Abstract
Most Alzheimer’s disease (AD)-associated genetic variants do not change protein coding sequence and thus likely exert their effects through regulatory mechanisms. RNA editing, the post-transcriptional modification of RNA bases, is a regulatory feature that is altered in AD patients that differs across ancestral backgrounds. Editing QTLs (edQTLs) are DNA variants that influence the level of RNA editing at a specific site. To study the relationship of DNA variants genome-wide, and particularly in AD-associated loci, with RNA editing, we performed edQTL analyses in self-reported individuals of African American (AF) or White (EU) race with corresponding global genetic ancestry averaging 82.2% African ancestry (AF) and 96.8% European global ancestry (EU) in the two groups, respectively. We used whole-genome genotyping array and RNA sequencing data from peripheral blood of 216 AD cases and 212 age-matched, cognitively intact controls. We identified 2144 edQTLs in AF and 3579 in EU, of which 1236 were found in both groups. Among these, edQTLs in linkage disequilibrium (r2 > 0.5) with AD-associated genetic variants in the SORL1, SPI1 and HLA-DRB1 loci were associated with sites that were differentially edited between AD cases and controls. While there is some shared RNA editing regulatory architecture, most edQTLs had distinct effects on the rate of RNA editing in different ancestral populations suggesting a complex architecture of RNA editing regulation. Altered RNA editing may be one possible mechanism for the functional effect of AD-associated variants and may contribute to observed differences in the genetic etiology of AD between ancestries.
Introduction
Alzheimer’s disease (AD) is the most common form of dementia (1), which places a large emotional and economic burden on patients and society. AD is highly heritable with an estimated 74% of liability explained by genetic factors (2,3). The most significant of genetic risk loci is the ε4 allele of apolipoprotein E (APOE) on chromosome 19q13 (4–8), but more than 30 genetic loci have been implicated in AD risk through large-scale genomic studies in diverse populations (9–14). These studies support the notion that the genetic architecture of AD partially overlaps but differs across ancestries, which is important as individuals of African ancestry (AF) are at increased risk for AD relative to individuals of European ancestry (EU) (15). Among several genetic risk factors with ancestry differences, one notable example is that the risk of AD associated with the APOEɛ4 allele is significantly higher when located on a European local ancestry haplotype relative to an African haplotype (16–18). Furthermore, there are African ancestry-specific risk variants in genes such as ABCA7 (12,19). Across ancestries, though, genetic loci associated with AD fall within or near genes that are enriched in molecular pathways including lipid metabolism (e.g. APOE, ABCA7), immune regulation (e.g. SORL1, TREM2) and endocytosis (e.g. BIN1), implicating these biological processes in the disease pathogenesis regardless of ancestry (9,11,13,14).
Despite the identification of associated variants, their functional effects are mostly unknown. They likely function by regulating gene expression (20), and several studies have identified transcriptional alterations in AD, including with expression differences specific to ancestral background (21–28). Beyond gene expression, gene function can also be impacted by post-transcriptional processes such as RNA editing. RNA editing is a process most commonly mediated by the ADAR family of enzymes by which individual bases in double-stranded mRNA are altered (29), which leads to downstream consequences including protein coding changes, altered splice sites, and the creation or disruption of microRNA (miRNA) binding sites (30,31). Importantly, altered editing levels at specific sites have been associated with various neurological disorders, such as Aicardi–Goutieres syndrome, amyotrophic lateral sclerosis, epilepsy, major depression, schizophrenia and AD (32–35), implicating this process broadly in maintaining appropriate neurological function. In AD specifically, studies have shown deficiencies of editing in hippocampal tissue in cases compared to controls at a few specifically tested sites (36,37), most prominently at the GluA2 QR site (37). Furthermore, we recently described transcriptome-wide AD-associated changes of RNA editing in a multi-ethnic cohort using RNA sequencing (RNA-seq) from whole blood (35). We found that genes that were differentially edited between AD cases and controls in AF and EU populations were enriched for biological processes related to immune regulation, inflammation and endocytosis (35).
In contrast to a rich literature on the role of DNA variants modulating gene expression across a variety of tissues as expression quantitative trait loci (eQTLs) (38), less work has been done to determine the role of DNA sequence variation in modulating post-transcriptional events such as RNA editing. Variation in DNA can influence the rate and location of editing through effects on RNA secondary structure (39,40), through variation of the sequence bound by ADAR enzymes (40) and likely other mechanisms. The genetic variants associated with the level of RNA editing at a specific site are known as RNA editing quantitative trait loci (edQTLs). Importantly, the role of disease-associated variants in modifying RNA editing rates in the context of diseases, including AD, is largely unknown. Furthermore, the effect of edQTLs modulating RNA editing from individuals of different ancestral backgrounds has been studied in lymphoblast cell lines, indicating that widespread cis variation of RNA editing may potentially contribute to phenotypic diversity across human populations (39). However, the overall role of ancestral background in edQTL effects remains to be further elucidated.
To address the potential disease risk contribution and ancestral differences in edQTLs, herein, we conducted a genome-wide screen for edQTLs in whole blood from AD cases and cognitively intact controls in AF and EU populations. Since there is evidence of ancestry-specific genetic risk factors in AD as well as ancestry-specific RNA editing signatures, here we tested whether edQTLs generalized across ethnicities or are population-specific. We hypothesized that the differences in RNA editing rates we previously observed between AD cases and controls (35) may be partially explained by genetic factors in the form of edQTLs. Furthermore, we tested whether edQTLs in AD-associated regions functionally contribute to the observed genetic signals. Our results bring to light the functional genomic impact of variants in AD-associated loci across ancestral backgrounds.
Results
Genome-wide edQTL identification
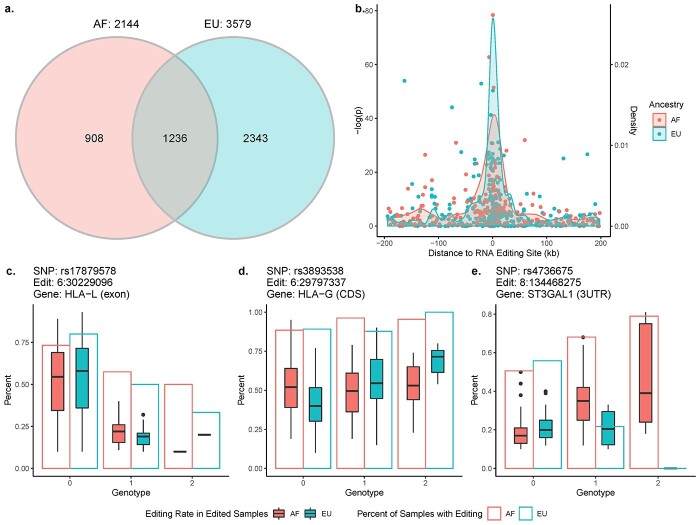
From our matched dataset of genotypes at 641 790 markers and RNA editing levels at 44985 individual sites in whole blood, we identified 2144 unique edQTL-edited site pairs in AF and 3579 in EU (FDR ≤ 0.01). Among them, 1236 edQTL-site pairs were significant in both ancestry groups (Fig. 1B). There was greater strength of association for single-nucleotide polymorphisms (SNPs) that were closer to the RNA editing site (Fig. 1B).
Figure 1.
Overview of edQTLs. (A) Venn diagram of edQTL-edited site pairs identified in African ancestry (AF) individuals and European ancestry (EU) individuals. (B) Distance between SNP and edited site versus the –log of the P-value of the association between the genotype and editing level (y-axis) with density plot showing frequency of significant edQTLs by distance (x-axis). Examples of edQTLs with shared (C), ancestry-specific (D) and reverse effects (E) across ancestral populations.
edQTL effects across ancestries
We observed four categories of edQTL effects when comparing across ancestry groups. First, 1138 edQTLs had shared effects across ethnicities: a SNP that was significantly associated with editing at the same site in both populations and had the same direction of effect (Fig. 1C). Among these edQTLs, only 85 (7.6%) were significantly associated with percent European ancestry, suggesting independent effects in each ancestry group. Second, we identified 2531 edQTLs with ancestry-specific effects, of which 535 were AF-specific and 1996 EU-specific. These were SNPs that were only significantly associated with editing in one population but not the other despite having a minor allele frequency (MAF) of at least 0.05 in both populations (Fig. 1D). Third, we found 720 edQTLs with ancestry-specific alleles (373 in AF, 347 in EU). These are SNPs that were significantly associated with editing in only one population and were too rare in the other population to measure effects (MAF < 0.05). Finally, we identified 98 edQTLs, the rarest category, with reverse effects: a SNP that was significantly associated with editing in both populations but with a different direction of effect (Fig. 1E).
edQTLs in AD-associated loci
Of the markers that were included in our genotyping data, 796 markers in 21 loci were both within 200 kb of an editing site and in linkage disequilibrium (LD) with an EU AD-associated signal from Kunkle et al. (13) using a minimum r2 of 0.5. We identified significant edQTLs in four AD-associated loci: SORL1, SPI1, TREM2 and HLA-DRB1 (Table 1, Supplementary Material, Table S3). These included an association between rs11218343 (in an intron of SORL1) and editing of an intronic site in SORL1, which was significant in both AF and EU. SNPs in the HLA-DRB1 region were associated with editing levels at multiple sites in both populations and included AF-specific, EU-specific and shared edQTLs. Conversely, the intronic variant rs116887820 in SPI1, which is in LD with rs3740688 in an adjacent intron of SPI1, was associated with editing at a nearby site in the same intron as the edQTL in AF alone, while rs62397402, which lies upstream of CCND3 and is in LD with rs75932628 in an exon of TREM2, was associated with editing in an intron of CCND3 in EU alone.
Table 1.
edQTLs in LD with AD-associated GWAS markers, defined as variants within an LD block with an r2 ≥0.5 of at least one of the top 25 genome-wide signals from Kunkle et al. (13) and the top 13 signals from Kunkle et al. (14)
| GWAS ancestral population | GWAS allele | GWAS gene | LD Block coordinates | edQTL | Edited site | Edited gene | Ancestry | Slope | 95% CI | P-value | FDR |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AF | rs184179037 | WDR70 | 5:37008690–5:37861883 | GSA-rs7716549 | 5:36968777 | NIPBL | EU | −0.446 | −0.7376, −0.1557 | 2.47E−03 | 4.66E−03 |
| EU | rs75932628 | TREM2 | 6:40706366–6:41365821 | GSA-rs62397402 | 6:41983812 | CCND3 | EU | 3.034 | 0.4203, 5.6472 | 2.21E−02 | 3.45E−02 |
| EU | rs3740688 | SPI1 | 11:47372377–11:47466790 | GSA-rs116887820 | 11:47389577 | SPI1 | AF | −0.927 | −1.4009, −0.454 | 1.12E−04 | 2.57E−04 |
| EU | rs11218343 | SORL1 | 11:121433926–11:121461593 | rs11218343 | 11:121403448 | SORL1 | AF | −2.774 | −3.6555, −1.8944 | 1.44E−09 | 5.06E−09 |
| EU | −2.405 | −3.1935, −1.6175 | 3.87E−09 | 1.45E−08 |
For edQTLs in the HLA-DRB1 gene region, see Supplementary Material, Table S2.
In addition, 510 markers in eight gene regions were within 200 kb of an editing site and in LD (r2 > 0.5) with an AD-associated signal from a genome-wide association study (GWAS) of an African-American cohort (14). Among these, we discovered an edQTL in WDR70, which had an effect in EU but not in AF (Table 1). No AF-specific edQTLs were discovered in LD with one of the AF-specific GWAS signals.
edQTLs associated with sites differentially edited in AD versus controls
We also evaluated edQTLs that were associated with an RNA editing site that was differentially edited between AD cases and controls. In a previous study, we found 449 sites in 254 genes in AF and 723 sites in 371 genes in EU that were differentially edited between AD cases and controls (35). We identified 460 edQTLs for 148 of these differentially edited sites with between 1 and 37 SNPs mapped to each edited site outside the HLA region and 209 SNPs mapping to the 4 sites within the HLA region (Supplementary Material, Table S4). Using a Fisher’s exact test, we found that differentially edited sites were significantly more likely to be associated with an edQTL than sites that were not differentially edited between cases and controls (P-value < 2.2 × 10−16, odds ratio = 7.23, 95% confidence interval: 5.97–8.72). Differentially edited sites with associated edQTLs included several AD-associated genes, including in SORL1, SPI1 and members of the HLA and SLC gene families. In addition, we found evidence for genetic regulation of RNA editing in genes involved in neuronal apoptosis and phagocytosis, such as GAS6 and AOAH.
Of note, edQTLs associated with differentially edited sites in HLA-DRB1, SORL1 and SPI1 were also in LD with GWAS markers. A formal colocalization analysis found strong evidence of the edQTL-GWAS pair influencing both differential RNA editing and AD status in SORL1 (posterior probability = 0.89) and SPI1 (posterior probability = 0.91). In HLA-DRB1, the posterior probability was 0.57 that both traits are associated and share a single causal variant, while the probability was 0.43 that both traits are associated but with different causal variants.
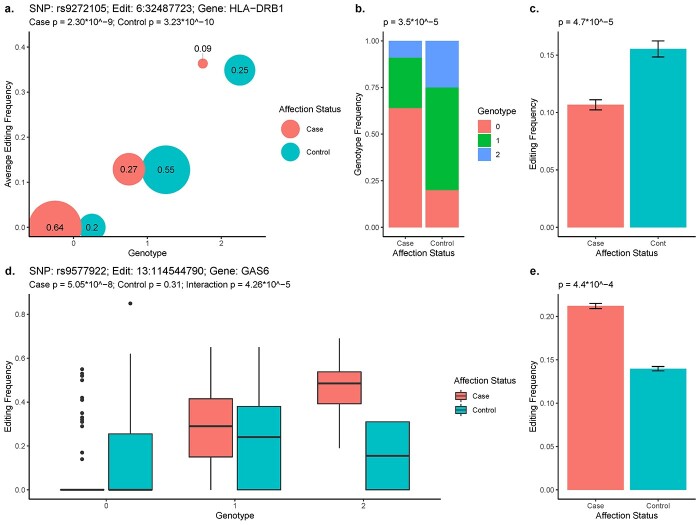
Among differentially edited sites with associated edQTLs, we then sought to test whether the edQTLs occurred at different genotype frequencies in cases versus controls. We found 11 edQTLs associated with differentially edited sites (four unique to AF, five unique to EU and two shared) where the edQTL had significantly different minor allele frequencies between AD cases and controls (FDR < 0.05, Fisher’s exact) (Table 2). Of note, rs2747454 in HLA-H was significantly associated with editing at 6:29856483 in HLA-H in both ancestral groups but only the EU cohort showed a significant difference in the allele frequency of rs2747454 across cases and controls and, in turn, a significant difference in the RNA editing rate. Also notable, a site in SORL1 and a site in HLA-DRB1 were both differentially edited in both AF and EU, both had a corresponding edQTL with different allele frequencies between cases and controls in both AF and EU, and were both in LD with an AD-associated marker (Fig. 2A).
Table 2.
edQTLs with frequency differences between cases and controls that are associated with differentially edited sites, as identified in Gardner et al. (35)
| SNP | Edited site | Edited gene | Ancestry | edQTL slope | edQTL 95% CI | edQTL P-value | edQTL FDR | MAF | Fisher FDR |
|---|---|---|---|---|---|---|---|---|---|
| rs7994900 | 13:114544790 | GAS6 | African | 1.14057 | 0.9902, 1.291 | 7.02E−35 | 5.16E−29 | 0.233696 | 0.016582 |
| rs1862514 | 19:7684449 | XAB2 | African | 0.73301 | 0.4978, 0.9682 | 2.07E−09 | 1.14E−05 | 0.154891 | 0.041413 |
| rs2747454 | 6:29856483 | HLA-H | European | 0.445014 | 0.24, 0.65 | 1.43E−05 | 0.023978 | 0.256667 | 0.045863 |
| rs4723537 | 7:36577129 | AOAH | African | 0.493498 | 0.262, 0.725 | 1.97E−05 | 0.020752 | 0.328804 | 0.009453 |
| rs74798237 | 4:119635931 | - | European | −1.18637 | −1.6945, −0.6782 | 3.60E−06 | 0.008451 | 0.026667 | 0.00903 |
| rs200290730 | 4:120327079 | LINC01061 | European | 0.876502 | 0.7099, 1.0431 | 6.21E−21 | 3.29E−16 | 0.336667 | 0.024092 |
| rs3094055 | 6:31166315 | HCG27 | African | 0.576924 | 0.3058, 0.8481 | 2.04E−05 | 0.021266 | 0.144022 | 0.005556 |
| rs75388473 | 10:120867016 | FAM45A | European | −1.03071 | −1.531, −0.5305 | 3.44E−05 | 0.045278 | 0.026667 | 0.045113 |
| rs35742971 | 15:32760892 | - | European | 0.468504 | 0.2693, 0.6677 | 3.10E−06 | 0.007524 | 0.436667 | 0.036915 |
| rs28366321 | 6:32487723 | HLA-DRB5 | African | 0.511273 | 0.3002, 0.7224 | 1.71E−06 | 0.003133 | 0.133152 | 0.018226 |
| European | 0.824663 | 0.5911, 1.0582 | 2.12E−11 | 2.51E−07 | 0.136667 | 0.005158 | |||
| rs11218343 | 11:121403448 | SORL1 | African | −2.77498 | −3.6555, −1.8944 | 1.44E−09 | 8.48E−06 | 0.002717 | 0.0025 |
| European | −2.40548 | −3.1935, −1.6175 | 3.87E−09 | 2.58E−05 | 0.003333 | 0.012 |
Figure 2.
edQTLs associated with differentially edited sites. (A) A bubble plot showing a characteristic example with average frequency of editing by genotype for AD cases (red) and controls (blue), combined across both AF and EU populations. The size of the bubble is proportional to the genotype frequency. (B) Genotype frequencies in cases and controls for rs9272105, shown in a. (C) Editing frequency in cases and controls for site at 6:32487723, shown in A. (D) A characteristic example of an interaction effect between genotype and case–control status on frequency of editing. (E) Editing frequency in cases and controls for site at 13:114544790, shown in D.
In addition to edQTLs at different frequencies between cases and controls, we also tested for edQTLs that have a stronger effect size in cases compared to controls and vice versa in an interaction analysis. We identified 40 differentially edited sites where the underlying edQTL had a significantly increased effect in cases (26 in AF alone, 11 in EU alone, 3 in both) and 26 sites with a significantly decreased effect of the edQTL in cases (17 in AF alone, 9 in EU alone). One site with a significantly increased edQTL effect in cases was located in an intron of GAS6 (Fig. 2B). Overall, more than half of the edQTLs that were associated with differentially edited sites either appeared at significantly different frequencies in cases versus controls or had a significant interaction effect with case–control status.
Discussion
In this study, we found a total of 4487 genetic variants that acted as edQTLs in whole blood. SNPs that were close to the RNA editing site were more strongly associated with editing rates than those further away, which is consistent with findings from previous studies of edQTLs (39,40) and with the hypothesis that edQTLs can act by altering RNA secondary structure near the editing target site. Among the identified edQTLs, 1138 edQTLs showed a significant effect in the same direction in both the AF and EU populations, and percent global ancestry did not appear to be strongly associated with the vast majority of shared signals (92.4%). Of the remaining edQTLs, 720 had a MAF > 0.05 in only one population, and 2531 had MAF > 0.05 in both populations but showed ancestry-specific effects. This is consistent with previous studies that also observed substantial differences in edQTLs across populations, most noticeably between African and European ancestries (39). In an edQTL study of lymphoblastoid cell lines from four European populations (CEU-Utah, FIN-Finland, GBR-Britain, TSI-Italy) and one African population (YRI-Nigeria), nearly half of the edQTLs identified in YRI cell lines were unique to that population (39). Our study finds similar differences, despite higher levels of admixture, further highlighting the need to conduct functional genomic studies in genetically distinct populations. Further, our study has a broader scope and uses whole blood samples rather than cell lines. The use of whole blood versus cell lines provided the additional benefit of larger and more diverse sample sizes that are more representative of the broader population and allow a deeper investigation of RNA editing in the context of disease. In addition, whole blood has the added benefit of easy accessibility for potential future applications as a biomarker or expansions into additional underrepresented populations.
Interestingly, some of the edQTLs we identified were in LD with an AD-associated SNP reported in at least one of two recent genome-wide association studies in a European cohort and an African-American cohort, respectively (13,14). These included SNPs in SORL1, SPI1 and HLA-DRB1 that were associated with differential editing. In particular, SORL1 influences differential sorting of APP and regulation of amyloid-β production (41–43), and alterations to the gene interfere with APP trafficking by weakening the interaction of the SORL1 protein with full-length APP (44). Transcriptome regulatory functions may further disrupt SORL1 activity and contribute to disease pathogenesis. The HLA-DRB1 region was also heavily enriched for edQTLs, which may be attributable to the high levels of RNA editing in this region as RNA editing is thought to impact the downstream processing of HLA transcripts and to increase the variability and diversity of HLAs (45,46). Interestingly, an edQTL in WDR70, a gene region associated with AD in populations with African ancestry, was significantly associated with editing in EU but not AF. Notably, a lack of editing at a site may be as influential as the presence of editing; for example, blocking editing in GluR-B in mice leads to intractable seizures (47). Thus, disrupted RNA editing is one possible mechanism for the functional effect of AD-risk variants and may contribute to observed differences in the genetic etiology of AD between ancestral populations.
In addition to edQTLs in AD-associated regions, we found edQTLs associated with 148 unique sites that were differentially edited between AD cases and controls. Our previous work showed that differential editing was not necessarily associated with differential expression, but many differentially edited sites were predicted to have downstream consequences including disrupting miRNA binding and inducing deleterious coding changes, including in genes associated with AD-related pathways (35). Of the edQTLs associated with differentially edited sites, 11 had significant differences in genotype frequencies between cases and controls, indicating that differences in genotype frequency may be driving downstream differences in editing, including in some functional candidate genes such as GAS6. GAS6 overexpression has been shown to rescue cortical neurons from amyloid beta-induced apoptosis (48) and to inhibit Toll-like receptor-mediated inflammatory pathways in microglia (49), but human omics data linking GAS6 to AD have been limited. Another 66 differentially edited sites were associated with edQTLs that had different effect sizes in cases compared to controls. Previous evidence suggests that the pathological changes of AD alter RNA processing, including changes to the processing of non-coding RNAs and mRNA translation associated with the pathological features of AD (50,51) or increased processing of SINE Alu RNAs in the human brain associated with amyloid beta pathology (52). Consequently, it is possible that the intracellular and pathological features associated with the AD disease state may influence RNA secondary structure and thus change the activity of edQTLs, though further work is needed to assess this hypothesis. While replication of the reported findings is needed, this study suggests that analyses of RNA editing and edQTLs may provide new tools for candidate gene discovery and functional genomics work to provide additional evidence for existing candidate genes.
While this study offers new insights into post-transcriptional changes associated with AD, several limitations apply. Notably, this study focused on discovery and hypothesis generation, and thus further work is needed to replicate the findings reported here with particular emphasis on novel signals outside a priori hypotheses loci. While great efforts were taken to mitigate the differences in sex composition between the AF and EU groups, additional work is needed to explore the effects of biological sex on RNA editing both in health and disease. This study emphasized the importance of including ancestrally diverse populations in genomic and transcriptomic studies, but additional work should continue to expand the diversity of study samples beyond those included here. Furthermore, while several publications have suggested that the pathological changes seen in AD are associated with changes in RNA processing, the limits of our study do not allow us to establish a causal relationship between AD status and RNA editing, and a two-direction Mendelian randomization test would be helpful in future studies to further strengthen the link between AD, AD-risk alleles and RNA processing.
This paper offers the first genome-wide view of RNA editing regulation in AD and importantly in individuals of non-European ancestry. As such, this work complements ongoing large-scale efforts in genomics and functional genomics in the Alzheimer community including genome-wide association studies (13,14,17), whole exome and genome sequencing studies (53–55) and functional genomic studies including bulk and single cell transcriptomics (28,27). Importantly, these functional studies have identified differing gene expression profiles in whole blood (27) and in specific brain cell types (28) depending on ancestry supporting this important extension of research. Future analysis of RNA editing in larger datasets, including using whole-genome sequencing data rather than genotyping arrays, and expanding the ancestral backgrounds of individuals will provide a more complete picture of the potential role of RNA editing and its regulatory variants in AD.
Overall, these findings indicate that a substantial portion of AD-associated changes in RNA editing levels may be mediated through underlying genetic architecture and that altered editing may be a functional outcome of some variants in AD-associated regions. Furthermore, we found significant differences between edQTLs in AF and EU, suggesting that ancestry-specific background effects of edQTLs may contribute to observed differences in the genetic etiology of AD across ethnicities.
Materials and Methods
Sample collection
All participants in this study provided informed consent or the immediate next of kin or legal representative provided written consent on the behalf of the participant prior to their inclusion with oversight by the University of Miami Institutional Review Board protocol #20070307. Participants include 428 individuals (AF—105 with AD, 105 cognitively intact; EU—111 with AD, 107 cognitively intact) ascertained by the John P. Hussman Institute for Human Genomics (HIHG) at the University of Miami Miller School of Medicine (Miami, FL), Wake Forest University (Winston-Salem, NC) and Case Western Reserve University (Cleveland, OH) (Supplementary Material, Table S1). Each of the participants was ascertained using the protocol approved by the proper institutional review boards (IRBs). Patients were collected for this study over the course of 10 years, with IRB protocols and amendments being approved at each stage.
All participants underwent rigorous phenotyping; diagnostic criteria followed the previously described criteria of the National Institute of Neurological and Communicative Disorders and Stroke—Alzheimer’s Disease and Related Disorders Association (12,56,57). The cognitive status of controls was measured with either the Mini-Mental State Examination (58) or the Modified Mini-Mental State (59) combined with the Clinical Dementia Rating Scale, which assesses functional decline (60). All individuals included in this study resided in the USA and self-identified their race and ethnicity using the format of the 1990 US census (12). Specifically, individuals in the AF cohort self-identified as ‘Black or African American’ and individuals in the EU cohort self-identified as ‘White’.
Genotyping
Genome-wide SNP genotyping was performed on the Illumina Infinium Global Screening Array (Illumina, San Diego, CA). Quality control procedures were performed with PLINK v.2 (61). All samples had a call rate >95% and sex concordance using X-chromosome markers were consistent. All samples were confirmed to be unique and unrelated by using identity by descent estimation. SNPs with the call rate <97% were eliminated from further analysis. Given the challenges of accurate imputation in diverse and admixed populations (62), we limited our study to only the genotyped SNPs to maximize the confidence in our edQTL mapping.
Global genetic ancestry was determined using a model-based clustering algorithm using genotyping data implemented in the ADMIXTURE software (63). Supervised ADMIXTURE analysis was performed at K = 3 by including three reference populations (African, Amerindian and European) from the Human Genome Diversity Project as reference populations (64). Global ancestry of AF individuals ranged from 99.4% African and 0.6% European to 37.1% African and 61.2% European ancestry with a mean of 82.2% (± 12.4%) African ancestry, while EU individuals ranged from 33.4% African and 66.2% European to 0% African and 99.8% European with a mean of 96.8% (± 7.0%) European ancestry (Supplementary Material, Fig. S1). Among the 218 individuals in the EU cohort, only 1 had African ancestry between 10 and 20% and 1 had African ancestry >20%. By contrast, of the 210 individuals in the AF cohort, 69 had European ancestry between 10 and 20% and 62 had European ancestry >20%. Only three individuals in the AF cohort had European ancestry >50%.
Identification of editing sites
The RNA editing data used in this study were previously described in Gardner et al. (35). Briefly, RNA was isolated from whole blood collected in PAXgene RNA tubes (PreAnalytiX, Hombrechtikon, Switzerland) utilizing automation on the QIAsymphony instrument (Qiagen, Germantown, MD), quantified on the 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA), prepped for sequencing using the NuGEN Universal Plus mRNA-Seq with globin and ribosomal depletion (NuGEN, San Carlos, CA), and sequenced on paired end 100 bp reactions to generate at least 40 million reads on the Illumina HiSeq3000. Raw FASTQ reads were processed through an in-house bioinformatics pipeline. We then used the REDItools Denovo software package, version 1.0.4, to create tables of every potential RNA editing site (65) and used standard filtration criteria.
Editing frequency at an individual site is given by the number of edited reads divided by the total number of reads mapped to that specific site. Individual sites that were differentially edited between cases and controls, as well as their downstream molecular effects, were previously reported in Gardner et al. These differentially edited sites were identified using a logistic regression model with case–control status as the outcome variable and the RNA editing frequency as the independent variable, correcting for covariates, in R software version 3.5.1 (https://www.r-project.org/) (35).
Power analysis
Statistical power analysis was performed for sample size estimation, based on data from a published edQTL study by Park et al. (N = 445), comparing genotype to editing frequency (39). With a Bonferroni-corrected alpha = 0.05, power = 0.80, the number of SNPs tested = 641 790, an estimated slope of 0.13 and MAF = 0.1, the projected sample size needed is approximately N = 189 for this simple linear regression (Package ‘powerEQTL’ version 0.1.3, R statistical software, version 3.5.1) (66). Thus, our proposed sample size of 210 for AF and 218 for EU is adequate for the main objective of this study and allows for expected attrition and additional objectives of controlling for possible mediating/moderating factors and subgroup analysis.
Assessment of blood cell type heterogeneity
The blood cell type composition in AF and EU was assessed using CIBERSORTx with default parameters using the PMBC single cell RNAseq data as training data (https://cibersortx.stanford.edu/, Access Date: 1/6/2022) (67,68). For each cell type, a logistic regression with ancestry as the outcome variable and cell type percentage as the independent variable, adjusting for sex, age and batch, was used to test for differences in cell type across ancestral groups. No statistically significant differences were identified between groups (Supplementary Material, Table S2).
RNA editing QTL mapping
We performed edQTL mapping for all edited sites using a hurdle model (Package ‘pscl’ version 1.5.2, R statistical software, version 3.5.1) (69). The hurdle model is a two-part model for zero-inflated data. By using this model, we were able to test for edQTLs that are associated with not only the rate of RNA editing but also the probability of editing at a specific site. The probability of editing at a site refers to whether an individual has any edited transcripts mapping to a specific site versus none, a binary outcome, whereas the rate of RNA editing refers to the percent of transcripts that are edited in individuals who have any editing mapping to that site, a continuous outcome. Additional covariates included in the model were age, RNA sequencing batch, sample collection center, sex and overall RNAseq read depth. We applied our edQTL analysis to both AF and EU samples separately to explore how ancestral background influences RNA editing across populations. Given the high degree of admixture in some individuals in the AF cohort, we further investigated the contribution of European ancestry to overlapping or shared signals between the AF and EU cohorts by testing these edQTLs for association with percent global European ancestry within AF individuals using a hurdle model as above with the addition of percent European ancestry.
Our input data included genotyping data at 641790 markers using the Global Screening Array and 44 985 editing events previously generated from RNAseq data (35). In keeping with parameters previously established in a study of edQTLs across multiple populations, we limited our tests to SNPs within 200 kb of an editing site (39), since RNA editing is largely regulated in cis as edQTLs often act through altering the hairpin loop secondary structure of RNA (70), and edQTLs identified outside of this region are less likely to be true positives (39). We then adjusted for multiple testing using false discovery rate (FDR), and considered edQTLs with FDR < 0.05 and a MAF > 0.05 to be statistically significant. FDR was used because the power of the FDR method is uniformly larger than Bonferroni methods, especially as the number of hypothesis tests increases (71). Given the hypothesis generating nature of this study, our goal was to maximize the power of the discovery phase to identify candidate for further evaluation.
Identification of edQTLs in AD-associated regions
To identify edQTLs in AD-associated regions, we used the top 25 associated markers from European (13) and the top 13 signals from a African-American AD genome-wide association studies (14) and identified LD blocks for each signal with an r2 ≥0.5 using the LDlink R package (version 1.0.1) (72). LD blocks were calculated independently for African and European ancestry individuals, and for each query allele, we used the larger LD block from the ancestry-specific results.
Identification of differentially edited sites influenced by genetic variants
We examined edQTLs that influence the rate of editing at sites that we previously identified as differentially edited between AD cases and controls in AF and EU populations (35). We used a Fisher’s exact test to examine whether differentially edited sites were more likely to be associated with an edQTL than non-differentially edited sites. We then used a Fisher’s exact test to test whether alleles appear in cases and controls at different rates. Interaction effects between affection status and genotype on editing levels were assessed using a hurdle regression model using the same covariates as were used for QTL mapping above. For both the Fisher’s exact test and the interaction test, we required an FDR < 0.05 for significance.
For edQTLs that were both in LD with a GWAS marker and associated with a differentially edited site, we conducted a formal colocalization analysis for the regions of interest using the coloc.abf() function with default parameters from the coloc package in R (version 5.1.0) (73). A posterior probability >0.75 was considered strong evidence that the variant in question was associated with both AD status and differential RNA editing.
Supplementary Material
Acknowledgements
We thank the community members who graciously agreed to participate in the study and made this research possible.
Conflict of Interest statement. The authors have no conflicts of interest to disclose.
Contributor Information
Olivia K Gardner, John P. Hussman Institute for Human Genomics, Miller School of Medicine, University of Miami, Miami, FL 33136, USA.
Derek Van Booven, John P. Hussman Institute for Human Genomics, Miller School of Medicine, University of Miami, Miami, FL 33136, USA.
Lily Wang, John P. Hussman Institute for Human Genomics, Miller School of Medicine, University of Miami, Miami, FL 33136, USA; Department of Public Health Sciences, Miller School of Medicine, University of Miami, Miami, FL 33136, USA.
Tianjie Gu, John P. Hussman Institute for Human Genomics, Miller School of Medicine, University of Miami, Miami, FL 33136, USA.
Natalia K Hofmann, John P. Hussman Institute for Human Genomics, Miller School of Medicine, University of Miami, Miami, FL 33136, USA.
Patrice L Whitehead, John P. Hussman Institute for Human Genomics, Miller School of Medicine, University of Miami, Miami, FL 33136, USA.
Karen Nuytemans, John P. Hussman Institute for Human Genomics, Miller School of Medicine, University of Miami, Miami, FL 33136, USA; Maya Angelou Center for Health Equity, Wake Forest University, Winston-Salem, NC 27101, USA.
Kara L Hamilton-Nelson, John P. Hussman Institute for Human Genomics, Miller School of Medicine, University of Miami, Miami, FL 33136, USA.
Larry D Adams, John P. Hussman Institute for Human Genomics, Miller School of Medicine, University of Miami, Miami, FL 33136, USA.
Takiyah D Starks, Maya Angelou Center for Health Equity, Wake Forest University, Winston-Salem, NC 27101, USA.
Michael L Cuccaro, John P. Hussman Institute for Human Genomics, Miller School of Medicine, University of Miami, Miami, FL 33136, USA; Department of Human Genetics, Dr. John T Macdonald Foundation, Miller School of Medicine, University of Miami, Miami, FL 33136, USA.
Eden R Martin, John P. Hussman Institute for Human Genomics, Miller School of Medicine, University of Miami, Miami, FL 33136, USA; Department of Human Genetics, Dr. John T Macdonald Foundation, Miller School of Medicine, University of Miami, Miami, FL 33136, USA.
Jeffery M Vance, John P. Hussman Institute for Human Genomics, Miller School of Medicine, University of Miami, Miami, FL 33136, USA; Department of Human Genetics, Dr. John T Macdonald Foundation, Miller School of Medicine, University of Miami, Miami, FL 33136, USA.
William S Bush, Department of Population & Quantitative Health Sciences, Case Western Reserve University, Cleveland, OH 44106, USA; Cleveland Institute for Computational Biology, Cleveland, OH 44106, USA.
Goldie S Byrd, Maya Angelou Center for Health Equity, Wake Forest University, Winston-Salem, NC 27101, USA.
Jonathan L Haines, Department of Population & Quantitative Health Sciences, Case Western Reserve University, Cleveland, OH 44106, USA; Cleveland Institute for Computational Biology, Cleveland, OH 44106, USA.
Gary W Beecham, John P. Hussman Institute for Human Genomics, Miller School of Medicine, University of Miami, Miami, FL 33136, USA; Department of Human Genetics, Dr. John T Macdonald Foundation, Miller School of Medicine, University of Miami, Miami, FL 33136, USA.
Margaret A Pericak-Vance, John P. Hussman Institute for Human Genomics, Miller School of Medicine, University of Miami, Miami, FL 33136, USA; Department of Human Genetics, Dr. John T Macdonald Foundation, Miller School of Medicine, University of Miami, Miami, FL 33136, USA.
Anthony J Griswold, John P. Hussman Institute for Human Genomics, Miller School of Medicine, University of Miami, Miami, FL 33136, USA; Department of Human Genetics, Dr. John T Macdonald Foundation, Miller School of Medicine, University of Miami, Miami, FL 33136, USA.
Funding
National Institutes of Health (NIH)/National Institute on Aging (NIA) (grants AG028786, AG027944, AG070935, AG070864, AG072547 and an administrative supplement to NIH/NIA grant U01AG052410-01).
References
- 1. Alzheimer's Association (2014) 2014 Alzheimer’s disease facts and figures. Alzheimers Dement., 10, e47–e92. [DOI] [PubMed] [Google Scholar]
- 2. Gatz, M., Pedersen, N.L., Berg, S., Johansson, B., Johansson, K., Mortimer, J.A., Posner, S.F., Viitanen, M., Winblad, B. and Ahlbom, A. (1997) Heritability for Alzheimer’s disease: the study of dementia in Swedish twins. J. Gerontol. A Biol. Sci. Med. Sci., 52A, M117–M125. [DOI] [PubMed] [Google Scholar]
- 3. Pericak-Vance, M.A., Yamaoka, L.H., Haynes, C.S., Speer, M.C., Haines, J.L., Gaskell, P.C., Hung, W.Y., Clark, C.M., Heyman, A.L., Trofatter, J.A.et al. (1988) Genetic linkage studies in Alzheimer’s disease families. Exp. Neurol., 102, 271–279. [DOI] [PubMed] [Google Scholar]
- 4. Farrer, L.A., Cupples, L.A., Haines, J.L., Hyman, B., Kukull, W.A., Mayeux, R., Myers, R.H., Pericak-Vance, M.A., Risch, N. and vanDuijn, C.M. (1997) Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA, 278, 1349–1356. [PubMed] [Google Scholar]
- 5. Pericak-Vance, M.A., Bebout, J.L., Gaskell PC Jr, Yamaoka, L.H., Hung, W.Y., Alberts, M.J., Walker, A.P., Bartlett, R.J., Haynes, C.A. and Welsh, K.A. (1991) Linkage studies in familial Alzheimer disease: evidence for chromosome 19 linkage. Am. J. Hum. Genet., 48, 1034–1050. [PMC free article] [PubMed] [Google Scholar]
- 6. Strittmatter, W.J., Weisgraber, K.H., Huang, D.Y., Dong, L.M., Salvesen, G.S., Pericak-Vance, M., Schmechel, D., Saunders, A.M., Goldgaber, D. and Roses, A.D. (1993) Binding of human apolipoprotein E to synthetic amyloid beta peptide: isoform-specific effects and implications for late-onset Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A., 90, 8098–8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Corder, E.H., Saunders, A.M., Strittmatter, W.J., Schmechel, D.E., Gaskell, P.C., Small, G.W., Roses, A.D., Haines, J.L. and Pericak-Vance, M.A. (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science, 261, 921–923. [DOI] [PubMed] [Google Scholar]
- 8. Corder, E.H., Saunders, A.M., Risch, N.J., Strittmatter, W.J., Schmechel, D.E., Gaskell, P.C.J., Rimmler, J.B., Locke, P.A., Conneally, P.M., Schmader, K.E.et al. (1994) Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat. Genet., 7, 180–184. [DOI] [PubMed] [Google Scholar]
- 9. Lambert, J.C., Ibrahim-Verbaas, C.A., Harold, D., Naj, A.C., Sims, R., Bellenguez, C., DeStafano, A.L., Bis, J.C., Beecham, G.W., Grenier-Boley, B.et al. (2013) Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet., 45, 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jun, G.R., Chung, J., Mez, J., Barber, R., Beecham, G.W., Bennett, D.A., Buxbaum, J.D., Byrd, G.S., Carrasquillo, M.M., Crane, P.K.et al. (2017) Transethnic genome-wide scan identifies novel Alzheimer’s disease loci. Alzheimers Dement., 13, 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. International Genomics of Alzheimer's Disease Consortium (IGAP) (2015) Convergent genetic and expression data implicate immunity in Alzheimer’s disease. Alzheimers Dement., 11, 658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reitz, C., Jun, G., Naj, A., Rajbhandary, R., Vardarajan, B.N., Wang, L.-S., Valladares, O., Lin, C.-F., Larson, E.B., Graff-Radford, N.R.et al. (2013) Variants in the ATP-binding cassette transporter (ABCA7), apolipoprotein E 4,and the risk of late-onset Alzheimer disease in African Americans. JAMA, 309, 1483–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kunkle, B.W., Grenier-Boley, B., Sims, R., Bis, J.C., Damotte, V., Naj, A.C., Boland, A., Vronskaya, M., van derLee, S.J., Amlie-Wolf, A.et al. (2019) Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet., 51, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kunkle, B.W., Schmidt, M., Klein, H.-U., Naj, A.C., Hamilton-Nelson, K.L., Larson, E.B., Evans, D.A., De Jager, P.L., Crane, P.K., Buxbaum, J.D.et al. (2021) Novel Alzheimer disease risk loci and pathways in African American individuals using the African genome resources panel: a meta-analysis. JAMA Neurol., 78, 102–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tang, M.X., Cross, P., Andrews, H., Jacobs, D.M., Small, S., Bell, K., Merchant, C., Lantigua, R., Costa, R., Stern, Y.et al. (2001) Incidence of AD in African-Americans, Caribbean Hispanics, and Caucasians in northern Manhattan. Neurology, 56, 49–56. [DOI] [PubMed] [Google Scholar]
- 16. Tang, M.X., Stern, Y., Marder, K., Bell, K., Gurland, B., Lantigua, R., Andrews, H., Feng, L., Tycko, B. and Mayeux, R. (1998) The APOE-epsilon 4 allele and the risk of Alzheimer disease among African Americans, whites, and Hispanics. JAMA, 279, 751–755. [DOI] [PubMed] [Google Scholar]
- 17. Rajabli, F., Feliciano, B.E., Celis, K., Hamilton-Nelson, K.L., Whitehead, P.L., Adams, L.D., Bussies, P.L., Manrique, C.P., Rodriguez, A., Rodriguez, V.et al. (2018) Ancestral origin of ApoE ε4 Alzheimer disease risk in Puerto Rican and African American populations. PLoS Genet., 14, e1007791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Blue, E.E., Horimoto, A.R.V.R., Mukherjee, S., Wijsman, E.M. and Thornton, T.A. (2019) Local ancestry at APOE modifies Alzheimer’s disease risk in Caribbean Hispanics. Alzheimers Dement., 15, 1524–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cukier, H.N., Kunkle, B.W., Vardarajan, B.N., Rolati, S., Hamilton-Nelson, K.L., Kohli, M.A., Whitehead, P.L., Dombroski, B.A., Van Booven, D., Lang, R.et al. (2016) ABCA7 frameshift deletion associated with Alzheimer disease in African Americans. Neurol. Genet., 2, e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Naj, A.C., Schellenberg, G.D. and for the Alzheimer's Disease Genetics Consortium (ADGC) (2017) Genomic variants, genes, and pathways of Alzheimer’s disease: an overview. Am. J. Med. Genet. B Neuropsychiatr. Genet., 174, 5–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Allen, M., Zou, F., Chai, H.S., Younkin, C.S., Crook, J., Pankratz, V.S., Carrasquillo, M.M., Rowley, C.N., Nair, A.A., Middha, S.et al. (2012) Novel late-onset Alzheimer disease loci variants associate with brain gene expression. Neurology, 79, 221–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Martiskainen, H., Viswanathan, J., Nykanen, N.-P., Kurki, M., Helisalmi, S., Natunen, T., Sarajarvi, T., Kurkinen, K.M.A., Pursiheimo, J.-P., Rauramaa, T.et al. (2015) Transcriptomics and mechanistic elucidation of Alzheimer’s disease risk genes in the brain and in vitro models. Neurobiol. Aging, 36, 1221.e15–1221.e28. [DOI] [PubMed] [Google Scholar]
- 23. Karch, C.M., Jeng, A.T., Nowotny, P., Cady, J., Cruchaga, C. and Goate, A.M. (2012) Expression of novel Alzheimer’s disease risk genes in control and Alzheimer’s disease brains. PLoS One, 7, e50976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Humphries, C., Kohli, M.A., Whitehead, P., Mash, D.C., Pericak-Vance, M.A. and Gilbert, J. (2015) Alzheimer disease (AD) specific transcription, DNA methylation and splicing in twenty AD associated loci. Mol. Cell. Neurosci., 67, 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stopa, E.G., Tanis, K.Q., Miller, M.C., Nikonova, E.V., Podtelezhnikov, A.A., Finney, E.M., Stone, D.J., Camargo, L.M., Parker, L., Verma, A.et al. (2018) Comparative transcriptomics of choroid plexus in Alzheimer’s disease, frontotemporal dementia and Huntington’s disease: implications for CSF homeostasis. Fluids Barriers CNS, 15, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rangaraju, S., Dammer, E.B., Raza, S.A., Gao, T., Xiao, H., Betarbet, R., Duong, D.M., Webster, J.A., Hales, C.M., Lah, J.J.et al. (2018) Quantitative proteomics of acutely-isolated mouse microglia identifies novel immune Alzheimer’s disease-related proteins. Mol. Neurodegener., 13, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Griswold, A.J., Sivasankaran, S.K., Van Booven, D., Gardner, O.K., Rajabli, F., Whitehead, P.L., Hamilton-Nelson, K.L., Adams, L.D., Scott, A.M., Hofmann, N.K.et al. (2020) Immune and inflammatory pathways implicated by whole blood transcriptomic analysis in a diverse ancestry Alzheimer’s disease cohort. J. Alzheimers Dis., 76, 1047–1060. [DOI] [PubMed] [Google Scholar]
- 28. Griswold, A.J., Celis, K., Bussies, P.L., Rajabli, F., Whitehead, P.L., Hamilton-Nelson, K.L., Beecham, G.W., Dykxhoorn, D.M., Nuytemans, K., Wang, L.et al. (2021) Increased APOE ε4 expression is associated with the difference in Alzheimer’s disease risk from diverse ancestral backgrounds. Alzheimers Dement., 17, 1179–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gott, J.M. and Emeson, R.B. (2000) Functions and mechanisms of RNA editing. Annu. Rev. Genet., 34, 499–531. [DOI] [PubMed] [Google Scholar]
- 30. Bazak, L., Haviv, A., Barak, M., Jacob-Hirsch, J., Deng, P., Zhang, R., Isaacs, F.J., Rechavi, G., Li, J.B., Eisenberg, E.et al. (2014) A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res., 24, 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang, L., Yang, C.S., Varelas, X. and Monti, S. (2016) Altered RNA editing in 3′ UTR perturbs micro RNA-mediated regulation of oncogenes and tumor-suppressors. Sci. Rep., 6, 23226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maas, S., Kawahara, Y., Tamburro, K.M. and Nishikura, K. (2006) A-to-I RNA editing and human disease. RNA Biol., 3, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tariq, A. and Jantsch, M.F. (2012) Transcript diversification in the nervous system: A to I RNA editing in CNS function and disease development. Front. Neurosci., 6, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Slotkin, W. and Nishikura, K. (2013) Adenosine-to-inosine RNA editing and human disease. Genome Med., 5, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gardner, O.K., Wang, L., vanBooven, D., Whitehead, P.L., Hamilton-Nelson, K.L., Adams, L.D., Starks, T.D., Hofmann, N.K., Vance, J.M., Cuccaro, M.L.et al. (2019) RNA editing alterations in a multi-ethnic Alzheimer disease cohort converge on immune and endocytic molecular pathways. Hum. Mol. Genet., 28, 3053–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Khermesh, K., D’Erchia, A.M., Barak, M., Annese, A., Wachtel, C., Levanon, E.Y., Picardi, E. and Eisenberg, E. (2016) Reduced levels of protein recoding by A-to-I RNA editing in Alzheimer’s disease. RNA, 22, 290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gaisler-Salomon, I., Kravitz, E., Feiler, Y., Safran, M., Biegon, A., Amariglio, N. and Rechavi, G. (2014) Hippocampus-specific deficiency in RNA editing of GluA2 in Alzheimer’s disease. Neurobiol. Aging, 35, 1785–1791. [DOI] [PubMed] [Google Scholar]
- 38. GTEx Consortium (2020) The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science, 369, 1318–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Park, E., Guo, J., Shen, S., Demirdjian, L., Wu, Y.N., Lin, L. and Xing, Y. (2017) Population and allelic variation of A-to-I RNA editing in human transcriptomes. Genome Biol., 18, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ramaswami, G., Deng, P., Zhang, R., Anna Carbone, M., Mackay, T.F. and Billy Li, J. (2015) Genetic mapping uncovers cis-regulatory landscape of RNA editing. Nat. Commun., 6, 8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Andersen, O.M., Schmidt, V., Spoelgen, R., Gliemann, J., Behlke, J., Galatis, D., McKinstry, W.J., Parker, M.W., Masters, C.L., Hyman, B.T.et al. (2006) Molecular dissection of the interaction between amyloid precursor protein and its neuronal trafficking receptor SorLA/LR11. Biochemistry, 45, 2618–2628. [DOI] [PubMed] [Google Scholar]
- 42. Rogaeva, E., Meng, Y., Lee, J.H., Gu, Y., Kawarai, T., Zou, F., Katayama, T., Baldwin, C.T., Cheng, R., Hasegawa, H.et al. (2007) The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat. Genet., 39, 168–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Offe, K., Dodson, S.E., Shoemaker, J.T., Fritz, J.J., Gearing, M., Levey, A.I. and Lah, J.J. (2006) The lipoprotein receptor LR11 regulates amyloid beta production and amyloid precursor protein traffic in endosomal compartments. J. Neurosci., 26, 1596–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cuccaro, M.L., Carney, R.M., Zhang, Y., Bohm, C., Kunkle, B.W., Vardarajan, B.N., Whitehead, P.L., Cukier, H.N., Mayeux, R., St George-Hyslop, P.et al. (2016) SORL1 mutations in early- and late-onset Alzheimer disease. Neurol. Genet., 2, e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mannion, N.M., Greenwood, S.M., Young, R., Cox, S., Brindle, J., Read, D., Nellaker, C., Vesely, C., Ponting, C.P. and McLaughlin, P.J. (2014) The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep., 9, 1482–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liddicoat, B.J., Hartner, J.C., Piskol, R., Ramaswami, G., Chalk, A.M., Kingsley, P.D., Sankaran, V.G., Wall, M., Purton, L.E., Seeburg, P.H.et al. (2016) Adenosine-to-inosine RNA editing by ADAR1 is essential for normal murine erythropoiesis. Exp. Hematol., 44, 947–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brusa, R., Zimmermann, F., Koh, D.S., Feldmeyer, D., Gass, P., Seeburg, P.H. and Sprengel, R. (1995) Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science, 270, 1677–1680. [DOI] [PubMed] [Google Scholar]
- 48. Yagami, T., Ueda, K., Asakura, K., Sakaeda, T., Nakazato, H., Kuroda, T., Hata, S., Sakaguchi, G., Itoh, N., Nakano, T.et al. (2002) Gas6 rescues cortical neurons from amyloid beta protein-induced apoptosis. Neuropharmacology, 43, 1289–1296. [DOI] [PubMed] [Google Scholar]
- 49. Gilchrist, S.E., Goudarzi, S. and Hafizi, S. (2020) Gas6 inhibits Toll-like receptor-mediated inflammatory pathways in mouse microglia via Axl and Mer. Front. Cell. Neurosci., 14, 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ashraf, G.M., Ganash, M. and Athanasios, A. (2019) Computational analysis of non-coding RNAs in Alzheimer’s disease. Bioinformation, 15, 351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ghosh, A., Mizuno, K., Tiwari, S.S., Proitsi, P., Gomez Perez-Nievas, B., Glennon, E., Martinez-Nunez, R.T. and Giese, K.P. (2020) Alzheimer’s disease-related dysregulation of mRNA translation causes key pathological features with ageing. Transl. Psychiatry, 10, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cheng, Y., Saville, L., Gollen, B., Veronesi, A.A., Mohajerani, M., Joseph, J.T. and Zovoilis, A. (2021) Increased Alu RNA processing in Alzheimer brains is linked to gene expression changes. EMBO Rep., 22, e52255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vardarajan, B.N., Barral, S., Jaworski, J., Beecham, G.W., Blue, E., Tosto, G., Reyes-Dumeyer, D., Medrano, M., Lantigua, R., Naj, A.et al. (2018) Whole genome sequencing of Caribbean Hispanic families with late-onset Alzheimer’s disease. Ann. Clin. Transl. Neurol., 5, 406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bis, J.C., Jian, X., Kunkle, B.W., Chen, Y., Hamilton-Nelson, K.L., Bush, W.S., Salerno, W.J., Lancour, D., Ma, Y., Renton, A.E.et al. (2020) Whole exome sequencing study identifies novel rare and common Alzheimer’s-associated variants involved in immune response and transcriptional regulation. Mol. Psychiatry, 25, 1859–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ma, Y., Jun, G.R., Zhang, X., Chung, J., Naj, A.C., Chen, Y., Bellenguez, C., Hamilton-Nelson, K., Martin, E.R., Kunkle, B.W.et al. (2019) Analysis of whole-exome sequencing data for Alzheimer disease stratified by APOE genotype. JAMA Neurol., 76, 1099–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. McKhann, G., Drachman, D., Folstein, M., Katzman, R., Price, D. and Stadlan, E.M. (1984) Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology, 34, 939–944. [DOI] [PubMed] [Google Scholar]
- 57. McKhann, G.M., Knopman, D.S., Chertkow, H., Hyman, B.T., Jack, C.R.J., Kawas, C.H., Klunk, W.E., Koroshetz, W.J., Manly, J.J., Mayeux, R.et al. (2011) The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement., 7, 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Folstein, M.F., Folstein, S.E. and McHugh, P.R. (1975) ‘Mini-mental state’. A practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res., 12, 189–198. [DOI] [PubMed] [Google Scholar]
- 59. Teng, E.L. and Chui, H.C. (1987) The Modified Mini-Mental State (3MS) examination. J. Clin. Psychiatry, 48, 314–318. [PubMed] [Google Scholar]
- 60. Morris, J.C. (1993) The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology, 43, 2412–2414. [DOI] [PubMed] [Google Scholar]
- 61. Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M.A.R., Bender, D., Maller, J., Sklar, P., deBakker, P.I.W., Daly, M.J.et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Huang, L., Li, Y., Singleton, A.B., Hardy, J.A., Abecasis, G., Rosenberg, N.A. and Scheet, P. (2009) Genotype-imputation accuracy across worldwide human populations. Am. J. Hum. Genet., 84, 235–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Alexander, D.H., Novembre, J. and Lange, K. (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res., 19, 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cavalli-Sforza, L.L. (2007) Human evolution and its relevance for genetic epidemiology. Annu. Rev. Genomics Hum. Genet., 8, 1–15. [DOI] [PubMed] [Google Scholar]
- 65. Picardi, E., D'Erchia, A.M., Montalvo, A. and Pesole, G. (2015) Using REDItools to detect RNA editing events in NGS datasets. Curr. Protoc. Bioinformatics, 49, 12.12.1–12.12.15. [DOI] [PubMed] [Google Scholar]
- 66. Dong, X., Li, X., Chang, T.-W., Scherzer, S.W., Weiss, S.T. and Qiu, W. (2021) powerEQTL: an R package and shiny application for sample size and power calculation of bulk tissue and single-cell eQTL analysis. Bioinformatics, 37, 4269–4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Newman, A.M., Liu, C.L., Green, M.R., Gentles, A.J., Feng, W., Xu, Y., Hoang, C.D., Diehn, M. and Alizadeh, A.A. (2015) Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods, 12, 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Newman, A.M., Steen, C.B., Liu, C.L., Gentles, A.J., Chaudhuri, A.A., Scherer, F., Khodadoust, M.S., Esfahani, M.S., Luca, B.A., Steiner, D.et al. (2019) Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol., 37, 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zeileis, A., Kleiber, C. and Jackman, S. (2008) Regression models for count data in R. J. Stat. Softw. Artic., 27, 1–25. [Google Scholar]
- 70. Walkley, C.R. and Li, J.B. (2017) Rewriting the transcriptome: adenosine-to-inosine RNA editing by ADARs. Genome Biol., 18, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Benjamini, Y. and Hochberg, Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B, 57, 289–300. [Google Scholar]
- 72. Machiela, M.J. and Chanock, S.J. (2015) LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics, 31, 3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Giambartolomei, C., Vukcevic, D., Schadt, E.E., Franke, L., Hingorani, A.D., Wallace, C. and Plagnol, V. (2014) Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet., 10, e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.