Abstract
Accumulation of inappropriately phosphorylated tau into neurofibrillary tangles is a defining feature of Alzheimer’s disease, with Tau pT231 being an early harbinger of tau pathology. Previously, we demonstrated that expressing a single genomic copy of human phosphomimetic mutant tau (T231E) in Caenorhabditis elegans drove age-dependent neurodegeneration. A critical finding was that T231E, unlike wild-type tau, completely and selectively suppressed oxidative stress-induced mitophagy. Here, we used dynamic imaging approaches to analyze T231E-associated changes in mitochondria and mitolysosome morphology, abundance, trafficking, and stress-induced mitophagy as a function of mitochondrial fission mediator dynamin-related protein 1, which has been demonstrated to interact with hyper phosphorylated tau and contribute to Alzheimer’s disease pathogenesis, as well as Pink1, a well-recognized mediator of mitochondrial quality control that works together with Parkin to support stress-induced mitophagy. T231E impacted both mitophagy and mitolysosome neurite trafficking with exquisite selectivity, sparing macroautophagy as well as lysosome and autolysosome trafficking. Both oxidative-stress-induced mitophagy and the ability of T231E to suppress it were independent of drp-1, but at least partially dependent on pink-1. Organelle trafficking was more complicated, with drp-1 and pink-1 mutants exerting independent effects, but generally supported the idea that the mitophagy phenotype is of greater physiologic impact in T231E. Collectively, our results refine the mechanistic pathway through which T231E causes neurodegeneration, demonstrating pathologic selectivity for mutations that mimic tauopathy-associated post-translational modifications, physiologic selectivity for organelles that contain damaged mitochondria, and molecular selectivity for dynamin-related protein 1-independent, Pink1-dependent, perhaps adaptive, and mitophagy.
Keywords: Alzheimer’s disease, tau phosphorylation, Caenorhabditis elegans, mitochondria, Drp1, Pink1, mitophagy
Introduction
Alzheimer’s disease (AD) is the most common progressive neurodegenerative disorder (DeTure and Dickson 2019). A prominent pathological hallmark of AD is the intraneuronal neurofibrillary tangles (NFTs), primarily comprised of fibrillar tau that is modified with a myriad of post-translational modifications (PTMs) which accumulate stoichiometrically on tau as the disease progresses (Neddens et al. 2018; Wesseling et al. 2020). Phosphorylation of tau at specific residues occurs early in the disease process, leading to an attenuation in its normal microtubule binding function (Sengupta et al. 1998; Ding et al. 2006; Qiang et al. 2018) concurrent with an increased propensity to self-associate (Lasagna-Reeves et al. 2012; Carroll et al. 2021). Indeed, the progressive phosphorylation of tau correlates with the evolution of cognitive impairment in AD, clearly indicating the importance of the phosphorylation of tau at key sites in the disease process (Wesseling et al. 2020; Arnsten et al. 2021).
In the past decades, transgenic animal models of tau toxicity have been developed in organisms such as Caenorhabditis elegans (Kraemer et al. 2003; Brandt et al. 2009; Miyasaka et al. 2018), Drosophila melanogaster (Chee et al. 2006; Steinhilb et al. 2007; Kosmidis et al. 2010), and mice (Santacruz et al. 2005; Yoshiyama et al. 2007). Toxicity in these model organisms has generally been achieved by over-expressing various tau species. Though these approaches have undoubtedly led to new insights, overexpression has the potential to mask subtle modifiers in a multifactorial, complex disease process.
Mitochondria play a complex role in the cell—they not only generate most of the energy needed to support the various neuronal functions (Safiulina and Kaasik 2013), but also are mediators of homeostatic processes that are necessary for neuronal health (Mattson et al. 2008). Mitochondrial dysfunction is a prominent feature of many neurodegenerative diseases, including AD (Flippo and Strack 2017; Hu et al. 2017; Murphy and Hartley 2018; Swerdlow 2018). Pathological accumulation of tau in animal models is associated with mitochondrial reactive oxygen species (ROS) production and impacts oxidative phosphorylation (David et al. 2005; Li et al. 2016; Perez et al. 2018), morphology and turnover (Cummins et al. 2019; Fang, Hou, Palikaras, et al. 2019), and trafficking (Flannery and Trushina 2019), though the precise mechanisms are still not well understood.
To better understand the role of gatekeeper tau PTMs in the context of AD relevant mitochondrial dysfunction, we developed transgenic, single-copy C. elegans models expressing wild-type human tau in a defined set of 6 mechanosensory (mec) neurons (Guha, Fischer, et al. 2020). Clustered regularly interspaced short palindromic repeats (CRISPR)–CRISPR associated protein 9 genome editing was used to introduce a phosphomimetic T-to-E mutation at T231, a site associated with early progression of tau to a toxic form in AD (Alonso et al. 2010; Neddens et al. 2018; Aragao Gomes et al. 2021). Although tau is phosphorylated at many sites as AD progresses, this progression is stereotypical, with modification of early sites such as T231 being a harbinger of selective vulnerability (Wesseling et al. 2020; Carroll et al. 2021). It is important to note that because tau is not normally expressed in worms, these were by definition still overexpression models, albeit less so than previous models given the use of single-copy gene insertion techniques and limited targeting repertoire of 6 neurons.
A combination of behavioral assays and dynamic imaging of fluorescent biosensors clearly demonstrated a selective and age-dependent effect of T231E on sensory neuron function, morphology, and, perhaps most importantly, on mitochondrial autophagy (mitophagy) elicited by oxidative stress (Guha, Fischer, et al. 2020)—phenotypes generally lacking in the strain expressing wild-type tau. Hence, this model confers the ability to discern between the pathological consequences of individual tau mutants and the age-dependence of mito-centric phenotypes with unprecedented precision.
Dynamin-related protein 1 (Drp1) is a GTPase that regulates mitochondrial morphology through membrane fission (Imoto et al. 1998; Smirnova et al. 1998). In addition to its contribution to canonical mitophagy (Dagda et al. 2009), Drp1 has also been shown to selectively interact with hyper-phosphorylated forms of tau (Manczak and Reddy 2012; Kandimalla et al. 2021) and to influence the pathogenesis of AD (Bera et al. 2022).
Phosphatase and tensin homolog-induced kinase 1 (PINK1) is a serine/threonine kinase that is mutated in hereditary early-onset Parkinson’s disease (PD) and which upon mitochondrial depolarization can be stabilized on the mitochondrial membrane to initiate PINK1-Parkin-dependent mitophagy [for review, see Pickrell and Youle (2015)]. There are numerous studies reporting altered levels of PINK1 in the context of AD pathology, and PINK1 overexpression has been shown to alleviate phenotypic outputs in a mouse AD model (Jiang et al. 2021). However, the specific impact of PINK1 on tau-driven pathology as well as pathological forms of tau on PINK1-dependent mitophagy remain largely underexplored.
With this in mind, here we set out to examine the intersection of tau pT231, mitophagy, and organelle trafficking in promoting neurodegeneration using our single-copy transgenic tau mutant strains together with established C. elegans loss of function mutants in drp-1 (Labrousse et al. 1999; Breckenridge et al. 2008) and pink-1 (Sämann et al. 2009; Bess et al. 2012; Chikka et al. 2016).
The results of our study provide clear in vivo evidence that phosphomimetic T231E tau elicits profound and selective effects on stress-induced mitophagy and mitolysosomal trafficking starting in early adulthood, with more subtle effects occurring on mitochondria themselves. We further demonstrate that the molecular mechanism underlying these effects is pink-1 dependent but drp-1 independent. Our observations provide focus on adaptive mitophagy as an unanticipated target of toxic tau, and may hence be worth exploring as a potential therapeutic target for tau pathology in AD.
Methods
Caenorhabditis elegans cultivation
Worms were maintained using standard techniques (Stiernagle 2006) at 20°C on nematode growth media (NGM) plates made with Bacto Agar (BD Biosciences) and seeded with live E. coli OP50-1 bacterial strain (cultured overnight at 37°C in Luria Bertani Miller Broth). For experimental assays, fourth larval stage (L4) hermaphrodites were generated by synchronization using standard alkaline-bleach treatment to isolate embryos. The synchronized animals were moved to plates containing 25 µM 2′-deoxy-5-fluorouridine to suppress fertility and assayed as adults either 3 or 10 days later.
To induce oxidative stress, N,N′-dimethyl-4,4′-bipyridinium dichloride, commonly known as paraquat (PQT; Sigma-Aldrich, St. Louis, MO) was added to a final concentration of 8 mM in OP50-seeded NGM agar plates, and animals were maintained on these plates at 20°C overnight prior to analysis.
Strains and reporters
A list of strains and reporter transgenes used in this study is provided in Supplementary Table 1. Single copy tau strains have been described (Guha, Fischer, et al. 2020). Reporters included mitochondrial localization sequence (MLS)::green fluorescent protein (GFP) to measure mitochondrial morphology and dynamics (Mondal et al. 2012), mito-mKeima to measure mitophagy and ML trafficking (Katayama et al. 2011; Suzuki et al. 2017), Atg8/LGG-1::mCherry::GFP to measure macroautophagy and autophagosome/autolysosome (AP/AL) trafficking (Chang et al. 2017), and LMP-1::mScarlet to measure lysosome trafficking (S. Swords and B. Grant, Rutgers University). All of these reporters were expressed under the control of a touch-cell specific mec promoter. In addition, strains were used that contain integrated reporters expressing pan-neuronal fluorescent mitochondria-targeted transgenes such as Y45G12B.3 (C. elegans L-2-hydroxyglutarate dehydrogenase; S. Pena and K. Nehrke) and outer membrane protein TOMM-20 (Traa et al. 2021). Standard C. elegans genetic techniques were used to cross reporters into single-copy tau strains and/or the drp-1 (tm1108)IV and pink-1 (ok3538)II mutants. In all cases, “control” refers to reporters in an N2 background. Reporters were followed under a standard fluorescent dissecting microscope. Single worm genomic PCR was used to discern the presence of the tau expression cassettes in the Mos ttTi5605 locus on Chr II, as well as for drp-1 (tm1108) and pink-1 (ok3538) genotyping.
Primers used for drp-1 genotyping are as follows:
drp-1 geno F1 5′-ccagacttcgatgccgtgcga-3′
drp-1 geno R1 5′-acgagcacctccgcatagctca-3′
drp-1 geno F2 (Int) 5′-cctgcactgaaagcccgcgt-3′
Primers used for pink-1 genotyping are as follows:
pink geno Fwd 5′-tgtcgtggaaatggagcctt-3′
pink geno Rev 5′-aacgattcggaaaagcagca-3′
pink geno Int 5′-gctgaaggctcggatgatga-3′
Primers for tau genotyping have been described (Guha, Fischer, et al. 2020). T231A and T231E tau mutations were confirmed in all final transgenic strains by PCR sequencing, as described (Guha, Fischer, et al. 2020).
Behavioral assay
The sensory response to light touch (using an eyelash) was assessed as described by Hobert et al. (1999) and modified according to Chen and Chalfie (2014). Individual worms were visualized under a conventional dissecting scope on OP50-1 seeded NGM-agar plates and touched gently behind the terminal bulb of the pharynx a total of 10 times. Touches were separated by a minimum of 10 s to prevent habituation. Positive responses were scored if an omega turn or locomotory reversal was observed. Worms that crawled off the food were censored. The experimentalists used a mec-4 mutant with a selective deficit in its response to light touch to calibrate their technique, thus ensuring that alternative harsh touch response pathways that occur through separate genetically and physiologically distinguishable circuits were not inadvertently activated (O'Hagan et al. 2005).
Fluorescent imaging
Mitochondrial morphology—Animals expressing touch-cell MLS::GFP were mounted on 3% agarose pads on glass slides and anesthetized with 10 mM tetramisole hydrochloride. Imaging of the soma was performed using a Nikon Eclipse TE2000 inverted microscope coupled to a 6-channel LED light source (Intelligent Imaging Innovation, Denver, CO), an ORCA-Flash4.0 V2 Digital CMOS camera (Hamamatsu Photonics, Bridgewater Township, NJ) and Slidebook6 software (Intelligent Imaging Innovation). The PLM cell body was brought into focus under visible light using DIC contrast, a 100x oil immersion objective, and a 1.5x magnifier. Once in focus, DIC was removed, and epifluorescence image acquisition was performed using 488-nm excitation, a 515-nm dichroic, and a 535/25 emission filter.
Freehand tracing of the mitochondria in ImageJ was used to calculate mitochondrial area and the proportion of somatic area occupied by mitochondria, which we termed density (1 pixel = 0.0019 µm2). Two experimentalists independently analyzed subsets of images and compared results to ensure the reproducibility of the analysis.
Mitophagy—Animals expressing touch-cell specific mito-mKeima were immobilized and imaged essentially as described above using dual excitation fluorescence ratiometry (ex. 550-nm/440-nm.; em. 600-nm+), with background subtraction, thresholding, and quantitation performed using Slidebook6 (Guha, Johnson, et al., 2020).
Autophagy—Animals expressing touch-cell specific LGG-1::GFP::mCherry were immobilized as described above and imaged using a Nikon A1R HD scanning confocal microscope running NIS-Elements version 5.11 software. Images were acquired through a 60x oil objective using the resonance scanner mode with 488-nm or 561-nm laser excitation at 2.5% power, 2x zoom, 4x averaging, and a pixel size of 0.14 µm, and emissions were monitored through a 525/50 or 595/50 filter for the green and red channels, respectively. The PMT gain was HV = 5 for the red channel and HV = 10 for the green channel, and the z-interval was 0.6 µm. Maximum projection composite images were generated using ImageJ software and used to assess AP (green and red) and AL (red only) puncta.
Motility assays—Time-lapse recordings were acquired with a Nikon A1R HD Laser scanning confocal microscope (Eclipse Ti2) at 60× oil objective using NIS-Elements version 5.11 software with parameters similar to those describe above. Mitochondria were captured using 488-nm excitation and a 525/50 emission window, whereas lysosome, AL, and ML movements were captured using 561-nm excitation and a 595/50 emission window. The capture period was 5 min with 30 s/frame interval. Particle dynamics were analyzed using the Imaris x6.2 version software. Run length was obtained as the distance between the starting and last stopping points of a motile particle. Other parameters considered were % motility and respective speed as described (Morsci et al. 2016).
Statistical analysis
All statistical analyses were conducted using Prism 8.0 (GraphPad Software), with alpha-error level of P < 0.05 considered to be significant. Data sets were first tested for normalcy, and in most cases group differences were analyzed with either 1-way or 2-way ANOVA depending upon the variables. Data sets which did not fit normalcy were analyzed by a Kruskal–Wallis test. The sample sizes were based on those found previously in the laboratory to provide appropriate power for discerning phenotypic differences among genotypes. Graphs were plotted in Prism 8.0.
Results
Phosphomimetic tau T231E, but not wild-type tau, exacerbates an age-dependent reduction of mitochondrial density in PLM neurons and sensitizes them to oxidative stress
Recently, we developed an AD model where single-copy transgenes coding for tau fused to the fluorescent protein Dendra2 were expressed specifically in C. elegans mec touch neurons (Guha, Fischer, et al. 2020). In this work, we showed that strains expressing phosphomimetic mutant human tau T231E (T231E hereafter), but not wild-type tau (0N4R or TauT4 hereafter) or a phosphoablation mutant T231A (T231A hereafter) exhibited mild fragmentation of the PLM cell mitochondrial network along with progressive morphologic and sensory defects. However, the relatively small size of the neurons and difficulty in skeletonizing/segmenting the mitochondria network confounded use of conventional metrics to describe morphology. Hence, fragmentation in the original work was assessed categorically.
Due to the modest effect size, we repeated this work here using a more quantitative metric, recapitulating an approach taken to define morphologic remodeling with age (Morsci et al. 2016). The mitochondria of touch neurons were visualized using transgenic matrix-targeted GFP driven by a touch neuron specific mec-7 promoter. The proportion of soma area taken up by mitochondria was calculated as “density,” which reflects morphology, distribution, and abundance in aggregate. In order to assess the impact of age, we performed identical analyses in young adults and old adults at Days 3 and 10 following the onset of reproductive maturity—with the normal worm adult lifespan being ∼2 weeks. Reproduction during adulthood was inhibited by the addition of 5′-fluorodeoxyuridine (FUdR) to the growth media. Although FUdR is routinely used for this purpose, it has been noted to impact mitochondrial phenotypes (Rooney et al. 2014), and we acknowledge the potential for synthetic interactions.
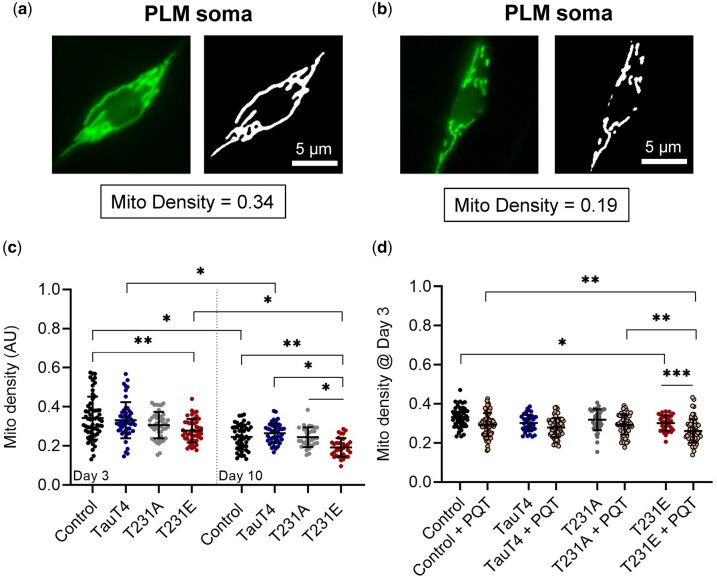
Representative images from the upper and lower quartiles of our combined datasets illustrate the range of natural variation in wild-type animals (Fig. 1, a and b). Indeed, most neurons exhibited relatively normal tubulo-reticular networks within the defined range of natural variation independent of tau genotype (Fig. 1c). However, T231E was statistically different than the control (MLS::GFP reporter strain) at day 3 of adulthood, and by Day 10 exhibited a slight reduction in mitochondrial density as compared to all other strains (Fig. 1c). This was reminiscent of age-dependent morphologic decline shown to occur in extremely old animals (Morsci et al. 2016; Coppa et al. 2020).
Fig. 1.
Mitochondrial morphology in PLM soma as a function of single-copy Tau mutants. An MLS::GFP reporter expressed via the mec-7 promoter was used to image mitochondria in the soma of PLM neurons. (a, b) Images on the left are the original fluorescent micrographs obtained from wild-type animals with mitochondrial networks that differ within the scope of our observations, representing natural variation and falling in the upper and lower quartile of the density metric, with values as shown. Images on the right represent a binary mask used to derive density. Scale bar: 5 µm. (c) Quantitative analysis of mitochondrial density at day 3 and 10 as a function of single-copy tau genotype. (d) Density as a function of 24 h treatment with 8 mM PQT starting at day 2 of adulthood. Data were collected via 3 independent biological replicates where all relevant genotypes were compared in parallel. Individual data points are values from single PLM cells from separate animals, with the mean ± SD plotted in the background (N = 33–71). Statistical analyses were 2-way ANOVA with Tukey’s multiple comparison test, with ***P < 0.001, **P < 0.01, *P < 0.05 when comparing bracketed samples.
Age is associated with the accumulation of oxidative damage (Chaudhuri et al. 2018; Hou et al. 2019) and chronic mitochondrial stress is likely to be a factor in neurodegenerative diseases including AD (Galindo et al. 2010; Weidling and Swerdlow 2019). PQT is a redox cycler that stimulates ROS production in mitochondria (Cochemé and Murphy 2008). We have previously shown that PQT stimulates mitophagy in PLM neurons, and that T231E selectively suppresses this effect (Guha, Fischer, et al. 2020). Surprisingly, we found here that PQT—at the low concentration used—had very little impact on PLM mitochondrial density in control, T231A, and TauT4 Day 3 adults, but caused a significant reduction of density in T231E (Fig. 1d). The most parsimonious conclusion is that T231E exacerbates morphologic remodeling such as occurs in extreme aged animals (Morsci et al. 2016; Coppa et al. 2020).
T231E selectively reduces neurite mitochondrial abundance and motility in older animals
The above results motivated us to examine other physiologic processes that require mitochondrial morphologic remodeling. Mitochondria are dynamic organelles, both with respect to morphology as well as to movement through neuronal processes (Flippo and Strack 2017). Abnormal mitochondrial morphology, distribution, and axonal transport have been observed in AD postmortem brains and in AD mouse models (Manczak et al. 2011; Reddy et al. 2012). Pathologic tau reduces mitochondrial mobility in cortical neurons treated with Aβ (Ebneth et al. 1998; Quintanilla et al. 2012) as well as in worm neurites expressing pro-aggregate forms of tau (Fatouros et al. 2012).
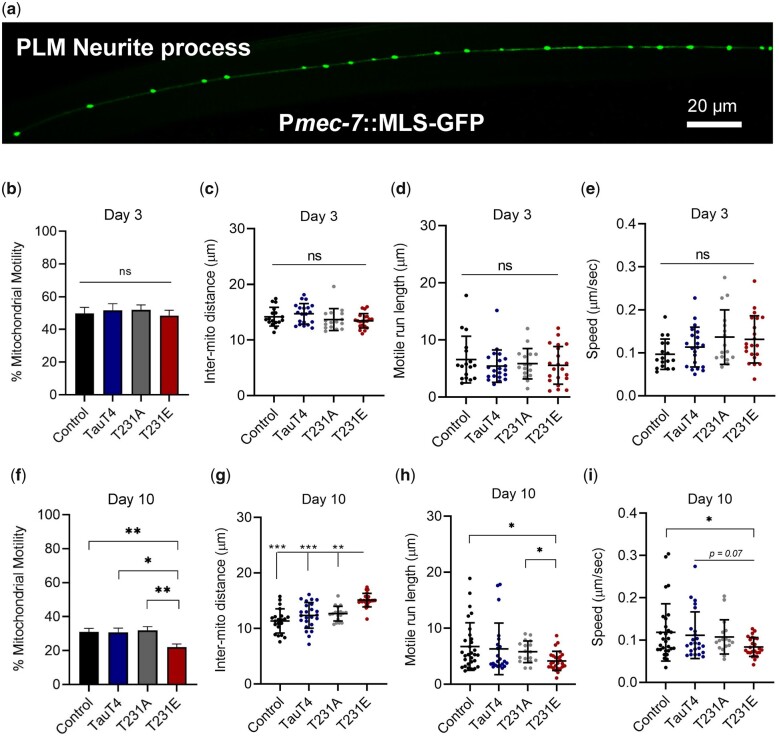
GFP-labeled mitochondria in PLM neuronal processes (Fig. 2a) appear as sphere and rod-shaped organelles of varying size distributed at specific intervals, similar to axonal mitochondria in mammalian neurons (Lin and Sheng 2015; Takihara et al. 2015). Given the established role of tau in facilitating axonal transport, we investigated the impact of TauT4, T231A, and T231E on several metrics that reflect the distribution of mitochondria. These include motility, spacing, and active trafficking dynamics such as motile run length and speed in the distal segment of PLM processes (∼300 µm).
Fig. 2.
Mitochondrial distribution and transport are reduced by pathological phosphomimetic Tau T231E. Mitochondrion were visualized through dynamic fluorescent imaging of an MLS::GFP reporter to track their movement in the PLM neuronal process of live worms. a) Representative confocal image of a PLM neurite from a wild-type worm expressing MLS::GFP. Scale bar: 20 µm. b, f) Mitochondrial motility (fraction of mitochondria that moved during imaging); c, g) intermitochondrial distance (distance between 2 consecutive mitochondria); d, h) run length (distance traversed on average by a healthy mitochondrion), and (e, i) speed, as a function of age and single-copy tau genotype. Motility metrics were limited to mitochondria that exhibited movement over the course of 5 min imaging. Two independent biological replicates were performed on different days, with all genotypes analyzed in parallel. Individual data points demarcate values from single PLM cells from separate animals, with the mean ± SD as shown (N = 20–26). Statistical analysis within day 3 and day 10 datasets was by 1-way ANOVA with Tukey’s multiple comparison test, with ***P < 0.001, **P < 0.01, *P < 0.05 when comparing bracketed samples.
Motile mitochondria represented ∼1/2 of the pool at day 3 (Fig. 2b), and overall fractional motility decreased by day 10, with T231E exhibiting a greater impact of age (Fig. 2f). The average distance between mitochondria was independent of tau genotype at day 3, but at day 10 the average inter-mitochondrial distance in T231E was significantly greater than all other 3 tau genotypes (Fig. 2, c and g). For trafficking metrics, immobile mitochondria were not used in calculating the values shown. As observed for somatic mitochondria, tau genotype had no significant impact on mean run length or speed at day 3 (Fig. 2, d and e), but T231E became selectively distinguishable by day 10 (Fig. 2, h and i). These results demonstrate that our model can detect subtle differences in mitochondrial number, distribution, and transport induced by toxic tau species associated with “disease” progression, where mitochondrial dysfunction may contribute to neurite deficits. These results motivated us to query the selectivity of T231E for mitochondria by examining the movement of other dynamic organelles.
Neurite transport of mitolysosomes is suppressed selectively by T231E in both old and young animals
Mitophagy is a type of cargo-selective autophagy (Hamacher-Brady and Brady 2016; Martinez-Vicente 2017), where damaged mitochondria are engulfed by APs and subsequently degraded by fusion with lysosomes to form a distinct type of AL termed mitolysosome (ML). We previously demonstrated a failure in oxidative-stress induced mitophagy in T231E but not TauT4 or T231A (Guha, Fischer, et al. 2020).
Here, we used the mitophagy reporter mito-mKeima to follow the movement of ML along PLM neurites (Fig. 3a). Mito-mKeima is a matrix-targeted variant of the genetically encoded pH biosensor Keima (Kogure et al. 2006). Upon fusion of the engulfed mitochondria with the acidic lysosome, mito-mKeima—which is resistant to degradation by acid proteases—exhibits a spectral shift in the excitation maxima from 440 to 550 nm. Mito-mKeima ratios have been widely used as a metric to assess mitophagy (Chang et al. 2017; Sun et al. 2017), but this property also permits focusing on ML movement through the neurite by selectively imaging the red channel.
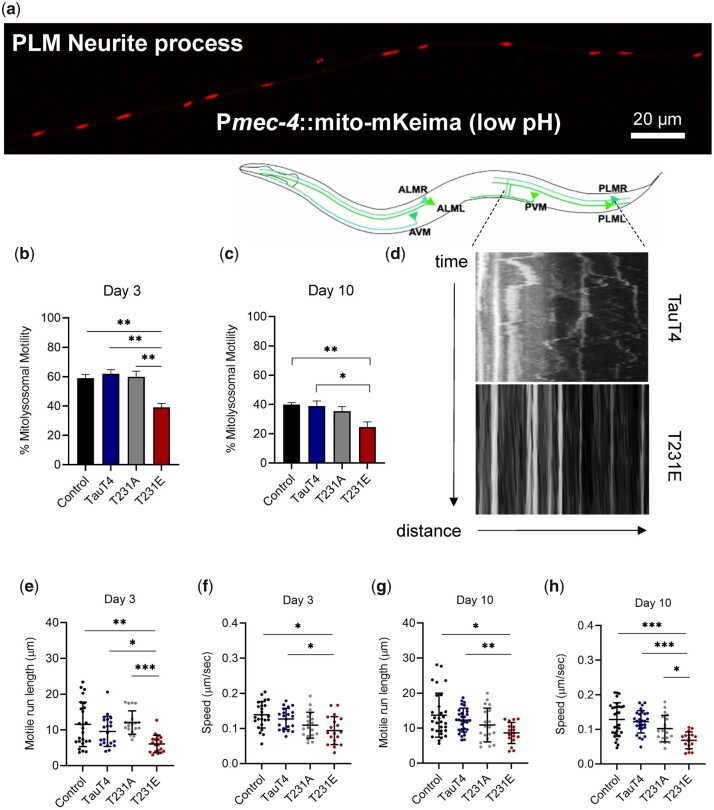
Fig. 3.
Pathological phosphomimetic tau T231E reduces mitolysosome motility. Mitolysosomes (MLs) were visualized through dynamic fluorescent imaging of mito-mKeima (550-nm excitation) to track their movement in the PLM neuronal process of live worms. a) Representative confocal image of a wild-type PLM neurite expressing mito-mKeima Scale bar: 20 µm. b, c) ML motility; e, g) run length and (f, h) speed, as a function of age and single-copy tau genotype. Motility metrics were limited to MLs that exhibited movement over the course of 5 min imaging. d) Diagram showing the C. elegans touch-sensitive mechanosensory neurons, including the PLM neuronal process (demarcated by dotted lines), with kymographs of ML movement in TauT4 and T231E, as labeled. The T231E example is extreme, but not unique. Two independent biological replicates were performed on different days. Individual data points demarcate values from single PLM neurites from separate animals, with the mean ± SD as shown (N = 22–30). Statistical analysis within day 3 and 10 datasets was by 1-way ANOVA with Tukey’s multiple comparison test, with ***P < 0.001, **P < 0.01, *P < 0.05 when comparing bracketed samples.
One caveat of this approach is that severe mitochondrial dysfunction can impact lysosomal activity (Fernandez-Mosquera et al. 2019), and under such circumstances lysosomes may not be properly acidified, hence suppressing the spectral shift in mito-mKeima even in the face of functional mitophagy. However, unlike most models of severe mitochondrial dysfunction, T231E has a modest, subtle phenotype that is only prominent following oxidative stress (Guha, Fischer, et al. 2020). Moreover, limiting imaging to the red channel, which is only visible at low pH, ensures that we are tracking acidified vesicles. However, we appreciate the potential interplay between mitochondrial dysfunction and lysosomal activity in regulating metabolism. We also note that new tools are being developed to interrogate those connections (Chen et al. 2020).
Unlike mitochondria, where the impact of T231E was not apparent until day 10, the fractional motility of ML in T231E was reduced by day 3 of adulthood (Fig. 3, b and c; Supplementary Videos 1 and 2). This was particularly apparent when comparing kymographs depicting bi-directional movement of individual ML over-time (Fig. 3d), with the T231E ML nearly stationary, albeit in an extreme example of the phenotype. Moreover, even though the total number of ML in the neurites remained constant across the genotypes (data not shown), the T231E run length and the average speed were also significantly lower at both day 3 and 10 (Fig. 3, e–h).
In order to address the selectivity of this transport deficit for ML, we used 2 approaches. First, we asked whether it applies generally to lysosomes by measuring their trafficking with a lysosomal-associated membrane protein 1 (LMP-1)::mScarlet fusion transgene, generated using miniMos single-copy insertion (Frøkjaer-Jensen et al. 2014). However, neither their motile run length nor their speed was impacted by T231E at either day 3 or day 10 of adulthood (Supplementary Fig. 1, a and b). We then narrowed our query by using a tandem mCherry-GFP-Atg8/LGG-1 reporter that labels touch neuron autophagic vesicles in both red and green, but is nonselective for cargo (Chang et al. 2017). Once again, our results suggested a lack of effect on autophagic vesicle trafficking with respect to tau genotype at either day 3 or day 10 (Supplementary Fig. 1, c–f). To summarize, pathological tau mutation T231E impacts ML movement in young and old adults, whereas its effects on mitochondria motility are limited to older animals, and neither lysosome nor AL motility are affected (Supplementary Fig. 1g).
Dysregulation of autophagy has been linked with neurodegenerative diseases, including AD (Nixon et al. 2005; Menzies et al. 2015; Bordi et al. 2016; Xu et al. 2021). Given the selective effect of T231E on ML motility, but not bulk AL motility, we asked whether there was a similarly selective impact on mitophagy over autophagy. Autophagy was triggered using food deprivation (Alirezaei et al. 2010) and measured as a function of tau genotype, with acidic quenching of GFP in the mCherry-GFP-Atg8/LGG-1 reporter as a means of distinguishing between AP and AL (Supplementary Fig. 1, h–n). In all cases, fasting induced an aging and genotype-independent increase in AL number (Supplementary Fig. 1, i–n). In addition, AL were more abundant than AP, particularly after fasting, suggesting that they turn over more slowly than AP. This validated the autophagy reporter, but also confirmed the selectivity of T231E in impairing mitophagy over bulk autophagy (given that overall lysosome and AL number were unchanged by tau mutant expression).
Finally, we explored whether PQT stimulation, which fails to induce mitophagy in the PLM soma of the T231E mutant (Guha, Fischer, et al. 2020), had a similar phenotype with respect to ML formation in the neuronal processes, and whether this in turn affected trafficking of this very specific cargo containing vesicle. Interestingly, while PQT treatment stimulated increased ML abundance in both control and TauT4 but not T231E (Supplementary Fig. 2a), it also suppressed ML speed and motility such that both TauT4 and T231A phenocopied T231E (Supplementary Fig. 2, b and c). We conclude that the effect of T231E is selective for ML formation and trafficking, though perhaps through different mechanisms.
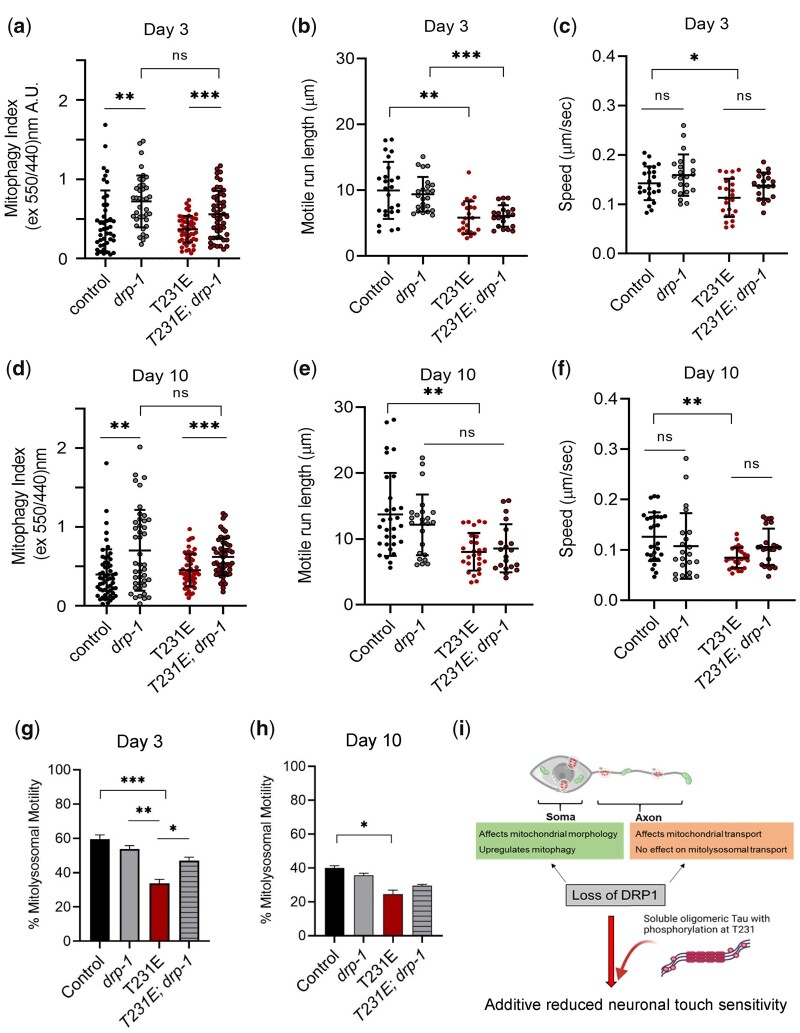
Loss of drp-1 causes mitochondrial blob morphology, independent of tau genotype
Alterations in mitochondrial dynamics are increasingly being recognized as early-stage pathological events in neurodegenerative diseases, including AD, Huntington’s disease, and PD (Reddy et al. 2012; Sheng et al. 2012; Malpartida et al. 2021). Regulation of mitochondrial morphology by fission is important for mitophagy (Chen and Chan 2009; Bhattacharya et al. 2021). The canonical fission mediator, Drp1, is a monomeric GTPase that oligomerizes at the mitochondrial outer membrane upon activation (Pernas and Scorrano 2016). Previous studies indicate that hyper-phosphorylated tau interacts directly with Drp1, likely increasing its activity and inducing mitochondrial fragmentation (Manczak and Reddy 2012). This led us to ask whether Drp1 contributes to the changes in mitochondrial morphology, trafficking or oxidative stress induced clearance that we observe in our model. Unlike the embryonic lethality observed in in Dnm1l knockout mice, loss of drp-1 is tolerated in C. elegans, and a tm1108 mutant has been well studied (Wakabayashi et al. 2009; Byrne et al. 2019).
Unexpectedly, in contrast to the normal tubulo-reticular mitochondrial network observed in wild-type C. elegans PLM cell body (Fig. 1a), PLM mitochondria in tm1108 presented as swollen blobs (Fig. 4a), reminiscent of worms expressing dominant interfering K40A DRP-1 (Labrousse et al. 1999). This blob-like phenotype was observed independent of tau genotype (Fig. 4a), with multiple biosensors (see mito-mKeima images in Fig. 6 as well as L-2-HGDH::mRuby3 and TOMM20::mScarlet in Supplementary Fig. 3), and in at least some other neurons (Supplementary Fig. 3). In general, both the qualitative and quantitative effect of tm1108 on mitochondrial density were disproportionally greater than tau genotype (Fig. 4, b and d), and we were unable to interpret the biological relevance of the subtle, albeit significant, differences due to tau genotype.
Fig. 4.
Drp-1 has extreme effects on mitochondrial morphology and motility, but only subtle effects on touch sensitivity. Mitochondria were visualized through time-lapse fluorescent imaging of an MLS::GFP reporter to measure mitochondrial morphology in the soma and to track their movement in the PLM neuronal process of live worms. a) Representative binary images used to quantitate morphology in the genotypes indicated. Note that the mitochondria appear as blobs in the drp-1(tm1108) mutant, irrespective of the tau genotype and age. Scale bar: 5 µm. b, d) Quantitative analysis of mitochondrial density in the distal PLM cell bodies at day 3 and day 10 as a function of drp-1 and tau genotype. Control indicates data points from MLS::GFP reporter. Individual data points demarcate values from single PLM cells from separate animals (N = 30–75). c, e) Responsiveness to touch was plotted as a function of drp-1 and T231E at day 3 and 10. The data are from 3 independent biological replicates (N = 60–115). f, g) Representative confocal images of the PLM neurite from control and drp-1(tm1108) expressing MLS::GFP. Scale bar: 20 µm. Quantitative analysis of mitochondria (h, l) motility, (i, m) distribution, (j, n) run length, and (k, o) speed in distal PLM processes at day 3 or day 10 as a function of drp-1 or T231E, as indicated. Individual data points are from single PLM neurites from separate animals (N = 17–25). Data are the mean ± SD from 3 independent biological replicates. Statistical analysis for all datasets was by 1-way ANOVA with Tukey’s multiple comparison test, with ***P < 0.001, **P < 0.01, *P < 0.05 when comparing bracketed samples.
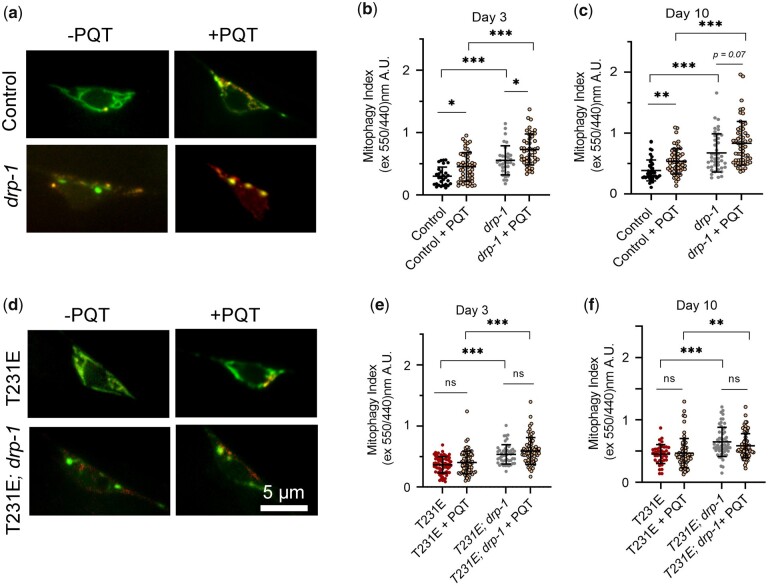
Fig. 6.
Oxidative-stress stimulated mitophagy and suppression by phosphomimetic Tau T231E are drp-1 independent. Mitophagy was measured using dual excitation ratio imaging of mito-mKeima expressed in C. elegans PLM neurons. Drp-1 refers to the drp-1(tm1108) allele. a, d) Representative merged images where 440-nm excitation was used to detect mitochondria (green) and 550-nm excitation was used to detect mitolysosomes (red) as a function of drp-1 genotype, T231E or 24 h PQT treatment, as indicated. Scale bar: 5 µm. b, c, e, f) Quantitative analysis of mitophagy as a function of age, drp-1 genotype, T231E or PQT. Data are the mean ± SD from 2 biological replicates performed on different days (N = 30–75). Statistical analysis was by 2-way ANOVA followed by Tukey’s post hoc test, with ***P < 0.001, **P < 0.01, *P < 0.05 denoting significance when comparing bracketed samples.
This motivated us to question the extent to which mitochondrial morphology per se affects neuronal function. Mec neurons such as PLM mediate the locomotion response to light touch (O'Hagan et al. 2005; Chen and Chalfie 2014). At day 3, tm1108 displayed a subtle, but not statistically significant touch defect (Fig. 4c), which achieved significance in day 10 mutants (Fig. 4e). As reported previously, T231E also exhibited an age-dependent functional decline in touch responsiveness (Fig. 4, c and e). Most interestingly, however, the T231E phenotype was additive to tm1108 at both day 3 and 10 (Fig. 4, c and e), suggesting that T231E and drp-1 may contribute to neuronal dysfunction through independent mechanisms.
We also examined the distribution and movement of GFP-labelled mitochondria in the PLM cell process (Fig. 4, f and g) via time-lapse fluorescent imaging. Fractional motility in tm1108 was significantly decreased at both day 3 and 10 (Fig. 4, h and l), and the decrease at day 10 was not additive to the decrease in T231E (Fig. 4l). Additionally, tm1108 exhibited increased inter mitochondrial distance (Fig. 4, l and m), and a reduction in both motile run length (Fig. 4, j and n) and speed (Fig. 4, k and o), regardless of T231E’s presence or absence. In short, tm1108 mitochondria in the cell body were dysmorphic, while in the nerve processes they were spaced further apart and did not move appreciably. The independent effect of T231E was comparatively small, making it difficult to interpret the non-additivity of mitochondrial parameters, given the disproportionate impact of tm1108. Nevertheless, it was striking that tm1108 could have such a profound influence on mitochondrial morphology and trafficking, but so little impact on touch sensitivity.
T231E selectively suppresses drp-1 independent mitophagy following oxidative stress
As reported previously (Guha, Fischer, et al. 2020), T231E had no impact on the baseline mitophagy index at either day 3 or day 10 (Fig. 5, a and b). In contrast, tm1108 exhibited a significant increase in the mitophagy index in both young and old animals, independent of T231E (Fig. 5, a and d). However, it is important to note that the extreme change in mitochondrial morphology could impact these calculations, rather than increased ML abundance per se.
Fig. 5.
The effect of T231E on baseline mitophagy and ML motility in a drp-1(tm1108) loss-of-function mutant. Mito-mKeima was used to measure mitophagy in the PLM soma and to visualize PLM neurite ML trafficking through dynamic fluorescent imaging. Drp-1 refers to the drp-1(tm1108) allele. a, d) Dual excitation fluorescence ratio imaging was used to derive a mitophagy index as a function of age, drp-1 genotype, and T231E, as indicated. Individual data points demarcate values from single PLM cells from separate animals (N = 40–58) collected by 2 independent researchers. b, c, e–h) Quantification of ML trafficking parameters (run length, speed, and motility) in the PLM cell neurites as a function of age, drp-1 genotype, and T231E, as indicated. Individual data points demarcate average values from single PLM cells from separate animals (N = 20–29). i) Cartoon illustrating the location of mitochondria (fibrils) and MLs (circles) in a neurite process. Drp-1 loss of function severely affects mitochondrial morphology and affects mitochondrial transport. The effects of T231E are independent and additive, leading to further loss of neuronal touch sensitivity. Data are the mean ± SD from 2 biological replicates performed on different days. Statistical analysis within day 3 and 10 datasets was by 1-way ANOVA with Tukey’s multiple comparison test, with ***P < 0.001, **P < 0.01, *P < 0.05.
As shown previously, fractional motility of ML was reduced by T231E, but somewhat surprisingly this reduction was partially suppressed by tm1108 at day 3, but not day 10 (Fig. 5, g and h). However, neither run length nor speed were impacted by tm1108, either in control or T231E (Fig. 5, b, c, e, f). While one might hypothesize that adaptive mechanisms in young tm1108 worms could mitigate the effects of T231E, it is difficult to reconcile the suppression of motility defects with the exacerbated touch response (Fig. 4c), if the 2 are truly related.
Finally, we measured oxidative-stress-induced mitophagy. As we have reported previously (Guha, Fischer, et al. 2020), PQT treatment increased the mitophagy index in the control at both day 3 and day 10 of adulthood (Fig. 6, a–c), and T231E suppressed this effect (Fig. 6, d–f). Mitophagy was likewise increased by PQT treatment in tm1108, and this increase was also suppressed by T231E (Fig. 6, d–f). To conclude, the drp-1 mutant mounted a robust mitophagy response to oxidative stress, despite the observed change in mitochondrial shape, which is consistent with mitochondrial fission occurring independent of the canonical Drp1 mediated pathway (Fig. 7a). In addition, the T231E phosphomimetic mutant tau continued to suppress oxidative stress-induced mitophagy in the drp-1(lf) background (Fig. 7b). This motivated us to test other potential genetic interactors.
Fig. 7.
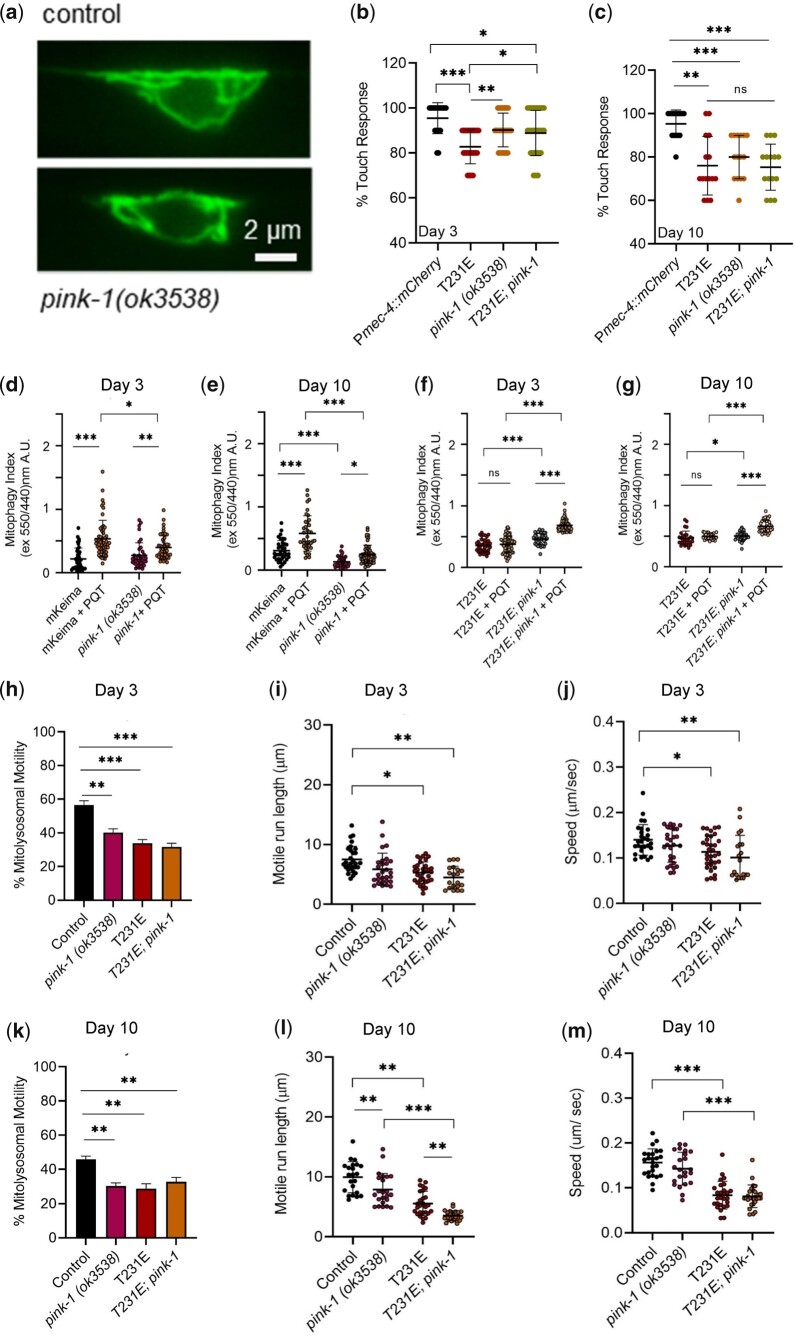
Tau T231E interacts with pink-1 to regulate mitophagy and behavior. a) Representative fluorescent images of mito-mKeima in day 3 control and pink-1(ok3538) animals. Scale bar: 2 µm. Mito-mKeima was also used to measure mitophagy in the PLM soma and to visualize PLM neurite ML trafficking through dynamic fluorescent imaging. Pink-1 refers to the ok3538 loss-of-function allele. b, c) Responsiveness to touch was plotted as a function of pink-1 and T231E at day 3 and 10 (N = 15–38 animals, touched 10 times apiece). d–g) Quantitative analysis of mitophagy using dual excitation ratio imaging of mito-mKeima as a function of age, pink-1 genotype, T231E, and PQT. Data are represented as the mean ± SD, with individual data points demarcating values from single PLM cells from separate animals (N = 30–65, from 2 biological replicates). h–m) Quantification of ML trafficking parameters (percentage motility, run length, and speed) in the distal PLM cell neurites at day 3 and 10 as a function of age, pink-1 genotype, and T231E, as indicated. Data are represented as the mean ± SD, with individual data points demarcating values from single PLM cells from separate animals (N = 21–34, from 2 biological replicates). Statistical analysis was by 1-way or 2-way ANOVA followed by Tukey’s post hoc test, with ***P < 0.001, **P < 0.01, and *P < 0.05 denoting significance when comparing bracketed samples.
pink-1 interacts genetically with T231E
PINK1 and Parkin work together to form one of the most recognizable mitochondrial damage recognition mechanisms, and together they support mitochondrial quality control (MQC) through the regulated induction of mitophagy (Goudarzi et al. 2021). In fact, mitophagy stimulation through PINK1/Parkin-dependent mechanisms has been shown to reduce tau and Aβ pathology in several AD models, including C. elegans (Fang, Hou, Palikaras, et al. 2019). Here, we examined mitophagy, ML motility, and the impact of T231E using a pink-1 loss-of-function mutant ok3538.
It has been reported that C. elegans pink-1 mutants accumulate dysfunctional mitochondria as they age (Cooper et al. 2017). While we observed that mitochondrial morphology was normal in ok3538 animals (Fig. 7a), the baseline mitophagy index was reduced in Day 10 worms compared with controls (Fig. 7e), a finding that was reproduced in pdr-1(gk488) mutants (data not shown), consistent with the previous observation. As might be predicted, the PQT-induced mitophagy index was also reduced in ok3538, but not eliminated entirely (Fig. 7, d and e). Strikingly, however, T231E was unable to suppress the smaller, but nevertheless significant PQT-induced mitophagy index increased observed in ok3538 (Fig. 7, f and g). This is consistent with induction of an alternate (and perhaps adaptive) mechanism that is refractory to the effect of T231E. Finally, contrary to our expectations, T231E actually suppressed the reduction in baseline mitophagy index in aged ok3538 (Fig. 7, d and e).
In the nerve processes, fractional ML motility in ok3538 phenocopied T231E (Fig. 7, h and k). In contrast, the speed of motile ML was not significantly different from controls in day 3 or day 10 ok3538 animals, whereas the motile run length was slightly reduced only in day 10 animals (Fig. 7, i and l). In the double mutants, T231E was the dominant effector (Fig. 7, j and m).
Finally, touch responsiveness suggests that ok3538 can partially suppress the detrimental effects of T231E in young animals (Fig. 7b)—coinciding with the appearance of a T231E resistant form of stress-induced mitophagy (Fig. 7f). However, the beneficial effect of ok3538 on T231E touch responsiveness was not recapitulated in older adults (Fig. 7c), consistent with the benefits of adaptation manifesting most strongly early in life (Supplementary Fig. 4d).
Overall, these data suggest that T231E interacts genetically with pink-1, which does not of course imply a physical interaction. However, the contrast between pink-1 and drp-1 mutants may be revealing. In ok3538, suppression of the T231E touch deficit aligns very well with its restoration of PQT-stimulated mitophagy—and strikingly does not align with neurite ML trafficking metrics. This is in comparison to tm1108, where mitochondria are relatively immobile, ML trafficking is partially restored, but the touch response deficit is exacerbated. We conclude that mitophagy appears to be of greater impact than organelle trafficking with regards to neurodegeneration, at least with respect to toxic T231E tau and in an organism that has very short neuronal processes (Supplementary Fig. 4).
Discussion
Tau accumulation is a cardinal feature of AD brain, with increasing PTMs as the disease progresses (Augustinack et al. 2002; Mi and Johnson 2006; Alquezar et al. 2020; Wesseling et al. 2020). While it is clear that tau is a necessary component of the pathologic pathways in AD (Roberson et al. 2007), the primary toxic entity is likely not the insoluble NFTs (Santacruz et al. 2005; Le Corre et al. 2006). Instead, toxicity is likely due to soluble or oligomeric forms of tau (Santacruz et al. 2005; Lasagna-Reeves et al. 2012; Ozcelik et al. 2016; Martinisi et al. 2021) that possess increased, disease-associated PTMs such as phosphorylation at specific residues that alter tau turnover and function (Alquezar et al. 2020; Guha, Johnson, et al. 2020). One such disease relevant residue is T231, where phosphorylation occurs early in the evolution of tau pathology (Augustinack et al. 2002; Luna-Muñoz et al. 2005; Schwalbe et al. 2015). Additionally, phosphorylation at this specific site precedes the formation of tau oligomers (Lasagna-Reeves et al. 2012), which likely contribute to tau toxicity (Shafiei et al. 2017), suggesting that this PTM may be, at least in part, causative for AD pathology. However, the precise cellular mechanism of how increased phosphorylation at this site negatively impacts neuronal health remains unclear.
The dynamic distribution of mitochondria via bidirectional trafficking in the neurite processes helps to maintain local energy demands in axons and dendrites (Vanhauwaert et al. 2019). Likewise, the turnover of damaged and dysfunctional mitochondria via mitophagy contributes to MQC and supports neuronal health (Sharma et al. 2019). These processes are facilitated by changes in mitochondrial size and shape, which are dynamically regulated through fusion and fission (Scott and Youle 2010; Dikov and Reichert 2011). Indeed, a major role of fission is to facilitate the clearance of dysfunctional mitochondria by mitophagy, and defective mitophagy is a prominent dysfunction in age-related diseases (Fivenson et al. 2017). Defective mitophagy likely occurs early in AD (Reddy and Oliver 2019), and this defect is thought to contribute to premature aging and neurodegeneration such as that observed in Werner’s syndrome patients (Fang, Hou, Lautrup, et al. 2019).
Another well-established role of fission is to create mitochondrial fragments that can be transported more easily along neuronal processes, and of course, tau is a mediator of microtubule dynamics (Baas and Qiang 2019). In fact, it has been shown that in hippocampal neurons, overexpression of a tau construct that mimics a caspase cleaved form significantly impairs mitochondrial transport in the neurites (Quintanilla et al. 2020). Indeed, many studies of tau, microtubules and trafficking have utilized tau overexpression models, and there is data that challenges the conclusion that tau disrupts neuronal transport mechanisms (Morfini et al. 2007; Yuan et al. 2008; Janning et al. 2014).
Here, we have demonstrated a defect in mitochondrial transport in our single copy T231E strain, consistent with many studies that have linked impaired mitochondrial transport with overexpression of tau (Ebneth et al. 1998; Trinczek et al. 1999), both in vitro and in vivo [for a review, see Cheng and Bai (2018)]. We have also shown that neurite transport of MLs is selectively and disproportionately impacted by T231E (Fig. 3). Indeed, our results suggest a relatively subtle effect on mitochondrial trafficking, and none at all on AP or lysosome trafficking (Fig. 2 and Supplementary Fig. 1). Hence, it is unclear that the exquisitely selective defect observed here in ML transport is comparable to the more wide-spread defects reported in the literature. It is interesting to speculate that defective mitochondria may prime adjacent lysosomes in a manner that allows them to be selectively recognized by transport machinery, as there is ample evidence supporting mitochondria-lysosome communication axes [for review, see Deus et al. (2020)].
More surprising to us was the observation that neither oxidative stress-induced mitophagy nor the deficit in ML trafficking responded appreciably to the loss of the fission mediator Drp1. Studies have shown that in AD patient brain samples, phospho-tau interacts with Drp1 (Manczak and Reddy 2012), which may lead to increased mitochondrial fragmentation and possibly contributing to cognitive decline (DuBoff et al. 2012; DuBoff et al. 2013). In C. elegans, drp-1(lf) is able to suppress certain forms of genetically induced mitochondrial fragmentation (Jiang et al. 2015), and very recent work has suggested that a reduction in mammalian Drp1 elevates mitophagy and may be beneficial in a symptomatic tau transgenic mice (Kandimalla et al. 2021), perhaps through a mechanism where APs remove larger-than-normal mitochondrial segments (Wrighton et al. 2021). While we did observe an increased mitophagy index at baseline in the drp-1(tm1108) mutants, it is important to appreciate that the severe alternation in tm1108 mitochondrial morphology could impact the baseline mitophagy index independent of actual mitophagy. We have confirmed this alternation using multiple sensors (Supplementary Fig. 3), which makes direct comparisons between tm1108 and control animals difficult.
Regardless of this, oxidative stress was capable of inducing robust levels of mitophagy in tm1108, which T231E suppressed entirely (Fig. 6). We interpret this to mean that mitophagy can be occur through at least 2 separate mechanisms: a canonical drp-1-dependent pathway and a second that is drp-1-independent, upregulated in response to oxidative stress, and suppressed by T231E (Supplementary Fig. 4). It is also important to note that the influence of drp-1 loss could occur through cell-nonautonomous or compensatory mechanisms, as there is good precedent for mitochondrial stress, which can be associated with changes in morphology, acting in this fashion (Durieux et al. 2011; Zhang et al. 2019; Haeussler et al. 2020).
We have also demonstrated that PQT-induced mitophagy occurs primarily, but not exclusively, through a pink-1 mediated process. Clear genetic interactions between T231E and ok3538 were observed for both touch responsiveness and mitophagy. It is intriguing that T231E suppressed decreased baseline mitophagy in aged ok3538, whereas ok3538 suppressed T231E’s effect on oxidative stress-induced mitophagy both early and later in life. Our data do not imply a direct mechanistic interaction, but should motivate a focused effort to examine whether T231E selectively impacts mitophagy through pink-1 and whether this interaction changes in response to age or the induction of compensatory mechanisms that support MQC. It is of specific interest to identify the mechanism through which PQT-induced mitophagy occurs in the pink-1 mutant, as it is refractory to the effect of T231E and would presumably represent an appropriate target for AD-centric therapeutic mitophagy stimulation.
Even with respect to mitophagy itself, there is precedent for alternative pathways compensating for the loss of the canonical pathway. Of particular interest are studies suggesting the importance of mitochondrial-derived vesicles (MDVs) (Soubannier, McLelland, et al. 2012), which can deliver cargo to the endolysosomal system through a Pink1/Parkin/Syntaxin 17-dependent pathway (McLelland et al. 2016), and may compensate for the loss of LC3-mediated mitophagy (Towers et al. 2021). Interestingly, MDV formation has been associated with oxidative stress (Soubannier, Rippstein, et al. 2012; Vasam et al. 2021).
Moving forward, it will be important to identify the molecular mechanism(s) through which oxidative stress-induced mitophagy occurs. Mitophagy signaling can involve multiple diverse recognition cues on the outer membrane, the most well-studied being the ubiquitin-dependent Pink1/Parkin pathway (Goudarzi et al. 2021), which we have examined here, as well as ubiquitin-independent mitophagy receptors including BNIP, NIX, and FUNDC1 (Narendra 2021), with the later implicated in hypoxic mitophagy in both mammals and worms (Liu et al. 2012; Lim et al. 2021). There is also emerging interest in what’s been termed “adaptive mitophagy” (Palikaras et al. 2018; Montava-Garriga and Ganley 2020), where proteins other than Drp1 have been shown to mediate mitochondrial removal. For example, Fis1 can regulate mitochondrial morphology independently of Drp1 (Onoue et al. 2013) and has been implicated in both homeostatic and adaptive signaling regiments that contribute to MQC through mitophagy (Shen et al. 2014; Pei et al. 2018; Xian et al. 2019; Chai et al. 2020). Additionally, femtosecond laser wounding triggers mitochondrial fragmentation in C. elegans and has also been shown to occur independent of DRP-1 (as well as other known regulators of mitochondrial morphology), and it instead requires the mitochondrial Rho GTPase MIRO-1 and cytosolic calcium signaling (Fu et al. 2020).
Finally, the mechanism by which tau T231E selectively suppresses mitophagy in a phosphomimetic-specific manner also remains to be established. One possible mechanism could be that phospho-tau inserts directly into the mitochondrial membrane, disrupting Pink1/Parkin-mediated mitophagy (Cummins et al. 2019; Fang, Hou, Palikaras, et al. 2019). Another possibility involves tau targeting one of the alternative receptors that function in stress-induced mitophagy. Whether the mechanism that impacts mitophagy is related to the ML motility deficit is similarly unknown, but it is intriguing to speculate that these may be conjoined at the level of mitochondrial–lysosomal communication. Moving forward, we anticipate being able to use our single-copy model to provide a genetic perspective on how tau modification impacts the molecular mechanisms through which mitochondria respond to stress in the context of aging and to further decipher how these processes contribute to neurodegeneration in AD.
Conclusions
It is apparent that tau toxicity in our single-copy gene insertion model is selective for the phosphomimetic T231E mutation. It is likewise apparent that there is additional selectivity for acidic organelles that contain damaged mitochondria, and that these organelles are derived from mitochondria by non-canonical processing that does not involve typical DRP-1 mediated fission, but does involve PINK1. Moving forward, we anticipate being able to use our model to provide a genetic perspective on how tau modification impacts the specific molecular mechanisms through which mitochondria respond to stress in the context of aging and to further decipher how these processes contribute to neurodegeneration in AD.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplementary Table 2 shows the means, SD, N-values, and biological replicate information for all data that were currently represented in graphical format.
Supplemental material is available at GENETICS online.
Supplementary Material
Acknowledgments
Technical assistance provided by Thomas Delgado, Duy Doan, and Ryunosuke Wajima was greatly appreciated. We thank all members of Dr Johnson’s lab, the Mitochondrial Research and Interest Group at the University of Rochester Medical Center, and members of the Western New York Worm Group for their valuable suggestions and helpful discussions. Authors would like to acknowledge Bio-render for providing an online paid subscription platform to create all the figures. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Funding
Research reported in this manuscript was supported by grants from the National Institute on Aging, National Institute of Neurological Disorders and Stroke, and National Institute of General Medical Sciences of the National Institutes of Health, including R01AG067617 (KN, GVWJ, SG, TC, AC), R01NS115906 (SAK), and F31NS117034 and R01GM135326 (SS).
Conflicts of interest
None declared.
Author contributions
SG, GVWJ, and KN conceived and designed the experiments. SG, AC, TC, DK, SAK, and SS generated reagents or performed the experiments. SG, AC, TC, DK, and SAK analyzed the data. SG, TC, GVWJ, and KN wrote the paper. All authors read, edited, and approved the final manuscript.
Contributor Information
Sanjib Guha, Department of Anesthesiology & Perioperative Medicine, University of Rochester, Rochester, NY 14642, USA.
Anson Cheng, Department of Anesthesiology & Perioperative Medicine, University of Rochester, Rochester, NY 14642, USA.
Trae Carroll, Department of Pathology and Laboratory Medicine, University of Rochester, Rochester, NY 14642, USA.
Dennisha King, Department of Neuroscience, University of Rochester, Rochester, NY 14642, USA.
Shon A Koren, Department of Anesthesiology & Perioperative Medicine, University of Rochester, Rochester, NY 14642, USA.
Sierra Swords, Department of Molecular Biology and Biochemistry, Rutgers University, New Brunswick, NJ 08901, USA.
Keith Nehrke, Department of Medicine, Nephrology Division, University of Rochester, Rochester, NY 14642, USA.
Gail V W Johnson, Department of Anesthesiology & Perioperative Medicine, University of Rochester, Rochester, NY 14642, USA.
Literature cited
- Alirezaei M, Kemball CC, Flynn CT, Wood MR, Whitton JL, Kiosses WB.. Short-term fasting induces profound neuronal autophagy. Autophagy. 2010;6(6):702–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso AD, Di Clerico J, Li B, Corbo CP, Alaniz ME, Grundke-Iqbal I, Iqbal K.. Phosphorylation of tau at Thr212, Thr231, and Ser262 combined causes neurodegeneration. J Biol Chem. 2010;285(40):30851–30860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alquezar C, Arya S, Kao AW.. Tau post-translational modifications: dynamic transformers of tau function, degradation, and aggregation. Front Neurol. 2020;11:595532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragão Gomes L, Uytterhoeven V, Lopez-Sanmartin D, Tomé SO, Tousseyn T, Vandenberghe R, Vandenbulcke M, von Arnim CAF, Verstreken P, Thal DR, et al. Maturation of neuronal AD-tau pathology involves site-specific phosphorylation of cytoplasmic and synaptic tau preceding conformational change and fibril formation. Acta Neuropathol. 2021;141(2):173–192. [DOI] [PubMed] [Google Scholar]
- Arnsten AFT, Datta D, Del Tredici K, Braak H.. Hypothesis: tau pathology is an initiating factor in sporadic Alzheimer's disease. Alzheimers Dement. 2021;17(1):115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustinack JC, Schneider A, Mandelkow EM, Hyman BT.. Specific tau phosphorylation sites correlate with severity of neuronal cytopathology in Alzheimer's disease. Acta Neuropathol. 2002;103(1):26–35. [DOI] [PubMed] [Google Scholar]
- Baas PW, Qiang L.. Tau: it’s not what you think. Trends Cell Biol. 2019;29(6):452–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bera A, Lavanya G, Reshmi R, Dev K, Kumar R.. Mechanistic and therapeutic role of Drp1 in the pathogenesis of Alzheimer's disease. Eur J Neurosci. 2022. doi: 10.1111/ejn.15611. [DOI] [PubMed] [Google Scholar]
- Bess AS, Crocker TL, Ryde IT, Meyer JN.. Mitochondrial dynamics and autophagy aid in removal of persistent mitochondrial DNA damage in Caenorhabditis elegans. Nucleic Acids Res. 2012;40(16):7916–7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Oliveira NK, Savitt AG, Silva VKA, Krausert RB, Ghebrehiwet B, Fries BC.. Low glucose mediated fluconazole tolerance in Cryptococcus neoformans. J Fungi (Basel). 2021;7:489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordi M, Berg MJ, Mohan PS, Peterhoff CM, Alldred MJ, Che S, Ginsberg SD, Nixon RA.. Autophagy flux in CA1 neurons of Alzheimer hippocampus: increased induction overburdens failing lysosomes to propel neuritic dystrophy. Autophagy. 2016;12(12):2467–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt R, Gergou A, Wacker I, Fath T, Hutter H.. A Caenorhabditis elegans model of tau hyperphosphorylation: induction of developmental defects by transgenic overexpression of Alzheimer's disease-like modified tau. Neurobiol Aging. 2009;30(1):22–33. [DOI] [PubMed] [Google Scholar]
- Breckenridge DG, Kang B-H, Kokel D, Mitani S, Staehelin LA, Xue D.. Caenorhabditis elegans drp-1 and fis-2 regulate distinct cell-death execution pathways downstream of ced-3 and independent of ced-9. Mol Cell. 2008;31(4):586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne JJ, Soh MS, Chandhok G, Vijayaraghavan T, Teoh J-S, Crawford S, Cobham AE, Yapa NMB, Mirth CK, Neumann B, et al. Disruption of mitochondrial dynamics affects behaviour and lifespan in Caenorhabditis elegans. Cell Mol Life Sci. 2019;76(10):1967–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll T, Guha S, Nehrke K, Johnson GVW.. Tau post-translational modifications: potentiators of selective vulnerability in sporadic Alzheimer's disease. Biology (Basel). 2021;10(10):1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai N, Haney MS, Couthouis J, Morgens DW, Benjamin A, Wu K, Ousey J, Fang S, Finer S, Bassik MC, et al. Genome-wide synthetic lethal CRISPR screen identifies FIS1 as a genetic interactor of ALS-linked C9ORF72. Brain Res. 2020;1728:146601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Kumsta C, Hellman AB, Adams LM, Hansen M.. Spatiotemporal regulation of autophagy during Caenorhabditis elegans aging. Elife. 2017;6:e18459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J, Bains Y, Guha S, Kahn A, Hall D, Bose N, Gugliucci A, Kapahi P.. The role of advanced glycation end products in aging and metabolic diseases: bridging association and causality. Cell Metab. 2018;28(3):337–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee F, Mudher A, Newman TA, Cuttle M, Lovestone S, Shepherd D.. Overexpression of tau results in defective synaptic transmission in Drosophila neuromuscular junctions. Biochem Soc Trans. 2006;34(Pt 1):88–90. [DOI] [PubMed] [Google Scholar]
- Chen H, Chan DC.. Mitochondrial dynamics–fusion, fission, movement, and mitophagy–in neurodegenerative diseases. Hum Mol Genet. 2009;18(R2):R169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Fang H, Shao X, Tian Z, Geng S, Zhang Y, Fan H, Xiang P, Zhang J, Tian X, et al. A dual-labeling probe to track functional mitochondria-lysosome interactions in live cells. Nat Commun. 2020;11(1):6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Chalfie M.. Modulation of C. elegans touch sensitivity is integrated at multiple levels. J Neurosci. 2014;34(19):6522–6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Bai F.. The association of tau with mitochondrial dysfunction in Alzheimer's disease. Front Neurosci. 2018;12:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikka MR, Anbalagan C, Dvorak K, Dombeck K, Prahlad V.. The mitochondria-regulated immune pathway activated in the C. elegans intestine is neuroprotective. Cell Rep. 2016;16(9):2399–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochemé HM, Murphy MP.. Complex I is the major site of mitochondrial superoxide production by paraquat. J Biol Chem. 2008;283(4):1786–1798. [DOI] [PubMed] [Google Scholar]
- Cooper JF, Machiela E, Dues DJ, Spielbauer KK, Senchuk MM, Van Raamsdonk JM.. Activation of the mitochondrial unfolded protein response promotes longevity and dopamine neuron survival in Parkinson's disease models. Sci Rep. 2017;7(1):16441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppa A, Guha S, Fourcade S, Parameswaran J, Ruiz M, Moser AB, Schlüter A, Murphy MP, Lizcano JM, Miranda-Vizuete A, et al. The peroxisomal fatty acid transporter ABCD1/PMP-4 is required in the C. elegans hypodermis for axonal maintenance: a worm model for adrenoleukodystrophy. Free Radic Biol Med. 2020;152:797–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins N, Tweedie A, Zuryn S, Bertran-Gonzalez J, Götz J.. Disease-associated tau impairs mitophagy by inhibiting Parkin translocation to mitochondria. EMBO J. 2019;38(3):e99360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagda RK, Cherra SJ, Kulich SM, Tandon A, Park D, Chu CT.. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009;284(20):13843–13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David DC, Hauptmann S, Scherping I, Schuessel K, Keil U, Rizzu P, Ravid R, Dröse S, Brandt U, Müller WE, et al. Proteomic and functional analyses reveal a mitochondrial dysfunction in P301L tau transgenic mice. J Biol Chem. 2005;280(25):23802–23814. [DOI] [PubMed] [Google Scholar]
- DeTure MA, Dickson DW.. The neuropathological diagnosis of Alzheimer's disease. Mol Neurodegener. 2019;14(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deus CM, Yambire KF, Oliveira PJ, Raimundo N.. Mitochondria-lysosome crosstalk: from physiology to neurodegeneration. Trends Mol Med. 2020;26(1):71–88. [DOI] [PubMed] [Google Scholar]
- Dikov D, Reichert AS.. How to split up: lessons from mitochondria. EMBO J. 2011;30(14):2751–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Matthews TA, Johnson GV.. Site-specific phosphorylation and caspase cleavage differentially impact tau-microtubule interactions and tau aggregation. J Biol Chem. 2006;281(28):19107–19114. [DOI] [PubMed] [Google Scholar]
- DuBoff B, Feany M, Gotz J.. Why size matters - balancing mitochondrial dynamics in Alzheimer's disease. Trends Neurosci. 2013;36(6):325–335. [DOI] [PubMed] [Google Scholar]
- DuBoff B, Gotz J, Feany MB.. Tau promotes neurodegeneration via DRP1 mislocalization in vivo. Neuron. 2012;75(4):618–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A.. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144(1):79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebneth A, Godemann R, Stamer K, Illenberger S, Trinczek B, Mandelkow E.. Overexpression of tau protein inhibits kinesin-dependent trafficking of vesicles, mitochondria, and endoplasmic reticulum: implications for Alzheimer's disease. J Cell Biol. 1998;143(3):777–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Hou Y, Lautrup S, Jensen MB, Yang B, SenGupta T, Caponio D, Khezri R, Demarest TG, Aman Y, et al. NAD(+) augmentation restores mitophagy and limits accelerated aging in Werner syndrome. Nat Commun. 2019;10(1):5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Hou Y, Palikaras K, Adriaanse BA, Kerr JS, Yang B, Lautrup S, Hasan-Olive MM, Caponio D, Dan X, et al. Mitophagy inhibits amyloid-beta and tau pathology and reverses cognitive deficits in models of Alzheimer's disease. Nat Neurosci. 2019;22(3):401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatouros C, Pir GJ, Biernat J, Koushika SP, Mandelkow E, Mandelkow E-M, Schmidt E, Baumeister R.. Inhibition of tau aggregation in a novel Caenorhabditis elegans model of tauopathy mitigates proteotoxicity. Hum Mol Genet. 2012;21(16):3587–3603. [DOI] [PubMed] [Google Scholar]
- Fernandez-Mosquera L, Yambire KF, Couto R, Pereyra L, Pabis K, Ponsford AH, Diogo CV, Stagi M, Milosevic I, Raimundo N, et al. Mitochondrial respiratory chain deficiency inhibits lysosomal hydrolysis. Autophagy. 2019;15(9):1572–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fivenson EM, Lautrup S, Sun N, Scheibye-Knudsen M, Stevnsner T, Nilsen H, Bohr VA, Fang EF.. Mitophagy in neurodegeneration and aging. Neurochem Int. 2017;109:202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannery PJ, Trushina E.. Mitochondrial dynamics and transport in Alzheimer's disease. Mol Cell Neurosci. 2019;98:109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flippo KH, Strack S.. Mitochondrial dynamics in neuronal injury, development and plasticity. J Cell Sci. 2017;130(4):671–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C, Davis MW, Sarov M, Taylor J, Flibotte S, LaBella M, Pozniakovsky A, Moerman DG, Jorgensen EM.. Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat Methods. 2014;11(5):529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Zhou H, Yu X, Xu J, Zhou J, Meng X, Zhao J, Zhou Y, Chisholm AD, Xu S, et al. Wounding triggers MIRO-1 dependent mitochondrial fragmentation that accelerates epidermal wound closure through oxidative signaling. Nat Commun. 2020;11(1):1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo MF, Ikuta I, Zhu X, Casadesus G, Jordán J.. Mitochondrial biology in Alzheimer's disease pathogenesis. J Neurochem. 2010;114(4):933–945. [DOI] [PubMed] [Google Scholar]
- Goudarzi S, Hosseini A, Abdollahi M, Haghi-Aminjan H.. Insights into Parkin-mediated mitophagy in Alzheimer's disease: a systematic review. Front Aging Neurosci. 2021;13(674071):674071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha S, Fischer S, Johnson GVW, Nehrke K.. Tauopathy-associated tau modifications selectively impact neurodegeneration and mitophagy in a novel C. elegans single-copy transgenic model. Mol Neurodegener. 2020;15:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha S, Johnson GVW, Nehrke K.. The crosstalk between pathological tau phosphorylation and mitochondrial dysfunction as a key to understanding and treating Alzheimer's disease. Mol Neurobiol. 2020;57(12):5103–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler S, Köhler F, Witting M, Premm MF, Rolland SG, Fischer C, Chauve L, Casanueva O, Conradt B.. Autophagy compensates for defects in mitochondrial dynamics. PLoS Genet. 2020;16(3):e1008638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamacher-Brady A, Brady NR.. Mitophagy programs: mechanisms and physiological implications of mitochondrial targeting by autophagy. Cell Mol Life Sci. 2016;73(4):775–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O, Tessmar K, Ruvkun G.. The Caenorhabditis elegans lim-6 LIM homeobox gene regulates neurite outgrowth and function of particular GABAergic neurons. Development (Cambridge, England). 1999;126(7):1547–1562. [DOI] [PubMed] [Google Scholar]
- Hou Y, Dan X, Babbar M, Wei Y, Hasselbalch SG, Croteau DL, Bohr VA.. Ageing as a risk factor for neurodegenerative disease. Nat Rev Neurol. 2019;15(10):565–581. [DOI] [PubMed] [Google Scholar]
- Hu H, Tan CC, Tan L, Yu JT.. A mitocentric view of Alzheimer's disease. Mol Neurobiol. 2017;54(8):6046–6060. [DOI] [PubMed] [Google Scholar]
- Imoto M, Tachibana I, Urrutia R.. Identification and functional characterization of a novel human protein highly related to the yeast dynamin-like GTPase Vps1p. J Cell Sci. 1998;111(10):1341–1349. [DOI] [PubMed] [Google Scholar]
- Janning D, Igaev M, Sündermann F, Brühmann J, Beutel O, Heinisch JJ, Bakota L, Piehler J, Junge W, Brandt R, et al. Single-molecule tracking of tau reveals fast kiss-and-hop interaction with microtubules in living neurons. Mol Biol Cell. 2014;25(22):3541–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H-C, Hsu J-M, Yen C-P, Chao C-C, Chen R-H, Pan C-L.. Neural activity and CaMKII protect mitochondria from fragmentation in aging Caenorhabditis elegans neurons. Proc Natl Acad Sci. 2015;112(28):8768–8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang XJ, Wu YQ, Ma R, Chang YM, Li LL, Zhu JH, Liu GP, Li G.. PINK1 alleviates cognitive impairments via attenuating pathological tau aggregation in a mouse model of tauopathy. Front Cell Dev Biol. 2021;9:736267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandimalla R, Manczak M, Pradeepkiran JA, Reddy PH. A partial reduction of Drp1 enhances mitophagy, autophagy, mitochondrial biogenesis, dendritic spines and synaptic activity in a transgenic Tau mouse model of Alzheimer disease . Human Mol Genet. 2022;31(11):1788–1805. [DOI] [PMC free article] [PubMed]
- Katayama H, Kogure T, Mizushima N, Yoshimori T, Miyawaki A.. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem Biol. 2011;18(8):1042–1052. [DOI] [PubMed] [Google Scholar]
- Kogure T, Karasawa S, Araki T, Saito K, Kinjo M, Miyawaki A.. A fluorescent variant of a protein from the stony coral Montipora facilitates dual-color single-laser fluorescence cross-correlation spectroscopy. Nat Biotechnol. 2006;24(5):577–581. [DOI] [PubMed] [Google Scholar]
- Kosmidis S, Grammenoudi S, Papanikolopoulou K, Skoulakis EMC.. Differential effects of Tau on the integrity and function of neurons essential for learning in Drosophila. J Neurosci. 2010;30(2):464–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer BC, Zhang B, Leverenz JB, Thomas JH, Trojanowski JQ, Schellenberg GD.. Neurodegeneration and defective neurotransmission in a Caenorhabditis elegans model of tauopathy. Proc Natl Acad Sci U S A. 2003;100(17):9980–9985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM.. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol Cell. 1999;4(5):815–826. [DOI] [PubMed] [Google Scholar]
- Lasagna-Reeves CA, Castillo-Carranza DL, Sengupta U, Sarmiento J, Troncoso J, Jackson GR, Kayed R.. Identification of oligomers at early stages of tau aggregation in Alzheimer's disease. FASEB J. 2012;26(5):1946–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Corre S, Klafki HW, Plesnila N, Hübinger G, Obermeier A, Sahagún H, Monse B, Seneci P, Lewis J, Eriksen J, et al. An inhibitor of tau hyperphosphorylation prevents severe motor impairments in tau transgenic mice. Proc Natl Acad Sci USA. 2006;103(25):9673–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-C, Hu Y, Wang Z-h, Luo Y, Zhang Y, Liu X-P, Feng Q, Wang Q, Ye K, Liu G-P, et al. Human wild-type full-length tau accumulation disrupts mitochondrial dynamics and the functions via increasing mitofusins. Sci Rep. 2016;6:24756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y, Berry B, Viteri S, McCall M, Park EC, Rongo C, Brookes PS, Nehrke K.. FNDC-1-mediated mitophagy and ATFS-1 coordinate to protect against hypoxia-reoxygenation. Autophagy. 2021;17(11):3389–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MY, Sheng ZH.. Regulation of mitochondrial transport in neurons. Exp Cell Res. 2015;334(1):35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14(2):177–185. [DOI] [PubMed] [Google Scholar]
- Luna-Muñoz J, García-Sierra F, Falcón V, Menéndez I, Chávez-Macías L, Mena R.. Regional conformational change involving phosphorylation of tau protein at the Thr231, precedes the structural change detected by Alz-50 antibody in Alzheimer's disease. J Alzheimer's Dis. 2005;8(1):29–41. [DOI] [PubMed] [Google Scholar]
- Malpartida AB, Williamson M, Narendra DP, Wade-Martins R, Ryan BJ.. Mitochondrial dysfunction and mitophagy in Parkinson's disease: from mechanism to therapy. Trends Biochem Sci. 2021;46(4):329–343. [DOI] [PubMed] [Google Scholar]
- Manczak M, Calkins MJ, Reddy PH.. Impaired mitochondrial dynamics and abnormal interaction of amyloid beta with mitochondrial protein Drp1 in neurons from patients with Alzheimer's disease: implications for neuronal damage. Hum Mol Genet. 2011;20(13):2495–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manczak M, Reddy PH.. Abnormal interaction between the mitochondrial fission protein Drp1 and hyperphosphorylated tau in Alzheimer's disease neurons: implications for mitochondrial dysfunction and neuronal damage. Hum Mol Genet. 2012;21(11):2538–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M. Neuronal mitophagy in neurodegenerative diseases. Front Mol Neurosci. 2017;10:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinisi A, Flach M, Sprenger F, Frank S, Tolnay M, Winkler DT.. Severe oligomeric tau toxicity can be reversed without long-term sequelae. Brain. 2021;144(3):963–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Gleichmann M, Cheng A.. Mitochondria in neuroplasticity and neurological disorders. Neuron. 2008;60(5):748–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLelland GL, Lee SA, McBride HM, Fon EA.. Syntaxin-17 delivers PINK1/parkin-dependent mitochondrial vesicles to the endolysosomal system. J Cell Biol. 2016;214(3):275–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies FM, Fleming A, Rubinsztein DC.. Compromised autophagy and neurodegenerative diseases. Nat Rev Neurosci. 2015;16(6):345–357. [DOI] [PubMed] [Google Scholar]
- Mi K, Johnson GV.. The role of tau phosphorylation in the pathogenesis of Alzheimer's disease. Curr Alzheimer Res. 2006;3(5):449–463. [DOI] [PubMed] [Google Scholar]
- Miyasaka T, Shinzaki Y, Yoshimura S, Yoshina S, Kage-Nakadai E, Mitani S, Ihara Y.. Imbalanced expression of tau and tubulin induces neuronal dysfunction in C. elegans models of tauopathy. Front Neurosci. 2018;12:415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal S, Ahlawat S, Koushika SP.. Simple microfluidic devices for in vivo imaging of C. elegans, Drosophila and zebrafish. J Vis Exp. 2012;67:3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montava-Garriga L, Ganley IG.. Outstanding questions in mitophagy: what we do and do not know. J Mol Biol. 2020;432(1):206–230. [DOI] [PubMed] [Google Scholar]
- Morfini G, Pigino G, Mizuno N, Kikkawa M, Brady ST.. Tau binding to microtubules does not directly affect microtubule-based vesicle motility. J Neurosci Res. 2007;85(12):2620–2630. [DOI] [PubMed] [Google Scholar]
- Morsci NS, Hall DH, Driscoll M, Sheng ZH.. Age-related phasic patterns of mitochondrial maintenance in adult Caenorhabditis elegans neurons. J Neurosci. 2016;36(4):1373–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MP, Hartley RC.. Mitochondria as a therapeutic target for common pathologies. Nat Rev Drug Discov. 2018;17(12):865–886. [DOI] [PubMed] [Google Scholar]
- Narendra DP. Managing risky assets—mitophagy in vivo. J Cell Sci. 2021;134:jcs240465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neddens J, Temmel M, Flunkert S, Kerschbaumer B, Hoeller C, Loeffler T, Niederkofler V, Daum G, Attems J, Hutter-Paier B, et al. Phosphorylation of different tau sites during progression of Alzheimer's disease. Acta Neuropathol Commun. 2018;6(1):52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM.. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64(2):113–122. [DOI] [PubMed] [Google Scholar]
- O'Hagan R, Chalfie M, Goodman MB.. The MEC-4 DEG/ENaC channel of Caenorhabditis elegans touch receptor neurons transduces mechanical signals. Nat Neurosci. 2005;8(1):43–50. [DOI] [PubMed] [Google Scholar]
- Onoue K, Jofuku A, Ban-Ishihara R, Ishihara T, Maeda M, Koshiba T, Itoh T, Fukuda M, Otera H, Oka T, et al. Fis1 acts as a mitochondrial recruitment factor for TBC1D15 that is involved in regulation of mitochondrial morphology. J Cell Sci. 2013;126(Pt 1):176–185. [DOI] [PubMed] [Google Scholar]
- Ozcelik S, Sprenger F, Skachokova Z, Fraser G, Abramowski D, Clavaguera F, Probst A, Frank S, Müller M, Staufenbiel M, et al. Co-expression of truncated and full-length tau induces severe neurotoxicity. Mol Psychiatry. 2016;21(12):1790–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palikaras K, Lionaki E, Tavernarakis N.. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat Cell Biol. 2018;20(9):1013–1022. [DOI] [PubMed] [Google Scholar]
- Pei S, Minhajuddin M, Adane B, Khan N, Stevens BM, Mack SC, Lai S, Rich JN, Inguva A, Shannon KM, et al. AMPK/FIS1-mediated mitophagy is required for self-renewal of human AML stem cells. Cell Stem Cell. 2018;23(1):86–100.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez MJ, Jara C, Quintanilla RA.. Contribution of Tau pathology to mitochondrial impairment in neurodegeneration. Front Neurosci. 2018;12:441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernas L, Scorrano L.. Mito-morphosis: mitochondrial fusion, fission, and cristae remodeling as key mediators of cellular function. Annu Rev Physiol. 2016;78:505–531. [DOI] [PubMed] [Google Scholar]
- Pickrell AM, Youle RJ.. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron. 2015;85(2):257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang L, Sun X, Austin TO, Muralidharan H, Jean DC, Liu M, Yu W, Baas PW.. Tau does not stabilize axonal microtubules but rather enables them to have long labile domains. Curr Biol. 2018;28(13):2181–2189.e4. [DOI] [PubMed] [Google Scholar]
- Quintanilla RA, Dolan PJ, Jin YN, Johnson GV.. Truncated tau and Aβ cooperatively impair mitochondria in primary neurons. Neurobiol Aging. 2012;33:619.e625–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintanilla RA, Tapia-Monsalves C, Vergara EH, Pérez MJ, Aranguiz A.. Truncated tau induces mitochondrial transport failure through the impairment of TRAK2 protein and bioenergetics decline in neuronal cells. Front Cell Neurosci. 2020;14:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PH, Oliver DM.. Amyloid beta and phosphorylated tau-induced defective autophagy and mitophagy in Alzheimer's disease. Cells. 2019;8(5):488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PH, Tripathi R, Troung Q, Tirumala K, Reddy TP, Anekonda V, Shirendeb UP, Calkins MJ, Reddy AP, Mao P, et al. Abnormal mitochondrial dynamics and synaptic degeneration as early events in Alzheimer's disease: implications to mitochondria-targeted antioxidant therapeutics. Biochim Biophys Acta. 2012;1822(5):639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson ED, Scearce-Levie K, Palop JJ, Yan F, Cheng IH, Wu T, Gerstein H, Yu G-Q, Mucke L.. Reducing endogenous tau ameliorates amyloid beta-induced deficits in an Alzheimer's disease mouse model. Science (New York, N.Y.). 2007;316(5825):750–754. [DOI] [PubMed] [Google Scholar]
- Rooney JP, Luz AL, González-Hunt CP, Bodhicharla R, Ryde IT, Anbalagan C, Meyer JN.. Effects of 5'-fluoro-2-deoxyuridine on mitochondrial biology in Caenorhabditis elegans. Exp Gerontol. 2014;56:69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safiulina D, Kaasik A.. Energetic and dynamic: how mitochondria meet neuronal energy demands. PLoS Biol. 2013;11(12):e1001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sämann J, Hegermann J, von Gromoff E, Eimer S, Baumeister R, Schmidt E.. Caenorhabditits elegans LRK-1 and PINK-1 act antagonistically in stress response and neurite outgrowth. J Biol Chem. 2009;284(24):16482–16491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santacruz K, Lewis J, Spires T, Paulson J, Kotilinek L, Ingelsson M, Guimaraes A, DeTure M, Ramsden M, McGowan E, et al. Tau suppression in a neurodegenerative mouse model improves memory function. Science (New York, N.Y.). 2005;309(5733):476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalbe M, Kadavath H, Biernat J, Ozenne V, Blackledge M, Mandelkow E, Zweckstetter M.. structural impact of tau phosphorylation at threonine 231. Structure. 2015;23(8):1448–1458. [DOI] [PubMed] [Google Scholar]
- Scott I, Youle RJ.. Mitochondrial fission and fusion. Essays Biochem. 2010;47:85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta A, Kabat J, Novak M, Wu Q, Grundke-Iqbal I, Iqbal K.. Phosphorylation of tau at both Thr 231 and Ser 262 is required for maximal inhibition of its binding to microtubules. Arch Biochem Biophys. 1998;357(2):299–309. [DOI] [PubMed] [Google Scholar]
- Shafiei SS, Guerrero-Munoz MJ, Castillo-Carranza DL.. Tau oligomers: cytotoxicity, propagation, and mitochondrial damage. Front Aging Neurosci. 2017;9:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Smith HJ, Yao P, Mair WB.. Causal roles of mitochondrial dynamics in longevity and healthy aging. EMBO Rep. 2019;20(12):e48395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Yamano K, Head BP, Kawajiri S, Cheung JTM, Wang C, Cho J-H, Hattori N, Youle RJ, van der Bliek AM, et al. Mutations in Fis1 disrupt orderly disposal of defective mitochondria. Mol Biol Cell. 2014;25(1):145–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng B, Wang X, Su B, Lee H-g, Casadesus G, Perry G, Zhu X.. Impaired mitochondrial biogenesis contributes to mitochondrial dysfunction in Alzheimer's disease. J Neurochem. 2012;120(3):419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM.. A human dynamin-related protein controls the distribution of mitochondria. J Cell Biol. 1998;143(2):351–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubannier V, McLelland G-L, Zunino R, Braschi E, Rippstein P, Fon EA, McBride HM.. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr Biol. 2012;22(2):135–141. [DOI] [PubMed] [Google Scholar]
- Soubannier V, Rippstein P, Kaufman BA, Shoubridge EA, McBride HM.. Reconstitution of mitochondria derived vesicle formation demonstrates selective enrichment of oxidized cargo. PLoS One. 2012;7(12):e52830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhilb ML, Dias-Santagata D, Fulga TA, Felch DL, Feany MB.. Tau phosphorylation sites work in concert to promote neurotoxicity in vivo. Mol Biol Cell. 2007;18(12):5060–5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T. Maintenance of C. elegans. WormBook; 2006. p. 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Malide D, Liu J, Rovira II, Combs CA, Finkel T.. A fluorescence-based imaging method to measure in vitro and in vivo mitophagy using mt-Keima. Nat Protoc. 2017;12(8):1576–1587. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Akamatsu W, Kisa F, Sone T, Ishikawa K-I, Kuzumaki N, Katayama H, Miyawaki A, Hattori N, Okano H, et al. Efficient induction of dopaminergic neuron differentiation from induced pluripotent stem cells reveals impaired mitophagy in PARK2 neurons. Biochem Biophys Res Commun. 2017;483(1):88–93. [DOI] [PubMed] [Google Scholar]
- Swerdlow RH. Mitochondria and mitochondrial Cascades in Alzheimer's disease. J Alzheimers Dis. 2018;62(3):1403–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takihara Y, Inatani M, Eto K, Inoue T, Kreymerman A, Miyake S, Ueno S, Nagaya M, Nakanishi A, Iwao K, et al. In vivo imaging of axonal transport of mitochondria in the diseased and aged mammalian CNS. Proc Natl Acad Sci USA. 2015;112(33):10515–10520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers CG, Wodetzki DK, Thorburn J, Smith KR, Caino MC, Thorburn A.. Mitochondrial-derived vesicles compensate for loss of LC3-mediated mitophagy. Dev Cell. 2021;56(14):2029–2042.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traa A, Machiela E, Rudich PD, Soo SK, Senchuk MM, Van Raamsdonk JM.. Identification of novel therapeutic targets for polyglutamine diseases that target mitochondrial fragmentation. Int J Mol Sci. 2021;22:13447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinczek B, Ebneth A, Mandelkow EM, Mandelkow E.. Tau regulates the attachment/detachment but not the speed of motors in microtubule-dependent transport of single vesicles and organelles. J Cell Sci. 1999;112(14):2355–2367. [DOI] [PubMed] [Google Scholar]
- Vanhauwaert R, Bharat V, Wang X.. Surveillance and transportation of mitochondria in neurons. Curr Opin Neurobiol. 2019;57:87–93. [DOI] [PubMed] [Google Scholar]
- Vasam G, Nadeau R, Cadete VJJ, Lavallée-Adam M, Menzies KJ, Burelle Y.. Proteomics characterization of mitochondrial-derived vesicles under oxidative stress. FASEB J. 2021;35(4):e21278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi J, Zhang Z, Wakabayashi N, Tamura Y, Fukaya M, Kensler TW, Iijima M, Sesaki H.. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol. 2009;186(6):805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidling I, Swerdlow RH.. Mitochondrial dysfunction and stress responses in Alzheimer's disease. Biology. 2019;8(2):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesseling H, Mair W, Kumar M, Schlaffner CN, Tang S, Beerepoot P, Fatou B, Guise AJ, Cheng L, Takeda S, et al. Tau PTM profiles identify patient heterogeneity and stages of Alzheimer's disease. Cell. 2020;183(6):1699–1713.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton PJ, Shwartz A, Heo JM, Quenzer ED, LaBella KA, Harper JW, Goessling W.. Quantitative intravital imaging in zebrafish reveals in vivo dynamics of physiological-stress-induced mitophagy. J Cell Sci. 2021;134:jcs256255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian H, Yang Q, Xiao L, Shen HM, Liou YC.. STX17 dynamically regulated by Fis1 induces mitophagy via hierarchical macroautophagic mechanism. Nat Commun. 2019;10(1):2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Propson NE, Du S, Xiong W, Zheng H.. Autophagy deficiency modulates microglial lipid homeostasis and aggravates tau pathology and spreading. Proc Natl Acad Sci USA. 2021;118:e2023418118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama Y, Higuchi M, Zhang B, Huang S-M, Iwata N, Saido TC, Maeda J, Suhara T, Trojanowski JQ, Lee VM-Y, et al. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron. 2007;53(3):337–351. [DOI] [PubMed] [Google Scholar]
- Yuan A, Kumar A, Peterhoff C, Duff K, Nixon RA.. Axonal transport rates in vivo are unaffected by tau deletion or overexpression in mice. J Neurosci. 2008;28(7):1682–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lanjuin A, Chowdhury SR, Mistry M, Silva-García CG, Weir HJ, Lee C-L, Escoubas CC, Tabakovic E, Mair WB, et al. Neuronal TORC1 modulates longevity via AMPK and cell nonautonomous regulation of mitochondrial dynamics in C. elegans. Elife. 2019;8:e49158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplementary Table 2 shows the means, SD, N-values, and biological replicate information for all data that were currently represented in graphical format.
Supplemental material is available at GENETICS online.