Abstract
Understanding gene expression and regulation requires insights into RNA transcription, processing, modification, and translation. However, the relationship between the epitranscriptome and the proteome under drought stress remains undetermined in poplar (Populus trichocarpa). In this study, we used Nanopore direct RNA sequencing and tandem mass tag-based proteomic analysis to examine epitranscriptomic and proteomic regulation induced by drought treatment in stem-differentiating xylem (SDX). Our results revealed a decreased full-length read ratio under drought treatment and, especially, a decreased association between transcriptome and proteome changes in response to drought. Epitranscriptome analysis of cellulose- and lignin-related genes revealed an increased N6-Methyladenosine (m6A) ratio, which was accompanied by decreased RNA abundance and translation, under drought stress. Interestingly, usage of the distal poly(A) site increased during drought stress. Finally, we found that transcripts of highly expressed genes tend to have shorter poly(A) tail length (PAL), and drought stress increased the percentage of transcripts with long PAL. These findings provide insights into the interplay among m6A, polyadenylation, PAL, and translation under drought stress in P. trichocarpa SDX.
Drought stress increases distal poly(A) site usage and promotes m6A, stimulating RNA degradation and reducing transcription and translation of secondary cell wall genes in Populus trichocarpa.
Introduction
Plants must adapt their physiology to the variable environment. Rapid regulation of gene expression for adapting to abiotic stresses occurs at different levels, including capping, splicing, polyadenylation, methylation, and translation (Floris et al., 2009; Hu et al., 2021). Drought is the major abiotic stress affecting the growth and productivity of plants; thus, various response mechanisms that involve reprogramming of gene expression have evolved to cope with drought stress (Ryan, 2011; Maud et al., 2016; Li et al., 2019; Zhang et al., 2019a, 2019b, 2019c). It is therefore extremely important to capture precise transcriptome and translation profiles for understanding the molecular mechanisms underlying the drought response.
N 6-Methyladenosine (m6A) is the most extensive dynamic RNA modification and is typically enriched near the stop codon and 3′-untranslated region (3′-UTR). Increasing evidence shows that m6A can affect mRNA in multiple ways, such as alternative polyadenylation (APA) and altered translation, degradation, and nuclear export of mRNAs (Shao et al., 2021). Epitranscriptome studies have been reported in maize (Zea mays) (Nichols, 1979), Arabidopsis (Arabidopsis thaliana; Zhong et al., 2008), tomato (Solanum lycopersicum; Zhou et al., 2019), rice (Oryza sativa japonica; Zhang et al., 2019a, 2019b, 2019c), and poplar (Populus trichocarpa; Lu et al., 2020; Gao et al., 2021). However, studies of m6A in response to stress response are almost exclusively focused on Arabidopsis (Martínez-Pérez et al., 2017; Anderson et al., 2018; Kramer et al., 2020; Hou et al., 2021; Song et al., 2021). For example, a reduction in m6A was recently associated with global changes in gene expression under salt stress conditions (Hu et al., 2021). Therefore, epitranscriptome regulation during abiotic stress should be further explored in plants.
APA generates mRNAs with distinct 3′-UTRs, which are implicated in stress responses (Yan et al., 2021). The key polyadenylation factor of the 30-kDa subunit of the Cleavage and Polyadenylation Stimulatory Factor (CPSF30) produces two splice isoforms. The longer isoform (CPSF30-L) binds m6A and functions as an m6A reader in regulating flowering and the abscisic acid (ABA) response (Song et al., 2021). The m6A reader CPSF30-L also modulates APA of nitrate signaling-related genes, such as nitrate transporter 1.1 (Li et al., 2017). The genome-wide profiling of APA sites using short-read sequencing greatly expanded our insights into recognition and selection of polyadenylation signals (Wu et al., 2011; Velten et al., 2015; Zhou et al., 2016; Hwang and Darnell, 2017; Routh et al., 2017; West et al., 2018; Ye et al., 2019; Chakrabarti et al., 2020). With the benefit of long-read sequencing, Nanopore direct RNA sequencing (DRS) offers a promising method for identifying authentic sites of mRNA cleavage and polyadenylation (Sherstnev et al., 2012; Parker et al., 2020), providing a great advantage for investigating the association between polyadenylation and stress.
The poly(A) tail is a homopolymeric stretch of adenosines at the 3′-end of most eukaryotic mRNAs. Poly(A) tail length (PAL) affects gene expression (Liu et al., 2021a, 2021b, 2021c, 2021d, 2021e), mRNA stability, translation, and nuclear export of mature mRNAs (Fuke and Ohno, 2008; Eckmann et al., 2011; Subtelny et al., 2014; Liu et al., 2021a, 2021b, 2021c, 2021d, 2021e). In particular, a recent study reported a connection between m6A and PAL (Liu et al., 2021a, 2021b, 2021c, 2021d, 2021e). High levels of variability in PAL are observed among transcripts of plant genes during heat shock, and PAL contributes to a swift posttranslational stress response (Wu et al., 2020). Endoplasmic reticulum stress-induced mRNAs harbor shorter poly(A) tails in RNA granules than in the cytoplasm (Woo et al., 2018). Full-length mRNA sequencing using Nanopore DRS allows us to estimate PAL (Krause et al., 2019). However, the extent of PAL regulation during stress remains largely unexplored in plants.
Transcription, mRNA degradation, translation, and protein degradation are the four fundamental cellular processes in gene regulation (Schwanhäusser et al., 2011). Translation is modulated in response to environmental stimuli (Bailey-Serres, 1999), and proteomic analysis strengthens our understanding of the biological processes of stress responses (Wei et al., 2018a, 2018b; Guan et al., 2019; Xiao et al., 2020). Tandem mass tag (TMT) labeling technology provides accurate quantification of peptides and proteins (Thompson et al., 2003). Although the association between the transcriptome and proteome has been investigated previously (Ideker et al., 2001), this association remains unknown under drought stress in plants.
In this study, we identified differential epitranscriptome and proteome changes under drought treatment in P.trichocarpa stem-differentiating xylem (SDX) using Nanopore DRS and TMT-based proteomic analysis, respectively. Our results revealed a low association between transcript level and protein abundance upon drought treatment. We investigated the dynamic profile of m6A in response to drought, finding that the m6A ratio associated with wood-formation genes was increased by drought stress, whereas transcript levels and translation levels were reduced. Moreover, we found that proximal poly(A) site usage shifted to distal poly(A) site usage under drought stress. Finally, our results revealed that highly expressed and well-translated transcripts have a strong bias toward short PAL. In summary, our work provides genome-wide profiles of the epitranscriptome and proteome in response to drought stress in P. trichocarpa SDX and their impact on gene regulation.
Results
Nanopore DRS of SDX transcriptome under drought treatment
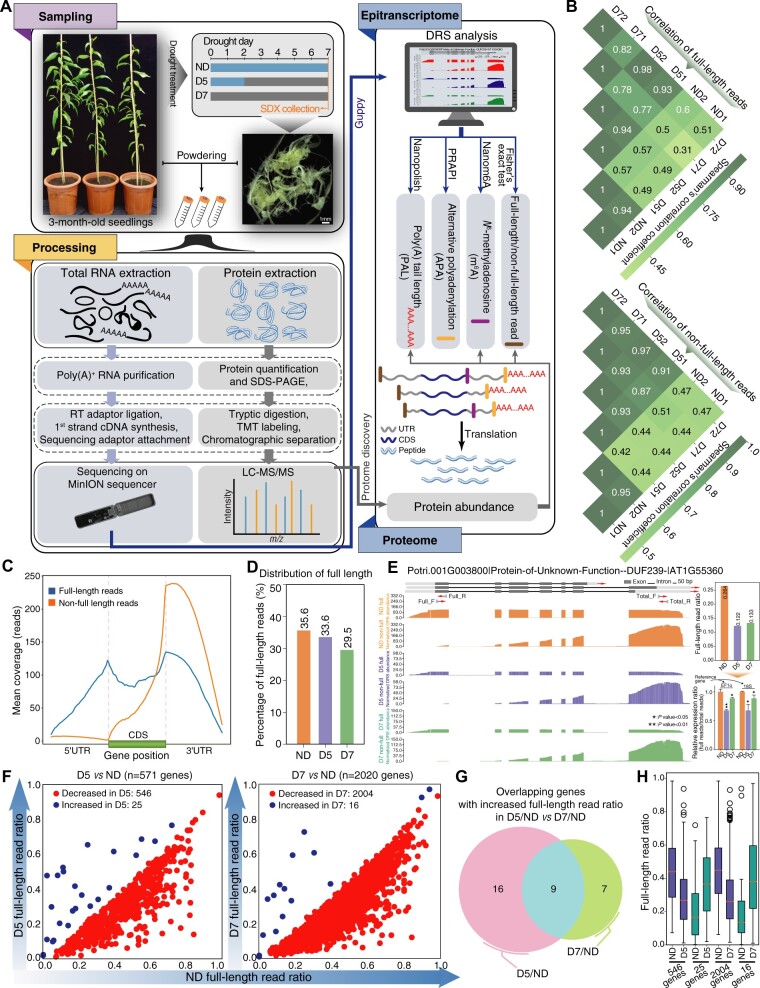
To capture the diversity and dynamics of the epitranscriptome in response to drought stress in P. trichocarpa SDX, we performed DRS of RNA from control (no drought, ND), 5-day (D5), and 7-day (D7) drought treatments using Nanopore MinION (Figure 1A). DRS libraries with two biological repeats were sequenced using the SQK-RNA002 kit protocol. A total of 9.9 million real-time single-molecule sequencing reads were generated from six MinION flow cells (FLO-MIN106) after basecalling with Guppy (version 3.6.1; Supplemental Table S1). After correction using LoRDEC (Salmela and Rivals, 2014), 99.6% DRS reads were successfully aligned to the P. trichocarpa version 3.0 genome using minimap2 (Li, 2018), suggesting the reliability of DRS reads.
Figure 1.
Nanopore DRS of RNA from SDX subjected to drought stress. A, Flowchart for analysis of epitranscriptome and proteome changes triggered by drought stress using Nanopore DRS and TMT labeling. Nanopolish, PRAPI, and Nanom6A are three pipelines for analysis of long reads. B, Heatmap showing Spearman’s correlation among ND, D5, and D7 samples for full-length and nonfull-length reads. C, Coverage of full-length and nonfull-length reads along transcripts. D, Histogram showing the dynamics of the full-length read ratio during drought stress. E, Wiggle plot, histogram, and RT-qPCR showing decreased full-length read ratio of DUF239 upon drought treatment. Significant difference was evaluated by t test and presented with asterisk rating system (*P < 0.05; **P < 0.01). Total_F/Total_R and Full_F/Full_R are the primer positions. F, Scatter plot showing differential full-length ratios upon drought treatment. G, Venn diagram showing overlap of genes with increased full-length read ratio in D5 versus ND and D7 versus ND comparisons. H, Boxplot showing the ratio distribution for genes with differential full-length read ratios in ND, D5, and D7 samples. The box limits represent 25th and 75th percentiles; center line, 50th percentiles; and whiskers, 10th and 90th percentiles. The circle represents outlier value.
Ratio of full-length transcripts decreases upon drought treatment
The ratio of full-length reads and nonfull-length reads is directly related to the balance between RNA biosynthesis and degradation. However, there are no reports on changes in this ratio in response to abiotic stress because it is hard to distinguish full-length reads from nonfull-length reads using short-read sequence platforms. Long-read sequencing technology solves this problem by reflecting features of biological RNA decay (Parker et al., 2020). Correlation analysis of both full-length and nonfull-length categories revealed high correlation between replicates (Figure 1B). With Reverse Transcription Adapter (RTA) ligation from the SQK-RNA002 kit protocol, DRS only detects transcripts with poly(A) tails. Thus, truncated reads starting from downstream of annotated transcription start sites and ending at the cleavage and polyadenylation sites show extreme 3′-bias and are distinct from full-length reads (Figure 1C). The full-length reads comprised about one-third of the total DRS reads in each sample, and the percentage of full-length reads decreased upon drought treatment (Figure 1D). For example, the full-length read ratio (full-length reads/total reads) of Potri.001G003800, a homolog of tRNA-splicing ligase (Domain of unknown function, DUF239) that shows altered expression during seed imbibition in Arabidopsis (Nakabayashi et al., 2005), was decreased under drought treatment compared with that under ND, which was validated by real time quantitative PCR (RT-qPCR) (Figure 1E).
We identified 546 and 2,004 genes (P < 0.005, Fisher’s exact test) with decreased full-length read ratio in D5 versus ND and D7 versus ND, respectively (Figure 1F;Supplemental Table S2). To our surprise, we observed few genes with increased full-length read ratio after drought treatment and only identified nine genes with increased full-length read ratio in both D5 versus ND and D7 versus ND comparisons (Figure 1G;Supplemental Table S3). For example, STAY-GREEN 1 (SGR1) has a crucial role in chlorophyll degradation in Arabidopsis (Ono et al., 2019; Zhang et al., 2019a, 2019b, 2019c). The full-length read ratio of SGR1 in P. trichocarpa was significantly increased under drought stress, which was consistent with the results of validated RT-qPCR (Supplemental Figure S1). Alteration of SGR1 might regulate stress-induced chlorophyll degradation. The average full-length read ratio was 35.6% in ND. Interestingly, genes with increased full-length read ratios in D5 versus ND and D7 versus ND had very low full-length read ratios (<20%) in ND (Figure 1H). Genes with decreased full-length read ratios in D5 versus ND and D7 versus ND showed a high full-length read ratios (>40%) in ND (Figure 1H).
Both endonuclease decay pathways and exonuclease decay pathways regulate mRNA decay (Schoenberg and Maquat, 2012). We did not observe obvious differential expression of 5′- to 3′-exoribonucleases (XRN2, XRN3, and XRN4) upon drought treatment (Supplemental Figure S2A). We also did not detect any differentially expressed protein (DEP) of 5′- to 3′-exoribonucleases (Supplemental Figure S2B), suggesting that the decreased full-length read ratio might not be caused by an alteration in exoribonuclease activity.
DEPs showed significant overlap with differentially expressed genes upon drought stress
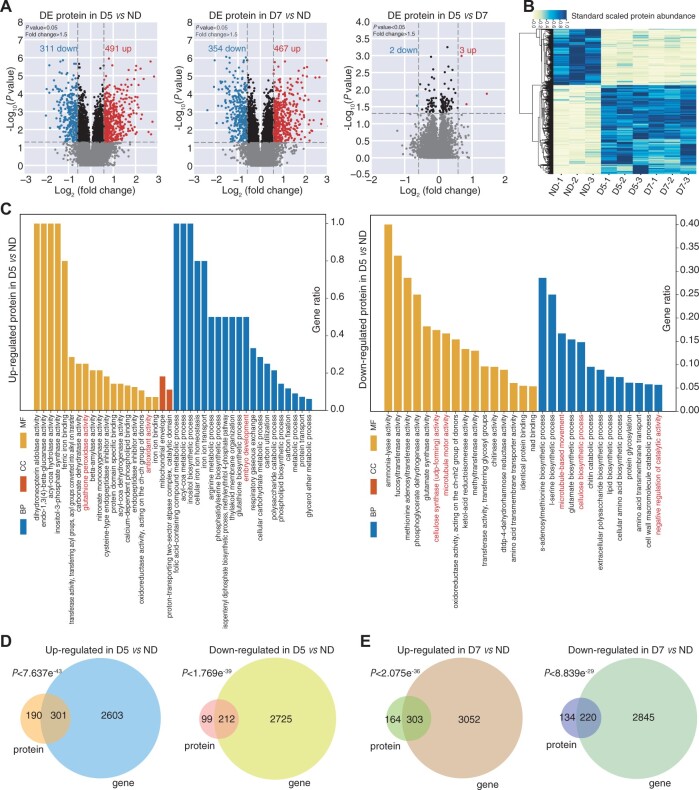
To create proteome profiles for ND, D5, and D7, we used TMT-labeling technology with three biological repeats (Figure 1A). A total of 86,904 peptide fragments corresponding to a total of 9,872 proteins were identified. Proteins with significant changes in abundance (P < 0.05) and greater than 1.5-fold change were considered DEPs (Figure 2A). Pairwise comparisons of ND, D5, and D7 samples led to the identification of 802 DEPs in D5 versus ND (491 upregulated and 311 downregulated) and 821 DEPs in D7 versus ND (467 upregulated and 354 downregulated), respectively (Supplemental Table S4). However, there was almost no difference in the number of DEPs (3 upregulated and 2 downregulated) between D5 and D7 (Figure 2, A and B), revealing that changes in protein abundance caused by drought treatment were already completed by the D5 stage.
Figure 2.
Mass spectrometry-based proteomics of SDX during drought stress. A, Volcano plots showing DEPs for D5 versus ND, D7 versus ND, and D5 versus D7, respectively. B, Heatmap of DEPs. Dark blue and light yellow represent high and low protein abundance, respectively. C, Pairwise comparison of D5 versus ND showing enriched GO terms among upregulated (left) and downregulated (right) proteins, respectively. D and E, Venn diagrams showing the overlap between DEPs and DEGs in D5 versus ND (D) and D7 versus ND (E).
To uncover the biological processes regulated in response to drought stress, we performed a gene ontology (GO) enrichment analysis of DEPs. Significantly enriched GO terms among the 491 upregulated DEPs (D5 versus ND) were “antioxidant activity” (GO:0016209, P = 0.004), “glutathione peroxidase activity” (GO:0004602, P = 0), and “embryo development” (GO:0009790, P = 0) (Figure 2C, left). For the 311 downregulated DEPs, significantly enriched GO terms included “microtubule-based movement” (GO:0007018, P = 0), “microtubule motor activity” (GO:0003777, P = 0), “negative regulation of catalytic activity” (GO:0043086, P = 0.001), “cellulose synthase activity” (GO:0016760, P = 0), and “cellulose biosynthetic process” (GO:0030244, P = 0) (Figure 2C, right), revealing that proteins related to cellulose biosynthesis were downregulated in response to drought stress. Among the downregulated proteins, we identified 22 associated with secondary cell wall genes: 7 cellulose synthases, 11 hemicellulose synthases, and 4 lignin synthases (Supplemental Table S5).
We further investigated “response to water deprivation” terms and identified 48 DEPs, including 36 DEPs in both D5 versus ND and D7 versus ND comparisons (Supplemental Table S6). In total, 81% of these DEPs were upregulated proteins, including ABI5-binding protein, aldehyde dehydrogenase, NAC domain-containing protein, Late embryogenesis abundant (LEA) family protein, CBL-interacting protein kinase, lipid transfer protein, β-amylase, and raffinose synthase family protein. NAC (Li et al., 2019) and LEA (Park et al., 2003) are reported to be strongly induced by drought, consistent with our results. The eight proteins downregulated during the drought response included PHE ammonia lyase, chitinase protein, and CHASE domain-containing histidine kinase protein.
PlantTFDB contains 428 annotated transcription factors (TFs) in P. trichocarpa (Jin et al., 2017). Among these, we identified 23 upregulated TFs and 4 downregulated TF proteins under drought stress, which could be classified into 16 families including NAC, basic leucine zipper (bZIP), and ERF (Supplemental Figures S3 and S4). A previous study revealed that ABA-responsive element binding (AREB) protein PtrAREB1-2 can bind to the promoter of PtrNAC genes to enhance H3K9ac for the development of drought tolerance (Li et al., 2019). We found that protein abundance of PtrAREB1-2 (Potri.002G125400) and PtrAREB1-4 (Potri.014G028200) from the bZIP family was upregulated under drought stress (Supplemental Figure S4, A and B).
ABA regulates guard cells in response to water deprivation. A large number of TFs are also regulated by ABA (Cutler et al., 2010). We identified 43 upregulated proteins and 2 downregulated proteins associated with “abscisic acid biosynthetic process” (GO:0009688), “abscisic acid metabolic process” (GO:0009687), and “response to abscisic acid” (GO:0009737) (Supplemental Table S7). There were more upregulated ABA-related proteins than downregulated ABA-related proteins in SDX, illustrating that the upregulated ABA proteins might increase ABA levels in response to water deprivation.
The association between mRNA and protein abundance varies widely among different organisms, cell types, and functions (Greenbaum et al., 2003; De Sousa Abreu et al., 2009; Vogel et al., 2010; Schwanhäusser et al., 2011; Ponnala et al., 2014). To investigate the consistency of the relationship between protein abundance and mRNA level in response to drought stress, we also identified differentially expressed genes (DEGs) in different samples. We identified 2,904 upregulated genes and 2,937 downregulated genes in D5 versus ND and 3,355 upregulated genes and 3,065 downregulated genes in D7 versus ND (Supplemental Table S8). Among these DEGs, 165 and 176 DEGs were associated with the “response to water deprivation” GO term in D5 and D7, respectively (Supplemental Table S6). The percentage of upregulated DEGs in this category was 74.55% (123/165) and 76.70% (135/176) for D5 and D7, respectively. Approximately 61.3% (301/491) and 68.2% (212/311) upregulated and downregulated proteins were identified as differentially expressed at the mRNA level in D5 versus ND (Figure 2D). For the comparison of D7 versus ND, ∼64.9% (303/467) and 62.1% (220/354) upregulated and downregulated proteins were detected as differentially expressed at the mRNA level (Figure 2E).
Association between protein abundance and mRNA expression decreased in response to drought stress
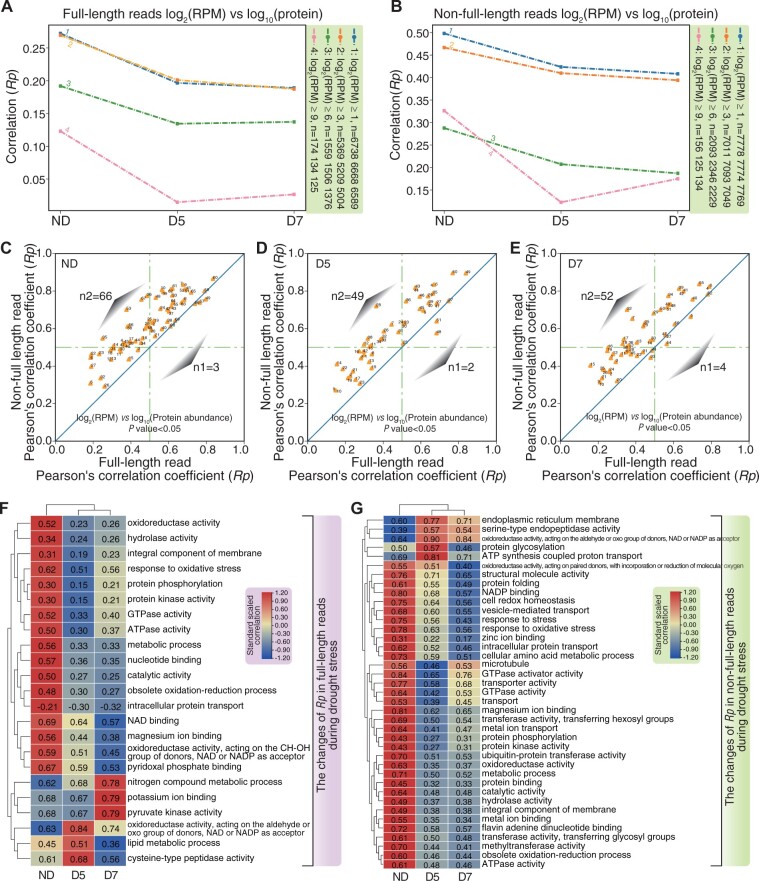
Previous studies analyzed mRNA and protein abundance in response to perturbation using RNA-Seq combined with proteomics (Vogel and Marcotte, 2012; Ponnala et al., 2014). Here, we took advantage of DRS and TMT-labeling technology to investigate the Pearson’s correlation coefficient between mRNA expression and protein abundance (Rp) of full-length and nonfull-length RNA during drought stress. Expression of mRNA was normalized using reads per million (RPM). TMT-labeled protein data were normalized using the sum of the total intensity of every report ion channel between samples. The median log10(protein) was ∼2.9 between samples (Supplemental Figure S5A), while the median log2(RPM) of full-length and nonfull-length RNA was ∼4.4 and 4.5, respectively (Supplemental Figure S5, B and C). With an increased threshold of normalized gene expression, the Rp of both full-length and nonfull-length RNA decreased (Figure 3, A and B). Our published PacBio Iso-Seq data showed a similar trend to the DRS data from this study (Liu et al., 2021a, 2021b, 2021c, 2021d, 2021e; Supplemental Figure S6). Furthermore, we also observed a decreased association between RNA and protein upon drought treatment, which revealed divergent changes between RNA expression and protein abundance during drought stress (Figure 3, A and B).
Figure 3.
Correlation between protein abundance and mRNA expression during drought stress. A\–B, Line plots of Rp with different RPM threshold levels for full-length (A) and nonfull-length RNA (B). C–E, Scatter plots showing different Rp based on GO terms between full-length RNA and nonfull-length RNA in ND (C), D5 (D), and D7 (E). F, Heatmap showing changes in Rp of full-length reads in response to drought treatment. The color of heatmpap (dark blue to red) indicates the correlation (low to high). G, Heatmap showing changes in Rp of nonfull-length reads in response to drought treatment.
To evaluate the Rp dynamics within different groups of genes, we estimated the Rp of gene clusters according to GO terms using full-length RNA (Supplemental Figure S7) and nonfull-length RNA (Supplemental Figure S8), respectively. For full-length RNA, the Rp of 83 GO terms revealed a significant correlation (P < 0.05) between RNA reads and protein abundance (Supplemental Figure S7; Supplemental Table S9). For example, the “oxidoreductase activity” GO term was associated with 206 genes in ND, and the correlation between protein abundance and mRNA expression was 0.52. The “microtubule activity” GO term was associated with 19 genes in ND, and the correlation between protein abundance and mRNA expression was 0.53. Interestingly, the Rp of “endoplasmic reticulum to Golgi vesicle-mediated transport” in D5 and “structural molecule activity” in D7 was ∼−0.69 and −0.53, respectively. It will be interesting to investigate whether these genes show distinct transcriptional or translational regulation (Supplemental Figure S9). For nonfull-length RNA, we found positive trends for Rp of 119 GO terms in the different samples (Supplemental Figure S8; Supplemental Table S9); no GO terms showed a negative correlation.
To compare the differences in Rp between full-length RNA and nonfull-length RNA, we used the significant GO terms for full-length and nonfull-length RNA for downstream analysis. Nonfull-length RNA had a higher Rp than full-length RNA in ND (Figure 3C), D5 (Figure 3D), and D7 (Figure 3E), respectively. To further investigate changes in Rp during drought stress, we compared overlapping GO terms in ND, D5, and D7, which revealed that the Rp of six GO terms was increased while that of 17 GO terms was decreased in full-length RNA (Figure 3F). Rp increased for terms involved in cysteine-type peptidase activity, and Rp decreased for terms associated with response to oxidative stress (Figure 3F). For nonfull-length RNA, the Rp of five GO terms increased while that of 34 GO terms decreased during drought stress (Figure 3G). Rp increased for terms with functions in protein glycosylation, and Rp decreased for terms with functions in the response to oxidative stress and other stresses (Figure 3G).
Dynamics of m6A under drought stress
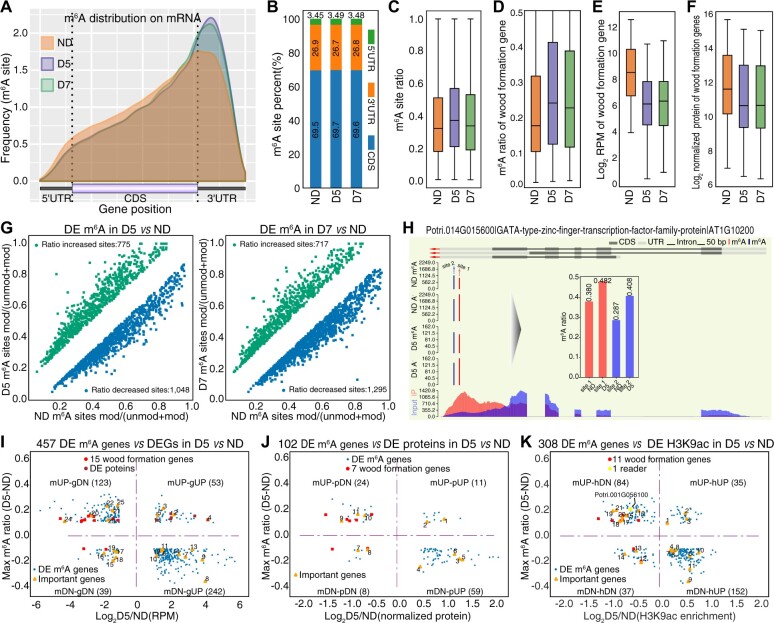
To investigate m6A dynamics upon drought stress, we analyzed the DRS data using our previously described method (Gao et al., 2021), which can detect RNA modification at single-base resolution. We detected a total of 86,313, 41,623, and 53,983 m6A sites associated with 13,069, 9,273, and 10,839 genes in ND, D5, and D7, respectively (Supplemental Figure S10, A and B). Transcriptome analysis of the m6A profiles revealed that m6A peaks were mainly distributed in the coding sequence and 3′-UTR in all samples (Figure 4, A and B). N6-adenosine methyltransferase MT-A70-like gene (MTA) is an RNA methyltransferase that enhances drought tolerance by regulating the development of trichomes and roots in poplar (Lu et al., 2020). Expression of PtrMTA was slightly increased under drought stress (Supplemental Figure S10C), with the protein level showing a similar trend (Supplemental Figure S10D). In addition to MTA, we found that m6A readers (ECT2 and ECT8) were significantly upregulated (P < 0.05, fold change >1.5) and an m6A eraser (ALBH8B) was significantly downregulated (P < 0.05, exact test for the negative binomial distribution) upon drought stress, which was further validated by RT-qPCR experiments (Supplemental Figure S11).
Figure 4.
Dynamics of m6A methylation in response to drought stress. A, Distribution of m6A methylation sites along transcripts. B, Histogram showing the percentage of m6A sites in the annotated gene region including UTR and CDS in ND, D5, and D7. C, Boxplot showing m6A ratio in ND, D5, and D7. D–F, Boxplots showing m6A ratio (D), gene expression (E), and protein level (F) of wood-formation genes in ND, D5, and D7. The box limits represent 25th and 75th percentiles; center line, 50th percentiles; and whiskers, 10th and 90th percentiles. G, Scatter plots showing differential m6A modification (mod) upon drought treatment in D5 (left) and D7 (right). H, Differential m6A modification in transcription factor WLIM1. I–K, Scatter plots from the pairwise comparison D5 versus ND showing the relationships of m6A ratio with gene expression (I), protein level (J), and H3K9ac level (K).
The average m6A ratio increased under drought stress (Figure 4C). The m6A ratio of wood-formation genes was higher in both D5 and D7 than in ND (Figure 4D) and was accompanied by decreased expression of mRNA (Figure 4E) and protein (Figure 4F), suggesting that m6A may play roles in repressing wood formation under stress. These wood-formation genes included CESA9 (Potri.002G066600 and Potri.005G194200, which function as cellulose synthase A9), HCT (Potri.003G183900, involved in the lignin biosynthetic process), BP1 (Potri.002G113300, functioning in xylem development), and IRX10 (Potri.001G068100, functioning in secondary cell wall biogenesis).
Furthermore, we identified differentially expressed m6A (DE m6A) sites. In total, 1,048 sites showed decreased m6A and 775 sites showed increased m6A in D5 versus ND (Figure 4G, left; Supplemental Table S10) while 1,295 sites showed decreased m6A and 717 sites showed increased m6A in D7 versus ND (Figure 4G, right; Supplemental Table S10) using Fisher’s exact test (P < 0.05). For example, WLIM1 (Potri.014G015600) is a TF that functions in actin bundle formation, and the m6A ratios of two m6A sites in WLIM1 increased under drought stress (Figure 4H). Genes exhibiting increased m6A ratios were enriched in protein-related process, RNA polyadenylation, and membrane coat functions (Supplemental Figure S12, A and C); genes exhibiting decreased m6A ratios were enriched in response to water, protein-related process, and metabolic process functions (Supplemental Figure S12, B and D). In particular, we found 10 DE m6A sites in eight m6A readers among the list of DE m6A sites (Supplemental Table S10).
We also integrated our transcriptome data (D5 versus ND and D7 versus ND) to analyze the relationship between DE m6A sites and gene expression. We identified 457 DEGs with DE m6A sites in D5 versus ND (Figure 4I;Supplemental Figure S13A; Supplemental Table S11). These were grouped in four quadrants: Quadrant I, mUP-gUP, increased m6A ratio and gene expression (53 genes); Quadrant II, mUP-gDN, increased m6A ratio and decreased gene expression (123 genes); Quadrant III, mDN-gDN, decreased m6A ratio and gene expression (39 genes); Quadrant IV, mDN-gUP, decreased m6A ratio and increased gene expression (242 genes). Quadrants II and IV had more genes than Quadrants I and III, which is consistent with m6A methylation generally being negatively associated with transcript abundance (Zhou et al., 2019). In the mUP-gDN category (Figure 4I, Quadrant II), the expression of 13 wood-formation genes was decreased by drought stress and the m6A ratio was increased. NAC genes present differential expression during drought stress (Li et al., 2019). We found that the expression of two NAC domain genes and one WRKY gene was downregulated while m6A modification was increased by drought stress. LEA (Potri.005G122400) and Dehydrin (Potri.013G062300) in the mDN-gUP category (Figure 4I, Quadrant IV) encode proteins that protect cells from abiotic stress and reduce the formation of reactive oxygen species, respectively. Senescence-associated genes (Potri.008G075200, Potri.001G112600, and Potri.004G174100) were also found in the mDN-gUP category.
Furthermore, we explored the connection between m6A ratio and protein abundance, observing a trend similar to that of gene expression (Figure 4J;Supplemental Figure 13B; Supplemental Table S11). In the mUP-pDN category (Figure 4J, Quadrant II), five wood-formation genes including CELLULOSE SYNTHASE-INTERACTIVE PROTEIN 1 (CSI1, Potri.005G080100) showed an increased m6A ratio and decreased protein abundance. A NAC domain protein (Potri.017G016700) and an RNA-binding family protein (Potri.006G015700) showed the same trend as wood-formation proteins. In the mDN-pUP category (Figure 4J, Quadrant IV), there were several stress-related genes, including dehydrin (Potri.013G062300), ERD7 (Potri.004G174100), and CIPK3 protein kinase (Potri.001G222600). ERD7 (Potri.004G174100) is an early-responsive gene to dehydration.
Recently, drought stress-responsive genes were shown to exhibit differential H3K9ac (Li et al., 2019), providing H3K9ac data for investigating the association between m6A and H3K9ac. Thus, we integrated the m6A ratio and H3K9ac level in SDX during drought stress (Figure 4K;Supplemental Figure S13C; Supplemental Table S11), which presented a similar trend to gene expression (Figure 4I) and protein abundance (Figure 4J). Nine wood-formation genes (Figure 4K, Quadrant II) and several translation-related genes including ribosome protein genes L34e (Potri.017G084500) and S8e (Potri.001G360500; Figure 4K, Quadrant IV) were identified in the mUP-hDN and mDN-hUP groups, respectively.
Transgenic expression of human RNA demethylase fat mass and obesity associated gene (FTO) induces transcriptional activation and increased drought tolerance in rice (O.sativa; Yu et al., 2021). However, it remains unknown whether m6A regulates the full-length read ratio (Figure 1D). Therefore, we used a DRS dataset of mutants defective in m6A writer, including vir-1 from Arabidopsis (Parker et al., 2020) and shMETTL3 from human HEK293T cells (Lorenz et al., 2020), to investigate the dynamics of full-length read ratio. Both vir-1 and shMETTL3 mutants showed an increased full-length read ratio (Supplemental Figure S14), suggesting a preference of m6A to promote RNA degradation. GO enrichment analysis of vir-1 genes with increased full-length ratio showed that their functions were enriched in photosynthesis and responses to different stresses (Supplemental Figure S15).
Drought stress promotes the usage of distal poly(A) sites
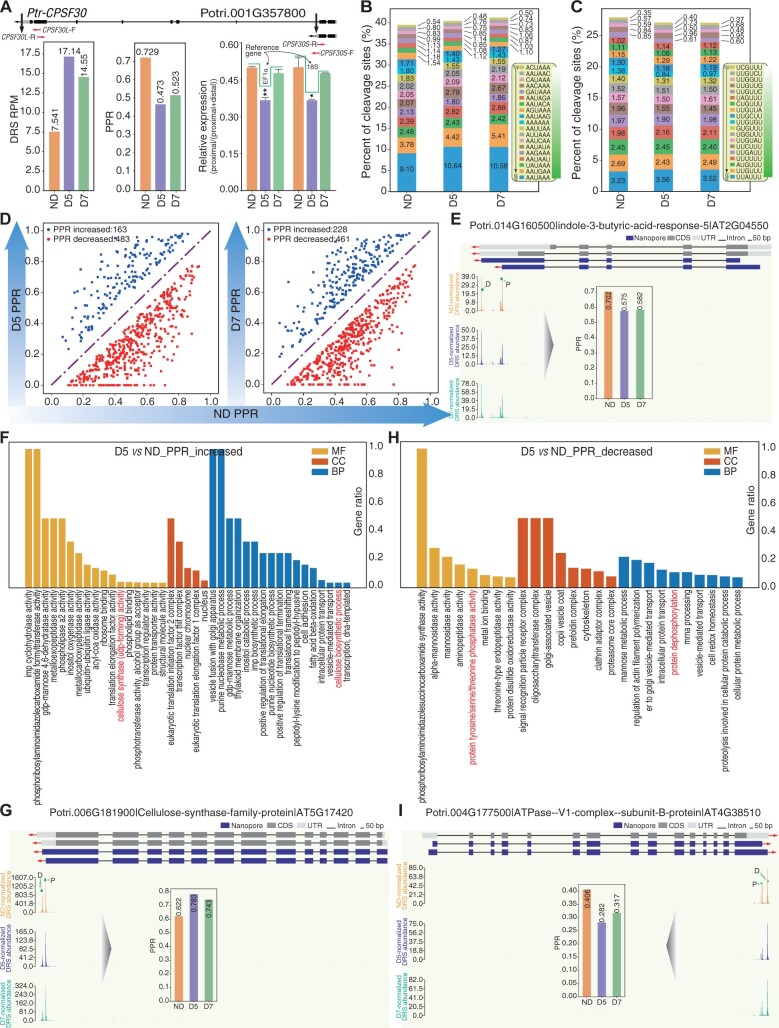
Recent studies showed that the m6A reader CPSF30-L affects nitrate signaling by controlling APA (Hou et al., 2021) and recognizes m6A-modified far-upstream element signals to control poly(A) site choice in Arabidopsis (Song et al., 2021). We observed an m6A peak in the 3′-UTR of CPSF30-L based on our recent MeRIP-seq data (Supplemental Figure S16; Gao et al., 2021). APA is extensively involved in plant stress responses (Ye et al., 2019; Chakrabarti et al., 2020). CPSF30 is a core component of the CPSF complex and controls APA site choices genome wide (Thomas et al., 2012). We found that expression of Ptr-CPSF30 (Potri.001G357800) was upregulated (D5 versus ND: P = 0.05, D7 versus ND: P = 0.006) during drought stress (Figure 5A). We further investigated the poly(A) site usage shift of Ptr-CPSF30 during drought stress; the proximal poly(A) site usage ratio (proximal/distal and proximal, PPR) of Ptr-CPSF30 was downregulated under drought stress, which was further validated by RT-qPCR (Figure 5A). A previous study showed that the most common motif associated with preferred cleavage site is AAUAAA and also identified another U-rich motif, UUGUUU, in Arabidopsis (Sherstnev et al., 2012). In this study, we found that the poly(A) signal ratio of AAUAAA and 1-nt variations was slightly increased (from 39.52% in ND to 41.27% in D7; Figure 5B), while that of UCGUUU and 1-nt variations was slightly decreased, upon drought stress (Figure 5C).
Figure 5.
Dynamics of poly(A) site usage in response to drought stress. A, Histogram showing increased expression and decreased PPR of CPSF30 validated by RT-qPCR under drought treatment. Significant difference was evaluated by t test and presented with asterisk rating system (*P < 0.05; **P < 0.01). B and C, Histograms showing the percentage of AAUAAA (and 1-nt variations) (B) and UCGUUU (and 1-nt variations) (C) hexamer motifs under drought stress. D, Scatter plots showing changes in PPR in response to drought stress. E, Wiggle plot and histogram showing decreased PPR in IBR5 (Potri.014G160500). F/H, GO enrichment analysis of genes exhibiting increased PPR (F) and decreased PPR (H), respectively. G/I, Wiggle plots, and histograms showing CESA7 and VAB2 as examples of genes with increased PPR (G) and decreased PPR (I).
To identify poly(A) site usage upon drought stress, we quantified poly(A) site usage using Fisher’s test (P < 0.005). We identified 163 genes with increased PPR and 483 genes with decreased PPR in D5 versus ND (Figure 5D, left; Supplemental Table S12) and 228 genes with increased PPR and 461 genes with decreased PPR in D7 versus ND (Figure 5D, right; Supplemental Table S12), revealing that distal poly(A) site usage increased upon drought stress. For example, Ptr-IBR5 (Potri.014G160500), a key component of the leaf-serration regulatory machinery (Kong et al., 2019), preferred to use the distal poly(A) site under drought stress (Figure 5E). Among these differential APA events, we investigated genes related to the “response to water deprivation” GO term, comprising 10 genes exhibiting differential APA in D5 versus ND (Supplemental Table S6) and 17 genes exhibiting differential APA in D7 versus ND. We then further explored the functions of genes with differential APA induced by drought. Genes with increased PPR were enriched in “cellulose synthase activity” (P-value = 0.002) and “cellulose biosynthetic process” (P = 0.006; Figure 5F), such as Ptr-CESA7 (Potri.006G181900 and Potri.018G103900) (Figure 5G;Supplemental Figure S17). Genes with decreased PPR were enriched in “protein dephosphorylation” (P = 0) and “protein tyrosine/serine/threonine phosphatase activity” (P = 0; Figure 5H), such as Ptr-IBR5 (Potri.014G160500; Figure 5E) and Ptr-VAB2 (Potri.004G177500; Figure 5I).
APA can generate different isoforms with different 3′-termini including or excluding microRNA sites (Zhang et al., 2021). We, therefore, extracted the alternative regions from the proximal poly(A) to the distal poly(A) sites for genes with decreased PPR to predict miRNA targets using psRNATarget (Dai et al., 2018). We identified 18 genes with poly(A) switch regions including miRNA target sites (Supplemental Table S13). For example, CesA8-B (Potri.004G059600) preferred to use the distal poly(A) site to create a long isoform including the miR172h-5p target site upon drought treatment. It will be interesting to investigate whether the long isoform of this cellulose synthase is regulated by miRNA cleavage or translation inhibition of miR172. APA can also generate different isoforms with poly(A) switch regions including or excluding m6A sites. We identified 465 and 158 differential poly(A) switch regions including m6A sites in D5 versus ND and D7 versus ND, respectively. m6A sites were obviously enriched upstream of proximal poly(A) sites (Supplemental Figure S18).
Differential PAL is induced by drought stress
During nuclear pre-mRNA processing, a poly(A) tail is added to the 3′-end of mature RNA transcripts (Bardwell et al., 1990; Eckmann et al., 2011). PAL affects mRNA stability, translational efficiency, and transfer of RNA from the nucleus to the cytoplasm, and distinguishes between stable RNAs and unstable RNAs (Huang and Carmichael, 1996; Meyer et al., 2004; Beilharz and Preiss, 2007; Subtelny et al., 2014; Lima et al., 2017; Niazi et al., 2021). Taking advantage of the full-length poly(A) tail sequences produced by DRS, we determined that the median PAL in ND, D5, and D7 samples was 79, 80, and 75 nt, respectively (Figure 6A;Supplemental Table S14). The 10 GO terms with the longest median PAL possessed functions including “transcription coregulator activity” and “response to hormone,” while the 10 GO terms with the shortest PAL functioned in translation and ribosome composition (Figure 6B;Supplemental Figure S19). The five gene function groups showing the greatest increase in PAL during drought stress (D5 > ND) were all associated with protein process, while enzyme activity and response to hormones were the main functions in the five groups showing the greatest decrease in PAL during drought stress (D5 < ND; Figure 6C).
Figure 6.
Differential PAL in response to drought stress. A, Violin plot showing median PAL in ND, D5, and D7. Dashed horizontal lines presenting PAL of 80 nt. B, Box plot showing 10 GO terms with the longest PAL (orange) and 10 GO terms with the shortest PAL (blue) in ND. The box limits represent 25th and 75th percentiles; center line, 50th percentiles; and whiskers, 10th and 90th percentiles. Dashed horizontal lines presenting PAL of 80 nt. C, Box plot showing gene function groups presenting the greatest increase (left) and decrease in PAL (right) in D5 versus ND (top) and D7 versus ND (bottom). A, represents “kinase activity”; B, “nucleotide-excision repair”; C, “protein deubiquitination”; D, “protein metabolic process”; E, “G protein-coupled receptor signaling pathway”; F, “response to hormone”; G, “N-acetyltransferase activity”; H, “isoprenoid biosynthetic process”; I, “protein serine/threonine kinase activity”; J, “nucleobase-containing compound metabolic process”; K, “metal ion transmembrane transporter activity”; L, “large ribosomal subunit”; M, “protein phosphatase type 2A regulator activity”; N, “protein phosphatase type 2A complex”; O represents “microtubule-based movement”; P, “microtubule motor activity”; Q, “kinesin complex”. Dashed horizontal lines presenting PAL of 80 nt. D, Box plot showing PAL distribution for fully spliced (no_IR) and IR transcripts. The dash lines representing the median PAL in no_IR and IR transcripts. E and F, Scatter plots showing differential PAL for D5 versus ND (E) and D7 versus ND (F). G and H, Box plots showing drought stress-induced longer PAL (G) and shorter PAL (H) for Potri.008G102500 and Potri.013G034800, respectively.
A positive association between intron retention (IR) and PAL distribution is reported for Caenorhabditis elegans (Roach et al., 2020). In this study, we also found a similar trend of transcripts with IR possessing longer poly(A) tails (Figure 6D), and reads with retained introns, whether full-length or nonfull length, always had a longer PAL than fully spliced reads (Supplemental Figure S20).
Furthermore, we investigated differential PAL in response to drought stress. There were 150 genes with longer PAL and 118 genes with shorter PAL in D5 versus ND (Figure 6E;Supplemental Table S15). Genes with longer PAL in D5 were enriched in transcription activity, RNA polyadenylation, and translation-related events (Supplemental Figure S21), while genes with shorter PAL functioned in gmp synthase activity and the gmp biosynthetic process, among others (Supplemental Figure S21). In D7 versus ND, we identified 146 genes with longer PAL and 143 genes with shorter PAL (Figure 6F;Supplemental Table S15). Genes with longer PAL were enriched in “NADH dehydrogenase activity,” “translation,” and “RNA polyadenylation” GO categories (Supplemental Figure S22). Shorter genes were associated with transcription activator activity and ATP-dependent chromatin remodeling (Supplemental Figure S22). For example, the median PAL of Potri.008G102500 [encoding poly(A) polymerase], increased by 53 and 61 nt in D5 and D7 compared with ND, respectively (Figure 6G). A similar trend was observed for Potri.001G152200, Potri.001G169900, Potri.018G057200, and Potri.018G119200 (Supplemental Figure S23A). Decreased PAL during drought stress was observed in some genes including Potri.013G034800 (Figure 6H), Potri.016G023100, Potri.016G037400, Potri.008G172400, and Potri.010G252000 (Supplemental Figure S23B). Among these differential PAL events, we investigated genes related to the ‘response to water deprivation” GO term and identified three and eight genes exhibiting differential PAL in D5 versus ND and D7 versus ND, respectively (Supplemental Table S6).
PAL is associated with transcript level and translation
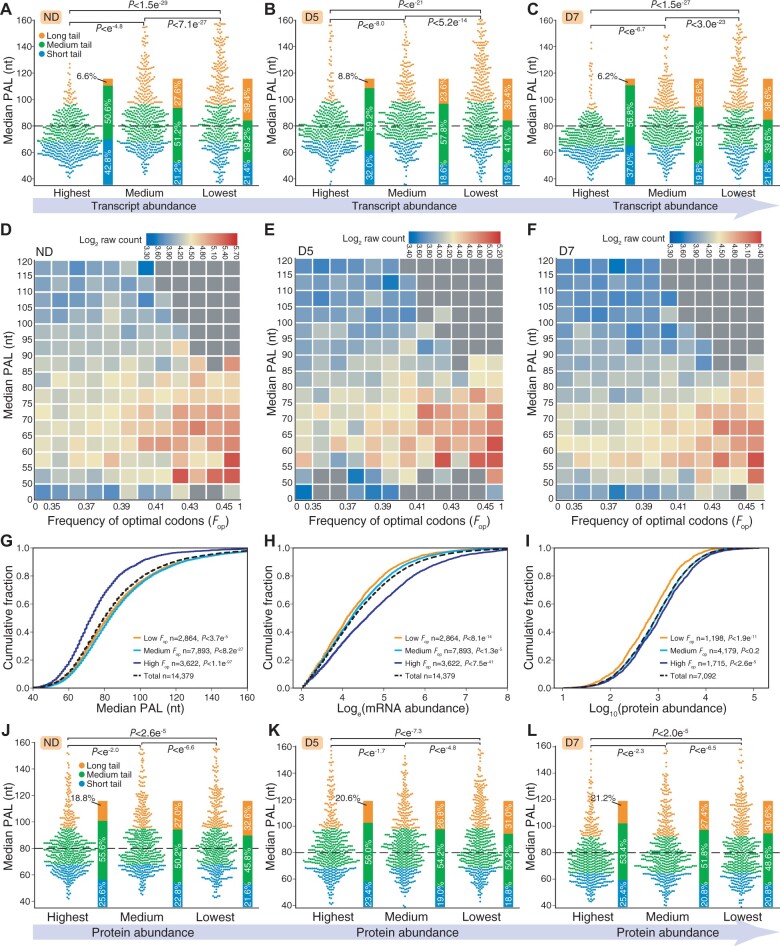
A previous study showed that the majority of highly expressed transcripts possessed short PAL to accommodate a single poly(A)-binding protein and undergo active degradation (Lima et al., 2017). We divided genes into three categories according to transcript abundance: high, medium, and low abundance (Figure 7, A–C; Supplemental Figure S24). Comparison of PALs among the three categories revealed that highly expressed genes tend to have short PAL, while genes with low expression show a preference for long PAL, which was in line with data for C. elegans (Lima et al., 2017). The percentage of different PAL (long, medium, and short) for genes highly expressed in ND did not present an obvious change in D5 or D7 (Supplemental Figure S25). However, for genes with medium transcript abundance in ND, the percentage of long PAL increased in both D5 and D7; for genes with low transcript abundance in ND, the percentage of long PAL only increased in D7 (Supplemental Figure S25).
Figure 7.
Highly expressed transcripts tend to have a short PAL. A–C, PAL distribution of genes in pools with different expression levels in ND (A), D5 (B), and D7 (C). Highest, medium, and lowest represent the 500 most highly expressed genes, the 500 genes with closest to median expression, and the 500 genes with lowest expression, respectively. P-value was calculated by Mann–Whitney U rank test. D–F, Heat maps representing the association between the frequency of optimal codons (Fop) and PAL in ND (D), D5 (E), and D7 (F). G–I, Cumulative sum plots showing association of translational efficiency (codon optimization) with PAL (G), mRNA abundance (H), and protein abundance (I). J–L, PAL distribution of genes in pools with different protein levels in ND (J), D5 (K), and D7 (L). P-value was calculated by Mann–Whitney U rank test.
Research has revealed that PAL is associated with translation in mouse GV oocytes (Liu et al., 2019), and that transcripts with optimal codons shape translational efficiency (Presnyak et al., 2015; Bazzini et al., 2016; Radhakrishnan et al., 2016; Lima et al., 2017). This inspired us to explore the association between PAL and translational efficiency in P. trichocarpa. We found that highly expressed genes tended to be enriched for optimal codons and show a strong bias toward short PAL in ND, D5, and D7, respectively (Figure 7, D–F). Using cumulative fraction analysis, we observed significant differences in the distribution of PAL (Figure 7G), mRNA abundance (Figure 7H), and protein abundance (Figure 7I) for transcripts with low, medium, and high levels of optimal codons. However, there was no substantial association between PAL and protein abundance in either ND or drought treatments (Figure 7, J–L). Taken together, these results indicate that highly expressed transcripts have a strong bias toward short PAL.
Exonuclease-mediated poly(A)+ RNA degradation begins with removal of the poly(A) tail by deadenylase complexes and a decapping complex (Dave and Chao, 2020). In this study, we observed that the PAL of full-length reads was longer than that of nonfull-length reads (Supplemental Figure S26). Interestingly, transcripts exhibiting IR also had longer PAL, consistent with results in humans and C. elegans (Workman et al., 2019; Roach et al., 2020). Recent studies found that the majority of nuclear transcripts have longer poly(A) tails than cytoplasmic transcripts (Liu et al., 2021a, 2021b, 2021c, 2021d, 2021e). Thus, IR or incompletely spliced RNAs might contribute to longer poly(A) tails in nuclear RNAs. Finally, we investigated the relationship between PAL dynamics and m6A modification. We classified transcripts into m6A-modified and unmodified transcripts for comparison of median PAL between these two categories. The number of genes exhibiting PALmod > PALunmod was approximately twice that displaying PALmod < PALunmod (Supplemental Figure S27), illustrating that m6A-modified transcripts tend to have longer PAL, which is consistent with MePAIso-seq2 results in mouse (Liu et al., 2021a, 2021b, 2021c, 2021d, 2021e).
Discussion
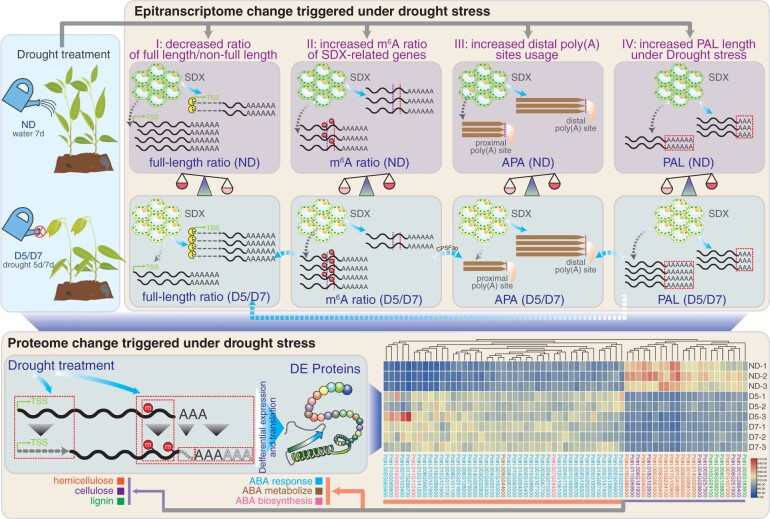
DRS sequencing technology has been used for predicting microRNA cleavage sites, suggesting that DRS reads can reflect RNA decay (Parker et al., 2020). In this study, we found that the full-length read ratio decreased in response to drought (Figure 8, Module I; Figure 1D). We did not observe differentially expressed exoribonucleases involved in either 5′–3′ or 3′–5′ decay upon drought treatment. PAL is related to mRNA stability (Eckmann et al., 2011; Weill et al., 2012; Liu et al., 2021a, 2021b, 2021c, 2021d, 2021e). Here, we observed that the PAL of full-length reads was longer than that of nonfull-length reads (Supplemental Figure S26). Future work should focus on other ribonucleolytic activities influencing mRNA decay, such as shortening of the poly(A) tail through deadenylation (Figure 8, Module IV). RNA decay is triggered by deadenylation, which is directed by adenine/uridine-rich elements (AREs) in the 3′-UTR (Chen and Shyu, 2011). It will be interesting to investigate whether ARE-mediated decay is regulated by APA (Figure 8, Module III), generating isoforms with distinct AREs.
Figure 8.
Model for drought response. Module I: Drought stress decreases full-length read ratio. Module II: Drought stress increases m6A ratio. Module III: Drought stress increases distal poly(A) site usage. Module IV: Drought stress increases PAL. The four modules are connected to each other. For example, PAL (Module IV) and m6A (Module II) might regulate full-length read ratio (Module I). APA (Module III) might interplay with m6A (Module II) via CPSF30, which can generate both m6A reader and polyadenylation factor.
In addition to deadenylation followed by 5′–3′ and 3′–5′ decay, several studies have revealed that m6A regulates steady-state levels of mRNA. A study in Arabidopsis demonstrates that m6A sites act as a mark to affect the stability of transcripts encoding salt response proteins (Anderson et al., 2018). Disruption of m6A methyltransferase FIONA1 reduces m6A, leading to slower degradation of PIF4, CRY2, and CO transcripts (Wang et al., 2022). Loss-of-function of m6A demethylase ALKBH10B affects floral transition because increased m6A of FT, SP3, and SPL9 accelerates mRNA decay of these transcripts in Arabidopsis (Duan et al., 2017). We also observed increased full-length read ratios in mutants of m6A writer components including shMETTL3 and vir-1 (Supplemental Figure S14), suggesting that m6A promotes mRNA decay. Interestingly, three m6A readers, Potri.008G100200, Potri.003G222700, and Potri.014G001000, showed significantly (P < 0.05, exact test) decreased full-length read ratios under drought stress (Figure 8, Module II; Supplemental Table S2). However, we did not observe this trend for m6A methyltransferase or demethylase.
In this study, we found that the average m6A ratio and that of wood-formation genes increased under drought stress (Figure 4, C and D), which was accompanied by reduced mRNA and abundance of proteins including 7 cellulose synthases, 11 hemicellulose synthases, and 4 lignin synthases (Figures 4, E, F, and 8; Supplemental Table S5). Thus, we infer that alteration of m6A methylation regulatory factors, such as PtrMTA (Supplemental Figure S10C) and PtrALKBH8B (Supplemental Figure S11), cause an increase in m6A, which promotes mRNA degradation and represses protein production from secondary cell wall genes to reduce ATP consumption for better survival under drought stress (Figure 8, Module I versus Module II). Downregulation of wood-formation genes upon drought is in sharp contrast to upregulation of 43 ABA-related proteins involved in the ABA biosynthetic process, ABA metabolic process, and response to ABA (Figure 8; Supplemental Table S7). As expected, ABA-related proteins were upregulated because of the expression of stress-responsive genes is regulated by ABA during plant abiotic and biotic stress (Cutler et al., 2010).
mRNA–protein associations have revealed that ∼60% of variation in protein concentration cannot be explained by mRNA abundance alone (Vogel and Marcotte, 2012). In this study, we also found modest Rp (˂50%), and mRNA–protein correlations were decreased in both D5 and D7 compared with ND (Figure 3, A and B), suggesting that regulation of protein translation is far more complex under drought. The lack of strong correlation between full-length RNAs and proteins can be partly explained by posttranscriptional regulation, such as m6A, APA, and PAL (Figure 8). It is clear from previous studies that protein biosynthesis and cap-independent translation initiation is promoted by two well-characterized m6A readers, YTHDF1 (Wang et al., 2015) and YTHDF2 (Zhou et al., 2015), respectively. In this study, we found increased expression of three m6A readers in response to drought stress (Supplemental Figure S11). However, it remains unclear whether m6A readers influence protein biosynthesis upon drought in P. trichocarpa.
Recent technological advances in Nanopore DRS provide an unprecedented opportunity for distinguishing full-length and nonfull-length RNAs. We empirically assumed that full-length RNAs were the main contributors to protein concentrations. Surprisingly, we found that the Rp of nonfull-length RNA was always higher than that of full-length RNA based on genome-scale measurements of DRS-based RNA and MS-based protein levels. Although the mechanism remains unclear, we summarize several potential explanations for the lack of association between full-length RNA and protein abundance, which should be explored in the future. First, mRNAs and proteins have different turnovers associated with regulation by degradation. The lifetime for mRNA and protein is minutes and hours, respectively (Payne, 2015). After translation, full-length RNAs are rapidly degraded into nonfull-length RNAs. Thus, proteins are more stable than RNAs, which might explain why we always observed a higher Rp for nonfull-length RNA than for full-length RNA. The correlation between RNA and protein can be more accurately quantified through intensive time-course measurements based on mRNA–protein time series. Specifically, future correlation analysis should combine transcription and translation rates with mRNA and protein degradation rates. Second, the delay between transcription of full-length RNA and translation of protein can cause a temporal delay in protein biosynthesis during drought stress since the translation of full-length RNAs takes some time (Liu et al., 2016a). Third, not all full-length RNAs are translated. Future work should focus on identifying full-length RNAs bound by ribosomes using global ribosome profiling-based methods (Ingolia, 2014). Such studies will reveal whether ribosome-engaged full-length RNAs correlate better with protein level.
In rice, ∼48% of expressed genes use alternative polyadenylation (APA) to generate transcriptomic and proteomic diversity (Fu et al., 2016). In particular, APA is involved in stress-responsive dynamics in rice (Ye et al., 2019). In this study, we directly detected poly(A) usage shifts during drought stress using Nanopore DRS, which offers a great advantage over other methods for the identification of APA dynamics induced by drought. We identified a global increase in distal poly(A) site usage in drought-treated SDX, with genes exhibiting drought-responsive APA dynamics mostly functioning in cellulose synthase activity and protein-related activity. CPSF30 regulates poly(A) site choice of most transcripts in Arabidopsis (Chakrabarti and Hunt, 2015), and recent studies show that the CPSF30-L isoform regulates APA globally and has a slight preference for distal poly(A) sites (Hou et al., 2021; Song et al., 2021). In this study, we found that expression of Ptr-CPSF30 was upregulated and exhibited a shift to the distal poly(A) site upon drought treatment. It will be interesting to investigate whether Ptr-CPSF30 is involved in global poly(A) switching under drought treatment. The mutant of VIR promotes lengthening of the 3′-UTR of ATAF1, GI, and GSTU17 transcripts via APA (Hu et al., 2021). Hypomethylated sites in mutants of the m6A methyltransferase FIONA1 are associated with APA shifting to usage of proximal poly(A) sites (Xu et al., 2022). Furthermore, we found an m6A peak/site in the 3′-UTR of CPSF30-L (Supplemental Figure S16). It will be interesting to investigate the functional regulation of these m6A sites in the 3′-UTR, which might reflect the interplay between RNA modification and polyadenylation (Figure 8, Module II versus Module III).
Nanopore DRS and RT-qPCR validation revealed several significant differences in m6A-related genes including ECT2, ECT8, ALKBH8B, and CPSF30 in response to drought stress. Future work to explore dynamic binding sites of these erasers and readers under stress will provide further insights into the drought-regulated epitranscriptome. Toward this goal, recently developed approaches such as formaldehyde cross-linking and immunoprecipitation, which was developed for identifying binding sites on transcripts targeted by ECT2 in Arabidopsis (Wei et al., 2018a, 2018b), and HyperTRIBE, adapted to identify targets of m6A reader proteins, will be of great use in furthering our understanding of the drought-regulated epitranscriptome (Arribas-Hernandez et al., 2021; ZHou et al., 2021).
In summary, our study provides a comprehensive profile of epitranscriptome and proteome changes triggered by drought stress in P. trichocarpa SDX. Under drought treatment, the full-length read ratio decreases and m6A stimulates RNA degradation in cellulose- and lignin-related genes. Moreover, drought stress promotes a shift to distal poly(A) site usage in many genes, including CPSF30. Finally, PAL is related to the ratio of full-length reads and m6A-modification. These findings greatly expand our understanding of the effects of drought on posttranscriptional aspects of gene expression.
Materials and methods
Drought treatment and sample collection
Poplar (P.trichocarpa) plants were grown in a 22°C–25°C greenhouse and subjected to drought treatment as described previously (Li et al., 2019; Gao et al., 2021). In short, P. trichocarpa plants were divided into three groups: control (ND), D5, and D7. SDX tissues were collected directly into liquid nitrogen and stored at −80°C for DRS and liquid chromatography-tandem mass spectrometer (LC-MS/MS).
Library construction and analysis for Nanopore DRS
For ND, D5, and D7 samples, total RNA was extracted using an RNAprep Pure Kit (polysaccharides and polyphenolics-rich) (Code no. DP441, Tiangen Co. Ltd, Beijing, China) following the manufacturer’s instructions and treated with DNase I to remove DNA. Libraries for DRS were prepared using the Oxford Nanopore Technologies SQK-RNA002 kit protocol. In brief, poly(A)+ RNA was isolated using a Dynabeads mRNA purification kit (Thermo Fisher Scientific, Waltham, MA, USA; 61006) and ligated to Nanopore RTA adapter; first-strand cDNA was synthesized using SuperScript III reverse transcriptase (Thermo Fisher; 18080093) and ligated to RNA adapter (RMX). A total of 75-μL library was loaded onto the flow cell (Gao et al., 2021). Six libraries (two biological repeats for ND, N5, and N7) were constructed and run on a MinION flow cell (FLO-MIN106) independently. Details of DRS libraries and read statistics are provided in Supplemental Table S1. DRS raw data was analyzed using Guppy (v3.6.1) with default parameters to generate FASTQ, which was then converted to FASTA format and corrected using LoRDEC (Salmela and Rivals, 2014) with Illumina short reads (PRJNA315705; Li et al., 2019).
Estimation of full-length and nonfull-length reads
Full-length reads were identified among Nanopore DRS data using a previously described method with minor modifications (Roach et al., 2020). Annotated translation start sites were used as a standard for estimating the completeness of long reads. The first 20 bp were considered error sites to avoid sequence bias at the 5′-end. Reads were treated as full length or nonfull length according to whether their 5′-ends fell within ±20 of an annotated transcription start site. The full-length read ratio for each gene was calculated using the following formula: number of full-length reads/(number of full-length + nonfull-length reads). Differential full-length read ratio (ND versus D5, ND versus D7) for each gene was identified using the Fisher’s test based on the number of full-length reads and nonfull-length reads using P < 0.005 as the cutoff.
Identification of DEGs
Corrected DRS reads were aligned to the P. trichocarpa v3.0 genome using minimap2 (version 2.10) (Li, 2018) with –secondary=no -ax splice -uf -k14 -t 40 parameters. BAM format files were then converted to bed12 format using BEDTools with the bamtobed -bed12 -split option (Quinlan and Hall, 2010). Reads corresponding to each gene were called using BEDTools with intersect -wo -a -b.
DEGs in ND versus D5 and ND versus D7 comparisons were identified using edgeR (Robinson et al., 2010) with fold change >2 and P < 0.005 as a cutoff. GO enrichment analysis of DEGs was performed with the Poplar Gene Web Server (Liu et al., 2016b) using the Chi-square statistical test and the Hochberg FDR multi-test adjustment method; the significance level was false discovery rate (FDR) < 0.05.
Identification of DE m6A
m6A sites were identified using the published Nanom6A (Gao et al., 2021) pipeline with default parameters, and the m6A ratio for each site in each gene was calculated using the following formula: number of modified reads/(number of modified reads + number of unmodified reads). Fisher’s extract test was used to identify differential m6A sites between ND and drought treatments using the number of m6A-modified and unmodified reads. P < 0.05 and differential m6A ratio >0.1 were used to identify DE m6A sites.
RT-qPCR validation
Total RNA was extracted using an RNAprep Pure Kit (polysaccharides and polyphenolics-rich) (Tiangen; no. DP441) following the manufacturer’s instructions and treated with DNase I to remove DNA. Total RNA (1 μg) from each sample was reverse transcribed into cDNA using a PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa, Shiga, Japan; RR047A) for validation of the m6A functional factors. Oligo (dT) primer was used for validating the full-length read ratios of DUF239 and SGR1, and the APA of CPSF30, according to the protocol of the HiScript II 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Vazyme, Nanjing, China; R212-01). All cDNAs were diluted 10×, and 1–2 μL of 10× cDNA was used as template in a 20-μL RT-qPCR system. EF1α and 18S rRNA were used as reference genes. RT-qPCR was performed using Hieff qPCR SYBR Green Master Mix (YEASEN, Shanghai, China; 11202ES08) on an Agilent M × 3005P Real-Time PCR System, following the manufacturers’ instructions. All RT-qPCR primers are detailed in Supplemental Table S16 and Supplemental Figure S28. For RT-qPCR analysis, expression relative to that under ND was firstly calculated using ΔΔCt and then using the full-length read ratio approximately equal to full-length read expression divided by total read expression; PPR relative expression was obtained by proximal/(proximal + distal). Lastly, an F.TEST was performed, and T.TEST was used to evaluate significant difference (*P < 0.05).
Identification of differentially cleaved poly(A) sites
The 3′-positions of mapped individual reads were obtained and allocated into 25-nt poly(A) site windows (PAWs). The depth of PAWs was calculated using in-house python scripts. The two most abundant PAWs were retained for identifying differentially cleaved poly(A) sites using Fisher’s exact test with P < 0.005 as the cutoff. A proximal poly(A) usage ratio of >10% was considered as a differential APA event.
Identification of differential PAL
PAL of individual reads was calculated using Nanopolish (version 0.11.1; Workman et al., 2019). Differential PAL was identified using edgeR (Robinson et al., 2010). Fold change >1.5 and P< 0.05 between ND and drought treatments were considered to represent differential PAL.
GO enrichment and heatmap
The Poplar Gene Web Server (Liu et al., 2016b) was used for GO enrichment analysis using the Chi-square statistical test and the Hochberg FDR multi-test adjustment method with FDR < 0.05 as a cutoff. Heatmaps were plotted using TBtools (Chen et al., 2020).
Protein extraction, digestion, and TMT labeling
Fresh SDX (0.3 g; three biological repeats for ND, N5, and N7) was ground in liquid nitrogen and suspended in 1-mL lysis buffer (10-mM Tris–HCl (pH 8.0), 5-mM EDTA, 1% (w/v), sodium dodecylsulfate (SDS), 8-M urea, and 20-mM dithiothreitol). Samples were vortexed and then incubated on ice for 30 min. After incubation at 4°C for 2 h with 20g, the lysate was centrifuged at 15,000g for 30 min at 4°C. The supernatant was aspirated to a new tube and centrifuged again for 15 min. The supernatant was then transferred to another new tube without touching any impurities then precipitated overnight at −20°C using six volumes of 10% (v/v) TCA/acetone (10% trichloroacetic acid dissolved in acetone). After centrifugation at 15,000g for 15 min at 4°C, pellets were washed 2 times with precooled acetone (flushed repeatedly using a pipette tip and gently poked several times). The supernatant was then removed, and pellets were air-dried for 2–10 min. Protein samples were dissolved in buffer (8-M urea, 25× Cocktail (Thermo Scientific; A32963), 100-mM Tris–HCl, pH 8.0) on ice, centrifuged 15,000g for 15 min at 4°C, and the supernatant was transferred to a new tube. The concentration of purified proteins was measured by bicinchoninic acid (BCA)protein assay (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions.
A total of 100 μg of each sample (three biological repeats for ND, N5, and N7) was digested using a filter-aided sample preparation procedure with minor modifications (Wiśniewski et al., 2009). Briefly, several volumes of 8-M urea were added to each sample to bring the final concentration of urea to >4 M; samples were then mixed with 1-M dithiothreitol to a final concentration of 20 mM and incubated at 37°C for 1 h. Next, 1-M iodoacetamide was added to a final concentration of 50–60 mM, incubated at room temperature in darkness for an additional 30 min, and centrifuged at 12,000g for 10 min. Subsequently, 100 μL 8-M urea was added, and samples were centrifuged again at 12,000g for 10 min; this step was repeated 2 times. A total of 100 μL 50-mM ammonium bicarbonate (ABC) or triethylammonium bicarbonate (TEAB) was added, and samples were centrifuged at 12,000g for 10 min. This step was repeated two to 5 times. The supernatant was collected in a new tube and digested using 100-μL trypsin buffer (trypsin in 100 μL 50-mM ABC or TEAB) with a ratio of 1:50 (trypsin buffer: protein sample) at 37°C for at least 8 h. Samples were then centrifuged at 12,000g for 10 min before adding 100-μL HPLC water, vortexing, and centrifuging again. The supernatant was collected and dried by vacuum freeze-drying and stored at −20°C until use.
The resulting peptide mixture was labeled using 10-plex TMT reagent according to the manufacturer’s instructions (Thermo Fisher Scientific, MA, USA). The peptide mixture was prefractionated using an Ultimate 3000 (Thermo Fisher Scientific, MA, USA), and MS analysis was performed following methods described in a previous study (Yu et al., 2019).
LC–MS/MS analysis
Raw data were searched against the UniProt protein database and all redundant sequences were removed using Proteome Discoverer version 2.2 software. For protein identification, the following options were used: MS1 tolerance = 10 ppm; MS/MS fragment tolerance = 0.02 Da; enzyme was set to trypsin with no more than two miss cleavages; fixed modification: carbamidomethyl (C), TMT 10 plex (N-term), TMT 10 plex (K); variable modification of oxidation (M) and acetylation (N-term); FDR < 0.01, and more than one unique peptide. Only unique peptides with report ion S/N > 15 and FDR < 0.01 were used for protein quantification; the signal error between different samples was normalized using the sum of the total intensity of every report ion channel. For the selection of DEPs, a 1.5-fold cutoff and P < 0.05 were used.
Accession numbers
All ONT FAST5 sequencing data have been deposited in NCBI under accession PRJNA672182. Mass spectrometry proteomics data were deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2018) partner repository with the dataset identifier PXD029406. Analysis and visualization were performed using Python scripts; all code is available by request from the corresponding author. Transcript sequences, coding sequences, and protein sequences of important genes mentioned in this study are listed in Supplemental Table S17. All data in this study are available at forestry.fafu.edu.cn/db/Ptr_Drought. The database provides modules for processing queries by gene ID or GO term and a Genome Browser to visualize Nanopore DRS reads.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Wiggle plot, histogram, and RT-qPCR show increased full-length read ratio of SGR1 upon drought treatment.
Supplemental Figure S2. Expression and translation of exonucleases in response to drought treatment.
Supplemental Figure S3. Heatmap of 23 upregulated TFs and 4 downregulated TFs in SDX under drought treatment.
Supplemental Figure S4. Transcription, posttranscription, and translation of PtrAREB1-2, PtrAREB1-4, PtrGT56, ERF76, ERF38, MYB010, BLH6a, and GRAS under drought stress.
Supplemental Figure S5. Frequency distribution plots for protein abundance, full-length RNA abundance (RPM), and nonfull-length RNA abundance (RPM) across all samples.
Supplemental Figure S6. Association between full-length transcript level and protein abundance from PacBio Iso-Seq.
Supplemental Figure S7. Rp of gene clusters in full-length reads according to GO term.
Supplemental Figure S8. Rp of gene clusters in nonfull-length reads according to GO term.
Supplemental Figure S9. Rp of “endoplasmic reticulum to Golgi vesicle-mediated transport” and “structural molecule activity” in full-length reads.
Supplemental Figure S10. m6A sites, m6A genes, MTA expression, and MTA protein abundance under drought stress.
Supplemental Figure S11. Expression profiles of genes encoding m6A functional factors and RT-qPCR validation.
Supplemental Figure S12. GO enrichment of genes with altered m6A ratio in D5 and D7.
Supplemental Figure S13. Scatter plots from the pairwise comparison D7 versus ND show the relationships of m6A ratio with gene expression, protein level, and H3K9ac level.
Supplemental Figure S14. Scatter plots showing full-length RNA ratio in Col-0 versus vir-1, VIRc versus vir-1, and WT versus shMETTL3.
Supplemental Figure S15. Histogram showing GO enrichment analysis of genes with increased full-length RNA ratio in vir-1.
Supplemental Figure S16. Wiggle plot showing an m6A peak in the 3′-UTR of CPSF30-L.
Supplemental Figure S17. Wiggle plot and histogram showing CESA7 (Potri.018G103900) as an example of a gene with increased PPR.
Supplemental Figure S18. Differential poly(A) switch regions with m6A sites showing enrichment at proximal poly(A) sites in D5 and D7.
Supplemental Figure S19. Box plots showing functions of 10 groups with the longest PAL (orange) and 10 groups with the shortest PAL (blue) in D5 and D7.
Supplemental Figure S20. Box plots showing PAL distribution for fully spliced (no_IR) and IR transcripts in full-length reads and nonfull-length reads.
Supplemental Figure S21. GO enrichment of genes with differential PAL in D5 versus ND.
Supplemental Figure S22. GO enrichment of genes with differential PAL in D7 versus ND.
Supplemental Figure S23. Box plots show that drought stress induces longer PAL and shorter PAL compared with ND.
Supplemental Figure S24. Distribution of median PAL per gene in ND, D5, and D7.
Supplemental Figure S25. PAL distribution in pools of genes with different expression levels in ND, D5, and D7.
Supplemental Figure S26. Full-length RNA has longer PAL than nonfull-length RNA.
Supplemental Figure S27. Scatter plot showing median PAL for m6A-modified and non-m6A-modified transcripts in ND, D5, and D7.
Supplemental Figure S28. Primer positions for m6A functional factors.
Supplemental Table S1. DRS library construction.
Supplemental Table S2. Genes with differential full-length read ratio in D5 versus ND and D7 versus ND.
Supplemental Table S3. Nine genes with increased full-length read ratio in both D5 versus ND and D7 versus ND comparisons.
Supplemental Table S4. DE proteins in D5 versus ND and D7 versus ND.
Supplemental Table S5. Dynamics of full-length read ratio, DEGs, DEPs, DE m6A, differential PAL, and differential APA associated with wood-formation genes under drought stress.
Supplemental Table S6. Dynamics of full-length read ratio, DEGs, DEPs, DE m6A, differential PAL, and differential APA based on the GO term “response to water deprivation” under drought stress.
Supplemental Table S7. Dynamics of full-length read ratio, DEGs, DEPs, DE m6A, differential PAL, and differential APA associated with ABA-related genes under drought stress.
Supplemental Table S8. DEGs in D5 versus ND and D7 versus ND.
Supplemental Table S9. Rp of full-length reads versus protein abundance and nonfull-length reads versus protein abundance.
Supplemental Table S10. DE m6A in ND versus D5 and ND versus D7.
Supplemental Table S11. DE m6A genes versus DEGs, DE m6A genes versus proteins, and DE m6A genes versus DE H3K9ac in D5 versus ND and D7 versus ND.
Supplemental Table S12. Differential APA in D5 versus ND and D7 versus ND.
Supplemental Table S13. MicroRNA target sites in genes with decreased PPR.
Supplemental Table S14. PAL of genes in ND, D5, and D7.
Supplemental Table S15. Differential PAL in ND versus D5 and ND versus D7.
Supplemental Table S16. RT-qPCR validation of m6A functional factors, full-length read ratios of DUF239 and SGR1, and the APA of CPSF30.
Supplemental Table S17. Names, transcript sequences, coding sequences, and protein sequences of important genes mentioned in this study.
Supplementary Material
Acknowledgments
We thank Liang Yang and Nan Yao for TMT-based proteomic experiments. The contents of this study are solely the responsibility of the authors.
Funding
This work was supported by the National Key Research and Development Program of China (2016YFD0600106), Fujian Forest Seedling Technology Project, a National Natural Science Foundation of China Grant (31970182), the Fund for scientific and technological innovation of Fujian Agriculture and Forestry University (CXZX2020093A), Undergraduate Training Program for Innovation and Entrepreneurship of China (202110389013), and Forestry Peak Discipline Construction Project from Fujian Agriculture and Forestry University.
Conflict of interest statement. None declared.
Contributor Information
Yubang Gao, College of Forestry, Basic Forestry and Proteomics Research Center, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
Xuqing Liu, College of Forestry, Basic Forestry and Proteomics Research Center, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
Yandong Jin, College of Forestry, Basic Forestry and Proteomics Research Center, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
Ji Wu, College of Forestry, Basic Forestry and Proteomics Research Center, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
Shuang Li, State Key Laboratory of Tree Genetics and Breeding, Northeast Forestry University, Harbin 150040, China.
Yaxing Li, Basic Forestry and Proteomics Research Center, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
Binqing Chen, Basic Forestry and Proteomics Research Center, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
Yaxin Zhang, College of Forestry, Basic Forestry and Proteomics Research Center, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
Linxiao Wei, College of Forestry, Basic Forestry and Proteomics Research Center, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
Wei Li, State Key Laboratory of Tree Genetics and Breeding, Northeast Forestry University, Harbin 150040, China.
Ruili Li, National Engineering Research Center of Tree Breeding and Ecological Restoration, College of Biological Sciences and Technology, Beijing Forestry University, Beijing 100083, China.
Chentao Lin, Department of Molecular, Cell and Developmental Biology, University of California, Los Angeles, California 90095, USA.
Anireddy S N Reddy, Department of Biology and Program in Cell and Molecular Biology, Colorado State University, Fort Collins, Colorado, USA.
Pankaj Jaiswal, Department of Botany and Plant Pathology, Oregon State University, Corvallis, Oregon, USA.
Lianfeng Gu, College of Forestry, Basic Forestry and Proteomics Research Center, Fujian Agriculture and Forestry University, Fuzhou 350002, China.
L.F.G. conceived and designed this project. Y.B.G. performed bioinformatics. X.Q.L., Y.D.J., J.W., S.L., and W.L. performed drought treatments, total RNA extraction, poly(A)+ RNA purification, and Nanopore DRS. X.Q.L., Y.X.L., and Y.X.Z. performed TMT-based proteomic experiments. X.Q.L., B.Q.C., and L.X.W. performed RT-qPCR validation. Y.B.G., X.Q.L., and L.F.G. wrote the manuscript. R.L.L., C.T.L., A.S.N.R., and P.J. discussed the research plan and revised the article. All authors read and approved the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is Lianfeng Gu (lfgu@fafu.edu.cn).
References
- Anderson SJ, Kramer MC, Gosai SJ, Yu X, Vandivier LE, Nelson AD, Anderson ZD, Beilstein MA, Fray RG, Lyons E (2018) N6-methyladenosine inhibits local ribonucleolytic cleavage to stabilize mRNAs in Arabidopsis. Cell Rep 25:1146–1157 e1143 [DOI] [PubMed] [Google Scholar]
- Arribas-Hernandez L, Rennie S, Koster T, Porcelli C, Lewinski M, Staiger D, Andersson R, Brodersen P (2021) Principles of mRNA targeting via the Arabidopsis m(6)A-binding protein ECT2. eLife 10: e72375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J (1999) Selective translation of cytoplasmic mRNAs in plants. Trend Plant Sci 4:142. [DOI] [PubMed] [Google Scholar]
- Bardwell VJ, Zarkower D, Edmonds M, Wickens M (1990) The enzyme that adds poly (A) to mRNAs is a classical poly (A) polymerase. Mol Cell Biol 10:846–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Del Viso F, Moreno-Mateos MA, Johnstone TG, Vejnar CE, Qin Y, Yao J, Khokha MK, Giraldez AJ (2016) Codon identity regulates mRNA stability and translation efficiency during the maternal-to-zygotic transition. EMBO J 35:2087–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilharz TH, Preiss T (2007) Widespread use of poly (A) tail length control to accentuate expression of the yeast transcriptome. RNA 13:982–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti M, de Lorenzo L, Abdel-Ghany SE, Reddy ASN, Hunt AG (2020) Wide-ranging transcriptome remodelling mediated by alternative polyadenylation in response to abiotic stresses in Sorghum. Plant J 102:916–930 [DOI] [PubMed] [Google Scholar]
- Chakrabarti M, Hunt AG (2015) CPSF30 at the interface of alternative polyadenylation and cellular signaling in plants. Biomolecules 5:1151–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13:1194–1202 [DOI] [PubMed] [Google Scholar]
- Chen CY, Shyu AB (2011) Mechanisms of deadenylation-dependent decay. Wiley Interdiscip Rev RNA 2:167–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61:651–679 [DOI] [PubMed] [Google Scholar]
- Dai X, Zhuang Z, Zhao PX (2018) psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res 46:W49–W54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave P, Chao JA (2020) Insights into mRNA degradation from single-molecule imaging in living cells. Curr Opin Struct Biol 65:89–95 [DOI] [PubMed] [Google Scholar]
- De Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C (2009) Global signatures of protein and mRNA expression levels. Mol BioSyst 5:1512–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan HC, Wei LH, Zhang C, Wang Y, Chen L, Lu Z, Chen PR, He C, Jia G (2017) ALKBH10B is an RNA N6-methyladenosine demethylase affecting Arabidopsis floral transition. Plant Cell 29:2995–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann CR, Rammelt C, Wahle E (2011) Control of poly (A) tail length. Wiley Interdiscip Rev RNA 2:348–361 [DOI] [PubMed] [Google Scholar]
- Floris M, Mahgoub H, Lanet E, Robaglia C, Menand B (2009) Post-transcriptional regulation of gene expression in plants during abiotic stress. Int J Mol Sci 10:3168–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Yang D, Su W, Ma L, Shen Y, Ji G, Ye X, Wu X, Li QQ (2016) Genome-wide dynamics of alternative polyadenylation in rice. Genome Res 26:1753–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuke H, Ohno M (2008) Role of poly (A) tail as an identity element for mRNA nuclear export. Nucleic Acids Res 36:1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Liu X, Wu B, Wang H, Xi F, Kohnen MV, Reddy AS, Gu L (2021) Quantitative profiling of N 6-methyladenosine at single-base resolution in stem-differentiating xylem of Populus trichocarpa using Nanopore direct RNA sequencing. Genom Biol 22:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum D, Colangelo C, Williams K, Gerstein M (2003) Comparing protein abundance and mRNA expression levels on a genomic scale. Genom Biol 4:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L, Zhao M, Qian Y, Yu H, Wu E (2019) Phenotypic analysis combined with tandem mass tags (TMT) labeling reveal the heterogeneity of strawberry stolon buds. BMC Plant Biol 19:505–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Sun J, Wu B, Gao Y, Nie H, Nie Z, Quan S, Wang Y, Cao X, Li S (2021) CPSF30-L-mediated recognition of mRNA m6A modification controls alternative polyadenylation of nitrate signaling-related gene transcripts in Arabidopsis. Mol Plant 14:688–699 [DOI] [PubMed] [Google Scholar]
- Hu J, Cai J, Park SJ, Lee K, Li Y, Chen Y, Yun JY, Xu T, Kang H (2021) N 6-Methyladenosine mRNA methylation is important for salt stress tolerance in Arabidopsis. Plant J 106:1759–1775 [DOI] [PubMed] [Google Scholar]
- Huang Y, Carmichael GG (1996) Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol Cell Biol 16:1534–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang HW, Darnell RB (2017) Comprehensive identification of mRNA polyadenylation sites by PAPERCLIP. mRNA Proc 1648:79–93 [DOI] [PubMed] [Google Scholar]
- Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L (2001) Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292:929–934 [DOI] [PubMed] [Google Scholar]
- Ingolia NT (2014) Ribosome profiling: new views of translation, from single codons to genome scale. Nat Rev Genet 15:205–213 [DOI] [PubMed] [Google Scholar]
- Jin J, Feng T, De-Chang Y, Yu-Qi M, Lei K, Luo J, Gao G (2017) PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res 45:D1040–D1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Huang G, Xiong Y, Zhao C, Wang J, Song X, Giri J, Zuo K (2019) IBR5 regulates leaf serrations development via modulation of the expression of PIN1. Int J Mol Sci 20:4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer MC, Janssen KA, Palos K, Nelson AD, Vandivier LE, Garcia BA, Lyons E, Beilstein MA, Gregory BD (2020) N6-methyladenosine and RNA secondary structure affect transcript stability and protein abundance during systemic salt stress in Arabidopsis. Plant Direct 4:e00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Niazi AM, Labun K, Cleuren YNT, Müller FS, Valen E (2019) tailfindr: alignment-free poly (A) length measurement for Oxford Nanopore RNA and DNA sequencing. RNA 25:1229–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2018) Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34:3094-3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Lin YJ, Wang P, Zhang B, Li M, Chen S, Shi R, Tunlaya-Anukit S, Liu X, Wang Z, et al. (2019) The AREB1 transcription factor influences histone acetylation to regulate drought responses and tolerance in Populus trichocarpa. Plant Cell 31:663–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang R, Gao Y, Wang C, Zhao L, Xu N, Chen KE, Qi S, Zhang M, Tsay YF (2017) The Arabidopsis CPSF30-L gene plays an essential role in nitrate signaling and regulates the nitrate transceptor gene NRT 1.1. New Phytologist 216:1205–1222 [DOI] [PubMed] [Google Scholar]
- Lima SA, Chipman LB, Nicholson AL, Chen YH, Yee BA, Yeo GW, Coller J, Pasquinelli AE (2017) Short poly (A) tails are a conserved feature of highly expressed genes. Nat Struct Mol Biol 24:1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, u Y, Beyer A, Aebersold R (2016a) On the dependency of cellular protein levels on mRNA abundance. Cell 165:535–550 [DOI] [PubMed] [Google Scholar]
- Liu Q, Ding C, Chu Y, Chen J, Zhang W, Zhang B, Huang Q, Su X (2016b) PoplarGene: poplar gene network and resource for mining functional information for genes from woody plants. Sci Rep 6:31356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Gao Y, Liao J, Miao M, Chen K, Xi F, Wei W, Wang H, Wang Y, Xu X (2021a) Genome-wide profiling of circular RNAs, alternative splicing, and R-loops in stem-differentiating xylem of Populus trichocarpa. J Integr Plant Biol 63:1294–1308 [DOI] [PubMed] [Google Scholar]
- Liu Y, Nie H, Liu H, Lu F (2019) Poly (A) inclusive RNA isoform sequencing (PAIso− seq) reveals wide-spread non-adenosine residues within RNA poly (A) tails. Nat Commun 10:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Nie H, Sun R, Wang J, Lu F (2021b) Enhancement of synthetic mRNA translation efficiency through engineered poly (A) tails. bioRxiv 10.1101/2021.08.30.458298 [DOI] [Google Scholar]
- Liu Y, Nie H, Zhang C, Hou Z, Wang J, Lu F (2021c) Poly(A) tail length is a major regulator of maternal gene expression during the mammalian oocyte-to-embryo transition. bioRxiv 10.1101/2021.08.29.458052 [DOI] [Google Scholar]
- Liu Y, Nie H, Zhang Y, Lu F, Wang J (2021d) Comprehensive analysis of mRNA poly(A) tail reveals complex and conserved regulation. bioRxiv 10.1101/2021.08.29.458068 [DOI] [Google Scholar]
- Liu Y, Zhang Y, Lu F, Wang J (2021e) Interactions between RNA m6A modification, alternative splicing, and poly (A) tail revealed by MePAIso-seq2. bioRxiv 10.1101/2021.08.29.458071 [DOI] [Google Scholar]
- Lorenz DA, Sathe S, Einstein JM, Yeo GW (2020) Direct RNA sequencing enables m6A detection in endogenous transcript isoforms at base-specific resolution. RNA 26:19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Zhang Y, He Q, Qi Z, Zhang G, Xu W, Yi T, Wu G, Li R (2020) MTA, an RNA m6A methyltransferase, enhances drought tolerance by regulating the development of trichomes and roots in poplar. Int J Mol Sci 21:2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Pérez M, Aparicio F, López-Gresa MP, Bellés JM, Sánchez-Navarro JA, Pallás V (2017) Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc Natl Acad Sci USA 114:10755–10760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maud V, Hazel K, Smith D, Cohen J, Dewoody H (2016) Adaptive mechanisms and genomic plasticity for drought tolerance identified in European black poplar (Populus nigra L). Tree Physiol 36:909–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Temme C, Wahle E (2004) Messenger RNA turnover in eukaryotes: pathways and enzymes. Critic Rev Biochem Mol Biol 39:197–216 [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41:697–709 [DOI] [PubMed] [Google Scholar]
- Niazi AM, Krause M, Valen E (2021) Transcript isoform-specific estimation of poly (A) tail length by Nanopore sequencing of native RNA. Methods Mol Biol (Clifton, NJ) 2284:543–567 [DOI] [PubMed] [Google Scholar]
- Nichols J (1979) ‘Cap’structures in maize poly (A)-containing RNA. Biochim Biophys Acta 563:490–495 [DOI] [PubMed] [Google Scholar]
- Ono K, Kimura M, Matsuura H, Tanaka A, Ito H (2019) Jasmonate production through chlorophyll a degradation by stay-green in Arabidopsis thaliana. J Plant Physiol 238:53–62 [DOI] [PubMed] [Google Scholar]
- Park JA, Cho SK, Kim JE, Chung HS, Hong JP, Hwang B, Hong CB, Kim WTJPS (2003) Isolation of cDNAs differentially expressed in response to drought stress and characterization of the Ca-LEAL1 gene encoding a new family of atypical LEA-like protein homologue in hot pepper (Capsicum annuum L. cv. Pukang). Plant Sci 165:471–481 [Google Scholar]
- Parker MT, Knop K, Sherwood AV, Schurch NJ, Simpson GG (2020) Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m6A modification. Elife Sci 9:e49658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne SH (2015) The utility of protein and mRNA correlation. Trends Biochem Sci 40:1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. (2018) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47:D442–D450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnala L, Wang Y, Sun Q, van Wijk KJ (2014) Correlation of mRNA and protein abundance in the developing maize leaf. Plant J 78:424–440 [DOI] [PubMed] [Google Scholar]
- Presnyak V, Alhusaini N, Chen YH, Martin S, Morris N, Kline N, Olson S, Weinberg D, Baker KE, Graveley BR (2015) Codon optimality is a major determinant of mRNA stability. Cell 160:1111–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A, Chen YH, Martin S, Alhusaini N, Green R, Coller J (2016) The DEAD-box protein Dhh1p couples mRNA decay and translation by monitoring codon optimality. Cell 167:122–132. e129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach NP, Sadowski N, Alessi AF, Timp W, Taylor J, Kim JK (2020) The full-length transcriptome of C. elegans using direct RNA sequencing. Genome Res 30:299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routh A, Ji P, Jaworski E, Xia Z, Li W, Wagner EJ (2017) Poly (A)-ClickSeq: click-chemistry for next-generation 3′-end sequencing without RNA enrichment or fragmentation. Nucleic Acids Res 45:e112–e112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MG (2011) Tree responses to drought. Tree Physiol 31:237–239 [DOI] [PubMed] [Google Scholar]
- Salmela L, Rivals E (2014) LoRDEC: accurate and efficient long read error correction. Bioinformatics 30:3506–3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg DR, Maquat LE (2012) Regulation of cytoplasmic mRNA decay. Nat Rev Genet 13:246–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M (2011) Global quantification of mammalian gene expression control. Nature 473:337–342 [DOI] [PubMed] [Google Scholar]
- Shao Y, Wong CE, Shen L, Yu H (2021) N6-methyladenosine modification underlies messenger RNA metabolism and plant development. Curr Opin Plant Biol 63:102047. [DOI] [PubMed] [Google Scholar]
- Sherstnev A, Duc C, Cole C, Zacharaki V, Hornyik C, Ozsolak F, Milos PM, Barton GJ, Simpson GG (2012) Direct sequencing of Arabidopsis thaliana RNA reveals patterns of cleavage and polyadenylation. Nat Struct Mol Biol 19:845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song P, Yang J, Wang C, Lu Q, Shi L, Tayier S, Jia G (2021) Arabidopsis N6-methyladenosine reader CPSF30-L recognizes FUE signals to control polyadenylation site choice in liquid-like nuclear bodies. Mol Plant 14:571–587 [DOI] [PubMed] [Google Scholar]
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP (2014) Poly (A)-tail profiling reveals an embryonic switch in translational control. Nature 508:66–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PE, Wu X, Liu M, Gaffney B, Ji G, Li QQ, Hunt AG (2012) Genome-wide control of polyadenylation site choice by CPSF30 in Arabidopsis. Plant Cell 24:4376–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A, Schäfer J, Kuhn K, Kienle S, Hamon C (2003) Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem 75:1895–1904 [DOI] [PubMed] [Google Scholar]
- Velten L, Anders S, Pekowska A, Järvelin AI, Huber W, Pelechano V, Steinmetz LM (2015) Single-cell polyadenylation site mapping reveals 3′ isoform choice variability. Mol Syst Biol 11:812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, de Sousa Abreu R, Ko D, Le SY, Shapiro BA, Burns SC, Sandhu D, Boutz DR, Marcotte EM, Penalva LO (2010) Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol Syst Biol 6:400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, Marcotte EM (2012) Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 13:227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Yang J, Song P, Zhang W, Lu Q, Yu Q, Jia G (2022) FIONA1 is an RNA N(6)-methyladenosine methyltransferase affecting Arabidopsis photomorphogenesis and flowering. Genome Biol 23:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C (2015) N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161:1388–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei LH, Song P, Wang Y, Lu Z, Tang Q, Yu Q, Xiao Y, Zhang X, Duan HC, Jia G (2018a) The m6A reader ECT2 controls trichome morphology by affecting mRNA stability in Arabidopsis. Plant Cell 30:968–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, Wang X, Jiang D, Dong S (2018b) Physiological and proteome studies of maize (Zea mays L.) in response to leaf removal under high plant density. BMC Plant Biol 18:378–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill L, Belloc E, Bava FA, Méndez R (2012) Translational control by changes in poly(A) tail length: recycling mRNAs. Nat Struct Mol Biol 19:577–585 [DOI] [PubMed] [Google Scholar]
- West SM, Mecenas D, Gutwein M, Aristizábal-Corrales D, Piano F, Gunsalus KC (2018) Developmental dynamics of gene expression and alternative polyadenylation in the Caenorhabditis elegans germline. Genome Biol 19:1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6:359–362 [DOI] [PubMed] [Google Scholar]
- Woo YM, Kwak Y, Namkoong S, Kristjánsdóttir K, Lee SH, Lee JH, Kwak H (2018) TED-seq identifies the dynamics of poly (A) length during ER stress. Cell Rep 24:3630–3641 e3637 [DOI] [PubMed] [Google Scholar]
- Workman RE, Tang AD, Tang PS, Jain M, Tyson JR, Razaghi R, Zuzarte PC, Gilpatrick T, Payne A, Quick J, et al. (2019) Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat Methods 16:1297–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Liu M, Downie B, Liang C, Ji G, Li QQ, Hunt AG (2011) Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc Natl Acad Sci USA 108:12533–12538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Wang J, Wu X, Hong Y, Li QQ (2020) Heat shock responsive gene expression modulated by mRNA poly (A) tail length. Front Plant Sci 11:1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Liu L, Zhang Y, Sun H, Zhang K, Bai Z, Dong H, Liu Y, Li C (2020) Tandem mass tag-based (TMT) quantitative proteomics analysis reveals the response of fine roots to drought stress in cotton (Gossypium hirsutum L.). BMC Plant Biol 20:1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Wu X, Wong CE, Fan S, Zhang Y, Zhang S, Liang Z, Yu H, Shen L (2022) FIONA1-mediated m(6) a modification regulates the floral transition in Arabidopsis. Adv Sci 9:e2103628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Wang Y, Lyu T, Hu Z, Ye N, Liu W, Li J, Yao X, Yin H (2021) Alternative Polyadenylation in response to temperature stress contributes to gene regulation in Populus trichocarpa. BMC Genom 22:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C, Zhou Q, Wu X, Ji G, Li QQ (2019) Genome-wide alternative polyadenylation dynamics in response to biotic and abiotic stresses in rice. Ecotoxicol Environ Saf 183:109485. [DOI] [PubMed] [Google Scholar]
- Yu Q, Liu S, Yu L, Xiao Y, Zhang S, Wang X, Xu Y, Yu H, Li Y, Yang J, et al. (2021) RNA demethylation increases the yield and biomass of rice and potato plants in field trials. Nat Biotechnol 39:1581–1588 [DOI] [PubMed] [Google Scholar]
- Yu X, Wang Y, Kohnen MV, Piao M, Tu M, Gao Y, Lin C, Zuo Z, Gu L (2019) Large scale profiling of protein isoforms using label-free quantitative proteomics revealed the regulation of nonsense-mediated decay in moso bamboo (Phyllostachys edulis). Cells 8:744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Zhang YC, Liao JY, Yu Y, Zhou YF, Feng YZ, Yang YW, Lei MQ, Bai M, Wu H (2019a) The subunit of RNA N6-methyladenosine methyltransferase OsFIP regulates early degeneration of microspores in rice. PLoS Genet 15:e1008120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Fan Y, Sun X, Chen L, Terzaghi W, Bucher E, Li L, Dai M (2019b) A large‐scale circular RNA profiling reveals universal molecular mechanisms responsive to drought stress in maize and Arabidopsis. Plant J 98:697–713 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu L, Qiu Q, Zhou Q, Ding J, Lu Y, Liu P (2021) Alternative polyadenylation: methods, mechanism, function, and role in cancer. J Exp Clin Cancer Res 40:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Chen LQ, Zhao YL, Yang CG, Roundtree IA, Zhang Z, Ren J, Xie W, He C, Luo GZ (2019c) Single-base mapping of m 6 A by an antibody-independent method. Sci Adv 5:eaax0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, Fray RG (2008) MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20:1278–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Niu R, Zhou Y, Luo M, Peng Y, Wang H, Wang Z, Xu G (2021) Proximity editing to identify RNAs in phase-separated RNA binding protein condensates. Cell Discov 7:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB (2015) Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 526:591–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Tian S, Qin G (2019) RNA methylomes reveal the m 6 A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening. Genom Biol 20:1–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Li R, Michal JJ, Wu XL, Liu Z, Zhao H, Xia Y, Du W, Wildung MR, Pouchnik DJ (2016) Accurate profiling of gene expression and alternative polyadenylation with whole transcriptome termini site sequencing (WTTS-Seq). Genetics 203:683–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.