Abstract
This study aimed to provide a further characterization of the lactic microbiota present in Minas artisanal cheese (MAC) from the Serro region by using culture-independent methods, as a complementary analysis of a previous study. The total DNA extracted from MAC samples (n = 55) was subjected to repetitive extragenic palindromic-PCR (rep-PCR) and PCR-denaturing gradient gel electrophoresis (PCR-DGGE). Rep-PCR analysis showed that core microbiota of Serro MAC was closely related, independent of the production town, farm size, or time of production. The sequencing of PCR-DGGE bands identified the prevalence of Lactococcus lactis in all samples, and Streptococcus salivarius was also identified. Thus, we conclude that when more accurate methods are unavailable, rep-PCR can be used as a culture-independent method to demonstrate if the microbiota is closely related or not among the samples. PCR-DGGE results also matched to the main findings of high-throughput sequencing, previously presented, confirming its confidence to detect the main microbial groups present in the raw milk cheeses.
Keywords: Artisanal cheese, Lactic microbiota, Rep-PCR, PCR-DGGE
Introduction
Recently, a study conducted by our research group have characterized by culture-dependent and culture-independent methods the microbiota signature of Minas artisanal cheeses (MAC) from Serro region, a traditional artisanal cheese-producing region in the state of Minas Gerais, Brazil [1]. The cheese samples were subjected to lactic acid bacteria (LAB) enumeration, and most of the obtained isolates were identified as lactobacilli; producer city, farm size, starter culture, and time of production had no influence on LAB counts and species distribution (P > 0.05) [1]. The total DNA from cheese samples was then subjected to high-throughput 16S rRNA sequencing, targeting V3–V4 region of the 16S rRNA, and the results revealed Lactococcus lactis as the predominant species in all samples, with no differences detected by alpha-diversity and beta-diversity analysis [1].
Minas artisanal cheeses are produced using raw milk, and the presence of LAB in this product is extremely desirable, once these microbial group confers typical sensory characteristics [2]. These cheeses are composed by complex ecosystems susceptible to abiotic and environmental factors, and changes in these factors have a direct influence within the microbiome community, affecting the organoleptic properties as well as the safety of the end products [3, 4].
The assessment of the microbial diversity of artisanal cheeses is very important to ensure their quality and safety, and that can be performed by culture-dependent and culture-independent methods. Culture-dependent methods are more laborious, consisting of homogenization of the sample, followed by dilution and plating in culture medium, selection of colonies, and phenotypic characterization [5, 6]. These methods have some limitations, since it is difficult to simulate the necessary nutrients and the conditions for many genera of bacteria to grow in vitro. On the other hand, the culture-independent methods are based on the analysis of genetic material extracted directly from the cheese. These methods have advantages for detection and differentiation among species and for identification of unculturable microbes [6, 7].
Considering the advantages and limitations of culture-dependent and culture-independent methods, combined approaches are the best way to characterize the microbial diversity of artisanal cheeses. Since there are many protocols available with this purpose, herein we continued a previous study of our group related to the characterization of the lactic microbiota of MAC from Serro region [1], by using further culture-independent methodologies.
Material and methods
The same Serro MAC samples (n = 55) obtained by Nero et al. [1] were considered in the present study, as well as the procedures for total DNA extraction. Samples were obtained from producers of different cities located in the Serro region, with distinct production characteristics as detailed previously [1].
Rep-PCR reactions of total DNA were performed according to Gevers et al. [8], using single primer (GTG)5 (5′-GTG GTG GTG GTG GTG-3′). PCR reactions contained 12.5 µL of GoTaq Master Mix 2X (Promega Corp., Madison, WI, USA), 1 mL of 50 pMol primer, 2 µL of DNA extracted from each cheese, and ultra-pure water (Promega) to complete a final volume of 25 µL. The PCR conditions were 95 °C for 5 min, 30 cycles at 95 °C for 30 s, 40 °C for 45 s, 65 °C for 8 min, and the final extension at 65 °C for 16 min. The PCR products were electrophoresed on agarose gels (1.5% w/v) in TBE buffer (0.5 × Tris/Borate/EDTA) at constant voltage (90 V) for 3 h. 1 kb DNA ladder (Sigma-Aldrich, St. Louis, MO, USA) was used as a molecular size marker. The obtained fingerprints were analyzed using BioNumerics 6.6 (Applied Maths NV, Sint-Martens-Latem, Belgium). The similarities between the profiles were calculated using the Dice coefficient and UPGMA method (tolerance 1.5%, optimization 5%).
To perform PCR-DGGE, the V3 variable region of the bacterial 16S rRNA gene was amplified according to Ampe et al. [9] by using primers 338F (5′-AC TCC TAC GGG AGG CAG CAG-3′) and 518R (5′-ATT ACC GCG GCT GCT GG-3′). A 40 bp GC-clamp (5′-CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGGG-3′) was attached to the 5′ end of the forward primer for DGGE analysis in order to ensure that the fragment of DNA remains partially double stranded and that the region screened is in the lowest melting domain, generating amplicons with a size of approximately 236 bp [10].
PCR reaction was performed in a final volume of 50 µL containing 1 × GoTaq® Flexi Reaction buffer (Promega), 1.5 mM MgCl2, 0.2 mM dNTPs, 0.2 µM of each primer, 1.25 U Taq pol DNA polymerase (Cellco Biotec, São Carlos, SP, Brazil), 2 µL of total DNA of cheese, and ultra-pure PCR water. PCR conditions were 10 min at 95 °C, 35 cycles of 1 min at 95 °C, 1 min at 42 °C, 2 min at 72 °C, and a final extension of 7 min at 72 °C. The PCR products were electrophoresed on agarose gels (1.5% w/v) in TBE buffer and stained with ethidium bromide.
The DCode™ universal mutation detection system (Bio-Rad Lab., Hercules, CA, USA) was used for DGGE analysis. Electrophoresis was performed in a 0.8-mm-thick polyacrylamide gel (8% w/v) acrylamide:bisacrylamide (37.5:1) with a denaturing gradient of urea and formamide ranging from 25 to 60% in 1 × TAE buffer (40 mM Tris base, 20 mM acetic acid, and 1 mM EDTA, pH 8) at constant voltage of 120 V for 6 h at 60 °C, stained (30 min) in 1 × TAE containing 1 × SYBR GOLD (Sigma-Aldrich). Ladders were included as reference in gels (1 kb, Kasvi, São José dos Pinhais, PR, Brazil). Gels were photographed and analyzed on a UV transilluminator with the photo documentation imaging system.
Representative bands of polyacrylamide gels were collected using sterile pipette tips, and their DNA eluted overnight in 50 µL of nuclease-free water at 4 °C. The DNA was then re-amplified using the same primer pair but without the GC-clamp by using the conditions described above. Amplicons were purified using the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) and sequenced on an Applied Biosystems 3500 Genetic Analyzer (Applied Biosystems, Waltham, MA, USA). Sequences were compared to those deposited in the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/genbank), using the Basic Alignment Search Tool (BLAST, http://blast.ncbi.nlm.nih.gov/Blast.cgi) software.
Results and discussion
In the previous study carried out by Nero et al. [1], lactobacilli were the predominant LAB identified by conventional plating and sequencing of amplified 16S rRNA. On the other hand, when the total DNA extracted from cheese samples was subjected to high-throughput 16S rRNA sequencing, Lactococcus lactis was clearly the dominant species. Considering the obtained data, further analysis was carried out to characterize the lactic microbiota of the same Serro MAC samples by alternative culture-independent methods. Then, results of these alternative approaches were compared to those presented by Nero et al. [1].
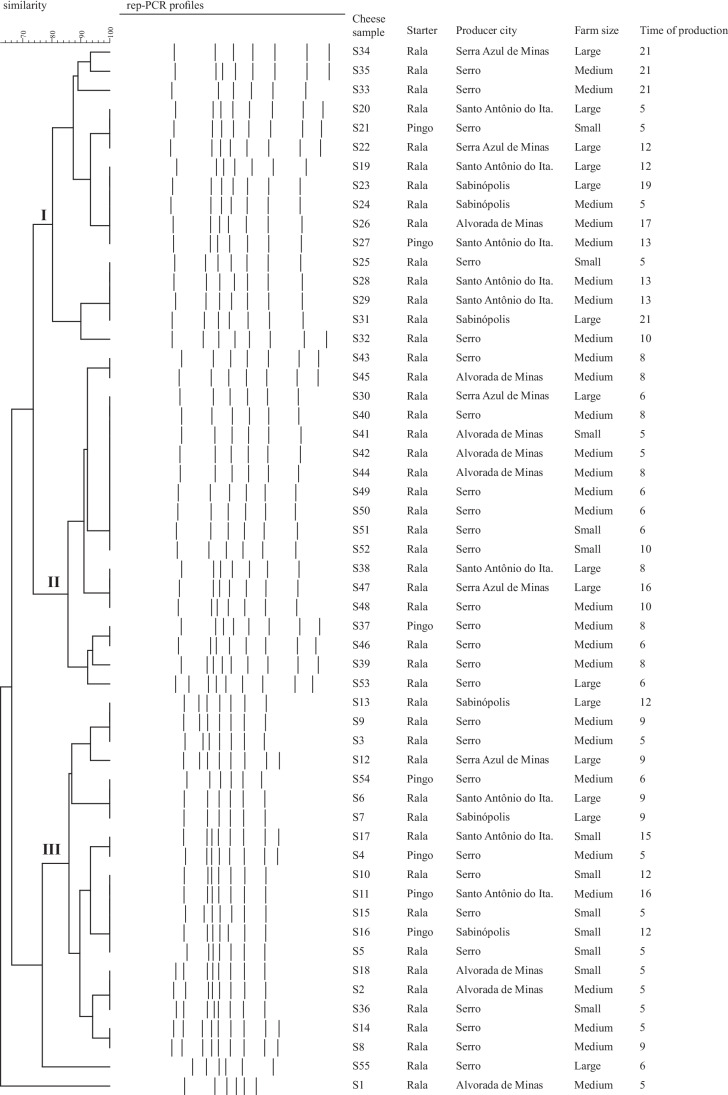
The rep-PCR profiles, cheese origin, and their production characteristics are presented in the Fig. 1. Considering a coefficient of similarity of 80%, three main clusters were obtained in the generated dendrogram. Rep-PCR is a subtyping method usually employed to differentiate LAB by grouping clusters of genetic-related isolates for subsequent selection of isolates and identification by sequencing [11]. However, this method may have other application, as to demonstrate the total microbiota of artisanal cheeses by grouping them based on their different geographical origins [12]. Another study conducted by Perin et al. [13] has demonstrated that rep-PCR can provide a clear evidence of the in situ interference caused by adding a bacteriocinogenic L. lactis subsp. lactis strain in Minas cheese produced with raw goat milk.
Fig. 1.
Cluster analysis of rep-PCR fingerprints obtained from Serro Minas artisanal cheese samples with different production characteristics (city; farm size—based on daily milk production—small, lower than 150 L; medium, 150–200 L; large, higher than 200 L; starter culture; and time of production, days). The similarities between the profiles were calculated by using Dice coefficient, and the dendrogram were constructed using unweighted pair group method with arithmetic mean (UPGMA), with tolerance of 1.5% and optimization of 5%
Interesting, in this study, the application of rep-PCR as culture-independent method suggested that the core microbiota of Serro MAC can be closely related independent of the production town, farm size, or maturation time (Fig. 1). This result makes sense once the towns where samples were collected compose a micro-region with particular climate and soil characteristics (terroirs), influencing the selection of cheese microbiota. Despite microbial groups present in each sample cannot be identified by rep-PCR, this method provided evidence that factors related to the production of Serro MAC did not have a major interference in the microbiota of the final products, as demonstrated in the first study by Nero et al. [1].
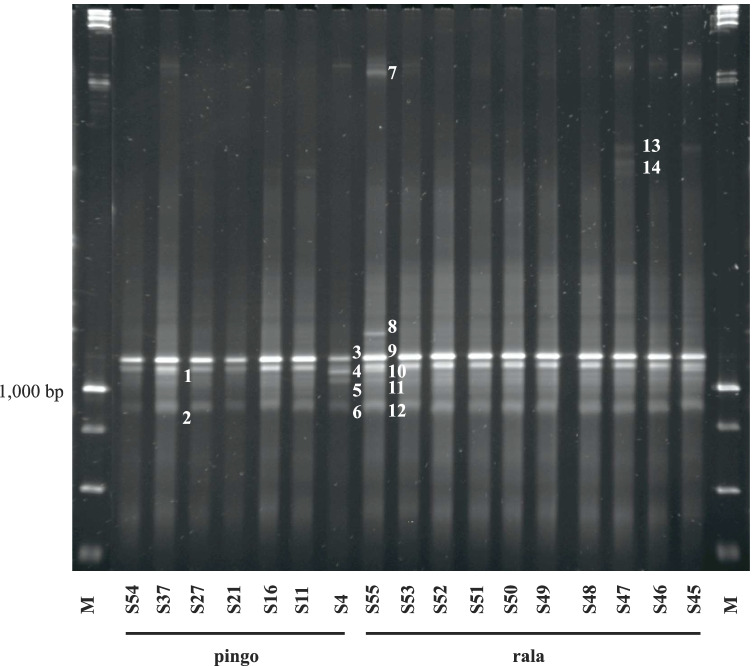
PCR-DGGE confirmed that LAB profile was similar in all 55 cheese samples, regardless of production characteristics. Based on denaturation profiles, 3 to 6 bands were observed as specific to all samples, and 14 bands were selected for sequencing and identification (Fig. 2). The sequencing of bands indicated the prevalence of L. lactis in all samples. In addition, L. lactis subsp. lactis, L. lactis sp., Streptococcus salivarius, and Streptococcus sp. were identified (Table 1). More than one genus was identified in bands 4 and 6, indicating the same denaturation profile (Table 1).
Fig. 2.
PCR-DGGE profiles of the bacterial ecology of Serro Minas artisanal cheese samples (17 out of 55) with different production characteristics (city; farm size—based on daily milk production—small, lower than 150 L; medium, 150–200 L; large, higher than 200 L; starter culture; and time of production, days). The lines indicate the identification of the 17 samples (S54, S37, S27, S21, S16, S11, S4, S55, S53, S52, S51, S50, S49, S48, S47, S46, and S45) and ladder markers of 1 kb (M). The numbers indicate the bands subjected to sequencing for molecular identification (results reported in Table 1)
Table 1.
Identification of bacterial species present in Serro Minas artisanal cheese samples based on sequencing of PCR-DGGE bands and BLAST sequence comparison in GenBank
| Banda | Closest relative | % of identityb | GenBank accession no |
|---|---|---|---|
| 1 | Lactococcus lactis | 99 | MT645510.1 |
| 2 | Lactococcus lactis | 98 | MT645510.1 |
| 3 | Lactococcus lactis | 98 | MT645510.1 |
| 4 | Lactococcus sp. | 97 | KJ804064.1 |
| Streptococcus sp. | 96 | MW045815.1 | |
| 5 | Streptococcus salivarius | 96 | MT512105.1 |
| 6 | Streptococcus sp. | 96 | MW045807.1 |
| Lactococcus lactis subsp. lactis | 94 | JQ973604.1 | |
| 7 | Streptococcus sp. | 97 | MT512026.1 |
| 8 | Lactococcus lactis | 96 | MT645510.1 |
| 9 | Lactococcus lactis | 99 | MT645510.1 |
| 10 | Lactococcus lactis | 99 | MT645510.1 |
| 11 | Lactococcus lactis | 99 | MT645510.1 |
| 12 | Lactococcus lactis | 99 | MT645510.1 |
| 13 | Lactococcus lactis | 96 | JQ754459.1 |
| 14 | Lactococcus lactis | 96 | MT115989.1 |
aThe numbers correspond to the band numbers in Fig. 2
bPercentage of similarity between the sequences obtained from the PCR-DGGE band and the sequence of the closest species in the GenBank database
PCR-DGGE proved to be useful for identification of the main species which compose the cheese microbiota, and our results agree to those observed in other studies [1, 2], where16S rRNA high-throughput sequencing was employed to study microbial signature of Serro MAC. However, PCR-DGGE was clearly much more limited when compared to high-throughput sequencing to study microbial diversity in complex food matrix.
Arcuri et al. [10] analyzed samples of MAC from Serro, Araxá, Cerrado, and Serra da Canastra regions by PCR-DGGE. The samples of Serro MAC also presented low microbial diversity and showed 4 or 5 bands as specific. One specific band was the most prominent, and it was present in all MAC samples, corresponding to Streptococcus thermophilus after sequencing; bands correspondent to L. lactis were also observed in nearly all MAC samples. However, this study included a very limited sampling. Van Hoorde et al. [14] evaluated the LAB diversity of Flemish artisan raw milk Gouda-type cheese by conventional culturing and PCR-DGGE. As results, lactobacilli were predominant by culture-dependent methodology, as well as by PCR-DGGE analysis of culturable fractions. However, L. lactis was predominant (present in all profiles) when PCR-DGGE was done using the total DNA extracted direct from cheeses. The authors emphasize the use of an integrated approach between culture-dependent and culture-independent techniques to cover as much as possible the taxonomic spectrum of LAB present in raw milk cheeses.
L. lactis is a highly relevant LAB for the dairy industry due its ability to acidify and grow at different concentrations of NaCl. This species has the ability to colonize and to adapt at different niches, being found naturally in a wide variety of environments, such as vegetables (grasses and silages) and animals, mainly in raw milk from different animals [15], explaining their prevalence in artisanal cheeses. However, our results suggests that L. lactis population may be underestimated by culture-dependent methods, as stated previously due to different factors related to laboratory conditions of cultivation and the viability of microorganisms conferred by adverse environmental conditions, such as effects of fermentation, nutrient depletion, and pH variation [16]. These factors can induce the microbial cells in a stressed state characterized by the inability to produce colonies on culture media even if they are still able to perform metabolic activity [16, 17].
Considering the observed results, culture-dependent methods do not fully represent the community and the actual microbial diversity present in raw milk cheeses. The conventional protocols used for isolation, identification, and characterization of LAB will always be relevant, especially for the selection of beneficial microbes that can be applied in the food industry. To overcome such limitation in studies focused on microbial diversity, nowadays the high-throughput sequencing is considered an excellent method, being applied to different food matrix [4, 18, 19]. However, it must be considered that by using this method, DNA of all microorganism present, including potentially dead, lysed, and in non-cultivable viable bacteria can be amplified [12, 20, 21].
Combining culture-dependent and culture-independent approaches is always the best for obtaining a complete view of the existing microbial ecosystem in complex food matrix. Among culture-independent methods, high-throughput sequencing emerged as a very powerful tool to study microbial diversity in fermented foods. But it is well know that the costs of high-throughput sequencing and technical support for data analysis can still be limiting factors. Considering the results presented by Nero et al. [1] and by this complementary study, we concluded that application of rep-PCR as culture-independent method in raw milk cheeses may provide evidence of factors related to the production when other methods are unavailable. In addition, the results of PCR-DGGE matched to the main findings of high-throughput sequencing, confirming its confidence to detect the main microbial groups present in the raw milk cheeses.
Acknowledgements
The authors are thankful to the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brasília, DF, Brazil, Finance code 001), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brasília, DF, Brazil), and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG, Belo Horizonte, MG, Brazil).
Author contribution
Leticia R. Ferreira: methodology, investigation, and writing (original draft and review). Milimani Andretta and Thaiza T. Almeida: methodology and investigation. Luana M. Perin and Anderson C. Camargo: supervising, methodology, formal analysis, and writing (review and editing). Antonio F. Carvalho and Luís A. Nero: conceptualization, validation, formal analysis, resources, data curation, writing (review and editing), supervision, project administration, and funding acquisition.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nero LA, Andretta M, Almeida TT, Ferreira LR, Camargo AC, Yamatogi RS, Carvalho AF, Call DR. Lactic microbiota of the minas artisanal cheese produced in the serro region, Minas Gerais. Brazil LWT. 2021;148:111698. doi: 10.1016/J.LWT.2021.111698. [DOI] [Google Scholar]
- 2.Kamimura BA, Magnani M, Luciano WA, Campagnollo FB, Pimentel TC, Alvarenga VO, Pelegrino BO, Cruz AG, Sant’Ana AS. Brazilian artisanal cheeses: an overview of their characteristics, main types and regulatory aspects. Compr Rev Food Sci Food Saf. 2019;18:1636–1657. doi: 10.1111/1541-4337.12486. [DOI] [PubMed] [Google Scholar]
- 3.Blaya J, Barzideh Z, LaPointe G. Symposium review: interaction of starter cultures and nonstarter lactic acid bacteria in the cheese environment1. J Dairy Sci. 2018;101:3611–3629. doi: 10.3168/JDS.2017-13345. [DOI] [PubMed] [Google Scholar]
- 4.Ferrocino I, Rantsiou K, Cocolin L. Investigating dairy microbiome: an opportunity to ensure quality, safety and typicity. Curr Opin Biotechnol. 2022;73:164–170. doi: 10.1016/J.COPBIO.2021.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Beresford TP, Fitzsimons NA, Brennan NL, Cogan TM. Recent advances in cheese microbiology. Int Dairy J. 2001;11:259–274. doi: 10.1016/S0958-6946(01)00056-5. [DOI] [Google Scholar]
- 6.Carraro L, Maifreni M, Bartolomeoli I, Martino ME, Novelli E, Frigo F, Marino M, Cardazzo B. Comparison of culture-dependent and -independent methods for bacterial community monitoring during Montasio cheese manufacturing. Res Microbiol. 2011;162:231–239. doi: 10.1016/J.RESMIC.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Al-Awadhi H, Dashti N, Khanafer M, Al-Mailem D, Ali N, Radwan S. Bias problems in culture-independent analysis of environmental bacterial communities: a representative study on hydrocarbonoclastic bacteria. Springerplus. 2013;2:1–11. doi: 10.1186/2193-1801-2-369/FIGURES/5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gevers D, Huys G, Swings J. Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol Lett. 2001;205:31–36. doi: 10.1111/J.1574-6968.2001.TB10921.X. [DOI] [PubMed] [Google Scholar]
- 9.Ampe F, Ben Omar N, Moizan C, Wacher C, Guyot JP. Polyphasic study of the spatial distribution of microorganisms in Mexican pozol, a fermented maize dough, demonstrates the need for cultivation-independent methods to investigate traditional fermentations. Appl Environ Microbiol. 1999;65:5464–5473. doi: 10.1128/AEM.65.12.5464-5473.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arcuri EF, El Sheikha AF, Rychlik T, Piro-Métayer I, Montet D. Determination of cheese origin by using 16S rDNA fingerprinting of bacteria communities by PCR–DGGE: preliminary application to traditional Minas cheese. Food Control. 2013;30:1–6. doi: 10.1016/J.FOODCONT.2012.07.007. [DOI] [Google Scholar]
- 11.Cocolin L, Dolci P, Rantsiou K. Biodiversity and dynamics of meat fermentations: the contribution of molecular methods for a better comprehension of a complex ecosystem. Meat Sci. 2011;89:296–302. doi: 10.1016/J.MEATSCI.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 12.Perin LM, Savo Sardaro ML, Nero LA, Neviani E, Gatti M. Bacterial ecology of artisanal Minas cheeses assessed by culture-dependent and -independent methods. Food Microbiol. 2017;65:160–169. doi: 10.1016/J.FM.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Perin LM, Dal Bello B, Belviso S, Zeppa G, de Carvalho AF, Cocolin L, Nero LA. Microbiota of Minas cheese as influenced by the nisin producer Lactococcus lactis subsp. lactis GLc05. Int J Food Microbiol. 2015;214:159–167. doi: 10.1016/J.IJFOODMICRO.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Van Hoorde K, Verstraete T, Vandamme P, Huys G. Diversity of lactic acid bacteria in two Flemish artisan raw milk Gouda-type cheeses. Food Microbiol. 2008;25:929–935. doi: 10.1016/J.FM.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Martins MC de F, Fusieger A, Freitas R de, Valence F, Nero LA, Carvalho AF (2020) Novel sequence types of Lactococcus lactis subsp. lactis obtained from Brazilian dairy production environments. LWT 124:109146 . 10.1016/J.LWT.2020.109146
- 16.Ruggirello M, Cocolin L, Dolci P. Fate of Lactococcus lactis starter cultures during late ripening in cheese models. Food Microbiol. 2016;59:112–118. doi: 10.1016/J.FM.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Ruggirello M, Giordano M, Bertolino M, Ferrocino I, Cocolin L, Dolci P. Study of Lactococcus lactis during advanced ripening stages of model cheeses characterized by GC-MS. Food Microbiol. 2018;74:132–142. doi: 10.1016/J.FM.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 18.de Almeida TT, Andretta M, Ferreira LR, de Carvalho AF, Nero LA. The complex microbiota of artisanal cheeses interferes in the performance of enumeration protocols for lactic acid bacteria and staphylococci. Int Dairy J. 2020;109:104791. doi: 10.1016/J.IDAIRYJ.2020.104791. [DOI] [Google Scholar]
- 19.Dolci P, Alessandria V, Rantsiou K, Cocolin L (2015) Advanced methods for the identification, enumeration, and characterization of microorganisms in fermented foods. Adv Fermented Foods Beverages Improv Qual Technol Heal Benefits 157–176.10.1016/B978-1-78242-015-6.00007-4
- 20.Cocolin L, Alessandria V, Dolci P, Gorra R, Rantsiou K. Culture independent methods to assess the diversity and dynamics of microbiota during food fermentation. Int J Food Microbiol. 2013;167:29–43. doi: 10.1016/J.IJFOODMICRO.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 21.Fusco V, Quero GM. Culture-dependent and culture-independent nucleic-acid-based methods used in the microbial safety assessment of milk and dairy products. Compr Rev Food Sci Food Saf. 2014;13:493–537. doi: 10.1111/1541-4337.12074. [DOI] [PubMed] [Google Scholar]