Schwann cell precursors (SCPs) are transient glial progenitors that are important for the formation of late neural crest derivatives, yet their heterogeneity and developmental potential remain incompletely understood. In this issue, Kastriti, Faure, von Ahsen et al (2022) use comprehensive single‐cell RNA sequencing analyses to identify a transient “hub” state common to SCPs and neural crest cells (NCCs), revealing a striking similarity of SCPs to late migrating NCCs. These results raise important questions about the potential role of such a state in adult tissue regeneration and tumourigenesis.
Subject Categories: Development, Neuroscience
Recent single‐cell transcriptome profiling of mouse embryonic development reveals an unexpected transient multipotent state shared between Schwann cell progenitors and neural crest cells.
Schwann cell precursors (SCPs) are embryonic glial progenitors that are closely associated with developing nerves of the peripheral nervous system, along which they migrate (Jessen & Mirsky, 2019). SCPs of the distal dorsal and ventral nerve roots are derived from neural crest cells (NCCs) that emigrate from the neural tube and migrate into the periphery. In contrast, SCPs of the proximal part of the dorsal and ventral nerve roots are derived from boundary cap cells that are a secondary transient multipotent cell type that also originates from NCCs and that can give rise to both neuronal and glial cells.
Lineage tracing experiments have previously shown that SCPs can generate, in addition to the Schwann cell lineage, various cell types such as endoneurial fibroblasts, melanocytes, chondrocytes and osteoblasts, as reviewed in Erickson et al (2022). Consequently, SCPs provide a transient multipotent cell population that is important for the formation of late neural crest derivatives. Given their broad developmental potential, SCPs have been referred to as nerve‐associated neural crest stem cells. However, SCPs are strongly dependent on axonal signalling for their survival, unlike neural crest stem cells. Moreover, although SCPs and NCCs share a number of expressed genes, SCPs express Schwann cell lineage‐specific genes and downregulate certain NCC‐specific genes. Importantly, in contrast to NCCs, the developmental potential and heterogeneity of SCPs have so far not been addressed thoroughly at the single‐cell level. In mice, clonal analyses have been performed for both cranial and trunk NCCs by means of genetic lineage tracing, revealing multipotency of at least some individual NCCs (Baggiolini et al, 2015; Kaucka et al, 2016). However, conventional lineage tracing technologies possess certain limitations, especially for migratory cells such as NCCs, since the assessment of their clonality may be difficult on tissue sections and the number of genetically traceable clones is restricted (Tang & Bronner, 2020). Therefore, clonality analysis has been complemented with transcriptomic approaches using single‐cell RNA sequencing (scRNAseq) of the NCC lineage, which allows a higher resolution than genetic lineage tracing. In particular, a recent study by Soldatov and colleagues yielded a thorough reconstruction of neural crest lineage progression and cell fate decisions in a spatiotemporal context (Soldatov et al, 2019).
In the current issue of this journal, Kastriti et al (2022) addressed both the similarity between NCCs and SCPs and the extent of SCP multipotency and heterogeneity in mouse embryos by an in‐depth scRNAseq approach. The authors showed that NCCs and SCPs pass through a “hub state” in which these two cell types intermingle and which appears to be associated with multipotency (Fig 1). Specifically, the authors determined that “hub” cells express not only both neural crest as well as SCP markers, but they also identified “hub” state‐specific genes such as Sox8. Moreover, analysis of transcriptional programmes (metaregulons) revealed a striking similarity between late migratory NCCs and early SCPs. Indeed, the onset of “hub” gene expression is already detectable in previously characterized multipotent migratory NCCs, gradually increases and was found to be the highest once SCPs associate with the developing nerves.
Figure 1.
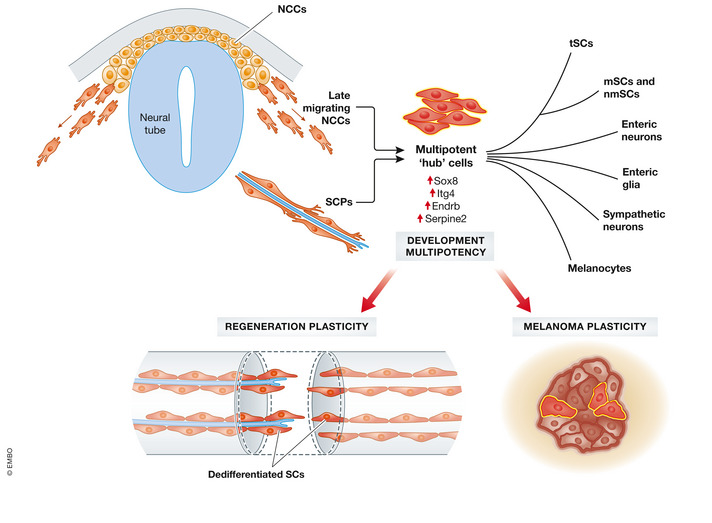
Neural crest cell / Schwann cell precursor “hub” cells contribute to diverse neural crest derivatives. Single‐cell RNA sequencing reports a multipotent “hub” cell state generated by late migrating neural crest cell (NCC) and Schwann cell precursors (SCPs) that express specific genes such as Sox8. NCC/SCP “hub” cells give rise to different NC‐derived cell populations. A state reminiscent of NCC/SCP “hub” cells re‐emerges in malignant tumours such as melanoma, although a potential contribution of the NCC/SCPs‐like state to tumorigenesis remains to be shown. Likewise, the NCC/SCP “hub” state might play a role during tissue regeneration, given that dedifferentiated Schwann cells were shown to emerge upon nerve injury and wounding of various tissues including the skin. NCC = Neural crest cells, SCPs = Schwann cell precursors, tSCs = terminal Schwann cells, mSCs = myelinating Schwann cells, nmSCs = non‐myelinating Schwann cells.
To clarify whether all the NCCs go through this “hub” state, the authors studied how “hub” markers emerge in the tissue. Based on this, the authors suggest three possible ways of NCC lineage transition: In the first scenario, NCCs pass through the “hub” state without nerve association; in the second, NCCs bypass the “hub” state; and in the third, NCCs converge into nerve‐associated SCPs. This emphasizes one caveat of scRNAseq: Despite the accurate identification of the internal states of cells, the predicted lineage trajectory does not necessarily reflect the actual clonal lineage. This is because fate restrictions could occur earlier or later than predicted by the transcriptional landscape. Clonal labelling studies will be required to determine the actual clonal lineage, complementary to the scRNAseq study presented in this study. Moreover, functional validation has to be performed to reveal the genes that drive fate selection and to distinguish those from genes that simply demarcate the decision‐making phases.
The study by Kastriti, Faure, von Ashen and colleagues provides an in‐depth source for the research community to better understand the molecular programmes defining neural crest‐derived lineages. Previously, work by the same authors and others suggested that SCPs generate a wide variety of cell types, including melanocytes and various mesenchymal derivatives (Erickson et al, 2022). However, whether all these predicted SCP derivatives emerge via a “hub” state could not be revealed. This could be due to technical limitations or also a lack of temporal resolution. Therefore, it will be important in the future to complement the knowledge gained by the present study with spatial transcriptomic approaches in order to take the tissue architecture into account and to refine the developmental stages to be investigated. Furthermore, analysis of the epigenome, for instance, by ATAC seq, of the “hub” state and downstream derivatives may provide further information on core transcriptional networks underlying the individual cell states (Klemm et al, 2019).
The notion that late migratory NCCs appear to form the “hub” cell state prior to nerve association and that this “hub” cell state then becomes more prominent upon association with developing peripheral nerves implies that the signal driving “hub” gene expression is already present in migrating NCCs. However, nerve association appears to reinforce the “hub” gene signature. These findings contrast with what has been observed in the mature peripheral nervous system, where axonal signalling/association is believed to maintain Schwann cell quiescence. The adult peripheral nerve is a highly quiescent tissue; however, Schwann cells retain the plasticity to dedifferentiate into a progenitor‐like state. Following nerve injury, all cell types appear to re‐enter the cell cycle (Stierli et al, 2018), and dedifferentiated Schwann cells without nerve association orchestrate the nerve regeneration process (Cattin & Lloyd, 2016). Likewise, Schwann cells induced to dedifferentiate upon injury promote tissue repair and regeneration also in other tissues, such as the heart and the skin (Parfejevs et al, 2018). Importantly, these previous studies revealed that dedifferentiated Schwann cells re‐differentiate within their lineage and apparently do not generate multiple cell types during tissue regeneration. However, whether dedifferentiated Schwann cells may harbour the potential for multi‐lineage differentiation, for instance, when exposed to pathogenic environments, remains to be shown. The signals responsible for maintaining Schwann cell quiescence and for driving Schwann cell dedifferentiation in the adult, as well as for supporting the embryonic “hub” state in SCPs are not known. It is open whether it is indeed axonal signalling that drive these processes and whether embryonic axonal signalling is different in contrast to mature axonal signalling. In addition to axonal association, SCPs are exposed to a very different microenvironment as opposed to mature Schwann cells. Moreover, in a Schwann cell‐derived tumour, neurofibroma, it was shown that the injury microenvironment is critical for tumour formation (Ribeiro et al, 2013). This implies that the tissue microenvironment, in addition to axonal signals, likely contributes to the different Schwann cell states. It will be intriguing to complement the knowledge gained with the present scRNAseq study with spatial transcriptomics to enhance spatio‐temporal information on “hub” cells and to reveal the cellular communication of nerve‐associated SCPs and their microenvironment. The latter could clarify whether it is nerve association that promotes the SCP “hub” state or whether it is the embryonic tissue microenvironment. Furthermore, comparing the transcriptome of dedifferentiated Schwann cells to the SCP “hub” cell state will be critical to determine the gene expression differences between dedifferentiated Schwann cells in the adult and SCPs in the embryo. Are dedifferentiated Schwann cells more similar to nerve‐associated “hub” cells or to NCCs that are not nerve associated? Similarly, it is well known that cancer cells are highly plastic and, interestingly, that cells with a NCC signature re‐emerge within malignant neural crest‐derived tumours (Diener & Sommer, 2021). Accordingly, malignant tumours, such as melanoma, contain tumour populations that map to the SCP/NCC “hub” state, as shown by Kastriti, Faure, von Ashen et al. Thus, it will be interesting to elucidate whether and how the “hub” state identified during embryonic development contributes to tumorigenesis and tissue regeneration in the adult (Fig 1).
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
The EMBO Journal (2022) 41: e111955
See also: ME Kastriti et al (September 2022)
References
- Baggiolini A, Varum S, Mateos JM, Bettosini D, John N, Bonalli M, Ziegler U, Dimou L, Clevers H, Furrer R et al (2015) Premigratory and migratory neural crest cells are multipotent in vivo . Cell Stem Cell 16: 314–322 [DOI] [PubMed] [Google Scholar]
- Cattin AL, Lloyd AC (2016) The multicellular complexity of peripheral nerve regeneration. Curr Opin Neurobiol 39: 38–46 [DOI] [PubMed] [Google Scholar]
- Diener J, Sommer L (2021) Reemergence of neural crest stem cell‐like states in melanoma during disease progression and treatment. Stem Cells Transl Med 10: 522–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson AG, Kameneva P, Adameyko I (2022) The transcriptional portraits of the neural crest at the individual cell level. Semin Cell Dev Biol 10.1016/j.semcdb.2022.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R (2019) Schwann cell precursors; multipotent glial cells in embryonic nerves. Front Mol Neurosci 12: 69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastriti ME, Faure L, von Ahsen D, Bouderlique GT, Boström J, Solovieva T, Jackson C, Bronner M, Meijer D, Hadjab S et al (2022) Schwann cell precursors represent a neural crest‐like hub state with biased multipotency. EMBO J 41: e108780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaucka M, Ivashkin E, Gyllborg D, Zikmund T, Tesarova M, Kaiser J, Xie M, Petersen J, Pachnis V, Nicolis SK et al (2016) Analysis of neural crest‐derived clones reveals novel aspects of facial development. Sci Adv 2: e1600060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm SL, Shipony Z, Greenleaf WJ (2019) Chromatin accessibility and the regulatory epigenome. Nat Rev Genet 20: 207–220 [DOI] [PubMed] [Google Scholar]
- Parfejevs V, Antunes AT & Sommer L (2018) Injury and stress responses of adult neural crest‐derived cells. Dev Biol 444(Suppl 1): S356–S365 [DOI] [PubMed] [Google Scholar]
- Ribeiro S, Napoli I, White IJ, Parrinello S, Flanagan AM, Suter U, Parada LF, Lloyd AC (2013) Injury signals cooperate with Nf1 loss to relieve the tumor‐suppressive environment of adult peripheral nerve. Cell Rep 5: 126–136 [DOI] [PubMed] [Google Scholar]
- Soldatov R, Kaucka M, Kastriti ME, Petersen J, Chontorotzea T, Englmaier L, Akkuratova N, Yang Y, Häring M, Dyachuk V et al (2019) Spatiotemporal structure of cell fate decisions in murine neural crest. Science 364: eaas9536 [DOI] [PubMed] [Google Scholar]
- Stierli S, Napoli I, White IJ, Cattin AL, Cabrejos AM, Calavia NG, Malong L, Ribeiro S, Nihouarn J, Williams R et al (2018) The regulation of the homeostasis and regeneration of peripheral nerve is distinct from the CNS and independent of a stem cell population. Development 145: dev170316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Bronner ME (2020) Neural crest lineage analysis: from past to future trajectory. Development 147: dev193193 [DOI] [PMC free article] [PubMed] [Google Scholar]


