Abstract
The detection and disclosure of secondary findings (SFs) is a preventive strategy for medically-actionable hereditary health conditions. Some countries have guidelines on management and disclosure of SFs, while others avoid detection and disclosure of SFs. This study is a review of clinical guidelines from six countries and the European Union to identify similarities and differences among SF guidelines. Evidence from this review supports harmonization of guidelines across countries to promote broad international collaborations on genomics and to benefit precision medicine. This study can serve as a reference for development of SF management guidelines in China by contributing evidence from other countries to the ethical and methodological challenges under debate.
Keywords: global guideline, secondary genomic finding, incidental finding, health policy precision medicine, health economics evaluation, discrete choice experiment
INTRODUCTION
Genome sequencing provides information about disease risk, prognosis, or treatment response, and is rapidly becoming an affordable and useful tool for patients (1-2). Sequencing occasionally identifies gene variants, called secondary findings (SFs), that are neither relevant to the primary intention of sequencing nor to the patient’s health condition. Disclosure of these SFs to patients is much debated (3-5). Some pathogenic variants are not medically actionable, and the disclosure of these variants may cause the patient anxiety (6), psychological harm (7), or information overload (8). However, non-disclosure of some SFs may miss opportunities to encourage patients to consider life-planning options or accept interventional recommendations (9-10).
The lack of consensus on SF management has led to differences in clinical guidelines across countries (11). For example, the American College of Medical Genetics and Genomics (ACMG) recommends returning SFs that are medically-actionable and have high penetrance, and has published a list of gene-condition pairs that should be returned to patients on a consent basis (12). As a result, disclosure of SFs becomes a preventive strategy to increase health outcomes from genomic interventions, such as population genetic screening and opportunistic screening. In contrast, Canada takes a different approach and recommends avoiding detection and return of SFs to patients (13).
In the era of precision medicine (succinctly described as providing tailored treatments to the right patients), maintenance of public health and development of healthcare technologies have benefited from the accumulation of vast amounts of genomic data worldwide (14). Since SFs contribute a large proportion of the available genomic data, discordance between countries’ SF management and disclosure guidelines may hinder the accumulation of genomic data and the development of precision medicine to address patient- and population-level health (15). In contrast, harmonized international guidelines on disclosing SFs would benefit genomic data accumulation and precision medicine research based on genomics. Therefore, it is useful to examine county-level SF guidelines and their concordance to inform the development of harmonized global guidelines. Such an examination can provide a global landscape of guidelines for stakeholders in China, where no guidelines on SF disclosure currently exist, and can serve as a reference for the establishment of national guidelines.
METHODS
Search Strategy
This scoping review compared clinical guidelines on disclosing SFs derived from genome sequencing. EMBASE, PubMed, MEDLINE, Web of Science, and HumGen, an international database of law and policies related to human genetics (16), were searched for published guidelines. The “grey literature” was searched using Google and Google Scholar. The search terms were “incidental findings,” “secondary findings,” “unanticipated findings,” “unsolicited findings,” “ancillary findings,” “opportunistic findings,” and “accessory findings” (3–4), accompanied by terms related to genome sequencing and communication of results.
Inclusion Criteria
The study included documents published by professional organizations, governments, and bioethical committees that were relevant to SF disclosure in a clinical setting. The review was limited to documents written in English and published after 2010, when genome sequencing was initially used. The selection process included two rounds: screening by title and abstract, followed by full-text review (Figure 1).
Figure 1.
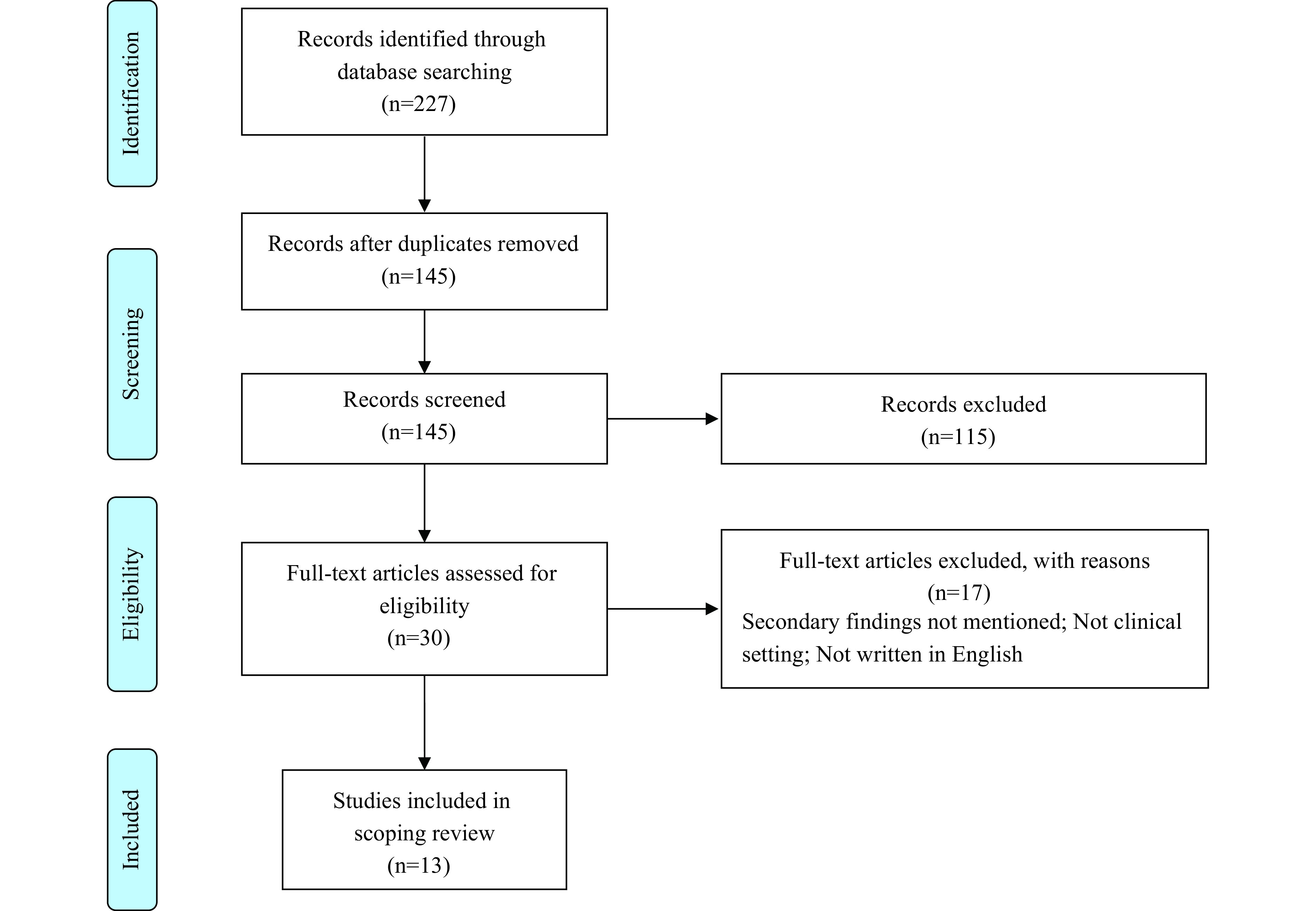
Article selection process.
Summary and Comparison
Characteristics and major contents of the guidelines were extracted from the original documents and included guideline title, source country, publication year, responsible professional organization, and major recommendations. Characteristics and recommendations were summarized by country, separately for adults and children.
To further compare guidelines, the major recommendations were summarized as succinct points, and the guidelines for each jurisdiction were checked for the presence or absence of these points. Results of the presence check were placed in a table to display similarities and differences of the guidelines.
RESULTS
Characteristics of Selected Articles
Thirteen articles were included in the review (Table 1) and covered the leading countries in genomics research and practice: the United States (US), the United Kingdom (UK), Canada, Australia, Germany, Denmark, and the European Union (EU). Contents of the clinical guidelines are shown in Table 2.
Table 1. Characteristics of included clinical guidelines.
| No. | Country (s) | Time | Organization | Targeted population | Title |
| Abbreviation: ACMG=American College of Medical Genetics and Genomics; AGNC=Association of Genetic Nurses and Counsellors; ASHG=American Society of Human Genetics; CCMG=Canadian College of Medical Geneticists; ESHG=European Society of Human Genetics; EU=European Union; HUGO=Human Genome Organization; P3G=Public Population Project in Genomics and Society; RCPA=Royal College of Pathologists of Australasia; UK=the United Kingdom; US=the United States. | |||||
| 1 | US | 2013 | Presidential Commission for the Study of Bioethical Issues | Adults; children and adolescents | Anticipate and communicate. Ethical management of incidental and secondary findings in the clinical, research, and direct-to-consumer contexts (17) |
| 2 | US | 2013 | ACMG | Adults; children and adolescents | Points to consider for informed consent for genome/exome sequencing (18) |
| 3 | US | 2013 | ACMG | Adults; children and adolescents | ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing (19) |
| 4 | US | 2014 | ACMG | Adults; children and adolescents | ACMG policy statement: updated recommendations regarding analysis and reporting of secondary findings in clinical genome-scale sequencing (20) |
| 5 | US | 2015 | ASHG | Children and adolescents | Ethical, legal, and psychosocial implications of genetic testing in children and adolescents (21) |
| 6 | US | 2017 | ACMG | Adults; children and adolescents | ACMG Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0) (22) |
| 7 | Canada | 2015 | CCMG | Adults; children and adolescents | Position statement of the Canadian college of medical geneticists (13) |
| 8 | EU | 2013 | ESHG | Adults; children and adolescents | Whole-genome sequencing in health care: recommendations of the European Society of Human Genetics (23) |
| 9 | EU | 2015 | P3G, ESHG, HUGO, and PHG Foundation | Children and adolescents | A statement on the continued importance of targeted approaches in newborn screening programmes (24) |
| 10 | UK | 2014 | AGNC | Adults; children and adolescents | Position statement on opportunistic genomic screening (25) |
| 11 | UK | 2014 | PHG Foundation | Adults | Realising genomics in clinical practice (26) |
| 12 | Germany | 2013 | German Ethics Council | Adults | The future of genetic diagnosis: from research to clinical practice (27) |
| 13 | Australia | 2014 | RCPA | Adults | Implementation of massively parallel sequencing in diagnostic medical genetic testing (28) |
Table 2. Contents of guidelines by country on the return of SFs in a clinical context.
| Guideline | Key points |
| Abbreviation: ACA=analytic validity, clinical significance, and actionability; ACMG=American College of Medical Genetics and Genomics; IFs=incidental findings; SFs=secondary findings; WES=whole exome sequencing; WGS=whole genome sequencing; UK=the United Kingdom; US=the United States. | |
| US, 2013 | • A minimum list of conditions, genes and variants should be routinely evaluated and reported to the ordering clinician who can place them into the context of that patient’s medical and family history, physical examination and other laboratory testing. |
| • Any findings included by the list should be reported by laboratories without seeking consents from the patient and family. | |
| • A clinician should be trained to communicate and interpret the results of incidental findings. | |
| US, 2014 | • Before performing the clinical sequencing, a written consent should be obtained from the patient, describing the interpretive uncertainty, privacy, possible impact on other family members, and the inevitable generation of data. |
| • Patients have the right to opt out of the analysis of the medically actionable gene list, so that they can choose not to receive the results of IFs. | |
| • Same policies apply to children as to adults. | |
| • Patients must decide whether to analyze the whole set of medically actionable genes (gene list) or none of them. It is infeasible to analyze a subset of the genes due to practical difficulties. | |
| US, 2015 | • The disclosure of IFs should occur only when the information has clear clinical utility. |
| • Informed consent should be obtained if there is a substantial likelihood of generating clinically relevant IFs. | |
| • Parents should be able to decline to receive IFs of their child. | |
| • In case of IFs that indicate urgent and serious implications for a child’s health, the results must be returned regardless of any preferences of the child and parents. | |
| • It is ethically acceptable, but not required, to search for IFs that are not relevant to the clinical indication for sequencing. | |
| US, 2017 | • ACMG setup a new process for accepting and evaluating nominations for updates to the secondary findings list. |
| • Based on the previous secondary findings list, 4 genes have been added and one gene been removed. A new updated secondary findings minimum list includes 59 medically actionable genes recommended for return in clinical genomic sequencing. | |
| Canada, 2003 | • Clinical genetics records should be maintained indefinitely except those that have minimal importance to future interaction in genetics with the patient or other family members. |
| Canada, 2015 | • Avoid the discovery of SFs, and focus on the primary indication of the test. |
| • Genome sequencing should only be considered when proved useful in the evaluation process and a selective filtering process is recommended. | |
| • The adult patient should have the right to receive or not to receive the results. | |
| • The results of the child must be returned to the parents if ACA criteria are satisfied. | |
| Canada and Europe, 2015 | • Targeted tests are recommended. |
| • If treatment or prevention is available during childhood and the variant indicates a serious problem, the IFs should be reported to parents. | |
| • When carrier status information is relevant to parents for reproductive choices, they could be offered this information. | |
| • Parents may also be offered information regarding unsolicited findings that are severe and clinically actionable relevant to their own health. | |
| UK, 2011 | • The possibility of incidental findings should be minimized. Targeted tests or analyses are preferred. |
| • Physicians have no obligation to provide IFs irrelevant with clinical question. | |
| • Informed consent is not sufficient to deal with IFs. | |
| • Where variants are of unknown significance, the responsibility to investigate might be less clear. | |
| UK, 2014 | • National Health Service (NHS) should adopt targeted tests using gene lists, which have greater clinical utility then WGS/WES. |
| • It should minimize the generation, interpretation and disclosure of IFs, because the potential harms are likely to outweigh the potential benefits. | |
| • Informed consent should be obtained from patients if it is necessary for WGS/WES, which should include the possible generation of IFs. | |
| • Disclosure decisions will be informed by clinical judgement, though patients should have opportunities to express views on disclosure. | |
| • Patients should be given the opportunity to opt out of re-contact. | |
| • Patients should be informed that the sequencing data would be stored in a secured, comprehensive, and accessible NHS database. | |
| • A multidisciplinary committee and many evidence-based studies are needed on the return of IFs. | |
| Denmark, 2012 | • Patients should be given a “degree of involvement in deciding whether, and to what extent, they are to receive feedback on any incidental findings.” |
| • Genetic testing should be accompanied by “impartial and comprehensive information as well as counseling, both before and after testing.” | |
| Europe, 2013 | • Targeted genetic tests are much preferred because they can avoid incidental findings that cannot be interpreted. |
| • Known genetic variants with limited or no clinical utility should be filtered out. | |
| • Patients’ claims to a right not to know about findings do not automatically override professional responsibilities. | |
| • Guidelines for testing minors should be established to balance the autonomy and interests of child and the parental rights of not receiving IFs of the child. | |
| Germany, 2013 | • The council refutes the idea that the doctor has the right to inform relatives of the patient who might also be affected by the genetic condition, or to recommend them to get genetic counseling. |
| • Prenatal genetic diagnosis should only be conducted if there is an increased risk of a genetic disease. | |
| • If the genetic information is irrelevant with the health of the born child, this finding should not be communicated to the pregnant woman. | |
| Australia, 2014 | • Genomic testing should have a sound evidence base before prescription. |
| • Targeted testing measures are recommended to avoid or minimize incidental findings. | |
| • Clinicians should use standard practices in deciding whether to return IFs given the patient has agreed to it. | |
Clinical Guidelines for Adults
US: In 2013, the US Presidential Commission for the Study of Bioethical Issues recognized the necessity for professional organizations to guide the management and disclosure of SFs (17). The Commission recommended that patients should be informed of the possibility of SFs, the scope of SFs that will be communicated, and the steps of discovering SFs, and that patient preference not to know SFs should be respected. In response, ACMG published two documents in 2013 (18-19). One document emphasized that patients should be informed of the possibility of finding SFs for which there may be interventions to prevent disease or reduce disease severity. The other document was specific to the reporting of SFs, and provided a list of reportable gene-condition pairs. ACMG recommended that listed variants should be reported regardless of the indication for sequencing and regardless of the age of the patient, including to minors. In 2014, ACMG updated its guidelines with two notable modifications: 1) patients should have the right to opt out of SF detection, and 2) once patients consent to the analysis, the entire list should be analyzed (20). In 2017 and again in 2021, ACMG updated the guidelines and extended the list of gene-disease pairs to additional SFs (12,21). ACMG also recognized that reporting SFs may introduce costs, impact individuals and families, and generate medical, legal, social, and economic ramifications that need to be assessed.
Canada: In 2015, the Canadian College of Medical Geneticists (CCMG) published guidelines that recommended a cautious approach towards the return of SFs to patients (13). Recognizing that reporting SFs is controversial, CCMG recommended focusing on the primary indication of the test, avoiding the discovery of SFs as much as possible. However, CCMG recommended that patients have the option to request and receive SFs.
EU: In 2013, the European Society of Human Genetics (ESHG) published guidelines recommending that SFs that are not interpretable or medically actionable should be avoided (22). However, if an SF indicates a serious health problem and is medically actionable, it should be reported to the patient, overriding the patient’s desire not to know.
UK: In 2014, the PHG Foundation published recommendations suggesting minimizing the generation, interpretation, and disclosure of SFs (23). In the same year, the Association of Genetic Nurses and Counsellors (AGNC) published a position paper stating that patients always have the right to opt-out of receiving SFs and that physicians should only disclose SFs if patients choose to receive them (24).
Germany: The German Ethical Council recognized the possibility of SFs with genome sequencing and recommended that the discovery of SFs should be avoided due to a lack of evidence on its benefits (25). The patient’s right not to know was emphasized.
Australia: In 2014, the Royal College of Pathologists of Australasia (RCPA) published guidelines and recommended that patients should be informed of the types of SFs that may be generated and reported, and have the option of not receiving SFs (26). Without a specific list, RCPA suggested that the return of SFs should be limited to variants that are pathogenic and deemed reportable.
Clinical Guidelines for Children
US: ACMG recommended the same policy for children as for adults. That is, once consent is obtained, SFs identified according to the ACMG SF list should be returned to the parents. Parents should have the option of opting out of the analysis for their child. In 2015, the American Society of Human Genetics (ASHG) published a specific position statement for children and adolescents, recommending that physicians should disclose SFs with clinical utility for the child and/or family members if the parents choose to receive them. If the SFs indicate a serious health problem for the child, disclosure should override parental preference.
Canada: CCMG recommended that physicians only return SFs to parents that indicate childhood-onset conditions with high penetrance and medical actionability. Variants that are associated with adult-onset conditions should not be reported, unless the parents want to receive them or disclosure could prevent serious harm to family members’ health.
EU: ESHG stated that benefits to the family members should be considered in a decision to disclose a child’s SF. In 2015, several professional organizations published a joint statement specific to newborn screening programs, recommending use of targeted sequencing to avoid irrelevant findings and focusing on the primary indications for testing (27).
UK: AGNC recommended that detection of children’s SFs associated with adult-onset conditions should be avoided.
Similarities and Differences in Guidelines
Table 3 shows similarities and differences between guidelines from the different countries. In terms of general attitude, the US guidelines take a more positive stance compared with other countries. For example, in the US, physicians have the right to search SFs with patient consent and they should return SFs to patients if the findings are on the ACMG SF list. Guidelines from other countries do not support physicians’ right to search SFs. Instead, they prioritize patients’ right to know or not to know any genomic findings. For children, the US uses the same guidelines used for adults, while the ESHG provides separate recommendations for children.
Table 3. Comparison of guidelines on returning SFs in clinical setting.
| Category | US | Canada | UK | Australia | Denmark | Germany | EU |
| Abbreviation: ACA=analytic validity, clinical significance, and actionability; SFs=secondary findings; All-or-none policy=patients could choose to receive all SFs or none of them; UK=the United Kingdom; US=the United States. | |||||||
| General attitude | |||||||
| Positive attitude | √ | ||||||
| Negative attitude | √ | √ | √ | √ | √ | ||
| Minimization of SFs | √ | ||||||
| No Harm principle | √ | ||||||
| For adults | |||||||
| ACA criteria | √ | ||||||
| Partial ACA criteria | √ | ||||||
| Informed consent | √ | √ | |||||
| Right to know | √ | √ | |||||
| Right not to know | √ | √ | √ | √ | |||
| All-or-none policy | √ | ||||||
| For clinicians | |||||||
| Must-return regardless of preference | √ | ||||||
| Obligation to disclose | √ | ||||||
| No obligation to disclose | √ | ||||||
| Right to search | √ | ||||||
| No obligation to search | √ | ||||||
| Case-by-case determination | √ | ||||||
| Decision by physicians | √ | √ | |||||
| For children | |||||||
| Specific guidelines for children | √ | ||||||
| Policy for children same as for adults | √ | ||||||
| For genomic data | |||||||
| Data protection | √ | √ | |||||
| Confidentiality | √ | √ | |||||
Lack of Evidence
The studied guidelines are discordant, reflecting three key differences. First, there is insufficient clinical evidence concerning the range of SFs that should be returned to patients. The ACMG guidelines recommend that only SFs associated with monogenic disorders be returned, and that SFs associated with disorders caused by structural variants, repeat expansions, or copy-number variations are insufficiently understood to be listed. Second, the health benefits of returning SFs are not clear. The CCMG guidelines recommend a cautious approach because the benefits of disclosure are not well established. The AGNC statement indicated that robust evidence for the benefits of SFs is needed. Third, there is a lack of evidence for the impact of disclosure on patients, physicians, and the healthcare system. For example, returning SFs that indicate disease risk will probably lead to confirmatory testing. If the disclosure becomes a routine practice for every patient, the downstream costs may become a financial burden for the healthcare system.
DISCUSSION
This study reviewed clinical guidelines of several countries concerning disclosure of SFs derived from genome sequencing. The review identified discordances between guidelines, as some countries take an open attitude towards disclosure, while others take a more cautious attitude. All guidelines value patient autonomy and the potential benefits of disclosing SFs with high penetrance and medical actionability. Generating new evidence for updating SF management guidelines is warranted.
Concordant Global Guidelines are Needed
SFs represent an important source of genomic data and are critically important for genomic research and precision medicine (28). Since the generation of valid evidence on the clinical implications of variants requires much larger datasets than do other medical research areas, it is necessary to collate clinical genome data internationally, including the retrieval of SFs. For example, recent studies usually employ genome data from nearly 300,000 participants (29). Ideally, international genome data should originate from multi-center, multidisciplinary, large-scale studies on a worldwide scale. Therefore, international collaboration on genome data is required.
Considering the significant role of SFs in the accumulated genomic data, globally concordant guidelines on detecting and disclosing SFs would greatly benefit the merging of genome data across countries and support large-scale international collaborations (11). Advances in genomic data accumulation may boost progress in genomic research and precision medicine, and the whole of society may be able to fully harness the benefits of genome sequencing.
Patient and Physician Preferences for SF Disclosure
Current guidelines recognize that some patients may want SFs returned and some patients may not. Understanding psychological motivations of wanting or not wanting to know SFs may provide important evidence for updating current guidelines. Methods such as discrete choice experiments (DCE) (30–31) and best-worst scaling (BWS) (32–33) would be appropriate tools to investigate psychological motivation (5,34).
Physician preferences on SFs disclosure are also fundamental for clinical guidelines. If physician views are not incorporated, the guidelines may lack consensus and generate much debate among physicians (35). Updated guidelines should consider the circumstances under which physicians decide to return SFs, irrespective of patient preference. It would be valuable to identify factors behind physicians’ decisions and simulate the effect of the factors (5). Regier et al. (2015) and Jiang et al. (2020) utilized DCE to elicit patient and physician preferences towards the return of SFs in Canada (5,36). Generation of more evidence on preferences for disclosure is warranted.
A Research Agenda to Inform Guidelines for China
Genomics-based precision medicine is increasingly being studied and used in China to advance healthcare technologies (37). For example, the China Precision Medicine Initiative (CPMI) is accelerating genomic research (38). The lack of guidelines on SF disclosure, however, indicates that precision medicine is at an early stage in China. Physicians and patients have no recommendations to follow when SFs are identified. Professional guidelines are needed to provide support and guidance to SF detection and disclosure in China. This review provides stakeholders in China with a landscape of international guidelines and their similarities and differences that can contribute to the development of guidelines in China.
Cost-effectiveness analysis (CEA) of disclosing SFs is important evidence supporting guideline development. CEA is an established method to inform decision-makers about the value for money of different healthcare technologies, interventions, or policies (39–40). However, CEAs that examine the value of returning SFs face methodological challenges due to the long-term impact of sequencing on quality-of-life and numerous downstream interventions after sequencing (41). Implementation of CEAs on returning SFs in China faces additional challenges, including measurement of quality-of-life, long-term financial impact, the use of modelling techniques to capture the impact of downstream interventions, and appropriate reimbursement thresholds (39,42–43). Researchers may need to overcome methodological difficulties to determine whether disclosing SFs is cost-effective in China. Such economic evidence is of critical importance for decision-makers developing guidelines on SF disclosure.
CONCLUSIONS
This study identified differences and similarities between several countries’ clinical guidelines on return of secondary genomic findings to patients. Evidence should be established on which SFs to return, the health benefits of disclosure, and the impact of disclosure on patients, physicians, and the healthcare system.
This study brings attention to the differences between clinical guidelines and supports calls for greater harmonization of guidelines across countries. Concordant guidelines would promote broader international collaboration on genomics and thus may help fully harness the benefits of genome sequencing for all.
This study provides a landscape of international guidelines for stakeholders to use as a reference for developing local guidelines on SF disclosure in China. More economic evaluations on the cost-effectiveness of returning SFs to patients are needed to facilitate the establishment of SF management and disclosure guidelines in China.
References
- 1.Biesecker LG. Opportunities and challenges for the integration of massively parallel genomic sequencing into clinical practice: lessons from the ClinSeq project. Genet Med 2012;14(4):393-8. https://www.nature.com/articles/gim201178.
- 2.Green RC, Rehm HL, Kohane IS. Clinical genome sequencing. In: Ginsburg GS, Willard HF, editors. Genomic and personalized medicine. Amsterdam: Elsevier. 2013; p. 102-22. https://www.genomes2people.org/wp-content/uploads/2012/07/Green-Rehm-Kohane-ClinGenomeSequencing-2012.pdf.
- 3.Mackley MP, Fletcher B, Parker M, Watkins H, Ormondroyd E. Stakeholder views on secondary findings in whole-genome and whole-exome sequencing: a systematic review of quantitative and qualitative studies. Genet Med 2017;19(3):283-93. https://www.nature.com/articles/gim2016109.
- 4.Delanne J, Nambot S, Chassagne A, Putois O, Pelissier A, Peyron C, et al. Secondary findings from whole-exome/genome sequencing evaluating stakeholder perspectives. A review of the literature. Eur J Med Genet 2019;62(6):103529. https://www.sciencedirect.com/science/article/pii/S1769721218301228?casa_token=Io3N7hZWhLkAAAAA:meFnUB7PfjIoSphlvfdzTknEa7r9HHLqhISuUIVdHnVofaG6xUPAlQCEV1EDpncU9MluAbXQHQ.
- 5.Jiang S, Anis AH, Cromwell I, Mohammadi T, Schrader KA, Lucas J, et al. Health-care practitioners’ preferences for the return of secondary findings from next-generation sequencing: a discrete choice experiment. Genet Med 2020;22(12):2011-9. https://www.nature.com/articles/s41436-020-0927-x.
- 6.Townsend A, Adam S, Birch PH, Lohn Z, Rousseau F, Friedman JM. “I want to know what's in Pandora’s Box”: comparing stakeholder perspectives on incidental findings in clinical whole genomic sequencing. Am J Med Genet Part A 2012;158A(10):2519-25. https://onlinelibrary.wiley.com/doi/full/10.1002/ajmg.a.35554?casa_token=39aw0eVQBnMAAAAA%3At1Rdqen1t9pkeewc6eWLJjhP2AoIkR7xnLAnAygaoBeT333PQzb6A9RdRXm0p2YjuuSxuWpkNlXiiw.
- 7.Downing NR, Williams JK, Daack-Hirsch S, Driessnack M, Simon CM. Genetics specialists’ perspectives on disclosure of genomic incidental findings in the clinical setting. Patient Educ Couns 2013;90(1):133-8. https://www.sciencedirect.com/science/article/pii/S0738399112003837?casa_token=giR11F1rz_wAAAAA:jt3cd371ip393YC4k31nyRLpBkdKZYKp38_eacO8grqVUI9NiXZpj3zRlYAuoiNmQT76dr2l6g.
- 8.Gray SW, Hicks-Courant K, Lathan CS, Garraway L, Park ER, Weeks JC Attitudes of patients with cancer about personalized medicine and somatic genetic testing. J Oncol Pract. 2012;8(6):329–35. doi: 10.1200/JOP.2012.000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mighton C, Carlsson L, Clausen M, Casalino S, Shickh S, McCuaig L, et al. Development of patient “profiles” to tailor counseling for incidental genomic sequencing results. Eur J Hum Genet 2019;27(7):1008-17. https://www.nature.com/articles/s41431-019-0352-2.
- 10.Hufnagel SB, Martin LJ, Cassedy A, Hopkin RJ, Antommaria AHM. Adolescents’ preferences regarding disclosure of incidental findings in genomic sequencing that are not medically actionable in childhood. Am J Med Genet Part A 2016;170(8):2083-8. https://onlinelibrary.wiley.com/doi/full/10.1002/ajmg.a.37730?casa_token=hngBVly5T5YAAAAA%3AbHiE-sUB0vnvDH7H3AJ-gi9lEHo4xeoYlgZcAw2DWqJKm_toHi23YtA95GKamehT5NQtDtUmbjyYTg.
- 11.Knoppers BM, Zawati MH, Sénécal K. Return of genetic testing results in the era of whole-genome sequencing. Nat Rev Genet 2015;16(9):553-9. https://www.nature.com/articles/nrg3960.
- 12.Miller DT, Lee K, Chung WK, Gordon AS, Herman GE, Klein TE, et al. ACMG SF v3.0 list for reporting of secondary findings in clinical exome and genome sequencing: a policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med 2021;23(8):1381-90. https://www.nature.com/articles/s41436-021-01172-3.
- 13.Boycott K, Hartley T, Adam S, Bernier F, Chong KR, Fernandez BA, et al. The clinical application of genome-wide sequencing for monogenic diseases in Canada: position statement of the Canadian college of medical geneticists. J Med Genet 2015;52(7):431-7. https://jmg.bmj.com/content/52/7/431.short.
- 14.Khoury MJ, Armstrong GL, Bunnell RE, Cyril J, Iademarco MF. The intersection of genomics and big data with public health: opportunities for precision public health. PLoS Med 2020;17(10):e1003373. https://journals.plos.org/plosmedicine/article?id=10.1371/journal.pmed.1003373.
- 15.Khoury MJ, Bowen MS, Clyne M, Dotson WD, Gwinn ML, Green RF, et al. From public health genomics to precision public health: a 20-year journey. Genet Med 2018;20(6):574-82. https://www.sciencedirect.com/science/article/pii/S1098360021010042.
- 16.Knoppers BM, Zawati MH, Senecal K Senecal K. Return of genetic testing results in the era of whole-genome sequencing. Nature Reviews Genetics. 2015;16(9):553–9. doi: 10.1038/nrg3960. [DOI] [PubMed] [Google Scholar]
- 17.Weiner C. Anticipate and communicate: ethical management of incidental and secondary findings in the clinical, research, and direct-to-consumer contexts (December 2013 report of the Presidential Commission for the Study of Bioethical Issues). Am J Epidemiol 2014;180(6):562-4. https://academic.oup.com/aje/article/180/6/562/2739282?login=true.
- 18.ACMG Board of Directors. Points to consider for informed consent for genome/exome sequencing. Genet Med 2013;15(9):748-9. https://pubmed.ncbi.nlm.nih.gov/23970068/.
- 19.Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 2013;15(7):565-74. https://pubmed.ncbi.nlm.nih.gov/23788249/.
- 20.ACMG Board of Directors. ACMG policy statement: updated recommendations regarding analysis and reporting of secondary findings in clinical genome-scale sequencing. Genet Med 2015;17(1):68-9. https://pubmed.ncbi.nlm.nih.gov/25356965/.
- 21.Kalia SS, Adelman K, Bale SJ, Chung WK, Eng C, Evans JP, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med 2017;19(2):249-55. https://pubmed.ncbi.nlm.nih.gov/27854360/.
- 22.Van El CG, Cornel MC, Borry P, Hastings RJ, Fellmann F, Hodgson SV, et al. Whole-genome sequencing in health care: recommendations of the European Society of Human Genetics. Eur J Hum Genet 2013;21(6):580-4. https://pubmed.ncbi.nlm.nih.gov/23676617/.
- 23.Hall A, Finnegan T, Alberg C. Realising genomics in clinical practice. 2014. https://www.phgfoundation.org/media/173/download/Realising-genomics-clinical-practice-executive-summary.pdf?v=1&inline=1. [2021-3-31].
- 24.Middleton A, Patch C, Wiggins J, Barnes K, Crawford G, Benjamin C, et al. Position statement on opportunistic genomic screening from the Association of Genetic Nurses and Counsellors (UK and Ireland). Eur J Human Genet 2014;22(8):955-6. https://www.nature.com/articles/ejhg2013301.
- 25.Ethikrat D. The future of genetic diagnosis—from research to clinical practice. 2013. https://www.ethikrat.org/fileadmin/Publikationen/Stellungnahmen/englisch/opinion-the-future-of-genetic-diagnosis.pdf. [2021-3-31].
- 26.Royal College of Pathologists of Australasia. Implementation of massively parallel sequencing in diagnostic medical genetic testing 2013. https://www.hgsa.org.au/hgsanews/guidelines-for-implementation-of-massively-parallel-sequencing.
- 27.Howard HC, Knoppers BM, Cornel MC, Clayton EW, Sénécal K, Borry P, et al. Whole-genome sequencing in newborn screening? A statement on the continued importance of targeted approaches in newborn screening programmes. Eur J Hum Genet 2015;23(12):1593-600. https://pubmed.ncbi.nlm.nih.gov/25626707/.
- 28.Biesecker LG. Incidental variants are critical for genomics. Am J Hum Genet 2013;92(5):648-51. https://pubmed.ncbi.nlm.nih.gov/23643378/.
- 29.Okbay A, Baselmans BML, De Neve JE, Turley P, Nivard MG, Fontana MA, et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet 2016;48(6):624-33. https://pubmed.ncbi.nlm.nih.gov/27089181/.
- 30.Lu J, Jiang S, Wang Y, Zhang N, Jiao G. PNS41 preferences of older patients with chronic diseases for medication review services in Shanxi, China: results from a discrete choice experiment. Value Heal Reg Issues 2020;22(S1):S88. https://www.valuehealthregionalissues.com/article/S2212-1099(20)30509-4/fulltext.
- 31.Liu P, Jiang S, Li SP. Systematic review of measuring the preference for targeted therapy in cancer patients by discrete choice experiment. Chin J Cancer Prev Treat 2021;28(4):318-22. https://d.wanfangdata.com.cn/periodical/ChlQZXJpb2RpY2FsQ0hJTmV3UzIwMjIwNzE5Eg9xbHpsenoyMDIxMDQwMTMaCDliamdqb2Rp. (In Chinese).
- 32.Zhang LF, Huang JJ, Wang H, Jiang S. An overview on theoretic and applied research of best-worst scaling. Stat Inf Forum 2019;34(3):24-30. https://d.wanfangdata.com.cn/periodical/ChlQZXJpb2RpY2FsQ0hJTmV3UzIwMjIwNzE5EhB0anl4eGx0MjAxOTAzMDA0GggxcXprd3Uyag%3D%3D. (In Chinese).
- 33.Jiang S, Lu J. PNS20 best-worst scaling in health economics in China: past, present and future. Value Health 2020;23(S1):S287. https://www.valueinhealthjournal.com/article/S1098-3015(20)31219-5/fulltext.
- 34.Jiang S, Gu YY, Yang F, Wu T, Wang H, Cutler H, et al. Tertiary hospitals or community clinics? An enquiry into the factors affecting patients' choice for healthcare facilities in urban China. China Econ Rev 2020;63:101538. https://www.sciencedirect.com/science/article/pii/S1043951X20301358?casa_token=PFothgKNYVAAAAAA:T_sTmyrOoqIfMW_w-jjVAIy2KRciCfVEtef52K2HrzcHXwZLr2IKCbC7HnFMvh0UBI2P3f28wQ.
- 35.Scheuner MT, Peredo J, Benkendorf J, Bowdish B, Feldman G, Fleisher L, et al. Reporting genomic secondary findings: ACMG members weigh in. Genet Med 2015;17(1):27-35. https://www.nature.com/articles/gim2014165.
- 36.Regier DA, Peacock SJ, Pataky R, Van Der Hoek K, Jarvik GP, Hoch J, et al. Societal preferences for the return of incidental findings from clinical genomic sequencing: a discrete-choice experiment. CMAJ 2015;187(6):E190-7. https://pubmed.ncbi.nlm.nih.gov/25754703/.
- 37.Liu XQ, Luo X, Jiang CY, Zhao H. Difficulties and challenges in the development of precision medicine. Clin Genet 2019;95(5):569-74. https://onlinelibrary.wiley.com/doi/abs/10.1111/cge.13511?casa_token=p4n6-XRcSmoAAAAA:jNU6e3HXO0IYXq1m3m89lLSa9YV-bK-i1a0wK_Ao8DCFWDVpZM1O3JnqPIeJwkGBQ2BnZLafg2hvqrG_.
- 38.Xue Y, Lameijer EW, Ye K, Zhang KL, Chang SH, Wang XY, et al. Precision medicine: what challenges are we facing? Genom Proteom Bioinform 2016;14(5):253-61. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5093857/.
- 39.Jiang S, Chen Z, Wu J, Zang X, Jiang YW. Addressing methodological and ethical issues in practicing health economic evaluation in China. J Global Health 2020;10(2):020322. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7561214/.
- 40.Chen Z, Zhou L, Jiang S, Haddix A. Identifying options of best value: use of economic evaluation in public health. China CDC Wkly 2020;2(5):75-8. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8393107/.
- 41.Phillips KA, Deverka PA, Marshall DA, Wordsworth S, Regier DA, Christensen KD, et al. Methodological issues in assessing the economic value of next-generation sequencing tests: many challenges and not enough solutions. Value Health 2018;21(9):1033-42. https://www.sciencedirect.com/science/article/pii/S1098301518322691.
- 42.Jiang S, Wang YT, Zhou JW, Jiang YW, Liu GGE, Wu J. Incorporating future unrelated medical costs in cost-effectiveness analysis in China. BMJ Global Heal 2021;6(10):e006655. https://gh.bmj.com/content/6/10/e006655.abstract.
- 43.Cai D, Shi S, Jiang S, Si L, Wu J, Jiang YW. Estimation of the cost-effective threshold of a quality-adjusted life year in China based on the value of statistical life. Eur J Health Econ 2022;23(4):607-15. https://link.springer.com/article/10.1007/s10198-021-01384-z.


