Abstract
Purpose:
SORAMIC is a randomized controlled trial in patients with advanced hepatocellular carcinoma (HCC) undergoing sorafenib ± selective internal radiation therapy (SIRT). We investigated the value of extracellular vesicle (EV)-based proteomics for treatment response prediction.
Experimental Design:
The analysis population comprised 25 patients receiving SIRT+sorafenib and 20 patients receiving sorafenib alone. Patients were classified as responders or nonresponders based on changes in AFP and imaging or overall survival. Proteomic analysis was performed on plasma EVs by LC/MS, followed by bioinformatics analysis. Clinical relevance of candidate EV proteins was validated by survival and receiver-operating characteristic analysis with bootstrap internal sampling validation. Origin of circulating EV was explored by IHC staining of liver and tumor tissues and transcriptomics of blood cells.
Results:
Proteomic analysis identified 56 and 27 EV proteins that were differentially expressed in plasma EVs between responders and nonresponders receiving SIRT+sorafenib and sorafenib alone, respectively. High EV-GPX3/ACTR3 and low EV-ARHGAP1 were identified as candidate biomarkers at baseline from the 13 responders to SIRT+sorafenib with statistically significant AUC = 1 for all and bootstrap P values 2.23 × 10−5, 2.22 × 10−5, and 2.23 × 10−5, respectively. These patients showed reduced abundance of EV-VPS13A and EV-KALRN 6 to 9 weeks after combined treatment with significant AUC and bootstrap P values. In reverse, low GPX3 and high ARHGAP1 demonstrated better response to sorafenib monotherapy with AUC = 0.9697 and 0.9192 as well as bootstrap P values 8.34 × 10−5 and 7.98 × 10−4, respectively. HCC tumor was the likely origin of circulating EVs.
Conclusions:
In this exploratory study, EV-based proteomics predicted response to SIRT+sorafenib and sorafenib-only treatment in patients with advanced HCC of metabolic origin.
Translational Relevance.
The SORAMIC trial showed no overall survival benefit of adding SIRT to sorafenib. However, analyses of the per-protocol population showed survival benefit in patient subgroups. Tumor biopsy is not commonly performed in HCC according to diagnostic guidelines. Liquid biopsy is easily accessible and extracellular vesicles (EV) have been widely studied and proposed to be potential liquid biopsy candidates as predictive or prognostic cancer biomarkers. This study showed that EVs with unique proteins predicted response to treatment in patients with advanced HCC of metabolic origin. Global EV profiles were highly similar among patients; however, specific EV proteins were associated with clinical outcomes according to either SIRT+sorafenib or sorafenib-only treatments. HCC tumors were proven as the origin of blood-circulating EV-GPX3 and EV-ACTR3. These EVs therefore represented predictive biomarkers. If confirmed in further validation studies, EV proteomics will evolve as a novel predictive biomarker in HCC.
Introduction
Hepatocellular carcinoma (HCC) accounts for 80% to 90% of all liver cancers, most often in association with chronic liver disease related to alcohol abuse, nonalcoholic steatohepatitis (NASH), or viral hepatitis (1). Only about 30% of patients are diagnosed early enough to benefit from potentially curative therapies (2, 3). For patients with inoperable, liver-confined intermediate-stage HCC, locoregional treatment by transarterial chemoembolization (TACE) is the recommended treatment of choice, supplemented by 90Y-radioembolization in recent European guidelines (4, 5). In advanced HCC, systemic therapy with sorafenib confers survival benefit (6). Recently, the IMbrave150 study showed that the combination of atezolizumab and bevacizumab in patients with advanced HCC was superior to sorafenib in first line, likely determining the future standard of care (7, 8).
In locoregional or systemic treatment of HCC, a significant proportion of patients do not achieve durable responses, and response rates vary (6, 7, 9). Clinical decision-making is currently generally based on staging, performance status, and basic liver function (9). An unmet medical need exists for biomarkers stratifying patients with HCC to therapeutic strategies.
EVs promote various biological processes such as intercellular communication, inflammation, immune response, and disease progression (10, 11). Their activity is mediated by proteins, nucleic acids, and lipids trapped in stabilizing lipid double-layers of the originating cells. In tumor-derived EVs, proteomic-based analyses have indicated the potential to unravel changes in protein expression under different clinical conditions. Therefore, EVs may be candidates as predictive or prognostic biomarkers (12, 13).
Sorafenib in combination with local microtherapy guided by gadolinium-EOB-DTPA–enhanced MRI (SORAMIC; EudraCT 2009-012576-27, NCT0112 6645) is a prospective study that comprised three substudies, including a cohort comparing the combination of SIRT with 90Y resin microspheres with sorafenib to sorafenib alone in patients with advanced HCC (14, 15). In the SORAMIC substudy presented herein, we sought to determine the role of EV proteins as predictive biomarkers of therapeutic response to sorafenib ± SIRT in advanced HCC.
Materials and Methods
SORAMIC is a prospective, phase II, open label, multicenter, randomized controlled trial. The study was conducted at 38 sites in 12 countries in Europe and Turkey. The post hoc analyses described herein are exploratory.
Ethical considerations
The institutional review boards of all participating centers approved the study prior to initiation of the trial, and written informed consent was obtained from all patients. Study procedures were performed in accordance with the protocol and ethical principles that have their origin in the Declaration of Helsinki and the International Council for Harmonisation-Good Clinical Practice.
Patient and public involvement
Details of inclusion and exclusion criteria of the SORAMIC study have been published elsewhere (14). Key selection criteria for the palliative treatment group were Barcelona Clinic Liver Cancer (BCLC) B (not eligible for TACE per investigator decision) and C, and Child Pugh ≤7. Patients were recruited between January 5, 2011, and April 19, 2016, and the de-identified samples were used in this translational and exploratory study.
Study objective
To assess the value of EV-based proteomics for response prediction in patients with advanced HCC receiving sorafenib ± SIRT.
Study design and experimental set up
Patient selection
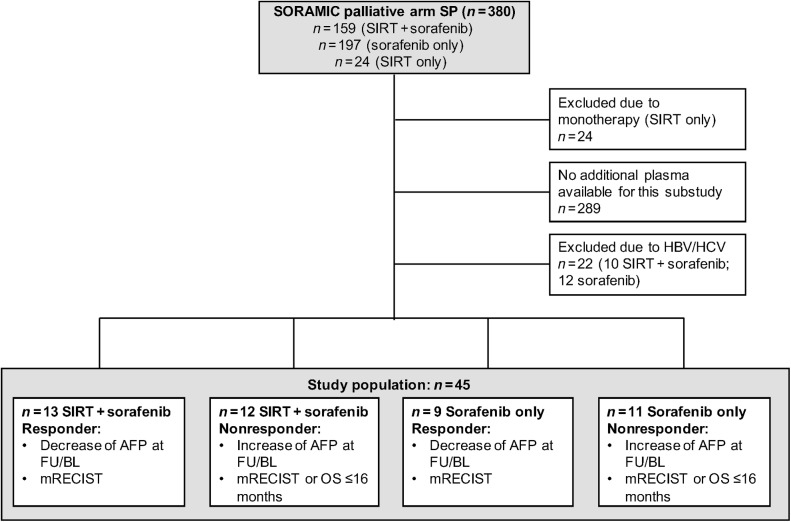
Our exploratory cohort was based on the SORAMIC safety population with patients undergoing SIRT+sorafenib or sorafenib alone (n = 380). Because of limited numbers of viral patients with HCC recruited in the European-based SORAMIC trials, we selected patients with nonviral etiology of the disease as a more homogeneous cohort for exploratory analysis. As a result, our study population comprised n = 25 and 20 patients receiving sorafenib±SIRT and sorafenib alone, respectively. For each patient, samples collected at two different time points (baseline and 6–9 weeks posttherapy) were included in the study. For details, see Fig. 1.
Figure 1.
CONSORT chart. AFP, alfa-fetoprotein; BL, baseline; FU, follow-up; SP, safety population.
Patients were classified as responders and nonresponders based on changes in alfa-fetoprotein (AFP) reduction at 6 to 9 weeks and overall survival (OS) assessment (>/≤16 months) or the mRECIST criteria if adjudication was necessary [complete response (CR) and partial response (PR) vs. stable disease (SD) and progressive disease (PD)]. In brief, in 42 of 45 patients, classification was based on changes in AFP and imaging. Baseline characteristics of all patients are displayed in Table 1 and the change in AFP and OS data are shown in Supplementary Fig. S1. Tumor tissue samples at baseline from 9 patients served for validation of the origin of EVs identified in blood. Only three of these tissues showed a minimal amount (<5%) of necrosis (Supplementary Fig. S2).
Table 1.
Baseline characteristics.
| P | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | SIRT+sorafenib responders | SIRT+sorafenib nonresponders | Sorafenib responders | Sorafenib nonresponders | ||||||
| Variables | (N = 45) | (N = 13) | (N = 12) | (N = 9) | (N = 11) | SIRT+sorafenib vs. sorafenib | Responders vs. nonresponders (overall) | Responders vs. nonresponders (SIRT+sorafenib) | Responders vs. nonresponders (sorafenib) | |
| Sex | 0.1058 | 0.1571 | 0.1775 | |||||||
| Female | 2 (4.4) | 2 (18.2) | ||||||||
| Male | 43 (95.6) | 13 (100) | 12 (100) | 9 (100) | 9 (81.8) | |||||
| Age (years) | 0.8691 | 0.5223 | 0.9472 | 0.1874 | ||||||
| Mean (SD) | 68.0 (7.4) | 67.8 (9.4) | 68.0 (7.7) | 70.2 (6.3) | 66.6 (5.4) | |||||
| Median (IQR) | 69.0 (11.0) | 70.0 (9.0) | 67.0 (12.0) | 71.0 (8.0) | 66.0 (9.0) | |||||
| Min–Max | 46.0–81.0 | 46.0–81.0 | 56.0–78.0 | 60.0–78.0 | 58.0–75.0 | |||||
| Age category | 0.2888 | 0.3990 | 0.3268 | 0.7952 | ||||||
| <65 years | 15 (33.3) | 4 (30.8) | 6 (50.0) | 2 (22.2) | 3 (27.3) | |||||
| ≥65 years | 30 (66.7) | 9 (69.2) | 6 (50.0) | 7 (77.8) | 8 (72.7) | |||||
| Liver cirrhosis | 0.3949 | 0.5498 | 0.3268 | 0.1561 | ||||||
| Yes | 41 (93.2) | 12 (92.3) | 12 (100) | 9 (100) | 8 (80.0) | |||||
| No | 3 (6.8) | 1 (7.7) | 2 (20.0) | |||||||
| Missing | 1 (2.2) | 1 (9.1) | ||||||||
| Alcohol etiology | 1.0000 | 0.0514 | 0.0221 | 0.7136 | ||||||
| Yes | 27 (60.0) | 5 (38.5) | 10 (83.3) | 5 (55.6) | 7 (63.6) | |||||
| No | 18 (40.0) | 8 (61.5) | 2 (16.7) | 4 (44.4) | 4 (36.4) | |||||
| Child Pugh | 0.7530 | 0.0569 | 0.0218 | 0.6676 | ||||||
| 5 | 29 (64.4) | 12 (92.3) | 5 (41.7) | 6 (66.7) | 6 (54.5) | |||||
| 6 | 9 (20.0) | 1 (7.7) | 4 (33.3) | 1 (11.1) | 3 (27.3) | |||||
| 7/8 | 7 (15.6) | 3 (25.0) | 2 (22.2) | 2 (18.2) | ||||||
| BCLC | 0.7872 | 0.0471 | 0.1654 | 0.1421 | ||||||
| BCLC B | 19 (42.2) | 4 (30.8) | 7 (58.3) | 2 (22.2) | 6 (54.5) | |||||
| BCLC C | 26 (57.8) | 9 (69.2) | 5 (41.7) | 7 (77.8) | 5 (45.5) | |||||
| Extrahepatic metastases | 0.2433 | 0.6732 | 0.9304 | 0.3534 | ||||||
| Yes | 5 (11.1) | 2 (15.4) | 2 (16.7) | 1 (9.1) | ||||||
| No | 40 (88.9) | 11 (84.6) | 10 (83.3) | 9 (100) | 10 (90.9) | |||||
| PVT | 0.3779 | 0.7628 | 0.7292 | 0.3428 | ||||||
| Yes | 21 (47.7) | 5 (38.5) | 5 (45.5) | 6 (66.7) | 5 (45.5) | |||||
| No | 23 (52.3) | 8 (61.5) | 6 (54.5) | 3 (33.3) | 6 (54.5) | |||||
| Missing | 1 (2.2) | 1 (8.3) | ||||||||
Abbreviations: BCLC, Barcelona Clinic Liver Cancer; PVT, portal vein thrombosis.
Experimental design
We performed a comprehensive proteomic analysis of proteins isolated from the EVs and evaluated their potential as predictive biomarkers for sorafenib±SIRT treatment. EVs were extracted from plasma and protein profiles were identified on the basis of LC/MS. Bioinformatics analysis, including differentially expressed protein (DEP) analysis and gene ontology (GO) enrichment analysis, was performed to discover candidates for outcome prediction. The top 100 commonly identified EV proteins from ExoCarta and Vesiclepedia were used to compare with the plasma EV proteome. The clinical relevance of the discovered proteins was analyzed by survival and receiver operating characteristic (ROC) analysis. Finally, the potential origin of the circulating EVs was investigated by transcriptomic analysis of blood cells and IHC analysis of HCC tissue samples. The complete experimental workflow is schematically described in Supplementary Fig. S3.
Sample collection
Peripheral venous blood was collected at baseline and at follow-up (6–9 weeks from radioembolization treatment), providing two samples per patient. Blood (5 mL) was drawn in EDTA tubes (Becton Dickinson) and processed immediately (centrifugation 1,300 × g, 5 minutes, 4°C) to isolate plasma, which was then aliquoted and stored at −80°C until use.
EV isolation, characterization, and MS analysis
In total, 90 plasma samples (100 μL) were analyzed by Tymora Analytical (West Lafayette) and used for enrichment of EVs using EVTrap magnetic beads, which were comprehensively characterized to show over 95% recovery yield and much better performance than traditional differential centrifugation with ultracentrifugation (16–18). EV extraction was validated by tunable resistive pulse sensing (TRPS) with 150 nm nanopore for size distribution and by Western blot analysis (19). The advantages of using EVTrap over traditional isolation methodologies were comprehensively described in previous publications by Tymora Analytical. The EVs were quantified and in total of 1 μg of peptides from each sample was subjected to MS analysis. Non-EV markers were not detected from the LC/MS analysis. The raw data of LC/MS were generated and processed according to published methodology (16). The abundance levels of all peptides and proteins were normalized to the spiked-in internal indexed retention time (iRT) standard. The details of the LC/MS sample preparation are shown in the Supplementary Materials and Methods.
DEP and GO enrichment analyses
The normalized MS data were processed and analyzed by Partek Genomics Suites version 7.19.1125 (Partek Incorporated). The value of 0.0001 was assigned to the proteins, which were below detection limit in the MS dataset, followed by log2 transformation. All protein ID was converted to matched gene ID prior to analysis. Principal component analysis (PCA) was first done and DEP analysis was then performed between responders and nonresponders in both treatments with the threshold of 2-fold change and FDR-adjusted P value <0.001, followed by hierarchical clustering. The lists of DEP were then subjected to BioVenn web-based Venn diagram analysis (20) and ShinyGO (v0.65) GO-enrichment analysis (21) with enrichment FDR-adjusted P value <0.05 and gene sets of GO biological process, GO cellular components, and GO molecular function.
Histologic and IHC analysis of liver tissue samples
Tumor blocks collected before therapy were available for a limited number of patients only. Two μm serial tissue sections were cut from formalin-fixed, paraffin-embedded tumor tissues, dewaxed, and rehydrated according to standard procedure (preheating at 60°C; deparaffinization in Neo-Clear, Merck KGaA; rehydration in graded series of ethanol and distilled water) and stained with hematoxylin and eosin for determination of areas of tumor necrosis. For IHC analysis, the primary antibodies anti-GPx3 (goat IgG polyclonal, dilution 5 μg/mL; R&D Systems; Catalog No. AF4199) and ACTR3 (rabbit monoclonal, clone JB33–44, dilution 1:200; Novus Biologicals; Catalog No. JB33–44) were applied overnight at 4°C, followed by incubation with secondary antibodies (for GPx3: Peroxidase AffiniPure Donkey Anti-Goat; Jackson ImmunoResearch Europe Ltd.; for ACTR3: Dako EnVision+System-HRP Anti-Rabbit; Carpinteria). DAB substrate kit (DAB Substrate Kit; Cell Signaling Technology) was used as chromogen. Sections were counterstained with hematoxylin, dehydrated, and mounted using Neo-Mount (Merck KGaA).
Transcriptomics analysis
Total RNA was extracted from frozen blood samples, containing lymphocytes, monocytes, granulocytes, and platelets, using AllPrep DNA/RNA/miRNA Universal Kit (Qiagen; Catalog No. 80224) as per manufacturer's instruction. The extracted RNA samples underwent library preparation using TruSeq Stranded Total RNA with Ribo-Zero Globin Kit, followed by sequencing with NovaSeq 6000 S4 Reagent Kit in a NovaSeq 6000 sequencer as per manufacturer's instruction (Illumina). In total, around 50 million read data were generated for each sample. The pipeline of bioinformatic analysis was done in the Partek Flow (Partek Incorporated). In brief, the raw data were aligned to whole genome of hg38 by STAR aligner version 2.7.3a and quantified to RefSeq Transcripts 97 - 2021–02–01 by Partek E/M model, followed by gene count normalization using method of counts per million (CPM) and adding 0.0001. The normalized count matrix file was then analyzed in Partek Genomics Suites version 7.19.1125.
Statistical analysis
Proteins in different groups were compared using the nonparametric Mann–Whitney U test. Bootstrap analysis with 1,000 randomization experiments of all detected EV proteins was performed as an internal resampling approach by Partek Genomics Suites version 7.19.1125 (Partek Incorporated). OS was calculated using the log-rank Kaplan–Meier method. All statistical analyses including the D'Agostino and Pearson normality test and ROC curve analysis were performed by GraphPad Prism Version 7.0 (GraphPad) and SAS Version 9.4 (SAS Institute Inc.).
Data availability statement
The data generated in this study are available within the article and its Supplementary Data files.
Results
EV isolation and characterization
EVs were extracted and isolated as described in Supplementary Fig. S4A. Examination by TRPS of the eluted post-EVTrap EVs demonstrated the expected diameter range between 100 and 200 nm (Supplementary Fig. S4B). EV characterization by Western blot analysis revealed that EV marker proteins, CD9, and newly identified pan-EV marker, Moesin (MSN; ref. 16), were present in the samples (Supplementary Fig. S4C). The proteomes of the plasma EVs from all samples were characterized according to the ISEV2018 recommendations (Supplementary Fig. S4D; ref. 22), showing CD9, MSN, and FLOT1/2 are expressed in all EV samples and AGO2 is under the detection limit. Contaminants such as ALB, APOA1/2, HSP90B1, and CANX are present evenly across different samples as background noise. Altogether, these results show that EVs were successfully isolated from the clinical plasma samples with minimal contaminants.
Proteomic profile of plasma EVs
A total of 26,810 unique peptides and 3,841 unique proteins were identified in the proteome profiles. To verify their identities and to determine the subcellular localization of the identified high-abundance proteins, we performed GO enrichment analysis. The detected proteins were classified according to the GO cellular components, the GO biological processes, and the GO molecular functions. For the GO cellular components, the proteins were strongly enriched in vesicle (enrichment P < 0.0001), extracellular organelle (enrichment P < 0.0001), extracellular exosome (enrichment P < 0.0001), and extracellular vesicle (enrichment P < 0.0001), confirming EV identity (Supplementary Fig. S5A). The GO biological processes analysis revealed that proteins were mainly involved in transport processes as well as in immune-related processes (Supplementary Fig. S5B), whereas the GO molecular functions analysis showed that the proteins were part of cell adhesion and molecular binding pathways (Supplementary Fig. S5C). By comparing with EV public domain databases, ExoCarta and Vesiclepedia, our isolated EVs are proved to express ∼90% of the top 100 most commonly reported EV proteins. In summary, EV characterization shown in Supplementary Figs. S4B to S4D and comparison to the EV database (Supplementary Fig. S5D) confirmed the identity of the isolated EVs. GO enrichment analysis further suggested abundance of proteins with functions in vesicle-mediated transport, immune regulation, and cell adhesion.
Analysis of the differential protein abundance of EVs
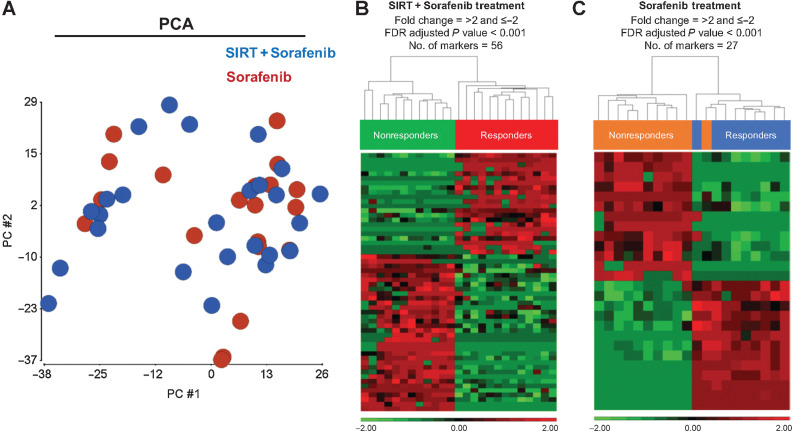
First, we sought to identify any variability of EV cargos between sorafenib ± SIRT patients at baseline. PCA was performed with the proteomic profiles of isolated EVs. Figure 2A indicates that there was little difference between the two treatment groups, suggesting that the global profiles of plasma EVs from both groups before treatment were highly similar. We then sought to identify unique high-abundance proteins that were specifically expressed in patients who received sorafenib±SIRT treatments and correlated with clinical outcomes. Supervised clustering with the proteome of the EVs isolated from baseline plasma samples was done subsequently. Among the patients receiving SIRT+sorafenib, we found 56 unique EV proteins which discriminated responders and nonresponders according to abundance and statistical significance (Fig. 2B; Supplementary Table S1), with bootstrap internal resampling validation showing statistical significance for all 56 EV proteins. In the patients receiving sorafenib only, 27 EV proteins were differentially expressed in responders and nonresponders, supported by bootstrap analysis (Fig. 2C; Supplementary Table S2). Statistical analysis showed significant DEP in responders and nonresponders in both treatment groups.
Figure 2.
Differential protein abundance analysis. A, PCA of the proteomics of plasma EVs at baseline time point in SIRT+sorafenib-treated patients (blue) vs. that of plasma EVs in sorafenib-treated patients (red). B, Heatmap of the differential level of 56 EV proteins in SIRT+sorafenib-treated patients. C, Heatmap of the differential level of 27 EV proteins in sorafenib-treated patients. Responders and nonresponders were clustered according to average linkage algorithm using Euclidean distance. Relative expression level was represented in red and green in the heatmap.
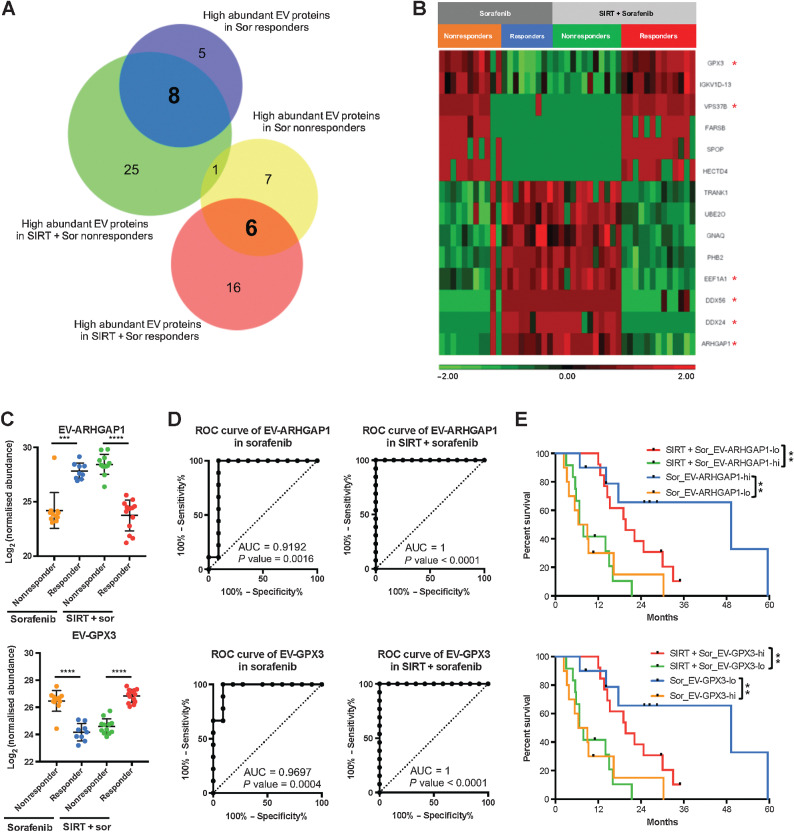
Identification of predictive biomarkers for both treatments
Next, we aimed to identify EV proteins which were present in both treatment groups, but were showing opposed trends in their relative abundance according to clinical response. On the basis of the Venn diagram analysis, 14 EV proteins were identified, of which eight were shared between responders receiving only sorafenib and nonresponders receiving SIRT+sorafenib (Fig. 3A). The other six proteins were shared between responders receiving SIRT+sorafenib and nonresponders receiving sorafenib only (Fig. 3A). On the basis of the statistical significance of the identified 14 EV proteins, 6 EV proteins were ranked as the most differentially expressed and statistically significant high-abundance EV proteins from both treatments: EV-ARHGAP1, EV-GPX3, EV-VPS37B, EV-EEF1A1, EV-DDX24, and EV-DDX56 (Fig. 3B and C; Supplementary Fig. S6). These EV proteins showed a strong correlation with high AUC values and survival outcomes (Fig. 3D and E; Supplementary Fig. S6). EV-GPX3 and EV-ARHGAP1 showed the highest AUC (AUC = 0.9697, P = 0.0004 and AUC = 0.9192, P = 0.0016, respectively, for the sorafenib arm and AUC = 1, P < 0.0001, both, for the SIRT+sorafenib arm; Fig. 3D). Patients with low level of EV-ARHGAP1 (log-rank P = 0.0015) and high level of EV-GPX3 (log-rank P = 0.001) showed improved response when receiving combined SIRT+sorafenib therapy, whereas patients with high level of EV-ARHGAP1 (log-rank P = 0.0028) and low level of EV-GPX3 (log-rank P = 0.003) showed a better response when treated with sorafenib alone (Fig. 3E). Similar results were found for the other four identified EV proteins, that is, EV-VPS37B, EV-EEF1A1, EV-DDX24, and EV-DDX56 (Supplementary Fig. S6).
Figure 3.
Differential abundance of 14 EV proteins correlating with treatment outcomes. A, Venn diagram of proteins detected from the EV samples. B, Heatmap of the differential abundance of the EV proteins identified in the four response groups. Relative expression levels are represented in red and green in the heatmap. C–E, Abundance, ROC analysis, and survival analysis of EV-ARHGAP1 and EV-GPX3; nonresponders (orange) and responders (blue) from the sorafenib group; nonresponders (green) and responders (red) from the SIRT+sorafenib group (**, P < 0.01; ***, P < 0.001).
Identification of predictive biomarkers before and after therapy
Next, we sought to identify distinct biomarkers for both treatments by analyzing the abundance of EV proteins with respect to the time point of plasma collection (baseline vs. follow-up). Only 7 EV proteins (EV-VPS13A, EV-SERPINH1, EV-KALRN, EV-DDX56, EV-SSH3, EV-VPS37B, and EV-SPOP) were identified as candidate biomarkers in patients receiving combination treatment (Supplementary Table S3). To further explore the utility of these EV protein candidates as biomarkers and to validate by internal resampling, a ROC analysis and bootstrap analysis with 1,000 randomization experiments were performed. As shown in Supplementary Fig. S7A and Supplementary Table S3, only EV-VPS13A and EV-KALRN showed statistically significant AUC values, that is, 0.7929 and 0.7396, with significant bootstrap P value respectively, in patients receiving the combination treatment. In contrast, two EV proteins (EV-STXBP5 and EV-LNPEP) were identified in patients receiving sorafenib only. However, only EV-LNPEP showed a statistically significant AUC value of 0.8182 with significant bootstrap P value (Supplementary Fig. S7B; Supplementary Table S3). These comparisons suggest that there was little change in EV cargo between baseline and follow-up, given similar EV proteome profiles with close to 4,000 EV proteins detected. EV proteome therefore was stable and not affected by therapeutic interventions within a time period of 6 to 9 weeks in our study. However, the two identified unique EV proteins, EV-VPS13A and EV-KALRN, demonstrating reduced abundance 6 to 9 weeks after SIRT+sorafenib treatment may represent potential biomarkers of therapeutic response and will require further studies.
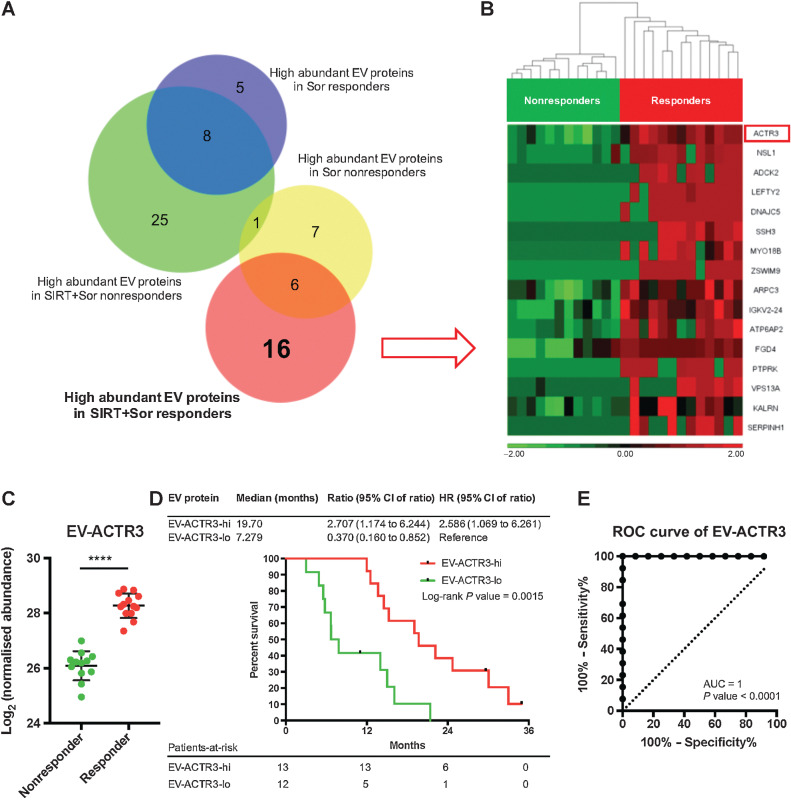
Identification of predictive biomarkers specifically for the treatment of SIRT+sorafenib
To identify unique predictive biomarkers for the treatment of SIRT+sorafenib, we performed Venn diagram analysis and clustered the EV proteins according to therapeutic response and therapy type (responders and nonresponders receiving sorafenib±SIRT treatments, respectively). We identified 16 proteins highly expressed only in responders and 25 proteins only in nonresponders receiving SIRT+sorafenib (Fig. 4A). In a similar way, five proteins showed much higher abundance only in responders and seven proteins only in nonresponders receiving sorafenib only (Fig. 4A). Among the 16 proteins identified in the responder group receiving SIRT+sorafenib, EV-ACTR3 showed the most significant false discovery rate-adjusted P value (Fig. 4B) and significant bootstrap P value of 2.22 × 10−5 (Supplementary Table S1). This protein was found to be significantly higher in the EVs isolated from responders compared with nonresponders (P < 0.0001; Fig. 4C) and Kaplan–Meier survival analysis showed that the much higher abundance of EV-ACTR3 significantly correlated with response (log-rank P = 0.0015; Fig. 4D). The AUC for EV-ACTR3 was 1 (P < 0.0001), indicating a strong predictive value (Fig. 4E). These results suggest that plasma EV-ACTR3 can differentiate between responders and nonresponders receiving SIRT+sorafenib therapy, but not in patients receiving sorafenib only (Supplementary Fig. S8). The abundance analysis and the ROC analysis of the other 15 EV proteins are shown in Supplementary Fig. S9.
Figure 4.
Identification of predictive EV proteins for SIRT+sorafenib treatment. A, Venn diagram of proteins detected from the EV samples. B, Heatmap of the differential abundance of the 16 EV proteins identified in the SIRT+sorafenib responders. Responders and nonresponders were clustered according to average linkage algorithm using Euclidean distance. Relative expression levels are represented in red and green in the heatmap. C, Abundance of EV-ACTR3 detected from responders and nonresponders from the SIRT+sorafenib group. Mann–Whitney test was used. D, Survival analysis of the patients based on the EV-ACTR3 abundance by log-rank (Mantel–Cox) test. E, ROC analysis of EV-ACTR3 for SIRT+sorafenib treatment.
Origin of circulating EVs
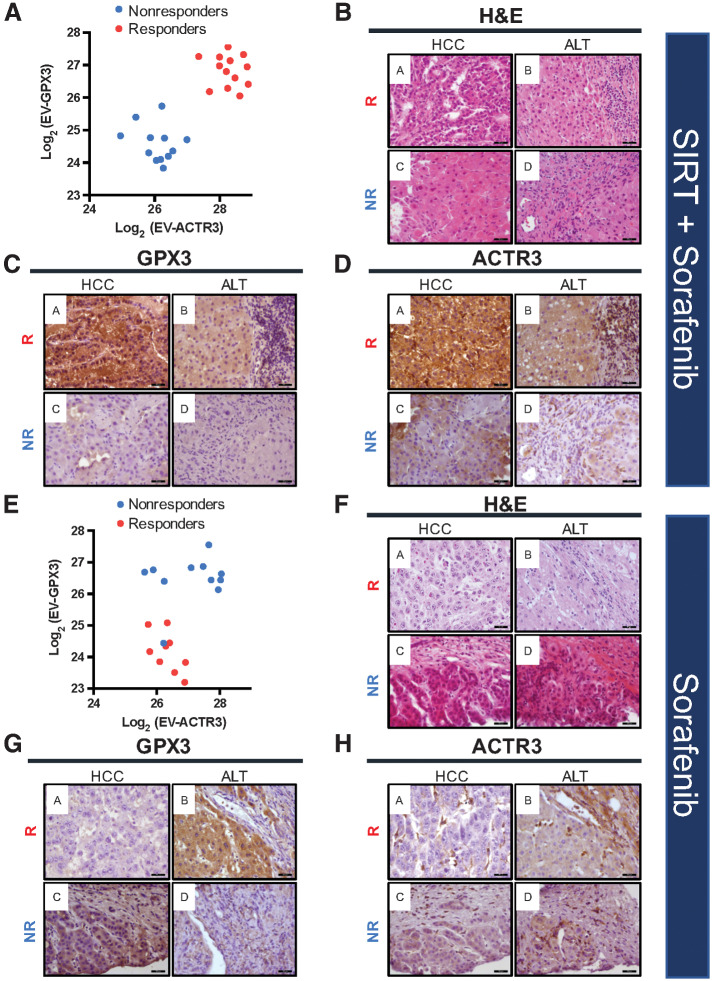
To identify the potential putative source of EV proteins in plasma, we performed IHC analysis of liver tissues including tumor and adjacent parenchyma as well as RNA sequencing of blood cell components (such as red blood cells, white cells, granulocytes, and platelets). First, to explore correlation of the unique EV profiles with protein expression in HCC tumors, histopathologic examination of tissues was performed to reveal the histology, distribution, and expression levels of GPX3 and ACTR3. These proteins were chosen as candidates since they were demonstrated to be highly abundant, most differentially expressed, and significantly different between the patient groups. The correlation analysis performed for the SIRT+sorafenib treatment group clearly indicated high EV-GPX3 and EV-ACTR3 levels in responders and low EV-GPX3 and EV-ACTR3 in nonresponders (Fig. 5A). Hematoxylin and eosin (H&E) staining was used to identify the regions of HCC tumor and adjacent liver tissue (ALT; Fig. 5B) and IHC analysis of the consecutive sections showed a strong cytoplasmic/membranous GPX3 and ACTR3 staining pattern in tumor from a patient responding to SIRT+sorafenib therapy (Fig. 5C and D, top row). On the contrary, a much weaker staining of both proteins was detected in a patient nonresponding to the therapy (Figs. 5C and D, bottom row).
Figure 5.
Levels of EV-GPX3 and EV-ACTR3 and corresponding protein expression in HCC and ALT prior to SIRT+sorafenib treatment (A–D) and sorafenib treatment (E–H) in responders (R) and nonresponders (NR). A, Correlation analysis of EV-GPX3 and EV-ACTR3 showing distinct clusters of responders and nonresponders in the SIRT+sorafenib treatment group. B, Representative images of hematoxylin and eosin (H&E) staining of HCC tissue and ALT prior to SIRT+sorafenib treatment. C, IHC staining of GPX3 in HCC tissue and ALT prior to SIRT+sorafenib treatment. D, IHC staining of ACTR3 in HCC tissue and ALT prior to SIRT+sorafenib treatment. E, Correlation analysis of EV-GPX3 and EV-ACTR3 in sorafenib treatment group. F, Representative images of H&E staining of HCC tissue and ALT prior to sorafenib treatment. G, IHC staining of GPX3 in HCC tissue and ALT prior to sorafenib treatment. H, IHC staining of ACTR3 in HCC tissue and ALT prior to sorafenib treatment. Magnification: 400×; scale bar: 50 μm.
In contrast, only the abundance of EV-GPX3 but not that of EV-ACTR3 could separate responders from nonresponders to sorafenib treatment alone (Fig. 5E). H&E staining and IHC analysis showed an undetectable GPX3 staining pattern in tumors of sorafenib-alone responders but a strong GPX3 staining pattern in the ALT of the same patients. The staining pattern of GPX3 in non-responders to sorafenib was the opposite to that in responders to the same treatment (Figs. 5F and G). As unraveled by DEP and correlation analyses, ACTR3 expression level did not differ significantly between tumoral and adjacent areas in both responders and nonresponders (Fig. 5H). However, stromal tissues including lymphocytes, endothelial cells, and macrophages showed a strong ACTR3 protein expression. Additional IHC analyses in responders (n = 4) receiving mono- and combination treatment is shown in Supplementary Fig. S10.
Next, whole-blood transcriptomic and EV proteomic profiles were compared. On the basis of the previously discovered 83 EV candidates (Figs. 2B and C), unsupervised clustering analysis (Supplementary Fig. S11A) and differentially expressed gene (DEG) analysis (Supplementary Fig. S11B) were performed. The difference among groups did not reach statistical significance (figure illustrates EV-GPX3 and EV-ACTR3). These results suggest that blood cells do not represent the origin of those unique plasma EV proteins.
In conclusion, these data establish a clear association between the levels of the proteins detected in plasma EVs and in tissues but not blood cells, suggesting tumor as the originating tissue.
Discussion
Biomarker-based decision-making is an unmet medical need in patients with HCC. The value of therapeutic decisions relies on prediction of best response to a specific therapy, along with high tolerability. Neither in systemic treatment of HCC by the tyrosine-kinase inhibitors sorafenib or lenvatinib, nor in recently approved immunotherapy, have predictive biomarkers yet been identified (23). As for many other clinical biomarkers, AFP was previously considered a prognosticator only (24). However, in the REACH-2 trial in patients with advanced HCC after previous sorafenib treatment, the human mAb (IgG1) ramucirumab led to a significant survival benefit in patients with AFP >400 ng/mL. To our knowledge, REACH-2 is the only positive phase III trial performed in a biomarker-selected patient population with HCC (25). Separately, recent study has suggested that patients with HCC with nonviral etiology, more specifically NASH HCC, responded less well to checkpoint blockade inhibitor (26), leading to the new hypothesis that targeted therapy and SIRT could still be useful for NASH patients with advanced HCC. It remains clinically relevant and scientifically important to explore the predictive biomarkers for using SIRT and/or sorafenib in advanced HCC. Therefore, studies for biomarkers of response, especially treatment for nonviral HCC, are urgently needed.
In locoregional techniques such as SIRT or TACE, no established biomarkers exist. Clinically, patient stratification into high- and low-risk groups follows tumor spread, Eastern Cooperative Oncology Group (ECOG) performance status, and basic liver function (27–29). In 1,000 consecutive HCC cases treated by SIRT, baseline albumin-bilirubin (ALBI) grade predicted median OS, with ALBI grades 1, 2, and 3 displaying 46.7, 19.1, and 8.8 months OS, respectively (30). Separately, our recent study showed that mRECIST has been found to predict survival in the SORAMIC cohort (31). For patients receiving SIRT+sorafenib, SORAMIC trial data proved that high baseline IL6 was the only independent prognostic factor for OS (HR, 2.35; 95% CI, 1.35–4.1; P = 0.002). However, multivariate analysis also confirmed IL6 (HR, 2.99; 95% CI, 1.22–7.3; P = 0.017) and IL8 (HR, 2.19; 95% CI, 1.02–4.7; P = 0.044) as independent predictors of OS in the control group of patients receiving sorafenib only (32).
EV activity including exosome-mediated signaling has gained increasing interest recently for understanding the tumor microenvironment, its impact on tumor behavior, and utility as biomarkers. EVs participate in multiple steps during neoplastic invasive processes and contribute to early steps involved in the development of metastasis (33). Several investigations have indicated participation in modeling extracellular matrix (34), pro-angiogenic signaling (35), communication with neighboring nontumor cells (36), and in epithelial-to-mesenchymal transition (EMT; ref. 37). Therefore, the nature of mechanisms mediated by EV indicates their potential to serve as prognostic or even predictive biomarkers. There are ongoing studies focusing on the clinical relevance and applications of EV-based biomarkers in multiple cancers including HCC (10, 38, 39) and so exploring EV-based biomarkers in a clinical trial setting would offer greater clinical insight toward the discovery of predictive and treatment biomarkers.
In this study, we sought to determine whether EV-based proteomics represents predictive biomarkers for response to SIRT+sorafenib or sorafenib alone in advanced HCC. The analysis was carried out in a subgroup of patients from the prospective SORAMIC trial. As a proof-of-concept study, we excluded HCC patients with viral etiology due to limited number of patients recruited in the trial in European countries. Our HCC cohort comprised patients with metabolic disease, NASH, or alcoholic liver disease, therefore forming a fairly homogeneous cohort with respect to tumor etiology. We identified a set of EV-based proteins with strong predictive value as proven by stratification of patients into responders and nonresponders to SIRT+sorafenib or sorafenib only. IHC performed on tissues suggested a tumor origin of these EVs, whereas transcriptomic analysis of blood cells excluded blood cells as the origin. In line with our proteomic results, a recent landmark study has confirmed that tumor-derived EVs can be detected in human plasma (19). More specifically, Arp2/3 complex-positive EVs (ACTR3+ and ARPC3+ EVs) have been proven to be of tumor origin in cancer patients. We therefore hypothesize that HCC tumor tissue is the origin of the detected plasma EVs. Furthermore, tumors with high expression of GPX3 and ACTR3 and low expression of ARHGAP1 benefit strongly from SIRT+sorafenib treatment as compared with those without. In addition, expression of GPX3 and ACTR3 in treatment-naïve HCC tumors did not correlate with OS, indirectly suggesting that EV-GPX3 and EV-ACTR3 are associated with clinical benefits of the treatment but are themselves not prognostic factors of treatment-naïve HCC (Supplementary Fig. S12).
Most of the EV proteins identified as markers of interest in our study are not fully understood yet in their biological relevance. Among the EV candidates, high plasma levels of EV-GPX3 and EV-ACTR3 and low levels of EV-ARHGAP1 were found in the patients responding to combination therapy, with improved OS and high AUC, whereas the opposite trend was found in patients receiving sorafenib only. Our results demonstrate a biological role for these proteins as predictive biomarkers in this therapeutic setting. Glutathione peroxidase 3 (GPX3) belongs to a class of antioxidant enzymes functioning as scavengers of reactive oxygen species (ROS; refs. 39–41). ARHGAP1, also known as Rho GTPase activating protein 1 (RhoGAP1) or p50-RhoGAP, is a positive regulator of p53 (42). ACTR3 (actin-related protein 3), part of the actin-related protein 2/3 (Arp2/3) complex (43), is involved in the formation of lamellipodia, cell migration, and cell invasion (44). One of the proposed mechanisms of action of sorafenib in HCC is the induction of ROS and p53 followed by cellular death (45). Assuming that plasma and tissue levels of the proteins are correlated, we speculate that tumors expressing low GPX3/ACTR3 and high ARHGAP1 are more sensitive to sorafenib monotherapy. The opposite effect with high GPX3/ACTR3 and low ARHGAP1 was observed in good responders to combination treatment including SIRT. Further in vitro and in vivo functional analyses with and without radiation/SIRT treatment would be needed to help delineate the functions of the tumor-secreting EV-GPX3, ACTR3, and ARHGAP1 locally within the tumor microenvironment among tumor cells, stromal cells, and adjacent noncancerous hepatocytes as well as systemically in the circulation.
One limitation of our study is that the cohort was limited to patients with HCC with nonviral etiology. Therefore, no conclusion can be drawn for patients with HCC associated with viral etiology. The small sample size and lack of external validation are another limitation of the study. In addition, the results of our study are limited as all patients received sorafenib, with some randomized to receiving additional SIRT. Therefore, it remains unknown whether the predictive value of the EV proteins derives from radiation or must be attributed to the combination of SIRT+sorafenib. Confirmation in a patient cohort receiving SIRT only is therefore mandatory. Finally, our findings should be explored further by molecular studies investigating the origin, mechanisms, and biology of the identified EVs and would trigger further initiatives in other forms of HCC treatments as well, specifically in alternative locoregional treatments such as TACE or its combination with systemic approaches.
In summary, in this post hoc analysis of the SORAMIC trial, we identified a panel of EV-based proteins predicting response to therapy in patients with nonviral advanced HCC undergoing SIRT+sorafenib or sorafenib-only treatments. To the best of our knowledge, this is the first report in any disease on the predictive role of EV-based proteomics with data derived from a prospective randomized controlled trial setting. Our findings suggest a distinct role of EV-based liquid biopsy profiling for tumor molecular classification and as a predictive biomarker. EVs have the potential to become a liquid biopsy–based predictive biomarker platform in patients with HCC.
Supplementary Material
Acknowledgments
Financial support by Sirtex Medical and Bayer Healthcare, Singapore National Medical Research Council (NMRC/OFLCG/003/2018), and National Cancer Centre Singapore Centre Block Grant.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
P. Malfertheiner reports grants from Bayer Healthcare and Sirtex Medical during the conduct of the study. M. Wildgruber reports personal fees from Bayer Oncology and Sirtex Medical Europe outside the submitted work. M. Pech reports grants from Sirtex and Bayer during the conduct of the study. B. Sangro reports personal fees from Adaptimmune, AstraZeneca, Bayer, Eisai, Eli Lilly, Ipsen, Roche, and Sirtex; and grants and personal fees from BMS and Boston Scientific outside the submitted work. J. Ricke reports grants and personal fees from Sirtex Medical and Bayer Healthcare during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
T.W.H. Shuen: Conceptualization, data curation, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, project administration. M. Alunni-Fabbroni: Conceptualization, formal analysis, supervision, writing–original draft, writing–review and editing. E. Öcal: Formal analysis, writing–original draft, writing–review and editing. P. Malfertheiner: Conceptualization, formal analysis, funding acquisition, writing–review and editing. M. Wildgruber: Conceptualization, formal analysis, writing–review and editing. R. Schinner: Data curation, formal analysis, writing–review and editing. M. Pech: Data curation, formal analysis, project administration, writing–review and editing. J. Benckert: Data curation, formal analysis, writing–review and editing. B. Sangro: Data curation, formal analysis, project administration, writing–review and editing. C. Kuhl: Data curation, formal analysis, project administration, writing–review and editing. A. Gasbarrini: Data curation, formal analysis, project administration, writing–review and editing. P.K.H. Chow: Data curation, formal analysis, project administration, writing–review and editing. H.C. Toh: Conceptualization, data curation, formal analysis, supervision, writing–original draft, project administration, writing–review and editing. J. Ricke: Conceptualization, data curation, formal analysis, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing.
References
- 1. Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol 2006;45:529–38. [DOI] [PubMed] [Google Scholar]
- 2. Mazzaferro V, Llovet JM, Miceli R, Bhoori S, Schiavo M, Mariani L, et al. Predicting survival after liver transplantation in patients with hepatocellular carcinoma beyond the Milan criteria: a retrospective, exploratory analysis. Lancet Oncol 2009;10:35–43. [DOI] [PubMed] [Google Scholar]
- 3. Villanueva A. Hepatocellular carcinoma. Reply. N Engl J Med 2019;381:e2. [DOI] [PubMed] [Google Scholar]
- 4. Vogel A, Martinelli E, ESMO Guidelines Committee. Updated treatment recommendations for hepatocellular carcinoma (HCC) from the ESMO clinical practice guidelines. Ann Oncol 2021;32:801–5. [DOI] [PubMed] [Google Scholar]
- 5. Reig M, Forner A, Rimola J, Ferrer-Fàbrega J, Burrel M, Garcia-Criado Á, et al. BCLC strategy for prognosis prediction and treatment recommendation: The 2022 update. J Hepatol 2021;S0168–8278:02223–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc J-F, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359:378–90. [DOI] [PubMed] [Google Scholar]
- 7. Finn RS, Qin S, Ikeda M, Galle PR, Ducreux M, Kim T-Y, et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med 2020;382:1894–905. [DOI] [PubMed] [Google Scholar]
- 8. Casak SJ, Donoghue M, Fashoyin-Aje L, Jiang X, Rodriguez L, Shen Y-L, et al. FDA approval summary: atezolizumab plus bevacizumab for the treatment of patients with advanced unresectable or metastatic hepatocellular carcinoma. Clin Cancer Res 2021;27:1836–41. [DOI] [PubMed] [Google Scholar]
- 9. European Association for Study of Liver, European Organisation for Research and Treatment of Cancer. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. Eur J Cancer 2012;48:599–641. [DOI] [PubMed] [Google Scholar]
- 10. Lee Y-T, Tran BV, Wang JJ, Liang IY, You S, Zhu Y, et al. The role of extracellular vesicles in disease progression and detection of hepatocellular carcinoma. Cancers (Basel) 2021;13:3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu Q, Zhou L, Lv D, Zhu X, Tang H. Exosome-mediated communication in the tumor microenvironment contributes to hepatocellular carcinoma development and progression. J Hematol Oncol 2019;12:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sorop A, Constantinescu D, Cojocaru F, Dinischiotu A, Cucu D, Dima SO. Exosomal microRNAs as biomarkers and therapeutic targets for hepatocellular carcinoma. Int J Mol Sci 2021;22:4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sasaki R, Kanda T, Yokosuka O, Kato N, Matsuoka S, Moriyama M. Exosomes and hepatocellular carcinoma: from bench to bedside. Int J Mol Sci 2019;20:E1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ricke J, Klümpen HJ, Amthauer H, Bargellini I, Bartenstein P, de Toni EN, et al. Impact of combined selective internal radiation therapy and sorafenib on survival in advanced hepatocellular carcinoma. J Hepatol 2019;71:1164–74. [DOI] [PubMed] [Google Scholar]
- 15. Ricke J, Steffen IG, Bargellini I, Berg T, Jaureguizar JIB, Gebaur B, et al. Gadoxetic acid-based hepatobiliary MRI for treatment decisons in hepatocellular carcinoma: a two-step non-inferiority/superiority trial. JHEP Reports 2020;In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Iliuk A, Wu X, Li L, Sun J, Hadisurya M, Boris RS, et al. Plasma-derived extracellular vesicle phosphoproteomics through chemical affinity purification. J Proteome Res 2020;19:2563–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu X, Li L, Iliuk A, Tao WA. Highly efficient phosphoproteome capture and analysis from urinary extracellular vesicles. J Proteome Res 2018;17:3308–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Andaluz Aguilar H, Iliuk AB, Chen I-H, Tao WA. Sequential phosphoproteomics and N-glycoproteomics of plasma-derived extracellular vesicles. Nat Protoc 2020;15:161–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoshino A, Kim HS, Bojmar L, Gyan KE, Cioffi M, Hernandez J, et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell 2020;182:1044–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 2008;9:488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ge SX, Jung D, Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020;36:2628–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 2018;7:1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roderburg C, Wree A, Demir M, Schmelzle M, Tacke F. The role of the innate immune system in the development and treatment of hepatocellular carcinoma. Hepat Oncol 2020;7:HEP17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Czauderna C, Schmidtmann I, Koch S, Pilz L, Heinrich S, Otto G, et al. High pretreatment static and dynamic alpha-fetoprotein values predict reduced overall survival in hepatocellular carcinoma. United European Gastroenterol J 2021. [DOI] [PubMed] [Google Scholar]
- 25. Zhu AX, Kang YK, Yen CJ, Finn RS, Galle PR, Llovet JM, et al. Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased alpha-fetoprotein concentrations (REACH-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2019;20:282–96. [DOI] [PubMed] [Google Scholar]
- 26. Pfister D, Núñez NG, Pinyol R, Govaere O, Pinter M, Szydlowska M, et al. NASH limits anti-tumour surveillance in immunotherapy-treated HCC. Nature 2021;592:450–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. European Association for the Study of the Liver. EASL clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol 2018;69:182–236. [DOI] [PubMed] [Google Scholar]
- 28. Nam JY, Lee YB, Lee J-H, Yu SJ, Kim H-C, Chung JW, et al. A prognostic prediction model of transarterial radioembolization in hepatocellular carcinoma: SNAP-HCC. Dig Dis Sci 2022;67:329–36. [DOI] [PubMed] [Google Scholar]
- 29. Spreafico C, Sposito C, Vaiani M, Cascella T, Bhoori S, Morosi C, et al. Development of a prognostic score to predict response to Yttrium-90 radioembolization for hepatocellular carcinoma with portal vein invasion. J Hepatol 2018;68:724–32. [DOI] [PubMed] [Google Scholar]
- 30. Antkowiak M, Gabr A, Das A, Ali R, Kulik L, Ganger D, et al. Prognostic role of albumin, bilirubin, and ALBI scores: analysis of 1000 patients with hepatocellular carcinoma undergoing radioembolization. Cancers (Basel) 2019;11:E879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Öcal O, Schinner R, Schütte K, de Toni EN, Loewe C, van Delden O, et al. Early tumor shrinkage and response assessment according to mRECIST predict overall survival in hepatocellular carcinoma patients under sorafenib. Cancer Imaging 2022;22:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Öcal O, Schütte K, Kupčinskas J, Morkunas E, Jurkeviciute G, de Toni EN, et al. Baseline Interleukin-6 and -8 predict response and survival in patients with advanced hepatocellular carcinoma treated with sorafenib monotherapy: an exploratory post hoc analysis of the SORAMIC trial. J Cancer Res Clin Oncol 2022;148:475–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell 2016;30:836–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sung BH, Ketova T, Hoshino D, Zijlstra A, Weaver AM. Directional cell movement through tissues is controlled by exosome secretion. Nat Commun 2015;6:7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. King HW, Michael MZ, Gleadle JM. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 2012;12:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Luga V, Zhang L, Viloria-Petit AM, Ogunjimi AA, Inanlou MR, Chiu E, et al. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 2012;151:1542–56. [DOI] [PubMed] [Google Scholar]
- 37. Tauro BJ, Mathias RA, Greening DW, Gopal SK, Ji H, Kapp EA, et al. Oncogenic H-ras reprograms Madin-Darby canine kidney (MDCK) cell-derived exosomal proteins following epithelial-mesenchymal transition. Mol Cell Proteomics 2013;12:2148–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Möller A, Lobb RJ. The evolving translational potential of small extracellular vesicles in cancer. Nat Rev Cancer 2020;20:697–709. [DOI] [PubMed] [Google Scholar]
- 39. Chang C, Worley BL, Phaëton R, Hempel N. Extracellular glutathione peroxidase GPx3 and its role in cancer. Cancers (Basel) 2020;12:E2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yi Z, Jiang L, Zhao L, Zhou M, Ni Y, Yang Y, et al. Glutathione peroxidase 3 (GPX3) suppresses the growth of melanoma cells through reactive oxygen species (ROS)-dependent stabilization of hypoxia-inducible factor 1-α and 2-α. J Cell Biochem 2019;120:19124–36. [DOI] [PubMed] [Google Scholar]
- 41. Reddy AT, Lakshmi SP, Banno A, Reddy RC. Role of GPx3 in PPARγ-induced protection against COPD-associated oxidative stress. Free Radic Biol Med 2018;126:350–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu J, Zhou X, Wang J, Li Z, Kong X, Qian J, et al. RhoGAPs attenuate cell proliferation by direct interaction with p53 tetramerization domain. Cell Rep 2013;3:1526–38. [DOI] [PubMed] [Google Scholar]
- 43. Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol 2006;7:713–26. [DOI] [PubMed] [Google Scholar]
- 44. Dimchev V, Lahmann I, Koestler SA, Kage F, Dimchev G, Steffen A, et al. Induced Arp2/3 complex depletion increases FMNL2/3 formin expression and filopodia formation. Front Cell Dev Biol 2021;9:634708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Coriat R, Nicco C, Chéreau C, Mir O, Alexandre J, Ropert S, et al. Sorafenib-induced hepatocellular carcinoma cell death depends on reactive oxygen species production in vitro and in vivo. Mol Cancer Ther 2012;11:2284–93. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available within the article and its Supplementary Data files.