Abstract
The acidity of soils significantly reduces the productivity of legumes mainly because of the detrimental effects of hydrogen ions on the legume plants, leading to the establishment of an inefficient symbiosis and poor biological nitrogen fixation. We recently reported the analysis of the fully sequenced genome of Rhizobium favelukesii LPU83, an alfalfa-nodulating rhizobium with a remarkable ability to grow, nodulate and compete in acidic conditions. To gain more insight into the genetic mechanisms leading to acid tolerance in R. favelukesii LPU83, we constructed a transposon mutant library and screened for mutants displaying a more acid-sensitive phenotype than the parental strain. We identified mutant Tn833 carrying a single-transposon insertion within LPU83_2531, an uncharacterized short ORF located immediately upstream from ubiF homolog. This gene encodes a protein with an enzymatic activity involved in the biosynthesis of ubiquinone. As the transposon was inserted near the 3′ end of LPU83_2531 and these genes are cotranscribed as a part of the same operon, we hypothesized that the phenotype in Tn833 is most likely due to a polar effect on ubiF transcription.
We found that a mutant in ubiF was impaired to grow at low pH and other abiotic stresses including 5 mM ascorbate and 0.500 mM Zn2+. Although the ubiF mutant retained the ability to nodulate alfalfa and Phaseolus vulgaris, it was unable to compete with the R. favelukesii LPU83 wild-type strain for nodulation in Medicago sativa and P. vulgaris, suggesting that ubiF is important for competitiveness. Here, we report for the first time an ubiF homolog being essential for nodulation competitiveness and tolerance to specific stresses in rhizobia.
Graphical abstract

Keywords: Acid tolerance, ubiF, Rhizobium favelukesii LPU83, Stress, Competitiveness
Introduction
Rhizobia are Gram-negative bacteria that live in soils and symbiotically associate with the roots of leguminous plants to fix atmospheric nitrogen and, in turn, receive carbon compounds derived from plant photosynthates. While, in general, the rhizobium-legume symbiosis shows different degrees of specificity according to the associated species, the nodulation of Medicago sativa (alfalfa) is known to be particularly restricted to Ensifer meliloti (formerly Sinorhizobium meliloti) and other few rhizobia [1–3] including the unusual and genetically closely related strains Rhizobium favelukesii Or191 (previously named Rhizobium sp. Or191) [4] and the Rhizobium favelukesii LPU83-type strain isolated in the USA and Argentina, respectively [5, 6]. However, neither of the latter strains is able to establish a full N2-fixing symbiosis on Medicago, Melilotus, Trigonella or Phaseolus vulgaris [6–8]. Some features of these alfalfa-nodulating rhizobia include acid tolerance [6]; extended host range for nodulation [9]; inefficiency of nitrogen fixation in M. sativa, Medicago truncatula and P. vulgaris; and their extremely low genetic diversity independent of their geographical origin [9, 10]. In addition, analysis of exopolysaccharides—necessary for the establishment of an effective symbiosis—in R. favelukesii LPU83 revealed an identical composition to those in E. meliloti [11]. All these characteristics, along with their remarkably successful competition with the N2-fixing symbionts for nodulation, point to this type of rhizobium as a detrimental agent in agricultural soils when it coexists with E. meliloti [5]. We have previously shown that under acidic conditions, the strain R. favelukesii LPU83 increased its competitiveness to nodulate alfalfa against the efficient symbiont E. meliloti [12]. Contrasting E. meliloti, the attachment of R. favelukesii LPU83 to alfalfa roots is almost not influenced by pH or Ca2+ concentration [13]. Although we have characterized the behaviour of these rhizobia in soil microcosms, we have, so far, no data available on the persistence and ecology of these bacteria in acidic fields, or on how the symbiosis between alfalfa and the efficient rhizobia is affected.
The low performance of alfalfa in acidic soils results from several conditions that affect the host plant, their rhizobia and the symbiotic interactions between them [14–17]. Acid tolerance of rhizobia has long been considered a phenotypic characteristic that would positively affect persistence and symbiosis under low pH. For this reason, the identification and manipulation of the genetic determinants of acid tolerance is a high-priority task towards the phenotypic improvement of rhizobial inoculants for acid soils. We and others have previously identified and characterized genes associated with tolerance to acidity (act genes) in alfalfa-nodulating rhizobia [18–23]. In addition, other factors like the production of exopolysaccharides have been pointed out as critical for tolerance to acid conditions and other stresses in E. meliloti [24, 25]. On the other hand, transcriptomic and proteomic analyses of R. favelukesii LPU83 under acid conditions revealed the differential expression of 1294 and 336 genes, respectively [11, 26]. The affected genes belonged to different categories, with a prominent number of those involved in cell envelope modification, cell permeability and reverse proton efflux. This response was consistent with the one observed in Rhizobium tropici when exposed to low pH [27] and seems to be a general bacterial response when exposed to acidic environments [28].
To gain more insight on the molecular basis of the acid tolerance in R. favelukesii LPU83, we performed transposon mutagenesis and selected acid-sensitive mutants. We focused on Tn833, which displayed a remarkable lower ability to grow on acid media. We find that the transposon was inserted within a short ORF located upstream an ubiF homolog and constructed mutants to characterize their acid tolerance, symbiosis capabilities and competitiveness in the nodulation of the host plant.
Materials and methods
Bacterial strains and media
The bacterial strains and plasmids used in this work are listed in Table 1. The rhizobia were grown at 28 °C on tryptone yeast medium [35] or yeast extract mannitol medium [36]. The screening of acid-sensitive mutants and the growth curves was performed on GS minimal medium [6]. Escherichia coli strains were grown at 37 °C on Luria–Bertani medium [37]. For the agarized media, 15 g of agar per litre of the medium was added. When required, the media were supplemented with 120 µg/ml neomycin (Nm), 400 µg/ml streptomycin (Str), 200 µg/ml ampicillin (Ap), 10 µg/ml tetracycline (Tc) for E. coli or 2 µg/ml for Rhizobium and 25 µg/ml kanamycin (Km). The plasmids were introduced into rhizobia from Escherichia coli S17-1 by biparental mating or from E. coli DH5α by triparental mating with pRK2013 as a helper plasmid.
Table 1.
Strains and plasmids used in this study
| Strains/plasmids | Description | Reference |
|---|---|---|
| Strains | ||
| R. favelukesii LPU83 | Wild-type, spontaneous Smr; is the specie’s type strain (= CECT 9014T = LMG 29160T); previously named Rhizobium sp. LPU83 | Del Papa et al. (1999) [6] |
| Tn833 | Tn5 mutant of wild-type LPU83, Smr Nmr | This study |
| LPU83_2531− | LPU83_2531 mutant in R. favelukesii LPU83, Smr Nmr | This study |
| ubiF− | ubiF mutant in R. favelukesii LPU83, Smr Nmr | This study |
| E. coli DH5 | recA1, lacU169 80dlacZ M15; host strain used in cloning experiments | Bethesda Research Laboratories |
| E. coli S17-1 | MM294, RP4-2-Tc::Mu-Km::Tn7 chromosomally integrated | Simon et al. (1991) [29] |
| E. coli TOP10 | F′ {lacIqTn10(TetR)} ncrA _(mrr-hsdRNS-ncrBC) _80lacZ_M15 _lacX74 recA1 | Invitrogen |
| Plasmids | ||
| Tn5B20 | Promoter probe transposon that carries the promoterless reporter genes lacZ and nptII as a selective marker | Simon et al. (1989) [30] |
| pCR-Topo | PCR-cloning vector Apr, Kmr, lacZ | Invitrogen |
| pK18mob | Kmr ori ColE1, Mob + , lacZ + used for directed insertional disruption | Schäfer et al. (1994) [31] |
| pK18mob-ORF1 | pK18mob with a 232-bp internal fragment of LPU83_2531 cloned as a blunt insert, Kmr | This study |
| pK18mob-ubiF | pK18mob with a 427-bp internal fragment of ubiF cloned as a blunt insert, Kmr | This study |
| pGEMT | Multicopy vector, Apr | Promega |
| pBBR1-MCS5 | Mobilizable vector, lacZalpha, Gmr | Kovach et al. (1994) [32] |
| pBBR1M CS5::ubiF | pBBR1MCS5 with ubiF cloned as a HindIII-EcoRI insert, Gmr | This study |
| pFAJ1708 | Cloning vector, Tcr | Dombrecht et al. (2001) [33] |
| pLow1 | Low-copy number plasmid, Cmr | Hansen et al. (1997) [34] |
| pLow1-833 | pLow1 plus 22-kbp KpnI fragment carrying Tn5 insertion of mutant Tn833 | This study |
Growth curves in acidic conditions
Starter cultures were grown in GS minimal medium. GS medium was supplemented with 20 mM MES buffer (2-[N-morpholino]ethanesulfonic acid) to adjust the pH to the range of 5.0 to 6.0. Twenty millimolar PIPES (piperazine-N,N′-bis(2-ethanesulfonic acid)) was added to the GS medium to maintain the pH within the range of 6.5 to 7.0. The bacterial cultures on the liquid medium were grown at 28 °C, with shaking at 250 rpm. The kinetics of bacterial growth were studied in the GS minimal medium supplemented with MES or PIPES depending on the pH of the experiment. The pH was approximated with HCl or KOH before autoclaving and adjusted more precisely after the addition of filter-sterilized vitamins and micronutrients. The pH of the cultures during bacterial growth was monitored each time that the optical density was measured.
DNA manipulations
Plasmid preparations and DNA cloning, bacterial transformations, restriction endonuclease digestions, agarose gel electrophoresis and molecular hybridization were carried out as described by Sambrook et al. [37]. Total genomic DNA was prepared as described by Simon et al. [29].
Screening for acid-sensitive mutants
Transposon mutagenesis on R. favelukesii LPU83 strain was performed using transposon Tn5-B20 [30], according to the protocol for Tn5 mutagenesis of Rhizobium sp. described by Rossbach et al. [38]. Mutants were screened for acid sensitivity by using toothpicks to streak individual colonies on GS plates at both acid and neutral pH levels. Transconjugants that grew poorly on GS medium at pH 5.0 but grew well at pH 7.0 were selected for further analysis.
PCR amplification
PCR amplifications were carried out in a total reaction volume of 25 µl containing PCR buffer (50 mM KCl, 20 mM Tris HCl, pH 8.0), 200 µM each of dNTPs, 3 mM MgCl2, 2 µl of DNA template and 1 U Taq DNA polymerase (Gibco, BRL). All amplifications were performed with an Idaho capillary air thermocycler under the following temperature conditions: an initial 1 min denaturation at 94 °C followed by 35 cycles of 94 °C for 10 s, 52 °C for 10 s and 72 °C for 30 s, and a final 1-min extension at 72 °C. PCRs were separated in 1.5% w/v agarose gels containing ethidium bromide (1 µg/ml).
DNA sequencing and sequence analysis
The characterization of the insertion site of transposon Tn5B20 was carried out by cloning a 22-kbp KpnI restriction fragment into the low-copy plasmid pLOW1 [34] digested with the same enzyme. The digestion mixtures were joined and electroporated in E. coli DH5α. The nucleotide sequence was obtained by a primer-walking strategy using specific deoxyoligonucleotides and plasmid DNA as the template. The reaction products were separated and analysed by an automatic laser–induced fluorescent DNA sequencer (Pharmacia). The sequence was compared to the complete genome of R. favelukesii strain LPU83 that is available with the following accession numbers: HG916852 (chromosome), HG916853 (pLPU83a), CBYB010000001-58 (pLPU83b), HG916854 (pLPU83c) and HG916855 (pLPU83d). All sequence data were downloaded from public databases. Sequence comparisons and alignments were performed through BLAST (www.ncbi.nlm.nih.gov/blast) and ClustalX [39] software, respectively.
Construction of R. favelukesii LPU83ubiF− and LPU83_2531− mutants
An internal region of ubiF or ORF LPU83_2531 was amplified by PCR with Pfx polymerase and DNA from strain LPU83 using primers UBIF-Fw (GGGCAATATCACGCGTTT) and UBIF-Rv (GAGTTGCGCACCTTCCTC), or orfF (GGCTTTGGCGTGCTGAT) and orfR (CCGCGAAGCTGTCGATAC) to amplify internal fragments of 427 bp and 232 bp, respectively. Each blunt PCR fragment corresponding to the gene internal sequence was cloned into vector pK18mob (KmR, lacZ) previously digested with SmaI [31]. The resulting plasmid was transferred by conjugation to strain LPU83 to yield mutants LPU83ubiF− and LPU83_2531− by site-specific insertional mutagenesis. All PCR amplifications were performed as described above except that the Pfx polymerase was used at its optimal elongation temperature of 68 °C.
All internal fragments cloned into pK18mob were confirmed by sequencing. The resulting recombinant plasmids were finally transferred by conjugation from E. coli S17-1 to strain R. favelukesii LPU83. Putative mutants that had been generated after the site-specific integration of the plasmid (single crossover) were selected by their expected Strr, Km-Nmr phenotype. After plasmid transfer from E. coli S17-1 to R. favelukesii LPU83 by conjugation, the correct plasmid integration was confirmed by PCR using M13 primers and specific oligonucleotides annealing to the flanking regions of the rhizobial genome.
Complementation of the LPU83ubiF− mutant
The ubiF gene from R. favelukesii LPU83 was PCR amplified using UBIFHindIII (TTTAAGCTTTGAAGGAGGAACGGTGCAATGAAGAC) and UBIFRevEcoRI (TTTTGAATTCATCCTGCTTGCGATCAAATC) primers and then digested with EcoRI and HindIII. The DNA fragment was ligated into the replicative vector pBBR1MCS5 (Gmr) [32] or pFAJ1708 (Tcr) [33], both having been previously digested with the same two enzymes. The resulting recombinant plasmids (pBBR1MCS5::ubiF or pFAJ1708::ubiF) were individually electroporated into E. coli DH5α. After several unsuccessful attempts to introduce the ubiF construction in E. coli cells, the DNA ligation mix was transferred by electroporation to LPU83ubiF− to not to use E. coli as a recipient strain and to search for the complemented strain, but we were not able to have positive clones.
Plant nodulation tests
Medicago sativa seeds (alfalfa, cv. Super Monarca, obtained from Instituto Nacional de Tecnología Agropecuaria, Argentina) were surface sterilized for 10 min with 20% v/v commercial bleach (NaClO concentration equivalent to 11 g active Cl2 per litre) followed by six washes with sterile distilled water. The sterilized seeds were germinated on 1.5% w/v water agar. Two- to four-day-old seedlings were transferred to gamma irradiation–sterilized plastic growth pouches (Mega Minneapolis International, Minneapolis, MN, USA) containing 10 ml of nitrogen-free Fåhraeus-modified mineral solution, pH 7.0 [40] or pH 5.6. Three days later, the primary roots were inoculated with 106 colony-forming units (CFU) of rhizobia by dripping 100 µl of a bacterial suspension onto the root from the tip towards the base. The rhizobia were obtained from log-phase yeast extract mannitol medium cultures. The plants were cultured in a growth chamber at 22 °C with a 16-h photoperiod. The CFU contained in the inoculum were estimated by plate counts. The competition was evaluated at 4 weeks post-inoculation.
Nodulation assays with common beans cv. Negro Jamapa were carried out in 200-ml plastic pots containing vermiculite and Fåhraeus mineral solution, pH 7.0 [41]. Surface-sterilized seeds were germinated on 1.5% w/v water agar, and two small seedlings were transferred to each pot. The seedlings were inoculated with ca. 107 rhizobia/pot. Plant roots were analysed for the presence of nodules 30 days after inoculation. The results are given as the average number of nodules per plant. Where present, the error bars indicate the standard deviation (σ/√n). Results are taken from a representative experiment among a set of three.
Sensitivity assays
For all assays, exponential cultures (OD600 = 1) grown in GS medium at pH 7.0 supplemented with antibiotics, when required, were centrifuged at 640 g and the cell pellet was resuspended in saline solution (0.89% w/v NaCl). An aliquot of cells was used to inoculate flasks containing GS either with or without the addition of 100 µM CuSO4, 10% w/v sucrose, 0.5 M NaCl, 500 µM ZnSO4 or 5 mM ascorbic acid. The cultures were then incubated at 28 °C and 250 rpm for 6 h or 24 h. After treatment, cultures were plated on TY media for CFU determination. CFU were compared to non-treated samples (control). Photosensitivity was tested as described by Nakahigashi et al. [42].
Results
Isolation of an acid-sensitive mutant and identification of an act gene in R. favelukesii LPU83
In order to identify novel genetic determinants of tolerance to acidity in alfalfa-nodulating rhizobia, random transposon mutagenesis was performed with transposon Tn5B20 on the acid-tolerant strain R. favelukesii LPU83. The collection of transposon mutants was selected in TY solid medium supplemented with Sm and Nm. More than 3000 mutants were streaked on GS solid medium at both pH levels 7.0 (control) and 5.0. Acid-sensitive mutants were expected to show either no growth or reduced growth in the minimal medium at pH 5.0, but near wild-type growth at pH 7.0. Mutants that had completely lost their ability to grow under acid conditions (pH 5.0) but had preserved normal growth at neutral pH were not found. We only found one mutant that displayed an impaired growth selectively under acidic conditions, Tn833, which was selected for further characterization. When mutant Tn833 was examined by Southern hybridization analysis utilizing an IS50 probe, single EcoRI- and KpnI-hybridizing signals were found that corresponded to fragments of 15 kb and 22 kb, respectively (data not shown).
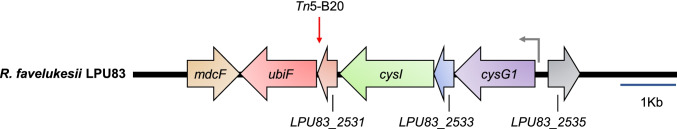
In order to identify the disrupted gene in mutant Tn833, the 22-kb KpnI restriction fragment that contained the Tn5B20 was cloned into the low-copy number plasmid pLOW1 and a 10-kb sequence of the rhizobial DNA bordering the transposon was obtained (Fig. 1). The Tn5B20 transposon was inserted close to the 3′ end of LPU83_2531, whose translation product corresponds to a hypothetical protein containing a domain of the DUF934 superfamily, identified in several rhizobia (Rhizobium leguminosarum bv. viciae, R. leguminosarum bv. trifolii, Rhizobium etli and E. meliloti, among others) and other bacteria. We identified a homolog of ubiF downstream LPU83_2531 gene. ubiF encodes a hydroxylase of 2-octaprenyl-6-methoxyphenol, an enzyme that catalyzes the sixth step in the biosynthesis of ubiquinones from the precursor chorismate under aerobic conditions [43]. Ubiquinone is required for ATP production through the electron transport chain, and its function is important to maintain proper cell respiration. ubiF is well conserved among rhizobial species, but its role in acid tolerance has not been yet investigated. The LPU83 ubiF ortholog presented the highest identity with the homologous genes present in R. etli CIAT 652 and in R. leguminosarum bv. viciae 3841, both bean-nodulating rhizobia. In R. favelukesii LPU83, LPU83_2531 and ubiF are present in a single copy in the genome. ubiF is the last gene of an operon comprising LPU83_2531 and three other genes located upstream that are cotranscribed from a single promoter (analysis performed using RNA-seq data from Nilsson et al. [44]. The genes in the operon are cysG1 that codifies for uroporphyrin III C-methyltransferase protein, cysI that codifies for putative sulfite reductase (NADPH) protein from the cysteine biosynthetic pathway, LPU83_2531 which is an open reading frame interrupted by Tn5B20, LPU83_2533 which is an open reading frame with unknown function and ubiF putative hydroxylase of 2-octaprenyl-6-methoxyphenol from the quinone biosynthetic pathway. Due to the well-known role of UbiF in proton transport and to the proximity of the transposon to the 5′ end of ubiF gene, we hypothesize that the phenotype observed in Tn833 is likely due to the disruption of this gene rather than LPU83_2531 or the disruption of both genes.
Fig. 1.
Genetic organization of the DNA region interrupted by transposon Tn5B20 insertion in the Tn833 mutant. The red arrow indicates the position where transposon was inserted (genome position 2,469,710), and the grey arrow shows the position of the predicted promoter of the operon containing LPU883_2531 (ORF1) and ubiF (genome position 2,473,610). The genome coordinates are relative to the reference genome of R. favelukesii LPU83. The genes in the region nearby are as follows: mdcF, malonate transport protein; ubiF, putative 2-polyprenyl-6-hydroxyphenol hydroxylase from the quinone biosynthetic pathway (UQ, coenzyme Q); LPU83_2531, open reading frame interrupted by Tn5B20; cysI, putative sulfite reductase (NADPH) protein from the cysteine biosynthetic pathway; cysG1, uroporphyrin III C-methyltransferase protein; LPU83_2535, unknown function
Mutants in LPU83_2531 and ubiF genes exhibit altered growth kinetics at low pH
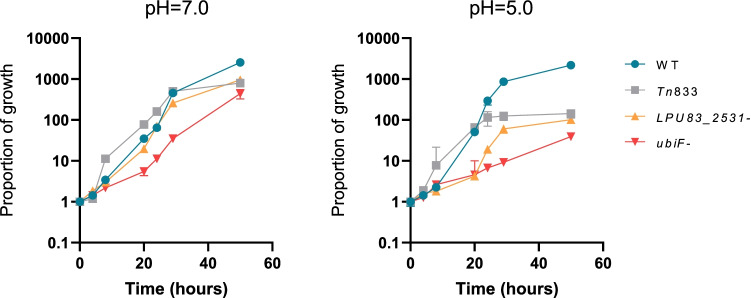
In order to evaluate the role of LPU83_2531 and UbiF in acid tolerance, mutants LPU83_2531− and ubiF−, respectively, were obtained by site-directed integration, as described in the “Materials and methods”. Both mutants displayed similar colony morphology to the WT strain in rich and minimal media. When grown in GS broth at pH 7.0, no significant differences between the WT and Tn833 and LPU83_2531 mutants were observed during the first 30 h, although ubiF− was considerably affected in this condition (Fig. 2). Incubation at pH 5.0 showed remarkable differences in growth kinetics for all mutants (Fig. 2). Interestingly, the mutants were not able to grow beyond 109 CFU/ml, and LPU83_2531− and ubiF− were also affected in log phase, being the last one more impacted (Fig. 2). Although cultures were buffered to maintain the initial pH, an increase in the pH of the extracellular medium of about 0.2 to 0.5 pH units was found under both pH conditions.
Fig. 2.
Disruption of LPU83_2531/ubiF impairs growth in acid media. Growth kinetics in neutral (pH = 7.0) or acid (pH = 5.0) GS medium for the WT and mutants are shown. The proportion of growth was calculated as the CFU at each time point relative to time zero. Error bars represent the average of at least two independent experiments. The variations in the pH throughout each experiment were no greater than 0.6 units
To complement the mutants with ubiF endogenous gene, we tried to clone the open reading frame from R. favelukesii LPU83 in different expression vectors. However, several attempts using different E. coli vectors failed, suggesting that the expression of ubiF from R. favelukesii LPU83 is toxic for E. coli. Attempts made at cloning ubiF in R. favelukesii LPU83 resulted in non-viable colonies.
LPU83_2531− and ubiF− mutants are affected in other stress conditions
Rhizobial mutants that express a higher sensitivity to hydrogen ions frequently show a lower tolerance to increased concentrations of heavy metals [45]. Such a common phenotypic pattern strongly suggests the presence of some related, but unknown yet, biochemical alterations in all mutants irrespective of their primary act genetic alteration. To investigate the response of the mutants to other stresses, rhizobia were exposed to the presence of heavy metals (100 mM Cu2+ and 0.5 mM Zn2+), osmotic stress (0.5 M NaCl or 10% w/v sucrose) and reducing conditions (5 mM ascorbic acid). Though no differences in cell viability between the mutants and the parental strain were observed in the presence of Cu2+, sucrose or NaCl (data not shown), there were differences in the other conditions. In presence of high concentrations of Zn2+, both mutants and the WT strain were importantly affected (Fig. 3). However, the impact was more severe in the mutants, particularly in ubiF−, where no CFU were detected after 24 h of incubation. On the other hand, while the WT was able to grow in the presence of ascorbic acid, the mutants were impaired to grow in this condition and CFU decreased dramatically after 24 h, being undetectable in the case of ubiF− after this time (Fig. 3). These results indicate that ubiF or LPU83_2531/ubiF genes are involved in tolerance to other stresses beyond acidity. Finally, concerning the effects of physical agents on ubi mutants, it is worth mentioning that the photosensitive phenotype described for ubiF mutants in E. coli [42] was not observed in our LPU83 derivatives.
Fig. 3.
Lack of LPU83_2531/ubiF impacts on the survival of R. favelukesii LPU83 in stressing conditions. Survival of ubiF− and LPU83_2531.− mutants and the WT strain was evaluated in the presence of high concentrations of ZnSO4 and ascorbic acid. CFU counting in medium at pH = 7.0 was used as a control. Log-phase cultures were diluted to a concentration of 107 CFU/ml and incubated at 28 °C and 280 RPM. For each condition, the number of CFU was determined after 6 h or 24 h. nd, not detected
UbiF is important for plant nodulation competitiveness
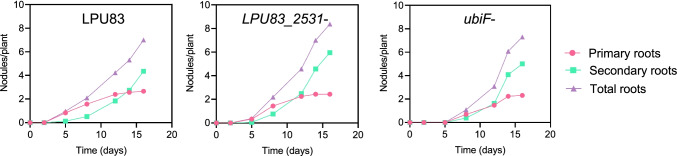
To determine if the observed decrease in acid tolerance has any effect on the rhizobial ability to associate with alfalfa, the symbiotic properties of the mutants at neutral and acidic pH levels were evaluated. We performed single-inoculation assays using alfalfa plants in a nitrogen-free medium at neutral pH and determined the number of nodules at different time points. We found that both LPU83_2531− and ubiF− achieved a similar number of nodules per plant than the WT. However, the nodulation kinetics of ubiF− were slightly different than the parental strain, showing a delay in the formation of primary roots (Fig. 4). Despite the number of nodules, a more important and desirable feature in soils is the ability to compete with other rhizobia and colonize roots. Thus, to evaluate the competitiveness of the mutants, we performed coinoculation assays infecting alfalfa and common bean plants with a similar CFU number of the WT and each of the mutant strains, as described in the “Materials and methods”. Interestingly, we found that LPU83_2531− and ubiF− were nearly fully outcompeted by the WT strain in M. sativa plants (Fig. 5) and completely outcompeted in P. vulgaris, where the entire bacterial recovery from those nodules corresponded to the WT. These results suggest that ubiF, and possibly LPU83_2531, plays an important role in competition in alfalfa and the common bean.
Fig. 4.
Alfalfa nodulation kinetics of R. favelukesii LPU83 and its derivative strains. The nodules from the principal roots (pink), secondary roots (green) and total roots (purple) were determined at different time points as indicated in the “Materials and methods”. The results, expressed as the average number of nodules per plant, are taken from a representative experiment within a set of three. The standard deviations throughout were lower than 20% in all cases
Fig. 5.
Loss of LPU83_2531/ubiF dramatically reduces the ability to compete with the WT strain. Nodulation competitiveness of LPU83_2531− (A) or ubiF− (B) mutants was evaluated against the R. favelukesii LPU83 WT strain in alfalfa plants. The results are presented as the mean value of the percentage of nodules occupied by each strain coinoculated from a 1:1 mixture. The occupancy of each strain in the nodules was determined by differential growth on TY solid plates supplemented with selective antibiotics
Discussion
Acid soils are seen as problematic in the matter of plant cultivation as acidity impacts both the plant and the associated rhizobia, having negative consequences in crop yield [17]. To address this issue, we and others have directed our research towards the identification of acid-tolerant rhizobia [6, 46–48]. On the other hand, the study of the genes and mechanisms underlying acid tolerance in these bacteria is critical to improve bioinoculants and to have better outcomes. In this regard, here, we performed random transposon mutagenesis on the acid-tolerant strain R. favelukesii LPU83 to identify novel determinants of acid tolerance. By screening on solid acid media, we found a mutant, Tn833, which was severely affected on its ability to grow at pH 5.0 but subtly disturbed in neutral conditions and further characterized its phenotype.
It is important to note that out of more than three thousand Tn5-screened mutants, only one clone appeared to be specifically affected in their growth at low pH without significant changes in the ability to grow in neutral culture media. Mutants that had completely lost their ability to grow under acidic conditions but grew well at neutral pH were not found, reinforcing the concept that tolerance to acidity in rhizobia is a multigenic phenotype that is related to the general bacterial physiology [16, 44, 49]. Although the transposon in Tn833 was inserted within the ORF of the unknown function LPU83_2531, the acid-tolerant phenotype in the mutant is probably due to the effect on the downstream gene, ubiF, cotranscribed with LPU83_2531 [44]. Since LPU83_2531− is the upstream gene in a probable operon, the polarity of a plasmid insertion in LPU83_2531− might decrease the expression of ubiF and this proposed inhibition could contribute to the phenotype of the both mutants (Tn833 and LPU83_2531−). UbiF is part of the aerobic biosynthetic pathway of ubiquinone, a key component of the electron transport chain that plays a key role in energy-generating processes in bacteria [50]. Then, it is reasonable to think that ubiquinone plays a role in acid tolerance, considering its implication in H+ transfer through the plasma membrane. Disruption of its function could lead to a malfunction of the electron transport chain and an imbalance of protons between the cytosol and the outer membrane space. In agreement with that, it was found that an ubiF mutant in E. coli was more susceptible to grow under acidic conditions and other stresses [51], supporting our findings.
As an alternative pathway for the biosynthesis of ubiquinone under anaerobic conditions, E. coli uses three other hydroxylases along with the oxygen atoms of water [52, 53]. In Rhizobium, neither the structure of the ubiquinone biosynthetic route nor the presence or absence of an alternative pathway has been determined. Because of the natural requirement of oxygen in rhizobia, a complete blockage of ubiquinone production should not be possible (only a few of rhizobia may use menaquinone as an electron carrier under symbiosis condition, as this quinone is characteristic of obligate anaerobes). The increased concentrations of hydrogen ions, heavy metals and osmotically active compounds in the extracellular medium are all factors that generate biochemical stress to the bacterial cell via different mechanisms. Proper preservation of the redox balance is crucial in rhizobia to adapt to the extracellular acidity [54]. Considering the phenotype of the mutant described in this work, these alterations are likely a result of primary changes in the biosynthetic route of ubiquinone. Although the quinone pool is often considered a simple group of reduction-equivalent carriers among membrane-protein complexes, these compounds are now clearly known to participate in defence mechanisms against oxidative stress [43] and are critical to preserving the regular activity of multiple cellular processes.
By using a transcriptomic approach, an increased expression of an ubiF homolog has been observed in S. meliloti in the presence of NO [55]. We show here that the alteration in ubiF strongly impacts bacterial behaviour under several stressful conditions and during the interaction with the host plant. Concerning the participation of quinones in the phenomena of tolerance to particular stress situations, a pleiotropic phenotype in ubiCA mutants has been observed, in which, for example, the tolerance to temperature is increased: interestingly enough, those mutants are likewise more resistant to phleomycin compounds and linoleic acid [43]. Consistent with our observations, the analysis of ubiCA mutants of E. coli also showed that these strains were highly sensitive to the action of the reducing agent dithiothreitol, possibly as a consequence of the incapacity of the altered respiratory chain to maintain the redox balance within the periplasm [43]. Interestingly, in our case, LPU83_2531− and ubiF− mutants did not show any significant difference in their tolerance to the oxidative stress generated by 400 µM H2O2 (data not shown). Such observation weakens the possibility that the low competitiveness for nodulation displayed by the mutants is due to a reduced antioxidant ability necessary to face the peroxides produced by the plant during rhizobial root infection [56–58]. However, it cannot be ruled out that during the development of symbiosis, rhizobia could be exposed to other stresses (i.e. low pH) for which the ubi mutants are known to be impaired.
The identification of ubiF as a novel act gene in the acid-tolerant strain R. favelukesii LPU83 presented here is an important contribution towards unravelling the mechanisms underlying acid tolerance in rhizobia. Future experiments should be directed to explore whether the stressing conditions that disturb the nodulation competitiveness of these mutants operate initially in the rhizosphere, subsequently in the rhizoplane, and/or after the bacterium enters the host during infection thread development and also to determine how low pH, Zn2+ and ascorbic acid sensitivity could be connected.
Acknowledgements
The authors would like to honour Prof. Gabriel Favelukes and Prof. Oscar Grau who made valuable contributions to the development of the Institute of Biotechnology and Molecular Biology.
Author contribution
MCM, WOD, MJL, MP and CV performed the experiments, and GATT carried out the molecular data analysis. AL and MFDP designed the experiments. MCM and MFDP wrote the manuscript. All authors revised and contributed to the manuscript.
Funding
This work was supported by the National Scientific and Technical Research Council of Argentina (Consejo Nacional de Investigaciones Científicas y Técnicas – CONICET, Argentina) (Grant/Award PIP2015-0700) and the Ministry of Science, Technology and Productive Innovation (Ministerio de Ciencia, Tecnolología e Innovación Productiva – MinCyT, Argentina) (Grants/Awards PICT2017-2833 and PICT2017-2371). MCM and CV were supported by fellowships from CONICET. GTT, WOD, ML, MP, AL and MFDP are researchers at CONICET.
Data Availability
Not applicable.
Code availability
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cloutier J, Serge L, Yves C, Hani A (1996) Characterization and mutational analysis of nodHPQ genes of Rhizobium sp. strain N33. Mol Plant Microbe Interact Vol 9, 720–728. 10.1094/mpmi-9-0720, https://pubmed.ncbi.nlm.nih.gov/8870271/ [DOI] [PubMed]
- 2.Barran LR, Bromfield ESP, Brown DCW. Identification and cloning of the bacterial nodulation specificity gene in the Sinorhizobium meliloti–Medicago laciniata symbiosis. Can J Microbiol. 2002;48:765–771. doi: 10.1139/w02-072. [DOI] [PubMed] [Google Scholar]
- 3.Villegas MDC, et al. Nitrogen-fixing sinorhizobia with Medicago laciniata constitute a novel biovar (bv. medicaginis) of S. meliloti. Syst Appl Microbiol. 2006;29:526–538. doi: 10.1016/j.syapm.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Tejerizo GT, et al. Rhizobium favelukesii sp. nov., isolated from the root nodules of alfalfa (Medicago sativa L) Int J System Evol Microbiol. 2016;66(11):4451–4457. doi: 10.1099/ijsem.0.001373. [DOI] [PubMed] [Google Scholar]
- 5.Eardly BD, David B Hannaway, Bottomley P (1985) Characterization of Rhizobia from ineffective alfalfa nodules: ability to nodulate bean plants [Phaseolus vulgaris (L.) Savi.] t. 50, 1422–1427. 10.1128/aem.50.6.1422-1427.1985 [DOI] [PMC free article] [PubMed]
- 6.MF, del P, et al. Isolation and characterization of alfalfa-nodulating rhizobia present in acidic soils of central argentina and uruguay. Appl Environ Microbiol. 1999;65:1420–1427. doi: 10.1128/AEM.65.4.1420-1427.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eardly B. Phylogenetic position of Rhizobium sp. strain Or 191, a symbiont of both Medicagosativa and Phaseolus vulgaris, based on partial sequences of the 16S rRNA and nifH genes. Appl Environ Microbiol. 1992;58:1809–1815. doi: 10.1128/aem.58.6.1809-1815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Del Papa MF, et al. Identification and characterization of a nodH ortholog from the alfalfa-nodulating Or191-like rhizobia. Mol Plant Microbe Interact. 2007;20:138–145. doi: 10.1094/MPMI-20-2-0138. [DOI] [PubMed] [Google Scholar]
- 9.Wegener C et al (2001) Genetic uniformity and symbiotic properties of acid-tolerant alfalfa-nodulating rhizobia isolated from dispersed locations throughout Argentina. Symbiosis 30, 141–162. https://dalspace.library.dal.ca/bitstream/handle/10222/77860/VOLUME%2030-NUMBERS%202-3-2001-PAGE%20141.pdf?sequence=1
- 10.Tabares-da RS, et al. Rhizobia inoculants for alfalfa in acid soils: a proposal for Uruguay. Inoculantes rizobianos para alfalfa en suelos ácidos: una propuesta para Uruguay. Agrociencia Uruguay. 2019;23:1–13. doi: 10.31285/agro.23.120. [DOI] [Google Scholar]
- 11.Castellani LG, et al. Exopolysaccharide characterization of Rhizobium favelukesii LPU83 and its role in the symbiosis with alfalfa. Front Plant Sci. 2021;12:1–17. doi: 10.3389/fpls.2021.642576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Del Papa MF, et al. A microcosm study on the influence of pH and the host-plant on the soil persistence of two alfalfa-nodulating rhizobia with different saprophytic and symbiotic characteristics. Biol Fertil Soils. 2003;39:112–116. doi: 10.1007/s00374-003-0690-6. [DOI] [Google Scholar]
- 13.Soto María, José; Pieter van Dillewijn, F. M. ınez-A. & José I. Jiménez-Zurdo, N. T Attachment to plant roots and nod gene expression are not affected by pH or calcium in the acid-tolerant alfalfa-nodulating bacteria. FEMS Microbiol Ecol. 2004;48:71–77. doi: 10.1016/j.femsec.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 14.Brockwell J, Pilka A, Holliday RA (1991) Soil pH is a major determinant of the numbers of naturally occurring Rhizobium meliloti in non-cultivated soils in central New South Wales. Aust J Exp Agric 1. 10.1071/EA9910211
- 15.Graham PH. Stress tolerance in Rhizobium and Bradyrhizobium, and nodulation under adverse soil conditions. Can J Microbiol. 1992;38:475–484. doi: 10.1139/m92-079. [DOI] [Google Scholar]
- 16.Glenn AR, Dilworth MJ. The life of root nodule bacteria in the acidic underground. FEMS Microbiol Lett. 1994;123:1–10. doi: 10.1111/j.1574-6968.1994.tb07193.x. [DOI] [Google Scholar]
- 17.Ferguson BJ, Lin M, Gresshoff PM. Regulation of legume nodulation by acidic growth conditions. Plant Signal Behav. 2013;8(3):1–5. doi: 10.4161/psb.23426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goss TJ, O’Hara GW, Dilworth MJ, Glenn AR. Cloning, characterization, and complementation of lesions causing acid sensitivity in Tn5-induced mutants of Rhizobium meliloti WSM419. J Bacteriol. 1990;172:5173–5179. doi: 10.1128/jb.172.9.5173-5179.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tiwari RP, Reeve WG, Gleenn AR. Mutations conferring acid sensitivity in the acid-tolerant strains Rhizobium meliloti WSM419 and Rhizobium leguminosarum biovar viciae WSM710. FEMS Microbiol Lett. 1992;100:107–112. doi: 10.1111/j.1574-6968.1992.tb14027.x. [DOI] [Google Scholar]
- 20.Tiwari RP, Reeve WG, Dilworth MJ, Glenn AR. Acid tolerance in Rhizobium meliloti strain WSM419 involves a two-component sensor-regulator system. Microbiology. 1996;142(Pt 7):1693–1704. doi: 10.1099/13500872-142-7-1693. [DOI] [PubMed] [Google Scholar]
- 21.Tiwari RP, Reeve WG, Dilworth MJ, Glenn AR. An essential role for actA in acid tolerance of Rhizobium meliloti. Microbiology. 1996;142:601–610. doi: 10.1099/13500872-142-3-601. [DOI] [PubMed] [Google Scholar]
- 22.Kiss E, Huguet T, Poinsot V, Batut J. The typA gene is required for stress adaptation as well as for symbiosis of Sinorhizobiummeliloti 1021 with certain Medicago truncatula lines. Mol Plant Microbe Interact. 2004;17:235–244. doi: 10.1094/MPMI.2004.17.3.235. [DOI] [PubMed] [Google Scholar]
- 23.Albicoro FJ, et al. The two-component system ActJK is involved in acid stress tolerance and symbiosis in Sinorhizobiummeliloti. J Biotechnol. 2021;329:80–91. doi: 10.1016/j.jbiotec.2021.01.006. [DOI] [PubMed] [Google Scholar]
- 24.Hawkins JP, Geddes BA, Oresnik IJ. Succinoglycan production contributes to acidic pH tolerance in Sinorhizobiummeliloti Rm1021. Mol Plant Microbe Interact. 2017;30:1009–1019. doi: 10.1094/MPMI-07-17-0176-R. [DOI] [PubMed] [Google Scholar]
- 25.Primo ED, et al. Exopolysaccharide production in Ensifer meliloti laboratory and native strains and their effects on alfalfa inoculation. Arch Microbiol. 2019 doi: 10.1007/s00203-019-01756-3. [DOI] [PubMed] [Google Scholar]
- 26.Nilsson JF, et al. Proteomic analysis of Rhizobium favelukesii LPU83 in response to acid stress. J Proteome Res. 2019 doi: 10.1021/acs.jproteome.9b00275. [DOI] [PubMed] [Google Scholar]
- 27.Guerrero-Castro J, Lozano L, Sohlenkamp C. Dissecting the acid stress response of Rhizobium tropici CIAT 899. Front Microbiol. 2018;9:846. doi: 10.3389/fmicb.2018.00846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guan N (2020) Microbial response to acid stress: mechanisms and applications. Appl Microbiol Biotechnol 51–65. 10.1007/s00253-019-10226-1 [DOI] [PMC free article] [PubMed]
- 29.Simon R, Hotte B, Klauke B, Kosier BOB. Isolation and characterization of insertion sequence elements from gram-negative bacteria by using new broad-host-range, positive selection vectors. J Bacteriol. 1991;173:1502–1508. doi: 10.1128/jb.173.4.1502-1508.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon R, Quandt J, Klipp W. New derivatives of transposon tn5 suitable for mobilization of replicons, generation of operon fusions and induction of genes in Gram-negative bacteria. Gene. 1989;80:161–169. doi: 10.1016/0378-1119(89)90262-X. [DOI] [PubMed] [Google Scholar]
- 31.Schäfer A, et al. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 32.Kovach ME, Phillips RW, Elzer PH, Roop RM, Peterson KM (1994) pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16, 800–802 . https://pubmed.ncbi.nlm.nih.gov/8068328/ [PubMed]
- 33.Dombrecht B, Vanderleyden J, Michiels J. Stable RK2-derived cloning vectors for the analysis of gene expression and gene function in gram-negative bacteria. Mol Plant-Microbe Interact. 2001;14:426–430. doi: 10.1094/MPMI.2001.14.3.426. [DOI] [PubMed] [Google Scholar]
- 34.Hansen LH, Sørensen SJ, Jensen LB. Chromosomal insertion of the entire Escherichia coli lactose operon, into two strains of Pseudomonas, using a modified mini-Tn5 delivery system. Gene. 1997;186:167–173. doi: 10.1016/S0378-1119(96)00688-9. [DOI] [PubMed] [Google Scholar]
- 35.Behringer JE. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- 36.Vincent JM (1970) A manual for the practical study of root nodule bacteria. (Blackwell, Oxford and Edinburgh. 10.1002/jobm.19720120524
- 37.Sambrook J, Fritsch ER, Maniatis T. Molecular cloning: a laboratory manual. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Rossbach S, Kulpa DA, Rossbach U, de Bruijn FJ. Molecular and genetic characterization of the rhizopine catabolism (mocABRC) genes of Rhizobium meliloti L5–30. MGG Mol Gen Genet. 1994;245:11–24. doi: 10.1007/BF00279746. [DOI] [PubMed] [Google Scholar]
- 39.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lodeiro Aníbal R, López-garcía SL, Vázquez TEE, Favelukes G. Stimulation of adhesiveness, infectivity, and competitiveness for nodulation of Bradyrhizobiumjaponicum by its pretreatment with soybean seed lectin. FEMS Microbiol Lett. 2000;188:177–184. doi: 10.1111/j.1574-6968.2000.tb09190.x. [DOI] [PubMed] [Google Scholar]
- 41.Fähraeus G. The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J Gen Microbiol. 1957;16:374–381. doi: 10.1099/00221287-16-2-374. [DOI] [PubMed] [Google Scholar]
- 42.Nakahigashi K, Miyamoto K, Nishimura K, Inokuchi H. Isolation and characterization of a light-sensitive mutant of Escherichia coli K-12 with a mutation in a gene that is required for the biosynthesis of ubiquinone. J Bacteriol. 1992;174:7352–7359. doi: 10.1128/jb.174.22.7352-7359.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Søballe B, Poole RK. Microbial ubiquinones: multiple roles in respiration, gene regulation and oxidative stress management. Microbiology. 1999;145:1817–1830. doi: 10.1099/13500872-145-8-1817. [DOI] [PubMed] [Google Scholar]
- 44.Nilsson JF et al (2021) Global transcriptome analysis of Rhizobium favelukesii LPU83 in response to acid stress. 1–16. 10.1093/femsec/fiaa235 [DOI] [PubMed]
- 45.Reeve WG, Tiwari RP, Wong CM, Dilworth MJ, Glenn AR. The transcriptional regulator gene phrR in Sinorhizobiummeliloti WSM419 is regulated by low pH and other stresses. Microbiology. 1998;144:3335–3342. doi: 10.1099/00221287-144-12-3335. [DOI] [PubMed] [Google Scholar]
- 46.Shamseldin A, Werner D. High salt and high pH tolerance of new isolated Rhizobium etli strains from Egyptian soils. Curr Microbiol. 2005;50:11–16. doi: 10.1007/s00284-004-4391-7. [DOI] [PubMed] [Google Scholar]
- 47.Bahena MHR, Salazar S, Velázquez E, Laguerre G, Peix A. Characterization of phosphate solubilizing rhizobacteria associated with pea (Pisum sativum L.) isolated from two agricultural soils. Symbiosis. 2015;67:33–41. doi: 10.1007/s13199-015-0375-6. [DOI] [Google Scholar]
- 48.Oliveira DP, et al. Acid tolerant Rhizobium strains contribute to increasing the yield and profitability of common bean in tropical soils. J Soil Sci Plant Nutr. 2017;17:922–934. doi: 10.4067/S0718-95162017000400007. [DOI] [Google Scholar]
- 49.Chen L, James LP, Helmann JD. Metalloregulation in Bacillus subtilis: isolation and characterization of two genes differentially repressed by metal ions. J Bacteriol. 1993;175:5428–5437. doi: 10.1128/jb.175.17.5428-5437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aussel L, et al. Biosynthesis and physiology of coenzyme Q in bacteria. Biochim Biophys Acta - Bioenerg. 2014;1837:1004–1011. doi: 10.1016/j.bbabio.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 51.Ma C, et al. Energy production genes sucB and ubiF are involved in persister survival and tolerance to multiple antibiotics and stresses in Escherichia coli. FEMS Microbiol Lett. 2010;303:33–40. doi: 10.1111/j.1574-6968.2009.01857.x. [DOI] [PubMed] [Google Scholar]
- 52.Alexander K, Young IG. Alternative hydroxylases for the aerobic and anaerobic biosynthesis of ubiquinone in Escherichia coli. Biochemistry. 1978;17:4750–4755. doi: 10.1021/bi00615a024. [DOI] [PubMed] [Google Scholar]
- 53.Kwon O, Hudspeth ME, Meganathan R. Anaerobic biosynthesis of enterobactin Escherichia coli: regulation of entC gene expression and evidence against its involvement in menaquinone (vitamin K2) biosynthesis. J Bacteriol. 1996;178:3252–3259. doi: 10.1128/jb.178.11.3252-3259.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riccillo PM, et al. Glutathione is involved in environmental stress responses in Rhizobium tropici, including acid tolerance. J Bacteriol. 2000;182:1748–1753. doi: 10.1128/JB.182.6.1748-1753.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meilhoc E, Cam Y, Skapski A, Bruand C. The response to nitric oxide of the nitrogen-fixing symbiont Sinorhizobiummeliloti. Mol Plant Microbe Interact. 2010;23:748–759. doi: 10.1094/MPMI-23-6-0748. [DOI] [PubMed] [Google Scholar]
- 56.Santos R, Herouart D, Sigaud S, Touati D, Puppo A. Oxidative burst in alfalfa-Sinorhizobiummeliloti symbiotic interaction. Mol Plant Microbe Interact. 2007;14:86–89. doi: 10.1094/MPMI.2001.14.1.86. [DOI] [PubMed] [Google Scholar]
- 57.Rubio MC, et al. Localization of superoxide dismutases and hydrogen peroxide in legume root nodules. Mol Plant Microbe Interact. 2004;17:1294–1305. doi: 10.1094/MPMI.2004.17.12.1294. [DOI] [PubMed] [Google Scholar]
- 58.Chang C, Damiani I, Puppo A, Frendo P. Redox changes during the legume–Rhizobium symbiosis. Mol Plant. 2009;2:370–377. doi: 10.1093/mp/ssn090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Not applicable.