Abstract
Crosses between Drosophila simulans females and Drosophila melanogaster males produce viable F1 sons and poorly viable F1 daughters. Unlike most hybrid incompatibilities, this hybrid incompatibility violates Haldane’s rule, the observation that incompatibilities preferentially affect the heterogametic sex. Furthermore, it has a different genetic basis than hybrid lethality in the reciprocal cross, with the causal allele in Drosophila melanogaster being a large species-specific block of complex satellite DNA on its X chromosome known as the 359-bp satellite, rather than a protein-coding locus. The causal allele(s) in Drosophila simulans are unknown but likely involve maternally expressed genes or factors since the F1 females die during early embryogenesis. The maternal haploid (mh) gene is an intriguing candidate because it is expressed maternally and its protein product localizes to the 359-bp repeat. We found that this gene has diverged extensively between Drosophila melanogaster and Drosophila simulans. This observation led to the hypothesis that Drosophila melanogaster mh may have coevolved with the 359-bp repeat and that hybrid incompatibility thus results from the absence of a coevolved mh allele in Drosophila simulans. We tested for the functional divergence of mh by creating matched transformants of Drosophila melanogaster and Drosophila simulans orthologs in both Drosophila melanogaster and Drosophila simulans strains. Surprisingly, we find that Drosophila simulans mh fully complements the female sterile phenotype of Drosophila melanogaster mh mutations. Contrary to our hypothesis, we find no evidence that adding a Drosophila melanogaster mh gene to Drosophila simulans increases hybrid viability.
Keywords: Drosophila, hybrid incompatibilities, speciation
Introduction
The evolution of reproductive isolation via hybrid incompatibilities can be complex, with multiple incompatibilities contributing to isolation within a single species pair. Two genetically distinct lethal hybrid incompatibilities exist between the sister species Drosophila melanogaster and Drosophila simulans (Sawamura, Watanabe, et al. 1993; Barbash 2010). When D. melanogaster females are crossed to D. simulans males, the F1 hybrid sons are invariably lethal, while the F1 daughters are generally fully viable, at least at lower temperatures (∼<25°C) (Sturtevant 1920; Barbash et al. 2000). Importantly, this pattern of lethality is not sex specific but rather caused by the presence of the D. melanogaster X chromosome. Experiments that can detect products of nondisjunction or use attached X chromosomes demonstrate that daughters inheriting both X chromosomes from their D. melanogaster mother are lethal while sons inheriting their X from their D. simulans father are viable (Barbash 2010). The lethality is caused by an incompatibility between the D. melanogaster allele of the gene Hmr on the D. melanogaster X, interacting with the D. simulans alleles of the autosomal genes Lhr and GFZF (Watanabe 1979; Hutter and Ashburner 1987; Brideau et al. 2006; Phadnis et al. 2015).
In contrast, the reciprocal cross of D. simulans females to D. melanogaster males produces viable F1 sons and poorly viable F1 daughters that die as early embryos (Sturtevant 1920). While the D. melanogaster X is again implicated in causing this embryonic lethality, Sawamura, Yamamoto, et al. (1993) showed that Hmr is not responsible for this F1 lethality. Instead, the lethal effect of the D. melanogaster X maps to the pericentromeric heterochromatin in a region called Zhr (Sawamura and Yamamoto 1993). The lethal effect of Zhr+ appears to be caused by mis-segregation during early embryogenesis of a multimillion base pair block of complex satellite DNA sequences (Ferree and Barbash 2009). These satellite sequences are known alternatively as the 359-bp or the 1.688-g/cm3 satellites (Lohe and Brutlag 1986; Sawamura and Yamamoto 1993; Ferree and Barbash 2009). While D. simulans contains some dispersed 359-bp repeats, it does not have the large X-linked block found in D. melanogaster (Lohe et al. 1993; Lima et al. 2020; Sproul et al. 2020). This extensive difference in abundance of the 359-bp satellite between the species suggests that D. melanogaster may contain allele(s) that have co-evolved with the X-linked 359-bp satellite block to help promote its proper mitotic segregation.
This logic further implies that the lack of the X-linked 359-bp satellite block in D. simulans would cause it to be unable to regulate this satellite block when inherited from a D. melanogaster parent, leading to hybrid incompatibility. Because the F1 hybrid female lethality occurs during early embryonic development, the allele(s) hypothesized to be missing from D. simulans are likely to be maternally expressed (Sturtevant 1920; Ferree and Barbash 2009). The penetrance of F1 hybrid female lethality is highly variable across strains, which has complicated efforts to identify the genes causing this lethality (Gérard and Presgraves 2012). Sawamura, Taira, et al. (1993) identified a strain of D. simulans called maternal hybrid rescue (mhr) that produces high viability of F1 hybrid daughters and mapped the causal locus (or loci) to the second chromosome. Orr also implicated the D. simulans second chromosome in contributing to hybrid lethality in this cross (Orr 1996). Another study directly tested the satellite-binding protein D1 as a candidate but found no evidence for a role in the incompatibility (Ferree and Barbash 2009). Further attempts to identify the genetic basis of the D. simulans maternal effect on hybrid viability led to the plausible conclusion that it is a polygenic effect (Gérard and Presgraves 2012). Others have suggested that the incompatibility may be caused by the absence in D. simulans of maternally deposited small RNAs homologous to the 359-bp repeat, rather than by protein-coding genes (Ferree and Barbash 2007). Several studies have shown that such RNAs are produced by heterochromatic satellites including the X-linked 359-bp satellite block (Usakin et al. 2007; Wei et al. 2021), though their potential role in hybrid lethality remains untested.
The X-linked gene maternal haploid (mh) is an intriguing candidate for contributing to this interspecific incompatibility. Mutations in mh were first identified based on its female sterility phenotype (Gans et al. 1975). Embryos from mh mutant mothers typically arrest within the first few nuclear cycles with condensation defects specific to the paternally inherited chromosomes, with a minority reaching late embryogenesis as lethal gynogenetic haploids (Zalokar et al. 1975; Loppin et al. 2001). The mh gene encodes a predicted metalloprotease, homologs of which are involved in DNA damage repair (Delabaere et al. 2014; Tang et al. 2017). Most relevant to this study, the Mh protein localizes to the 359-bp satellite during embryogenesis and the satellite shows aberrant segregation in the embryonic progeny of mh mutant mothers (Tang et al. 2017). These findings, as well as the evolutionary patterns described below, motivated us to test for functional divergence of mh orthologs between D. melanogaster and D. simulans and for possible effects on hybrid lethality.
In particular, we tested the hypothesis that hybrid female embryonic lethality results from the inability of the maternally expressed D. simulans Mh protein to properly interact with the paternally inherited X-linked D. melanogaster 359-bp satellite block. Under this hypothesis, we predict that adding mel-mh to D. simulans mothers would suppress hybrid lethality.
Materials and methods
Nomenclature
We use the abbreviations mel-mh and sim-mh to refer to the mh ortholog in D. melanogaster and D. simulans, respectively. We use phi{mel-mh-Gfp} and phi{sim-mh-Gfp} to designate phiC31-mediated transgenes containing Gfp fusions of mel-mh and sim-mh, respectively. As described in the Results, D. simulans has a tandem duplication of mh. The mh-p copy is more similar than the mh-d copy in sequence and structure to D. melanogaster mh. We therefore consider sim-mh-p to be the ortholog of D. melanogaster mh, and for simplicity, refer to it as sim-mh.
Drosophila stocks
D. melanogaster stocks w mh6/FM7a, P{sChRFP}1 and w, mh31/FM7, Gfp+ were kindly provided by Xiaona Tang and Yikang Rong. D. simulans stocks containing attP landing sites were kindly provided by David Stern (Stern et al. 2017).
Sequence analysis
Genome sequences were obtained from D. melanogaster release 6 and the second-generation D. simulans release (Hu et al. 2013; Hoskins et al. 2015). The sequences for both duplicates of mh from the D. simulans genome were taken from the genome sequence and aligned with the D. melanogaster ortholog to determine the consensus coding sequence (see Results). We then calculated DN/DS between the D. melanogaster mh and both D. simulans orthologs using MEGA 11 (Tamura et al. 2021). We compared these ratios to a genome-wide sample of DN/DS ratios from a previously published study to determine their percentile and relative rate of evolutionary change (Stanley and Kulathinal 2016). These data all consist of pairwise sequence comparisons between D. melanogaster and D. simulans orthologs. Iso-Seq sequences of mRNA transcripts from adult males were also analyzed to determine the reading frame and structure of the mh duplicates (Nouhaud, 2018). To do this, we ran a BLASTN search on the full length mh-d sequence, which, after manual filtering to remove duplicates and reads mapping to mh-p or other genes, turned up 4 unique reads mapping to mh-d. Out of these, only 2 reads mapped to mh-d upstream of exon 3, and neither of these reads mapped to potential exons 1 and 2.
mh transgene constructs
The mel-mh-Gfp transgene was kindly provided by Xiaona Tang and Yikang Rong and previously described as gfp-mh-pTV2gw (Tang et al. 2017). We added an attB site to this to create the plasmid gfp-mh-pTV2gw-attB by PCR-amplifying using oligos 502/503 from a plasmid with an attB site that derived from the pTA-attB plasmid (Groth et al. 2004). The PCR product was digested using NotI and inserted into gfp-mh-pTV2gw at its NotI site. All oligonucleotide sequences are listed in Supplementary Table 1.
The w+-attB-sim-mh-eGfp (p834) construct was designed to be parallel in structure to gfp-mh-pTV2gw-attB (Fig. 1 and Supplementary File 1) and was made in the following steps:
Fig. 1.
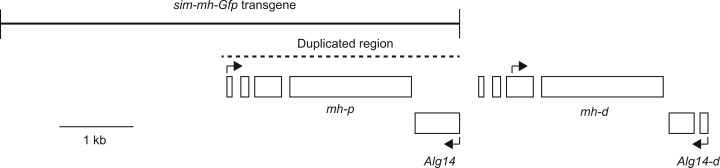
Map of mh region in D. simulans demonstrating the duplication event. The region in D. melanogaster has the same structure as the mh-p and Alg14 regions. The dashed line shows the region duplicated to form mh-d and Alg14-d. Black arrows show translation start sites (Chakraborty et al. 2021). The 2 boxes shown for Alg14-d represent a frameshift relative to Alg14. A full annotation of this region is shown in Supp File 1.
w+-attB-sim-mh. The D. simulans mh genomic region covering the coding region and ∼3000 bp upstream was PCR amplified from the strain w501 using oligos 2099/2100 and cloned into pCR-Blunt II-TOPO (Invitrogen). The sequence of sim-mh was confirmed by Sanger sequencing (using oligos 788, 823, 2078, 2079, 2080, 2081, 2082, 2083, 2084, and 2101). The insert was then released by XbaI digestion and ligated into the XbaI site of the plasmid w+-attB, a gift from Jeff Sekelsky (Addgene plasmid # 30326; http://n2t.net/addgene:30326; RRID: Addgene_30326). The resulting clone w+-attB-sim-mh was confirmed by checking the pattern of restriction enzyme digestion.
pCR-Blunt II-TOPO_sim-mh(partial)-eGfp. The coding sequence of eGfp was inserted into the coding sequence of sim-mh immediately after the start codon by Gibson assembly of fragments termed Dsmh1, eGfp, and Dsmh2. Dsmh1 and Dsmh2 were PCR amplified from D. simulans (strain w501) using oligo pairs 2016/2017 and 2020/2021, respectively, while eGfp was amplified using oligos 2018/2019 from pEGfp-attB (Drosophila Genomics Resource Center). A fusion product of Dsmh1- eGfp -Dsmh2 was PCR-amplified from the Gibson Assembly reaction mixture, using oligos 2080/2082 and cloned into pCR-Blunt II-TOPO. The sequence of the insert, i.e. the fusion of eGfp CDS and Dsmh (partial), was checked by Sanger sequencing using oligos 498, 788, 823, and 2081.
w+-attB-Dsmh-eGfp. The Dsmh(partial)_eGfp fragment was released from the pCR-Blunt II TOPO vector and ligated into w+-attB-sim-mh using double digestion (BsiWI-BstEII). The final construct w-attB-sim_mh-eGfp was confirmed by checking the restriction pattern of the construct.
mh transgenic lines
D. melanogaster transgenic lines were made by phiC31-mediated integration into the strain y w P{nos-phiC31\int.NLS}X; P{CaryP}attP40. D. simulans transformant lines were made by phiC31-mediated integration into the strains y w; pBac{3XP3::EYFP, attP}1048-2R and y w; pBac{3XP3::EYFP, attP}1029-3R (Stern et al. 2017). Microinjections were done by Rainbow Transgenic Flies, Inc.
Fertility tests
The mel-mh-Gfp and sim-mh-Gfp transgenes transformed into D. melanogaster were compared for their ability to complement the female sterility of mh null mutations. A small-scale pilot experiment was initially done at room temperature by crossing single virgin w mh6/w mh31; {mh-Gfp, w+}attP40/+ females to 2 DGRP-882 (wild-type) males, where mh-Gfp represents either of the 2 transgenes. Two sets were done: the first with females aged 3–4 days old before mating and the second aged 9–10 days. Vials were cleared after 5 days; if either the female or both males were dead, then the vial was discarded.
To generate F1 females to assay in a large-scale experiment, a parental cross was set up of w mh6; {mh-Gfp, w+}attP40 females and mh31/Y males, where mh-Gfp represents either of the 2 transgenes. Virgin F1 daughters of genotype w mh6/w mh31; {mh-Gfp, w+}attP40/+ were collected and aged for 3–5 days, followed by test crosses containing 1 virgin female and 2 Canton-S (wild type) males. After 4–5 days, parents were flipped to new vials and flipped again after another 4–5 days. At each flip, if either the female or both males were dead, then the vial was discarded. Otherwise, vials were kept for 16 (27°C) or 18 (25°C) days and all progeny counted. Three flips were performed at 27°C and 4 at 25°C; however, very few parents survived until the fourth flip at 25°C and, thus, we only report the first 3 flips in Fig. 3b. The overall time period across all 3 flips was 12 days at 25°C and 12–13 days at 27°C. Progeny per day are reported to normalize between flips that were 4 or 5 days.
Fig. 3.
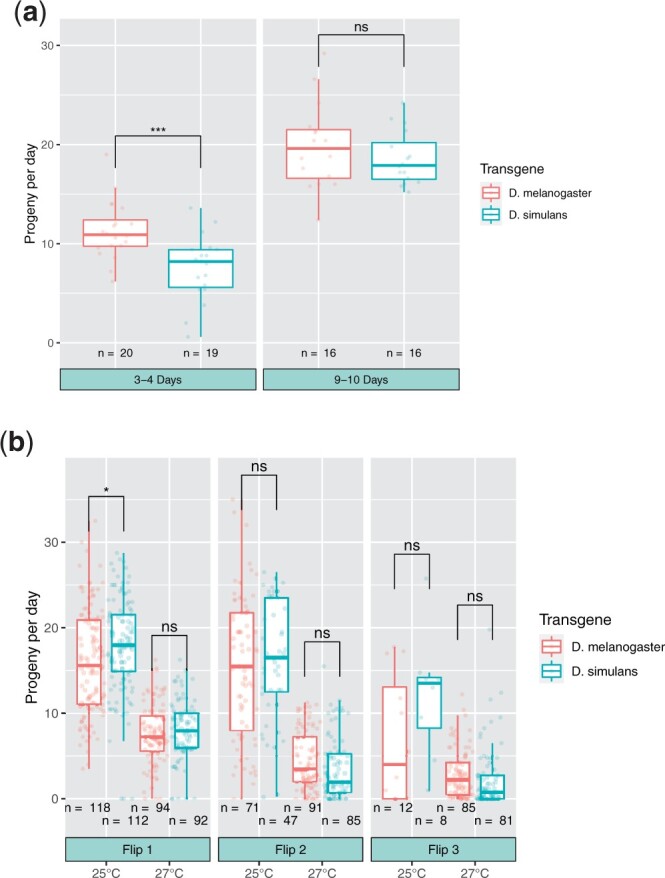
Comparison of fertility between D. melanogastermh mutant females carrying mel-mh-Gfp vs. sim-mh-Gfp transgenes (single w mh6/w mh31; {mel-mh-Gfp, w+}attP40/+ vs . w mh6/w mh31; {sim-mh-Gfp, w+}attP40/+). Statistical significance between genotypes was tested by a 2 sample unpaired t-test: *** = 0.0001 < P ≤ 0.001; * = 0.01 < P ≤ 0.05; ns = P > 0.05 (not significant). a) Pilot experiment at room temperature (∼20–22°C). Females were collected as virgins and aged for either 3–4 days (left) or 9–10 days (right) prior to mating and progeny collection for 5 days. Progeny are reported as “per-day” to normalize with (b). b) Two large-scale experiments, performed at 25 and 27°C. Virgin females were aged for 3–5 days prior to mating. Crosses were flipped to fresh vials after 4 or 5 days, and again after an additional 4 or 5 days, for a total collection period of 12–13 days; the exact length of each flip was recorded and is accounted for in calculating the “progeny per day.” Note that vials were discarded if the parents died, which is why N goes down between flips (see Materials and Methods).
Hybrid viability tests
The mel-mh-Gfp and sim-mh-Gfp transgenes transformed into D. simulans were assessed for their ability to rescue female viability in F1 D. simulans/D. melanogaster hybrids as compared to a matched control group lacking the transgenes. To generate F1 hybrids either carrying or not carrying a mh-Gfp transgene, for each transgenic D. simulans line, the following parental crosses were set; first, w501 virgin females were crossed to y w/Y; {mh-Gfp, w+} males, where mh-Gfp represents either of the 2 transgenes. Virgin daughters of the genotype w501/y w; {mh-Gfp, w+}/+ collected from this cross were subsequently mated to w501/Y males. For each set, 30–40 y? w/w501; {mh-Gfp, w+}/+ and y? w/w501 virgin daughters were separately collected, aged 0–1 days, and mated to 40–50 3–5-day-old D. melanogaster Canton-S virgin males. The crosses were kept at room temperature (19.4–22.0°C) and flipped every 2–4 days until they stopped producing progeny; the adult F1 hybrids were scored for sex.
Western blots
Young female virgin flies were fed yeast paste 2–3 days prior to dissection. Ovaries were dissected in 0.7% NaCl with 2 pairs of tweezers after flies were anesthetized by carbon dioxide. Ovaries were collected into 1.7-ml microcentrifuge tubes and flash-frozen in liquid nitrogen and then stored at −80°C until all dissections were completed.
To extract proteins, ovaries were ground in SDS sample buffer (62.5 mM Tris, pH 6.8, 2% SDS, 10% glycerol, 1% ß-mercaptoethanol, 0.05% bromophenol blue), boiled for 3 minutes, and centrifuged at 10,000 rpm for 5 min. Proteins were separated via 7.5% SDS-PAGE using a BioRad Mini-PROTEAN Vertical Electro Cell and transferred to polyvinylidene difluoride membrane using BioRad Mini Trans-Blot. The protein standard was Thermo Scientific PageRuler Prestained Protein Ladder, 10–180 kDa.
The protein-bound membrane was blocked in 5% skim milk in TBST (150 mM NaCl, 20 mM Tris, pH 7.5, 0.1% Tween-20) for 1 h at room temperature, followed by primary antibody incubation at 4°C for 16 h, and secondary antibody incubation at room temperature for 1 h. The membrane was washed 3 × 10 min in TBST after each antibody incubation. Primary antibodies used were: Anti-Gfp Rabbit Polyclonal Antibody (1/5,000, Rockland Immunochemical 600-401-215S) and Monoclonal Mouse Anti-α-Tubulin antibody (1/20,000, Sigma T9026), and secondary antibodies used were: HRP-Goat Anti-Rabbit IgG (H + L) (1/4,000, Jackson 111-035-003) and HRP-Goat Anti-Mouse IgG (H + L) (1/8,000, Jackson 115-035-003). Antibodies were diluted in either 5% skim milk or 5% BSA, in TBST. Signals were detected by applying ECL2 Western Blotting Substrate (Thermo Scientific 80197) to the membrane and exposing it to autoradiography film (VWR 490001-930).
Results
mh is duplicated in D. simulans
When attempting to identify the mh ortholog in D. simulans, we noticed 2 different regions with homology to mel-mh. The first region was a contig on the X with high similarity but that appeared to be incomplete and possibly contain partial duplications. While pursuing this analysis, a PacBio assembly of the D. simulans genome reported that mh is tandemly duplicated on the X along with the flanking gene alg14 (Chakraborty et al. 2021). Following Chakraborty et al. (2021), we refer to the D. simulans duplicates as mh-p (mh-proximal) and mh-d (mh-distal), though as noted below we suggest that mh-p can also be considered the parental copy and mh-d the daughter copy (Fig. 1).
We confirmed the duplication structure of D. simulans mh in an independent assembly of D. simulans created using Nanopore sequencing (Miller et al. 2018). The Nanopore and PacBio assemblies fully agree in structure, with the exception of a ∼25-bp insertion in the PacBio assembly immediately distal to the proximal copy (Supplementary File 1). The 2 genome assemblies also contain no differences in the coding sequences of either mh-p or mh-d.
We used Artemis and the Artemis Comparison Tool to compare and annotate the structures of the D. melanogaster and D. simulans mh regions (Carver et al. 2008; Supplementary File 1). We estimate that the duplicated region corresponds to approximately 3181 bp (Fig. 1). This leaves D. simulans mh-d having a duplication of the mh-p 3′ region as its 5′ region, which may be responsible for its novel testis-enriched expression pattern reported by Chakraborty et al. (2021). There is a 16-bp deletion in the coding region of what would correspond to exon 1 of mh-d, relative to mh-p and D. melanogaster mh. We confirmed that mh-d has this deletion in 5 D. simulans strains that were sequenced using Sanger sequencing (Begun et al. 2007). This deletion could change the coding potential of exon 1, or alternatively the transcript could potentially splice to exon 2 and restore the same reading frame as in mh-p. However, using Iso-Seq reads from adult male D. simulans testes, we did not find evidence that the potential exon 1 or exon 2 are expressed (see Materials and Methods), consistent with the conclusions of Chakraborty et al. (2021). This suggests that mh-d produces a truncated product relative to mh-p, with the apparent transcription start site being ∼50–80-bp upstream of exon 3. We have annotated the mh-d CDS as beginning at the first ATG in exon 3, with potential exons 1 and 2 also indicated (Fig. 1 and Supplementary File 1).
Regardless of this uncertainty regarding the N-terminal structure of mh-d, mh-p has greater similarity D. melanogaster mh in its structure, primary sequence, and expression pattern, suggesting that mh-p can be considered to be the parental copy and mh-d the daughter copy of the duplication. We thus define D. simulans mh-p as the ortholog of D. melanogaster mh.
The second region of lesser homology mapped to an intron of the tkv gene on chr 2. This region of chr 2 is annotated as the pseudogene CR14033 and was identified as producing siRNAs in testis (Czech et al. 2008; Okamura et al. 2008) homologous to a region of mh (Czech et al. 2008). This pseudogene is present in multiple Drosophila species including D. ananassae and D. pseudoobscura, but the region homologous of CR14033 homologous to mh is only present in D. melanogaster and its sister species D. simulans and D. sechellia (Sperry 2016).
mh coding sequences are rapidly evolving
We calculated pairwise divergence among the mh genes in D. melanogaster and D. simulans (Table 1). The DN/DS ratios are relatively high compared to a genome-wide sample of loci, indicating substantial nonsynonymous divergence. The comparison between mel-mh and sim-mh-p had a DN/DS ratio of 0.373, placing it in the top 10% of the genome-wide distribution. The comparison between mel-mh and sim-mh-d had a DN/DS ratio of 0.509, which is in the top 5% of the genome distribution. For reference, the hybrid incompatibility loci Hmr and Lhr are in the top 3% of the distribution. DN/DS is also higher between the paralogs sim-mh-p and sim-mh-d than between the orthologs mel-mh and sim-mh-p.
Table 1.
Divergence of mh genes between D. melanogaster and D. simulans.
| Gene 1 | Gene 2 | DN | DS | DN/DS | Percentile |
|---|---|---|---|---|---|
| mel_mh | sim-mh-p | 0.056 | 0.150 | 0.373 | 91.2 |
| mel_mh | sim-mh-d | 0.086 | 0.168 | 0.509 | 95.3 |
| sim-mh-p | sim-mh-d | 0.040 | 0.066 | 0.605 | n/a |
The numbers of nonsynonymous and synonymous substitutions per site were estimated using the Nei-Gojobori method as implemented in MEGA 11. Percentile refers to rank of DN/DS relative to 10,766 genes for which DN/DS was compared between D. melanogaster and D. simulans (Stanley and Kulathinal 2016). Note that mh was not included in the Stanley and Kulathinal (2016) gene set because the D. simulans ortholog was not identified at that time.
D. simulans mh complements D. melanogaster mh mutants
We constructed a transgene of D. simulans mh tagged with Gfp (called sim-mh-Gfp) to precisely match in structure the D. melanogaster mh-Gfp transgene of Tang et al. (2017) (Fig. 1; Materials and Methods). We transformed and integrated both transgenes into D. melanogaster at the same autosomal position. Western blots indicate that both transgenes express at similar levels (Fig. 2). We then crossed the transgenes into a mh6 mutant background. Stable stocks were established, indicating that both transgenes can complement the sterility of mh6. To quantitatively compare the activity of these transgenes, we performed fertility assays of females trans-heterozygous for 2 mh null alleles and heterozygous for a mh-Gfp transgene; that is w mh6/w mh31; {mel-mh-Gfp, w+}/+ compared to w mh6/w mh31; {sim-mh-Gfp, w+}/+. A small-scale pilot experiment found greater fertility among females carrying mel-mh-Gfp, but only among younger females (Fig. 3a). We then performed a more extensive experiment, examining fertility across a 12–13-day period at 2 different temperatures (Fig. 3b). No significant difference between the transgenic genotypes was observed at any time point, except for the first flip at 25°C, where females carrying the mel-mh-Gfp transgene had significantly fewer progeny than those carrying the sim-mh-Gfp transgene. We conclude that the D. melanogaster and D. simulans orthologs have not substantially diverged for the essential female fertility function of mh.
Fig. 2.
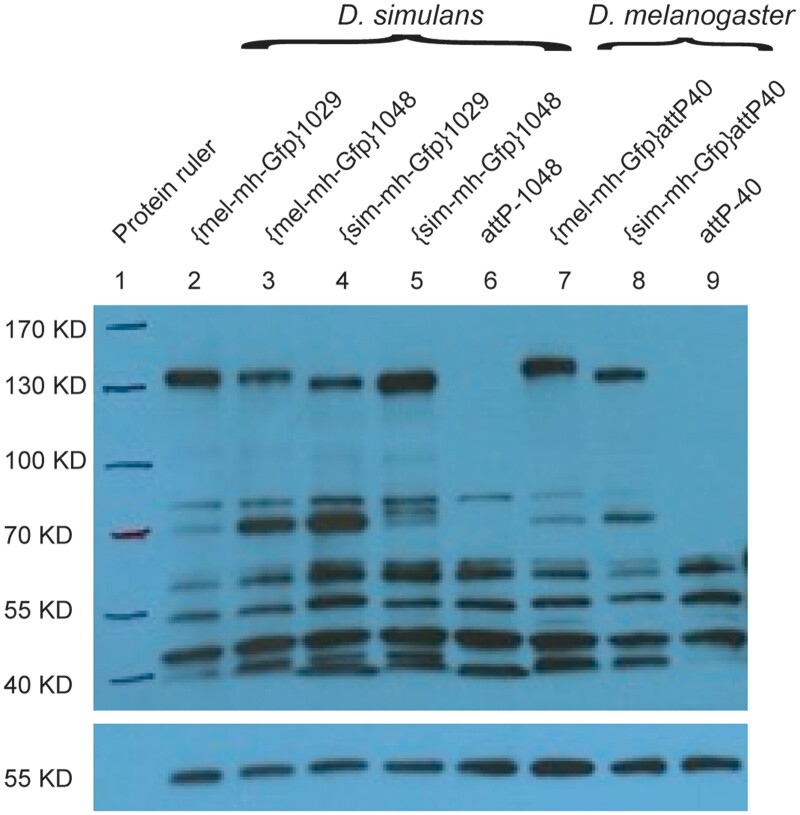
Western blot comparing Mh-Gfp protein accumulation in different transgenic genotypes. Western blot with an anti-Gfp antibody recognizes the Mh-eGfp protein in ovary extracts from females (top panel). Signals on the same membrane detected by an anti-α-Tubulin antibody served as a loading control (lower panel). Protein sizes of the prestained protein ladder were hand-marked on the membrane. Lanes 2–6 are from D. simulans extracts and lanes 7–9 from D. melanogaster extracts. Two independent transformants of the transgenes in D. simulans were analyzed, at attP sites 1029 and 1048. The attP-1048 and attP-40 samples are negative controls because they are the untransformed strains that carry the attP sites. The predicted molecular weights of mel-Mh-Gfp and sim-Mh-Gfp proteins are 108.6 and 109 kDa, respectively.
D. melanogaster mh does not affect hybrid viability
Having established that sim-mh-Gfp is functional within D. melanogaster, we turned to the primary question motivating this study, of whether mh is a hybrid incompatibility gene. We tested this by comparing the D. simulans and D. melanogaster mh transgenes for their ability to modulate F1 hybrid female viability. Specifically, we sought to determine whether the addition of a mh+ transgene to the mothers of these hybrids would increase viability of their daughters. We transformed these transgenes into 2 different attP sites in D. simulans, with Western blots with an anti-Gfp antibody demonstrating similar protein levels for both transgenes (Fig. 2).
We then generated F1 interspecific hybrids by crossing D. simulans females heterozygous for either mel-mh-Gfp or sim-mh-Gfp, along with sibling females not carrying a transgene as controls, to wild-type D. melanogaster males (Table 2). The viability of the control F1 daughters not carrying a transgene was quite high, ranging from ∼47% to 69% relative to F1 sons, which limited our ability to detect substantial increases in the progeny of interspecific hybrids from mothers carrying a transgene. This relatively high rescue may reflect the fact that the transgenes were outcrossed to the D. simulans w501 strain. We used this strain to be able to easily follow the transgenes in a white mutant background, but this strain is known to produce high hybrid viability in other crosses (Gérard and Presgraves 2012). Regardless, the results are opposite to our hypothesis. Both sets of crosses with the sim-mh-Gfp transgene showed increased female viability from transgenic mothers compared to control mothers, though only set D (with the 1048 insertion) was statistically significant. In contrast, crosses with the mel-mh-Gfp transgene showed essentially no differences in the relative viability of daughters between the transgenic and control genotypes. Within the resolution of our assay, we find no evidence suggesting that hybrid lethality results from the absence of mel-mh alleles in D. simulans.
Table 2.
Testing mh transgenes for modulation of interspecific F1 hybrid viability
| Set | Maternal genotype | # F1 hybrid daughters | # F1 hybrid sons | Ratio F1 daughters/sons | Relative ratio w/o and w transgene | P-Value |
|---|---|---|---|---|---|---|
| A | w | 760 | 1106 | 0.69 | ||
| w; {mel-mh-Gfp}1029/+ | 766 | 1105 | 0.69 | 0.99 | 0.895 | |
| B | w | 781 | 1144 | 0.68 | ||
| w; {mel-mh-Gfp}1048/+ | 830 | 1124 | 0.74 | 0.92 | 0.228 | |
| C | w | 144 | 211 | 0.68 | ||
| w; {sim-mh-Gfp}1029/+ | 84 | 88 | 0.95 | 0.71 | 0.0722 | |
| D | w | 248 | 529 | 0.47 | ||
| w; {sim-mh-Gfp}1048/+ | 461 | 693 | 0.67 | 0.70 | 0.000331 |
In the following descriptions, all flies are D. simulans unless noted otherwise. The designation {mh-Gfp, w+} refers to one of the 4 transgenic genotypes used in this table. The flies described in the “Maternal genotype” column were generated as follows: (1) w501 virgin females were crossed to y w/Y; {mh-Gfp, w+} males. (2) w501/y w; {mh-Gfp, w+}/+ virgin daughters were crossed to w501/Y males. (3) For each set, y? w/w501; {mh-Gfp, w+}/+ and y? w/w501 virgin daughters were separately collected. The females from step (3) were then mated to D. melanogaster Canton-S males. The full genotypes of the transgenes are noted in the Materials and Methods. P-Values were calculated using a 2 × 2 Chi-squared test.
Discussion
The mh gene is interesting based on its unusual mutant phenotype of producing gynogenetic haploid embryos. It also displays a relatively high rate of coding sequence evolution, has experienced a recent duplication in D. simulans, and has a pseudogene fragment in both D. melanogaster and D. simulans that indicates a possible second duplication in their common ancestor. Our major motivation in this study derived from the association of mel-Mh with the X-linked 359-bp satellite DNA block that is present in D. melanogaster but absent in D. simulans. We used transgenic constructs to compare the activity of mh orthologs from D. melanogaster and D. simulans in the background of both species. We designed the sim-mh-Gfp transgene to be parallel in structure to a mel-mh-Gfp transgene previously shown to be functional. Our sim-mh-Gfp transgene complements D. melanogaster mh mutations, as evidenced by the ability to maintain a fertile mh6; sim-mh-Gfp stock.
In the course of this study, Brand and Levine (2022) independently published a study of the evolution of mh between D. melanogaster and D. simulans. They compared mh ortholog function by replacing the endogenous D. melanogaster mh locus with the D. simulans ortholog. They found that females of this replacement line have reduced fertility compared to the D. melanogaster mh control line and further found that this reduced fertility is dependent on an intact Zhr+ locus, which contains a large block of the 359-bp satellite. Below we summarize our results and compare them to those of Brand and Levine where appropriate.
Similar fertility function of mel-mh and sim-mh
We observed some reduction of fertility in D. melanogaster mh mutants carrying sim-mh-Gfp compared to mel-mh-Gfp controls in one of the 2 small scale initial experiments (Fig. 3a). However, follow-up experiments at much larger scale failed to find any reduction (with 1 out of the 6 comparisons showing a modest but significant increase in fertility of sim-mh-Gfp relative to mel-mh-Gfp females; Fig. 3b). We conclude that D. melanogaster and D. simulans mh orthologs are largely interchangeable for female fertility.
This contrasts with the result of Brand and Levine (2022), who report a more than 2-fold mean reduction in fertility of D. melanogaster females carrying sim-mh compared to mel-mh. One possible explanation for these differences is that the fertility of mh genotypes are inherently variable due to environmental (or other) variation, which is plausible for any genotype that may be subfertile but not completely sterile. Another possibility may be differences in experimental design of the fertility assays. We assayed females individually while Brand and Levine analyzed fertility in broods of 4 females. If a female (and/or the males they were mating with) died during the course of our experiment we could exclude that vial, while in the Brand and Levine design individual deaths may not have been recorded and would reduce the brood size and thus presumably the progeny count. We saw very high rates of death in one of our experiments (at 25°C) that varied by genotype: mel-mh-Gfp crosses dropped from 118 to 71 from flip 1to 2, while for sim-mh-Gfp the drop was much greater, from 112 to 47.
There are also significant differences in experimental design of the gene replacements between the 2 studies. Here, we have used transgenic constructs integrated into an autosomal site and crossed into mh null-allele backgrounds to “replace” mh and compare mel-mh and sim-mh. We designed our sim-mh-Gfp transgene to match a previously described mel-mh-Gfp transgene that was shown to provide wild type mh function (Tang et al. 2017). This introduces a potential position effect as mh is now in an autosomal location. It is also possible that sim-mh-Gfp does not express properly in a D. melanogaster background since it has its endogenous regulatory regions. We did, however, see robust protein expression from our transgenes. Brand and Levine (2022) used a CRISPR replacement strategy, replacing the endogenous mh locus in D. melanogaster with FLAG-tagged and codon-optimized mel-mh and sim-mh coding sequences. This means that the sim-mh allele is synthetic in the sense that the synonymous sites in the transgene are not native to D. simulans. But again, Western blots suggest that it is fully expressed. Among other differences in the 2 studies is that our fertility assays were done with 1 functional copy of mh in the mothers (that is heterozygous for the mh+ transgene), while the Brand and Levine (2022) design has 2 functional copies (that is, homozygous for the replaced locus). Brand and Levine (2022) found that D. melanogaster females with 1 mel-mh and 1 sim-mh allele are fully fertile, and other experiments support their conclusion that deleterious effects of sim-mh on fertility and ovarian morphology are dose dependent.
Lack of effect of mel-mh on hybrid viability
We found no evidence that the absence of mel-mh contributes to hybrid female lethality. In crosses of D. simulans females to D. melanogaster males, D. simulans mothers carrying a mel-mh-Gfp transgene produced the same ratio of female hybrids compared to control crosses without the transgene. Repeating our experiments in D. simulans genetic backgrounds that have a lower baseline of hybrid female viability might therefore reveal more subtle effects that we could not detect. We also note that the hybrids contain both endogenous expressed sim-mh as well as sim-mh-Gfp or mel-mh-Gfp expressed from the transgene. It remains possible that potential effects of mel-mh in hybrids could be masked by the presence of the endogenous sim-mh. Surprisingly, we did observe a moderate effect of increased hybrid female viability produced by mothers that carried the sim-mh-Gfp transgene. This finding suggests that increased mh dosage may reduce mis-segregation of the 359-bp satellite in hybrids but that such effects are not dependent on the mel-mh ortholog that has coevolved with the D. melanogaster 359-bp satellite block.
Whither the D. simulans maternal effect?
The identity of the D. simulans genes that are interacting with the D. melanogaster 359-bp satellite to cause hybrid lethality remain unknown. One approach would be to map the alleles that distinguish the rescuing D. simulans mhr strain from other D. simulans strains that do not rescue hybrid lethality. Mapping efforts could also be extended to include other D. simulans strains that produce high F1 hybrid daughter viability (Gérard and Presgraves 2012). Another approach could test alternative hypotheses, such as that hybrid lethality results from a lack in D. simulans of small RNAs derived from the 359-bp satellite (Ferree and Barbash 2007). This would require developing a transgenic expression system in D. simulans that drives the deposition of such small RNAs into the developing egg. If this turned out to represent the mechanistic basis of hybrid female lethality, it remains of high interest to understand why there is such substantial interstrain variation in D. simulans for this hybrid lethality.
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material is available at G3 online.
Supplementary Material
Acknowledgments
Plasmids were obtained from the Drosophila Genomics Resource Center (supported by NIH Grant 2P40OD010949). The w+-attB plasmid was a gift from Jeff Sekelsky (Addgene plasmid # 30326). We thank David Stern, Xiaona Tang, and Yikang Rong for sharing strains.
Funding
This work was supported by National Institutes of Health (NIH) grant R01-GM07473 to DAB. DMC was supported by an NIH NRSA fellowship 5F32GM120896.
Conflicts of interest
None declared.
Contributor Information
Dean M Castillo, Institute of Agriculture and Natural Resources, University of Nebraska, Lincoln, NE 68588, USA; Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY 14850, USA.
Benjamin McCormick, Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY 14850, USA.
Connor M Kean, Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY 14850, USA.
Sahana Natesan, Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY 14850, USA.
Daniel A Barbash, Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY 14850, USA.
Literature cited
- Barbash DA, Roote J, Ashburner M.. The Drosophila melanogaster hybrid male rescue gene causes inviability in male and female species hybrids. Genetics. 2000;154(4):1747–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash DA. Ninety years of Drosophila melanogaster hybrids. Genetics. 2010;186(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Holloway AK, Stevens K, Hillier LW, Poh Y-P, Hahn MW, Nista PM, Jones CD, Kern AD, Dewey CN, et al. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5(11):e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand CL, Levine MT.. Cross-species incompatibility between a DNA satellite and the Drosophila Spartan homolog poisons germline genome integrity. Curr Biol. 2022;32(13):2962-2971.e4. 10.1016/j.cub.2022.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brideau NJ, Flores HA, Wang J, Maheshwari S, Wang X, Barbash DA.. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science. 2006;314(5803):1292–1295. [DOI] [PubMed] [Google Scholar]
- Carver T, , BerrimanM, , TiveyA, , PatelC, , BöhmeU, , BarrellBG, , ParkhillJ, , Rajandream M-A.. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics. 2008;24(23):2672–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty M, Chang C-H, Khost DE, Vedanayagam J, Adrion JR, Liao Y, Montooth KL, Meiklejohn CD, Larracuente AM, Emerson JJ, et al. Evolution of genome structure in the Drosophila simulans species complex. Genome Res. 2021;31(3):380–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453(7196):798–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delabaere L, Orsi GA, Sapey-Triomphe L, Horard B, Couble P, Loppin B.. The Spartan ortholog maternal haploid is required for paternal chromosome integrity in the Drosophila zygote. Curr Biol. 2014;24(19):2281–2287. [DOI] [PubMed] [Google Scholar]
- Ferree PM, Barbash DA.. Distorted sex ratios: a window into RNAi-mediated silencing. PLoS Biol. 2007;5(11):e303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferree PM, Barbash DA.. Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol. 2009;7(10):e1000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gans M, Audit C, Masson M.. Isolation and characterization of sex-linked female-sterile mutants in Drosophila melanogaster. Genetics. 1975;81(4):683–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gérard PR, Presgraves DC.. Abundant genetic variability in Drosophila simulans for hybrid female lethality in interspecific crosses to Drosophila melanogaster. Genet Res (Camb). 2012;94(1):1–7. [DOI] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP.. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166(4):1775–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins RA, , CarlsonJW, , WanKH, , ParkS, , MendezI, , GalleSE, , BoothBW, , PfeifferBD, , GeorgeRA, , Svirskas R,. et al. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 2015;25(3):445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, , EisenMB, , ThorntonKR, , Andolfatto P.. A second-generation assembly of the Drosophila simulans genome provides new insights into patterns of lineage-specific divergence. Genome Res. 2013;23(1):89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter P, Ashburner M.. Genetic rescue of inviable hybrids between Drosophila melanogaster and its sibling species. Nature. 1987;327(6120):331–333. [DOI] [PubMed] [Google Scholar]
- Lima LGd, Hanlon SL, Gerton JL.. Origins and evolutionary patterns of the 1.688 satellite DNA family in Drosophila phylogeny. G3 (Bethesda). 2020;10(11):4129–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe AR, Brutlag DL.. Multiplicity of satellite DNA sequences in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1986;83(3):696–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe AR, Hilliker AJ, Roberts PA.. Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Genetics. 1993;134(4):1149–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loppin B, Berger F, Couble P.. Paternal chromosome incorporation into the zygote nucleus is controlled by maternal haploid in Drosophila. Dev Biol. 2001;231(2):383–396. [DOI] [PubMed] [Google Scholar]
- Miller DE, Staber C, Zeitlinger J, Hawley RS.. Highly contiguous genome assemblies of 15 Drosophila species generated using nanopore sequencing. G3 (Bethesda). 2018;8(10):3131–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouhaud P. Long-read based assembly and annotation of a Drosophila simulans genome. bioRxiv. 2018;425710. [Google Scholar]
- Okamura K, Balla S, Martin R, Liu N, Lai EC.. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat Struct Mol Biol. 2008;15(6):581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. The unexpected recovery of hybrids in a Drosophila species cross: a genetic analysis. Genet Res. 1996;67(1):11–18. [DOI] [PubMed] [Google Scholar]
- Phadnis N, Baker EP, Cooper JC, Frizzell KA, Hsieh E, de la Cruz AFA, Shendure J, Kitzman JO, Malik HS.. An essential cell cycle regulation gene causes hybrid inviability in Drosophila. Science. 2015;350(6267):1552–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamura K, Taira T, Watanabe TK.. Hybrid lethal systems in the Drosophila melanogaster species complex. I. The maternal hybrid rescue (mhr) gene of Drosophila simulans. Genetics. 1993;133(2):299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamura K, Watanabe TK, Yamamoto MT.. Hybrid lethal systems in the Drosophila melanogaster species complex. Genetica. 1993;88(2–3):175–185. [DOI] [PubMed] [Google Scholar]
- Sawamura K, Yamamoto MT.. Cytogenetical localization of Zygotic hybrid rescue (Zhr), a Drosophila melanogaster gene that rescues interspecific hybrids from embryonic lethality. Mol Gen Genet. 1993;239(3):441–449. [DOI] [PubMed] [Google Scholar]
- Sawamura K, Yamamoto MT, Watanabe TK.. Hybrid lethal systems in the Drosophila melanogaster species complex. II. The zygotic hybrid rescue (Zhr) gene of D. melanogaster. Genetics. 1993;133(2):307–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperry EG. Genetic analysis and the evolutionary study of a pseudogene, CR14033, and the corresponding parent gene, CG9203 [master’s thesis]. New Brunswick, Canada: University of New Brunswick; 2016.
- Sproul JS, Khost DE, Eickbush DG, Negm S, Wei X, Wong I, Larracuente AM.. Dynamic evolution of euchromatic satellites on the X chromosome in Drosophila melanogaster and the simulans clade. Mol Biol Evol. 2020;37(8):2241–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley CE Jr, Kulathinal RJ.. flyDIVaS: a comparative genomics resource for Drosophila divergence and selection. G3 (Bethesda). 2016;6(8):2355–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Crocker J, Ding Y, Frankel N, Kappes G, Kim E, Kuzmickas R, Lemire A, Mast JD, Picard S, et al. Genetic and transgenic reagents for Drosophila simulans, D. mauritiana, D. yakuba, D. santomea, and D. virilis. G3 (Bethesda). 2017;7(4):1339–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH. Genetic studies on Drosophila simulans. I. Introduction. Hybrids with Drosophila melanogaster. Genetics. 1920;5(5):488–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Kumar S.. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38(7):3022–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Cao J, Zhang L, Huang Y, Zhang Q, Rong YS.. Maternal haploid, a metalloprotease enriched at the largest satellite repeat and essential for genome integrity in Drosophila embryos. Genetics. 2017;206(4):1829–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usakin L, Abad J, Vagin VV, de Pablos B, Villasante A, Gvozdev VA.. Transcription of the 1.688 satellite DNA family is under the control of RNA interference machinery in Drosophila melanogaster ovaries. Genetics. 2007;176(2):1343–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe TK. A gene that rescues the lethal hybrids between Drosophila melanogaster and D. simulans. Jpn J Genet. 1979;54(5):325–331. [Google Scholar]
- Wei X, Eickbush DG, Speece I, Larracuente AM.. Heterochromatin-dependent transcription of satellite DNAs in the Drosophila melanogaster female germline. eLife. 2021;10:e62375. 10.7554/eLife.62375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalokar M, Audit C, Erk I.. Developmental defects of female-sterile mutants of Drosophila melanogaster. Dev Biol. 1975;47(2):419–432. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material is available at G3 online.