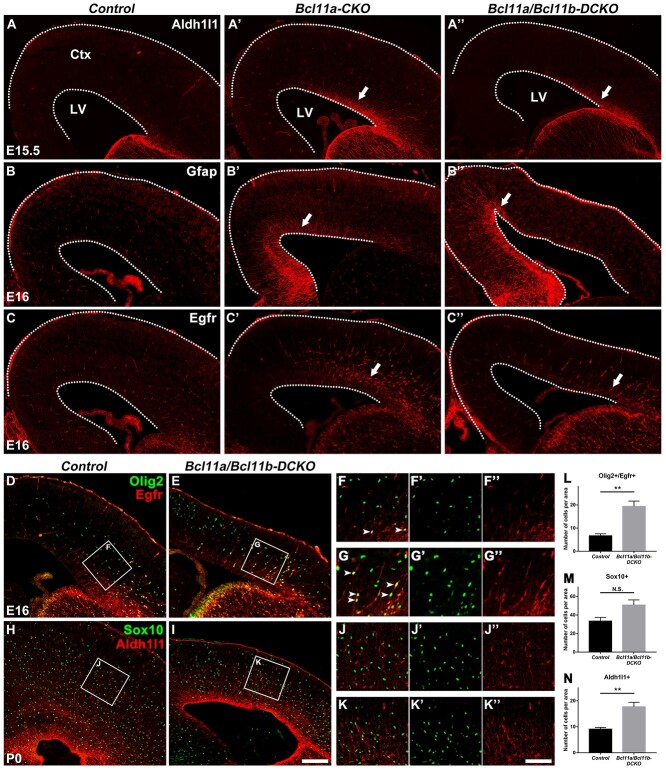
At the end of cortical neurogenesis, cortical RGCs undergo a major switch to generate cortical oligodendrocytes and astrocytes (Li et al. 2021). We investigated whether the neurogenesis to gliogenesis switch was affected in the knockout mice. During normal development, the expression of Aldh1l1 and Egfr, genes associated with gliogenesis, is detected in the cortical VZ/SVZ around E16.5 (Beattie et al. 2017; Zhang, Mennicke, et al. 2020; Li et al. 2021). Aldh1l1+ cells were observed in the cortical VZ/SVZ in the E15.5 Bcl11a-CKO and the Bcl11a/b-DCKO mice, whereas they were not observed in the controls at the same gestation age (Fig. 10A–A′′). Further, more Egfr+ cells were present in the cortical VZ/SVZ at E16 in the Bcl11a-CKO and the Bcl11a/b-DCKO mice (Fig. 10C–C′′). Another glial marker, Gfap, showed increased expression in the VZ/SVZ of both the Bcl11a-CKO and the Bcl11a/b-DCKO mice at E16 (Fig. 10B–B′′).
Figure 10.
Bcl11a and Bcl11b regulate the cortical RGC lineage progression. (A–A′′) The expression of astrocytic marker Aldh1l1 is markedly higher in Bcl11a-CKO and Bcl11a/b-DCKO than in control mice (arrows) at E15.5. (B–B″) The glial marker Gfap shows precocious expression in the cortices of Bcl11a-CKO and Bcl11a/b-DCKO mice (arrows) at E16. (C–C″) Glial progenitor marker Egfr shows significantly increased expression in progenitors of Bcl11a-CKO and Bcl11a/b-DCKO cortices (arrows) at E16. (D–G′) Olig2+/Egfr+ glial progenitors are increased in the Bcl11a/b-DCKO cortices compared to the control mice at E16 (arrows). (H–K′) Sox10+ oligodendrocytes, Aldh1l1+ astrocytes in the CP are increased in the cortices of Bcl11a/b-DCKO mice at P0. (L–N) Quantification of Olig2+/Egfr+, Sox10+, and Aldh1l1+ cells at E16 or P0. (Student’s t-test, N.S., not significant, **P < 0.01, n = 3, mean ± SEM) Ctx, cortex; LV, lateral ventricle. White dashed lines indicate the LV and cortex boundaries. Scale bars: 200 μm in I for A–C″, D–E, and H–I, 100 μm in K″ for F–G′′ and J–K″.
During early stage the cortical RGCs produce Eomes+ IPCs to give birth to projection neurons; during later stage they produce Olig2+/Egfr+ IPCs to produce olfactory bulb interneurons, cortical oligodendrocytes, and astrocytes. To further study the possible premature gliogenesis, we investigated the Olig2, Egfr, Sox10, Aldh1l1 expression in the Bcl11a/b-DCKO mice. We identified the glia population as follows: glial progenitors, Olig2+/Egfr+; oligodendrocytes, Sox10+; astrocytes, Aldh1l1+ cells in the CP. In the Bcl11a/b-DCKO mice, we observed increased Olig2+/Egfr+ IPCs at E16 (Fig. 10D–G′′,L), slightly but not significantly increased density of Sox10+ oligodendrocytes and significantly increased density of Aldh1l1+ astrocytes at P0 (Fig. 10H–K′′,M,N). These results indicate a precocious gliogenesis at late gestation stage in the Bcl11a-CKO and Bcl11a/b-DCKO mice.
Discussion
Bcl11a and Bcl11b each play individual and essential roles in regulating cortical neurogenesis (Arlotta et al. 2005; Wiegreffe et al. 2015; Woodworth et al. 2016). However, it remains unknown whether Bcl11a and Bcl11b share joint functions and act redundantly to regulate the proliferation and differentiation of cortical progenitor cells, or the subtype specification of cortical projection neurons. Here, we show that in addition to the postmitotic neurons, Bcl11a is expressed in cortical RGCs and intermediate progenitors, and it promotes proliferation and represses differentiation in progenitors in part through directly downregulating the expression of transcriptional factor Nr2f1. Our data also indicate that Bcl11a and Bcl11b function redundantly to promote the subtype specification of cortical projection neurons, especially for the deep-layer corticofugal neurons (Fig. 11). Furthermore, we find that Bcl11a (and possibly Bcl11b) prevents cortical progenitors from undergoing precocious neurogenesis to gliogenesis switch.
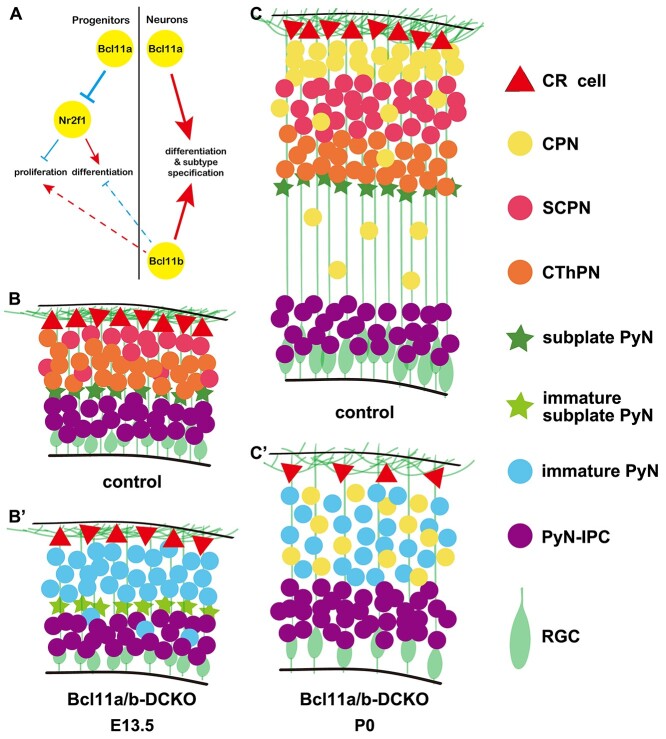
Figure 11.
Summary of Bcl11a and Bcl11b functions in cortical projection neuronal development. (A) An integrated diagram illustrating the genetic interactions of Bcl11a and Bcl11b play essential roles in different types of neurons. The red arrows indicate the positive regulation, while the blue arrows indicate the negative regulation. The dashed lines mean possible feedback regulations. (B–C′) Conditional deletion of both Bcl11a and Bcl11b results in depletion and premature differentiation of cortical RGCs, and blockage of cortical projection neuron differentiation at E13.5 and P0. This study showed that the proper intrinsic clock of proliferation and differentiation at early neurogenesis was disrupted in the absence of Bcl11a and Bcl11b, followed by precocious RGCs lineage switch. The projection neurons of the Bcl11a/b-DCKO mice failed to acquire distinct subtype identities at P0. CR cell, Cajal–Retzius cells; CPN, callosal projection neurons; SCPN, subcerebral neurons; CThPN, corticothalamic neurons; PyN, projection neurons; PyN-IPC, projection neuron intermediate progenitors; RGC, radial glial cells.
Bcl11a Regulates RGC Proliferation and Differentiation through Repressing Coup-TFI/Nr2f1 Expression
Previous studies reported that in the cerebral cortex, Bcl11a is only expressed in postmitotic cortical projection neurons and some cortical interneurons, and its expression is excluded from proliferating progenitors of the cortical VZ/SVZ (Wiegreffe et al. 2015). Our immunohistochemistry showed that the Bcl11a protein was detected in Pax6+ cells and Eomes+ intermediate progenitors at the onset of cortical neurogenesis (Fig. 1). Analysis of previously published single-cell RNA-seq data of E10.5 wild-type cortex confirmed the expression of Bcl11a in cortical progenitors (Di Bella et al. 2021). Similar expression pattern of BCL11A in the human cortex at mid-gestation week (GW17-GW18) was also observed (Supplementary Fig. S1). The expression of Bcl11a in cortical progenitors suggests that it may regulate the proliferation and differentiation of these cells. Indeed, analyses of the Bcl11a-CKO cortices revealed reduced proliferation of cortical RGCs, increased Eomes+ intermediate progenitors, and precocious production of cortical projection neurons at early stages of cortical neurogenesis. Interestingly, these defects were exacerbated in the Bcl11a/b-DCKO cortices, indicating that Bcl11a and Bcl11b function jointly to regulate the proliferation and differentiation of cortical progenitor cells.
Bcl11a and Bcl11b were shown to interact with members of Coup-Tfs subfamily (Avram et al. 2000, 2002). Nr2f1 (also known as Coup-TfI) is expressed in cortical progenitors and coordinately regulates cortical patterning, neurogenesis, and laminar fate (Faedo et al. 2008). Interestingly, similar to the Bcl11a-CKO and Bcl11a/b-DCKO mice, overexpression of Nr2f1 in the developing cortex led to reduced proliferation and increased neuronal differentiation (Faedo et al. 2008). We found that the expression of Nr2f1 is increased in the cortical progenitors of Bcl11a-CKO and Bcl11a/b-DCKO mice and that Bcl11a protein binds to a conserved sequence in the Nr2f1 gene. We speculate that Bcl11a promotes cortical RGC proliferation and prevents precocious neuronal differentiation through inhibiting high-level Nr2f1 expression.
The Redundant Functions of Bcl11 Genes in Specifying Distinct Cortical Projection Neuron Subtypes
At E14.5, Bcl11a and Bcl11b are co-expressed in the postmitotic projection neurons in the cortical plate (Supplementary Fig. S2). When the cortical projection neurons have reached their final destination, Bcl11a is expressed in projection neurons in all cortical layers, while Bcl11b shows highest expression in the layer V subcerebral neurons and low-level expression in the layer VI neurons. Consistent with the previous report (Woodworth et al. 2016), we observed increased Bcl11b expression in the layer VI cortical neurons in the Bcl11a-CKO mice. Further, we found that Bcl11a expression was increased in the cortical neurons of the Bcl11b-CKO mice. These results suggested the possibility that these two proteins may redundantly regulate the development of deep-layer cortical projection neurons. Previous studies have shown that in the Bcl11a-CKO mice, the numbers of layer V subcerebral neurons were increased at the expense of corticothalamic neurons and deep-layer callosal neurons (Woodworth et al. 2016) and that in the Bcl11b−/− mice, layer V subcerebral neurons, especially the corticospinal motor neurons, failed to extend axons at P0, when the mutant mice died (Arlotta et al. 2005). Our results confirmed these earlier results. In addition, we discovered that in the Bcl11a/b-DCKO mice, the deep-layer neurons showed much more severe defects in gene expression and axonal projections than the summation of the defects observed in the Bcl11a-CKO and those in the Bcl11b-CKO mice. Thus, besides the high expression of Bcl11a in corticothalamic, and deep-layer callosal neurons promotes these neurons to differentiate into their distinct subtypes and suppresses layer V subcerebral neuron identity, and Bcl11b promotes the extension and fasciculation of subcerebral axons, in particular the corticospinal tract axons in these neurons, our results indicate that these two genes might inhibit the high-level expression of each other in the differentiating neurons, and that they act redundantly to promote deep-layer neuron subtype identities.
Bcl11a and Bcl11b Regulate the Timing of Neurogenesis and Gliogenesis
While Bcl11b is not expressed in proliferating progenitors during normal development, there are two possible reasons for the more severe defects in the Pax6+ cells in the Bcl11a/b-DCKO cortices than the Bcl11a-CKO mice. The first possible reason is that in the absence of Bcl11a, Bcl11b expression might be upregulated in the cortical progenitors and partially compensated for the Bcl11a function in promoting progenitor cell proliferation and preventing precocious differentiation. The second possible reason is that mutant cortical projection neurons in the Bcl11a/b-DCKO mice might fail to send appropriate feedback signal to the RGCs to regulate their proliferation and differentiation. Indeed, previous studies have showed that intermediate progenitors and differentiating neurons release signaling molecules such as Delta, Jag, or Fgf9 that signal to the RGCs to regulate the cell fates of their progenies (Seuntjens et al. 2009; Parthasarathy et al. 2014; Wang et al. 2016). The Wnt7a and Wnt7b molecules that are expressed and secreted by the other radial glial cells and differentiating neurons, respectively, function in a paracrine fashion to promote the intermediate progenitors and newly generated neurons to express Delta and Jag, respectively, to activate Notch signaling in the RGCs (Wang et al. 2016). In the Bcl11a/b-DCKO cortices, the defective cortical projection neurons may fail to send the feedback signal to regulate RGC function.
At the end of cortical neurogenesis around E16.5, RGCs undergo a major lineage switch, concurrent with their gene expression change. For example, the cortical progenitors begin to express Aldh1l1 and Gfap. The cortical RGCs generate Egfr+ intermediate progenitors instead of projection neuron intermediate progenitors. The expression of Egfr in the cortex is a strong indication for the onset of cortical gliogenesis (Li et al. 2021). Surprisingly, we observed that the expression of Aldh1l1, Gfap, and Egfr in the Bcl11a-CKO and Bcl11a/b-DCKO cortices at an earlier stage than in the cortices of control mice (Fig. 10). This strongly suggests that cortical gliogenesis begins earlier in Bcl11a-CKO and Bcl11a/b-DCKO cortices. Thus, Bcl11a regulates the timing of neurogenesis-to-gliogenesis switch of cortical RGCs, and Bcl11b may play a joint role. However, currently we do not know how Bcl11a regulates this process.
Bcl11a/b Regulate Gene Expression through Chromatin Remodeling
Chromatin remodeling is an important mechanism to control gene expression. It can be carried out either by covalent modification of DNA and histones or by ATP-dependent moving, ejecting, or restructuring nucleosomes (Ho and Crabtree 2010; Sokpor et al. 2018). Previous studies suggested that Bcl11a/b were involved in both pathways to remodel chromatin. We will focus on the possible mechanisms underlying Bcl11a/b-regulated gene expression during cortical development.
Bcl11a/b could remodel chromatin through interacting with the ATP-dependent chromatin remodeling factors such as BAF complex. Bcl11a/b (also known as BAF100a/b, respectively) were identified as subunits of BRG1/BRM-associated factor (BAF) complex. BAF complex, the mammalian SWI/SNF complex, hydrolyzes ATP to mobilize nucleosomes to create nucleosome-free regions of DNA (Sokpor et al. 2018). BAF complex comprises 15 subunits, which can be classified into core subunits and variant subunits. The core subunits include the BRG1 or BRM catalytic ATPase subunit and several scaffolding proteins; the variant subunits are cell type-specific and exchangeable during lineage progression (Narayanan and Tuoc 2014). In the developing central neural system, it uses Actl6a/BAF45a/d/SS18 to assemble into the neural progenitor BAF (npBAF) complex and uses Actl6b/BAF45b/c/CREST to assemble into the neuron-specific BAF (nBAF) complex (Son and Crabtree 2014). Both npBAF and nBAF complex include a Bcl11a or Bcl11b subunit. To address the possibility of Bcl11a/b acting as BAF subunits, we compared the cortical developmental defects resulted from BAF subunits depletion or from Bcl11a/b depletion. Defects in proliferation and maintenance of neural stem cells after Bcl11a/b depletion were similarly reported after depletion of the core ATPase subunit Smarca4 (Brg1); however, reduction of cortical size seemed more severe in the Brg1 mutants; the misregulated Shh-driven ventralization of dorsal cortex in the Brg1 mutants (Zhan et al. 2011) was not observed in the Bcl11a and/or Bcl11b mutants. In addition, removing the npBAF subunit Actla6 (BAF53a) also showed reduced cortical size, premature differentiation of cortical RGCs into IPCs, and disrupted cortical layer formation; while deletion of Actla6 led to G2/M arrest of cortical RGCs (Braun et al. 2021), cell cycle progression seemed normal in the Bcl11a and/or Bcl11b mutants. Taken together, the similar effects of Bcl11a/b and BAF complex indicated that Bcl11a/b possibly control cortical development as subunits of BAF complex; the distinct effects of Bcl11a/b and BAF complexes suggested that they may also have unique functions in cortical development.
Both of Bcl11a/b contain the Nucleosome Remodeling Deacetylase (NuRD) interacting domains and Bcl11a/b possibly function as subunits of NuRD complex to repress target gene expression. The NuRD complex couples both ATP-dependent chromatin remodeling and histone deacetylase activities. It contains multiple subunits that include the histone deacetylase proteins HDAC1/2, the chromodomain-helicase-DNA-binding protein CHD3/4, and the metastasis-associated proteins MTA1/2/3 (Hoffmann and Spengler 2019). In T lymphocytes, Bcl11b associated with the NuRD complex to repress targeted genes (Cismasiu et al. 2005). In neuroblastoma, Bcl11a recruited NuRD complexes to the promoter region of Cdkn1c (also names p57KIP2, encodes a cyclin-dependent kinase inhibitor which negatively regulates cell proliferation) and suppressed its expression (Topark-Ngarm et al. 2006). During early mouse corticogenesis, deletion of Cdkn1c led to increased proliferation of RGCs and IPCs followed by increased deep layer neuron production (Hoffmann and Spengler 2019). Based on our RNA-seq data, Cdkn1c mRNA expression was upregulated after loss of Bcl11a/b function (Ctr vs. DCKO, e13.5, 41.6 vs. 65.5, P < 0.001; e15.5, 22.3 vs. 135.4, P < 0.001). It is possible that Bcl11a functions together with NuRD complex to restrict Cdkn1c expression from RGCs, which allows the normal proliferation of RGCs. In the other words, the increased Cdkn1c expression resulted from Bcl11a/b depletion could reduce the proliferation of neural progenitors and consequently reduce the cortical size.
Bcl11a/b could also remodel chromatin through covalently modifying histones. Bcl11a/b could interact directly with the histone deacetylase Sirtuin1 (Sirt1) which deacetylates proteins using NAD+ as a co-substrate. In vitro, Bcl11a recruited Sirt1 and deacetylated chromatin-associated histones H3/H4, which led to repression of the target genes in T cells (Senawong et al. 2003, 2005). But based on scRNA-seq data, Sirt1 mRNA was rarely expressed during cortical development. Thus, it is unlikely that Bcl11a/b regulated cortical development through recruiting Sirt1.
Taken together, Bcl11a/b could regulate target gene expression through ATP-dependent chromatin remodeling together with NuRD or BAF complex during cortical development.
Recent reports have revealed that Bcl11 mutations are associated with neurodevelopmental disorders in humans. BCL11A has been implicated in neuropsychiatric disorders, including intellectual disability and autism spectrum disorders, while BCL11B was required for the formation and maintenance of synapses during development as well as in adulthood, and its deficiency has been implicated in various neurodegenerative disorders (Bae and Kim 2017; Simon et al. 2020). Our study confirms previously reported unique functions of Bcl11a and Bcl11b in cortical neuron subtype specification and differentiation for distinct neuronal subtypes. We also show the novel function of Bcl11a in regulating cortical progenitor proliferation and differentiation, and the timing of neurogenesis to gliogenesis switch. In addition, we show that Bcl11a and Bcl11b function redundantly to promote the general subtype specification and differentiation of cortical projection neurons, especially for the deep-layer neuronal subtypes.
Author Contributions
H.D. performed experiments and data analysis. Z.W., R.G., L.Y., G.L., Z.Z., Z.X., and Y.T. helped conduct experiments and analyze the data. Z.Y., X.L., and B.C. guided the project and discussed some of results. Z.Y., X.L., H.D., and B.C. designed the experiments and analyzed the results. H.D., B.C., X.L., and Z.Y. wrote the paper.
Supplementary Material
Contributor Information
Heng Du, State Key Laboratory of Medical Neurobiology and MOE Frontiers Center for Brain Science, Institute of Pediatrics, Children’s Hospital of Fudan University, Institutes of Brain Science, Fudan University, Shanghai 200032, China.
Ziwu Wang, State Key Laboratory of Medical Neurobiology and MOE Frontiers Center for Brain Science, Institute of Pediatrics, Children’s Hospital of Fudan University, Institutes of Brain Science, Fudan University, Shanghai 200032, China.
Rongliang Guo, State Key Laboratory of Medical Neurobiology and MOE Frontiers Center for Brain Science, Institute of Pediatrics, Children’s Hospital of Fudan University, Institutes of Brain Science, Fudan University, Shanghai 200032, China.
Lin Yang, State Key Laboratory of Medical Neurobiology and MOE Frontiers Center for Brain Science, Institute of Pediatrics, Children’s Hospital of Fudan University, Institutes of Brain Science, Fudan University, Shanghai 200032, China.
Guoping Liu, State Key Laboratory of Medical Neurobiology and MOE Frontiers Center for Brain Science, Institute of Pediatrics, Children’s Hospital of Fudan University, Institutes of Brain Science, Fudan University, Shanghai 200032, China.
Zhuangzhi Zhang, State Key Laboratory of Medical Neurobiology and MOE Frontiers Center for Brain Science, Institute of Pediatrics, Children’s Hospital of Fudan University, Institutes of Brain Science, Fudan University, Shanghai 200032, China.
Zhejun Xu, State Key Laboratory of Medical Neurobiology and MOE Frontiers Center for Brain Science, Institute of Pediatrics, Children’s Hospital of Fudan University, Institutes of Brain Science, Fudan University, Shanghai 200032, China.
Yu Tian, State Key Laboratory of Medical Neurobiology and MOE Frontiers Center for Brain Science, Institute of Pediatrics, Children’s Hospital of Fudan University, Institutes of Brain Science, Fudan University, Shanghai 200032, China.
Zhengang Yang, State Key Laboratory of Medical Neurobiology and MOE Frontiers Center for Brain Science, Institute of Pediatrics, Children’s Hospital of Fudan University, Institutes of Brain Science, Fudan University, Shanghai 200032, China.
Xiaosu Li, State Key Laboratory of Medical Neurobiology and MOE Frontiers Center for Brain Science, Institute of Pediatrics, Children’s Hospital of Fudan University, Institutes of Brain Science, Fudan University, Shanghai 200032, China.
Bin Chen, Department of Molecular, Cell and Developmental Biology, University of California, Santa Cruz, CA 95064, USA.
Notes
We are grateful to Dr. Pengtao Liu for providing the Bcl11aFlox and Bcl11bFlox mice. Conflict of Interest: None declared.
Funding
National Key Research and Development Program of China (2018YFA0108000 to Z.Y.); National Natural Science Foundation of China (NSFC 31820103006 and 31630032 to Z.Y.); Shanghai Municipal Science and Technology Major Project (No. 2018SHZDZX01 to Z.Y.), ZJ Lab; Shanghai Center for Brain Science and Brain-Inspired Technology; National Natural Science Foundation of China (NSFC 32070971 to X.L.); NIH grants (R01 MH094589 and R01 NS089777 to B.C.).
References
- Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Farinas I, Grosschedl R, McConnell SK. 2008. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 57(3):364–377. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. 2005. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 45(2):207–221. [DOI] [PubMed] [Google Scholar]
- Avram D, Fields A, Pretty On Top K, Nevrivy DJ, Ishmael JE, Leid M. 2000. Isolation of a novel family of c(2)h(2) zinc finger proteins implicated in transcriptional repression mediated by chicken ovalbumin upstream promoter transcription factor (coup-tf) orphan nuclear receptors. J Biol Chem. 275(14):10315–10322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avram D, Fields A, Senawong T, Topark-Ngarm A, Leid M. 2002. Coup-tf (chicken ovalbumin upstream promoter transcription factor)-interacting protein 1 (ctip1) is a sequence-specific DNA binding protein. Biochem J. 368(Pt 2):555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae JR, Kim SH. 2017. Synapses in neurodegenerative diseases. BMB Rep. 50(5):237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balci TB, Sawyer SL, Davila J, Humphreys P, Dyment DA. 2015. Brain malformations in a patient with deletion 2p16.1: a refinement of the phenotype to bcl11a. Eur J Med Genet. 58(6–7):351–354. [DOI] [PubMed] [Google Scholar]
- Beattie R, Postiglione MP, Burnett LE, Laukoter S, Streicher C, Pauler FM, Xiao G, Klezovitch O, Vasioukhin V, Ghashghaei THet al. 2017. Mosaic analysis with double markers reveals distinct sequential functions of lgl1 in neural stem cells. Neuron. 94(3):517–533.e513. [DOI] [PubMed] [Google Scholar]
- Bedogni F, Hodge RD, Elsen GE, Nelson BR, Daza RA, Beyer RP, Bammler TK, Rubenstein JL, Hevner RF. 2010. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proc Natl Acad Sci U S A. 107(29):13129–13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borello U, Madhavan M, Vilinsky I, Faedo A, Pierani A, Rubenstein J, Campbell K. 2014. Sp8 and coup-tf1 reciprocally regulate patterning and fgf signaling in cortical progenitors. Cereb Cortex. 24(6):1409–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun SMG, Petrova R, Tang J, Krokhotin A, Miller EL, Tang Y, Panagiotakos G, Crabtree GR. 2021. Baf subunit switching regulates chromatin accessibility to control cell cycle exit in the developing mammalian cortex. Genes Dev. 35(5–6):335–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britanova O, Juan RC, Cheung A, Kwan KY, Schwark M, Gyorgy A, Vogel T, Akopov S, Mitkovski M, Agoston Det al. 2008. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 57(3):378–392. [DOI] [PubMed] [Google Scholar]
- Canovas J, Berndt FA, Sepulveda H, Aguilar R, Veloso FA, Montecino M, Oliva C, Maass JC, Sierralta J, Kukuljan M. 2015. The specification of cortical subcerebral projection neurons depends on the direct repression of tbr1 by ctip1/bcl11a. J Neurosci. 35(19):7552–7564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CM, Fulton J, Montiel-Duarte C, Collins HM, Bharti N, Wadelin FR, Moran PM, Mongan NP, Heery DM. 2013. A signature motif mediating selective interactions of bcl11a with the nr2e/f subfamily of orphan nuclear receptors. Nucleic Acids Res. 41(21):9663–9679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Schaevitz LR, McConnell SK. 2005. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc Natl Acad Sci U S A. 102(47):17184–17189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Wang SS, Hattox AM, Rayburn H, Nelson SB, McConnell SK. 2008. The fezf2-ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc Natl Acad Sci U S A. 105(32):11382–11387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cismasiu VB, Adamo K, Gecewicz J, Duque J, Lin Q, Avram D. 2005. Bcl11b functionally associates with the NuRD complex in T lymphocytes to repress targeted promoter. Oncogene. 24(45):6753–6764. [DOI] [PubMed] [Google Scholar]
- Di Bella DJ, Habibi E, Stickels RR, Scalia G, Brown J, Yadollahpour P, Yang SM, Abbate C, Biancalani T, Macosko EZet al. 2021. Molecular logic of cellular diversification in the mouse cerebral cortex. Nature. 595(7868):554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias C, Estruch SB, Graham SA, McRae J, Sawiak SJ, Hurst JA, Joss SK, Holder SE, Morton JE, Turner Cet al. 2016. Bcl11a haploinsufficiency causes an intellectual disability syndrome and dysregulates transcription. Am J Hum Genet. 99(2):253–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckler MJ, Larkin KA, McKenna WL, Katzman S, Guo C, Roque R, Visel A, Rubenstein JL, Chen B. 2014. Multiple conserved regulatory domains promote fezf2 expression in the developing cerebral cortex. Neural Dev. 9:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faedo A, Tomassy GS, Ruan Y, Teichmann H, Krauss S, Pleasure SJ, Tsai SY, Tsai MJ, Studer M, Rubenstein JL. 2008. Coup-tfi coordinates cortical patterning, neurogenesis, and laminar fate and modulates mapk/erk, akt, and beta-catenin signaling. Cereb Cortex. 18(9):2117–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, Jones KR. 2002. Cortical excitatory neurons and glia, but not gabaergic neurons, are produced in the emx1-expressing lineage. J Neurosci. 22(15):6309–6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. 2013. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 14(11):755–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig LC, Woodworth MB, Greppi C, Macklis JD. 2016. Ctip1 controls acquisition of sensory area identity and establishment of sensory input fields in the developing neocortex. Neuron. 90(2):261–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Liu G, Du H, Wen Y, Wei S, Li Z, Tao G, Shang Z, Song X, Zhang Zet al. 2019. Dlx1/2 are central and essential components in the transcriptional code for generating olfactory bulb interneurons. Cereb Cortex. 29(11):4831–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Kwan KY, Shim S, Lam MM, Shin Y, Xu X, Zhu Y, Li M, Sestan N. 2011. Tbr1 directly represses fezf2 to control the laminar origin and development of the corticospinal tract. Proc Natl Acad Sci U S A. 108(7):3041–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Tucciarone J, Lee S, Nigro MJ, Kim Y, Levine JM, Kelly SM, Krugikov I, Wu P, Chen Yet al. 2016. Strategies and tools for combinatorial targeting of gabaergic neurons in mouse cerebral cortex. Neuron. 91(6):1228–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Shi L, Justice N, Hsueh Y, Sheng M, Smiga S, Bulfone A, Goffinet AM, Campagnoni AT, Rubenstein JL. 2001. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 29(2):353–366. [DOI] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. 2010. Chromatin remodelling during development. Nature. 463(7280):474–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Spengler D. 2019. Chromatin remodeling complex nurd in neurodevelopment and neurodevelopmental disorders. Front Genet. 10:682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juric-Sekhar G, Hevner RF. 2019. Malformations of cerebral cortex development: molecules and mechanisms. Annu Rev Pathol. 14:293–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, Ahmad K, Henikoff S. 2019. Cut&tag for efficient epigenomic profiling of small samples and single cells. Nat Commun. 10(1):1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Lam MM, Krsnik Z, Kawasawa YI, Lefebvre V, Sestan N. 2008. Sox5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc Natl Acad Sci U S A. 105(41):16021–16026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai T, Jabaudon D, Molyneaux BJ, Azim E, Arlotta P, Menezes JR, Macklis JD. 2008. Sox5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron. 57(2):232–247. [DOI] [PubMed] [Google Scholar]
- Leid M, Ishmael JE, Avram D, Shepherd D, Fraulob V, Dolle P. 2004. Ctip1 and ctip2 are differentially expressed during mouse embryogenesis. Gene Expr Patterns. 4(6):733–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon MJ, Jones SP, Lovelace MD, Guillemin GJ, Brew BJ. 2016. Bcl11b: a new piece to the complex puzzle of amyotrophic lateral sclerosis neuropathogenesis? Neurotox Res. 29(2):201–207. [DOI] [PubMed] [Google Scholar]
- Lennon MJ, Jones SP, Lovelace MD, Guillemin GJ, Brew BJ. 2017. Bcl11b-a critical neurodevelopmental transcription factor-roles in health and disease. Front Cell Neurosci. 11:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK. 2008. The determination of projection neuron identity in the developing cerebral cortex. Curr Opin Neurobiol. 18(1):28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessel D, Gehbauer C, Bramswig NC, Schluth-Bolard C, Venkataramanappa S, Gassen KLI, Hempel M, Haack TB, Baresic A, Genetti CAet al. 2018. Bcl11b mutations in patients affected by a neurodevelopmental disorder with reduced type 2 innate lymphoid cells. Brain. 141(8):2299–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang C, Zhang Z, Wen Y, An L, Liang Q, Xu Z, Wei S, Li W, Guo Tet al. 2018. Transcription factors sp8 and sp9 coordinately regulate olfactory bulb interneuron development. Cereb Cortex. 28(9):3278–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Burke S, Wang J, Chen X, Ortiz M, Lee SC, Lu D, Campos L, Goulding D, Ng BLet al. 2010. Reprogramming of t cells to natural killer-like cells upon bcl11b deletion. Science. 329(5987):85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu G, Yang L, Li Z, Zhang Z, Xu Z, Cai Y, Du H, Su Z, Wang Zet al. 2021. Decoding cortical glial cell development. Neurosci Bull. 37(4):440–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Hargreaves VV, Zhu Q, Kurland JV, Hong J, Kim W, Sher F, Macias-Trevino C, Rogers JM, Kurita Ret al. 2018. Direct promoter repression by bcl11a controls the fetal to adult hemoglobin switch. Cell. 173(2):430–442 e417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. 2003. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 13(3):476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Zhang Z, Lindtner S, Li Z, Xu Z, Wei S, Liang Q, Wen Y, Tao G, You Yet al. 2019. Sp9 regulates medial ganglionic eminence-derived cortical interneuron development. Cereb Cortex. 29(6):2653–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynoga B, Morrison H, Price DJ, Mason JO. 2005. Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev Biol. 283(1):113–127. [DOI] [PubMed] [Google Scholar]
- McConnell SK. 1989. The determination of neuronal fate in the cerebral cortex. Trends Neurosci. 12(9):342–349. [DOI] [PubMed] [Google Scholar]
- McConnell SK. 1995. Constructing the cerebral cortex: neurogenesis and fate determination. Neuron. 15(4):761–768. [DOI] [PubMed] [Google Scholar]
- McKenna WL, Betancourt J, Larkin KA, Abrams B, Guo C, Rubenstein JL, Chen B. 2011. Tbr1 and fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J Neurosci. 31(2):549–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna WL, Ortiz-Londono CF, Mathew TK, Hoang K, Katzman S, Chen B. 2015. Mutual regulation between satb2 and fezf2 promotes subcerebral projection neuron identity in the developing cerebral cortex. Proc Natl Acad Sci U S A. 112(37):11702–11707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. 2005. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 47(6):817–831. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. 2007. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 8(6):427–437. [DOI] [PubMed] [Google Scholar]
- Narayanan R, Tuoc TC. 2014. Roles of chromatin remodeling baf complex in neural differentiation and reprogramming. Cell Tissue Res. 356(3):575–584. [DOI] [PubMed] [Google Scholar]
- Parthasarathy S, Srivatsa S, Nityanandam A, Tarabykin V. 2014. Ntf3 acts downstream of sip1 in cortical postmitotic neurons to control progenitor cell fate through feedback signaling. Development. 141(17):3324–3330. [DOI] [PubMed] [Google Scholar]
- Polioudakis D, Torre-Ubieta L, Langerman J, Elkins AG, Shi X, Stein JL, Vuong CK, Nichterwitz S, Gevorgian M, Opland CKet al. 2019. A single-cell transcriptomic atlas of human neocortical development during mid-gestation. Neuron. 103(5):785–801 e788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, Murtha MT, Bal VH, Bishop SL, Dong Set al. 2015. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron. 87(6):1215–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senawong T, Peterson VJ, Avram D, Shepherd DM, Frye RA, Minucci S, Leid M. 2003. Involvement of the histone deacetylase sirt1 in chicken ovalbumin upstream promoter transcription factor (coup-tf)-interacting protein 2-mediated transcriptional repression. J Biol Chem. 278(44):43041–43050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senawong T, Peterson VJ, Leid M. 2005. Bcl11a-dependent recruitment of sirt1 to a promoter template in mammalian cells results in histone deacetylation and transcriptional repression. Arch Biochem Biophys. 434(2):316–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seuntjens E, Nityanandam A, Miquelajauregui A, Debruyn J, Stryjewska A, Goebbels S, Nave KA, Huylebroeck D, Tarabykin V. 2009. Sip1 regulates sequential fate decisions by feedback signaling from postmitotic neurons to progenitors. Nat Neurosci. 12(11):1373–1380. [DOI] [PubMed] [Google Scholar]
- Shim S, Kwan KY, Li M, Lefebvre V, Sestan N. 2012. Cis-regulatory control of corticospinal system development and evolution. Nature. 486(7401):74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Brylka H, Schwegler H, Venkataramanappa S, Andratschke J, Wiegreffe C, Liu P, Fuchs E, Jenkins NA, Copeland NGet al. 2012. A dual function of bcl11b/ctip2 in hippocampal neurogenesis. EMBO J. 31(13):2922–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Wiegreffe C, Britsch S. 2020. Bcl11 transcription factors regulate cortical development and function. Front Mol Neurosci. 13:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Henikoff JG, Henikoff S. 2018. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc. 13(5):1006–1019. [DOI] [PubMed] [Google Scholar]
- Skene PJ, Henikoff S. 2017. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife. 6:e21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soblet J, Dimov I, Graf von Kalckreuth C, Cano-Chervel J, Baijot S, Pelc K, Sottiaux M, Vilain C, Smits G, Deconinck N. 2018. Bcl11a frameshift mutation associated with dyspraxia and hypotonia affecting the fine, gross, oral, and speech motor systems. Am J Med Genet A. 176(1):201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokpor G, Castro-Hernandez R, Rosenbusch J, Staiger JF, Tuoc T. 2018. Atp-dependent chromatin remodeling during cortical neurogenesis. Front Neurosci. 12:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son EY, Crabtree GR. 2014. The role of baf (mswi/snf) complexes in mammalian neural development. Am J Med Genet C Semin Med Genet. 166C(3):333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Yao Z, Graybuck LT, Smith KA, Nguyen TN, Bertagnolli D, Goldy J, Garren E, Economo MN, Viswanathan Set al. 2018. Shared and distinct transcriptomic cell types across neocortical areas. Nature. 563(7729):72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topark-Ngarm A, Golonzhka O, Peterson VJ, Barrett B Jr, Martinez B, Crofoot K, Filtz TM, Leid M. 2006. Ctip2 associates with the NuRD complex on the promoter of p57kip2, a newly identified ctip2 target gene. J Biol Chem. 281(43):32272–32283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of rna-seq experiments with tophat and cufflinks. Nat Protoc. 7(3):562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsyporin J, Tastad D, Ma X, Nehme A, Finn T, Huebner L, Liu G, Gallardo D, Makhamreh A, Roberts JMet al. 2021. Transcriptional repression by fezf2 restricts alternative identities of cortical projection neurons. Cell Rep. 35(12):109269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Jossin Y, Chai G, Lien WH, Tissir F, Goffinet AM. 2016. Feedback regulation of apical progenitor fate by immature neurons through wnt7-celsr3-fzd3 signalling. Nat Commun. 7:10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhou F, Dou Y, Tian X, Liu C, Li H, Shen H, Chen G. 2018. Melatonin alleviates intracerebral hemorrhage-induced secondary brain injury in rats via suppressing apoptosis, inflammation, oxidative stress, DNA damage, and mitochondria injury. Transl Stroke Res. 9(1):74–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegreffe C, Simon R, Peschkes K, Kling C, Strehle M, Cheng J, Srivatsa S, Liu P, Jenkins NA, Copeland NGet al. 2015. Bcl11a (ctip1) controls migration of cortical projection neurons through regulation of sema3c. Neuron. 87(2):311–325. [DOI] [PubMed] [Google Scholar]
- Woodworth MB, Greig LC, Liu KX, Ippolito GC, Tucker HO, Macklis JD. 2016. Ctip1 regulates the balance between specification of distinct projection neuron subtypes in deep cortical layers. Cell Rep. 15(5):999–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Nakashima M, Okanishi T, Kanai S, Fujimoto A, Itomi K, Morimoto M, Saitsu H, Kato M, Matsumoto Net al. 2018. Identification of novel bcl11a variants in patients with epileptic encephalopathy: expanding the phenotypic spectrum. Clin Genet. 93(2):368–373. [DOI] [PubMed] [Google Scholar]
- Yu Y, Wang J, Khaled W, Burke S, Li P, Chen X, Yang W, Jenkins NA, Copeland NG, Zhang Set al. 2012. Bcl11a is essential for lymphoid development and negatively regulates p53. J Exp Med. 209(13):2467–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Shi X, Zhang Z, Chen Y, Wu JI. 2011. Dual role of brg chromatin remodeling factor in sonic hedgehog signaling during neural development. Proc Natl Acad Sci U S A. 108(31):12758–12763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Mennicke CV, Xiao G, Beattie R, Haider MA, Hippenmeyer S, Ghashghaei HT. 2020. Clonal analysis of gliogenesis in the cerebral cortex reveals stochastic expansion of glia and cell autonomous responses to egfr dosage. Cells. 9(12):2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu G, Guo T, Liang XG, Du H, Yang L, Bhaduri A, Li X, Xu Z, Zhang Zet al. 2020. Cortical neural stem cell lineage progression is regulated by extrinsic signaling molecule sonic hedgehog. Cell Rep. 30(13):4490–4504 e4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.