Abstract
During sporulation, Bacillus thuringiensis produces intracellular, crystalline inclusions comprised of a mixture of protoxins active on insect larvae. A major class of these protoxin genes, designated cry1, is transcribed from two overlapping promoters (BtI and BtII) utilizing RNA polymerase containing sporulation sigma factors ςE and ςK, respectively. Fusions of these promoters to lacZ were constructed in order to analyze transcription patterns. Mutations within the −10 region of the BtII promoter (within the spacer region of the BtI promoter) which departed from the consensus −10 sequence for either ςE or ςK resulted in inactivation of transcription from BtII and a fivefold stimulation of transcription from BtI. In contrast, transcription from both promoters was inhibited with a change to the ςE consensus. One of the “promoter-up” mutations was fused to the cry1Ac1 gene, and enhanced transcription was confirmed by Northern blotting. There was an increase in the accumulation of Cry1Ac antigen at early but not later times in sporulation in the mutant. This shift was due to the rapid turnover of much of the excessively accumulated protoxin at the early times as measured by pulse-chase labeling. As a result of the turnover and the inactivation of the BtII promoter, the mutant produced smaller inclusions which contained two- to threefold-less protoxin than inclusions from the wild type. Promoter overlap is a mechanism for modulating protoxin synthesis, thus ensuring the efficient packaging of these protoxins into inclusions.
Bacillus thuringiensis is unique in its capacity to produce, primarily during sporulation, large, intracellular, crystalline inclusions comprised of protoxins active on insect larvae (4, 20, 38). Most B. thuringiensis subspecies contain multiple, plasmid-borne protoxin genes (4, 20). Related protoxins are present in the same inclusion, but the relative amounts differ as determined from steady-state mRNA levels (3) and protoxin contents (27, 28). Since each of these toxins has a unique specificity profile, usually for a subset of insects from within a particular order (20), the presence of several protoxins in different amounts is important for the overall toxicity profile of an isolate. There may also be synergistic interactions between specific toxins (23, 42).
In addition to the differential regulation of the various protoxin genes, there must be mechanisms to ensure the synthesis of very large amounts of the protoxins and their orderly deposition into a crystalline inclusion. The latter process involves extensive intermolecular disulfide cross-linking (9), and there are probably enzymes to aid this process as well as chaperones and/or protective proteins (2, 8) for the initial assembly process.
A very abundant class of protoxin genes designated cry1 all have very similar overlapping promoters, designated BtI and BtII, which function primarily at different times during sporulation (2, 8, 20). Transcription from the former is dependent upon a form of RNA polymerase containing a sigma factor with 88% identity to Bacillus subtilis ςE, whereas transcription from BtII utilizes a sigma factor with 85% identity to ςK from B. subtilis (1).
In B. subtilis, these sigma factors are necessary for the transcription of sporulation genes expressed in the mother cell (15, 26). There are examples of tandem promoters in sporulating B. subtilis cells which ensure extended transcription during sporulation (17) or during both growth and sporulation (12, 13). A few sporulation genes contain overlapping promoters (21, 43), but the significance, if any, of such an arrangement has not been studied. In B. thuringiensis, ςE and ςK must function for the transcription of mother cell sporulation genes as well as for the protoxin genes, a conclusion supported by in vitro studies (1).
Since this prevalent class of cry1 protoxin genes contains two promoters that overlap, the function of such a unique arrangement in regulation was examined by constructing fusions of wild-type and mutated promoter regions to lacZ. Mutations of the −10 region of the BtII promoter (within the spacer region of the BtI promoter) which departed from the consensus sequence resulted in stimulation of transcription from BtI, indicating a modulating function for this promoter arrangement. This regulation was critical for the rate of accumulation of protoxins and ultimately their deposition in an inclusion.
MATERIALS AND METHODS
Bacterial strains and growth.
Escherichia coli DH5α was the host for plasmid constructs, and E. coli CJ236 was the host for the preparation of uracil-containing DNA and for the propagation of helper phage R408 (Promega). B. thuringiensis strain 80-21 was used for the introduction of the lacZ fusions by electroporation (39). This strain had spontaneously lost the 44-mDa plasmid containing the cry1Ab3 gene from Bacillus thuringiensis subsp. kurstaki HD1 but still contained the cry1Aa1, cry1Ac1, cry2Aa1, and cry2Ab1 genes (5). B. thuringiensis subsp. kurstaki HD73 contains only the cry1Ac1 gene and was used for reverse transcriptase mapping.
A clone of the cry1Ac1 gene in pHT3101 (3, 24) was electroporated into strain CryB (an acrystalliferous, plasmid-cured derivative of B. thuringiensis subsp. kurstaki HD1) (41) to form strain CryB/pHT3101-1Acwt (wild type). CryB/pHT3101 was the control strain. The 242-bp NsiI promoter fragment from mutant 272 (see below) was cloned into pHT3101-1Acwt which had been digested with NsiI to remove the wild-type promoters. The resulting plasmid, pHT3101-1Ac272, contained the mutant promoters as demonstrated by the additional SpeI site (see below). The orientation of the promoters was established by digestion with HpaI, and the promoter region was sequenced to confirm the construct. This plasmid was also electroporated into strain CryB to create CryB/pHT3101-1Ac272.
E. coli was grown at 37°C in Luria-Bertani medium (35) with 50 μg of ampicillin per ml for selection. B. thuringiensis strains were grown at 30°C in G-Tris medium (6). This medium was supplemented with 7 μg of chloramphenicol per ml for selection of cells containing lacZ fusions and 10 μg of erythromycin per ml for those containing the cry1Ac1 clones. Growth was monitored in a Klett colorimeter (660-nm-wavelength filter). Cells clump at the end of growth (T0) due to the accumulation of organic acids in the medium, and phase-dull endospores are usually visible about 3 h later (T3), with phase-white endospores present 2 to 3 h after this time (T6).
Plasmid constructs.
A 242-bp NsiI fragment extending from nucleotides −150 to +91 relative to the start site of transcription for BtI contained the promoter region of the cry1Ab3 gene (7). This fragment was cloned into the NsiI site in plasmid pGEM-llZf(+) from Promega (Fig. 1) for site-directed mutagenesis (36). It should be noted that this region of the cry1Ab3 gene is identical in sequence to that of the cry1Ac1 gene (M. Geiser, personal communication). The latter was chosen for measuring protoxin synthesis because the protoxin is more stable (31). The 251-bp XbaI/HindIII fragment from this clone was also subcloned into M13 for mutagenesis. Single-stranded DNA was produced in E. coli CJ236 with helper phage R408 (Promega) for pGEM. The −10 region of each promoter was mutagenized by employing gel-purified and phosphorylated primer 271 (3′TACTCAGTCGACACAATTTAAC) for BtI and primer 272 (3′TAAAAAAGGCTTCTGATCAGTATA) for BtII by using pGEM. The following primers were all used with M13: primer 1076 (3′CGTAAAAAAGTACTCTACTCAGTA) for BtII, primer 1077 (3′CGTAAAAAAGTATGTTACTCAGTA) for BtII, and primer 1078 (3′GTATTCTACTGAGTATACAA) and primer 1079 (3′CGTAAAATAGTATTCTAC) for changes in the BtI spacer outside of the −10 region. The mutations are summarized in Table 1.
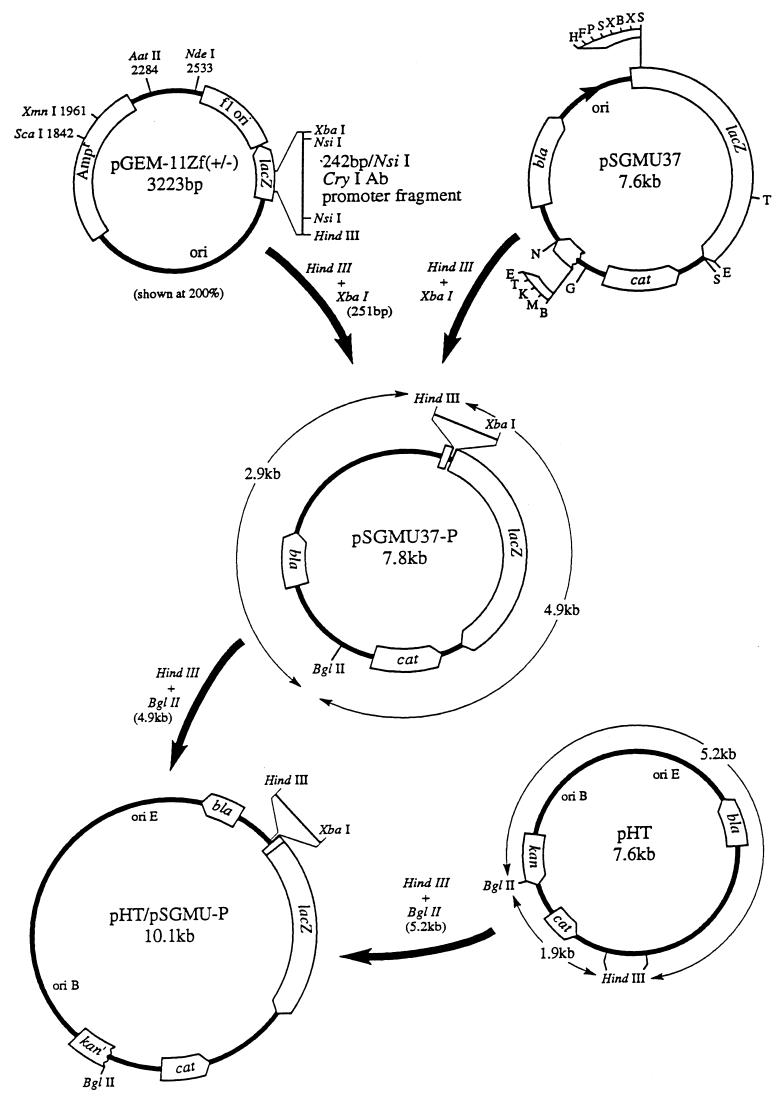
FIG. 1.
Construction of an E. coli-B. thuringiensis shuttle vector with a fragment containing the cry1A BtI and BtII promoters inserted upstream of the lacZ gene. See Materials and Methods for further discussion. Letters for pSGMU37 are restriction enzyme sites as described previously (14).
TABLE 1.
Effects of mutations in the spacer region of the overlapping promoters on transcription of lacZ fusions
| Region or mutation | Sequencea
|
Maximum units of β-galactosidase activityb | ||
|---|---|---|---|---|
| BtI (−35) | BtII (−10) | BtI (−10) | ||
| Region | ||||
| Promoter | GCATTTT T | T CATAAGATG AGT | CATATGTT | |
| B. subtilis ςEc | G/TC/AATATT | CATACA-T | ||
| B. subtilis ςKc | CATA-------Ta | |||
| Mutationd | ||||
| Wild type (both) | 1,100 | |||
| 271 (only BtII) | CGCATGTG | 300 | ||
| 272 (only BtI) | CGCAAGACT | 5,400 | ||
| 1076 (only BtI) | CATGAGATG | 5,200 | ||
| 1077 (both) | CATACAATG | 100 | ||
| 1078 (both) | CATAAGATG AC | 950 | ||
| 1079 (both) | A | T CATAAGATG | 900 | |
Twelve nanomoles of single-stranded DNA template was mixed with 32 pmol of each of the mutagenic primers and 3.5 pmol of each of the M13 primers. Following incubation at 80°C for 5 min and cooling to 27°C for 20 min, 4 U of Klenow fragment and 5 U of T4 DNA ligase were added for 20 h at 16°C. The DNA was precipitated with 2 volumes of ethanol and dissolved in 5 μl of Tris-EDTA (TE) buffer for transformation of E. coli DH5α. Clones with the desired mutations were identified by the introduction of unique restriction sites for the primer 271 (PvuII), 272 (SpeI), and 1076 (BspHI) oligonucleotide-primed reactions. All of the mutated promoters were sequenced (37) to verify the changes as shown in Table 1 and Fig. 2.
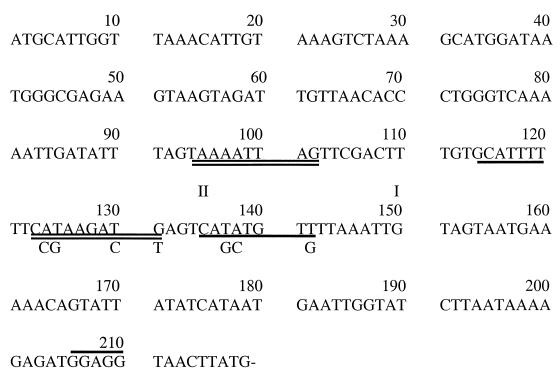
FIG. 2.
Sequence of the region upstream of the cry1Acl gene from the NsiI site to the ATG initiation codon. The −10 and −35 regions of the dual overlapping promoters are singly (BtI) or doubly (BtII) underlined, with the base substitutions in the −10 regions for the 271 (BtI) and 272 (BtII) oligonucleotides indicated below each. The start sites of transcription are marked (I and II), and the ribosome binding site is overlined. The sequence of an additional 900 bp upstream of the cry1A genes is available (GenBank accession number AF039908).
The 251-bp HindIII/XbaI wild-type and mutant promoter regions were excised and purified from low-melting-point agarose with GELase TM (Epicentre Technologies). These fragments were then subcloned into pSGMU37 (Fig. 1), a vector containing a promoterless lacZ gene (15). An E. coli-B. thuringiensis shuttle vector containing the promoter-lacZ fusions was then constructed by ligating purified HindIII/BglII fragments from pSGMU37-P and pHT to form pHT/pSGMU-P. Plasmid pHT is a shuttle vector consisting of plasmid pBD12 (from a gram-positive organism) with a nonessential PvuII fragment deleted (19) ligated to PvuII-digested pUC18 (H. Tsai and A. Aronson, unpublished results).
Enzyme assays and primer extension.
B. thuringiensis strains containing lacZ fusions were grown at 30°C in G-Tris plus 7 μg of chloramphenicol per ml. The optical density was monitored (Klett colorimeter with a 660-nm-wavelength filter), and 1-ml samples were removed periodically during growth and sporulation. Stages of sporulation were monitored in the phase microscope and the percent phase-white endospores was recorded. Cells were pelleted by centrifugation for 5 min in an Eppendorf microcentrifuge, and the pellets were frozen at −70°C. Samples were thawed and assayed in duplicate for β-galactosidase (18), and the average values (+/−6%) are reported as Miller units (30).
RNA was prepared (7) at various times during sporulation from cells grown as described above. For primer extension of promoters fused to lacZ, a 19-mer (3′CGGCGTAGATCTCAGCTGG) complementary to nucleotides 34 to 52 in the lacZ gene was purified on a Sephadex G-25 column. Oligonucleotide 5′GGTTACTTAAACAATTATAAGG (complementary to nucleotides 34 to 55 in the cry1Ac1 gene) was used for primer extension of cry1Ac1 RNA from strain 80-21 and B. thuringiensis subsp. kurstaki HD73. The oligonucleotides were labeled with [32P]ATP (6,000 Ci mmol−1) using T4 polynucleotide kinase (36). The labeled primers were mixed with 30 μg of RNA, annealed at 37°C, and extended with reverse transcriptase (16, 36). These primers were also used for the sequencing reactions.
For Northern hybridizations (36), 20 μg of RNA was electrophoresed in a sodium dodecyl sulfate (SDS)–6% polyacrylamide gel. The gel was examined under UV light to establish that the 16S and 23S rRNAs were intact. The RNA was then transferred to a polyvinylidene difluoride membrane and hybridized with a 32P-oligonucleotide specific to the cry1Ac1 gene (3), 5′TACCCCAATTAACGTTGAGGTGAATCGGGG (nucleotides 1609 to 1638 from the ATG codon of cry1Ac1). Following washing and exposure to an X-ray film, the oligonucleotide was scanned in a phosphorimager.
Measurements of protoxin synthesis.
Strains CryB/pHT3101, CryB/pHT3101-1Acwt, and CryB/pHT3101-1Ac272 were grown at 30°C in 30 ml of G-Tris medium in a New Brunswick shaker. As mentioned above, a marker for the commencement of sporulation is cell clumping, so 5-ml samples were removed approximately 1 h prior to this time, 1 h later, and at two subsequent times during sporulation (as monitored with the phase microscope). The remainder of the culture was incubated for 24 h in order to obtain free spores and inclusions.
The cells were washed once with 1 M KCl–5 mM EDTA, pH 8.0, and twice with 50 mM Tris–5 mM EDTA–2 mM phenylmethylsulfonyl fluoride, pH 7.6. The pellets were suspended in 6 M urea–1% SDS 2 mM phenylmethylsulfonyl fluoride–50 mM dithiothreitol, pH 9.6 (UDS), sonicated (Branson 45 with a microtip) for 40 s, and then heated to 90°C for 3 min. Aliquots of the cell extracts were precipitated with 10% trichloroacetic acid. The pellets were dissolved in 0.2 ml of 0.2 N NaOH for protein determinations with the bicinchoninic acid reagent (Pierce Chemical Co.).
Spores plus inclusions from the 24-h cultures were harvested, washed as described above, resuspended in 2 ml of deionized water, and sonicated for 12 s to remove clumps. A 0.2-ml sample was taken, and following centrifugation for 5 min in an Eppendorf microcentrifuge, the pellet was extracted twice with 40 μl of UDS at 37°C for 20 min each time and the supernatants were pooled.
A trace amount of purified 14C-inclusions (5,000 cpm) was added to the remaining spore-plus-inclusion suspensions in order to monitor inclusion recovery. The labeled inclusions were prepared by incubating 10 ml of sporulating cells with 5 μCi of [14C]isoleucine until sporulation was completed. These inclusions were purified as described below.
The suspensions were layered on step gradients of Renografin-76 (66% diatrizoate meglumine plus 10% diatrizoate sodium; Squibb) comprised of 1 ml of 60% and 3 ml each of 50 and 40% Renografin. The tubes were centrifuged for 1 h at 8,000 rpm in a Sorvall HB4 swinging-bucket rotor. The band of inclusions was removed with a syringe, diluted 10-fold with water, and collected by centrifugation at 10,000 rpm for 20 min in a Sorvall centrifuge. The pellets were examined in the phase microscope for spore contamination and recentrifuged through a second Renografin gradient if >5% of the initial spores were present. Samples of purified inclusions from the wild type, the mutant, and a mixture were photographed with a Nikon Eclipse phase microscope using the oil immersion objective (2,000×). Digital images were converted with Adobe Photoshop and printed.
The purified inclusions were dissolved in 50 μl of UDS by incubation for 10 min at 37°C. This step was repeated, and 10 μl of the pooled supernatants was precipitated with 10% trichloroacetic acid. The precipitates were collected on Whatman glass fiber filters, and the radioactivity was determined in a Beckman scintillation counter.
Fifty micrograms of cell extract protein, the extract from an equal volume of the spore-inclusion mixture, or an equal quantity of counts per minute from the purified inclusions (indicative of inclusion recovery) was diluted in UDS, heated at 90°C for 3 min, and electrophoresed in SDS–10% polyacrylamide. Immunoblotting was done as described previously by employing a Cry1Ac1 polyclonal antibody (32). Staining was done with the Pierce Gelcode blue stain reagent.
The stability of the Cry1Ac1 protoxin antigen was determined in pulse-chase experiments. Five milliliters from a 30-ml culture grown as described above was removed when the cells started to clump and at hourly intervals thereafter (five total samples). These subcultures were incubated with 5 μCi of [14C]isoleucine for 7 min. At that time, a 2-ml sample was pipetted into a tube on ice containing 1 mg of chloramphenicol. Unlabeled isoleucine was added to 200 μg/ml to the remaining culture, which was sampled as described above after an additional 60 min of incubation. Following centrifugation at 10,000 rpm for 10 min in a Sorvall SS34 rotor, the pellets were washed twice with 10 ml (each time) of 0.05 M Tris–0.15 M NaCl–1.0% Nonidet P-40 (pH 8.0), suspended in 0.8 ml of this buffer, and sonicated on ice three times for 40 s each time. The suspensions were centrifuged for 2 min in an Eppendorf microcentrifuge and the supernatants were stored at −70°C. This pulse-chase protocol was repeated at hourly intervals for at least four additional hours.
The radioactive and protein contents of aliquots of each sample were determined, and 150 μg of protein was incubated at 0°C for 1 h with preimmune serum plus 2 μg of purified, unlabeled Cry1Ac1 protoxin. A 50% suspension of protein A-Sepharose CL4B (Pharmacia) was added to 25% of the volume, and the suspensions were incubated at 4°C for 40 min. Following centrifugation for 2 min in an Eppendorf microcentrifuge, the supernatants were carefully removed and anti-Cry1Ac1 antibody (32) was added followed by protein A-Sepharose CL4B as described above. These suspensions were centrifuged, and the pellets were washed three times with 0.2 ml of the above buffer each time. The final pellets were suspended in 50 μl of UDS and heated at 90°C for 3 min, and one-half was electrophoresed by SDS–10% PAGE. The gels were dried and exposed to X-ray film and the 130-kDa bands were quantitated in a phosphorimager.
RESULTS
Transcription patterns.
Initially, the transcription pattern of the cry1Ac1 gene in a strain (80-21) which contained this gene as one of several protoxin genes (5) and in a strain in which this was the only protoxin gene (B. thuringiensis subsp. kurstaki HD73) was established by reverse transcriptase mapping (Fig. 3). The results confirmed the reverse transcriptase mapping of the cry1Ba1 gene (11) in that there was transcription from both BtI and BtII at different although somewhat overlapping times and to approximately the same extent. A fairly constant rate of transcription starting at T2 and continuing throughout much of sporulation was also found with the fusion of the wild-type cry1A promoters to lacZ (Fig. 4).
FIG. 3.
Reverse transcriptase mapping of the transcription of the cry1Ac1 gene in strain 80-21 (A) and B. thuringiensis subsp. kurstaki HD73 (B). In both cases, transcription from BtI started at about T2 and sporulation was completed 11 to 12 h after T0.
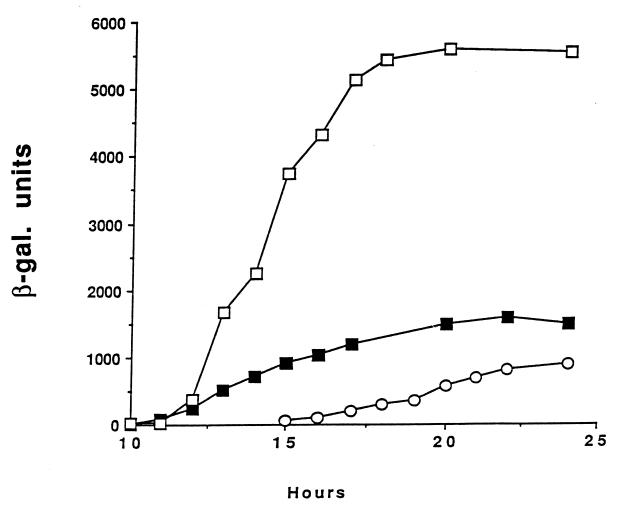
FIG. 4.
β-Galactosidase synthesis in B. thuringiensis 80-21 containing plasmids with the cry1A gene promoter region fused to lacZ. The fusions contained the wild-type promoter (■), the 272 mutant BtII promoter (□), and the 271 mutant BtI promoter (○). The cells were grown and sporulated in G-Tris medium plus 7 μg of chloramphenicol per ml and sampled as described in Materials and Methods. In all cases, exponential growth ended at about 10 h.
Mutagenesis of the promoter region.
Initially, each of the promoters was inactivated by introducing several mutations into conserved bases in the −10 regions (mutations 271 and 272 [Table 1; Fig. 2]). Loss of promoter function was determined by reverse transcriptase mapping using RNA prepared from sporulating cells of strain 80-21 transformed with each of the lacZ fusion plasmids (Fig. 5). At the sampling time for the wild type, there was a strong transcript initiating at BtI and a weaker one at BtII. There was also a transcript initiating 14 bp downstream from BtI which was not seen in the reverse transcriptase mapping of the cry1Ac1 gene (Fig. 3) or in the mutant 271 and 272 lanes. The lack of transcription from the BtII promoter in mutant 272 was confirmed by sampling at several times later in sporulation. There was a weak transcript from the BtI promoter with RNA prepared from the 271 mutant which was not enhanced by sampling at earlier times in sporulation (data not shown).
FIG. 5.
Reverse transcriptase mapping of the start sites of transcription from the wild-type (Wt) and mutant (271 and 272) promoters in plasmid pHT/pSGMU-P in B. thuringiensis 80-21. The start sites for BtI and BtII are shown on the right. The Wt RNA was prepared from cells at the equivalent of 14 to 15 h in Fig. 4, the RNA for the 271 lane was prepared from cells at the equivalent of 16 to 17 h in Fig. 4, and the RNA for the 272 lanes (separate gel) was prepared from cells at the equivalent of 16 and 17 h in Fig. 4. All of the sequencing lanes are in the order AGCT as indicated in the panel on the right side. Note that these sequencing lanes start six bases lower than the Wt and 271 sequencing lanes and that this sequence is compressed at the top.
Similar reverse transcriptase mapping was done with the other mutants listed in Table 1. Transcription from both promoters in mutants 1077, 1078, and 1079 was established by preparing RNA at T3 and T6. Transcription was barely detectable in mutant 1077 even with five times more RNA.
As previously mentioned, strain 80-21, containing a fusion of the wild-type promoters to lacZ, synthesized β-galactosidase over about 10 h commencing at T2 of sporulation (Fig. 4). Expression from a construct containing only a functional BtII promoter (mutation 271) began later and at a slightly lower rate. The rates at very late times were somewhat variable because of sporulation asynchrony and lysis of sporulated cells with the subsequent inactivation of the β-galactosidase.
Mutation within the −10 region of the BtII promoter (mutation 272) resulted in a ca. fivefold increase in both the initial rate and final amount of β-galactosidase. This stimulation was confirmed by making a single mutation within the −10 region of BtII which inactivated this promoter (mutation 1076 [Table 1]). Both the 272 and 1076 mutations resulted in further deviations from the B. subtilis −10 consensus sequence for ςK (and ςE) (Table 1). When the −10 region was changed to the ςE consensus (mutation 1077), there was transcription from both promoters but at a greatly reduced rate. Single base pair changes within the spacer region but outside the BtII −10 sequence (mutations 1078 and 1079) had no effect (Table 1).
Effect of a promoter-up mutation on mRNA and protoxin accumulation.
The cry1Ac1 gene containing the 272 promoter-up mutation was cloned and electroporated into strain CryB. Early in sporulation (equivalent to about 13 h in Fig. 4), there was 2.2-fold more cry1Ac1 mRNA in this strain than in a strain containing the cry1Ac1 gene with the wild-type promoters (CryB/pHT3101-1Acwt) (Fig. 6). At a later time (>70% phase-white endospores; equivalent to 18-20 h in Fig. 4), there was 30% less cry1Ac1 mRNA in the mutant. This decrease was due to the lack of transcription from the BtII promoter and decay of the earlier-transcribed mRNA.
FIG. 6.
Northern hybridizations of 20 μg of RNA prepared from CryB/pHT3101-1Acwt (lanes 1 and 4) and CryB/pHT3101-1Ac272 (lanes 2 and 3) at T2.5 (lanes 1 and 2) and T6 (lanes 3 and 4) of sporulation. Hybridization and quantitation of both bands were done as described in Materials and Methods. The upper bands in each lane are the expected size for cry1Ac1 mRNA; the lower, less prevalent bands are apparently stable degradation products.
The accumulation of Cry1Ac antigen in sporulating cells was determined over a 3-h period from about hour 13 to 16 in Fig. 4, when there was transcription from the BtI promoter. There was 2.5 times more antigen in the mutant than the wild type at the first sampling time and about 40% more 90 min later; the values were about equal after an additional 90 min (Fig. 7A).
FIG. 7.
(A) Immunoblot of cell extracts of strains CryB/pHT3101-1Acwt (lanes 1 to 3) and CryB/pHT3101-1Ac272 (lanes 4 to 6) sampled at T1.5 (lanes 1 and 4), T3.0 (lanes 2 and 5), and T4.5 (lanes 3 and 6). Electrophoresis and treatment with a Cry1Ac antibody were done as described in Materials and Methods. (B) Stained SDS–10% PAGE of crude spore-inclusion mixtures (lanes 1 and 2) and of purified inclusions (lanes 3 and 4) from CryB/pHT3101-1Acwt (lanes 1 and 3) and CryB/pHT3101-1Ac272 (lanes 2 and 4). Standards (STD), from top to bottom, are 116, 97, 66, 45, and 29 kDa.
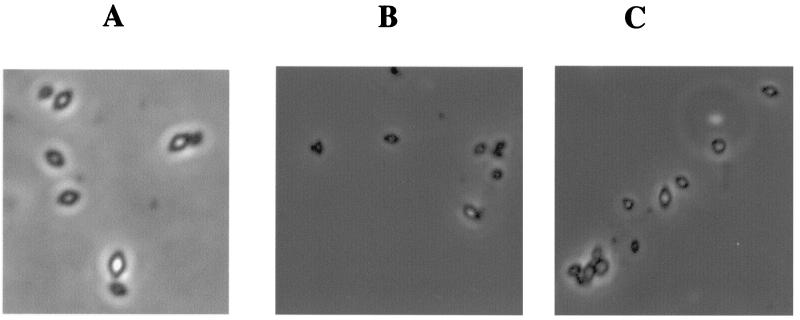
Following completion of sporulation, protoxin was extracted from the spore-plus-inclusion mixtures and from purified inclusions. The amount of protoxin antigen in spore-plus-inclusion extracts from the mutant was 50% higher than that in extracts from the wild type, but the amount in purified inclusions was 2.5-fold less (Fig. 7B). On the whole, inclusions produced by CryB/pHT3101-1Ac272 were much smaller than those produced by strain CryB/pHT3101-1Acwt (Fig. 8B and C).
FIG. 8.
Phase-contrast micrographs of purified inclusions from a mixture (A), CryB/pHT3101-1Ac272 (B), and CryB/pHT3101-1Acwt (C), examined with oil immersion (2,000×) with an additional ×2.5 magnification when photographed. Panel A shows three inclusions from the wild type with three from the mutant immediately adjacent.
Despite the hyperactivity of the BtI promoter in the mutant, there was an impairment in protoxin accumulation, especially in inclusions. Since both transcription and translation of the cry1Ac1 gene were enhanced in the mutant at early times, the lack of protoxin accumulation was most likely due to turnover. This possibility was examined by pulse-chase immunoprecipitation experiments as described in Materials and Methods (Fig. 9). There was a twofold-enhanced synthesis of 14C-Cry1Ac protoxin in the mutant during the earliest 7-min labeling, consistent with the protoxin antigen accumulation results in Fig. 7. In the mutant, however, almost all of the protoxin turned over, in contrast to its stability in the wild type (Fig. 9, compare lanes 1 and 2 with lanes 7 and 8). Even an hour later, at least half the 14C-protoxin in the mutant was unstable. This unstable fraction decreased over an additional 3 h until the switch in transcription from BtI to BtII. At this time, there was very little additional synthesis of protoxin in the mutant.
FIG. 9.
Autoradiogram of SDS-PAGE of immunoprecipitated Cry1Ac antigen from CryB/pHT3101-1Acwt (lanes 1 to 4) and CryB/pHT3101-1Ac272 (lanes 7 to 10) from a pulse-chase experiment (see Materials and Methods). Five-milliliter aliquots of the cultures were removed and incubated with [14C]isoleucine for 7 min at clumping (about T0.5) (lanes 1 and 7) and then chased with an excess of [12C]isoleucine for 1 h (lanes 2 and 8). At this time (T1.5), another 5-ml sample of cells was incubated with [14C]isoleucine for 7 min (lanes 3 and 9) and chased (lanes 4 and 10). Lanes 5 and 6 contained extracts, like lanes 1 and 2, but they were treated only with preimmune serum.
DISCUSSION
Fusions of the cry1A gene promoters to lacZ were used to establish the pattern of transcription of this class of protoxin genes. The timing and relative transcription from each of the cry1Ac1 promoters (Fig. 3) were independent of the presence of other cry1A genes and consistent with the reverse transcriptase mapping results obtained with the cry1Ba1 gene (11).
The −10 region of the BtII promoter within the spacer region of the BtI promoter is critical for regulation. Mutations which departed from the consensus ςE or ςK sequence resulted in stimulation of transcription (Fig. 4 and 6; Table 1), whereas a change to the ςE consensus was inhibitory (mutation 1077 [Table 1]). These results imply that ςE or some form of RNA polymerase containing this subunit is involved in the regulation. Transcription from BtI occurs primarily before there is functional ςK (although pro-ςK may be present), so this sigma subunit is unlikely to be involved. In addition, transcription of the cry1Aa1 gene in a strain lacking ςK was unaltered at early times, with no evidence for enhanced expression (10). Unproductive binding of ςE RNA polymerase to the BtII promoter could modulate transcription from BtI.
In general, promoter function is most severely affected by mutations in the −10 and −35 regions, but some within the spacer region which alter promoter bending (35) or perhaps other properties, such as conformation (29, 35, 45), can be detrimental. Promoter-up mutations in this region are rare, although a regulatory region analogous to the one described here is present in the spacer region of a DNA replication gene promoter in Caulobacter (46). While the sequence of this regulatory region was very similar to that of the −10 region, there was no evidence for overlapping promoters.
Related protoxins (such as the Cry1 types) are packaged into single inclusions (3). The packaging involves extensive intermolecular disulfide bond formation (9), so rate-limiting steps involving chaperones (2, 8) and disulfide interchange reactions (9) are likely. In such a multistep assembly process, efficient packaging over a prolonged time during sporulation may depend upon a mechanism for controlling the rate of protoxin synthesis. The results reported here indicate that this control can be achieved by the overlap of two mother cell sporulation promoters. Additional regulatory mechanisms are required for the differential transcription of the multiple protoxin genes (44) and to balance protoxin synthesis with that of mother cell spore components.
While there is a small amount of transcription of some cry genes at the end of growth (8, 47), most synthesis does not occur until stage II of sporulation, when ςE RNA polymerase becomes functional. Parenthetically, this timing coincides with the conditions necessary for inclusion assembly. Toxins active on Lepidoptera and Diptera form inclusions consisting of protoxins cross-linked by disulfide bonds and thus require the more oxidative conditions found in sporulating cells (40). In contrast, inclusions comprised of the Cry3Aa1 protoxin probably do not contain disulfide bonds (2), and this protoxin is readily solubilized at a lower pH in the absence of a reducing agent (reflecting conditions in the midgut of susceptible coleopteran larvae). Transcription of this gene is not dependent upon sporulation, and there is a novel ςA-like promoter which functions in late exponential-phase cells (2). Differences in the time of expression of protoxin genes may in part reflect the intracellular conditions needed for inclusion assembly.
It was possible to substantially increase the size of inclusions comprised of the Cry3Aa1 toxin either by expressing a cloned gene in a spo0A deletion strain (25) or by using a hybrid promoter (34). In the latter case, a 12.7-fold enhancement of Cry3Aa1 protein accumulation over that of the wild-type strain was found, but the toxicities were comparable. Apparently, these large inclusions either were not readily ingested or were not solubilized. In contrast, only a twofold increase in total Cry1 protoxin was obtained by introducing an additional gene, cry1Ca1, into the chromosome of B. thuringiensis subsp. kurstaki HD73 (22). The regulated expression of the cry1 genes by overlapping promoters as well as other regulatory steps in inclusion assembly could account for the less extensive enhancement of Cry1 protoxin accumulation.
ACKNOWLEDGMENTS
This research was supported by Public Health Service grant GM34035 from the National Institutes of Health and grant MCB-9600584 from the National Science Foundation.
The technical assistance of Lan Wu is appreciated. Dr. Chris Staiger was most helpful with the phase microscopy.
REFERENCES
- 1.Adams L F, Brown K I, Whiteley H R. Molecular cloning and characterization of two genes encoding sigma factors that direct transcription from a Bacillus thuringiensis crystal protein gene promoter. J Bacteriol. 1991;173:3846–3854. doi: 10.1128/jb.173.12.3846-3854.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agaisse H, Lereclus D. How does Bacillus thuringiensis produce so much insecticidal crystal protein? J Bacteriol. 1995;177:6027–6032. doi: 10.1128/jb.177.21.6027-6032.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aronson A. The protoxin composition of Bacillus thuringiensis insecticidal inclusions affects solubility and toxicity. Appl Environ Microbiol. 1995;61:4057–4060. doi: 10.1128/aem.61.11.4057-4060.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aronson A I. The two faces of Bacillus thuringiensis: insecticidal proteins and post-exponential survival. Mol Microbiol. 1993;7:489–496. doi: 10.1111/j.1365-2958.1993.tb01139.x. [DOI] [PubMed] [Google Scholar]
- 5.Aronson A I. Flexibility in the protoxin composition of Bacillus thuringiensis. FEMS Microbiol Lett. 1994;117:21–28. doi: 10.1111/j.1574-6968.1994.tb06737.x. [DOI] [PubMed] [Google Scholar]
- 6.Aronson A I, Angelo N, Holt S C. Regulation of extracellular protease production in Bacillus cereus T: characterization of mutants producing altered amounts of protease. J Bacteriol. 1971;106:1016–1025. doi: 10.1128/jb.106.3.1016-1025.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arvidson H, Dunn P E, Strnad S, Aronson A I. Specificity of Bacillus thuringiensis for lepidopteran larvae: factors involved in vivo and in the structure of a purified protein. Mol Microbiol. 1989;3:1533–1543. doi: 10.1111/j.1365-2958.1989.tb00139.x. [DOI] [PubMed] [Google Scholar]
- 8.Baum J A, Malvar T. Regulation of insecticidal crystal protein production in Bacillus thuringiensis. Mol Microbiol. 1995;18:1–12. doi: 10.1111/j.1365-2958.1995.mmi_18010001.x. [DOI] [PubMed] [Google Scholar]
- 9.Bietlot H P L, Vishmulhatla J, Carey P R, Pozsgay M, Kaplan H. Characterization of the cysteine residues and disulphide linkages in the protein crystal of Bacillus thuringiensis. Biochem J. 1990;267:309–315. doi: 10.1042/bj2670309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bravo A, Agaisse H, Salamitou S, Lereclus D. Analysis of cry1Aa expression in sigE and sigK mutants of Bacillus thuringiensis. Mol Gen Genet. 1996;250:734–741. doi: 10.1007/BF02172985. [DOI] [PubMed] [Google Scholar]
- 11.Brizzard B L, Schnepf H E, Kronstad J W. Expression of the cry1B crystal protein gene of Bacillus thuringiensis. Mol Gen Genet. 1991;231:59–64. doi: 10.1007/BF00293822. [DOI] [PubMed] [Google Scholar]
- 12.Carter H L, III, Wang L-F, Doi R H, Moran C P., Jr rpoD operon promoter used by ςH-RNA polymerase in Bacillus subtilis. J Bacteriol. 1988;170:1617–1621. doi: 10.1128/jb.170.4.1617-1621.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chibazakura T, Kawamura F, Takahashi H. Differential regulation of spo0A transcription in Bacillus subtilis: glucose represses promoter switching at the initiation of sporulation. J Bacteriol. 1991;173:2625–2632. doi: 10.1128/jb.173.8.2625-2632.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Errington J. A general method for fusion of the Escherichia coli lacZ gene to chromosomal genes in Bacillus subtilis. J Gen Microbiol. 1986;132:2953–2966. doi: 10.1099/00221287-132-11-2953. [DOI] [PubMed] [Google Scholar]
- 15.Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrari E, Henner D J, Perego M, Hoch J A. Transcription of Bacillus subtilis subtilisin and expression of subtilisin in sporulating mutants. J Bacteriol. 1988;170:289–295. doi: 10.1128/jb.170.1.289-295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foulger D, Errington J. Sequential activation of dual promoters by different sigma factors maintains spoVJ expression during successive developmental stages of Bacillus subtilis. Mol Microbiol. 1991;5:1363–1373. doi: 10.1111/j.1365-2958.1991.tb00783.x. [DOI] [PubMed] [Google Scholar]
- 18.Giacomini A, Corich V, Ollero F J, Squarlini H, Nuti M P. Experimental conditions may affect reproducibility of the β-galactosidase assay. FEMS Microbiol Lett. 1992;100:87–90. doi: 10.1111/j.1574-6968.1992.tb14024.x. [DOI] [PubMed] [Google Scholar]
- 19.Gryczan T, Shivakumar A G, Dubnau D. Characterization of chimeric plasmid cloning vehicles in Bacillus subtilis. J Bacteriol. 1980;141:246–253. doi: 10.1128/jb.141.1.246-253.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Höfte H, Whiteley H R. Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol Rev. 1989;53:242–255. doi: 10.1128/mr.53.2.242-255.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson C W, Moran C P, Jr, Losick R. Two RNA polymerase sigma factors from Bacillus subtilis discriminate between overlapping promoters for a developmentally regulated gene. Nature. 1983;302:800–804. doi: 10.1038/302800a0. [DOI] [PubMed] [Google Scholar]
- 22.Kalman S, Kiehne K L, Cooper N, Reynoso M S, Yamamoto T. Enhanced production of insecticidal proteins in Bacillus thuringiensis strains carrying an additional crystal protein gene in their chromosome. Appl Environ Microbiol. 1995;61:3063–3068. doi: 10.1128/aem.61.8.3063-3068.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee M K, Curtiss A, Alcantara E, Dean D H. Synergistic effect of the Bacillus thuringiensis toxins CryIAa and CryIAc on the gypsy moth, Lymantria dispar. Appl Environ Microbiol. 1996;62:583–586. doi: 10.1128/aem.62.2.583-586.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lereclus D, Arantes O, Chaufaux J, Lecadet M-M. Transformation and expression of a cloned δ-endotoxin gene in Bacillus thuringiensis. FEMS Microbiol Lett. 1989;60:211–218. doi: 10.1016/0378-1097(89)90511-9. [DOI] [PubMed] [Google Scholar]
- 25.Lereclus D, Agaisse H, Gominet M, Chaufaux J. Overproduction of encapsulated insecticidal crystal proteins in a Bacillus thuringiensis Spo0A mutant. Bio/Technology. 1995;13:67–71. doi: 10.1038/nbt0195-67. [DOI] [PubMed] [Google Scholar]
- 26.Losick R, Stragier P J. Crisscross regulation of cell-type specific gene expression during development in Bacillus subtilis. Nature. 1992;355:601–604. doi: 10.1038/355601a0. [DOI] [PubMed] [Google Scholar]
- 27.Masson L, Prefontaine G, Peloquin L, Lau P C K. Comparative analysis of the individual protoxin constituents in P1 crystals of Bacillus thuringiensis subsp. kurstaki isolates NRD12 and HD1. Biochem J. 1989;269:507–512. doi: 10.1042/bj2690507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masson L, Erlandson M, Puzstai-Carey M, Brousseau R, Juarez-Perez V, Frutos R. A holistic approach for determining the entomopathogenic potential of Bacillus thuringiensis strains. Appl Environ Microbiol. 1998;64:4782–4788. doi: 10.1128/aem.64.12.4782-4788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mellies J, Brems R, Villarejo M. The Escherichia coli proU promoter element and its contribution to osmotically signaled transcription activation. J Bacteriol. 1994;176:3638–3645. doi: 10.1128/jb.176.12.3638-3645.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 31.Minnich S A, Aronson A I. Regulation of protoxin synthesis in Bacillus thuringiensis. J Bacteriol. 1984;158:447–454. doi: 10.1128/jb.158.2.447-454.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohammed S I, Johnson D E, Aronson A I. Altered binding of the Cry1Ac toxin to larval membranes but not to the toxin-binding protein in Plodia interpunctella selected for resistance to different Bacillus thuringiensis isolates. Appl Environ Microbiol. 1996;62:4168–4173. doi: 10.1128/aem.62.11.4168-4173.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moran C P., Jr . RNA polymerase and transcription factors. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C.: American Society for Microbiology; 1993. pp. 653–667. [Google Scholar]
- 34.Park H-W, Ge B, Bauer L S, Federici B A. Optimization of Cry3A yields in Bacillus thuringiensis by use of sporulation-dependent promoters in combination with the STAB-SD mRNA sequence. Appl Environ Microbiol. 1998;64:3932–3938. doi: 10.1128/aem.64.10.3932-3938.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pérez-Martín J, Rojo F, de Lorenzo V. Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol Rev. 1994;58:268–290. doi: 10.1128/mr.58.2.268-290.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler D R, Dean D H. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev. 1998;62:775–806. doi: 10.1128/mmbr.62.3.775-806.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schurter W, Geiser M, Mathe D. Efficient transformation of Bacillus thuringiensis and B. cereus via electroporation: transformation of acrystalliferous strains with a cloned delta-endotoxin gene. Mol Gen Genet. 1989;218:177–181. doi: 10.1007/BF00330581. [DOI] [PubMed] [Google Scholar]
- 40.Setlow B, Setlow P. Levels of acetyl coenzyme A, reduced and oxidized coenzyme A, and coenzyme A in disulfide linkage to protein in dormant and germinated spores and growing and sporulating cells of Bacillus megaterium. J Bacteriol. 1977;132:444–452. doi: 10.1128/jb.132.2.444-452.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stahly C P, Dingman D W, Bulla L A, Jr, Aronson A I. Possible origin and function of the parasporal crystals in Bacillus thuringiensis. Biochem Biophys Res Commun. 1978;84:581–588. doi: 10.1016/0006-291x(78)90745-3. [DOI] [PubMed] [Google Scholar]
- 42.Tabashnik B E. Evaluation of synergism among Bacillus thuringiensis toxins. Appl Environ Microbiol. 1992;58:3343–3346. doi: 10.1128/aem.58.10.3343-3346.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tatti K M, Moran C P., Jr Utilization of one promoter by two forms of RNA polymerase from Bacillus subtilis. Nature. 1985;314:190–192. doi: 10.1038/314190a0. [DOI] [PubMed] [Google Scholar]
- 44.Walter T, Aronson A. Specific binding of the E2 subunit of pyruvate dehydrogenase to the upstream region of Bacillus thuringiensis protoxin genes. J Biol Chem. 1998;274:7901–7906. doi: 10.1074/jbc.274.12.7901. [DOI] [PubMed] [Google Scholar]
- 45.Warne S E, deHaseth P L. Promoter recognition by Escherichia coli RNA polymerase. Effects of single base pair deletions and insertions in the spacer DNA separating the −10 and −35 regions are dependent on spacer DNA sequence. Biochemistry. 1993;32:6134–6140. doi: 10.1021/bi00075a003. [DOI] [PubMed] [Google Scholar]
- 46.Winzeler E, Shapiro L. A novel promoter motif for Caulobacter cell cycle-controlled DNA replication genes. J Mol Biol. 1996;264:412–425. doi: 10.1006/jmbi.1996.0650. [DOI] [PubMed] [Google Scholar]
- 47.Yoshisue H, Ihara K, Nishimoto T, Sakai H, Komano T. Expression of the genes for insecticidal crystal proteins in Bacillus thuringiensis: cryIVA, not cryIVB, is transcribed by RNA polymerase containing ςH and that containing ςE. FEMS Microbiol Lett. 1995;127:65–67. doi: 10.1111/j.1574-6968.1995.tb07451.x. [DOI] [PubMed] [Google Scholar]