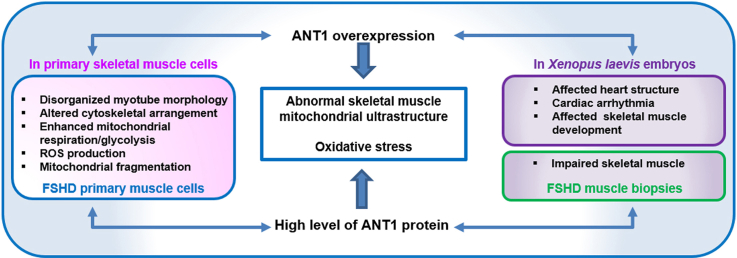
Abstract
Facioscapulohumeral muscular dystrophy (FSHD) is an autosomal dominant disorder characterized by progressive muscle weakness. Adenine nucleotide translocator 1 (ANT1), the only 4q35 gene involved in mitochondrial function, is strongly expressed in FSHD skeletal muscle biopsies. However, its role in FSHD is unclear. In this study, we evaluated ANT1 overexpression effects in primary myoblasts from healthy controls and during Xenopus laevis organogenesis. We also compared ANT1 overexpression effects with the phenotype of FSHD muscle cells and biopsies.
Here, we report that the ANT1 overexpression-induced phenotype presents some similarities with FSHD muscle cells and biopsies. ANT1-overexpressing muscle cells showed disorganized morphology, altered cytoskeletal arrangement, enhanced mitochondrial respiration/glycolysis, ROS production, oxidative stress, mitochondrial fragmentation and ultrastructure alteration, as observed in FSHD muscle cells. ANT1 overexpression in Xenopus laevis embryos affected skeletal muscle development, impaired skeletal muscle, altered mitochondrial ultrastructure and led to oxidative stress as observed in FSHD muscle biopsies. Moreover, ANT1 overexpression in X. laevis embryos affected heart structure and mitochondrial ultrastructure leading to cardiac arrhythmia, as described in some patients with FSHD.
Overall our data suggest that ANT1 could contribute to mitochondria dysfunction and oxidative stress in FSHD muscle cells by modifying their bioenergetic profile associated with ROS production. Such interplay between energy metabolism and ROS production in FSHD will be of significant interest for future prospects.
Keywords: Adenine nucleotide translocase type 1 (ANT1); Facioscapulohumeral muscular dystrophy (FSHD); Primary muscle cells; Xenopus laevis, Mitochondrial function; Metabolism; Oxidative stress; Muscle morphological abnormalities
Graphical abstract
Highlights
-
•
ANT1 overexpression leads to a disorganized myotube phenotype, as observed in FSHD myotubes.
-
•
Oxidative phosphorylation and glycolytic activity are increased in ANT1-overexpressing and FSHD cells.
-
•
ANT1 overexpressing and FSHD cells and also X. laevis embryos present abnormal morphology of muscle mitochondria.
-
•
ANT1 overexpressing and FSHD cells and also X. laevis embryos exhibit oxidative stress.
-
•
ANT1 overexpression in X. laevis embryos affects muscle and heart development.
1. Introduction
Facioscapulohumeral muscular dystrophy (FSHD), the third most common inherited neuromuscular disorder, is characterized by progressive weakness and atrophy of facial [1] and shoulder girdle muscles that can spread also to the lower extremity muscles [[1], [2], [3]]. In many patients, muscle involvement is asymmetric [2]. FSHD severity shows considerable inter- and intra-familial variability, from almost asymptomatic to wheelchair dependency [1,3,4]. This highly heterogeneous clinical spectrum suggests the involvement of multiple factors, including epigenetic regulators [2].
FSHD type 1 (FSHD1), which concerns more than 95% of patients, is associated with a reduction of the D4Z4 macrosatellite repeat array (from 1 to 10 D4Z4 units) on chromosome 4 (4q35) [5,6]. It is thought that in FSHD1, epigenetic derepression of double homeobox 4 (DUX4), a retrogene within the D4Z4 repeat, causes the disease through a gain-of-function mechanism [7]. It has been proposed that D4Z4 repeat array reduction results in the local relaxation of the DNA structure, leading to upregulated transcription of local genes [8]. DUX4 misexpression in skeletal muscle initiates a transcription deregulation cascade, leading to muscle atrophy, muscle differentiation defects, oxidative stress and ultimately cell death which are key features of FSHD [[9], [10], [11], [12]]. A recent study demonstrated that DUX4 expression in FSHD myocytes is increased by oxidative stress through a DNA damage response signaling pathway [13].
Despite major progress in understanding FSHD genetic basis, the mechanisms that contribute to the initial muscle mass loss and weakness are not fully understood. Besides DUX4, other genes in the 4q35 region [e.g. double homeobox 4 centromeric (DUX4c), adenine nucleotide translocator 1 (ANT1), FSHD region gene 1 and 2 (FRG1, FRG2)] could be derepressed and act as modifiers [7,[14], [15], [16], [17]] thus contributing to FSHD pathogenesis, either independently or synergistically. However, mRNA expression studies of all these genes including ANT1 in FSHD are controversial. Gabellini et al. reported a 10 fold higher ANT1 transcript levels in FSHD than in control skeletal muscle biopsies using 3 FSHD muscle biopsies [18]. Conversely, using real-time PCR, no consistent difference in steady-state ANT1 mRNA levels normalized to those for 18 S rRNA was observed when comparing FSHD and analogous disease-control muscle biopsy samples from patients with unrelated neuromuscular disease involving myopathic changes or denervation atrophy [19]. In the study of Klooster et al. [20], using quantitative RT-PCR, no significant difference in relative mRNA expression of the gene ANT1 was observed between the 10 FSHD and 10 control muscle biopsies and between 4 controls and 4 FSHD primary myoblasts and myotubes [20]. Conversely, Tsumagari et al. observed that ANT1 mRNA was upregulated 2-fold in FSHD vs. control myoblasts, although not in myotubes [21]. These discrepancies in the transcriptional level of ANT1 in FSHD skeletal muscle were proposed to partly be explained by the different technological approaches and variation in source of RNA (muscle pathology, duration of disease, etc.) [22]. At protein level, only limited data are available for ANT1 gene. In the study of Klooster et al. [20], ANT1 protein level could not be reliably detected in both FSHD muscle biopsies (n = 10) and cell cultures (n = 4). However, in our previous study [23], ANT1 protein levels were higher in 17 FSHD muscle biopsies compared with 16 healthy controls and this result was supported by the work of Macaione et al. performed in 8 FSHD and 8 control muscle biopsies [16]. The discrepancy between these results is not entirely clear but may partly be explained by the protein extract preparation, the choice of antibodies etc. Moreover, the work of Kim et al. brings a new evidence of ANT1 upregulation in FSHD [24]. A new enhancer element in the 4qA allele was proposed to regulate the expression of the FRG1 and ANT1 genes specifically in FSHD cells through a direct interaction with the respective gene promoters. This hypothesis was supported by the finding that (1.5–3 fold) up-regulation of these two genes in FSHD patients is consistent with the relatively weak effect of the 4qA enhancer in the luciferase assay [24]. This study reveals the first evidence of 4qA β-satellite repeats (BSR) implication in the transcriptional regulation of the 4q35 locus and precisely, in ANT1 gene transcriptional activity [24]. Furthermore, the zinc-finger protein 555 (ZNF555) binding to a D4Z4 4qA located enhancer was shown to play a critical role in regulating ANT1 promoter activity, particularly in FSHD [24]. Therefore, a new working model of the functional role of the 4qA allele in the transcriptional control of the ANT1 gene in FSHD patients was proposed where the cooperative 4qA β-satellite repeats (BSR) binding with the ZNF555 transcriptional factor could be a critical step in the formation of a transcriptionally productive complex [24]. These results were in general agreement with the study of abnormal chromatin conformation changes in FSHD myoblasts [21,25,26] leading to the direct interactions between the 4qA region and the long distance genes in 4q35 [27]. They are also consistent with the current model of interaction between distantly located enhancer and promoter through formation of the DNA loop enabling their physical interaction [28,29]. Finally, transgenic mice muscle-specifically overexpressing ANT1 do not seem to develop muscular dystrophy [30]. However, the absence of ANT1 levels evaluation in ANT1 overexpressing mouse was a considerable limitation given that ANT1 is highly abundant in muscle mitochondria [31]. Thus, whether Ant1 overexpression does, or does not, cause muscle pathology remains undetermined. Recently, the establishment of Ant1-transgenic mice using the Ant1 genomic locus and its native promoter support the idea that moderate overloading of ANT1 induces muscle atrophy [32].
ANT1 is predominantly expressed in post-differentiated tissues, such as heart and skeletal muscle, and catalyses ADP/ATP exchange across the mitochondrial inner membrane [33]. ANT1 mutations or altered expression have been associated with a growing list of human diseases. Moreover, we previously showed that the reduced physical performance of patients with FSHD is associated with mitochondrial dysfunction and oxidative stress [34], and can be improved by antioxidant supplementation [35]. The organization of mitochondria is altered in skeletal muscle biopsies from patients with FSHD. Large mitochondrial pools in the intermyofibrillar and subsarcolemmal compartments [34] and abnormal mitochondrial aggregation near blood capillaries [36] can be observed. The mitochondrial ultrastructure also is altered with some badly formed cristae and apparent swelling [34]. Ex vivo experiments showed that FSHD primary myoblasts are more sensitive to exogenous pro-oxidants [37,38]. Moreover, control myoblasts fuse to form branched myotubes with aligned nuclei. Conversely, FSHD primary myoblasts fuse to form thin and branched myotubes (atrophied myotubes) with aligned nuclei, or large myotubes with randomly distributed nuclei (disorganized myotubes) [37]. DUX4 expression induces atrophic myotubes and associated FSHD markers [12], and its silencing normalizes the FSHD atrophic myotube phenotype, but not the disorganized phenotype [12]. Since high levels of ANT1 are observed in FSHD muscle biopsies, and FSHD is associated with mitochondrial dysfunction and oxidative stress [16,23,[34], [35], [36], [37], [38]]. ANT1 remains an attractive candidate gene and could be involved in the pathogenesis of FSHD [16,18,21,23,[24], [25], [26], [27], [28], [29]]. In this study, we thoroughly evaluated the effects of ANT1 overexpression in primary myoblasts from healthy controls and during Xenopus laevis organogenesis. We also compared ANT1 overexpression effects with the phenotype of FSHD muscle cells and biopsies (age-matched with the healthy controls).
2. Materials and methods
2.1. Quadriceps biopsies and human primary cells
The following quadriceps biopsies and derived human primary myoblast cultures used in this study: FSHD8 (39-year-old man; 6 D4Z4 units), FSHD10 (20-year-old woman; 4 D4Z4 units), FSHD 14 (25-year-old man; 4 D4Z4 units) and 3 age- and sex-matched healthy controls (C: C1: 20-year-old woman; C2: 29- and C3: 43-year-old men) were provided by CHU Montpellier biobank (DC-2008-594). Upon differentiation, these FSHD cell cultures showed a disorganized myotubes morphology as previously described [37].
2.2. Human primary cell cultures
Primary myoblasts were cultured on poly-lysine coated coverslips in the presence of 1/1000 collagen, as previously described [37]. Cells were cultured at 37 °C in a humidified atmosphere of 5% CO2 in proliferation medium: Dulbecco's modified Eagle's medium (DMEM; Sigma Aldrich, Saint Quentin-Fallavier, France) supplemented with glutamine (Sigma Aldrich), 20% foetal bovine serum (FBS) (Eurobio), 2% Ultroser G (Biosepra, Cergy Saint Christophe, France), 50 μg/ml gentamycin (Sigma Aldrich). In 80% confluent myoblasts, myogenic differentiation was induced by switching to differentiation medium (DMEM, 2% FBS, 50 μg/ml gentamycin) for 3 days before analysis. According to our previous study [37], the proliferation rates of all FSHD and healthy control myoblasts were comparable. The mean doubling time of FSHD and healthy control myoblasts was very similar (41.7 ± 6.5 and 41 ± 3.6, respectively).
2.3. ANT1 overexpression in primary myoblasts
Primary myoblast cultures from 3 healthy control primary myoblast cultures (C: C1: 20-year-old woman; C2: 29- and C3: 43-year-old men) were used for the transfection experiments. Primary myoblasts were plated (2 × 105 cells per well) in six-well plates for 24 h before transfection (1 μg/well) of a pCMV6 plasmid expressing human ANT1 or empty vector (CMV promoter-derived mammalian expression vector from Origene) with Turbofectin (Origene Technologies Gmbh, herford, Germany) [39] (standard ratio of 3:1), according to the manufacturer's recommendations. Four conditions were studied: healthy control cells without Turbofectin (C), cells incubated only with Turbofectin (T), cells transfected with Turbofectin and the empty vector (V-) or the ANT1 plasmid (V+). Myoblasts were harvested 48 h after transfection and myotubes after 3 days of differentiation. The mean doubling time of C, T, V-, V+ was very similar (C: 41 h ± 3.6, T: 40.6 h ± 4; V-: 39 h ± 4; V+: 40.3 h ± 2.5).
2.4. ANT1 overexpression in Xenopus laevis embryos
2.4.1. X. laevis care and in vitro fertilization
Adult wild type X. laevis females were purchased from Xenopus Express Farm (Vernassal, France). Testicles were isolated from adult wild type X. laevis males from the institute. All experiments were performed according to the animal welfare regulations of the District Government of Lower Franconia. In vitro fertilization was performed as described [40]. Embryos were staged according to the Normal Table of X. laevis [41] and cultured in modified Ringer's solution [42] in charcoal agar plates with 0.5% sulfadiazine at 20 °C. Animal care and experimental procedures were approved by the Würzburg institutional animal care and use committee in accordance with the University of Würzburg ethical guidelines (agreement number 55.2-2532-2-604).
2.4.2. Plasmids
The human ANT1 cDNA sequence (SLC25A4) in the pHRTK-P229 plasmid was amplified with primers including the restriction sites Xbal and SalI (ANT1 Forward: 5′-GTCGACATGGGTGATCACGCTTGGAGC-3′, Reverse: 5′-TCTAGATTAGACATATTTTTTGATCTC-3′, Biomers, Ulm, germany) and the Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific). Cycling conditions were: initial denaturation at 94 °C for 2 min, 25 cycles of 94 °C for 30 s, 52 °C for 30 s, 72 °C for 90 s, followed by a final extension at 72 °C for 10 min. The target cDNA band was recovered and purified with the NucleoSpin® Extract II Kit (Macherey-Nagel GmbH, Düren, Germany) and cloned in the pCMV-Sport6 vector (Invitrogen, Thermo Fisher Scientific), following the manufacturer's instructions. After plasmid linearization, both SP6 and T7 promoters could be used for in vitro synthesis of capped mRNA.
2.4.3. mRNA synthesis and microinjection
In vitro synthesis of capped ANT1 mRNA and of the antisense ANT1 mRNA were performed using the mMESSAGE mMACHINE® SP6 and T7 Transcription Kits, respectively (Thermo Fisher Scientific), according to the manufacturer's instructions. After synthesis, mRNAs were purified with the RNeasy-Mini-Kit (Qiagen, Courtaboeuf, France). Approximatively 50 ng of capped ANT1 mRNA was injected in one blastomere of each 2-cell stage X. laevis embryo, as described [43,44]. As control, embryos were injected with antisense ANT1 mRNA, and the non-injected side of each embryo was used as internal control.
2.5. RNA isolation and quantitative PCR analysis of human muscle cells
Total RNA from muscle cells was prepared using the RNeasy® kit (Qiagen) and quantitated using NanoDrop spectrophotometer (Thermo Fischer Scientific). First strand cDNA was synthetized using the SuperScript III Kit (Invitrogen) following the manufacturer's instructions. ANT1, MFN1, MFN2, DRP1, OPA1, MnSOD relative expression was normalized to ribosomal protein, large, P0 (RPLP0) mRNA levels (Supplementary Table 1). The reaction mixtures were prepared according to the instructions of the SYBR Green Kit (Roche Diagnostics, Meylan, France) and real time quantitative reverse transcription PCR (RT-PCR) was performed using a Roche Lightcycler 480 system (Roche Diagnostics) and the following program: 95 °C for 2 min, followed by 37 cycles at 95 °C for 10 s, and 60 °C for 40 s. The relative expression levels were calculated using the 2 −ΔΔCt method.
2.6. Protein extraction and western blotting from muscle biopsies and cells and X. laevis
Protein extracts from human quadriceps biopsies (20 μg) and from cells (40 μg) were obtained as previously described [23,37]. Briefly, muscle powder from muscle biopsy specimens was homogenized in 1 ml buffer A [20 mM hydroxyethylpiperazine ethanesulfonic acid pH 7.5, 1 mM EDTA, 1 mM dithiothreitol, 1 mM MgCl2, 400 mM NaCl, 20% (v/v) glycerol, 0.75 mM spermidine, 0.15 mM spermine, 0.1% (v/v) Nonidet P40] with protease inhibitors. Primary muscle cells were lysed in hypertonic buffer containing 50 mM Tris pH7, 50 mM NaCl, 0.1% NP40, anti-proteases and 1 mM DTT. Samples were separated by SDS-PAGE and immunoblotted with an anti-ANT1 antibody (sc-9300, 1:1000, Santa Cruz Biotechnology, Le Perray-en-Yvelines, France), as previously reported [23]. Densitometry analyses were performed using Image J (NIH, Bethesda, MD) and values were normalized to loading controls (cytochrome c (muscle biopsies, sc-7159, 1:500, Santa Cruz Biotechnology), tubulin (primary muscle cells, T9026, 1:10000, Sigma Aldrich)). Protein extraction and western blotting were performed in X. laevis as previously described [45]. Vitellogenin was used as loading control.
2.7. Muscle cell immunocytochemistry
For immunofluorescence staining, cells were fixed with 4% paraformaldehyde (Electron Microscopy Science) and incubated with PBS/0.5%Triton X-100. Cells were then incubated at RT for 1 h with the following antibodies: anti-ANT1 antibody (sc-9300, Santa Cruz Biotechnology), AB2 (MS-1372 P1, Interchim, Montluçon, France), Troponin T (T6277, Sigma Aldrich), or Alexa Fluor 488 conjugated phalloidin (ab176757, Abcam). After incubation with Alexa 488 conjugated secondary antibody at RT for 1 h, cells were washed in PBS 3 times, and coverslips were mounted using Fluoroshied (F6057, Sigma Aldrich). Fluorescence was visualized using a Zeiss microscope.
2.8. Whole mount immunostaining
For whole mount X. laevis experiments, 3-day-old X. laevis tadpoles were fixed in Dent's fixative (80% methanol, 20% dimethyl sulfoxide) at −20 °C overnight, followed by bleaching and labelling, as previously described [44]. The anti-desmin primary antibody (1:13, Sigma-Aldrich) was followed by anti-mouse peroxidase-conjugated IgG (1:3500, Dianova), and antibody interaction visualization using diaminobenzidine.
2.9. Myotube fusion
To analyse the myotube phenotype, cell staining was performed as previously described [37]. After 3 days of differentiation, myotubes were incubated with anti-troponin T or -phalloidin antibodies and DAPI to visualize myotubes and nuclei. The Myogenic Fusion Index (MFI) was determined by dividing the number of nuclei in multi-nucleated myotubes by the total number of nuclei in a given microscopic field [37]. The Deformed Myotube Index (DMI) was calculated as the proportion of myotubes with deformed morphology characterized by abnormal repartition of nuclei [37].
2.10. Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) in muscle cells
OCR, an indicator of mitochondrial respiration [46] and ECAR, a measure of lactic acid levels formed during the conversion of glucose to lactate during glycolysis [47], were measured using a XF-96 Analyzer (Agilent technologies, Seahorse Bioscience Europe, Copenhagen, Denmark). OCR and ECAR measurement were performed in myoblasts and myotubes at the MÉTAMONTP facility, Montpellier, France. Myoblasts were grown in proliferation medium. Then, 24 h after plating, medium was switched to differentiation medium for 3 days to obtain myotubes. Healthy control (C) and FSHD myoblasts were plated (5000 cells/well for myoblasts study and 10,000 cells/well for myotube study) on XF Cell Culture 96-well plates (# 101085–004, Agilent) to obtain a cell monolayer as previously reported [48]. For overexpression experiments, after plating (5000 cells/well for myoblasts study and 10,000 cells/well for myotube study), healthy control myoblasts (C: C1: 20-year-old woman; C2: 29- and C3: 43-year-old men) were transfected with Turbofectin alone (T), empty vector (V-), or ANT1 expression plasmid (V+). Myoblasts and myotubes from V+, FSHD and controls (C, T, V-) were studied on separate XF Cell Culture 96-well plates.
For the mitochondrial stress test, cell medium was replaced by the assay medium (Seahorse Bioscience) supplemented with 1 mM pyruvate, 10 mM glucose, and 2 mM glutamine at 37 °C for 30 min, followed by measurements with the XF Cell Mito Stress Kit (Agilent). OCR (pmol (O2)/min/μg protein) and ECAR (pH/min/μg protein) were evaluated in the same well at baseline and after sequential addition of inhibitors, as described in the Seahorse Agilent protocol. Briefly, baseline rates were measured and then mitochondrial function inhibitors (2 mM oligomycin, 2 mM carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP), and 1 mM rotenone/0.5 μM antimycin A) were sequentially added. Five mitochondrial bioenergetic parameters were calculated from the profile (Supplementary Fig. 1a): basal respiration (basal cellular respiration minus non-mitochondrial respiration), maximal respiratory capacity (maximal uncoupled respiration minus non-mitochondrial respiration), ATP-linked OCR (basal respiration minus oligomycin-inhibited respiration), proton leak (oligomycin-inhibited respiration minus non-mitochondrial respiration), and NMOC (rotenone + antimycin A-inhibited respiration). OCR/ECAR ratios were calculated at baseline and after FCCP injection. Finally, baseline ECAR and OCR measurements were converted to ATP synthesis rates (JATP production) to allow the direct comparison of the glycolytic (JATPgly) and oxidative (JATPox) ATP production rates [49] and the conversion to the percentage of total ATP production. These calculations are reliable only for myoblasts [50].
For the glycolysis stress test, cells were incubated in the assay medium (Seahorse Bioscience) with 2 mM glutamine without glucose at 37 °C for 30 min before measurements with the Glycolytic Stress Test Kit (Seahorse Bioscience). ECAR and the corresponding OCR were measured in the same wells at baseline and after sequential addition of 10 mM glucose, and pharmacological inhibitors of OXPHOS (2 μM oligomycin) and glycolysis (100 mM 2-DG) as described by the manufacturer. Four parameters were calculated from the profile (Supplementary Fig. 1b): glycolysis (ECAR rate following the addition of glucose minus non-glycolytic acidification), glycolytic capacity (maximal ECAR rate following the addition of oligomycin minus non-glycolytic acidification), glycolytic reserve (glycolytic capacity minus glycolysis) and non-glycolytic acidification (2-DG-inhibited ECAR). Results were expressed as mpH/min/mg protein. After each experiment, the protein content per well was quantified using the BCA assay kit (23227, Pierce, Thermo Fisher Scientific), and used for normalization.
2.11. Lactate measurement in myoblasts and myotubes and in the corresponding supernatants
Lactate was measured using Ultra Performance Liquid Chromatography coupled to a triple-quadrupole mass spectrometer (Acquity UPLC – XEVO TQD, Waters Corporation). Calibrators, controls and samples (100 μL/each) were treated with 100 μL of 10% trichloroacetic acid/1 mM [2H2]-3-hydroxybutyrate as internal standard. After mixing and centrifugation (14000×g, 4 °C, 15 min), supernatants were transferred to Acquity vials and placed in the sample manager (injection volume = 10 μL). Samples were analysed on an Acquity Premier CSH Phenyl-Hexyl column, 100 mm × 2.1 mm, 1.7 μm (Waters Corporation). The chromatographic mobile phase was constituted of 0.1% formic acid (v/v) in water and 0.1% formic acid (v/v) in acetonitrile delivered at a flow rate of 0.4 mL/min at 50 °C. Lactate ionization was performed using negative electrospray ionization and the XevoTQD monitored the following transitions: m/z 88.7 to m/z 43.0 and m/z 104.8 to m/z 60.0 for lactate and [2H2]-3-hydroxybutyrate, respectively.
2.12. Transmission electron microscopy (TEM) of muscle biopsies and cells and X. laevis embryos
Muscle quadriceps biopsies were fixed and embedded as previously described [34]. TEM of muscle quadriceps biopsies was carried out at the Centre Regional d’Imagerie Cellulaire (CRIC) of Montpellier (France).
V+ and FSHD and control myoblasts (C, T, V-) were fixed, embedded in Epon and processed for TEM according to standard procedures (for details see Ref. [51]). Control (uninjected and antisense mRNA-injected) and ANT1 mRNA-injected X. laevis embryos were fixed and embedded as described [52], and ultrathin sections were cut using a Leica EM UC7 microtome. Semi-thin sections were stained with methylene blue Azur II, rinsed with water, and air-dried. Then, samples from primary myoblasts and X. laevis embryos sections were analysed using a JEOL JEM-2100 transmission electron microscope equipped with a TemCam F416 4 k x 4 k (Tietz video and Imaging Processing Systems, Gauting, Germany).
2.13. Mitochondrial mass of myoblasts
2.13.1. MitoTracker staining and imaging
Mitochondrial mass was monitored using the MitoTracker green probes [53] in V+ and control (C, T, V-) myoblasts. After incubation in pre-warmed growth medium containing 100 nM of probe at 37 °C for 30 min, cells were immediately washed twice in cold PBS and analysed using a Zeiss microscope. The fluorescence intensity of MitoTracker relative to the cell volume was quantified using Image J.
2.13.2. Citrate synthase activity
Mitochondrial mass was monitored using citrate synthase activity (citrate synthase activity assay kit (Ab119692)) [54] in FSHD and C myoblast samples. Citrate synthase activity was measured spectrophotometrically in myoblast samples at 30 °C using a Beckman spectrophotometer. Citrate synthase activity was monitored by recording the changes in absorbance at 412 nm for 150 sec and normalized to protein concentration.
2.14. Mitochondrial membrane potential of myoblasts
Mitochondrial membrane potential was detected in myoblasts with the Muse™ Cell Analyzer (Millipore, Billerica, MA, USA). V+, FSHD and control (C, T and V-) myoblasts were suspended in the working solution of the Muse MitoPotential Kit (MCH100110, Merck Millipore) and incubated at 37 °C for 25 min, according to the manufacturer's recommendations. Then, cells were incubated with 5 μl of Muse 7-aminoactinomycin D (7-AAD) at RT for 5 min to monitor cell death. The analysis was performed using the Muse Cell Soft V1.4.0.0 Analyser Assay (Merck Millipore). Cells incubated with 100 μM H2O2 for 5 min were used as positive control to validate the assay results. For this cytometry assay, a single assay result was derived from 3000 individual fluorescence measurements to decrease statistical error.
2.15. Mitochondrial dynamics and motility in FSHD myoblasts using the MitoSelect® technology
MitoSelect® analyses of intact living primary myoblasts were performed by Innovative Concepts in Drug Development investigators (ICDD, Gemenos, France), blind to the donor status (C/FSHD cells with disorganized phenotype). The Mitoselect® technology is a high content analysis of mitochondrial behaviour using 4D live-cell imaging to identify mitochondrial phenotypes associated with disease states [55]. It quantifies patterns of mitochondrial behaviour along four dimensions: mitochondrial dynamics, motility, organization and permeability with the captures of 48 descriptors. Mitochondria were visualized using MitoTracker green (Thermo Fisher) on a Zeiss Axioplan microscope, equipped with a temperature-controlled stage to maintain the sample at 37 °C, and the Slidebook 4.2 image acquisition software (Intelligent Imaging Innovation). In each imaging session, 3 to 5 cells were recorded allowing the analysis of motility and dye retention in 70–80 individual mitochondria/sample, and the analysis of mitochondrial dynamics and organization in 300–700 mitochondria/sample. Image analysis was performed with the Micv1.1-Mitostream® image analysis software (ICDD). Descriptors measuring the mitochondrial fusion-fission balance (scale), mitochondrial velocity (μm/min), mitochondrial organization (in ATP-demanding hubs, such as the microtubule-organizing centre, perinuclear area, and focal adhesion points along the plasma membrane), and dye retention rate as a surrogate measure of mitochondrial permeability were studied. Raw data (captured images) and quantification from image analysis (database) were stored in their original format.
2.16. Reactive oxygen species (ROS) production and oxidative stress
2.16.1. Measurement of intracellular ROS production by dihydroethidium in muscle cells
In V+ and FSHD and control (C, T and V-) myoblasts, intracellular ROS production was measured by cytometry using the Muse Oxidative Stress Kit (MCH100111, Merck Millipore). Briefly, cell suspensions were incubated in the working solution of Muse® reagent at 37 °C for 30 min, and then intracellular ROS production (percentage of ROS-positive cells) was detected with the Muse™ Cell Analyzer (Merck Millipore). For cytometry assay, a single assay result was derived from 3000 individual fluorescence measurements to decrease statistical error. The analysis was performed using the Muse Cell Soft V1.4.0.0 Analyser Assay (Merck Millipore).
In V+ and FSHD and control (C, T and V-) myotubes, intracellular superoxide activity was detected using dihydroethidium (DHE) (Invitrogen). Myotubes were incubated in medium containing 2 μM DHE in the dark at 37 °C for 30 min, according to the manufacturer's instructions. Images were recorded with a fluorescent Zeiss microscope and a 590-nm long-pass filter. DHE fluorescence signal was quantified in nuclei and averaged per images and per cell culture. DHE fluorescence signal was analysed using Image J software.
2.16.2. Measurement of mitochondrial superoxide production with MitoSOX in myoblasts
Mitochondrial superoxide levels were assessed by incubating V+ and FSHD and control (C, T and V-) myoblasts with MitoSOX (5 μM) at 37 °C for 30 min. Following incubation, myoblasts were washed twice and mounted for fluorescence assessment using a Zeiss microscope. The fluorescence intensity of MitoSOX per cell was measured using Image J. Experiments were performed in C, T, V-, V+ and in FSHD and C myoblasts. MitoSOX fluorescence signal was quantified in cells and averaged per images and experiments.
2.16.3. Ascorbyl free radicals (AFR) measurement by electron paramagnetic resonance spectroscopy in FSHD and control cells
Myoblasts and myotubes from healthy controls (C) and patients with FSHD were cultured in proliferation and differentiation medium, respectively, at 37 °C in a humidified atmosphere of 5% CO2. Supernatants were collected after 48 h of proliferation (myoblasts) and 72 h of differentiation (myotubes), placed in cryotubes and quickly frozen in liquid nitrogen [56]. AFR was measured after addition of dimethyl sulfoxide (DMSO) (AFR/DMSO) (1:1) to evaluate ascorbate concentration released in supernatants. Upon reacting with free radicals, ascorbate is oxidized to ascorbyl radical anion (also referred to as AFR). AFR/DMSO content is considered as a good marker of the cellular oxidative stress level. The method is detailed in Ref. [56]. EPR spectra of AFR were recorded at 9.8 GHz from supernatant, dimethyl sulfoxide (1:1) mixtures by averaging 4 scans with instrument parameters: microwave power, 10 mW; modulation frequency, 100 kHz; modulation amplitude, 0.883 G; time constant, 81.92 ms; receiver gain, 5 × 105 and sweep rate, 1.07 G/s.
2.16.4. Protein carbonylation in muscle cells and in X. laevis embryos
The Oxyblot Protein Oxidation Detection Kit was used to quantify the presence of carbonyl groups in V+ and FSHD and control (C, T and V-) myoblasts and myotubes as well as in 3-day-old X. laevis control (uninjected or antisense mRNA-injected) and ANT1 mRNA-injected embryos (according to the manufacturer's instructions. Signals were revealed using enhanced chemiluminescence (Roti®Lumin and Carl Roth Alpha Innotech Corporation, San Leandro, CA) for X. laevis embryos and C and FSHD myoblasts and myotubes and a Licor Odyssey scanner (Licor) for C, T, V-, V+ myoblasts and myotubes. Protein carbonyl content was calculated by adding the integrated density of individual protein bands. Normalization was performed using GAPDH [57] for C, T, V-, V+ myoblasts and tubulin for C, T, V-, V+ myotubes and C and FSHD myoblasts and myotubes. Protein concentration was used to normalize protein carbonyl content in X. laevis samples.
2.17. Data availability
All data supporting the findings described in this manuscript are available in the article and from the corresponding author upon reasonable request.
2.18. Statistical analysis
All analyses were performed using the Sigma Stat program (Jandel). N are indicated in the figure legends. The Shapiro-Wilks test was used to confirm data normality. Parametric continuous data were presented using mean and standard deviation (SD); non-parametric data using median and interquartile range. Sample sizes for studies was chosen based on observed effect sizes and standard errors. In order to verify the effect size in the comparisons of averages between control and FSHD samples, Cohen's d was calculated and the value of d was considered small if 0.20 ≤ d < 0.5 and large when d ≥ 0.80. Statistical significance was determined using the paired Student's t-test, unpaired Student's t-test for two group analyses. One-way Analysis of Variance (ANOVA) (with Holm-Sidak multiple comparison or Tukey test) was used to compare more than two groups. When considering categorical data contingency analyses were performed to determine the probability that the frequency distribution differ across groups using a Chi-square analysis. Statistical significance was set at p ≤ 0.05. *P < 0.05, **P < 0.01 and ***P < 0.001.
3. Results
3.1. ANT1 overexpression leads to a disorganized myotube phenotype, as observed in FSHD myotubes
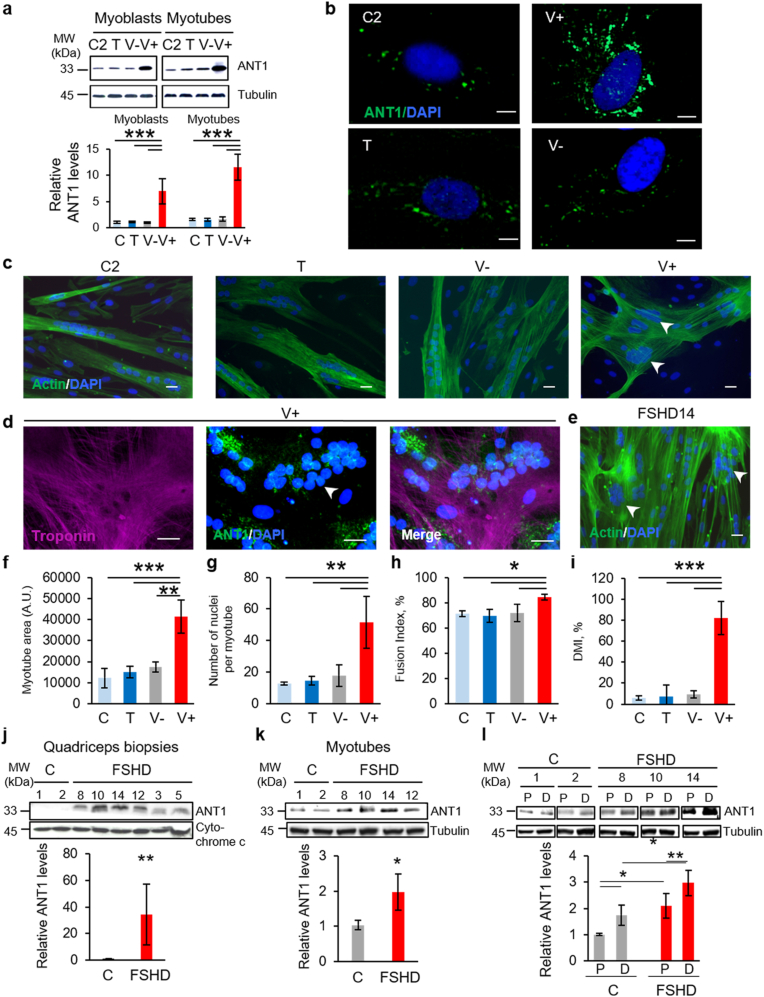
To evaluate the impact of ANT1 overexpression, we incubated each healthy control myoblast (C: C1: 20-year-old woman; C2: 29- and C3: 43-year-old men) culture with the transfection reagent Turbofectin alone (T) or together with the ANT1 construct (V+), or empty vector (V-). We confirmed ANT1 overexpression in V+ myoblasts and myotubes, compared with control cells without transfection reagent (C), T, and V- cells by RT-qPCR (Supplementary Fig. 2) and by Western blot analysis (Fig. 1a). Moreover, immunofluorescence analysis showed that ANT1 staining was stronger in V+ than in C, T and V- myoblasts (Fig. 1b).
Fig. 1.
ANT1 overexpression leads to a disorganized myotube phenotype, as observed in FSHD myotubes.
(a) Representative Western blot of the levels of ANT1 in C2, T, V-, V+ myoblasts and myotubes. The bar graphs show the summary of 3 independent cultures of each condition in duplicate as the mean ± SD of the intensity of ANT1 band over tubulin; *P = 0.05, ***P < 0.001 [Shapiro-Wilk test for normality, one-way ANOVA followed by Holm-Sidak method (F:99.080, DF:23, P < 0.001)]. (b) Representative immunofluorescent staining for ANT1 (green) and DAPI (blue) in control (C2, T, V-) and V+ myoblasts. For all conditions, the contrast range was set at the same level. (c) Myotube phenotype in C2, T, V- and V+ cultures at day 3 of differentiation: representative immunofluorescent staining for actin (phalloidin, green) in V+ and control myotubes (C2, T, V-) (c) and (d) for ANT1 (green) and troponin T (magenta) in V+ myotubes. (e) Myotube phenotype in FSHD (FSHD14) culture at day 3 of differentiation: representative immunofluorescent staining for actin (phalloidin, green) and DAPI (blue) in FSHD (FSHD14) myotubes. White arrowheads show clusters of nuclei (DAPI, blue) in V+ and FSHD myotubes. Scale bars: 10 μm. (f) Myotube area (arbitrary unit (A.U), (g) Number of nuclei per myotube, (h) Myogenic fusion index and (i) Deformed myotube index (DMI, %) in C, T, V- and V+ samples (6 randomly chosen fields per condition from 3 independent cultures of each condition in duplicate); *P = 0.05, **P = 0.01, ***P < 0.001 [one-way ANOVA (Holm-Sidak): (f) F:21.280, DF:11, P < 0.001; (g) F:12.487, DF:11, P = 0.002; (h) F:6.370, DF:11, P = 0.016; (i) F:44.690, DF:11, P < 0.001]. Data are presented as mean ± SD. (j, k, l) Representative Western blot of the levels of ANT1 (j) in myotubes, (k) in the corresponding quadriceps biopsies from 3 patients with FSHD (FSHD8, FSHD10, FSHD 14) and 2 age-matched healthy controls (C1, C2) and (l) in FSHD (FSHD8, FSHD10 and FSHD14) and control (C1, C2) muscle cells in proliferation (P) and after differentiation (D). (j, l) The bar graphs show the summary of 3 independent control (C) and FSHD cultures in duplicate as the mean ± SD of the intensity of ANT1 band over tubulin. (k) The bar graphs show the summary of 3 independent control and FSHD muscle biopsies in duplicate as the mean ± SD of the intensity of ANT1 band over cytochrome c. (j) *P = 0.010 (two tailed Student's t-test), Cohen's d = 2.33; (k) **P = 0.009 (two tailed Student's t-test), Cohen's d = 2.56; (l) *P = 0.05, **P < 0.01 [ANOVA (Holm-Sidak) (F: 19,027, DF:11, P < 0.001)]. Molecular weight (MW) markers are indicated in kilodaltons (kDa). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
After 3 days of differentiation, C, T and V- myoblasts fused to form branched myotubes with aligned nuclei and the actin was organized in a filamentous network parallel to the long axis of the syncytium (Fig. 1c). In contrast, V+ myotubes appeared larger and distorted with an abnormal distribution of nuclei (Fig. 1c) and the actin network was disorganized (Fig. 1c and d). The phenotype induced by ANT1 overexpression was reminiscent of the disorganized phenotype observed in primary FSHD myotubes (Fig. 1e): larger and distorted myotubes with an increased number of nuclei [37,58]. Indeed, phenotype analysis showed that myotube area and number of nuclei were significantly higher in V+ than in C, T and V- myotubes (Fig. 1f and g) as observed in FSHD myotubes (Supplementary Figs. 3a and b). Similarly, the myogenic fusion index (MFI) [37], to assess fusion competence, was significantly higher in V+ than in C, T and V- myotubes (Fig. 1h) as well as the deformed myotube index (DMI) [37] (57–100% in V+ versus 10% in C, T and V- myotubes) (Fig. 1i) as observed in FSHD myotubes (Supplementary Figs. 3c and d).
As ANT1 overexpression-induced phenotype was reminiscent of the disorganized phenotype observed in primary FSHD myotubes [37,58], we investigated ANT1 protein levels in FSHD primary myotubes and in the corresponding muscle biopsies. Quantification of the Western blot results showed that ANT1 protein levels were higher in FSHD primary myotubes and as previously reported [23] in muscle biopsies compared with age-matched healthy controls (C) (Fig. 1j and k). ANT1 protein levels were higher also in FSHD than in C myoblasts (Fig. 1l). Moreover, ANT1 protein levels were higher in myotubes than in myoblasts both in C and FSHD samples (Fig. 1l). We confirmed these results by immunofluorescence analysis of C and FSHD myoblasts and myotubes (Supplementary Figs. 3e and f).
3.2. Oxidative phosphorylation is increased in ANT1-overexpressing and FSHD myoblasts
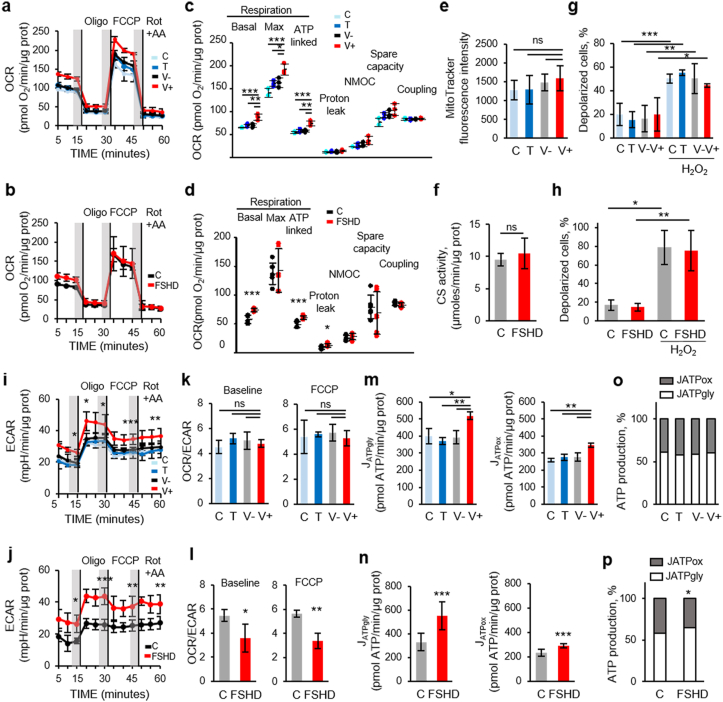
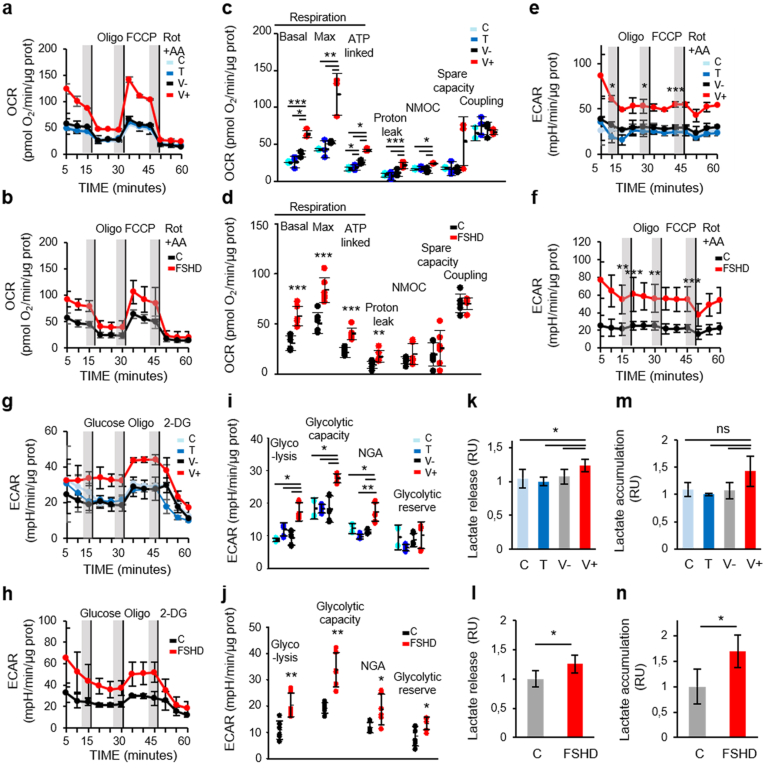
As ANT1 regulates mitochondrial metabolism, we then characterized the bioenergetic status of V+ and FSHD myoblasts compared with their respective controls using the Seahorse Cell Mito Stress Test (see schematic of the mitochondrial stress test in Supplementary Fig. 1a). In this system, mitochondrial respiration (oxygen consumption rate, OCR) is used to measure oxidative phosphorylation (OXPHOS). Based on the OCR profiles (Fig. 2a and b), we first established the “resting” OCR (basal OCR), which accounts for the initial respiration rate in myoblasts after removing the contribution from non-mitochondrial oxygen consumption (NMOC). Basal OCR was significantly higher in V+ than in C, T and V- myoblasts (Fig. 2c). Similarly, basal OCR was also significantly higher in FSHD than in C myoblasts (Fig. 2d). Quantification of MitoTracker green staining [53] showed no difference between V+ and control (C, T and V-) myoblasts (Fig. 2e), suggesting no change in mitochondrial mass. Similarly, citrate synthase activity, as an index of total mitochondrial mass [54], was similar in FSHD and C myoblasts (Fig. 2f). Upon addition of oligomycin that decreases the electron flow through the electron transport chain (ETC) by inhibiting adenosine triphosphate (ATP) synthase, mitochondrial respiration was reduced and this allowed identifying the fraction of basal OCR attributable to ATP production. Oxygen consumption used to produce mitochondrial ATP (ATP-linked respiration) was higher in V+ than in C, T and V- myoblasts (Fig. 2c). Similarly, oxygen consumption was also higher in FSHD than in C myoblasts (Fig. 2d). We also measured the energy dissipation, as proton leak, and found that it was similar in V+ and control (C, T and V-) myoblasts (Fig. 2c). Conversely, proton leak was higher in FSHD than in C myoblasts (Fig. 2d). Coupling efficiency was comparable in C, T, V- and V+ myoblasts (Fig. 2c). Similarly, no difference in coupling efficiency between FSHD and C myoblasts was observed (Fig. 2d). Then, addition of the mitochondrial uncoupler FCCP, to shuttle protons across the inner membrane, led to ETC maximum activity. The resulting effect was the maximum OCR value (maximal respiration) to maintain the proton motive force at the highest levels. The maximal respiratory capacity was significantly higher in V+ than in C, T and V- myoblasts (Fig. 2c), but no difference between FSHD and C myoblasts was observed (Fig. 2d). The maximal respiration curve allowed calculating the spare (or reserve) respiratory capacity (i.e., the difference between the FCCP-stimulated and resting OCR) and showing that it was comparable in C, T, V- and V+ myoblasts (Fig. 2c) and also in FSHD and C myoblasts (Fig. 2d). Finally, rotenone and antimycin A (Rot + AA) were added to block complex I and III, respectively. Together, these agents fully inhibit the mitochondrial electron transfer, and the residual OCR measured can be attributed to non-mitochondrial oxidases (NMOC). NMOC was comparable in C, T, V- and V+ myoblasts (Fig. 2c) and also in FSHD and C myoblasts (Fig. 2d). Moreover, we did not observe any difference in mitochondrial membrane potential (Δψm) in C, T, V- and V+ myoblasts (Fig. 2g) and also in FSHD and C myoblasts (Fig. 2h).
Fig. 2.
Oxygen consumption rate (OCR) is increased in ANT1-overexpressing and FSHD myoblasts.
(a, b) The MitoStress test was used to quantify OCR in (a) C, T, V-, V+ (3 independent cultures of each condition in duplicate) and (b) FSHD (3 independent control (C) and FSHD cultures in duplicate). (a, b) Shaded regions define the steady state before sequential addition of inhibitors: oligomycin (Oligo, 2 mM), FCCP (2 mM), rotenone + antimycin A (1 mM Rot + 0.5 μM AA); vertical lines indicate the injection time. (c, d) Basal respiration, maximal respiratory capacity, ATP-linked OCR, proton leak, non-mitochondrial respiration (NMOC), spare capacity and coupling in (c) C, T, V-, V+ myoblasts and in (d) FSHD and C myoblasts were calculated from the profiles (a, b). (c) C, T, V-, V+ myoblasts: *P < 0.05, **P < 0.01, ***P < 0.001 [one-way ANOVA (Holm-Sidak)], (d) FSHD and C myoblasts: *P < 0.05, ***P < 0.001 (two-tailed unpaired Student's t-test) (Basal respiration Cohen's d: 2.97, maximal respiratory capacity Cohen's d: 0.18, ATP-linked OCR Cohen's d: 2.75, proton leak Cohen's d: 1.51, NMOC Cohen's d: 0.49, spare capacity Cohen's d: 0.34 and coupling Cohen's d: 0.69). (e) Mitochondrial mass was based on MitoTracker fluorescence intensity per cell (A.U) in C, T, V-, V+ myoblasts (ANOVA, P = 0.465). Each histogram represents the mean MitoTracker fluorescence intensity per cell from 3 independent cultures of each condition in duplicate. (f) Mitochondrial mass was based on citrate synthase (CS) activity in FSHD and C myoblasts (3 independent control (C) and FSHD cultures in duplicate), P = 0.524, Cohen's d = 0.57, ns: non-significant. Each histogram represents the mean ± SD. (g, h) Mitochondrial membrane potential (g) in C, T, V-, V+ myoblasts (3 independent cultures of each condition in duplicate) ***P < 0.001 [one-way ANOVA (Holm-Sidak) (F:20.836, DF:23, P < 0.001)], and (h) in FSHD and C myoblasts (3 independent control (C) and FSHD cultures in duplicate) *P < 0.05, **P < 0.01 [one-way ANOVA (Holm-Sidak) (F:34.817, DF:11, P < 0.001), Cohen's d = 2.58] by flow cytometry using the MitoPotential Kit. Positive control: incubation with hydrogen peroxide (H2O2,100 μM for 5 min). (i, j) The corresponding ECAR of the OCR: representative ECAR profile (i) in C, T, V-, V+ and (j) in FSHD and C myoblasts. (k, l) OCR/ECAR ratios at baseline and after FCCP addition (k) in V+ and control myoblasts (C, T, V-) (ns: non-significant) and (l) in FSHD and C myoblasts (*P < 0.05, Cohen's d = 1.75); **P < 0.01 (Cohen's d = 1.76)). (m, n) Glycolytic and oxidative ATP production rates (JATPgly and JATPox, respectively) (m) in C, T, V-, V+ myoblasts (**P < 0.01, one-way ANOVA (Holm-Sidak) JATPgly: F:12.902, DF:11, P = 0.003; JATPox: F:13.447, DF:10, P = 0.003) and (n) in FSHD and C myoblasts (two-tailed unpaired Student's t-test: JATPgly: ***P < 0.001 (Cohen's d = 2.29), JATPox: ***P < 0.001 (Cohen's d = 2.78)). (o, p) JATPgly and JATPox percentages of the total JATP production were calculated respectively (o) in V+ and control (C, T, V-) myoblasts (non-significant) and (p) in FSHD and C myoblasts (two tailed Student's t-test: %JATPgly: *P < 0.05 (Cohen's d = 1.748), %JATPox: *P < 0.05 (Cohen's d = 1.748).
Concomitantly, we also assessed the glycolytic function in V+ and FSHD myoblasts and their respective controls by measuring the extracellular acidification rate (ECAR) that indicates the lactic acid levels generated by glycolysis (Fig. 2i and j). ECAR curve was similar to the OCR curve; however, while addition of oligomycin reduced OCR, ECAR was increased in both V+ and FSHD myoblasts compared with their respective controls. This indicated that glycolysis was upregulated in response to the inhibition of mitochondrial oxygen consumption (Fig. 2i and j), suggesting that compared with their respective controls, V+ and FSHD myoblasts more readily switch to glycolysis when ATP synthase and mitochondrial respiration are inhibited. Then, we calculated the OCR/ECAR ratio at baseline and at maximal respiration (upon FCCP addition) to assess the relative contribution of glycolysis and OXPHOS to energy generation. In both conditions, the OCR/ECAR ratios were comparable in C, T, V- and V+ myoblasts (Fig. 2k), whereas they were lower in FSHD than in C myoblasts (Fig. 2l). Then, we converted the baseline ECAR and OCR values to rates of ATP synthesis (JATP production) to directly compare the glycolytic (JATPgly) and oxidative (JATPox) ATP production rates [49]. Glycolytic and oxidative ATP production were significantly increased in V+ and FSHD myoblasts compared with their respective controls (Fig. 2m and n). Conversion to percentage of ATP production showed comparable glycolytic and mitochondrial ATP production percentages in C, T, V- and V+ myoblasts (Fig. 2o). Conversely, glycolytic ATP production was increased by 12% and mitochondrial ATP production was decreased by 16% in FSHD myoblasts compared with C (Fig. 2p).
3.3. Glycolytic activity is increased in ANT1-overexpressing and FSHD myoblasts
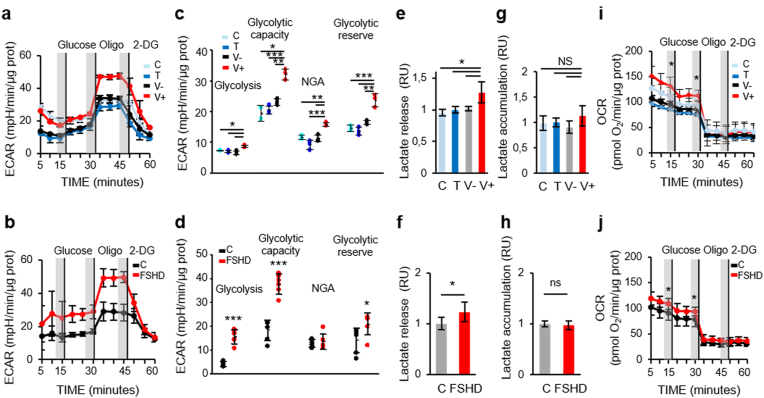
As myoblasts rely primarily on glycolysis for metabolic needs, we evaluated also the glycolytic activity in ANT1 and FSHD myoblasts compared with their respective controls using the Seahorse Glycolysis Stress Test (Supplementary Fig. 1b). This test starts with ECAR measurement in starved cells (Fig. 3a and b). The normal glycolysis rate was higher in V+ (Fig. 3c) than in control myoblasts (C, T and V-). Similarly, glycolysis rate was higher in FSHD than in C myoblasts (Fig. 3d). The baseline lactate release was significantly increased in the culture medium of V+ (by 28% compared with C, T and V- myoblasts) (Fig. 3e) and also of FSHD myoblasts (by 20% compared with C myoblasts) (Fig. 3f). Conversely, lactate accumulation was not different (Fig. 3g and h). Addition of oligomycin to stimulate the maximal ECAR, which shuts down ATP-dependent OCR, shifted metabolism to glycolysis more effectively in V+ (Fig. 3c) and FSHD (Fig. 3d) than in their respective control myoblasts. The glycolytic reserve capacity (i.e. the difference between maximal and basal ECAR) was higher in V+ (Fig. 3c) and FSHD (Fig. 3d) than in their respective control myoblasts. The test ended by total inhibition of glycolysis using the glucose analogue 2-DG, leading to the ECAR decrease to its non-glycolytic level. All myoblast samples were sensitive to glycolysis inhibition by 2-DG (Fig. 3a and b). Non-glycolytic acidification (NGA) might correspond to the conversion of respiratory CO2 generated in the TCA cycle to HCO3- and H+. NGA (i.e. acidification not due to lactate production) was higher in V+ than in C, T and V- myoblasts (Fig. 3c), but not in FSHD myoblasts (Fig. 3d). Concomitant extracellular flux analysis to assess OXPHOS in culture supernatants showed that after addition of 10 mM glucose, OCR was higher in V+ and FSHD than in their respective control myoblasts (Fig. 3i and j).
Fig. 3.
Extracellular acidification rate (ECAR) is increased in ANT1-overexpressing and FSHD myoblasts.
(a, b) The glycolytic stress test was used to quantify ECAR (a) in C, T, V-, V+ myoblasts (3 independent cultures of each condition in duplicate) and (b) in FSHD and control (C) myoblasts (3 independent control (C) and FSHD cultures in duplicate). After serial addition of glucose, oligomycin (Oligo) and 2-deoxy-d-glucose (2-DG), glycolysis (basal ECAR), glycolytic capacity, non-glycolytic acidification (NGA), and glycolytic reserve were calculated (c) in C, T, V-, V+ myoblasts and (d) in FSHD and C myoblasts from their corresponding profiles (a, b). (c) C, T, V-, V+: *P < 0.05, **P < 0.01, ***P < 0.001 (one-way ANOVA (Holm-Sidak)), (d) FSHD and C myoblasts (two-tailed unpaired Student's t-test) *P < 0.05, ***P < 0.001 (glycolysis Cohen's d: 5.17, glycolytic capacity Cohen's d: 4.56, NGA Cohen's d: 0.26, glycolytic reserve Cohen's d: 1.39). (e, f) Lactate release in the culture medium of (e) C, T, V-, V+ myoblasts and (f) of FSHD and C myoblasts, and (g, h) lactate accumulation (g) in C, T, V-, V+ myoblasts (3 independent cultures of each condition in duplicate) and (h) in FSHD and C myoblasts (3 independent control (C) and FSHD cultures in duplicate). Difference between C, T, V-, V+ myoblasts:(e) *P < 0.05 [one-way ANOVA (Holm-Sidak) (F:7.132, DF:11, P = 0.016)], (g) ns: non-significant; FSHD and C myoblasts: *P < 0.05 (two-tailed unpaired Student's t-test) and in (f) Cohen’ d:1.59 and (h) Cohen’ d:1.71. (i, j) The corresponding OCR of the ECAR: Representative profile of OCR simultaneously measured to ECAR (i) in C, T, V-, V+ myoblasts and (j) in FSHD and C myoblasts. Significant difference between V+ and control (C, T, V-) myoblasts (*P < 0.05 [one-way ANOVA (Holm-Sidak)], FSHD and C myoblasts *P < 0.05. (two-tailed unpaired Student's t-test).
3.4. Oxidative phosphorylation and glycolytic activity are increased also in ANT1-overexpressing and FSHD myotubes
We then determined the basal and maximal OCR, mitochondrial ATP production, spare respiratory capacity and proton leak in V+, FSHD and in their respective control myotubes. Myotubes are highly metabolically active cells that rely heavily on OXPHOS. We found that the OCR profile was higher in V+ and FSHD (Fig. 4a and b) than in their respective control myotubes. Specifically, basal respiration, maximal respiration, ATP production, and proton leak were significantly higher in V+ than C, T and V- myotubes (Fig. 4c). Similarly, ATP production was significantly higher in FSHD than in C myotubes (Fig. 4d). Conversely, NMOC was significantly increased only in V+ but not in FSHD myotubes (Fig. 4c and d) compared with their respective controls. Respiratory spare capacity and coupling efficiency were comparable in V+ and C, T, V- myotubes (Fig. 4c) and also in FSHD and C myotubes (Fig. 4d). Finally, the OCR increase was associated with higher basal ECAR in V+ and FSHD myotubes compared with their respective controls (Fig. 4e and f). Analysis of the glycolytic function using the glycolysis stress test showed that ECAR was higher in V+ and FSHD than in their respective control myotubes (Fig. 4g and h). Specifically, glycolysis, glycolytic capacity, and NGA were significantly increased in V+ myotubes compared with controls (Fig. 4i). Glycolysis also was higher in FSHD than C myotubes as well as the other respiratory parameters (Fig. 4j), except the glycolytic reserve higher in FSHD myotubes compared with C (Fig. 4j). The baseline lactate released in supernatant was significantly increased in V+ (by 28%) and FSHD (by 35%) samples compared with their respective controls (Fig. 4k and l). Conversely, baseline lactate accumulation was significantly increased only in FSHD myotubes compared with C, but not in V+ myotubes (Fig. 4m and n).
Fig. 4.
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) are increased in ANT1-overexpressing and FSHD myotubes.
(a, b) The MitoStress test was used to quantify OCR (a) in C, T, V-, V+ myotubes (3 independent cultures of each condition in duplicate) and (b) in FSHD and C myotubes (3 independent control (C) and FSHD cultures in duplicate) after sequential addition of oligomycin (2 mM), FCCP (2 mM) and rotenone + antimycin A (1 mM Rot + 0.5 μM AA). Each inhibitor was injected at the times indicated by the vertical lines. (c, d) The indicated mitochondrial bioenergetic parameters (c) in C, T, V-, V+ and (d) in FSHD and C myotubes were calculated from the profiles (a, b). Dot plots indicated individual experiments and the means ± SD are shown. (c) C, T, V-, V+ myotubes: *P < 0.05, **P < 0.01 [one-way ANOVA (Holm-Sidak)], (d) FSHD and C myotubes *P < 0.05, ***P < 0.001 (two-tailed unpaired Student's t-test) (basal respiration Cohen's d: 3.16, maximal respiratory capacity Cohen's d:2.83, ATP-linked OCR Cohen's d: 3.487, proton leak Cohen's d: 1.72, NMOC Cohen's d: 0.81, spare capacity Cohen's d: 0.41 and coupling Cohen's d: 0.68). (e, f) The corresponding ECAR of the OCR: representative ECAR profile (e) in C, T, V-, V+ and (f) in FSHD and C myoblasts. C, T, V-, V+ myotubes: *P < 0.05, ***P < 0.001 [one-way ANOVA (Holm-Sidak)], (d) FSHD and C myotubes **P < 0.01, ***P < 0.001 (two-tailed unpaired Student's t-test). (g, h) The glycolytic stress test was used to quantify ECAR (g) in C, T, V-, V+ myotubes (3 independent cultures of each condition in duplicate) and (h) in FSHD and control (C) myotubes (3 independent control (C) and FSHD cultures in duplicate). After serial addition of glucose, oligomycin (Oligo), and 2-deoxy-d-glucose (2-DG), the indicated parameters (i) in C, T, V-, V+ myotubes and (j) in FSHD and C myotubes were calculated from their corresponding profiles (g, h). Dot plots indicated individual experiments and the means ± SD are shown. (i) C, T, V-, V+ myotubes: *P < 0.05, **P < 0.01 [one-way ANOVA (Holm-Sidak)]; (j) FSHD and C myotubes: *P < 0.05, **P < 0.01, ***P < 0.001 (two-tailed unpaired Student's t-test). (glycolysis Cohen's d: 2.39, glycolytic capacity Cohen's d: 3.01, NGA Cohen's d: 1.59, glycolytic reserve Cohen's d: 1.57). (k, l) Lactate release in the culture medium and (m, n) lactate accumulation: (k, m) in C, T, V-, V+ myotubes (3 independent cultures of each condition in duplicate) and (l, n) in FSHD and C myotubes (3 independent control (C) and FSHD cultures in duplicate). *P < 0.05 [one-way ANOVA (Holm-Sidak) (F: 7.538, DF:11, P = 0.010), ns: non-significant] for C, T, V-, and V+ myotubes, two-tailed unpaired Student's t-test for FSHD and C myotubes (l) Cohen’ d: 1.63 and (n) Cohen’ d: 1.82.
3.5. The mitochondrial network is disturbed in ANT1-overexpressing and FSHD primary muscle cells
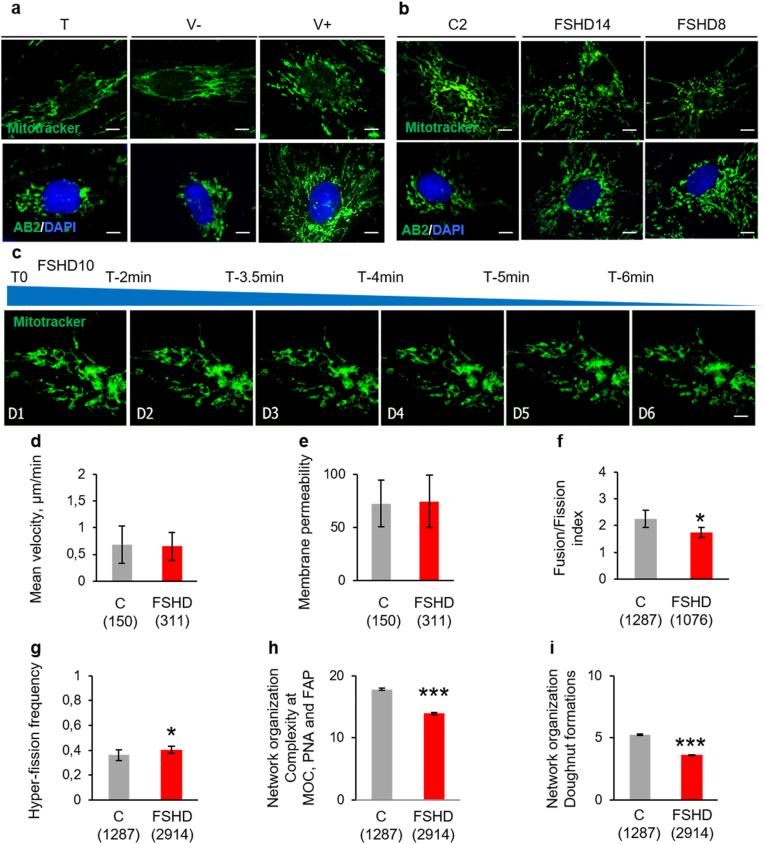
Many evidences show that mitochondrial bioenergetics and mitochondrial dynamics influence each other [59]. MitoTracker Green staining showed mitochondria with normal elongated-tubular structures in T, V- myoblasts (Fig. 5a, upper panels) and control myoblasts (C2) (Fig. 5b, upper panels). Conversely, in V+ myoblasts, mitochondria displayed a punctuated structure (Fig. 5a upper panel). We obtained similar results by immunofluorescence with a mouse monoclonal antibody against the mitochondrial marker AB2 (Fig. 5a and b, lower panel). We observed similar changes in FSHD myoblasts compared with C2 using the MitoTracker Green probe (Fig. 5b, upper panel), the antibody against AB2 (Fig. 5b, lower panel), and time-lapse microscopy tracking using a novel 4-dimension live imaging (three spatial dimensions over time) technology (Fig. 5c): the mitochondrial shape varied from tubular to rounded or punctuated in FSHD myoblasts.
Fig. 5.
Mitochondrial network abnormalities are observed in ANT1-overexpressing and FSHD myoblasts.
(a, b) Representative mitochondrial network visualization with MitoTracker Green (upper panels) and with AB2 (green)/DAPI (blue) staining (lower panels) (a) in V+, T, V- and (b) C2 myoblasts, (b) in FSHD (FSHD14, FSHD8) and C2 myoblasts. Scale bars = 10 μm. (c) Representative time lapse images of FSHD myoblasts (FSHD10) labelled with MitoTracker green. Six representative frames are shown (time 0, 2, 3.5, 4, 5, 6 min). Scale bars = 10 μm. (d–i) Mitochondrial movement was analysed in myoblasts from 3 controls (C) and 3 patients with FSHD in 3 independent plating of 3–5 individual cells. (d) Mean velocity, (e) Retention rate of dye in control (C) and FSHD myoblasts (n = 150 mitochondria in C and n = 311 in FSHD myoblasts for d and e). (f) Fusion/fission index in C (n = 1287 mitochondria) and FSHD (n = 1076 mitochondria) myoblasts, (g) Mitochondrial hyper-fission frequency in C (n = 1287 mitochondria) and FSHD (n = 2914 mitochondria) myoblasts, (h, i) Mitochondrial network organization (h) in ATP demanding hubs (microtubule organizing centre (MOC), perinuclear area (PNA), and focal points at the plasma membrane (FAP)) and (i) in doughnut formations (n = 1287 mitochondria in C and 2914 in FSHD myoblasts). Values are the mean ± SEM. *P < 0.05, **P < 0.001, ***P < 0.001 (two-tailed Student's t-test). (Mean velocity Cohen's d: 0.11, membrane permeability Cohen's d: 0.07, Fusion/fission index Cohen's d: 1.85, Mitochondrial hyper-fission frequency Cohen's d: 2.5, Mitochondrial network organization Cohen's d: 24.827 and doughnut formations Cohen's d: 36.11). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Using live cell imaging and the Mitoselect® technology, we found that mitochondrial velocity (Fig. 5d) was comparable in FSHD and in C myoblasts. Similarly, we did not observe any difference in the dye retention rate, suggesting no impact of the disease on mitochondrial membrane permeability (Fig. 5e). Conversely, analysis of the mitochondrial dynamics showed a slight, but significant decrease in the fusion-fission index (Fig. 5f) reflected by a significantly increased probability of higher frequency of hyper-fission in FSHD than C myoblasts (p = 0.004) (Fig. 5g). Mitochondrial network organization was significantly perturbed in FSHD myoblasts compared with C. The abundance and complexity of the reticular mitochondrial network were drastically reduced in FSHD myoblasts compared with C (p < 0.001) in regions of high ATP demand: microtubule organizing centre (MOC), perinuclear area (PNA), and focal points at the plasma membrane (FAP) (Fig. 5h). Similarly, the formation of doughnut-shaped mitochondria, a feature necessary for intracellular oxidative stress management, was greatly reduced in FSHD myoblasts compared with C (Fig. 5i) (p < 0.001). The expression of genes involved in fission-fusion (DRP1, FIS1, MFN1, MFN2 and OPA1) was comparable in FSHD and in C myoblasts (Supplementary Fig. 4).
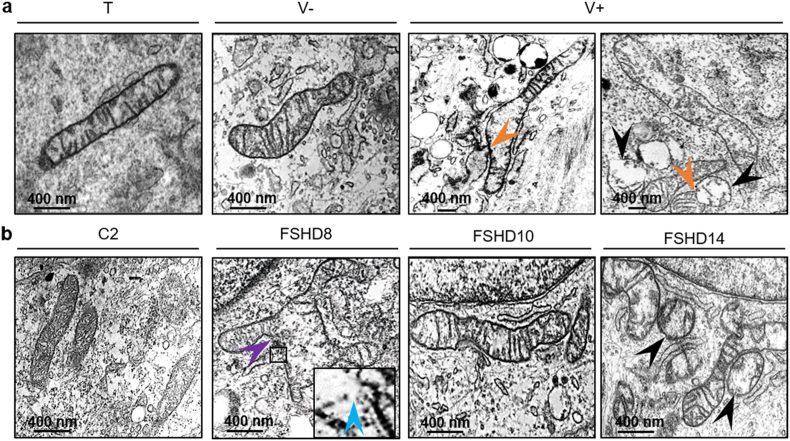
Transmission electron microscopy (TEM) highlighted morphological and shape abnormalities of mitochondria in V+ myoblasts compared with T, V- (Fig. 6a) and C2 (Fig. 6b): elongated tubular mitochondria and immature-like mitochondria with a round shape, absent or reduced number of cristae membranes, with empty matrix or enlarged matrix spaces, underdeveloped cristae (arrowhead), and partial damage of the outer mitochondrial membrane with blebs (arrowhead). In V+ myoblasts, mitochondrial fusion with a mitochondrion empty of cristae was observed (arrowhead). We observed similar changes also in FSHD myoblasts (FSHD8, FSHD10, FSHD 14) compared with C2 (Fig. 6b).
Fig. 6.
Mitochondrial ultrastructure abnormalities are observed in ANT1-overexpressing and FSHD myoblasts.
(a, b) Representative electron microscopy images of mitochondrial ultrastructure (a) in V+, T and V- myoblasts, (b) in FSHD (FSHD8, FSHD10, FSHD14) and C2 myoblasts. In V+ myoblasts, black arrowheads indicate the immature-like morphology of mitochondria with round-shaped morphology and under-developed cristae. In V+ myoblasts, orange arrowheads show mitochondrial fusion with a mitochondrion empty of cristae. In FSHD myoblasts, the purple arrow depicts an enlarged mitochondrion carrying aberrant cristae with total or partial absence of cristae, and the blue arrowhead (enlarged image) indicates partial damage of the surrounding membrane with blebs and membrane disruption. Scale bars: 400 nm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.6. ROS production is increased in ANT1-overexpressing and FSHD myoblasts and myotubes
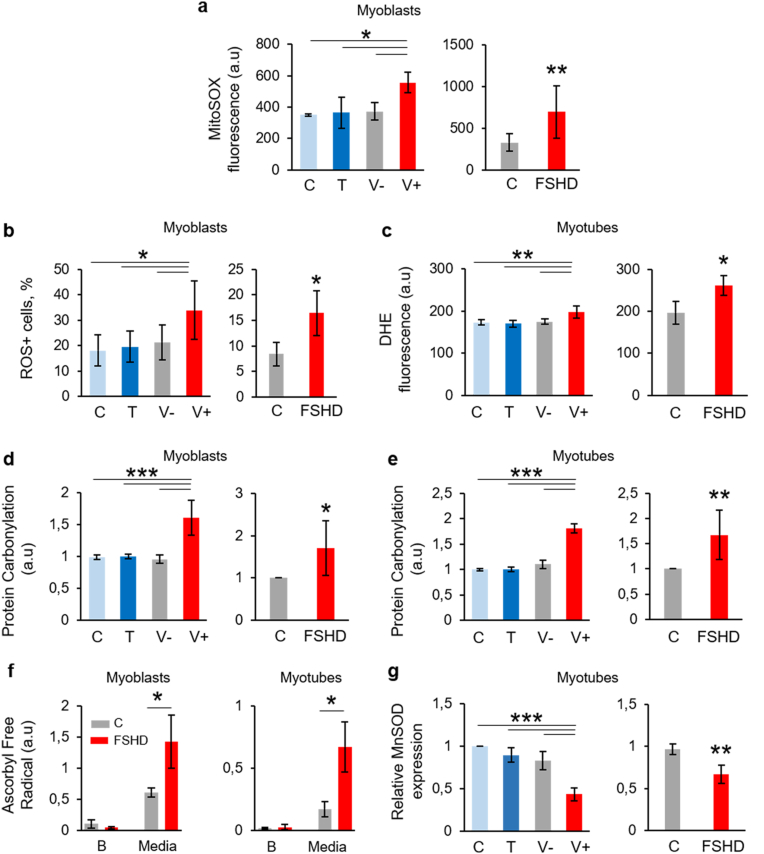
As increase in mitochondrial respiration may be associated with free radical formation, we evaluated mitochondrial ROS production using the MitoSOX probe (to detect mitochondrial superoxide production) in V + and FSHD and their respective control myoblasts. Mitochondrial superoxide production was significantly increased in V+ and FSHD myoblasts compared with their respective controls (Fig. 7a). Furthermore, using a DHE-based reagent (Muse Oxidative Stress kit), flow cytometry analysis showed that intracellular ROS levels were significantly higher in V+ and FSHD (Fig. 4c and d) than in their respective control myoblasts. Similarly, microscopy analysis of DHE fluorescence showed that intracellular ROS levels were significantly higher in V+ and FSHD (Fig. 4c and d) than in their respective control myotubes. Protein carbonylation levels also were higher in V+ and FSHD myoblasts (Fig. 7d) and myotubes (Fig. 7e) compared with their respective controls. Determination by electron spin resonance of ascorbyl free radical (AFR/DMSO) concentration showed that AFR release was significantly higher in supernatants from FSHD myoblasts (48 h) and myotubes (72 h) compared with C (Fig. 7f), suggesting that FSHD cells are exposed to free radical-induced oxidative stress. MnSOD, the essential mitochondrial antioxidant enzyme that detoxify superoxide generated by mitochondrial respiration was evaluated in V + and FSHD and their respective control myotubes. The expression of MnSOD gene was significantly decreased in V+ and FSHD myotubes compared with their respective controls (Fig. 7g).
Fig. 7.
Mitochondria contributes to the increased ROS production in ANT1-overexpressing and FSHD muscle cells.
(a) Mitochondrial superoxide production assessed with the MitoSOX Red fluorescent probe in V+, FSHD and their respective control myoblasts. Data are the mean ± SD. C, T, V-, V+: 3 independent cultures of each condition *P < 0.05 [one-way ANOVA (Holm-Sidak) F:6.505, DF:11, P = 0.015], FSHD and control (C) myoblasts: 3 independent control (C) and FSHD cultures in duplicate, **P < 0.01 (two-tailed unpaired Student's t-test, Cohen’ d: 4,17). (b) Percentage of myoblasts with high intracellular ROS production (ROS + cells) evaluated by cytometry and the Muse Oxidative Stress Kit in V+, FSHD, and their respective control myoblasts. Data are the mean ± SD. C, T, V-, V+: 3 independent cultures of each condition in duplicate, *P < 0.05 [one-way ANOVA (Holm-Sidak) (F = 7,671, DF:11, P = 0.018)]. FSHD and control myoblasts (C): 3 independent control and FSHD cultures in duplicate, *P < 0.05 (two tailed Student's t-test, Cohen’ d:3,53). (c) Intracellular ROS production using DHE fluorescence in C, T, V- and V+ myotubes (3 independent cultures of each condition) ***P < 0.001 [one-way ANOVA (Holm-Sidak) F = 13.5443 DF:11, P = 0.002] and in FSHD and C myotubes (3 independent control and FSHD cultures in duplicate, two tailed Student's t-test, *P = 0.015, Cohen’ d: 3.30). (d, e) Oxidative damage of proteins (protein carbonylation) was assessed with the Oxyblot kit in V+, FSHD and their respective control myoblasts (d) and myotubes (e). The bar graphs show the mean ± SD of 3 independent cultures of each condition in duplicate for C, T, V-, V+. *P < 0.05, **P < 0.01 [one-way ANOVA (Holm-Sidak) (Myoblasts: F = 44.99, DF:11, P < 0.001; Myotubes: F = 175.84, DF:11, P < 0.001). FSHD and C myoblasts and myotubes (3 independent control (C) and FSHD cultures in duplicate): *P < 0.05, **P < 0.05 (two tailed Student's t-test, for myoblasts Cohen’ d: 1.54, for myotubes Cohen’ d:1.97). (f) Levels of ascorbyl free radical - dimethyl sulfoxide (AFR/DMSO) in supernatant from FSHD and C cells (3 independent control (C) and FSHD cultures). Myoblasts and myotubes were cultured for 48 h and 72 h, respectively, and then supernatants were collected and analysed. Mean values (±SD) of AFR/DMSO from myoblast and myotube supernatants (media) and the respective backgrounds (B: culture media alone) are shown. *P < 0.05 [one-way ANOVA (Holm-Sidak)]. (g) Relative expression levels of MnSOD mRNA in V+, FSHD and their respective control myotubes were calculated relative to RPLPO mRNA levels. Values are expressed as relative to C myoblast levels. The bar graphs show the mean ± SD. C, T, V- and V+ myotubes (3 independent cultures of each condition) ***P < 0.001 [one-way ANOVA (Holm-Sidak) F = 30.526, DF:11, P < 0.001]. FSHD and C myotubes (3 independent control (C) and FSHD cultures in triplicate): ***P = 0.001 (two tailed Student's t-test, for myoblasts Cohen’ d: 1.84. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.7. ANT1 overexpression affects skeletal muscle development in X. laevis embryos
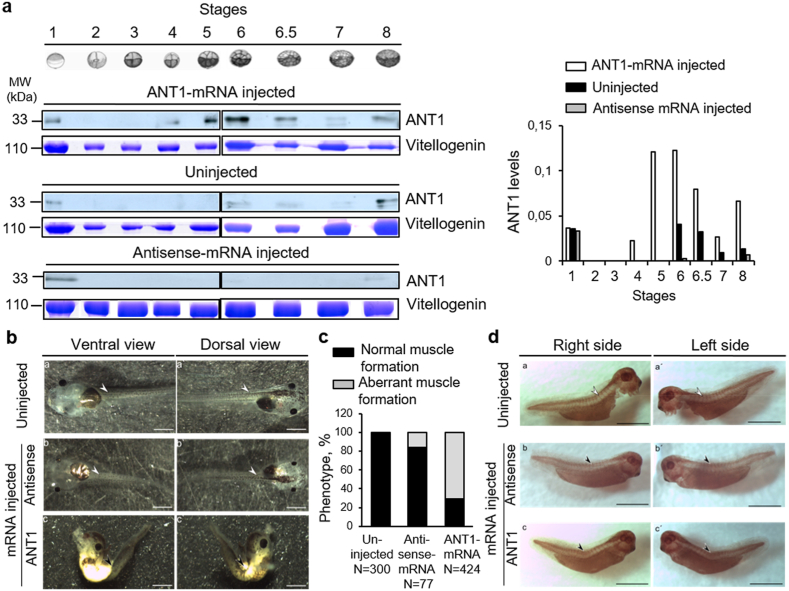
ANT1 is expressed primarily in the heart and skeletal muscles and encodes a carrier of ADP/ATP of the mitochondrial inner membrane [33]. In this study, we overexpressed ANT1 in primary myoblasts and studied the consequence on the muscle cell phenotype (disorganized myotube formation, modification of the energetic metabolism, ROS production, oxidative stress). Therefore, we developed a model of ANT1 overexpression to investigate the impact of ANT1 overexpression on X. laevis organ development after injection of in vitro synthetized human ANT1 mRNA in one blastomere of 2-cell-stage embryos (ANT1+) (Fig. 8a). The sequence of the human ANT1 gene (SLC2544) is 77% identical to that of X. laevis, and the amino acid sequences show 90.65% of identity and 95.6% of similarity (Supplementary Fig. 5). The injected ANT1 mRNA was successfully translated, leading to a significantly higher amount of ANT1 protein compared with controls (uninjected embryos and antisense mRNA-injected) (Fig. 8a lower panels). In controls, we detected ANT1 protein in unfertilized and antisense mRNA-injected eggs (Fig. 8a, stage 1), but not at stages 2–5, and weaker ANT1 expression at stages 6–8, compared with ANT1 mRNA-injected embryos (Fig. 8a). In ANT1+ embryos, we detected ANT1 from stage 4 (Fig. 8a). In 3-day-old ANT1+ embryos (stage 40/41) (Fig. 8b), the body was malformed, and this phenotype was explained by loss of somites and disorganization of the muscle structure along the tail in more than 70% of the ANT1+ embryos (Fig. 8c). Tail muscle development was normal in uninjected embryos and in embryos injected with antisense mRNA (Fig. 8b and c).
Fig. 8.
Xenopus laevis embryos that overexpress human ANT1 show altered phenotype.
(a) Western blots (left panel) and quantification (right panel) of ANT1 expression of experimental (injected with ANT1 mRNA) and control (uninjected or injected with antisense mRNA) embryos at different developmental stages. Total protein extracts from five not fertilized eggs (lane 1) and five X. laevis embryos (at different stages, lanes 2–8) after injection of ANT1 mRNA or antisense mRNA or uninjected (controls) were separated by SDS-PAGE and immunoblotted with an anti-ANT1 antibody. Vitellogenin expression as loading control was detected by Coomassie blue staining at each stage and was quantified by densitometry. (b) Microinjection of ANT1 mRNA leads to a spine developmental defect. Stereoscopic microscope images of control (uninjected or injected with antisense mRNA) and experimental (injected with ANT1 mRNA) 3-day-old embryos. In controls, spine shape was normal. In ANT1 mRNA-injected embryos, spine was malformed due to abnormal skeletal muscle formation. Scale bar: 1 mm. (c) Quantification of the percentage of normal and abnormal muscle formation in control (uninjected or injected with antisense mRNA) embryos and after injection of ANT1 mRNA. (d) Whole mount immunostaining with an anti-desmin antibody of uninjected (a, a’), injected with antisense mRNA (b, b’) or with ANT1 mRNA (c, c’) 3-day-old embryos. Controls (uninjected or injected with antisense mRNA) showed typical staining of normally developed somites: upper panels: right side (a, b), left side (a’, b’). Conversely, in ANT1 mRNA-injected animals, somite-specific staining was not observed: lower panels: right side (c), left side (c’). Scale bar: 1 mm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Whole mount immunostaining with an antibody against desmin, a somite marker [60] (Fig. 8d) showed that in uninjected embryos (a, a’) and antisense-mRNA-injected embryos (b, b’), somites developed normally. Conversely, in ANT1+ embryos (c, c'), desmin staining was absent in the curved tail, indicating the absence of somites.
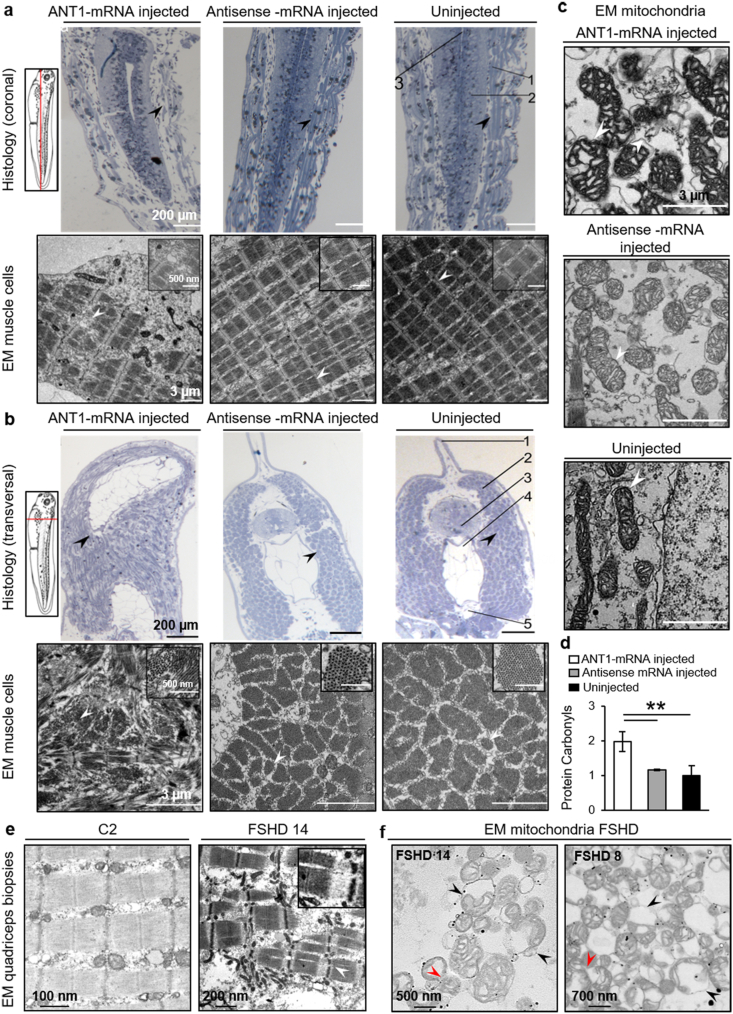
3.8. In ANT1-overexpressing X. laevis embryos, skeletal muscle and mitochondrial ultrastructure are abnormal
Staining of 3-day-old embryo sections with methylene blue showed that somite density was reduced and they were fragmented and disorganized (arrowhead) in the tail of ANT1+ embryos compared with control (uninjected and antisense mRNA-injected) embryos (arrowhead) (Fig. 9a). In transversal histological sections (Fig. 9b, top panels), the tails of ANT1+ embryos were curved compared with the straight tail of uninjected and antisense mRNA-injected embryos (arrowheads). TEM analysis confirmed that muscle tissues were severely disorganized in ANT1+ embryos (Fig. 9a and b, lower panels). Sarcomeres were disorganized and formed a “keel” (arrowhead). Conversely, in control embryos, myofibrils in the tail were well organized and tightly arranged (Fig. 9a, lower panels, arrowhead). In ANT1+ embryos, the morphology of skeletal muscle mitochondria also was abnormal. Compared with control embryos, the mitochondria of ANT1+ embryos showed abnormal cristae structure and partial damage of the outer membrane with blebs and disruption (arrowhead) (Fig. 9c). Moreover, protein carbonylation was higher in ANT1+ embryos than in controls (Fig. 9d). We observed sarcomere disorganization and abnormal mitochondrial morphology also in FSHD quadriceps muscle biopsies (Fig. 9e and f).
Fig. 9.
ANT1+ X. laevis embryos show abnormal skeletal muscle and mitochondria ultrastructure, and increased protein carbonylation.
Tail sections from ANT1 mRNA-injected and control (uninjected and antisens mRNA-injected) X. laevis 3-day-old embryos were stained with methylene blue (upper panels) (a) Coronal sections (upper panels): Scale bar: 200 μm. Lower panels: Electron microscopic (EM) images of tail somites. Scale bar: 3 μm; Scale bar of the inset: 500 nm. 1: Somites; 2: Spinal cord; 3: Notochord. (b) Transversal tail sections (upper panels). Scale bar: 200 μm. Lower panels: EM images of sarcomeres in the tail. Scale bar: 3 μm; Scale bar of the inset: 500 nm. 1: Dorsal fin; 2: Sarcomere; 3: Spinal cord; 4: Notochord; 5: Dorsal aorta. (c) Morphological analysis of the mitochondria in the tail somites from ANT1 mRNA-injected and control (uninjected and antisense mRNA-injected) X. laevis 3-day-old embryos. White arrowheads indicate the mitochondria outer membrane with blebs in ANT1 mRNA-injected X. laevis embryos. Scale bar: 3 μm. (d) Analysis of carbonylated proteins using the Oxyblot Kit and protein extracts from five ANT1 mRNA-injected and control (antisense mRNA-injected and uninjected) X. laevis 3-day-old embryos. Protein concentration was used to normalize protein carbonyl content in X. laevis samples. The bar graphs show the mean ± SD of each condition in duplicate. **P < 0.01 [one-way ANOVA (Holm-Sidak)]. Representative transmission electron microscopy micrographs of quadriceps muscle sarcomeres from healthy control (C2) and patient with FSHD (FSHD14) (e) and (e, f) mitochondria from healthy control (C2) and patients with FSHD (FSHD14, FSHD8). The insets show a higher magnification of FSHD sarcomere. Black arrowheads indicate rounded mitochondria with loss of cristae. Red arrowheads indicate mitochondria during fission where diffused membranes could not be observed between mitochondria. Scale bars: (e) 100 nm in control, and 200 nm in FSHD, (f) 500 nm and 700 nm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
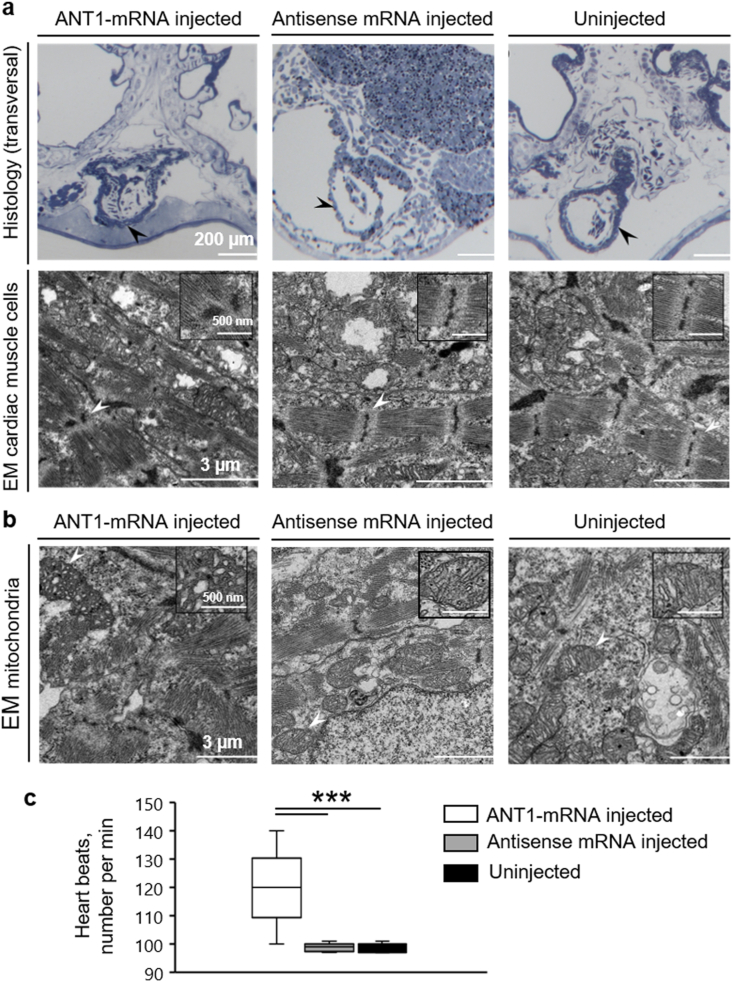
3.9. ANT1 overexpression in X. laevis embryos affects cardiac muscle structure
As ANT1 is mainly expressed in cardiac tissues, we analysed heart development and function in ANT1+ embryos. The heart morphology in semi-thin sections stained with methylene blue was comparable among groups (Fig. 10a). Conversely, TEM analysis showed fewer and disorganized sarcomeres and abnormal z-discs in cardiac muscle cells of ANT1+ embryos compared with controls (Fig. 10a lower panel, arrowhead). Moreover, we observed abnormal structure of the cristae and partial damage of the outer membrane in mitochondria in the heart of ANT1+ embryos compared with controls (Fig. 10b). Last, video analysis of beating hearts in 3-day-old X. laevis tadpoles revealed higher number of heart beats per min (Fig. 10c) and severe arrhythmia (Video in Supplementary Video 1) in ANT1+ compared with control animals (uninjected X. laevis embryos), although the positioning of ventricles and atria was not similar in the two groups.
Fig. 10.
In the heart of ANT1+ X. laevis embryos, sarcomeres are disorganized and mitochondrial ultrastructure is abnormal.
(a) Morphological analysis of cardiac muscle cells. Upper panels: heart sections of ANT1 mRNA-injected and control (uninjected and antisense mRNA-injected) 3-day-old X. laevis embryos were stained with methylene blue. Scale bar: 200 μm. Lower panels: electron microscopy (EM) micrographs showing sarcomeres in the heart of ANT1 mRNA-injected and control (uninjected and antisense mRNA-injected) 3-day-old X. laevis embryos. Scale bar: 3 μm; Scale bar of each inset: 500 nm. (b) Morphological analysis of the mitochondria in the heart of ANT1 mRNA-injected and control (uninjected and antisense mRNA-injected) 3-day-old X. laevis embryos. The insets show a higher magnification of a mitochondrion. Scale bar: 3 μm; Scale bar of each insert: 500 nm. (c) Analysis of the number of heart beats per min in ANT1 mRNA injected and control (antisense mRNA-injected and uninjected) 3-day-old embryos (n = 20/each). ***P < 0.001 [one-way ANOVA (Tukey test)], median and range are presented. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Here, we showed that ANT1 overexpression leads to large myotubes with randomly distributed nuclei, as observed in FSHD myotubes [37,58] where ANT1 protein levels are increased (myoblasts and myotubes) compared with healthy control myotubes. ANT1 overexpression may modify the energetic metabolism and ROS production in time and space, thus affecting muscle development and phenotype. In agreement, ANT1-overexpressing X. laevis embryos showed somite defects and disorganized muscle structure along the tail.
By OCR analysis, we found that the basal respiration and ATP-coupled respiration were increased in ANT1-overexpressing and FSHD myoblasts, indicating a higher oxygen requirement for ATP synthesis at baseline. The spare capacity, which is an estimate of the cell potential bioenergetics capacity to meet increased ATP demands, was similar in all groups. Non-mitochondrial respiration (e.g. ROS production) also was similar in ANT1-overexpressing, FSHD and control myoblasts. Moreover, we did not observe any deficit in ATP coupling efficiency or Δ impairment, indicating that ANT1-overexpressing and FSHD myoblasts rely partially on OXPHOS to produce ATP. However, FSHD myoblasts presented some additional changes not observed in ANT1-overexpressing cells. First, the increased proton leak may be used as a mechanism to regulate mitochondrial ATP production and to maintain the fraction of respiration coupled to ATP synthesis [61]. Second, FSHD myoblasts could not maintain the proton motive force at the highest level, suggesting an impaired capacity to stimulate the respiratory chain to operate at maximum capacity to meet energy demand.
Besides the mitochondrial respiration rate increase, glycolysis was increased in both ANT1-overexpressing and FSHD myoblasts compared with controls in basal conditions and upon oligomycin exposure, suggesting that ATP supply for glycolysis is higher in ANT1-overexpressing and FSHD myoblasts. Unlike in ANT1-overexpressing myoblasts, the baseline OCR/ECAR ratio was in favour of glycolysis in FSHD myoblasts, suggesting a significantly higher reliance of FSHD myoblasts on glycolytic metabolism for energy production. Although glycolytic and oxidative ATP production were significantly increased in ANT1-overexpressing and FSHD myoblasts, the proportion of glycolytic ATP production was increased and mitochondrial ATP production was decreased only in FSHD myoblasts.
The finding that ANT1-overexpressing and FSHD myoblasts may present a higher glycolytic capacity than controls may be reflected by higher glycolytic flux or stimulation. NGA was increased only in ANT1-overexpressing myoblasts, but not in FSHD myoblasts, indicating higher release of protons. Adding glucose triggered an increased glycolytic flux in ANT1-overexpressing and FSHD myoblasts compared with controls, suggesting a higher lactate production in these cells. The subsequent addition of oligomycin further increased ECAR in ANT1-overexpressing and FSHD myoblasts, suggesting a higher capacity of lactate production in these cells. At baseline, intracellular lactate levels were comparable in all groups, but lactate release in the supernatant was higher in ANT1-overexpressing and FSHD myoblasts, consistent with the increased glycolytic flux. Interestingly, incubation of C2C12 cells with lactate leads to a disorganized phenotype (increased myotube diameter and number of nuclei compared with controls [62]), similar to the phenotype observed in ANT1-overexpressing and FSHD myotubes. Our results suggest that extracellular lactate might participate in the activation of anabolic signals, leading to the increased diameter of ANT1-overexpressing and FSHD myotubes [63]. Moreover, the ANT1 overexpression-induced phenotype was reminiscent of the disorganized phenotype observed in DUX4c-overexpressing primary muscle cells [64], suggesting that ANT1 and DUX4c may both contribute to FSHD pathogenesis, either independently or synergistically. Moreover, OCR and ECAR were increased in ANT1-overexpressing and FSHD myotubes compared with controls. Specifically, basal respiration, maximal respiration, ATP production and proton leak were higher in ANT1-overexpressing and FSHD myotubes than in controls. Proton leak is modulated by several mechanisms, including the adenine translocator and by specific mitochondrial inner membrane proteins such as uncoupling proteins (UCPs) [65], a subfamily of the mitochondrial solute carrier family proteins that mediate transport of various metabolites across the inner mitochondrial membrane. UCP3 is predominantly expressed in skeletal muscle [66]. UCP3 expression may facilitate mitochondrial fatty acid uptake suggesting a role of UCP3 in lipid metabolism [67]. Furthermore, UCP3 is also known to play a role in the regulation of glucose homeostasis [68]. Therefore, we cannot exclude that UCP3 could be involved in the increase of proton leak. UCP3 is also a target of μ-Crystallin (Crym), a thyroid hormone binding protein [69,70] which was abnormally up-regulated in deltoid muscle but not in triceps and tibialis anterior muscles [71], where patients with FSHD are preferentially affected [72]. Inactivation of Crym increased UCP3 levels in muscle cells [73] and high levels of Crym are associated with a shift toward genes associated with oxidative type I muscle fibres where UCP3 expression is lower [74,75] than in the more glycolytic type II fibres. Moreover, mitochondria isolated from the skeletal muscle of UCP3 null mice were found to have reduced proton leak and increased ATP/ADP ratio [76] but these results were not confirmed in another study [77]. Nevertheless, further investigations are needed to evaluate the content of UCP3 in FSHD primary cells derived from quadriceps. Additionally, the basal and oligomycin-stimulated glycolytic fluxes were increased in both ANT1-overexpressing and FSHD myotubes, but the glycolytic reserve was larger only in FSHD myotubes, indicating that they did not function at the maximal glycolytic capacity. Similarly, we observed lactate accumulation only in FSHD myotubes. Lactate accumulation is also supported by the increased expression of lactate dehydrogenase in FSHD muscle observed in vitro and in vivo [78,79]. Our results suggest that ANT1 overexpression may modify the bioenergetic profile in FSHD cells and promote ATP production primarily via the existence of a larger glycolytic reserve (rather than mitochondrial capacity), reflecting an excess capacity of glycolytic reactions in these cells. This may lead to conditions of increased bioenergetic stress (ATP consumption) where a substantial amount of ATP production is via glycolysis in addition to mitochondrial phosphorylation [80].
We then found that mitochondrial and cytosolic superoxide production were significantly increased in ANT1-overexpressing myoblasts and myotubes and also in FSHD cells. In ANT1-overexpressing cells, increased superoxide production may be explained by increased oxidative phosphorylation. On the other hand, in FSHD myotubes, lactate accumulation [81] also may contribute to ROS production [82] and may facilitate HIF-1α stabilization through ROS production [83]. HIF-1α might be involved in FSHD [84]. Protein carbonylation, a measure of oxidative damage, was significantly increased in ANT1-overexpressing and FSHD cells. Superoxide anions can be detoxified via endogenous antioxidant enzymes such as manganese superoxide dismutase (MnSOD) which acts on superoxide in the mitochondrial matrix to form hydrogen peroxide [85]. However, MnSOD mRNA levels are downregulated in ANT1 overexpressing myotubes. Interestingly, a down-regulation of MnSOD was observed in ANT1 overexpressing Hela cells [86,87]. In Hela cells, overexpression of ANT1 was shown to specifically induce apoptosis and to potentiate the recruitment of NF-κB by reducing the nuclear NF-κB DNA binding activity and consequently decreasing MnSOD expression [86,87]. Interestingly, MnSOD mRNA levels are also downregulated in FSHD myotubes, corroborating our previous study performed in FSHD skeletal muscle biopsies showing that the only antioxidant downregulated was MnSOD [34]. An increased of ANT1 protein levels and a stimulation of the receptor for advanced glycation end products (RAGE)–NF–κB pathway were observed in FSHD muscle biopsies [16]. However, moderate activation of the nuclear NF-κB DNA binding activity was found [16]. As oxidative cellular damage was observed in FSHD muscle biopsies, the engagement of RAGE, which triggers ROS generation was proposed to be not balanced by an efficient activation of the nuclear NF-κB DNA binding activity [16]. Furthermore, owing to the increased apoptosis of FSHD muscle cells, NF-κB was proposed to be less active in FSHD cells [84]. Further studies are necessary to investigate RAGE–NF–κB pathway in ANT1 overexpressing and FSHD primary muscle cells.
Studies on mitochondrial dynamics identified an intriguing link between the energy demand/supply balance and mitochondrial architecture [59]. In ANT1-overexpressing and FSHD myoblasts, we observed a shift in mitochondrial shape from a tubular network to a fragmented morphology. Mitochondrial fission is physiologically important for mitochondrial quality control; however, excessive fragmentation of the mitochondrial network (due to increased mitochondrial fission and/or reduced mitochondrial fusion) results in accumulation of dysfunctional mitochondria. Our analysis of the mitochondrial dynamics in intact living myoblasts showed that motility was comparable in FSHD and control cells, but that hyper-fission was increased in FSHD myoblasts, leading to a decrease in the fusion/fission ratio. The increased hyper-fission frequency in FSHD myoblasts could be explained by a possible mitophagy defect in the context of deregulated expression of genes regulating fusion-fission dynamics [88]. Additional experiments are needed to confirm this hypothesis. We did not observe any expression change in DRP1 and FIS1 (two genes that regulate mitochondrial fission) and in MFN1/2 and OPA1 (three genes that regulate mitochondrial fusion) in FSHD myoblasts, consistent with a previous study [89]. However, DRP1 expression might be modulated also by post-transcriptional modifications [90] (e.g. S-nitrosylation, ubiquitination), independently of its expression levels, leading to mitochondrial fragmentation [91]. Furthermore, OPA1 and the recently identified mitochondrial contact site and cristae organizing system (MICOS) complex regulate the mitochondrial inner membrane morphology and remodelling [92] in a redox-dependent manner. We did not observe any defect in mitochondrial membrane permeability in FSHD myoblasts consistent with increased mitochondrial respiration, which may maintain the mitochondrial membrane potential [93]. Further experiments are needed to confirm the increased fission levels and expression of genes involved in fusion and fission in ANT1-overexpressing cells.
Mitochondrial morphology may adapt to the respiratory activity [94] and to the redox capability of the mitochondrial reticular network [95,96]. In FSHD myoblasts, we detected a drastic reorganization of the mitochondrial reticular network in the intracellular space, consistent with the observed modulation of mitochondrial bioenergetics, particularly in cell areas with high energy demand (microtubule-organizing centre, perinuclear area, focal adhesion points). The finding that reduction in doughnut-shaped mitochondria formation is associated with oxidative stress in FSHD myoblasts strongly suggests that oxidative stress management in these cells is impaired [97]. TEM analysis showed marked swelling, loss or fragmented cristae, and size variations of mitochondria in ANT1-overexpressing and FSHD myoblasts and also in ANT1+ X. laevis embryos. This suggests an adaptation to the bioenergetic metabolism because mitochondrial respiratory state and ATP production rate may be involved in cristae morphology [94]. Similarly, the presence of inner membrane blebbing in mitochondria of ANT1+ X. laevis embryos and FSHD muscle biopsies and myoblasts also could be related to a response to metabolic changes [98].
On the long term, lactate might affect adaptations in lactate transport via redox and ROS-generating mechanisms. Hogan et al. demonstrated that lactate accumulation decreases muscle tension by a direct effect on the contractile sites [99]. The increased lactate production, accumulation, and release in FSHD myotubes suggest a dysregulated lactate shuttling. Besides lactate, excessive oxidation of myofibrillar contractile proteins, such as actin and myosin heavy chain, also might contribute to sarcomere disorganization and muscle dysfunction [100,101].
In ANT1+ X. laevis embryos, sarcomere organization was altered in skeletal muscle and heart. We obtained similar results in FSHD skeletal muscle biopsies. Additionally, ROS and calcium are mutually interconnected. Oxidative stress during cardiac pathologies, such arrhythmia, is commonly associated with increased sarcoplasmic reticulum calcium leak through the hyperactive ryanodine receptor 2 [102]. ANT1-overexpressing Xenopus laevis embryos present cardiac arrhythmia and in FSHD, arrhythmia seems to be detected more frequently than expected [103,104]. Although cardiac screening and surveillance is not currently recommended in patients without cardiac symptoms [105,106]. Cardiac abnormalities have been proven in several case reports and cohort studies of genetically proven FSHD patients. Both structural and conduction abnormalities can occur in FSHD [107]. Incomplete right branch block (RBBB) is the most common cardiac abnormality detected in 23–33% of patients [105,108]. Supraventricular tachyarrhythmias were reported in approximatively 10% of FSHD patients [103,104]. Ventricular arrhythmias, cardiac conduction system abnormalities such as interventricular conduction delay and atrioventricular block, focal myocardial fibrosis or fatty infiltration with preserved ejection fraction, or a hypertrophic cardiomyopathy phenotype [103,104,[109], [110], [111]] were also observed in some patients with FSHD. Moreover, the results of a FSHD multicenter study indicate the susceptibility to supraventricular arrhythmia as a possible feature of FSHD [111]. Together with the finding that ANT inhibition or anti-oxidant strategy prevented sarcoplasmic reticulum calcium leak in a ROS-dependent pathway [112] may justify to assess calcium handling in FSHD models. Nevertheless, the deleterious effects of ANT1 overexpression on X. laevis whole embryos heart rate are inconsistent with the cardioprotective effects of ANT1 overexpression in heart diseases, such as hypertension-induced hypertrophic cardiomyopathy, diabetic or ischemic heart disease [[113], [114], [115], [116]]. Although the results seem enough plausible, the matters may be more complex. The work of Lynn et al. showed that transient upregulation of PGC-1alpha leading to ANT1 upregulation diminished cardiac ischemia tolerance [117]. Furthermore, siRNA knockdown of ANT1 abolished the deleterious effects of PGC-1α suggesting that the adverse effects of ischemia could be mediated, in part, by PGC-1α induced upregulation of ANT1 [117]. Moreover, in lung ischemia-reperfusion (I/R) injury, the protective administration of cyclosporine-A would depend on the decreased expression levels of ANT1 and VDAC1 [118]. Furthermore, ANT1 overexpression was deleterious in various cell types [86,87,119]. ANT1 up-regulation via the overexpression of regulator of calcineurin 1 isoform 1 was detrimental in human neuroblastoma SH‐SY5Y cells, leading to abnormal mitochondria [119].
Finally, in our study, we used primary myoblasts isolated from quadriceps muscle biopsies. Despite the observations that FSHD has a characteristic descending pattern of weakness, starting in the face, scapular girdle, and arms, followed later by the distal, then the proximal lower extremity muscles, it is considered that lower extremity involvement may be more common than previously appreciated [120]. It is interesting that the pattern of progression of pathologic changes in the vastus lateralis correlates with disease duration [121] and the grade of pathology of quadriceps could be a useful marker of the disease severity in FSHD [122]. In our previous studies [23,34], quadriceps muscle was chosen because a less clinically affected muscle would be more likely to show FSHD-specific changes at the pathological and molecular level than a muscle more clinically affected. Vastus lateralis was chosen to reduce the variability due to sampling from different muscles. In our previous studies [23], quadriceps muscle biopsies from patients with FSHD showed that proteins involved in mitochondrial function and protection from oxidative stress are specifically modified in all FSHD muscles, including clinically unaffected muscles such vastus lateralis and deltoid. Moreover, the highest levels of ANT1 protein were detected in FSHD deltoid and quadriceps muscles clinically non-affected [23]. This result is also consistent with the high level of ant1 observed in muscle biopsies with normal appearance and without sign of necrosis, inflammation or regenerating fibres [16]. Moreover, while MRI analysis showed normal appearance and comparable total volume of the thigh muscles in control and patient with FSHD, reduced quadriceps specific force was observed [36]. Consistent with these observations, it has been proposed that muscle weakness in FSHD is not only caused by a reduced amount of available contractile muscle tissue, but also by reduced of the intrinsic properties of the residual muscle tissue. Reduced muscle quality could be the consequence of several factors including oxidative stress which could affect the intrinsic properties of the muscle by acting on contractile proteins [100,101] and excitation/contraction coupling [102]. Together, our data open the possibility that ANT1 could participate to the decline of muscle quality by contributing to the mitochondrial dysfunction and oxidative stress. However, further investigations are necessary to better understand causes of muscle weakness.
In summary, our study suggests that ANT1 could contribute to mitochondria dysfunction and oxidative stress in FSHD muscle cells by modifying their bioenergetic profile associated with ROS production. Using in vitro (ANT1 overexpression in muscle cells), in vivo (ANT1-overexpressing X. laevis embryos) and ex vivo (muscle cells from patients with FSHD) models, we showed that the ANT1 overexpression-induced phenotype presents striking similarities with FSHD muscle cells and biopsies: skeletal muscle structure abnormalities, enhanced mitochondrial respiration/glycolysis, ROS production, oxidative stress, mitochondrial reticular network organization and ultrastructure alteration in skeletal muscle. However, our data needs to be replicated in more cell cultures. Moreover, ANT1-overexpressing Xenopus laevis embryos also present cardiac muscle structure and mitochondria ultrastructure abnormalities associated with arrhythmia observed more frequently than expected in patients with FSHD. However, a more thorough investigation of the heart of ANT1 X. Laevis is needed to decipher the link between mitochondrial function, structural abnormalities and cardiac arrhythmia.
Authors' contributions
Conceptualization: DLC, MCD; Methodology: DLC, MCD, SA, NC, TS, SP; Investigation: DLC, SA, HK, CF, NC, TS, FP, JPC, SP, NH, DMAM, JB, SF; Writing – original draft: SA, CF, MCD, DLC; All authors reviewed the manuscript and approved the final version. The authors had full access to data and full control of the decision to publish. Funding acquisition: DLC, MCD; Resources: DLC, MCD, JM, JPC, NC, SP; Supervision: DLC, MCD.
Author disclosure statement
The authors declare that they have no competing financial interest.
Funding information
This work was supported by AFM-Telethon (Association Française contre les Myopathies, France) and Amis FSH Europe Association.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank the MÉTAMONTP facility funded by FEDER-Région Occitanie-Plan Cancer. We thank E. Andermarcher for critical reading of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102450.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Pandya S., King W.M., Tawil R. Facioscapulohumeral dystrophy. Phys. Ther. 2008;88:105–113. doi: 10.2522/ptj.20070104. [DOI] [PubMed] [Google Scholar]
- 2.Schatzl T., Kaiser L., Deigner H.P. Facioscapulohumeral muscular dystrophy: genetics, gene activation and downstream signalling with regard to recent therapeutic approaches: an update. Orphanet J. Rare Dis. 2021;16:129. doi: 10.1186/s13023-021-01760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tawil R., Van Der Maarel S.M. Facioscapulohumeral muscular dystrophy. Muscle Nerve. 2006;34:1–15. doi: 10.1002/mus.20522. [DOI] [PubMed] [Google Scholar]
- 4.Richards M., Coppee F., Thomas N., Belayew A., Upadhyaya M. Facioscapulohumeral muscular dystrophy (FSHD): an enigma unravelled? Hum. Genet. 2012;131:325–340. doi: 10.1007/s00439-011-1100-z. [DOI] [PubMed] [Google Scholar]
- 5.van Deutekom J.C., Wijmenga C., van Tienhoven E.A., Gruter A.M., Hewitt J.E., Padberg G.W., van Ommen G.J., Hofker M.H., Frants R.R. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum. Mol. Genet. 1993;2:2037–2042. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- 6.Wijmenga C., Winokur S.T., Padberg G.W., Skraastad M.I., Altherr M.R., Wasmuth J.J., Murray J.C., Hofker M.H., Frants R.R. The human skeletal muscle adenine nucleotide translocator gene maps to chromosome 4q35 in the region of the facioscapulohumeral muscular dystrophy locus. Hum. Genet. 1993;92:198–203. doi: 10.1007/BF00219692. [DOI] [PubMed] [Google Scholar]
- 7.Lemmers R.J., van der Vliet P.J., Klooster R., Sacconi S., Camano P., Dauwerse J.G., Snider L., Straasheijm K.R., van Ommen G.J., Padberg G.W., Miller D.G., Tapscott S.J., Tawil R., Frants R.R., van der Maarel S.M. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329:1650–1653. doi: 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van der Maarel S.M., Frants R.R. The D4Z4 repeat-mediated pathogenesis of facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2005;76:375–386. doi: 10.1086/428361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixit M., Ansseau E., Tassin A., Winokur S., Shi R., Qian H., Sauvage S., Matteotti C., van Acker A.M., Leo O., Figlewicz D., Barro M., Laoudj-Chenivesse D., Belayew A., Coppee F., Chen Y.W. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc. Natl. Acad. Sci. U. S. A. 2007;104:18157–18162. doi: 10.1073/pnas.0708659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geng L.N., Yao Z., Snider L., Fong A.P., Cech J.N., Young J.M., van der Maarel S.M., Ruzzo W.L., Gentleman R.C., Tawil R., Tapscott S.J. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell. 2012;22:38–51. doi: 10.1016/j.devcel.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma V., Harafuji N., Belayew A., Chen Y.W. DUX4 differentially regulates transcriptomes of human rhabdomyosarcoma and mouse C2C12 cells. PLoS One. 2013;8 doi: 10.1371/journal.pone.0064691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanderplanck C., Ansseau E., Charron S., Stricwant N., Tassin A., Laoudj-Chenivesse D., Wilton S.D., Coppee F., Belayew A. The FSHD atrophic myotube phenotype is caused by DUX4 expression. PLoS One. 2011;6 doi: 10.1371/journal.pone.0026820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sasaki-Honda M., Jonouchi T., Arai M., Hotta A., Mitsuhashi S., Nishino I., Matsuda R., Sakurai H. A patient-derived iPSC model revealed oxidative stress increases facioscapulohumeral muscular dystrophy-causative DUX4. Hum. Mol. Genet. 2018;27:4024–4035. doi: 10.1093/hmg/ddy293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bosnakovski D., Xu Z., Gang E.J., Galindo C.L., Liu M., Simsek T., Garner H.R., Agha-Mohammadi S., Tassin A., Coppee F., Belayew A., Perlingeiro R.R., Kyba M. An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J. 2008;27:2766–2779. doi: 10.1038/emboj.2008.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsen M., Rost S., El Hajj N., Ferbert A., Deschauer M., Walter M.C., Schoser B., Tacik P., Kress W., Muller C.R. Diagnostic approach for FSHD revisited: SMCHD1 mutations cause FSHD2 and act as modifiers of disease severity in FSHD1. Eur. J. Hum. Genet. 2015;23:808–816. doi: 10.1038/ejhg.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macaione V., Aguennouz M., Rodolico C., Mazzeo A., Patti A., Cannistraci E., Colantone L., Di Giorgio R.M., De Luca G., Vita G. RAGE-NF-kappaB pathway activation in response to oxidative stress in facioscapulohumeral muscular dystrophy. Acta Neurol. Scand. 2007;115:115–121. doi: 10.1111/j.1600-0404.2006.00724.x. [DOI] [PubMed] [Google Scholar]
- 17.Rijkers T., Deidda G., van Koningsbruggen S., van Geel M., Lemmers R.J., van Deutekom J.C., Figlewicz D., Hewitt J.E., Padberg G.W., Frants R.R., van der Maarel S.M. FRG2, an FSHD candidate gene, is transcriptionally upregulated in differentiating primary myoblast cultures of FSHD patients. J. Med. Genet. 2004;41:826–836. doi: 10.1136/jmg.2004.019364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabellini D., Green M.R., Tupler R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110:339–348. doi: 10.1016/s0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- 19.Jiang G., Yang F., van Overveld P.G., Vedanarayanan V., van der Maarel S., Ehrlich M. Testing the position-effect variegation hypothesis for facioscapulohumeral muscular dystrophy by analysis of histone modification and gene expression in subtelomeric 4q. Hum. Mol. Genet. 2003;12:2909–2921. doi: 10.1093/hmg/ddg323. [DOI] [PubMed] [Google Scholar]
- 20.Klooster R., Straasheijm K., Shah B., Sowden J., Frants R., Thornton C., Tawil R., van der Maarel S. Comprehensive expression analysis of FSHD candidate genes at the mRNA and protein level. Eur. J. Hum. Genet. 2009;17:1615–1624. doi: 10.1038/ejhg.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsumagari K., Chang S.C., Lacey M., Baribault C., Chittur S.V., Sowden J., Tawil R., Crawford G.E., Ehrlich M. Gene expression during normal and FSHD myogenesis. BMC Med. Genom. 2011;4:67. doi: 10.1186/1755-8794-4-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Maarel S.M., Frants R.R., Padberg G.W. Facioscapulohumeral muscular dystrophy. Biochim. Biophys. Acta. 2007;1772:186–194. doi: 10.1016/j.bbadis.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Laoudj-Chenivesse D., Carnac G., Bisbal C., Hugon G., Bouillot S., Desnuelle C., Vassetzky Y., Fernandez A. Increased levels of adenine nucleotide translocator 1 protein and response to oxidative stress are early events in facioscapulohumeral muscular dystrophy muscle. J. Mol. Med. (Berl.) 2005;83:216–224. doi: 10.1007/s00109-004-0583-7. [DOI] [PubMed] [Google Scholar]
- 24.Kim E., Rich J., Karoutas A., Tarlykov P., Cochet E., Malysheva D., Mamchaoui K., Ogryzko V., Pirozhkova I. ZNF555 protein binds to transcriptional activator site of 4qA allele and ANT1: potential implication in Facioscapulohumeral dystrophy. Nucleic Acids Res. 2015;43:8227–8242. doi: 10.1093/nar/gkv721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrov A., Pirozhkova I., Carnac G., Laoudj D., Lipinski M., Vassetzky Y.S. Chromatin loop domain organization within the 4q35 locus in facioscapulohumeral dystrophy patients versus normal human myoblasts. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6982–6987. doi: 10.1073/pnas.0511235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu X., Tsumagari K., Sowden J., Tawil R., Boyle A.P., Song L., Furey T.S., Crawford G.E., Ehrlich M. DNaseI hypersensitivity at gene-poor, FSH dystrophy-linked 4q35.2. Nucleic Acids Res. 2009;37:7381–7393. doi: 10.1093/nar/gkp833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pirozhkova I., Petrov A., Dmitriev P., Laoudj D., Lipinski M., Vassetzky Y. A functional role for 4qA/B in the structural rearrangement of the 4q35 region and in the regulation of FRG1 and ANT1 in facioscapulohumeral dystrophy. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulaeva O.I., Nizovtseva E.V., Polikanov Y.S., Ulianov S.V., Studitsky V.M. Distant activation of transcription: mechanisms of enhancer action. Mol. Cell Biol. 2012;32:4892–4897. doi: 10.1128/MCB.01127-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petrascheck M., Escher D., Mahmoudi T., Verrijzer C.P., Schaffner W., Barberis A. DNA looping induced by a transcriptional enhancer in vivo. Nucleic Acids Res. 2005;33:3743–3750. doi: 10.1093/nar/gki689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gabellini D., D'Antona G., Moggio M., Prelle A., Zecca C., Adami R., Angeletti B., Ciscato P., Pellegrino M.A., Bottinelli R., Green M.R., Tupler R. Facioscapulohumeral muscular dystrophy in mice overexpressing FRG1. Nature. 2006;439:973–977. doi: 10.1038/nature04422. [DOI] [PubMed] [Google Scholar]
- 31.Brand M.D., Pakay J.L., Ocloo A., Kokoszka J., Wallace D.C., Brookes P.S., Cornwall E.J. The basal proton conductance of mitochondria depends on adenine nucleotide translocase content. Biochem. J. 2005;392:353–362. doi: 10.1042/BJ20050890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X., Middleton F.A., Tawil R., Chen X.J. Cytosolic adaptation to mitochondria-induced proteostatic stress causes progressive wasting. iSience. 2022;25 doi: 10.1016/j.isci.2021.103715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kokoszka J.E., Waymire K.G., Levy S.E., Sligh J.E., Cai J., Jones D.P., MacGregor G.R., Wallace D.C. The ADP/ATP translocator is not essential for the mitochondrial permeability transition pore. Nature. 2004;427:461–465. doi: 10.1038/nature02229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turki A., Hayot M., Carnac G., Pillard F., Passerieux E., Bommart S., Raynaud de Mauverger E., Hugon G., Pincemail J., Pietri S., Lambert K., Belayew A., Vassetzky Y., Juntas Morales R., Mercier J., Laoudj-Chenivesse D. Functional muscle impairment in facioscapulohumeral muscular dystrophy is correlated with oxidative stress and mitochondrial dysfunction. Free Radic. Biol. Med. 2012;53:1068–1079. doi: 10.1016/j.freeradbiomed.2012.06.041. [DOI] [PubMed] [Google Scholar]
- 35.Passerieux E., Hayot M., Jaussent A., Carnac G., Gouzi F., Pillard F., Picot M.C., Bocker K., Hugon G., Pincemail J., Defraigne J.O., Verrips T., Mercier J., Laoudj-Chenivesse D. Effects of vitamin C, vitamin E, zinc gluconate, and selenomethionine supplementation on muscle function and oxidative stress biomarkers in patients with facioscapulohumeral dystrophy: a double-blind randomized controlled clinical trial. Free Radic. Biol. Med. 2015;81:158–169. doi: 10.1016/j.freeradbiomed.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 36.Wilson V.D., Thomas C., Passerieux E., Hugon G., Pillard F., Andrade A.G., Bommart S., Picot M.C., Pincemail J., Mercier J., Arbogast S., Laoudj-Chenivesse D. Impaired oxygen demand during exercise is related to oxidative stress and muscle function in facioscapulohumeral muscular dystrophy. JCSM Rapid Commun. 2018;1:1–13. [Google Scholar]
- 37.Barro M., Carnac G., Flavier S., Mercier J., Vassetzky Y., Laoudj-Chenivesse D. Myoblasts from affected and non-affected FSHD muscles exhibit morphological differentiation defects. J. Cell Mol. Med. 2010;14:275–289. doi: 10.1111/j.1582-4934.2008.00368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winokur S.T., Barrett K., Martin J.H., Forrester J.R., Simon M., Tawil R., Chung S.A., Masny P.S., Figlewicz D.A. Facioscapulohumeral muscular dystrophy (FSHD) myoblasts demonstrate increased susceptibility to oxidative stress. Neuromuscul. Disord. 2003;13:322–333. doi: 10.1016/s0960-8966(02)00284-5. [DOI] [PubMed] [Google Scholar]
- 39.Neuhuber B., Huang D.I., Daniels M.P., Torgan C.E. High efficiency transfection of primary skeletal muscle cells with lipid-based reagents. Muscle Nerve. 2002;26:136–140. doi: 10.1002/mus.10171. [DOI] [PubMed] [Google Scholar]
- 40.Wolf D.P., Hedrick J.L. A molecular approach to fertilization. 3. Development of a bioassay for sperm capacitation. Dev. Biol. 1971;25:360–376. doi: 10.1016/0012-1606(71)90037-6. [DOI] [PubMed] [Google Scholar]
- 41.Nieuwkoop P.D., Faber J., editors. Normal Table of Xenopus laevis (Daudin) North-Holland Publishing Company; Amsterdam: 1967. [Google Scholar]
- 42.Newport J., Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- 43.Benavente R., Krohne G., Franke W.W. Cell type-specific expression of nuclear lamina proteins during development of Xenopus laevis. Cell. 1985;41:177–190. doi: 10.1016/0092-8674(85)90072-8. [DOI] [PubMed] [Google Scholar]
- 44.Klymkowsky M.W., Hanken J. Whole-mount staining of Xenopus and other vertebrates. Methods Cell Biol. 1991;36:419–441. doi: 10.1016/s0091-679x(08)60290-3. [DOI] [PubMed] [Google Scholar]
- 45.Reil M., Dabauvalle M.C. Essential roles of LEM-domain protein MAN1 during organogenesis in Xenopus laevis and overlapping functions of emerin. Eur. J. Cell Biol. 2013;92:280–294. doi: 10.1016/j.ejcb.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 46.Divakaruni A.S., Paradyse A., Ferrick D.A., Murphy A.N., Jastroch M. Analysis and interpretation of microplate-based oxygen consumption and pH data. Methods Enzymol. 2014;547:309–354. doi: 10.1016/B978-0-12-801415-8.00016-3. [DOI] [PubMed] [Google Scholar]
- 47.Ferrick D.A., Neilson A., Beeson C. Advances in measuring cellular bioenergetics using extracellular flux. Drug Discov. Today. 2008;13:268–274. doi: 10.1016/j.drudis.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 48.Kang J.G., Wang P.Y., Hwang P.M. Cell-based measurements of mitochondrial function in human subjects. Methods Enzymol. 2019;542:209–221. doi: 10.1016/B978-0-12-416618-9.00011-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mookerjee S.A., Gerencser A.A., Nicholls D.G., Brand M.D. Quantifying intracellular rates of glycolytic and oxidative ATP production and consumption using extracellular flux measurements. J. Biol. Chem. 2017;292:7189–7207. doi: 10.1074/jbc.M116.774471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Louie M.C., Ton J., Brady M.L., Le D.T., Mar J.N., Lerner C.A., Gerencser A.A., Mookerjee S.A. Total cellular ATP production changes with primary substrate in MCF7 breast cancer cells. Front. Oncol. 2020;10(1703) doi: 10.3389/fonc.2020.01703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benavente R., Krohne G. Involvement of nuclear lamins in postmitotic reorganization of chromatin as demonstrated by microinjection of lamin antibodies. J. Cell Biol. 1986;103:1847–1854. doi: 10.1083/jcb.103.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schoft V.K., Beauvais A.J., Lang C., Gajewski A., Prufert K., Winkler C., Akimenko M.A., Paulin-Levasseur M., Krohne G. The lamina-associated polypeptide 2 (LAP2) isoforms beta, gamma and omega of zebrafish: developmental expression and behavior during the cell cycle. J. Cell Sci. 2003;116:2505–2517. doi: 10.1242/jcs.00450. [DOI] [PubMed] [Google Scholar]
- 53.Kogot-Levin A., Saada A., Leibowitz G., Soiferman D., Douiev L., Raz I., Weksler-Zangen S. Upregulation of mitochondrial content in cytochrome c oxidase deficient fibroblasts. PLoS One. 2016;11 doi: 10.1371/journal.pone.0165417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Larsen S., Nielsen J., Hansen C.N., Nielsen L.B., Wibrand F., Stride N., Schroder H.D., Boushel R., Helge J.W., Dela F., Hey-Mogensen M. Biomarkers of mitochondrial content in skeletal muscle of healthy young human subjects. J. Physiol. 2012;590:3349–3360. doi: 10.1113/jphysiol.2012.230185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Compagnone N.A. 2010. Method to Predict Toxicity Using the Analysis of Dynamic Organelle Behaviour. [Google Scholar]
- 56.Culcasi M., Benameur L., Mercier A., Lucchesi C., Rahmouni H., Asteian A., Casano G., Botta A., Kovacic H., Pietri S. EPR spin trapping evaluation of ROS production in human fibroblasts exposed to cerium oxide nanoparticles: evidence for NADPH oxidase and mitochondrial stimulation. Chem. Biol. Interact. 2012;199:161–176. doi: 10.1016/j.cbi.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 57.Koechlin C., Maltais F., Saey D., Michaud A., LeBlanc P., Hayot M., Prefaut C. Hypoxaemia enhances peripheral muscle oxidative stress in chronic obstructive pulmonary disease. Thorax. 2005;60:834–841. doi: 10.1136/thx.2004.037531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tassin A., Leroy B., Laoudj-Chenivesse D., Wauters A., Vanderplanck C., Le Bihan M.C., Coppee F., Wattiez R., Belayew A. FSHD myotubes with different phenotypes exhibit distinct proteomes. PLoS One. 2012;7 doi: 10.1371/journal.pone.0051865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benard G., Bellance N., James D., Parrone P., Fernandez H., Letellier T., Rossignol R. Mitochondrial bioenergetics and structural network organization. J. Cell Sci. 2007;120:838–848. doi: 10.1242/jcs.03381. [DOI] [PubMed] [Google Scholar]
- 60.Debus E., Weber K., Osborn M. Monoclonal antibodies to desmin, the muscle-specific intermediate filament protein. EMBO J. 1983;2:2305–2312. doi: 10.1002/j.1460-2075.1983.tb01738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jastroch M., Divakaruni A.S., Mookerjee S., Treberg J.R., Brand M.D. Mitochondrial proton and electron leaks. Essays Biochem. 2010;47:53–67. doi: 10.1042/bse0470053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohno Y., Oyama A., Kaneko H., Egawa T., Yokoyama S., Sugiura T., Ohira Y., Yoshioka T., Goto K. Lactate increases myotube diameter via activation of MEK/ERK pathway in C2C12 cells. Acta Physiol (Oxf) 2018;223 doi: 10.1111/apha.13042. [DOI] [PubMed] [Google Scholar]
- 63.Ohno Y., Ando K., Ito T., Suda Y., Matsui Y., Oyama A., Kaneko H., Yokoyama S., Egawa T., Goto K. Lactate stimulates a potential for hypertrophy and regeneration of mouse skeletal muscle. Nutrients. 2019;11 doi: 10.3390/nu11040869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vanderplanck C., Tassin A., Ansseau E., Charron S., Wauters A., Lancelot C., Vancutsem K., Laoudj-Chenivesse D., Belayew A., Coppee F. Overexpression of the double homeodomain protein DUX4c interferes with myofibrillogenesis and induces clustering of myonuclei. Skeletal Muscle. 2018;8(2) doi: 10.1186/s13395-017-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parker N., Vidal-Puig A., Brand M.D. Stimulation of mitochondrial proton conductance by hydroxynonenal requires a high membrane potential. Biosci. Rep. 2008;28:83–88. doi: 10.1042/BSR20080002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Faraut B., Giannesini B., Matarazzo V., Marqueste T., Dalmasso C., Rougon G., Cozzone P.J., Bendahan D. Downregulation of uncoupling protein-3 in vivo is linked to changes in muscle mitochondrial energy metabolism as a result of capsiate administration. Am. J. Physiol. Endocrinol. Metab. 2007;292:E1474–E1482. doi: 10.1152/ajpendo.00292.2006. [DOI] [PubMed] [Google Scholar]
- 67.Hesselink M.K., Keizer H.A., Borghouts L.B., Schaart G., Kornips C.F., Slieker L.J., Sloop K.W., Saris W.H., Schrauwen P. Protein expression of UCP3 differs between human type 1, type 2a, and type 2b fibers. Faseb. J. 2001;15:1071–1073. [PubMed] [Google Scholar]
- 68.Schrauwen P., Hesselink M.K., Blaak E.E., Borghouts L.B., Schaart G., Saris W.H., Keizer H.A. Uncoupling protein 3 content is decreased in skeletal muscle of patients with type 2 diabetes. Diabetes. 2001;50:2870–2873. doi: 10.2337/diabetes.50.12.2870. [DOI] [PubMed] [Google Scholar]
- 69.Gong D.W., He Y., Karas M., Reitman M. Uncoupling protein-3 is a mediator of thermogenesis regulated by thyroid hormone, beta3-adrenergic agonists, and leptin. J. Biol. Chem. 1997;272:24129–24132. doi: 10.1074/jbc.272.39.24129. [DOI] [PubMed] [Google Scholar]
- 70.Solanes G., Pedraza N., Calvo V., Vidal-Puig A., Lowell B.B., Villarroya F. Thyroid hormones directly activate the expression of the human and mouse uncoupling protein-3 genes through a thyroid response element in the proximal promoter region. Biochem. J. 2005;386:505–513. doi: 10.1042/BJ20041073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reed P.W., Corse A.M., Porter N.C., Flanigan K.M., Bloch R.J. Abnormal expression of mu-crystallin in facioscapulohumeral muscular dystrophy. Exp. Neurol. 2007;205:583–586. doi: 10.1016/j.expneurol.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 72.Tawil R., van der Maarel S.M., Tapscott S.J. Facioscapulohumeral dystrophy: the path to consensus on pathophysiology. Skeletal Muscle. 2014;4(12) doi: 10.1186/2044-5040-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seko D., Ogawa S., Li T.S., Taimura A., Ono Y. mu-Crystallin controls muscle function through thyroid hormone action. Faseb. J. 2016;30:1733–1740. doi: 10.1096/fj.15-280933. [DOI] [PubMed] [Google Scholar]
- 74.Meex R.C., Schrauwen-Hinderling V.B., Moonen-Kornips E., Schaart G., Mensink M., Phielix E., van de Weijer T., Sels J.P., Schrauwen P., Hesselink M.K. Restoration of muscle mitochondrial function and metabolic flexibility in type 2 diabetes by exercise training is paralleled by increased myocellular fat storage and improved insulin sensitivity. Diabetes. 2010;59:572–579. doi: 10.2337/db09-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Phielix E., Meex R., Moonen-Kornips E., Hesselink M.K., Schrauwen P. Exercise training increases mitochondrial content and ex vivo mitochondrial function similarly in patients with type 2 diabetes and in control individuals. Diabetologia. 2010;53:1714–1721. doi: 10.1007/s00125-010-1764-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cline G.W., Vidal-Puig A.J., Dufour S., Cadman K.S., Lowell B.B., Shulman G.I. In vivo effects of uncoupling protein-3 gene disruption on mitochondrial energy metabolism. J. Biol. Chem. 2001;276:20240–20244. doi: 10.1074/jbc.M102540200. [DOI] [PubMed] [Google Scholar]
- 77.Cadenas S., Echtay K.S., Harper J.A., Jekabsons M.B., Buckingham J.A., Grau E., Abuin A., Chapman H., Clapham J.C., Brand M.D. The basal proton conductance of skeletal muscle mitochondria from transgenic mice overexpressing or lacking uncoupling protein-3. J. Biol. Chem. 2002;277:2773–2778. doi: 10.1074/jbc.M109736200. [DOI] [PubMed] [Google Scholar]
- 78.Kowaljow V., Marcowycz A., Ansseau E., Conde C.B., Sauvage S., Matteotti C., Arias C., Corona E.D., Nunez N.G., Leo O., Wattiez R., Figlewicz D., Laoudj-Chenivesse D., Belayew A., Coppee F., Rosa A.L. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul. Disord. 2007;17:611–623. doi: 10.1016/j.nmd.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 79.Winokur S.T., Chen Y.W., Masny P.S., Martin J.H., Ehmsen J.T., Tapscott S.J., van der Maarel S.M., Hayashi Y., Flanigan K.M. Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum. Mol. Genet. 2003;12:2895–2907. doi: 10.1093/hmg/ddg327. [DOI] [PubMed] [Google Scholar]
- 80.Hill B.G., Benavides G.A., Lancaster J.R., Jr., Ballinger S., Dell'Italia L., Jianhua Z., Darley-Usmar V.M. Integration of cellular bioenergetics with mitochondrial quality control and autophagy. Biol. Chem. 2012;393:1485–1512. doi: 10.1515/hsz-2012-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hashimoto T., Hussien R., Oommen S., Gohil K., Brooks G.A. Lactate sensitive transcription factor network in L6 cells: activation of MCT1 and mitochondrial biogenesis. Faseb. J. 2007;21:2602–2612. doi: 10.1096/fj.07-8174com. [DOI] [PubMed] [Google Scholar]
- 82.Ali M.A., Konishi T. Enhancement of hydroxyl radical generation in the Fenton reaction by alpha-hydroxy acid. Biochem. Mol. Biol. Int. 1998;46:137–145. doi: 10.1080/15216549800203642. [DOI] [PubMed] [Google Scholar]
- 83.Kozlov A.M., Lone A., Betts D.H., Cumming R.C. Lactate preconditioning promotes a HIF-1alpha-mediated metabolic shift from OXPHOS to glycolysis in normal human diploid fibroblasts. Sci. Rep. 2020;10(8388) doi: 10.1038/s41598-020-65193-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Banerji C.R., Knopp P., Moyle L.A., Severini S., Orrell R.W., Teschendorff A.E., Zammit P.S. beta-Catenin is central to DUX4-driven network rewiring in facioscapulohumeral muscular dystrophy. J. R. Soc. Interface. 2015;12 doi: 10.1098/rsif.2014.0797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boden M.J., Brandon A.E., Tid-Ang J.D., Preston E., Wilks D., Stuart E., Cleasby M.E., Turner N., Cooney G.J., Kraegen E.W. Overexpression of manganese superoxide dismutase ameliorates high-fat diet-induced insulin resistance in rat skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2012;303 doi: 10.1152/ajpendo.00577.2011. E798-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zamora M., Merono C., Vinas O., Mampel T. Recruitment of NF-kappaB into mitochondria is involved in adenine nucleotide translocase 1 (ANT1)-induced apoptosis. J. Biol. Chem. 2004;279:38415–38423. doi: 10.1074/jbc.M404928200. [DOI] [PubMed] [Google Scholar]
- 87.Bauer M.K., Schubert A., Rocks O., Grimm S. Adenine nucleotide translocase-1, a component of the permeability transition pore, can dominantly induce apoptosis. J. Cell Biol. 1999;147:1493–1502. doi: 10.1083/jcb.147.7.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Van der Bliek A.M., Sedensky M.M., Morgan P.G. Cell biology of the mitochondrion. Genetics. 2017;207:843–871. doi: 10.1534/genetics.117.300262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cheli S., Francois S., Bodega B., Ferrari F., Tenedini E., Roncaglia E., Ferrari S., Ginelli E., Meneveri R. Expression profiling of FSHD-1 and FSHD-2 cells during myogenic differentiation evidences common and distinctive gene dysregulation patterns. PLoS One. 2011;6 doi: 10.1371/journal.pone.0020966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cho D.H., Nakamura T., Fang J., Cieplak P., Godzik A., Gu Z., Lipton S.A. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Willems P.H., Rossignol R., Dieteren C.E., Murphy M.P., Koopman W.J. Redox homeostasis and mitochondrial dynamics. Cell Metabol. 2015;22:207–218. doi: 10.1016/j.cmet.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 92.Stephan T., Bruser C., Deckers M., Steyer A.M., Balzarotti F., Barbot M., Behr T.S., Heim G., Hubner W., Ilgen P., Lange F., Pacheu-Grau D., Pape J.K., Stoldt S., Huser T., Hell S.W., Mobius W., Rehling P., Riedel D., Jakobs S. MICOS assembly controls mitochondrial inner membrane remodeling and crista junction redistribution to mediate cristae formation. EMBO J. 2020;39 doi: 10.15252/embj.2019104105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Garcia O., Almeida A., Massieu L., Bolanos J.P. Increased mitochondrial respiration maintains the mitochondrial membrane potential and promotes survival of cerebellar neurons in an endogenous model of glutamate receptor activation. J. Neurochem. 2005;92:183–190. doi: 10.1111/j.1471-4159.2004.02851.x. [DOI] [PubMed] [Google Scholar]
- 94.Hackenbrock C.R. Ultrastructural bases for metabolically linked mechanical activity in mitochondria. I. Reversible ultrastructural changes with change in metabolic steady state in isolated liver mitochondria. J. Cell Biol. 1966;30:269–297. doi: 10.1083/jcb.30.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Iqbal S., Hood D.A. Oxidative stress-induced mitochondrial fragmentation and movement in skeletal muscle myoblasts. Am. J. Physiol. Cell Physiol. 2014;306:C1176–C1183. doi: 10.1152/ajpcell.00017.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Valera-Alberni M., Canto C. Mitochondrial stress management: a dynamic journey. Cell Stress. 2018;2:253–274. doi: 10.15698/cst2018.10.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ahmad T., Aggarwal K., Pattnaik B., Mukherjee S., Sethi T., Tiwari B.K., Kumar M., Micheal A., Mabalirajan U., Ghosh B., Sinha Roy S., Agrawal A. Computational classification of mitochondrial shapes reflects stress and redox state. Cell Death Dis. 2012;4 doi: 10.1038/cddis.2012.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mannella C.A. Consequences of folding the mitochondrial inner membrane. Front. Physiol. 2020;11:536. doi: 10.3389/fphys.2020.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hogan M.C., Gladden L.B., Kurdak S.S., Poole D.C. Increased [lactate] in working dog muscle reduces tension development independent of pH. Med. Sci. Sports Exerc. 1995;27:371–377. [PubMed] [Google Scholar]
- 100.Brooke M.H., Kaiser K.K. Three “myosin adenosine triphosphatase” systems: the nature of their pH lability and sulfhydryl dependence. J. Histochem. Cytochem. 1970;18:670–672. doi: 10.1177/18.9.670. [DOI] [PubMed] [Google Scholar]
- 101.Sweeney H.L., Bowman B.F., Stull J.T. Myosin light chain phosphorylation in vertebrate striated muscle: regulation and function. Am. J. Physiol. 1993;264:C1085–C1095. doi: 10.1152/ajpcell.1993.264.5.C1085. [DOI] [PubMed] [Google Scholar]
- 102.Dobrev D., Wehrens X.H. Role of RyR2 phosphorylation in heart failure and arrhythmias: controversies around ryanodine receptor phosphorylation in cardiac disease. Circ. Res. 2014;114:1311–1319. doi: 10.1161/CIRCRESAHA.114.300568. discussion 1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Laforet P., de Toma C., Eymard B., Becane H.M., Jeanpierre M., Fardeau M., Duboc D. Cardiac involvement in genetically confirmed facioscapulohumeral muscular dystrophy. Neurology. 1998;51:1454–1456. doi: 10.1212/wnl.51.5.1454. [DOI] [PubMed] [Google Scholar]
- 104.Trevisan C.P., Pastorello E., Armani M., Angelini C., Nante G., Tomelleri G., Tonin P., Mongini T., Palmucci L., Galluzzi G., Tupler R.G., Barchitta A. Facioscapulohumeral muscular dystrophy and occurrence of heart arrhythmia. Eur. Neurol. 2006;56:1–5. doi: 10.1159/000094248. [DOI] [PubMed] [Google Scholar]
- 105.van Dijk G.P., van der Kooi E., Behin A., Smeets J., Timmermans J., van der Maarel S., Padberg G., Voermans N., van Engelen B. High prevalence of incomplete right bundle branch block in facioscapulohumeral muscular dystrophy without cardiac symptoms. Funct. Neurol. 2014;29:159–165. [PMC free article] [PubMed] [Google Scholar]
- 106.Tawil R., Kissel J.T., Heatwole C., Pandya S., Gronseth G., Benatar M. Evidence-based guideline summary: evaluation, diagnosis, and management of facioscapulohumeral muscular dystrophy: report of the guideline development, dissemination, and implementation subcommittee of the American academy of neurology and the practice issues review panel of the American association of neuromuscular & electrodiagnostic medicine. Neurology. 2015;85:357–364. doi: 10.1212/WNL.0000000000001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ducharme-Smith A., Nicolau S., Chahal C.A.A., Ducharme-Smith K., Rehman S., Jaliparthy K., Khan N., Scott C.G., St Louis E.K., Liewluck T., Somers V.K., Lin G., Brady P.A., Milone M. Cardiac involvement in facioscapulohumeral muscular dystrophy (FSHD) Front. Neurol. 2021;12 doi: 10.3389/fneur.2021.668180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Labombarda F., Maurice M., Simon J.P., Legallois D., Guyant-Marechal L., Bedat-Millet A.L., Merle P., Saloux E., Chapon F., Milliez P. Cardiac abnormalities in type 1 facioscapulohumeral muscular dystrophy. J. Clin. Neuromuscul. Dis. 2017;18:199–206. doi: 10.1097/CND.0000000000000144. [DOI] [PubMed] [Google Scholar]
- 109.Galetta F., Franzoni F., Sposito R., Plantinga Y., Femia F.R., Galluzzi F., Rocchi A., Santoro G., Siciliano G. Subclinical cardiac involvement in patients with facioscapulohumeral muscular dystrophy. Neuromuscul. Disord. 2005;15:403–408. doi: 10.1016/j.nmd.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 110.Goselink, R. J. M.; Voermans, N. C.; Okkersen, K.; Brouwer, O. F.; Padberg, G. W.; Nikolic, A.; Tupler, R.; Dorobek, M.; Mah, J. K.; van Engelen, B. G. M.; Schreuder, T. H. A.; Erasmus, C. E. Early onset facioscapulohumeral dystrophy - a systematic review using individual patient data. Neuromuscul. Disord. 27:1077-1083. [DOI] [PubMed]
- 111.Blaszczyk E., Grieben U., von Knobelsdorff-Brenkenhoff F., Kellman P., Schmacht L., Funk S., Spuler S., Schulz-Menger J. Subclinical myocardial injury in patients with Facioscapulohumeral muscular dystrophy 1 and preserved ejection fraction - assessment by cardiovascular magnetic resonance. J. Cardiovasc. Magn. Reson. 2019;21:25. doi: 10.1186/s12968-019-0537-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Roussel J., Thireau J., Brenner C., Saint N., Scheuermann V., Lacampagne A., Le Guennec J.Y., Fauconnier J. Palmitoyl-carnitine increases RyR2 oxidation and sarcoplasmic reticulum Ca2+ leak in cardiomyocytes: role of adenine nucleotide translocase. Biochim. Biophys. Acta. 1852:749–758. doi: 10.1016/j.bbadis.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 113.Dorner A., Lynetskiy O., Euler G., Landmesser U., Schluter K.D., Heger J. Mitochondria isolated from hearts subjected to ischemia/reperfusion benefit from adenine nucleotide translocase 1 overexpression. Membranes (Basel) 2021;11 doi: 10.3390/membranes11110836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Klumpe I., Savvatis K., Westermann D., Tschope C., Rauch U., Landmesser U., Schultheiss H.P., Dorner A. Transgenic overexpression of adenine nucleotide translocase 1 protects ischemic hearts against oxidative stress. J. Mol. Med. (Berl.) 2016;94:645–653. doi: 10.1007/s00109-016-1413-4. [DOI] [PubMed] [Google Scholar]
- 115.Walther T., Tschope C., Sterner-Kock A., Westermann D., Heringer-Walther S., Riad A., Nikolic A., Wang Y., Ebermann L., Siems W.E., Bader M., Shakibaei M., Schultheiss H.P., Dorner A. Accelerated mitochondrial adenosine diphosphate/adenosine triphosphate transport improves hypertension-induced heart disease. Circulation. 2007;115:333–344. doi: 10.1161/CIRCULATIONAHA.106.643296. [DOI] [PubMed] [Google Scholar]
- 116.Wang Y., Ebermann L., Sterner-Kock A., Wika S., Schultheiss H.P., Dorner A., Walther T. Myocardial overexpression of adenine nucleotide translocase 1 ameliorates diabetic cardiomyopathy in mice. Exp. Physiol. 2009;94:220–227. doi: 10.1113/expphysiol.2008.044800. [DOI] [PubMed] [Google Scholar]
- 117.Lynn E.G., Stevens M.V., Wong R.P., Carabenciov D., Jacobson J., Murphy E., Sack M.N. Transient upregulation of PGC-1alpha diminishes cardiac ischemia tolerance via upregulation of ANT1. J. Mol. Cell. Cardiol. 2010;49:693–698. doi: 10.1016/j.yjmcc.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li J., Yan Z., Fang Q. A mechanism study underlying the protective effects of cyclosporine-A on lung ischemia-reperfusion injury. Pharmacology. 2017;100:83–90. doi: 10.1159/000458760. [DOI] [PubMed] [Google Scholar]
- 119.Jiang H., Zhang C., Tang Y., Zhao J., Wang T., Liu H., Sun X. The regulator of calcineurin 1 increases adenine nucleotide translocator 1 and leads to mitochondrial dysfunctions. J. Neurochem. 2016;140:307–319. doi: 10.1111/jnc.13900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.group T.F.-D. A prospective, quantitative study of the natural history of facioscapulohumeral muscular dystrophy (FSHD): implications for therapeutic trials. The FSH-DY Group. Neurology. 1997;48:38–46. doi: 10.1212/wnl.48.1.38. [DOI] [PubMed] [Google Scholar]
- 121.Olsen D.B., Gideon P., Jeppesen T.D., Vissing J. Leg muscle involvement in facioscapulohumeral muscular dystrophy assessed by MRI. J. Neurol. 2006;253:1437–1441. doi: 10.1007/s00415-006-0230-z. [DOI] [PubMed] [Google Scholar]
- 122.Statland J.M., Shah B., Henderson D., Van Der Maarel S., Tapscott S.J., Tawil R. Muscle pathology grade for facioscapulohumeral muscular dystrophy biopsies. Muscle Nerve. 2015;52:521–526. doi: 10.1002/mus.24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings described in this manuscript are available in the article and from the corresponding author upon reasonable request.
Data will be made available on request.