Abstract
Rapalogs (everolimus and temsirolimus) are allosteric mTORC1 inhibitors and approved agents for advanced clear cell renal cell carcinoma (ccRCC), although only a subset of patients derive clinical benefit. Progress in genomic characterization has made it possible to generate comprehensive profiles of genetic alterations in ccRCC; however, the correlations between recurrent somatic mutations and rapalog efficacy remain unclear. Here, we demonstrate by using multiple patient-derived ccRCC cell lines that compared to PTEN-proficient cells, PTEN-deficient cells exhibit hypersensitivity to rapalogs. Rapalogs inhibit cell proliferation by inducing G0/G1 arrest without inducing apoptosis in PTEN-deficient ccRCC cell lines. Using isogenic cell lines generated by CRISPR/Cas9, we validate the correlation between PTEN loss and rapalog hypersensitivity. In contrast, deletion of VHL or chromatin-modifying genes (PBRM1, SETD2, BAP1, or KDM5C) fails to influence the cellular response to rapalogs. Our mechanistic study shows that ectopic expression of an activating mTOR mutant (C1483F) antagonizes PTEN-induced cell growth inhibition, while introduction of a resistant mTOR mutant (A2034V) enables PTEN-deficient ccRCC cells to escape the growth inhibitory effect of rapalogs, suggesting that PTEN loss generates vulnerability to mTOR inhibition. PTEN-deficient ccRCC cells are more sensitive to the inhibitory effects of temsirolimus on cell migration and tumor growth in zebrafish and xenograft mice, respectively. Of note, PTEN protein loss as detected by immunohistochemistry is much more frequent than mutations in the PTEN gene in ccRCC patients. Our study suggests that PTEN loss correlates with rapalog sensitivity and could be used as a marker for ccRCC patient selection for rapalog therapy.
Keywords: clear cell renal cell carcinoma, mTOR, PTEN, rapalogs, recurrent mutations, sensitivity
Introduction
Renal cell carcinoma (RCC) encompasses a heterogeneous group of cancers derived from renal tubular epithelial cells. Clear cell renal cell carcinoma (ccRCC) is the most common histological subtype and accounts for the most RCC-specific deaths [1]. Surgery is the only curative treatment for localized RCC, while systemic treatment is necessary for patients with metastatic RCC. ccRCC is usually resistant to chemoradiotherapy. Targeted therapies, including inhibitors of vascular endothelial growth factor receptor (VEGFR) and its ligand or inhibitors of mammalian target of rapamycin (mTOR) signaling pathways, are standard treatment options for patients with metastatic ccRCC [2, 3].
The mTOR serine/threonine kinase exerts its effects mainly through mTOR complex 1 and 2 (mTORC1/2), defined by distinct protein partners. Rapamycin and its analogs everolimus and temsirolimus (so-called rapalogs) are inhibitors of the mTOR pathway, acting through FK506-binding protein 12 (FKBP12) to inhibit the mTORC1 complex [4]. Large, randomized clinical trials on mTOR inhibitors in the metastatic RCC setting led to Food and Drug Administration approval of temsirolimus and everolimus [5, 6].
The Cancer Genome Atlas (TCGA) data revealed that the genetic changes underlying ccRCC include alterations in genes controlling cellular oxygen sensing (e.g., VHL), the maintenance of chromatin states (e.g., PBRM1, SETD2, BAP1, and KDM5C), and the PI3K/AKT/mTOR pathway (e.g., PIK3CA, MTOR, and PTEN) [7]. Paradoxically, targeted treatments such as rapalogs are applied without assessing the genomic profile of tumors in individual ccRCC patients [8]. It is reasonable to anticipate that ccRCC patients with high-level activation of mTORC1 resulting from activating mutations in either MTOR or PIK3CA or inactivating mutations or loss of TSC1, TSC2 or PTEN are likely more sensitive to rapalogs. Remarkably, previous studies have shown controversial results regarding the potential correlations between alterations in mTOR pathway genes and the response to rapalog therapy in ccRCC [9–13]. Furthermore, whether other recurrent mutations affect the cellular response to rapalogs remains obscure [9, 10, 13]. Here, we sought to clarify the associations between recurrent mutations (VHL, PBRM1, SETD2, BAP1, KDM5C, and PTEN) and the response to rapalog treatment by using patient-derived ccRCC cell lines and isogenic cell models generated by gene editing techniques. Our study demonstrates that PTEN loss confers sensitivity to rapalogs in cell models and in xenografted zebrafish and mouse models.
Materials and methods
Cell culture
HEK293A cells were kindly provided by Dr. Kun-Liang Guan (University of California, San Diego). A498 and OS-RC2 cells were obtained from Cellcook Biotech Co., Ltd. (Guangzhou, China). A704 and HEK293T cells were available from ATCC (Beijing Zhongyuan Limited, Beijing, China). Caki-1, 769P, and 786O cells were obtained from the Cell Resource Center of the Chinese Academy of Sciences (Shanghai, China). KMRC1, KMRC20, VMRC-RCW, and VMRC-RCZ cells were obtained from the Japanese Collection of Research Bioresources (Tokyo, Japan). UT16, UT33a, and UT44 cells were obtained from the Chinese Academy of Medical Sciences (Beijing, China). A498, VMRC-RCW, and VMRC-RCZ cells were maintained in MEM (Thermo Fisher Scientific); Caki-1 cells were maintained in McCoy’s 5A medium (Thermo Fisher Scientific); HEK293A, HEK293T, KMRC1 and KMRC20 cells were maintained in DMEM (Thermo Fisher Scientific); UT16, UT33a and UT44 cells were maintained in DMEM/F12 (Thermo Fisher Scientific); and other cell lines were maintained in RPMI-1640 medium (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (Gemini) and 1% penicillin/streptomycin (Invitrogen). Cells were maintained at 37 °C in a humidified incubator with 5% CO2. No mycoplasma (Thermo Fisher Scientific) contamination was detected.
Gene deletion by the CRISPR/Cas9 system
The CRISPR/Cas9 system was used to delete genes of interest. The single guide RNA (sgRNA) sequences were cloned into the plasmids px459 v2 (Addgene #62988) or LentiCRISPR V2 (Addgene #52961). The sgRNA sequences targeting individual genes were as follows (5′-3′): CGCGCGTCGTGCTGCCCGTA (VHL #1), CCCGTATGGCTCAACTTCGA (VHL #2), GAAACCACTTCATAATAGTC (PBRM1 #1), TTACTCACAGGATCTACAGT (PBRM1 #2), TTAAAGAACCAGTTGATACG (SETD2 #1), TGTAACATCCAGGCCACTGC (SETD2 #2), ACCGAAATCTTCCACGAGCA (BAP1 #1), ACCCACCCTGAGTCGCATGA (BAP1 #2), GGTTTACCCCCCGAATCCAG (KDM5C #1), AGATTCCCAATGTAGAACGG (KDM5C #2), ACAGATTGTATATCTTGTAA (PTEN #1), and ACGCCTTCAAGTCTTTCTGC (PTEN #2). The constructed px459 plasmids were transfected into HEK293A cells. Twenty-four hours after transfection, HEK293A cells were treated with puromycin (1 μg/mL) for 3 days and were then sorted into 96-well plates with only one cell in each well. The clones were screened by Western blot analysis with gene-specific antibodies, and at least two independent clones for each gene deletion were used for each experiment described. For analysis of genome modification by CRISPR/Cas9 in knockout (KO) clones, PCR primers were designed to span the Cas9 cleavage site. Indel mutations were identified through Sanger sequencing of PCR products. PTEN was deleted in KMRC20 and 769P cells by a lentiviral CRISPR/Cas9 system. Lentiviral particles were generated using HEK293T cells as previously described [14]. Lentiviral supernatant was filtered through a 0.45 μm filter, supplemented with 5 μg/mL polybrene, and used for infection. The polyclonal population was used for the subsequent experiments shortly after puromycin selection (1 μg/mL, 3 days).
Viral infection
786O and OS-RC2 cells stably expressing empty vector, PTEN, or mTOR were generated by retroviral and lentiviral infection. HEK293T packaging cells were transfected with empty vector, (pQCXIH hygro) PTEN, (pQCXIH hygro) FKBP12, or (pLVX puro) mTOR constructs (Youbio Technologies). Forty-eight hours after transfection, retroviral and lentiviral supernatants were filtered through a 0.45 μm filter, supplemented with 5 μg/mL polybrene, and used for infection. Forty-eight hours after infection, cells were selected with hygromycin (500 μg/mL) or puromycin (1 μg/mL).
Immunoblot analysis and immunoprecipitation
Equal amounts of protein samples were separated by SDS–PAGE. Antibodies against mTOR (2983P, 1:1000), H2AK119ub1 (8240S, 1:200,000), HA (2367S, 1:1000), PTEN (9188S, 1:500), p-AKT (Thr308) (4056S, 1;1000), p-S6K (Thr389) (9234S, 1:1000), HIF-1α (36169T, 1:1000), H3K36me3 (8173S, 1:3000), H3K4me3 (9751S, 1:10,000) and VHL (68547S, 1:1000) were obtained from Cell Signaling Technology; those against BAP1 (sc-28383, 1:100) and GAPDH (sc-25778, 1:10,000) were obtained from Santa Cruz; those against PBRM1 (A301-591A, 1:1000) and KDM5C (A301-034A-M, 1:1000) were obtained from Bethyl Laboratories; the antibody against SETD2 (A3194, 1:1000) was obtained from ABclonal; and the antibody against Flag (F1804, 1:1000) was obtained from Sigma–Aldrich. For immunoprecipitation, cells were lysed with a buffer containing 0.05 M Tris-HCl (pH 7.5), 0.15 M NaCl, 0.5% Triton X-100, protease inhibitor cocktail (11873580001, Roche) and phosphatase inhibitor cocktail (88667, Thermo Fisher Scientific). Primary antibodies were added to the lysates and incubated with rotation overnight at 4 °C. Magnetic protein A/G beads (88802, Thermo Fisher Scientific) were added and incubated for an additional 2 h. The immunoprecipitates were washed three times with lysis buffer and were then subjected to immunoblot analysis.
Immunohistochemistry
ccRCC tissue arrays (OD-CT-UrKid01-002) were purchased from Shanghai Outdo Biotech Co., Ltd. Paraffin sections were deparaffinized and rehydrated with xylene and alcohol gradients. Sections were then subjected to heat-induced antigen retrieval using 10 mM sodium citrate buffer followed by 3% H2O2 for 30 min to quench endogenous peroxidase activity. Sections were incubated overnight at 4 °C with antibodies against PTEN (9188S, 1:500, Cell Signaling Technology) or Ki67 (9449S, 1:200, Cell Signaling Technology), and antibodies were detected using a Vectastain ABC kit and DAB peroxidase substrate kit (ZSGB-BIO). The intensity and extent of staining in the tumor cells were evaluated, and the cases were categorized as PTEN- or Ki67-positive/negative according to the following criteria. PTEN expression levels were assessed by scoring the staining intensity in the whole cell as 0 (absent), 1 (weak), 2 (moderate) or 3 (strong); PTEN-negative tumors were those with an intensity score of 0/1. Ki67 expression was assessed by scoring the intensity in the nucleus as negative, weakly positive or strongly positive; tumors with positive Ki67 expression were identified as those with weakly/strongly positive staining.
MTT cell viability assay
Cells were seeded in 96-well plates 1 day prior to treatment with everolimus (T1784, TargetMol) or temsirolimus (T2145, TargetMol) for 6 days. MTT (0793, VWR International) reagent was then added. The absorbance was measured with a spectrophotometer (Bio–Rad Laboratories) at a wavelength of 570 nm. Relative cell viability was calculated as the percentage of viable treated cells relative to vehicle control (DMSO) cells.
Colony formation assay
786O (800 cells per well) and OS-RC2 (1500 cells per well) cells were seeded in six-well plates. Colonies were stained using 0.1% crystal violet and counted 10–14 days after plating. The colony number and the sum of colony area were calculated by ImageJ software.
Cell cycle analysis
Cells were treated with temsirolimus at the indicated concentrations for 48 h. The cells were then fixed with ice-cold 75% ethanol in PBS and stained using propidium iodide. The distribution of cells in different phases of the cell cycle was analyzed by flow cytometry (BD FACSCanto II).
Annexin V staining for apoptosis analysis
Cells were treated with temsirolimus at the indicated concentrations for 6 days. The cells were collected and stained with Annexin V-APC and propidium iodide (Annexin V-APC/PI Apoptosis Detection Kit, KGA1030-100T, KeyGEN BIOTECH) according to the manufacturer’s protocol. Flow cytometry (BD LSRFortessa X-20) was used to analyze the stained cells within 1 h.
Wound healing assay
Cells were serum starved overnight, after which the cell layer in the six-well plate was scratched using a 100 μL pipette tip and was then cultured in serum-free medium with or without temsirolimus at the indicated concentrations. The wound area was measured at 0 and 48 h. Quantification of wound healing was performed using ImageJ. The wound area was used to determine the extent of wound healing, and the cell migration ability was evaluated using the “relative area of wound closed”.
Whole-exome and whole-genome sequencing
Genomic DNA was extracted from UT16, UT33a, UT44, and VMRC-RCZ cells using a PureLink Genomic DNA Mini Kit (Thermo Fisher Scientific). DNA was then sent to a sequencing company (Novogene) for whole-exome and whole-genome sequencing.
RNA sequencing and bioinformatics analysis
Total RNA was extracted with an RNAsimple Total RNA Kit (RE-03113, FOREGENE) from HEK293A cells, and PTEN KO#2 cells treated with vehicle control (DMSO) or 1000 nM temsirolimus for 24 h. Three replicates for each sample were established and analyzed. RNA was sent to a sequencing company (Novogene) for sequencing. Bioinformatics analysis was performed as described previously [15]. Genes with adjusted P < 0.05 and absolute log2 fold change >0.585 were considered significantly differentially expressed genes (DEGs). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of DEGs was performed using the online database KOBAS 3.0.
Animal work
All animal procedures were carried out according to protocols approved by the Institutional Animal Care and Use Committee of Southern Medical University (2015-0056).
Wild-type AB zebrafish were obtained from the China Zebrafish Resource Center (Wuhan, China). Zebrafish embryos at 48 h post fertilization (hpf) were dechorionated prior to cell injection. Approximately 100 CM-DiI (Thermo Fisher Scientific)-labeled cells were injected into the embryonic yolk sac using a micropipette and pump (World Precision Instruments). Engrafted embryos were sorted to remove falsely injected embryos. Then, the embryos were exposed to DMSO (solvent control) or temsirolimus at the indicated concentrations. The exposed embryos were maintained at 28 °C for 1 h and were then placed in an incubator at 34 °C for 6 days. The drug-containing medium was replaced every day. At 6 days post injection (6 dpi), engrafted zebrafish embryos were examined using an Olympus MVX10 Zoom Fluorescence Macro System Microscope.
Male nude mice (6 weeks old) purchased from Guangdong Medical Laboratory Animal Center (Guangzhou, China) were used to evaluate the anticancer efficacy of temsirolimus against A498 and OS-RC2 cells. A498 (5 × 106) and OS-RC2 (5 × 106) cells were grafted subcutaneously into the right back flanks of nude mice. When the tumors were just palpable, the mice were randomly assigned to the vehicle control (5% ethanol + 5% Tween 80 + 5% PEG 300 in water) or temsirolimus group (n = 10 per group). Temsirolimus was administered intraperitoneally at 20 mg/kg body weight once daily. Tumor height and width were measured with a caliper every other day to calculate tumor volume (=width2 × height × 0.5). Mice were sacrificed after 15 days of treatment. Tumor xenografts were excised, fixed with 4% paraformaldehyde, and embedded in paraffin.
Statistical analysis
All data analysis was performed using GraphPad Prism version 6.0c for Mac (GraphPad Software, CA).
Results
PTEN-deficient ccRCC cells are hypersensitive to mTORC1 inhibition
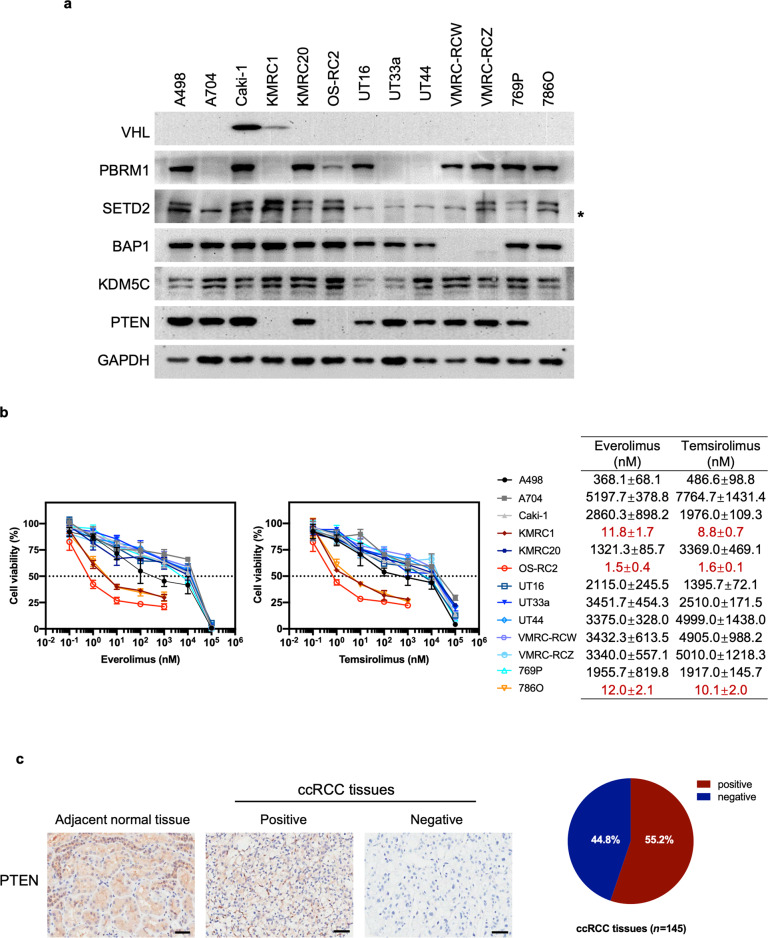
Genetically, ccRCC is characterized by early loss of the von Hippel-Lindau tumor suppressor gene (VHL) followed by alterations in chromatin remodeling genes and PI3K/AKT/mTOR pathway genes, as depicted in Supplementary Fig. S1. We checked the mutation status of VHL, PBRM1, SETD2, BAP1, KDM5C, MTOR, PTEN, PIK3CA, TSC1, and TSC2 in a panel of 13 cell lines established from ccRCCs. Whole-exome sequencing was performed in the UT16, UT33a, and UT44 cell lines, and whole-genome sequencing was applied to the VMRC-RCZ cell line. Whole-exome sequencing data of the remaining nine ccRCC cell lines were obtained from the Cell Model Passports website (https://cellmodelpassports.sanger.ac.uk/) [16]. Twelve of the 13 cell lines showed genetic alterations in VHL, while only Caki-1 cells did not carry VHL mutations. Six cell lines (A704, KMRC1, KMRC20, OS-RC2, UT33a, and UT44) harbored PBRM1 mutations, and six cell lines (A498, A704, UT16, UT33a, UT44, and VMRC-RCW) had SETD2 mutations. BAP1 mutations were present in four cell lines (KMRC20, VMRC-RCW, VMRC-RCZ, and 769P). None of the 13 cells lines carried KDM5C mutations. Six cell lines showed PTEN (KMRC1, OS-RC2, and 786O), MTOR (KMRC20), TSC1 (UT33a), and TSC2 (769P) mutations in a mutually exclusive manner. PIK3CA mutations were not detected in any of the cell lines (Supplementary Table S1).
We next evaluated the protein expression of VHL, PBRM1, SETD2, BAP1, KDM5C, and PTEN by Western blotting. Loss of VHL protein expression was observed in the majority of the cell lines. A704, KMRC1, UT33a, and UT44 cells did not express PBRM1 protein. SETD2 protein expression was absent in A704, UT16, UT33a, UT44, and VMRC-RCW cells. VMRC-RCW and VMRC-RCZ cells did not express BAP1 protein. Lack of PTEN protein expression was observed in KMRC1, OS-RC2, and 786O cells. KDM5C protein expression was present in all cell lines tested (Fig. 1a). In addition to genetic mutations (e.g., missense, nonsense, insertion or deletion), other genetic alterations (e.g., loss of heterogeneity) and epigenetic alterations (e.g., methylation and noncoding RNA) have also been reported for the tested genes in ccRCC [3]. According to copy number variation analysis, loss of heterogeneity could contribute to the absence of VHL, PBRM1, SETD2, and BAP1 protein expression (Supplementary Table S1). In addition, VHL loss could also be attributed to promoter hypermethylation (Supplementary Fig. S2). In contrast, the lack of PTEN expression is unlikely due to loss of heterogeneity or promoter methylation, suggesting that other epigenetic changes might be involved in addition to genetic mutations (Supplementary Table S1 and Supplementary Fig. S2). Collectively, these findings indicate that genetic and epigenetic changes led to the lack of specific protein expression in individual cell lines.
Fig. 1. PTEN-deficient ccRCC cell lines are more sensitive to everolimus and temsirolimus than PTEN-proficient ccRCC cell lines.
a Immunoblots showing relevant protein expression in 13 ccRCC cell lines. * indicates nonspecific bands. b Viability assay in ccRCC cell lines after treatment with everolimus or temsirolimus at the indicated concentrations for 6 days. The data are presented as the mean ± SD (n = 3) of one representative experiment out of three (left). The EC50 values for each cell line are presented as the mean ± SD (n = 3) of three separate experiments (right). c Representative immunohistochemical (IHC) images of PTEN expression in paraffin-embedded tissues from patients with ccRCC and adjacent normal tissues are shown. Scale bar: 50 μm (left). The percentage of PTEN-positive and PTEN-negative specimens among 145 ccRCC samples (right).
We then tested the sensitivity of the 13 ccRCC cell lines to the rapalogs everolimus and temsirolimus in vitro. PTEN-deficient ccRCC cell lines demonstrated dose-dependent sensitivity to everolimus with a half-maximal effective concentration (EC50) of 1–20 nM. Similar results were obtained for temsirolimus. However, PTEN-proficient ccRCC cell lines were largely insensitive to rapalogs, with an EC50 > 300 nM for everolimus and EC50 > 400 nM for temsirolimus in A498 cells and an EC50 > 1000 nM for both rapalogs in the other cell lines (Fig. 1b). These findings suggest an association of PTEN loss with cellular sensitivity to rapalogs.
We conducted immunohistochemical (IHC) analysis of ccRCC specimens. Positive PTEN staining in the adjacent normal tissues was detected. In contrast, negative PTEN staining was observed in 44.8% (65 of 145) of the tumors (Fig. 1c). This percentage is much higher than that expected for PTEN mutations (4% in the TCGA PanCancer ccRCC cohort) (Supplementary Fig. S1), implying that additional regulatory mechanisms contribute to the loss of PTEN protein expression in ccRCC patients.
PTEN deletion confers sensitivity to rapalogs
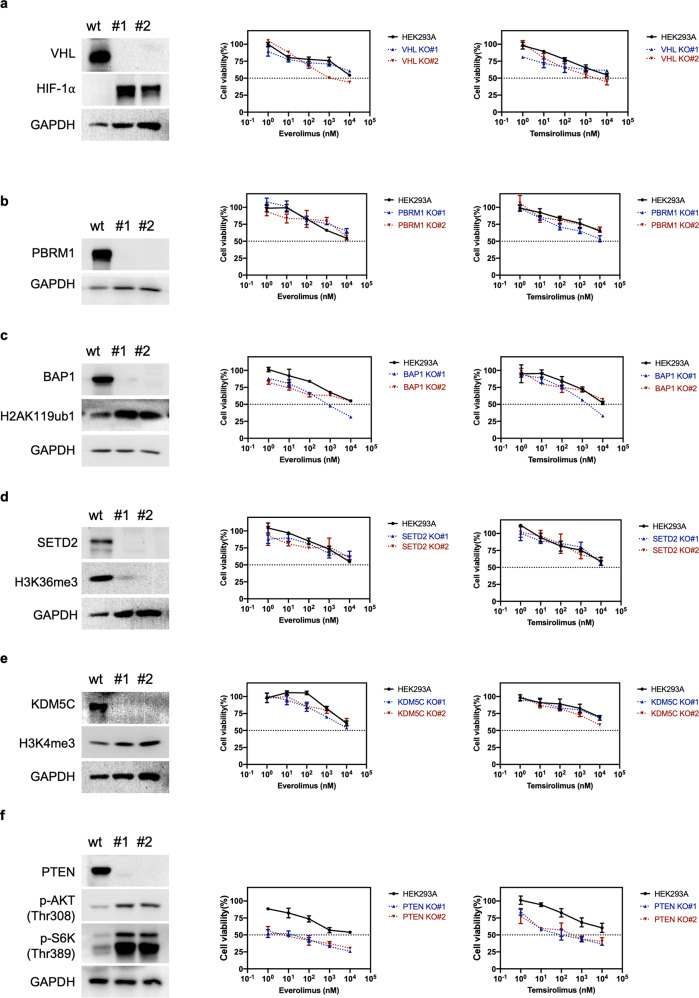
Most reported somatic mutations in VHL, PBRM1, BAP1, SETD2, KDM5C, and PTEN in ccRCC result in loss of function [3]; thus, we chose to delete each gene for phenotypic analysis. To comprehend the role of each individual gene in cellular sensitivity to rapalogs, we performed knockout (KO) of VHL, PBRM1, BAP1, SETD2, KDM5C, and PTEN in human HEK293A cells, a model cell line that can be efficiently transfected and is thus particularly amenable to genome editing. The isogenic mutant clones were generated by CRISPR/Cas9 and validated by Sanger sequencing and Western blotting. Two different clones generated by two independent CRISPR guide sequences were used for this study. Sanger sequencing confirmed the frameshift insertions and deletions (indels) within the targeted genes in both alleles in all selected KO clones except VHL KO#2, which showed an out-of-frame deletion (8 bp) of one allele and a large in-frame insertion of another allele, which resulted in the deletion of Phe91 and insertion of 32 amino acids (p.Phe91delins (32)) (Supplementary Table S2). The insertion was located in the beta domain, which is required for prolyl-hydroxylated HIF-α recognition [17]. VHL is the substrate recognition subunit of an E3 ligase known to target the α subunits of the heterodimeric transcription factor HIF for ubiquitin-mediated degradation under normoxic conditions [18]. VHL KO clones were verified by the lack of VHL protein expression and elevated HIF-1α level (Fig. 2a). Interestingly, the VHL protein was also undetectable in the partial VHL deletion clone (KO#2), suggesting that the insertion might lead to protein misfolding and degradation. VHL deletion failed to influence cellular sensitivity to everolimus and temsirolimus (Fig. 2a). PBRM1 encodes BRG1-associated factor (BAF)180, the defining subunit of the polybromo BAF (BRAF) SWI/SNF complex. SWI/SNF chromatin remodeling complexes utilize ATP to mobilize nucleosomes and thereby modulate chromatin structure [19]. PBRM1 deletion did not affect cellular sensitivity to rapalogs (Fig. 2b). BAP1 deletion was verified by the absence of BAP1 expression and increase in monoubiquitination of histone H2A at lysine 119 (H2AK119ub1), which is a physiological substrate of BAP1 [20]. The BAP1 KO clones did not exhibit altered rapalog sensitivity (Fig. 2c). SETD2 is the enzyme that trimethylates lysine 36 on histone H3 (H3K36me3), while KDM5C removes di- and trimethyl groups from lysine 4 on histone H3 (H3K4) [1, 3]. The SETD2 and KDM5C KO clones were functionally validated by the decreased H3K36me3 and increased H3K4me3 levels, respectively (Fig. 2d and e). Neither SETD2 deletion nor KDM5C deletion changed cellular sensitivity to rapalogs (Fig. 2d and e). As expected, PTEN deletion increased the levels of p-AKT (Thr308) (an effector downstream of PTEN) and p-S6K (Thr389) (one mTORC1 substrate). Interestingly, PTEN deletion conferred sensitivity to rapalogs (Fig. 2f). In summary, PTEN loss made HEK293A cells more sensitive to rapalogs, whereas loss of VHL, PBRM1, BAP1, SETD2, or KDM5C exerted minimal or no effects on cellular sensitivity to rapalogs.
Fig. 2. PTEN deletion renders HEK293A cells more sensitive to rapalogs.
VHL (a) PBRM1 (b), BAP1 (c), SETD2 (d), KDM5C (e), and PTEN (f) knockout clones were validated by Western blotting. Cells were treated with temsirolimus or everolimus at the indicated concentrations for 6 days, and an MTT assay was used to determine cell viability. The data are presented as the mean ± SD (n = 3) of one representative experiment out of three.
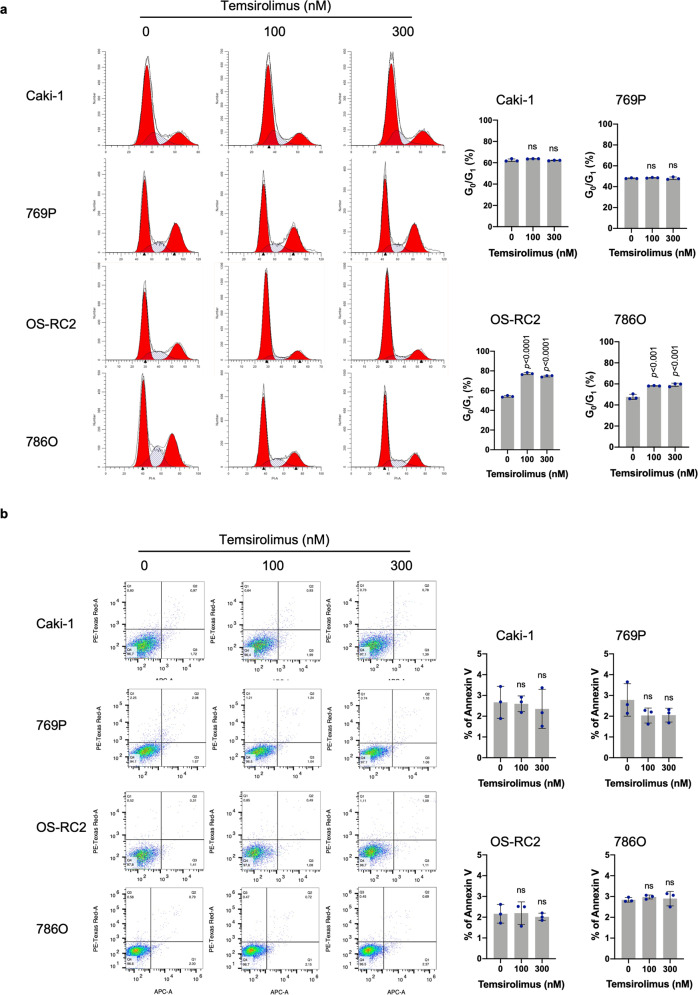
Temsirolimus induces G0/G1-phase cell cycle arrest in PTEN-deficient but not PTEN-proficient ccRCC cells
Temsirolimus significantly increased the percentage of PTEN-deficient OS-RC2 and 786O cells in G0/G1 phase. In contrast, temsirolimus did not alter the percentage of PTEN-proficient Caki-1 and 769P cells in G0/G1 phase (Fig. 3a). Fluorescence-activated cell sorting (FACS) analysis of Annexin V- and PI-stained cells after temsirolimus treatment did not indicate significant changes in apoptosis levels in PTEN-proficient Caki-1 and 769P cells or in PTEN-deficient OS-RC2 and 786O cells (Fig. 3b). These data suggest a cytostatic effect rather than a proapoptotic effect of temsirolimus on PTEN-deficient ccRCC cells.
Fig. 3. Temsirolimus induces G0/G1-phase cell cycle arrest in PTEN-deficient ccRCC cells.
a Cells were treated with temsirolimus at the indicated concentrations for 48 h, and their DNA content was analyzed by flow cytometry. The data are the mean ± SD (n = 3) of one representative experiment out of three. Significance was assessed using ANOVA followed by Tukey’s test. b Cells were treated with temsirolimus at the indicated concentrations for 6 days. Cellular apoptosis was assessed by phosphatidylserine externalization and binding of Annexin V-APC. The data are presented as the mean ± SD (n = 3) of one representative experiment out of three. Significance was assessed using ANOVA followed by Tukey’s test.
PTEN modulates cellular sensitivity to rapalogs by regulating the AKT/mTOR pathway
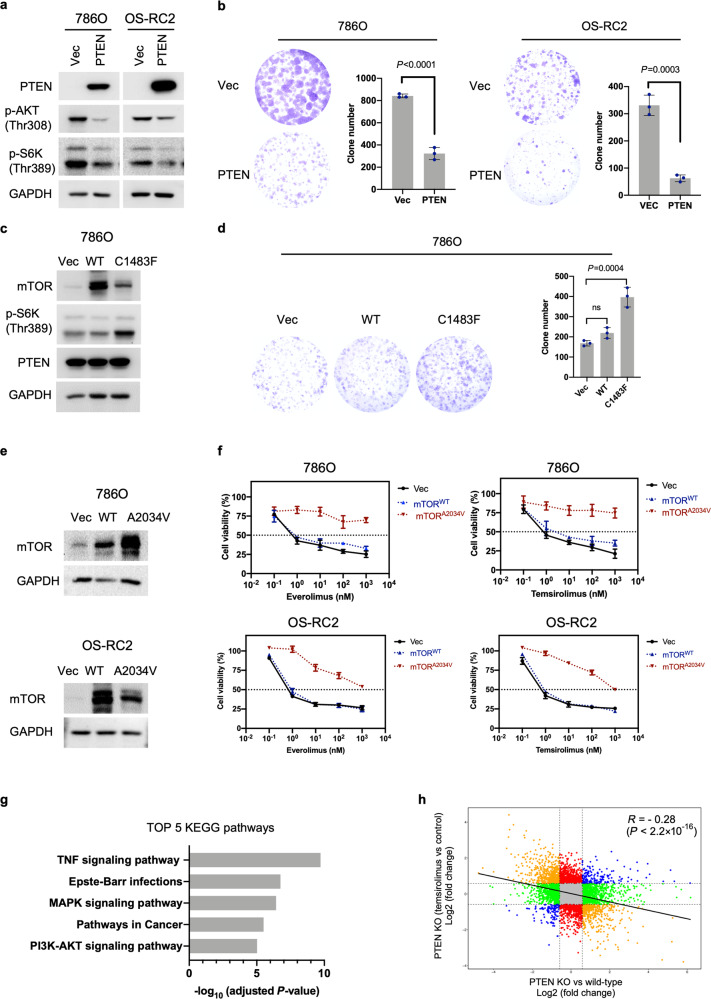
PTEN exerts much of its activity as a lipid phosphatase antagonizing the prosurvival and oncogenic PI3K/AKT/mTOR pathway; it also performs PI3K/AKT/mTOR-independent and protein phosphatase-dependent activities and functions within the nucleus [21]. Previous studies have demonstrated that cancer cell lines with hyperactivating MTOR mutations display heightened sensitivity to rapamycin both in culture and in in vivo xenografts, suggesting that such mutations confer mTOR pathway dependency [22]. Thus, we speculate that PTEN’s role in the PI3K/AKT/mTOR pathway is pivotal for conferring hypersensitivity to rapalogs. To test this hypothesis, we first determined whether PTEN regulates mTORC1 activity via AKT. HEK293A PTEN KO cells exhibited higher p-AKT (Thr308) and p-S6K (Thr389) levels than wild-type cells. Notably, inhibition of AKT with an allosteric inhibitor (miransertib) prevented AKT and S6K phosphorylation in both wild-type and PTEN KO HEK293A cells, indicating that PTEN loss activates mTORC1 via AKT (Supplementary Fig. S3). Next, we determined whether mTOR could counter PTEN’s effect on cell proliferation. Ectopic expression of PTEN in PTEN-deficient 786O or OS-RC2 cells decreased p-AKT (Thr308) and p-S6K (Thr389) levels and significantly reduced the colony-forming capacity, consistent with its role as a tumor suppressor (Fig. 4a and b). We established 786O cells stably expressing empty vector, wild-type (wt) mTOR, or mutant mTOR (C1483F) by lentiviral transduction. The mTORC1 activating mutation (C1483F) located in the FAT domain of mTOR reduces the mTOR-Deptor interaction, leading to increased mTOR pathway activity [22, 23]. Of note, mTOR mutations in the C1483 cluster are also particularly prevalent in kidney cancer [22, 24]. We next introduced PTEN into the aforementioned stable cells via retroviral vectors. Notably, ectopic expression of mutant mTOR (C1483F) but not wt mTOR elevated the p-S6K (Thr389) level (Fig. 4c). Concordantly, mutant mTOR (C1483F) significantly reversed the inhibitory effect of PTEN on colony formation. The same trend was observed for wild-type mTOR, although the difference was not statistically significant (Fig. 4d). These data suggest that PTEN suppresses cell proliferation primarily by modulating mTOR pathway activity.
Fig. 4. PTEN affects cell proliferation and rapalog sensitivity through the mTOR pathway.
a 786O and OS-RC2 cells were stably transduced with empty vector (pQCXIH-vec) or PTEN (pQCXIH-PTEN) retroviruses. PTEN, p-AKT (Thr308) and p-S6K (Thr389) protein levels were determined by immunoblot analysis. b Images and corresponding quantification of colony formation by 786O and OS-RC2 cells stably expressing vec or PTEN. The data are presented as the mean ± SD (n = 3) of one representative experiment out of three. Significance was assessed using Student’s t test. c 786O cells were stably transduced with empty vector (pLVX-vec), wt (pLVX-mTORwt) or mutant (pLVX-mTORC1483F) lentiviruses. The established stable cells were next infected with retroviruses encoding PTEN (pQCXIH-PTEN). The immunoblots show the expression of the indicated proteins. d Images and corresponding quantification of colony formation by the stable cells generated in (c). The data are presented as the mean ± SD (n = 3) of one representative experiment out of three. Significance was assessed using ANOVA followed by Tukey’s test. e 786O and OS-RC2 cells were stably transduced with empty vector (pLVX-vec), wt (pLVX-mTORwt) or mutant (pLVX mTORA2034V) lentiviruses. The immunoblots show the expression of mTOR. f Viability assay of the stable cells generated in (e) after treatment with everolimus or temsirolimus at the indicated concentrations for 6 days. The data are presented as the mean ± SD (n = 3) of one representative experiment out of three. g KEGG pathway enrichment analysis of the DEGs identified between PTEN KO#2 and parental HEK293A cells. The bars indicate the enrichment scores, −log10 (adjusted P values). h Comparison of the transcriptional changes induced by PTEN loss versus temsirolimus treatment (1000 nM, 24 h). The blue dots represent genes that were either upregulated or downregulated in both comparisons; the orange dots represent genes that were significantly upregulated in one comparison and downregulated in the other comparison. The green dots represent genes significantly changed only upon PTEN deletion in HEK293A cells, while the red dots represent genes significantly changed upon temsirolimus treatment in HEK293A PTEN KO#2 cells. The R value was calculated for the correlation; Pearson’s correlation coefficient.
To determine whether the mTOR pathway is directly responsible for the rapalog hypersensitivity phenotype observed in PTEN-deficient ccRCC cells, we introduced empty vector, wt, or mutant mTOR (A2034V) into 786O and OS-RC2 cells and then measured the efficacy of rapalogs (Fig. 4e). The A2034V mutation is located in the FRB (FKBP12-rapamycin binding) domain and confers rapamycin resistance by disrupting the interaction of mTOR with the FKBP12-rapamycin complex. In cells carrying the A2034V mutant, rapalogs failed to affect the phosphorylation of the normally rapamycin-sensitive sites in S6K (Thr389) and S6 (Ser240/244 and Ser235/236) [25]. Similarly, our immunoprecipitation (IP) assay showed that FKBP12-rapamycin interacted with wt mTOR but not with A2034 mTOR (Supplementary Fig. S4a). Moreover, treatment with either rapamycin or rapalogs abolished S6K (Thr389) phosphorylation in 786O and OS-RC2 cells expressing empty vector or wt mTOR, whereas ectopic expression of mutant mTOR (A2034V) substantially attenuated the inhibitory effects of rapamycin and rapalogs on S6K (Thr389) phosphorylation (Supplementary Fig. S4b). Concordantly, ectopic expression of wt mTOR did not affect the cellular response to everolimus and temsirolimus, whereas ectopic expression of the A2034V mutant reversed rapalog hypersensitivity in 786O and OS-RC2 cells (Fig. 4f). These findings suggest that PTEN mutation confers rapalog sensitivity due to elevated mTOR pathway activity.
We performed RNA-seq to analyze the transcriptomes of parental HEK293A and PTEN KO#2 cells. Using the resulting mRNA expression profiles, we identified 803 differentially expressed genes (DEGs; see “Materials and methods”) between PTEN KO#2 and wild-type cells (Supplementary Table S3). To understand cellular processes that were altered by PTEN deletion, we performed Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis for the identified DEGs. KEGG analysis revealed enrichment in a variety of biological processes ranging from broad terms such as pathways in cancer to more specific terms such as TNF signaling pathway, Epstein–Barr virus infection, MAPK signaling pathway and PI3K-AKT signaling pathway (Fig. 4g and Supplementary Table S4). Indeed, multiple levels of crosstalk between the PI3K/AKT and MAPK signaling pathways have been demonstrated [26]. Moreover, the PI3K/AKT pathway has been reported to be involved in the TNF signaling [27] and Epstein–Barr virus infection pathways [28]. These findings confirmed the role of PTEN in regulating the PI3K/AKT pathway. We next assessed transcriptional changes in HEK293A PTEN KO#2 cells in response to temsirolimus treatment (1000 nM, 24 h). We identified 593 DEGs between temsirolimus-treated and vehicle-treated HEK293A PTEN KO#2 cells (Supplementary Table S3). KEGG analysis showed that the identified DEGs are mainly involved in processes related to protein synthesis (Supplementary Fig. S5 and Supplementary Table S4), consistent with the reported function of mTORC1 [29]. Interestingly, we found a negative correlation between PTEN loss-induced transcriptional changes and temsirolimus-induced transcriptional changes, yielding a Pearson correlation coefficient of −0.28 (P < 2.2 × 10−16) (Fig. 4h), which is reminiscent of the opposite roles of PTEN loss and rapalogs in regulating mTORC1 activity.
PTEN-deficient ccRCC cells are more vulnerable to the antimigratory effect of temsirolimus than PTEN-proficient ccRCC cells
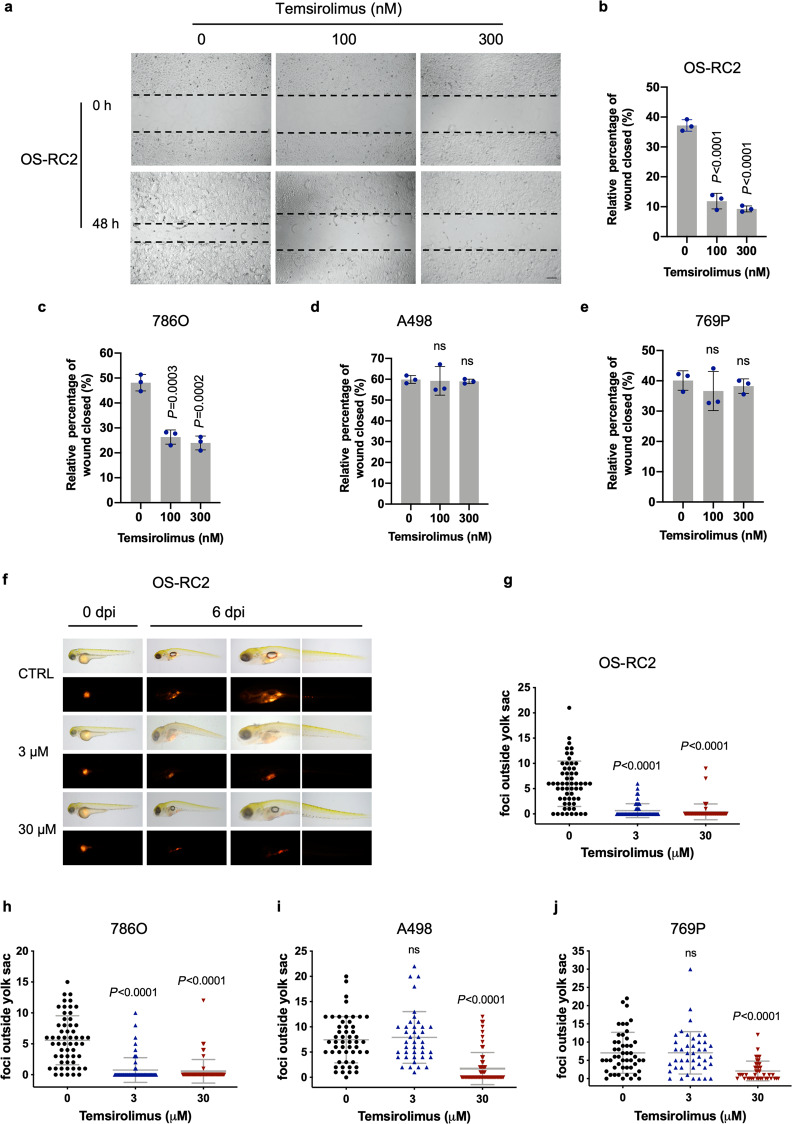
Inhibition of mTOR has been reported to regulate cell migration [30–32]. We explored whether rapalogs affect cell migration through wound healing assays. The wound healing assays showed that temsirolimus significantly inhibited the migration of PTEN-deficient OS-RC2 (Fig. 5a and b) and 786O (Fig. 5c, Supplementary Fig. S6) cells, while this compound had no significant effects (or only a weak inhibitory effect) on the migration of PTEN-proficient A498 (Fig. 5d, Supplementary Fig. S6) and 769P (Fig. 5e, Supplementary Fig. S6) cells.
Fig. 5. Temsirolimus exhibits a more potent inhibitory effect on migration in PTEN-deficient ccRCCs than in PTEN-proficient ccRCC cells.
a Wound healing assays were performed to examine the effects of temsirolimus on the migration of OS-RC2 cells. Representative pictures of OS-RC2 cells at 0 h (time of wounding) and after 48 h of exposure to the indicated concentrations of temsirolimus. Scale bar, 200 μm. Quantification of wound healing, shown as the wound closure percentage in OS-RC2 (b), 786O (c), A498 (d), and 769P (e) cells. The data are presented as the mean ± SD (n = 3, five independent fields for each sample) of one representative experiment out of three. Significance was assessed using ANOVA followed by Tukey’s test. f Representative images of zebrafish embryos engrafted with OS-RC2 cells at 0 and 6 days post injection (dpi). Zebrafish embryos were treated with temsirolimus at the indicated concentrations. Quantification of the dissemination of OS-RC2 (g), 786O (h), A498 (i), and 769P (j) cells. Each dot represents the number of foci outside the yolk sac in a single embryo. The mean ± SD of three independent experiments is shown. Significance was assessed using ANOVA followed by Tukey’s test.
We next determined whether PTEN deletion in ccRCC cell lines can affect cellular sensitivity to rapalogs. Unlike the HEK293A cell line, which is an easily transfected cell model, we found that it was challenging to achieve a high transfection efficiency in the ccRCC cell lines used in our study. Therefore, we chose to use a lentiviral CRISPR/Cas9 system to delete PTEN. Among the multiple tested ccRCC cell lines, high knockout efficiency was achieved in two cell lines (KMRC20 and 769P). The PTEN knockout efficiency of the tested single guide RNAs (g1 and g2) compared to that of empty vector (vec) in the polyclonal populations of KMRC20 and 769P cells following puromycin selection was validated by Western blotting (Supplementary Fig. S7a). Similar to the above results, PTEN deletion made cells more sensitive to the temsirolimus-induced effects on cell growth inhibition, G0/G1-phase cell cycle arrest and migration suppression (Supplementary Fig. S7b–d). These data suggest that PTEN loss occurring in patient tumors or generated by gene editing renders cells more sensitive to rapalogs.
The transparent and immunoprivileged nature of zebrafish embryos allows direct visualization of the migration, metastatic intravasation, and extravasation of single human cancer cells in vivo. Cell migration was observed on Day 6 after cell injection in 2-day-old zebrafish embryos that were exposed to 0, 3, and 30 μM temsirolimus. Dissemination of tumor cells from the original injection site (yolk sac) to distal regions (head and trunk) was measured. Compared to the vehicle control, temsirolimus significantly suppressed OS-RC2 (Fig. 5f and g) and 786O (Fig. 5h and Supplementary Fig. S8a) cell dissemination away from the yolk sac at concentrations of 3 and 30 μM. In contrast, temsirolimus significantly decreased A498 (Fig. 5i and Supplementary Fig. S8b) and 769P (Fig. 5j and Supplementary Fig. S8c) cell migration at a high concentration (30 μM) but showed no effect at a low concentration (3 μM). These findings suggest that PTEN-deficient ccRCC cells are more vulnerable to the antimigratory effect of temsirolimus than PTEN-proficient ccRCC cells.
PTEN-deficient xenografts are more sensitive to temsirolimus than PTEN-proficient xenografts
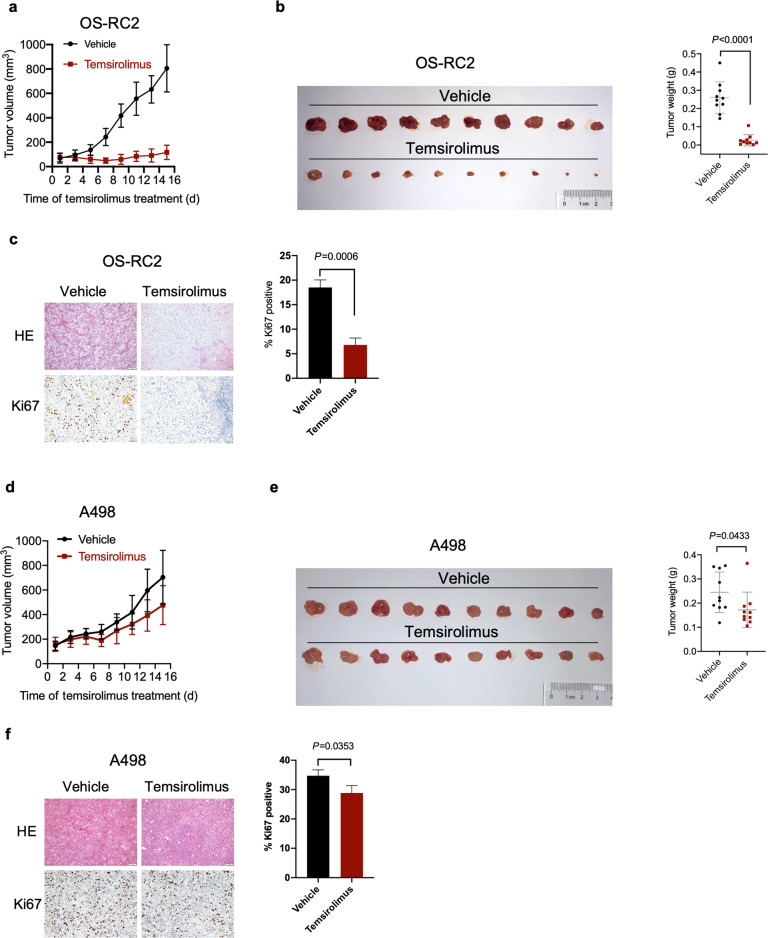
To assess the effects of temsirolimus on the growth of PTEN-deficient and PTEN-proficient tumors, we performed xenograft experiments in nude mice. Temsirolimus robustly reduced tumor growth in nude mice xenografted with PTEN-deficient OS-RC2 cells (Fig. 6a). Concordantly, temsirolimus significantly decreased the tumor weight (Fig. 6b) and percentage of cells positive for Ki67 (a proliferation marker) (Fig. 6c). In contrast, PTEN-proficient tumors were less responsive to temsirolimus. Temsirolimus marginally reduced the growth of PTEN-proficient A498 xenografts (Fig. 6d). Moreover, the inhibitory effects of temsirolimus on tumor weight (Fig. 6e) and the percentage of Ki67-positive cells (Fig. 6f) were modest. These data indicate that PTEN-deficient ccRCC tumors are more sensitive to temsirolimus.
Fig. 6. Temsirolimus exerts more potent tumor growth inhibitory activity in PTEN-deficient tumors than in PTEN-proficient tumors.
a Size of tumors derived from OS-RC2 cells subcutaneously implanted into nude mice and treated with either vehicle or temsirolimus. The data are presented as the mean ± SD values (n = 10). b Tumors derived from OS-RC2 cells were harvested and weighed on day 15 post treatment. The data are presented as the mean ± SD values (n = 10). Significance was assessed using Mann–Whitney U test. c IHC staining for Ki67 in representative OS-RC2 tumor tissues. Scale bar, 100 μm. The data are presented as the mean ± SD of three tumors (ten independent fields for each tumor). Significance was assessed using Student’s t test. d Size of tumors derived from A498 cells subcutaneously implanted into nude mice and treated with either vehicle or temsirolimus. The data are presented as the mean ± SD values (n = 10). e Tumors derived from A498 cells were harvested and weighed on day 15 post treatment. The data are presented as the mean ± SD values (n = 10). Significance was assessed using Mann–Whitney U test. f IHC staining for Ki67 in representative A498 tumor tissues. Scale bar, 100 μm. The data are presented as the mean ± SD of three tumors (ten independent fields for each tumor). Significance was assessed using Student’s t test.
Discussion
In this study, we found that the mutation status of VHL, PBRM1, SETD2, BAP1, or KDM5C is unrelated to the rapalog response in a panel of ccRCC cell lines. In sharp contrast, we established a causal link between PTEN loss and rapalog hypersensitivity. A mechanistic study showed that PTEN loss promotes rapalog sensitivity primarily by activating the mTOR pathway, suggesting that PTEN deficiency confers mTOR dependency.
Despite multiple attempts to investigate the potential association between mTOR pathway mutations and the rapalog response in ccRCC, the link remains unclear, and contrasting results have been reported in the literature. Some studies in ccRCC cases reported that mutations in MTOR, TSC1, and TSC2 correlated with the rapalog response [9, 12]. However, other studies failed to validate this association in ccRCC patients [11, 13]. It has been demonstrated that some ccRCC-derived mTOR missense mutations are activating mutations, while some are passenger mutations [24]. Thus, the heterogeneous functional effects of mTOR missense mutations on mTORC1 activity may contribute to the discrepancy between these studies. Remarkably, although studies considering only PTEN mutations found no association with the rapalog response in ccRCC [10, 11, 13], loss of PTEN expression (detected by immunohistochemistry) has been proposed as a marker of the therapeutic outcome of rapalog treatment in patients with ccRCC [11, 12]. The latter two referenced studies showed that negative PTEN IHC staining was detected in 50% [11] and 58% [12] of tumors. Our study showed a similar proportion of negative PTEN immunohistochemistry in ccRCC in a Chinese population (44.8%, 65 of 145). Of note, the percentage of tumors with PTEN loss as assessed by IHC staining was much higher than the PTEN mutation (point mutations, insertions, and deletions) frequency in ccRCC (4%) (Supplementary Fig. S1), implying the importance of alternative mechanisms of PTEN inactivation in ccRCC. We analyzed the copy number data of PTEN from the TCGA ccRCC dataset, which showed that a substantial fraction of samples (62 of 352) exhibited copy number loss (shallow or deep deletion) (Supplementary Fig. S9a). In addition to genetic alterations that partially or fully inactivate a given PTEN allele, accumulating evidence shows that PTEN expression is also suppressed via epigenetic mechanisms [21]. Notably, analysis of the TCGA ccRCC dataset revealed that PTEN pseudogene 1 (PTENP1) mRNA expression was strongly correlated with PTEN mRNA expression by Spearman correlation analysis (r = 0.88) (Supplementary Fig. S9b). PTEN and PTENP1 share extensive sequence homology, and thus PTENP1 mRNA can function as a decoy for PTEN-targeting microRNAs (miRNAs), counteracting miRNA-mediated repression and consequently boosting PTEN expression [21]. Thus, the competitive endogenous RNA (ceRNA) mechanism could be an important regulator of PTEN expression in ccRCC. Owing to a multilayered mode of PTEN regulation, we speculate that PTEN immunohistochemistry may constitute a straightforward and reliable way to screen for ccRCC patients with PTEN deficiency who are likely to benefit from rapalog therapy.
Inactivation of VHL occurs in essentially all ccRCCs via VHL mutations or epigenetic silencing owing to promoter methylation [3]. Previous studies have shown that the response to rapalog treatment is not dependent on the presence or absence of VHL alterations [9, 10]. In line with these findings, our data show that VHL mutation status is unrelated to rapalog efficacy in ccRCC. In contrast, multiple studies have reported conflicting data regarding the associations between mutations in chromatin remodeling genes (e.g., PBRM1, SETD2, BAP1, and KDM5C) and the response to rapalog therapy in ccRCC. Some studies failed to identify a relationship between mutations in these genes [9, 12] and rapalog response. However, the RECORD-3 phase III clinical trial showed that PBRM1 and BAP1 mutations were associated with longer and shorter PFS (progression-free survival) times, respectively, for ccRCC patients treated with everolimus [10]. Our study shows that rapalogs display similar cytostatic effects on ccRCC cell lines with or without mutations in PBRM1, SETD2, or BAP1. Furthermore, deletion of PBRM1, SETD2, BAP1, or KDM5C did not affect the cellular response to rapalogs compared to that of isogenic parental wild-type cells. Indeed, our findings are in line with the observation that PBRM1 downregulation by shRNA had no effect on growth inhibition by rapamycin in ccRCC cell lines [33]. Thus, our study suggests that the observed favorable or unfavorable clinical outcomes may not derive from the direct cytostatic effect of rapalogs on tumor cells carrying mutations in chromatin remodeling genes. The potential link between mutations in chromatin remodeling genes and the rapalog response and the underlying mechanism warrant further investigation.
Previous studies have shown that in vitro exposure to rapalogs usually arrests cell proliferation without inducing apoptosis [4]. Several mechanisms have been proposed to explain this phenotype. For example, rapalogs block the S6K1-IRS1 negative feedback loop, leading to the activation of PI3K/AKT prosurvival signals [30] or stimulate eIF4E phosphorylation via Mnk-dependent mechanisms, leading to apoptosis resistance [31]. Largely in line with these studies, rapalogs failed to induce apoptosis in PTEN-deficient ccRCC cells at concentrations that induced significant G0/G1 arrest in our study. It has been reported that inhibition of mTOR attenuates cell migration through diverse mechanisms, including epithelial-mesenchymal transition (EMT) inhibition [32], decreases in oncometabolite levels [34], or autophagy activation [35]. We also observed antimigratory effects of rapalogs in ccRCC, and this effect was more pronounced in PTEN-deficient cells than in PTEN wild-type cells. Although PTEN immunohistochemistry showed the presence of PTEN in the cytoplasm, membrane and nucleus in ccRCC samples with positive staining, our data support that PTEN’s role in mTOR regulation contributes to the observed hypersensitivity to rapalogs. In this regard, ectopic expression of an activating mTOR mutant (C1483F) attenuated PTEN-induced cell growth inhibition, while the introduction of a resistant mTOR mutant (A2034V) enabled PTEN-deficient ccRCC cells to escape the growth inhibitory effect of rapalogs. Of note, temsirolimus appeared to oppose PTEN loss-induced transcriptional changes. We speculate that the opposite roles of PTEN loss and rapalog treatment in regulating mTORC1 activity might contribute to this transcriptional regulation pattern. Interestingly, mTOR mutations and negative PTEN immunohistochemistry are not mutually exclusive events in ccRCC patients [12]. The co-occurrence of mTOR-activating mutations and loss of PTEN expression may lead to additive effects on the rapalog response.
Here, we demonstrate that PTEN loss rather than alterations in VHL or chromatin-modifying genes (PBRM1, SETD2, BAP1, and KDM5C) confers hypersensitivity to rapalogs in ccRCC, suggesting that patients with PTEN deficiency are more likely to benefit from rapalog therapy than patients with functional PTEN. A mechanistic study showed that PTEN loss creates vulnerabilities related to mTOR pathway dependency. Future studies could apply more sophisticated models of ccRCC, including patient-derived xenografts or organoids, to validate the observed correlation. Rapalogs are approved for clinical use in tumor types in addition to ccRCC, such as pancreatic neuroendocrine tumors and advanced breast cancer [4, 36]. In addition, multiple clinical trials have been conducted to evaluate the efficacy of rapalogs in treating a broad range of human cancers (clinicaltrials.gov). Given that PTEN is one of the most frequently somatically mutated genes in cancer [37], it would thus be of clinical significance to explore whether PTEN loss similarly heightens sensitivity to rapalogs in other cancer types. Elucidation of this issue would help patient select and optimize the clinical outcomes of rapalogs.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81973357), Guangdong Basic and Applied Basic Research Foundation (2020A1515010027), Guangzhou Basic and Applied Basic Research Foundation (4001616289), Wu Jieping Medical Foundation (3206750.2020-10-120), Bethune Charitable Foundation Research Programme (B-19-H-20200622) and HUI LAN Public Welfare (2021-HLJJ0328). We thank Dr. Kun-Liang Guan (University of California, San Diego) for critically discussing and revising the manuscript.
Author contributions
XLL and GMZ substantially contributed to designing the study, performing experiments, and analyzing the data. WHS, LXY, and SSH performed experiments and analyzed the data. ZLR analyzed the data. JJZ and SWL provided experimental resources. LY and YLL conceived and designed the study and experiments, assembled and interpreted the data, wrote and revised the manuscript, and approved the manuscript. All authors read and approved the final manuscript.
Data availability
The RNA-seq data have been deposited to the National Center for Biotechnology Information (NCBI) via the Sequence Read Archive (SRA) database repository under accession code PRJNA777372. All other relevant data supporting the findings of this study are available upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Xiao-lian Liu, Gui-ming Zhang
Contributor Information
Le Yu, Email: yulezy@smu.edu.cn.
Yi-lei Li, Email: lei@smu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41401-022-00862-1.
References
- 1.Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L, Schmidinger M, et al. Renal cell carcinoma. Nat Rev Dis Prim. 2017;3:17009. doi: 10.1038/nrdp.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiao GF, Yan X, Chen Z, Zhang RJ, Liu TZ, Hu WL. Identification of a novel immune-related prognostic biomarker and small-molecule drugs in clear cell renal cell carcinoma (ccRCC) by a merged microarray-acquired dataset and TCGA database. Front Genet. 2020;11:810. doi: 10.3389/fgene.2020.00810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dizman N, Philip EJ, Pal SK. Genomic profiling in renal cell carcinoma. Nat Rev Nephrol. 2020;16:435–51. doi: 10.1038/s41581-020-0301-x. [DOI] [PubMed] [Google Scholar]
- 4.Hua H, Kong Q, Zhang H, Wang J, Luo T, Jiang Y. Targeting mTOR for cancer therapy. J Hematol Oncol. 2019;12:71. doi: 10.1186/s13045-019-0754-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hudes G, Carducci M, Tomczak P, Dutcher J, Figlin R, Kapoor A, et al. Temsirolimus, interferon alfa, or both for advanced renal-cell carcinoma. N Engl J Med. 2007;356:2271–81. doi: 10.1056/NEJMoa066838. [DOI] [PubMed] [Google Scholar]
- 6.Motzer RJ, Escudier B, Oudard S, Hutson TE, Porta C, Bracarda S, et al. Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet. 2008;372:449–56. doi: 10.1016/S0140-6736(08)61039-9. [DOI] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013; 499: 43-9. [DOI] [PMC free article] [PubMed]
- 8.Ho TH, Choueiri TK, Wang K, Karam JA, Chalmers Z, Frampton G, et al. Correlation between molecular subclassifications of clear cell renal cell carcinoma and targeted therapy response. Eur Urol Focus. 2016;2:204–9. doi: 10.1016/j.euf.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Kwiatkowski DJ, Choueiri TK, Fay AP, Rini BI, Thorner AR, de Velasco G, et al. Mutations in TSC1, TSC2, and MTOR Are associated with response to rapalogs in patients with metastatic renal cell carcinoma. Clin Cancer Res. 2016;22:2445–52. doi: 10.1158/1078-0432.CCR-15-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lei Y, Yildiz S, Chen M. Re: James J. Hsieh, David Chen, Patricia I. Wang, et al. Genomic biomarkers of a randomized trial comparing first-line everolimus and sunitinib in patients with metastatic renal cell carcinoma. Eur Urol. 2017;71:405–14. [DOI] [PMC free article] [PubMed]
- 11.Voss MH, Chen D, Reising A, Marker M, Shi J, Xu J, et al. PTEN Expression, not mutation status in TSC1, TSC2, or mTOR, correlates with the outcome on everolimus in patients with renal cell carcinoma treated on the randomized RECORD-3 trial. Clin Cancer Res. 2019;25:506–14. doi: 10.1158/1078-0432.CCR-18-1833. [DOI] [PubMed] [Google Scholar]
- 12.Roldan-Romero JM, Beuselinck B, Santos M, Rodriguez-Moreno JF, Lanillos J, Calsina B, et al. PTEN expression and mutations in TSC1, TSC2 and MTOR are associated with response to rapalogs in patients with renal cell carcinoma. Int J Cancer. 2020;146:1435–44. doi: 10.1002/ijc.32579. [DOI] [PubMed] [Google Scholar]
- 13.Nassar AH, Hamieh L, Gray KP, Thorner AR, Fay AP, Lasseter KD, et al. Mutations and response to rapalogs in patients with metastatic renal cell carcinoma. Mol Cancer Ther. 2020;19:690–6. doi: 10.1158/1535-7163.MCT-19-0642. [DOI] [PubMed] [Google Scholar]
- 14.Yu L, Zhou D, Zhang G, Ren Z, Luo X, Liu P, et al. Co-occurrence of BAP1 and SF3B1 mutations in uveal melanoma induces cellular senescence. Mol Oncol. 2021; 10.1002/1878-0261.13128. [DOI] [PMC free article] [PubMed]
- 15.Xu YY, Ren ZL, Liu XL, Zhang GM, Huang SS, Shi WH, et al. BAP1 loss augments sensitivity to BET inhibitors in cancer cells. Acta Pharmacol Sin. 2021; 10.1038/s41401-021-00783-5. [DOI] [PMC free article] [PubMed]
- 16.van der Meer D, Barthorpe S, Yang W, Lightfoot H, Hall C, Gilbert J, et al. Cell model passports-a hub for clinical, genetic and functional datasets of preclinical cancer models. Nucleic Acids Res. 2019;47:D923–d9. doi: 10.1093/nar/gky872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stebbins CE, Kaelin WG, Jr, Pavletich NP. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science. 1999;284:455–61. doi: 10.1126/science.284.5413.455. [DOI] [PubMed] [Google Scholar]
- 18.Maranchie JK, Vasselli JR, Riss J, Bonifacino JS, Linehan WM, Klausner RD. The contribution of VHL substrate binding and HIF1-alpha to the phenotype of VHL loss in renal cell carcinoma. Cancer Cell. 2002;1:247–55. doi: 10.1016/S1535-6108(02)00044-2. [DOI] [PubMed] [Google Scholar]
- 19.Nargund AM, Pham CG, Dong Y, Wang PI, Osmangeyoglu HU, Xie Y, et al. The SWI/SNF protein PBRM1 restrains VHL-loss-driven clear cell renal cell carcinoma. Cell Rep. 2017;18:2893–906. doi: 10.1016/j.celrep.2017.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campagne A, Lee MK, Zielinski D, Michaud A, Le Corre S, Dingli F, et al. BAP1 complex promotes transcription by opposing PRC1-mediated H2A ubiquitylation. Nat Commun. 2019;10:348. doi: 10.1038/s41467-018-08255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee YR, Chen M, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat Rev Mol Cell Biol. 2018;19:547–62. doi: 10.1038/s41580-018-0015-0. [DOI] [PubMed] [Google Scholar]
- 22.Grabiner BC, Nardi V, Birsoy K, Possemato R, Shen K, Sinha S, et al. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov. 2014;4:554–63. doi: 10.1158/2159-8290.CD-13-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jewell JL, Russell RC, Guan KL. Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol. 2013;14:133–9. doi: 10.1038/nrm3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu J, Pham CG, Albanese SK, Dong Y, Oyama T, Lee CH, et al. Mechanistically distinct cancer-associated mTOR activation clusters predict sensitivity to rapamycin. J Clin Invest. 2016;126:3526–40. doi: 10.1172/JCI86120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodrik-Outmezguine VS, Okaniwa M, Yao Z, Novotny CJ, McWhirter C, Banaji A, et al. Overcoming mTOR resistance mutations with a new-generation mTOR inhibitor. Nature. 2016;534:272–6. doi: 10.1038/nature17963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aksamitiene E, Kiyatkin A, Kholodenko BN. Cross-talk between mitogenic Ras/MAPK and survival PI3K/Akt pathways: a fine balance. Biochem Soc Trans. 2012;40:139–46. doi: 10.1042/BST20110609. [DOI] [PubMed] [Google Scholar]
- 27.Burow ME, Weldon CB, Melnik LI, Duong BN, Collins-Burow BM, Beckman BS, et al. PI3-K/AKT regulation of NF-kappaB signaling events in suppression of TNF-induced apoptosis. Biochem Biophys Res Commun. 2000;271:342–5. doi: 10.1006/bbrc.2000.2626. [DOI] [PubMed] [Google Scholar]
- 28.Dawson CW, Tramountanis G, Eliopoulos AG, Young LS. Epstein-Barr virus latent membrane protein 1 (LMP1) activates the phosphatidylinositol 3-kinase/Akt pathway to promote cell survival and induce actin filament remodeling. J Biol Chem. 2003;278:3694–704. doi: 10.1074/jbc.M209840200. [DOI] [PubMed] [Google Scholar]
- 29.Kim J, Guan KL. mTOR as a central hub of nutrient signalling and cell growth. Nat Cell Biol. 2019;21:63–71. doi: 10.1038/s41556-018-0205-1. [DOI] [PubMed] [Google Scholar]
- 30.O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–8. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun SY, Rosenberg LM, Wang X, Zhou Z, Yue P, Fu H, et al. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res. 2005;65:7052–8. doi: 10.1158/0008-5472.CAN-05-0917. [DOI] [PubMed] [Google Scholar]
- 32.Zong H, Yin B, Zhou H, Cai D, Ma B, Xiang Y. Inhibition of mTOR pathway attenuates migration and invasion of gallbladder cancer via EMT inhibition. Mol Biol Rep. 2014;41:4507–12. doi: 10.1007/s11033-014-3321-4. [DOI] [PubMed] [Google Scholar]
- 33.Hamieh L, Choueiri TK, Ogórek B, Khabibullin D, Rosebrock D, Livitz D, et al. Mechanisms of acquired resistance to rapalogs in metastatic renal cell carcinoma. PLoS Genet. 2018;14:e1007679. doi: 10.1371/journal.pgen.1007679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hujber Z, Petővári G, Szoboszlai N, Dankó T, Nagy N, Kriston C, et al. Rapamycin (mTORC1 inhibitor) reduces the production of lactate and 2-hydroxyglutarate oncometabolites in IDH1 mutant fibrosarcoma cells. J Exp Clin Cancer Res. 2017;36:74. doi: 10.1186/s13046-017-0544-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tuloup-Minguez V, Hamaï A, Greffard A, Nicolas V, Codogno P, Botti J. Autophagy modulates cell migration and β1 integrin membrane recycling. Cell Cycle. 2013;12:3317–28. doi: 10.4161/cc.26298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roskoski R., Jr Properties of FDA-approved small molecule protein kinase inhibitors: a 2020 update. Pharmacol Res. 2020;152:104609. doi: 10.1016/j.phrs.2019.104609. [DOI] [PubMed] [Google Scholar]
- 37.Ngeow J, Eng C. PTEN in hereditary and sporadic cancer. Cold Spring Harb Perspect Med. 2020;10:a036087. doi: 10.1101/cshperspect.a036087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data have been deposited to the National Center for Biotechnology Information (NCBI) via the Sequence Read Archive (SRA) database repository under accession code PRJNA777372. All other relevant data supporting the findings of this study are available upon reasonable request.