Abstract
Escherichia coli thioredoxin 1 has been characterized in vivo and in vitro as one of the most efficient reductants of disulfide bonds. Nevertheless, under some conditions, thioredoxin 1 can also act in vivo as an oxidant, promoting formation of disulfide bonds in the cytoplasm (E. J. Stewart, F. Åslund, and J. Beckwith, EMBO J. 17:5543–5550, 1998). We recently showed that when a signal sequence is attached to thioredoxin 1 it is exported to the periplasm, where it can also act as an oxidant, replacing the normal periplasmic catalyst of disulfide bond formation, DsbA, in oxidizing cell envelope proteins (L. Debarbieux and J. Beckwith, Proc. Natl. Acad. Sci. USA 95:10751–10756, 1998). Here we report pulse-chase studies of the efficiency of disulfide bond formation in strains exporting thioredoxin 1 and more-oxidizing variants of it. While the exported thioredoxin 1 itself substantially speeds up the kinetics of disulfide bond formation, a version of this protein containing the DsbA active site exhibits kinetics that are indistinguishable from those of the DsbA protein itself. Further, we confirm the findings of Jonda et al. (S. Jonda, M. Huber-Wunderlich, R. Glockshuber, and E. Mössner, EMBO J. 18:3271–3281, 1999), who found that DsbB is responsible for the oxidation of exported thioredoxin 1, and we report the detection of a disulfide-bonded DsbB-thioredoxin 1 complex. Finally, we have found that under conditions of high-level expression of exported thioredoxin 1, the protein can act as both an oxidant and a reductant.
The thioredoxin superfamily is a set of enzymes that share two common characteristics: a folded structure, first observed in the three-dimensional structure of thioredoxin 1; and, within this conserved fold, a consensus active site composed of 2 cysteine residues separated by 2 amino acids residues, the CXXC motif (15). One feature of these enzymes is that via the use of this common motif embedded within a common structure, different members of the family can efficiently carry out diametrically opposite reactions: reduction of disulfide bonds or oxidation of cysteine thiols. Within the cell, these two antagonistic reactions usually occur in different subcellular compartments: reduction occurs in the cytoplasm, and oxidation occurs in extracytoplasmic environments (e.g., the eukaryotic endoplasmic reticulum and the gram-negative bacterial periplasmic space).
In Escherichia coli, genetic studies of disulfide bond formation and isomerization in the cell envelope have resulted in the identification of four proteins dedicated to these processes (18). DsbA, a periplasmic protein, is a very efficient oxidative enzyme. It is actively maintained in an oxidized active form by the cytoplasmic membrane protein DsbB. This reoxidation process involves the formation of a mixed disulfide bond between DsbA cysteine 30 and DsbB cysteine 104 (10, 12). DsbB is in turn reoxidized by a mechanism involving components of the respiratory chain (2, 13, 14). DsbC is a periplasmic enzyme required for the isomerization of disulfide bonds incorrectly formed during the oxidation process. The active-site cysteines of DsbC are maintained in a reduced state by DsbD, a cytoplasmic membrane protein (19). The DsbD active site is itself maintained in the reduced state via electrons transferred from the cytoplasmic thioredoxins 1 and 2 (20, 22).
We previously reported that the reductant thioredoxin 1 can catalyze the formation of disulfide bonds in proteins when it is placed in an oxidizing environment such as the periplasm. By fusing a signal sequence to thioredoxin 1, we showed that the protein can be exported to the periplasm, where it is able to partially complement the defect in disulfide bond formation of a dsbA strain (5). This ability of thioredoxin 1 to perform active oxidation in vivo is consistent with other studies from this laboratory, in which we demonstrated that cytoplasmic thioredoxin 1 can act as an oxidant when the redox environment of the cytoplasm is altered (6, 22). We also showed that the amplitude of the complementation is dependent at least on two factors: the efficiency of thioredoxin 1 export, and whether the DsbC and DsbD proteins were present in the cell envelope. These two reductive proteins lessen the ability of exported thioredoxin 1 to complement a dsbA mutant. We presume that the reductive activity of these two proteins interferes with the oxidation performed by exported thioredoxin 1. Using different thioredoxin 1-signal sequence fusions, we showed a correlation between the amount of thioredoxin 1 exported and the level of complementation of disulfide bond formation observed (5).
Here we report further studies of the function of thioredoxin 1 as an oxidant in the periplasm. By examining the in vivo kinetics of disulfide bond formation in the cell envelope protein OmpA, we measured the efficiencies of different versions of the exported thioredoxin 1. These results demonstrate directly that thioredoxin 1 carrying its native Cys-Gly-Pro-Cys motif exhibits a strong stimulus to periplasmic disulfide bond formation in a dsbA dsbC dsbD triple-mutant strain. Surprisingly, the kinetics of disulfide bond formation with a version of exported thioredoxin 1 containing the DsbA Cys-Pro-His-Cys sequence are indistinguishable from those of a wild-type (DsbA+) strain. During the course of this work, we learned that in contrast to our previous findings, Jonda et al. (11) found that DsbB is responsible for the oxidation of their version of exported thioredoxin 1. Here we show that their conclusion is correct and that our previous results were based on a mistaken strain specification. Based on these results, we examined the mechanism of the oxidation of exported thioredoxin 1 by DsbB and identified a mixed disulfide-bonded complex of thioredoxin 1 and DsbB comparable to that normally formed by DsbA and DsbB. Finally, we present results suggesting that in a dsbB mutant strain, in which exported thioredoxin 1 is no longer oxidized, this enzyme acts as a reductant. The same reduction is also observed when a large amount of thioredoxin 1 accumulates in the periplasm.
MATERIALS AND METHODS
Strains and media.
Strains used in this study are listed in Table 1. Strains were grown at 37°C either in NZ medium (NaCl, 8 g/liter; NZ amine, 10 g/liter; yeast extract, 5 g/liter) or in M63 minimal medium supplemented with vitamin B1, glucose as a carbon source, and all amino acids except methionine and cysteine. When necessary, ampicillin was added at 200 μg/ml. Cells were grown to an optical density at 600 nm of approximately 0.5. IPTG (isopropyl-β-d-thiogalactopyranoside) was used to induce the expression of the DsbA signal sequence (DsbAss)-TrxA fusion.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| MC1000 | araD139 Δ(araABC-leu)7679 galE galK Δ(lac)X74 rpsL thi | Laboratory collection |
| RI89 | MC1000 phoR Δara714 leu+ | 20 |
| RI90 | RI89 dsbA::Kanr | 20 |
| RI249 | RI89, ΔdsbC::CamrdsbA::KanrdsbD::mini-Tn10Cam1 cadC1::Tn10 | Laboratory collection |
| RI317 | RI89, dsbB::Kanr | 20 |
| RI342 | RI89, ΔtrxA::Kanr | Laboratory collection |
| LMD244 | RI89, dsbA::Kanrzij::Tn10 dsbB::Kanr | This work |
| Plasmids | ||
| pLMD60 | APss-6aa-TrxA fusion | |
| pLMD104 | TrxA under the control of the phoA promoter | |
| pLMD173 | pLMD60—DsbA active site | This work |
| pLMD186 | pLMD104—DsbA active site | This work |
| pLMD225 | pLMD60 Cys35Tyr | This work |
| pLMD256 | pLMD104 Cys35Tyr | This work |
| pTrx-Sec | DsbAss-TrxA fusion | 11 |
Plasmid constructions.
pLMD173 was constructed by cloning an EcoRI- and XbaI-cleaved PCR product into pLMD60 cut by the same enzymes. This PCR product was obtained with primers 1 and 24 (listed below), using pBAD33-TrxA (DsbA active site [17]) as a template.
pLMD186 was constructed by cloning a HindIII- and EcoRI-cleaved PCR product into pLMD173 cut by the same enzymes. This PCR product was obtained with primers 2 and 15, using pLMD104 as a template.
pLMD225 was constructed by subcloning a product obtained from two subsequent PCRs. First, primers 24 and 31 were used with pLMD60 as a template. The PCR product obtained was purified and employed as a primer for a second PCR using primer 1 and pLMD60 as a template. The final PCR product was subcloned into pLMD60, using XbaI and EcoRI restriction sites.
pLMD256 was constructed by subcloning the EcoRI-XbaI fragment containing the trxA gene from pLMD225 into pLMD104 cut by the same enzymes.
The primers used for plasmid construction were as follows: no. 1, 5′-GGGTCTAGATTACGCCAGGTTAGCGTC-3′; no. 2, 5′-ATCATCGATAAGCTTTAATGCGG-3′; no. 15, 5′-CACTTTGAATTCTCCATGTACAAATAC-3′; no. 24, 5′-TCCCGGAATTCACCATGAGCGATAAAATTATTCACCTG-3′; and no. 30, 5′-GAATCGGGGCGATCATTTTGTACGGACCGCACCACTCTGCC-3′.
Every plasmid construct was subjected to sequence analysis. T4 DNA ligase and all restriction enzymes were used in accordance with the manufacturer's recommendations (New England Biolabs).
AP assay.
Alkaline phosphatase (AP) assays were performed in triplicate as described previously (4, 19). The strains used to make these assays lack the phoR gene, which results in constitutive expression of both AP and APss-TrxA fusions.
Immunoblotting.
Cells extracts grown in rich medium were separated in 14% nonreducing polyacrylamide gels and transferred to a nitrocellulose membrane by the use of a semidry apparatus from Bio-Rad. Membranes were probed with a polyclonal antiserum to thioredoxin 1 (a gift from C. Richardson), DsbB (kindly provided by J. C. B. Bardwell), or AP.
RESULTS
The studies of exported thioredoxin 1 reported here were done with a construct expressing a polypeptide with the APss at its amino terminus, a 6-amino-acid linker resulting from the cloning approach, and the mature sequence of thioredoxin 1 retaining its amino-terminal methionine. Our previous findings showed that it is the processed version of this protein, containing the 6-amino-acid linker attached to thioredoxin 1, that is the active molecule in the periplasm.
In vivo kinetics of disulfide bond formation by exported thioredoxin 1.
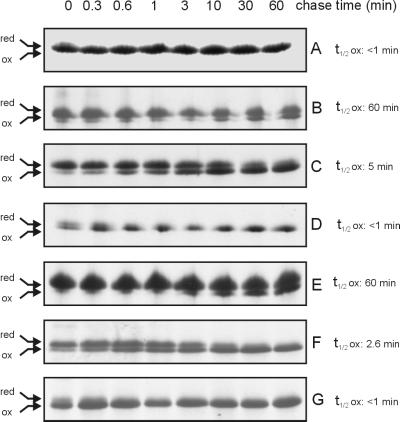
By using phenotypic properties of cells that are dependent on disulfide bond formation, such as motility on low-agar-concentration plates and AP enzymatic activities, we have previously concluded that exported thioredoxin 1 acts as an oxidant. To directly demonstrate in vivo disulfide bond formation in these studies, and to determine its efficiency, we examined the rate of oxidation of OmpA in pulse-chase experiments. OmpA is an outer membrane protein that contains one disulfide bond. Bardwell et al. (3) showed that more than 50% of the OmpA in a wild-type strain is oxidized within a labeling time of 1 min while in a dsbA strain it takes 60 min to oxidize 50% of the OmpA. We repeated these experiments, obtaining the same results for wild-type and dsbA strains as Bardwell et al., and further showed that a dsbA dsbC dsbD triple-mutant strain exhibits the same kinetics as a dsbA strain (Fig. 1). As a control for experiments with exported versions of thioredoxin 1, we used strains containing a plasmid overexpressing cytoplasmic thioredoxin 1. When we expressed the exported version of thioredoxin 1, we determined that 50% of the OmpA was oxidized in 5 min in a dsbA strain and in 2.6 min in a dsbA dsbC dsbD triple-mutant strain (Fig. 1). Thus, the exported thioredoxin 1 results in a 10- to 20-fold reduction in the half-time for disulfide bond formation, depending on the genetic background.
FIG. 1.
Rate of oxidation of OmpA catalyzed by exported thioredoxin 1. After labeling of cells with [35S]methionine (200 μCi/ml) for 1 min, chase point samples were obtained and incubated in the presence of 100 mM iodoacetamide in order to alkylate free thiol residues (0.1% final concentration of cold methionine was used for the chase). An OmpA antiserum was used to immunoprecipitate this protein (see reference 7 for details), and the samples were separated on a nonreducing sodium dodecyl sulfate-polyacrylamide gel. The OmpA oxidation rate was determined in a wild-type strain (A), a dsbA strain (B to D), and a dsbA dsbC dsbD triple-mutant strain (E to G) expressing cytoplasmic thioredoxin 1 (A, B, and E), exported thioredoxin 1 (C and F), or exported thioredoxin 1 with a DsbA active site (D and G). Values for 50% oxidation time (t1/2ox) were determined after the quantitation of reduced (red) and oxidized (ox) forms of OmpA by phosphorimager analysis (Bio-Rad).
We also expressed in both dsbA single-mutant and dsbA dsbC dsbD triple-mutant strains a version of exported thioredoxin 1 in which its active site (Cys-Gly-Pro-Cys) was changed to the DsbA active site (Cys-Pro-His-Cys). This thioredoxin 1 version is a more efficient oxidant in vitro (11). With this variant of thioredoxin 1, we found that the rate of formation of disulfide bonds in OmpA exhibited kinetics of disulfide bond formation that were indistinguishable from those of the wild-type (DsbA+) control (Fig. 1).
The dramatic changes in the kinetics of OmpA oxidation are mirrored by the levels of AP activity found in the different strains (Table 2). AP, a periplasmic protein, requires the formation of two disulfide bonds to achieve its final active conformation. In a dsbA dsbC dsbD triple-mutant strain, the amount of AP accumulated was approximately 50% of that found in wild-type strains when the exported thioredoxin 1 contained its wild-type active site. This activity rose to approximately 100% when the thioredoxin 1 with a DsbA Cys-Pro-His-Cys active site was exported. These activities represent approximately 10- and 20-fold increases over the control strain overexpressing only a cytoplasmic thioredoxin 1. These data show that the highly effective reductant thioredoxin 1, when exported to the periplasm with its own active site, is a surprisingly efficient oxidant. Furthermore, changing only 2 amino acids in this protein (in the active site) results in a thioredoxin 1 whose activity is indistinguishable from that of DsbA in vivo.
TABLE 2.
AP activities, mirroring disulfide bond formation
| TrxA typea | Mean AP activity ± SD for strain (or relevant genotype):
|
||||
|---|---|---|---|---|---|
| Wild type | dsbA | dsbB | dsbA dsbB | dsbA dsbC dsbD | |
| Cyt. | 850 ± 50 | 22 ± 6 | 20 ± 8 | 24 ± 10 | 35 ± 11 |
| Exp. | 850 ± 55 | 80 ± 8 | 5 ± 4 | 6 ± 3 | 350 ± 20 |
| Exp. (DsbA) | 750 ± 45 | 500 ± 10 | 8 ± 4 | 10 ± 4 | 1,000 ± 52 |
Cyt., cytoplasmic thioredoxin 1; exp., exported thioredoxin 1; exp. (DsbA), exported thioredoxin 1 with DsbA active site.
Mechanism of oxidation of exported thioredoxin 1 by DsbB.
In contrast to our previous report, Jonda et al. (11) showed that DsbB is essential for the oxidation of exported thioredoxin 1. We had compared a dsbA strain with a dsbA dsbB strain. To reanalyze the effect of DsbB on exported thioredoxin 1, we examined the properties of the following three strain backgrounds: dsbA single mutant, dsbB single mutant, and dsbA dsbB double mutant (we reconstructed the dsbA dsbB double mutant after finding that the strain used previously did not have the correct genotype). The results now confirm that DsbB is required for exported thioredoxin 1 to act as an oxidant in the periplasm (Table 2).
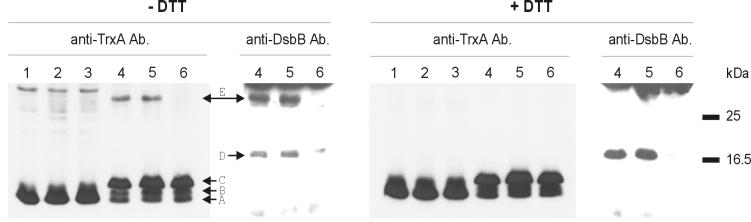
The mechanism of oxidation of DsbA, until now the only known substrate of DsbB, was clarified as the result of studies with dominant-negative mutants of DsbA (10, 12). A mutation in which the second cysteine residue of the DsbA active site is replaced by a tyrosine residue interferes with wild-type DsbA function (10). This dominant-negative effect is due to the accumulation of a disulfide-bonded complex of DsbB and the DsbA mutant protein that is formed between DsbA(Cys30) and DsbB(Cys104). The complex revealed by this approach is believed to represent an intermediate in the oxidation of DsbA by DsbB (10, 12). To determine whether the oxidation of exported thioredoxin 1 by DsbB occurs by the same mechanism, we introduced into thioredoxin 1 an analogous mutation changing the second active-site cysteine residue to a tyrosine residue (Cys35Tyr). In parallel, we constructed versions of exported thioredoxin 1 in which the complete active site was replaced by either the DsbA dominant-negative site or the DsbA wild-type site (as a control). Using thioredoxin 1 antibodies, we compared immunoblots of extracts of cells expressing cytoplasmic thioredoxin 1 Cys35Tyr and exported thioredoxin 1 Cys35Tyr. These blots exhibited a band of ∼32 kDa only when thioredoxin 1 was exported (Fig. 2, left panel, lanes 4 and 5). This band was not present in a dsbB mutant strain (Fig. 2, left panel, lane 6). This band also was not present when samples were reduced (Fig. 2, right panel, lanes 4 and 5), indicating that it contained a disulfide-bonded complex. Using antibodies to DsbB, we identified this band as a mixed disulfide complex of DsbB and thioredoxin 1 (Fig. 2, left panel, right part, lanes 4 and 5). The gels show that perhaps a majority of the DsbB is tied up in such a complex. Identical results have been observed with the thioredoxin 1 in which the active site was replaced by the dominant-negative Cys-Pro-His-Tyr sequence of DsbA (data not shown). We also showed that by using a version of DsbB in which the cysteine 104 residue was replaced by alanine, the mixed disulfide bond thioredoxin 1-DsbB complex was not present (data not shown). These data indicate that exported thioredoxin 1 interacts directly with DsbB in the same way as DsbA does.
FIG. 2.
Detection of a mixed disulfide bond between exported thioredoxin 1 and DsbB. Equal amounts of proteins from a wild-type (lanes 1 and 4), a dsbA (lanes 2 and 5), and a dsbB (lanes 3 and 6) strain expressing cytoplasmic thioredoxin 1-Cys35⧫Tyr (lanes 1 to 3) or exported thioredoxin 1-Cys35⧫Tyr (lanes 4 to 6) were separated on a nonreducing (minus dithiothreitol [−DTT]) and a reducing (plus dithiothreitol [+DTT]) sodium dodecyl sulfate-polyacrylamide gel and transferred to two nitrocellulose membranes. The left part of each of the membranes was incubated with TrxA antiserum (anti-TrxA Ab.), and the right part was incubated with DsbB antiserum (anti-DsbB Ab.). Prestained molecular mass markers were loaded between the left and the right panels so that the two pieces of membrane could be aligned. Band A is the cytoplasmic thioredoxin 1, band B is the 6 amino acids-thioredoxin 1 processed form, band C is the precursor APss-6 amino acids-thioredoxin 1, band D is DsbB, and band E is the mixed disulfide complex of exported thioredoxin 1 Cys35⧫Tyr and DsbB.
Exported thioredoxin 1 can also act as a reductant.
An examination of the data in Table 2 shows that not only does the dsbB mutation eliminate the ability of exported thioredoxin 1 to act as an oxidant, but also in this strain background the export of thioredoxin 1 actually reduces the amount of AP below the background level normally found in a dsbB mutant. This finding raised the possibility that exported thioredoxin 1, when not oxidized by DsbB, actually reduces disulfide bonds in cell envelope proteins. We considered that this finding might explain the results of Jonda et al. (11), who failed to detect any oxidative ability of exported thioredoxin 1 when it contained its native active site. In those experiments, the thioredoxin was expressed at such high levels that it may have saturated the capacity of DsbB to oxidize it, thus resulting in substantial amounts of the reduced form of the protein.
To test this explanation, we used the plasmid described by Jonda et al. (11) in which thioredoxin 1 is fused to the DsbAss. The expression of this construct is under the control of an IPTG-inducible promoter, and the product is processed more efficiently than ours (11). To determine the redox activity of exported thioredoxin 1, we monitored the enzymatic activity of AP in a dsbA dsbC dsbD triple-mutant strain in the presence of increasing concentrations of IPTG. Results presented in Table 3 show that at low concentrations of IPTG (0 to 20 μM), exported thioredoxin 1 is able to promote disulfide bond formation, but at higher concentrations of IPTG (40 to 200 μM), the level of AP activity progressively decreases. These results suggest that exporting a large amount of thioredoxin 1 into the periplasm changes the balance between reduced and oxidized thioredoxin 1 in favor of the reduced form, which may then reduce substrate proteins such as AP. We determined the level of expression induced with different concentrations of IPTG by immunoblotting, using both thioredoxin 1 and AP antibodies (data not shown). As expected at steady state, the level of thioredoxin 1 increased with the increase in IPTG concentration while the level of AP slightly decreased (the steady-state levels of AP protein decrease because the protein is rapidly degraded when not disulfide bonded [3]). Finally, pulse-chase experiments showed that neither the level of expression nor the secretion of AP was affected by the overexpression of the thioredoxin construct (data not shown).
TABLE 3.
AP activities of a dsbA dsbC dsbD strain expressing different levels of the DsbAss-TrxA fusion
| IPTG concn (μM) | AP activity (mean ± SD) |
|---|---|
| 0 | 50 ± 10 |
| 5 | 206 ± 16 |
| 10 | 220 ± 14 |
| 20 | 224 ± 20 |
| 40 | 153 ± 15 |
| 80 | 110 ± 14 |
| 200 | 95 ± 12 |
DISCUSSION
The formation of disulfide bonds in the periplasmic space of E. coli is normally a very rapid process. For example, in the case of OmpA, pulse-labeling studies show that in vivo the majority of full-length protein is fully oxidized within 30 s to 1 min. This efficient process is dependent on DsbA, a 23-kDa protein containing a thioredoxin-like domain. Here we report studies of the efficiency with which cytoplasmic thioredoxin 1, when exported to the periplasm, can substitute for DsbA. We showed that 50% of the OmpA is oxidized within 2 to 3 min by exported thioredoxin 1 in a dsbA dsbC dsbD triple-mutant strain. Previously, our laboratory found that a combination of oxidized thioredoxins 1 and 2, also with a half-time of 2 to 3 min, promotes disulfide bond formation in the cytoplasm (22). In the latter case, the oxidized thioredoxins accumulated in the cytoplasm of a mutant lacking thioredoxin reductase. Together, these findings show that despite its extremely low redox potential (−270 mV [1]), thioredoxin 1 can still act as a strong catalyst of oxidation in vivo.
However, and perhaps more striking, the introduction of the Cys-Pro-His-Cys DsbA active-site sequence into exported thioredoxin 1 resulted in a thioredoxin 1 oxidation capability that appears to be the same in vivo as that of DsbA. The kinetics of disulfide bond formation in OmpA protein are indistinguishable in the two cases. These results are somewhat surprising because DsbA has, along with its thioredoxin domain, additional polypeptide sequences amounting to approximately the same molecular weight as the thioredoxin portion. These extra sequences have been thought to serve functions necessary for an enzyme that catalyzes disulfide bond formation, such as binding of substrate polypeptides as well as interacting with DsbB (8, 9, 21). Our findings raise the possibility that these additional sequences in DsbA serve some function in the periplasm other than disulfide bond formation. Alternatively, they may contribute to a slight increase in efficiency not detectable in pulse-chase experiments, which, nevertheless, provides a selective advantage. For example, with some of the substrates other than OmpA, DsbA may be more effective than the altered thioredoxin 1 in promoting disulfide bond formation. Such differences in substrate reactivities could be due to differing redox potentials of the cysteines in those proteins or to differences in substrate accessibility.
We also report that our previous results suggesting that a cell envelope component different from DsbB is responsible for oxidizing exported thioredoxin 1 were incorrect. By reexamining the levels of complementation provided by exported thioredoxin 1 in a single dsbA, a single dsbB, and a reconstructed dsbA dsbB double-mutant strain, we confirmed that DsbB is essential to this complementation, as reported by Jonda et al. (11). The fact that DsbB can efficiently recognize thioredoxin 1, essentially a paired-down version of DsbA, in vivo as well as in vitro (11) raises interesting issues about the specificity of DsbB discussed by Jonda et al. (11).
Strains expressing dominant-negative mutants of the dsbA gene in which the second cysteine of the active site, Cys-33, is replaced by another amino acid accumulate a disulfide-bonded DsbB-DsbA complex (10, 12). These findings have been taken to indicate that this complex represents an intermediate in the oxidative pathway. We showed here that by expressing a mutant of exported thioredoxin 1 in which the analogous cysteine was altered (Cys-35), we could detect an analogous DsbB-thioredoxin 1 complex. As in the DsbB-DsbA complex, the disulfide bond is formed between the remaining cysteine of thioredoxin 1 and cysteine 104 of DsbB. This result indicates, not surprisingly, that exported thioredoxin 1 is oxidized by DsbB in the same way that DsbA is. However, our findings also raise the possibility that in general, protein-protein intermediates in such oxidation pathways will be detectable by employing mutant versions of one of the potential protein partners in which the appropriate cysteine is altered. In the cases described here, the proteins were mutated so that the exposed more-reactive cysteine of the Cys-X-X-Cys motif is left intact.
It has been known for some time that low background levels of disulfide bond formation still occur in dsbA dsbB and dsbA dsbB double mutants (19). There is currently no information to explain this background activity. Here, we report that exported thioredoxin 1 actually decreases this background in a dsbB mutant and a dsbA dsbB double mutant, but not in a dsbA mutant. This result is most easily explained by the fact that thioredoxin 1 accumulates in the reduced (and reducing) form in dsbB backgrounds and is, therefore, able to interfere with disulfide bond formation. This interference may occur either by direct reduction of disulfide bonds in substrate proteins or by interference with the source of oxidizing potential.
This last finding allows us to advance an explanation for a puzzling contradiction between one aspect of the studies of Jonda et al. (11) and our own. The former group reported, in contrast to our results, that exported thioredoxin 1, with its Cys-Gly-Pro-Cys active site intact, is incapable of promoting disulfide bond formation in the periplasm because of its low redox potential. As described above, we found a relatively efficient thioredoxin 1 oxidizing activity in the periplasm. Jonda et al. (11) expressed their thioredoxin 1 construct from a very-high-copy-number pUC plasmid and grew cells either in rich NYT medium or in M63 medium. It is likely that the optimized expression system used in their study, coupled with a high export efficiency, allowed the accumulation of much larger amounts of thioredoxin 1 in the periplasm when cells were grown in rich medium (a rich medium is more likely to contain traces of inducer). They actually reported that their expression system was very leaky under such conditions. In this situation, thioredoxin 1 may greatly exceed the reoxidation capacity of DsbB, resulting in a much higher level of reduced than oxidized thioredoxin 1. Given our findings suggesting that reduced thioredoxin 1 can interfere with disulfide bond formation, the absence of an oxidative effect of thioredoxin 1 under these conditions is not surprising. On the other hand, when cells are grown in M63 medium, such an expression system would be much less leaky, thereby not allowing the exportation of enough thioredoxin 1 for a complementation of disulfide bonds in a dsbA strain to be observed, as reported by Jonda et al. (11). We previously showed that a dsbA dsbC dsbD triple-mutant strain was more effective than a dsbA strain in the detection of complementation of disulfide bond defects by a weakly exported thioredoxin 1. We have obtained support for these explanations by showing that the use of the construct of Jonda et al. (11) in a dsbA dsbC dsbD triple-mutant strain grown in M63 medium led to the formation of disulfide bonds at low levels of induction and a decrease in formation of disulfide bonds when the level of induction was further increased.
The studies reported here suggest a surprisingly functional interchangeability of thioredoxin and DsbA, two oxidoreductases well characterized as carrying out opposing redox reactions. These and other findings (16, 20) suggest that in examining the function of members of the thioredoxin superfamily it is important to take into consideration the interaction of these proteins in vivo with partner proteins for which they are substrates. Such partners of thioredoxin 1 and DsbA are normally thioredoxin reductase and DsbB, respectively. These latter enzymes determine what kind of reactions the proteins will carry out, reactions which are to an unexpected extent independent of their redox potentials. This analysis suggests that in a reverse form of these experiments, expression of DsbA into the cytoplasm may compensate for a thioredoxin defect in the cytoplasmic disulfide reduction pathway. It appears that if DsbA evolved from a thioredoxin, it did not have to diverge very far to become a strong oxidant as long as there existed a protein such as DsbB which could receive electrons from it.
Despite this interchangeability of members of the thioredoxin superfamily, the redox potentials of these proteins are usually indicative of their in vivo function (17). Nevertheless, beyond laboratory manipulations, members of the thioredoxin family may switch their function in a biologically significant way.
ACKNOWLEDGMENTS
We thank R. Glockshuber for the thioredoxin 1 expression plasmid and J. C. A. Bardwell for antiserum to DsbB. We gratefully acknowledge members of the Beckwith laboratory particularly Hongping Tian and Daniel Ritz, for helpful discussions.
This work was supported by the National Institute of Health (grant GM41883 to J.B.).
REFERENCES
- 1.Åslund F, Berndt K D, Holmgren A. Redox potentials of glutaredoxins and other thiol-disulfide oxidoreductases of the thioredoxin superfamily determined by direct protein-protein redox equilibria. J Biol Chem. 1997;272:30780–30786. doi: 10.1074/jbc.272.49.30780. [DOI] [PubMed] [Google Scholar]
- 2.Bader M, Muse W, Ballou D P, Gassner C, Bardwell J C. Oxidative protein folding is driven by the electron transport system. Cell. 1999;98:217–227. doi: 10.1016/s0092-8674(00)81016-8. [DOI] [PubMed] [Google Scholar]
- 3.Bardwell J C A, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 4.Brickman E, Beckwith J. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and φ80 transducing phages. J Mol Biol. 1975;96:307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- 5.Debarbieux L, Beckwith J. The reductive enzyme thioredoxin 1 acts as an oxidant when it is exported to the Escherichia coli periplasm. Proc Natl Acad Sci USA. 1998;95:10751–10756. doi: 10.1073/pnas.95.18.10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derman A I, Prinz W A, Belin D, Beckwith J. Mutations that allow disulfide bond formation in the cytoplasm of Escherichia coli. Science. 1993;262:1744–1747. doi: 10.1126/science.8259521. [DOI] [PubMed] [Google Scholar]
- 7.Froshauer S, Green G N, Boyd D, McGovern K, Beckwith J. Genetic analysis of the membrane insertion and topology of MalF, a cytoplasmic membrane protein of Escherichia coli. J Mol Biol. 1988;200:501–511. doi: 10.1016/0022-2836(88)90539-6. [DOI] [PubMed] [Google Scholar]
- 8.Guddat L W, Bardwell J C, Martin J L. Crystal structures of reduced and oxidized DsbA: investigation of domain motion and thiolate stabilization. Structure. 1998;6:757–767. doi: 10.1016/s0969-2126(98)00077-x. [DOI] [PubMed] [Google Scholar]
- 9.Guddat L W, Bardwell J C, Zander T, Martin J L. The uncharged surface features surrounding the active site of Escherichia coli DsbA are conserved and are implicated in peptide binding. Protein Sci. 1997;6:1148–1156. doi: 10.1002/pro.5560060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guilhot C, Jander G, Martin N L, Beckwith J. Evidence that the pathway of disulfide bond formation in Escherichia coli involves interactions between the cysteines of DsbB and DsbA. Proc Natl Acad Sci USA. 1995;92:9895–9899. doi: 10.1073/pnas.92.21.9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jonda S, Huber-Wunderlich M, Glockshuber R, Mössner E. Complementation of DsbA deficiency with secreted thioredoxin variants reveals the crucial role of an efficient dithiol oxidant for catalyzed protein folding in the bacterial periplasm. EMBO J. 1999;18:3271–3281. doi: 10.1093/emboj/18.12.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kishigami S, Kanaya E, Kikuchi M, Ito K. DsbA-DsbB interaction through their active site cysteines. Evidence from an odd cysteine mutant of DsbA. J Biol Chem. 1995;270:17072–17074. doi: 10.1074/jbc.270.29.17072. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi T, Ito K. Respiratory chain strongly oxidizes the CXXC motif of DsbB in the Escherichia coli disulfide bond formation pathway. EMBO J. 1999;18:1192–1198. doi: 10.1093/emboj/18.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi T, Kishigami S, Sone M, Inokuchi H, Mogi T, Ito K. Respiratory chain is required to maintain oxidized states of the DsbA-DsbB disulfide bond formation system in aerobically growing Escherichia coli cells. Proc Natl Acad Sci USA. 1997;94:11857–11862. doi: 10.1073/pnas.94.22.11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin J L. Thioredoxin—a fold for all reasons. Structure. 1995;3:245–250. doi: 10.1016/s0969-2126(01)00154-x. [DOI] [PubMed] [Google Scholar]
- 16.Missiakas D, Georgopoulos C, Raina S. The Escherichia coli dsbC (xprA) gene encodes a periplasmic protein involved in disulfide bond formation. EMBO J. 1994;13:2013–2020. doi: 10.1002/j.1460-2075.1994.tb06471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mössner E, Huber-Wunderlich M, Rietsch A, Beckwith J, Glockshuber R, Åslund F. Importance of redox potential for the in vivo function of the cytoplasmic disulfide reductant thioredoxin from Escherichia coli. J Biol Chem. 1999;274:25254–25259. doi: 10.1074/jbc.274.36.25254. [DOI] [PubMed] [Google Scholar]
- 18.Rietsch A, Beckwith J. The genetics of disulfide bond metabolism. Annu Rev Genet. 1998;32:163–184. doi: 10.1146/annurev.genet.32.1.163. [DOI] [PubMed] [Google Scholar]
- 19.Rietsch A, Belin D, Martin N, Beckwith J. An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:13048–13053. doi: 10.1073/pnas.93.23.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rietsch A, Bessette P, Georgiou G, Beckwith J. Reduction of the periplasmic disulfide bond isomerase, DsbC, occurs by passage of electrons from cytoplasmic thioredoxin. J Bacteriol. 1997;179:6602–6608. doi: 10.1128/jb.179.21.6602-6608.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schirra H J, Renner C, Czisch M, Huber-Wunderlich M, Holak T A, Glockshuber R. Structure of reduced DsbA from Escherichia coli in solution. Biochemistry. 1998;37:6263–6276. doi: 10.1021/bi980136y. [DOI] [PubMed] [Google Scholar]
- 22.Stewart E J, Åslund F, Beckwith J. Disulfide bond formation in the Escherichia coli cytoplasm: an in vivo role reversal for the thioredoxins. EMBO J. 1998;17:5543–5550. doi: 10.1093/emboj/17.19.5543. [DOI] [PMC free article] [PubMed] [Google Scholar]