Abstract
Arabidopsis (Arabidopsis thaliana) FLOWERING LOCUS C (FLC) acts as a key flowering regulator by repressing the expression of the floral integrator FLOWERING LOCUS T (FT). Prolonged exposure to cold (vernalization) induces flowering by reducing FLC expression. The long noncoding RNAs (lncRNAs) COOLAIR and COLDAIR, which are transcribed from the 3′ end and the first intron of FLC, respectively, are important for FLC repression under vernalization. However, the molecular mechanism of how COOLAIR and COLDAIR are transcriptionally activated remains elusive. In this study, we found that the group-III WRKY transcription factor WRKY63 can directly activate FLC. wrky63 mutant plants display an early flowering phenotype and are insensitive to vernalization. Interestingly, we found that WRKY63 can activate the expression of COOLAIR and COLDAIR by binding to their promoters.WRKY63 therefore acts as a dual regulator that activates FLC directly under non-vernalization conditions but represses FLC indirectly during vernalization through inducing COOLAIR and COLDAIR. Furthermore, genome-wide occupancy profile analyses indicated that the binding of WRKY63 to vernalization-induced genes increases after vernalization. In addition, WRKY63 binding is associated with decreased levels of the repressive marker Histone H3 Lysine 27 trimethylation (H3K27me3). Collectively, our results indicate that WRKY63 is an important flowering regulator involved in vernalization-induced transcriptional regulation.
WRKY63 acts as a dual regulator that activates FLC directly under non-vernalization conditions but represses FLC indirectly after vernalization through activating COOLAIR/COLDAIR.
Introduction
The flowering time of Arabidopsis (Arabidopsis thaliana) is regulated by complex gene regulation networks associated with different regulation pathways, including photoperiod, autonomous and vernalization pathways (Hepworth and Dean, 2015). FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1) are two key integrators perceiving these pathways. In the autonomous and vernalization pathways, the MADS-box transcription factor FLOWERING LOCUS C (FLC) acts as a repressor of FT and SOC1 (Michaels and Amasino, 2001; Hepworth et al., 2002; Searle et al., 2006). The repression of FLC is mediated by the conserved chromatin-modifying complex Polycomb Repression Complex 2 (PRC2) (Gendall et al., 2001; De Lucia et al., 2008; Kim and Sung, 2013). Prolonged exposure to cold triggers the increased enrichment of PRC2 and increased levels of the repressive histone mark Histone H3 Lysine 27 trimethylation (H3K27me3) at FLC chromatin (De Lucia et al., 2008; Kim and Sung, 2013). Two Arabidopsis long noncoding RNAs (lncRNAs), COLD INDUCED LONG ANTISENSE INTRAGENIC RNA (COOLAIR) and COLD ASSISTED INTRONIC NONCODING RNA (COLDAIR), are originated from the 3′ end and the first intron of FLC, respectively. The expression of both COOLAIR and COLDAIR is induced by vernalization. Furthermore, COOLAIR and COLDAIR repress FLC transcription by increasing the H3K27me3 level via interacting with the PRC2 complex (Swiezewski et al., 2009; Heo and Sung, 2011; Marquardt et al., 2014; Kim et al., 2017). In addition, COOLAIR and COLDAIR are also associated with DNA folding and the loop structure of FLC to repress FLC expression (Sun et al., 2013; Kim et al., 2017). These findings demonstrate the importance of COOLAIR and COLDAIR in FLC expression. However, the molecular mechanism of how COOLAIR and COLDAIR are transcriptionally regulated remains elusive.
The WRKY protein family is one of the largest groups of transcription factors in plants (Llorca et al., 2014). All WRKY transcription factors contain zinc-finger-like motifs and one or two WRKY domains (Eulgem et al., 2000). The WRKY domain is a DNA-binding region of approximately 60 amino acids composed of the conserved amino acid sequence WRKYGQK (Rushton et al., 1995). The WRKY transcription factors containing two WRKY domains are classified as group I, whereas most WRKY transcription factors containing one WRKY domain are categorized as group II (Eulgem et al., 2000). Both of these groups have the C2H2-type zinc-finger motif (Eulgem et al., 2000). There is a small group of WRKY transcription factors containing the C2HC-type zinc-finger motif with one WRKY domain that are classified as group III (Eulgem et al., 2000). Four members of Arabidopsis group-IIIa WRKY transcription factors, namely WRKY63, WRKY64, WRKY66 and WRKY67, share two highly conserved domains on the N-terminus and the WRKY domain on the C-terminus (Kalde et al., 2003). The biological function of these group-IIIa WRKY transcription factors remains largely unknown. Although members of the WRKY protein family are involved in biotic/abiotic stress responses, vegetative growth, and reproductive development, only a few of them have been reported to regulate flowering time (Li et al., 2016b; Yu et al., 2016; Zhang et al., 2018). Members of the group-IIc WRKY transcription factors in Arabidopsis, including WRKY12, WRKY13, WRKY71 and WRKY75, are involved in flowering (Li et al., 2016b; Yu et al., 2016; Zhang et al., 2018). WRKY71 promotes flowering by directly activating FT and LEAFY (LFY), accelerating transition from shoot apical meristems to floral meristems (Yu et al., 2016). WRKY12, WRKY13, and WRKY75 are involved in GA signaling and modulate flowering time through interacting with DELLA proteins (Li et al., 2016b; Zhang et al., 2018). In addition, the group-I WRKY34 can repress FLC indirectly by activating ubiquitin-E3 ligase CULLIN3A (CUL3A) to enhance degradation of the FLC activator FRIGIDA (FRI) (Hu et al., 2014).
Arabidopsis WRKY63, also named as ABA OVERLY SENSITIVE 3 (ABO3), is a member of group-IIIa WRKY transcription factors. Previous studies have demonstrated that WRKY63 plays a role in ABA response and drought tolerance by activating ABSCISIC ACID RESPONSIVE ELEMENTS-BINDING FACTOR 2 (ABF2) (Ren et al., 2010). In addition, WRKY63 can also regulate stress responses of mitochondrial target proteins by activating ALTERNATIVE OXIDASE 1a (AOX1a) (Van Aken et al., 2013). In this study, we found that WRKY63 functions as a transcriptional activator of FLC. Furthermore, WRKY63 is involved in vernalization-induced flowering regulation by activating the expression of the lncRNAs COOLAIR and COLDAIR.
Results
WRKY63 is a transcriptional activator involved in flowering time regulation
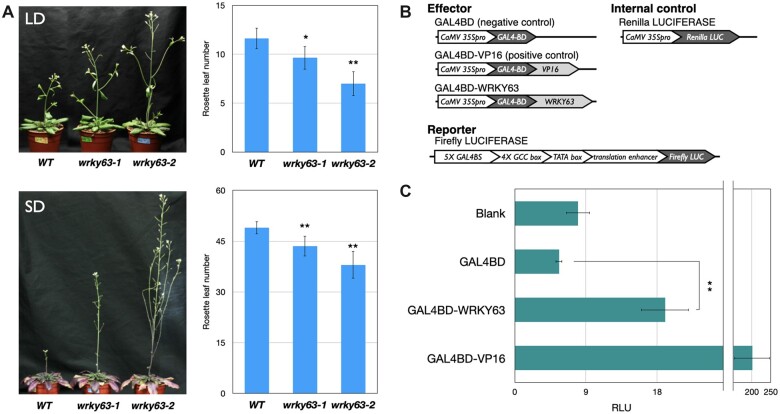
Previous studies indicated that WRKY63 is involved in ABA and stress responses by regulating ABF2 and AOX1a (Ren et al., 2010; Kim et al., 2017). To further investigate the biological function of WRKY63, the two wrky63 mutant lines wrky63-1 and wrky63-2 (abo3) were obtained from The Arabidopsis Information Resource (TAIR; www.arabidopsis.org). wrky63-1 and wrky63-2 have T-DNA inserted in the promoter and third exon of the WRKY63 locus, respectively (Supplemental Figure S1, A and B). Homozygous wrky63-1 and wrky63-2 lines were identified by reverse transcription quantitative PCR (RT-qPCR) analysis. Although the WRKY63 transcript was detected in both wrky63-1 and wild-type (WT), the expression of WRKY63 was decreased in wrky63-1, indicating that it is a WRKY63 knock-down line (Supplemental Figure S1, C and D). By contrast, the WRKY63 transcript was absent in wrky63-2, indicating that it is a WRKY63 knockout line (Supplemental Figure S1, C and D). Both wrky63-1 and wrky63-2 displayed an early flowering phenotype in both long-day (LD, 16 h light and 8 h dark) and short-day (SD, 8 h light and 16 h dark) conditions (Supplemental Figure 1, A and E), suggesting that WRKY63 is involved in flowering time regulation.
WRKY63 has been identified to be a transcriptional activator by using transcriptional activity assays in yeast (Ren et al., 2010). To further confirm whether WRKY63 can act as a transcriptional activator in planta, we conducted transient transcriptional activity assays in Arabidopsis protoplasts. WRKY63 fussed with the GAL4 DNA binding domain at the N-terminus (GALBD-WRKY63) was analyzed for whether it can activate the Firefly LUCIFERASE reporter with the GAL4 binding sites (GAL4BS) in the promoter. The GAL4 DNA binding domain fused with the VP16 activation domain (GAL4BD-VP16) was used as a positive control (Figure 1B). We found that the LUCIFERASE reporter can be activated by GAL4BD-WRKY63 (Figure 1C), indicating that WRKY63 can act as a transcriptional activator in Arabidopsis.
Figure 1.
Arabidopsis WRKY63 is a transcriptional activator and is involved in flowering regulation. A, The flowering phenotypes and rosette leaf numbers of WT and wrky63 mutants under LD and SD. **P < 0.001, *P < 0.01 (Student’s t test). Values are means ±sd. The experiments were performed with three biological replicates showing comparable results. B, Schematic structure of the effectors, reporter, and internal control used in the transient transcriptional activity assays. C, Transient transcriptional activity assays of WRKY63. GAL4BD-WRKY63 activated the expression of firefly LUCIFERASE (firefly LUC). Blank: no effector was added. GAL4BD was used as a negative control. GAL4BD fused with the activation domain VP16 (GAL4BD-VP16) was used as a positive control. Values are means ±sd. **P < 0.001 (Student’s t test). The experiments were performed with three biological replicates showing comparable results. RLU, relative luciferase activity.
WRKY63 is a transcriptional activator of FLC
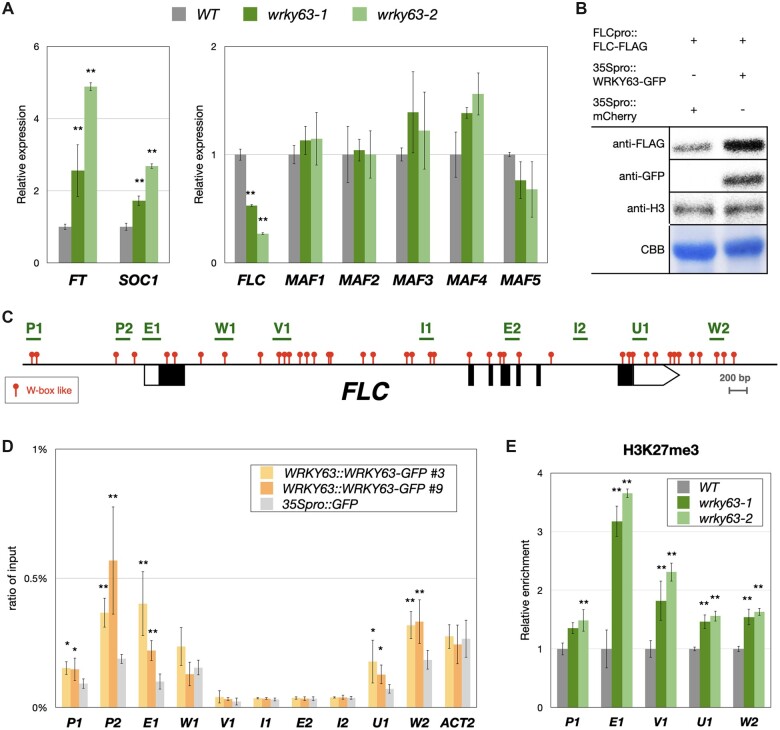
To investigate the function of WRKY63 in flowering time regulation, we analyzed expression of the core floral regulator genes FT, SOC1, and FLC in wrky63 mutants. The expression of FT and SOC1 was significantly increased, but the expression of FLC was decreased in both wrky63-1 and wrky63-2 compared to WT (Figure 2A), suggesting that WRKY63 acts upstream of FT, SOC1, and FLC (Figure 2A). To analyze the genetic interaction of WRKY63 with FT and FLC in flowering time control, we generated the wrky63/ft and wrky63/flc double mutants. wrky63/ft showed a late flowering phenotype similar to ft (Supplemental Figure S2), suggesting that WRKY63 acts upstream of FT in flowering time control. Compared to WT, wrky63/flc showed an early flowering phenotype similar to wrky63-2 and flc (Supplemental Figure S3).
Figure 2.
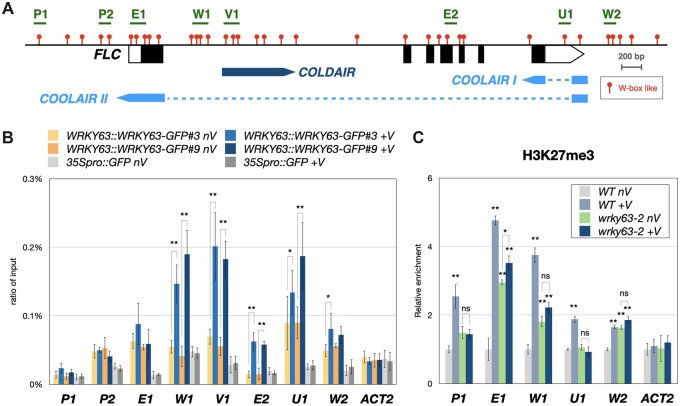
WRKY63 can activate FLC by direct targeting. A, The relative expression of FT, SOC1, FLC, MAF1, MAF2, MAF3, MAF4, and MAF5 in wrky63-1, wrky63-2, and WT Arabidopsis. B, Western blot of FLCpro::FLC-FLAG in Agrobacterium-infiltrated Nicotiana benthamiana leaves transformed with 35Spro::WRKY-GFP or the 35Spro::mcherry control. Western blots were performed with indicated antibodies, H3 and total proteins stained with CBB were used as the loading control. CBB, coomassie brilliant blue. C, The schematic structure of the FLC genome. The red dots indicate W-boxes like motifs (TGAC), and the regions analyzed for ChIP-qPCR are marked with green lines. D, Binding of WRKY63-GFP to FLC identified by ChIP-qPCR assays. Fourteen-day-old WRKY63pro::WRKY63-GFP transgenic plants were used for ChIP assays using an anti-GFP antibody. The 35Spro::GFP transgenic plants were used as a control. The amount of immunoprecipitated DNA was quantified by qPCR. Values represent the average immunoprecipitation efficiencies (%) against the total input DNA, and ACT2 was used as a negative control. E, ChIP analysis of H3K27me3 levels on the FLC locus. Fourteen-day-old plants were used for ChIP assays. The amounts of DNA after ChIP were quantified by qPCR and normalized to TA3. The values of these qPCR assays are means ±sd. These experiments were performed with three biological replicates showing comparable results. **P < 0.001, *P < 0.01 (Student’s t test).
To further investigate whether WRKY63 can directly activate FLC expression, we generated the FLCpro::FLC-FLAG construct containing the FLC coding sequence (CDS) fused with 3×FLAG driven by the FLC native promoter. FLCpro::FLC-FLAG was co-expressed with 35Spro::WRKY63-GFP or 35Spro::mcherry control in Nicotiana benthamiana leaves by using Agrobacterium-mediated infiltration. Compared to the negative control, the protein level of FLC-FLAG was increased when FLCpro::FLC-FLAG was co-expressed with 35Spro::WRKY63, suggesting that WRKY63 can induce FLC expression (Figure 2B).
We also generated WRKY63pro::WRKY63-GFP transgenic lines, in which the WRKY63 genome sequence containing its native promoter fused with the GFP epitope tag was transformed into wrky63-2. The early flowering phenotype of wrky63-2 was rescued by expressing WRKY63pro::WRKY63-GFP (Supplemental Figure S4), suggesting that WRKY63-GFP is functional. The binding of WRKY63 to FLC was analyzed by chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR). ChIP assays were performed with the anti-GFP antibody using WRKY63pro::WRKY63-GFP transgenic seedlings. We found that WRKY63 can be enriched in the FLC promoter and the transcription starting site regions, indicating that WRKY63 can activate FLC by directly targeting (Figure 2, C and D). Previous studies indicate that the expression of FLC is highly associated with H3K27me3 level changes (Gendall et al., 2001; De Lucia et al., 2008; Kim and Sung, 2013). We further investigated whether WRKY63 affects the level of H3K27me3 on the FLC locus. Compared to WT, we found that the H3K27me3 level of FLC was increased in wrky63 (Figure 2E). Collectively, these results indicated that WRKY63 can directly target FLC and activate FLC expression. Furthermore, WRKY63-induced FLC expression is associated with decreased H3K27me3.
WRKY63 is involved in vernalization-induced flowering
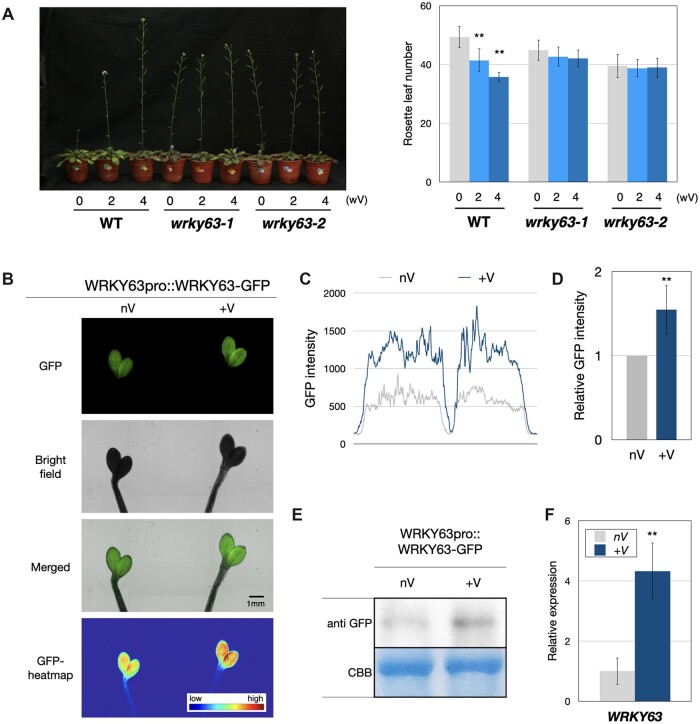
Previous studies indicated that FLC is involved in vernalization-induced flowering (De Lucia et al., 2008; Swiezewski et al., 2009; Heo and Sung, 2011; Kim and Sung, 2013). To investigate whether WRKY63 is involved in vernalization-induced flowering, wrky63 mutant plants were treated with 0, 2, and 4 weeks of vernalization. The flowering time of WT plants was decreased after vernalization. However, the flowering time of wrky63 mutants was not reduced after the vernalization treatment (Figure 3A; Supplemental Figure S5), indicating that wrky63 mutants are insensitive to vernalization.
Figure 3.
WRKY63 is induced by vernalization and is involved in vernalization-induced flowering. A, The flowering phenotypes and rosette leaf numbers of WT and wrky63 mutants under 0, 2, and 4 weeks of vernalization treatments in SD. At least 20 plants of each line were counted for the rosette leaf numbers at flowering. Values are means ±sd. **: P < 0.001 (Student’s t test). The experiments were repeated 3 times with comparable results. wV, weeks of vernalization. B, GFP fluorescence signal in WRKY63pro::WRKY63:GFP transgenic plants without vernalization (nV) or with 4 weeks of vernalization (+V). The experiments were performed with three biological replicates showing comparable results. Bar = 1 mm. C and D, Quantification of GFP fluorescent intensity (C) and the relative GFP fluorescent intensity (D) in WRKY63pro::WRKY63:GFP transgenic plants without vernalization (nV) or with 4 weeks of vernalization (+V). The X-axis represents the detected section of cotyledons, Y-axis fluorescence indicates the value of GFP intensity. Values of relative GFP intensity are normalized means ±sd. **P < 0.001. At least 10 leaves for each treatment were counted for the GFP intensity. E, Western blot of WRKY63pro::WRKY63:GFP in transgenic plants without vernalization (nV) or with 4 weeks of vernalization (+V). The western blots were performed with the indicated antibody, and the total protein stained with CBB was used as the loading control. F, The relative expression of WRKY63 in WT Arabidopsis without vernalization (nV) or with 4 weeks of vernalization (+V). **P < 0.001 (Student’s t test). Values are means ±sd. The experiments were performed with three biological replicates showing comparable results. CBB, coomassie brilliant blue.
We further analyzed whether the expression of WRK63 is affected by vernalization. WRKY63pro::WRKY63:GFP transgenic seedlings were analyzed using a fluorescent microscope. We found that the GFP signal was presented in the leaves and root-tips of the transgenic plants (Figure 3B; Supplemental Figure S6). Interestingly, the fluorescence intensity of GFP was increased after vernalization treatment (Figure 3, B–D; Supplemental Figure S6). Western-blot analysis indicated that the protein level of WRKY63 was increased after vernalization (Figure 3E). In addition, the transcript of WRKY63 was also increased after vernalization (Figure 3F). These results indicated that the expression of WRKY63 can be induced by vernalization. Collectively, our results suggested that WRKY63 is involved in vernalization-induced flowering.
WRKY63 is a transcriptional activator of the lncRNAs COOLAIR and COLDAIR
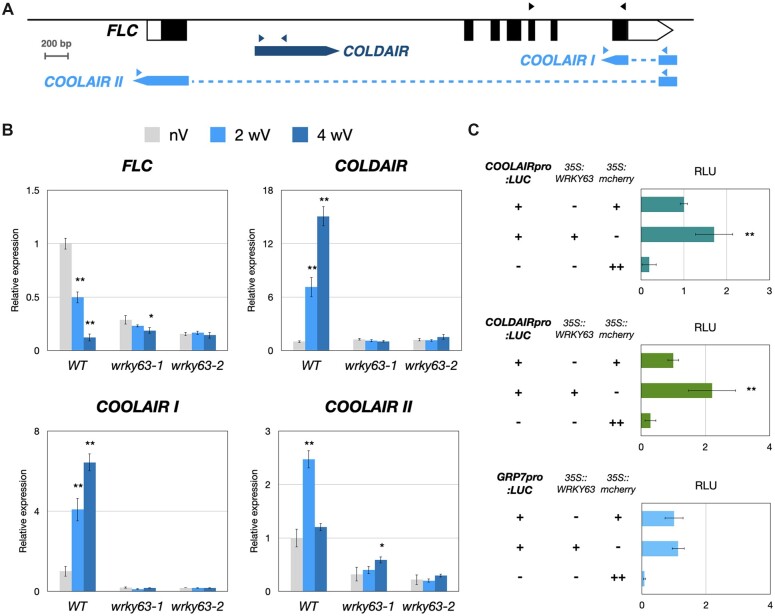
To further investigate how WRKY63 is involved in vernalization-induced flowering, we analyzed the expression of FLC after the vernalization treatment. After vernalization, FLC expression was decreased in WT but not in the wrky63 mutants (Figure 4, A and B), suggesting that vernalization-repressed expression of FLC is dependent on WRKY63. The Arabidopsis lncRNAs COOLAIR and COLDAIR can be induced by vernalization, and both of them can repress the expression of FLC (Swiezewski et al., 2009; Heo and Sung, 2011; Marquardt et al., 2014; Kim et al., 2017). Interestingly, we found that the expression of COOLAIR and COLDAIR was strongly increased in WT after vernalization treatment, but not in the wrky63 mutants (Figure 4, A and B). These results suggested that vernalization-induced COOLAIR/COLDAIR expression is dependent on WRKY63. To further confirm whether WRKY63 can activate COOLAIR/COLDAIR expression, COOLAIR/COLDAIR promoter-driven LUCIFERASE constructs (COOLAIRpro::LUC/COLDAIRpro::LUC) were co-expressed with 35Spro::WRKY63 or the 35Spro::mcherry control. We found that the expression of COOLAIRpro::LUC and COLDAIRpro::LUC was significantly increased when co-expressed with WRKY63, but not with the 35Spro::mcherry control (Figure 4C). By contrast, the GLYCINE RICH PROTEIN 7 (GRP7) promoter-driven LUCIFERASE construct (GRP7pro::LUC), which was used as a control, did not show increased expression when co-expressed with WRKY63 (Figure 4C). Together, these data indicated that WRKY63 is a transcriptional activator of COOLAIR and COLDAIR.
Figure 4.
The expression of COOLAIR and COLDAIR is activated by WRKY63. A, The FLC structure and the fragments used for the RT-qPCR assay. Arrows indicate the primer sets in RT-qPCR. The blocks represent transcribed regions of FLC, COLDAIR and COOLAIR. The dashed lines indicate intron regions. B, The relative expression of FLC, COLDAIR, Class I COOLAIR (COOLAIR I), and Class II COOLAIR (COOLAIR II) in wrky63-1, wrky63-2, and WT treated with 0, 2, or 4 weeks of vernalization (nV, 2wV, and 4wV). C, Transient transcriptional activity assays of COOLAIRpro::LUC, COLDAIRpro::LUC, and GRP7pro::LUC. The firefly LUCIFERASE (LUC) driven by the COOLAIR promoter, COLDAIR promoter, or GRP7 promoter were co-transformed with the effector 35S::WRKY63 and the 35Spro::mcherry control. RLU represents firefly LUCIFERASE normalized by co-expressed 35Spro::Renilla luciferase. The value of above assays represents means ±sd. These experiments were performed with three biological replicates showing comparable results. **P < 0.001, *P < 0.05 (Student’s t test). RLU, relative light units.
We further tested the wrky63 mutant for its vernalization response by the introgression of wrky63 into the FRIcol (Col-0 with a functional FRI allele) genetic background (Gazzani et al., 2003). We found that FRI/wrky63 was also insensitive to vernalization (Supplemental Figure S7, A and B). The gene expression pattern was also analyzed in FRIcol and FRI/wrky63. We found that the expression of FLC was significantly decreased in FRI/wrky63 compared to FRIcol (Supplemental Figure S7C). In addition, we also found that the expression of COOLAIR was decreased in FRI/wrky63 compared to FRIcol (Supplemental Figure S7C). These results indicated that vernalization-induced COOLAIR expression is dependent on WRKY63.
The binding of WRKY63 to FLC under vernalization (V) or non-vernalization (nV) treatment was further analyzed by ChIP-qPCR using WRKY63pro::WRKY63:GFP transgenic seedlings. We found that the binding of WRKY63 on the first intron (W1 and V1) and down-stream regions (U1 and W2) of FLC was significantly increased after vernalization treatment (Figure 5, A and B). Interestingly, two WRKY63 binding sites, W2 and W1, are located in the 5′ upstream regions of the transcription start sites (TSS) of COOLAIR and COLDAIR, respectively (Figure 5A). Collectively, these results suggested that WRKY63 can directly target the FLC locus to activate FLC under non-vernalization conditions. Furthermore, vernalization increases the binding of WRKY63 to the COOLAIR/COLDAIR promoter, which activates COOLAIR/COLDAIR transcription and suppresses the expression of FLC.
Figure 5.
Binding of WRKY63 and the H3K27me3 level on the FLC locus in vivo. A, The schematic structure of the FLC genome. The blocks represent transcribed regions of FLC, COLDAIR and COOLAIR. The dashed lines indicate intron regions. B, Binding of WRKY63pro::WRKY63:GFP to FLC identified by ChIP-qPCR assays. Fourteen-day-old WRKY63pro::WRKY63:GFP transgenic plants without vernalization (nV) or with 4 weeks of vernalization (+V) were used for ChIP assays using an anti-GFP antibody. The 35Spro::GFP transgenic plants were used as a control. The amount of immunoprecipitated DNA was quantified by qPCR. Values represent the average immunoprecipitation efficiencies (%) against the total input DNA, and ACT2 was used as a negative control. C, ChIP analysis of H3K27me3 levels on the FLC locus. Plants without vernalization (nV) or with 4 weeks of vernalization (+V) were used for ChIP assays. The amounts of DNA after ChIP were quantified by qPCR and normalized to TA3. Error bars indicate sd from three biological replicates. *P < 0.01, **P < 0.001, ns: no significant difference (Student’s t test).
Previous studies have revealed that the vernalization-repressed expression of FLC is associated with PRC2-mediated H3K27me3 enrichment (De Lucia et al., 2008; Swiezewski et al., 2009; Heo and Sung, 2011; Kim and Sung, 2013). To further investigate whether WRKY63 is involved in the regulation of histone modifications, we also analyzed the H3K27me3 and H3 acetylation (H3Ac) levels in wrky63 treated with vernalization (+V) or non-vernalization (nV). The H3K27me3 level of FLC in wrky63 was increased compared to WT before vernalization (Figure 5C). Furthermore, the H3K27me3 level of FLC was significantly increased after vernalization in WT. By contrast, the H3K27me3 level of FLC in wrky63 was not changed after vernalization (Figure 5C). In addition, the H3Ac level of FLC was not substantially affected by vernalization in both wrky63 and WT (Supplemental Figure S8). These results indicated that WRKY63-induced gene expression is associated with decreased H3K27me3 levels.
Genome-wide occupancy profiles of WRKY63
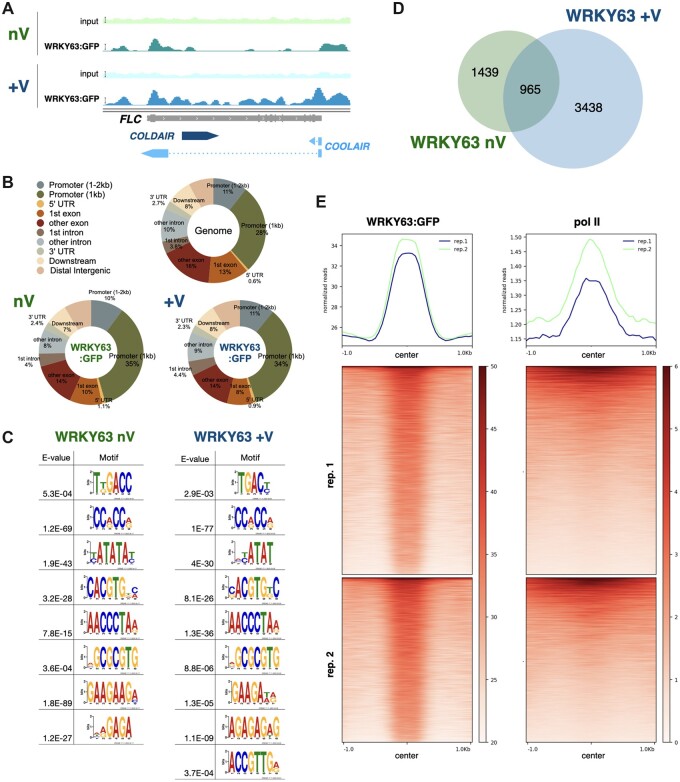
To investigate the genome-wide function of WRKY63 in gene regulation, we mapped the genome-wide occupancy of WRKY63. Chromatin immunoprecipitation followed by sequencing (ChIP-seq) was carried out to identify the WRKY63-targeted genomic regions using WRKY63pro::WRKY63:GFP transgenic plants under vernalization or non-vernalization treatment. Similar to the ChIP-qPCR data, the genome browser views of WRKY63 ChIP-Seq data also show that WRKY63 can target the FLC promoter and first exon, and vernalization increased the binding of WRKY63 on the first intron and downstream regions (Figure 6A). Compared to the Arabidopsis genomic region distribution, we found that the enrichment of WRKY63 was higher in the 1-kb promoter regions, but lower in the exon regions (Figure 6B). The genomic distribution ratio of WRKY63 was similar among vernalization and non-vernalization treatments (Figure 6B). In addition, we identified many cis-elements that were enriched within the WRKY63 binding sites, including TGACC/T, CCRCCR, TATA-box, G-box (CACGTG), AACCCTA and AG/AAG sequences (Figure 6C). The enriched cis-element TGACC/T is the W-box like motif, which is found to be targeted by many WRKY family transcription factors (Eulgem et al., 2000; Maeo et al., 2001). We found that the WRKY63 enriched cis-elements of vernalization and non-vernalization treatments were similar (Figure 6C). Taken together, these results suggested that the genomic distribution ratio and the DNA binding motifs of WRKY63 are not affected by vernalization.
Figure 6.
Genome-wide occupancy profiles of WRKY63. A, IGV showing the binding of WRKY63 on the FLC locus without (nV) or with (+V) vernalization. Bars: normalized reads = 25. B, Pie charts showing the distribution of WRKY63 annotated genic and intergenic regions in the genome. C, The DNA binding motifs significantly enriched in the WRKY63 binding sites. D, A Venn diagram showing the overlap between WRKY63 targeted genes among vernalization (WRKY63 +V) and non-vernalization (WRKY63 nV) treatments. E, Heat map representation of the occupancy of WRKY63 and RNA polymerase II (pol II) in the WRKY63 occupied genomic region. Each horizontal line represents a WRKY63 binding region, and the signal intensity is shown for the occupancy of WRKY63 and pol II (center). Columns show the genomic region surrounding each WRKY63 peak. Signal intensity is indicated by the shade of heatmap.
However, we also found that the number of WRKY63-targeted genes is increased after vernalization (Figure 6D). Many of the WRKY63-targeted genes under vernalization were not targeted by WRKY63 without vernalization (Figure 6D), suggesting that WRKY63 binds to a subset of genes only after vernalization. These results indicated that WRKY63 might be involved in the vernalization-induced gene activation. We further classified the WRKY63 target genes into three different groups, namely the genes specific for vernalization, those specific for non-vernalization, and those for both vernalization and non-vernalization. These WRKY63-targeted genes were further analyzed according to Gene Ontology Biological Processes (GO-BP). We found that the WRKY63 target genes are involved in different types of hormone responses, stress responses, and development pathways (Supplemental Figure S9). Interestingly, we found that the GO term “vernalization response (GO:0010048)” was only enriched in the genes specifically targeted by WRKY63 after vernalization (Supplemental Figure S9). By contrast, the genes specifically targeted under non-vernalization were more associated with the light response and circadian rhythm, but less associated with developmental responses (Supplemental Figure S9). These results suggested that WRKY63 may be involved in different regulation processes under vernalization and non-vernalization.
We also compared the WRKY63-occupied genomic regions with the previously published ChIP-Seq data of RNA polymerase II (pol II) (Liu et al., 2018). The plotprofile and plot heatmap analyses indicate that the center of WRKY63-binding regions was highly associated with the enrichment of pol II (Figure 6E). These results support that WRKY63 can act as a transcriptional activator and may be associated with the regulation of transcription initiation.
WRKY63 is associated with vernalization-induced gene activation and decreased H3K27me3
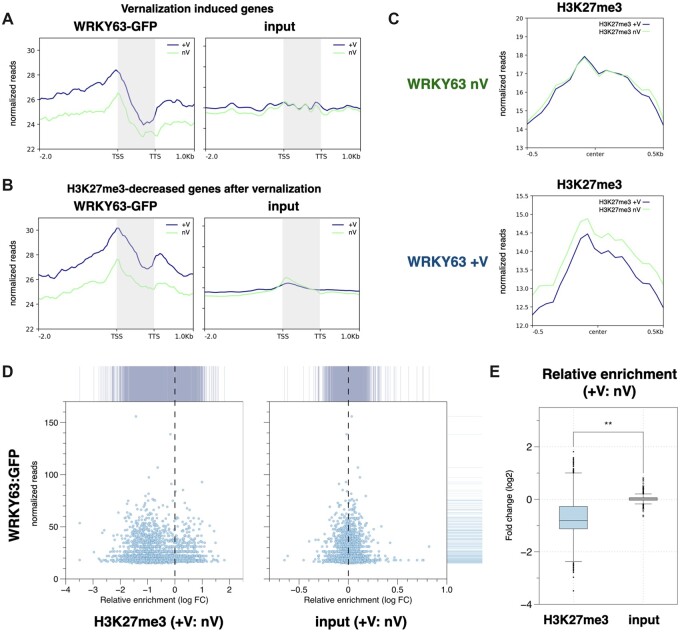
We further investigated the function of WRKY63 in the vernalization-induced gene activation. A subset of vernalization-induced genes was previously identified by the RNA-sequencing data of Arabidopsis treated with vernalization (Gibbs et al., 2018). We found that the binding of WRKY63 was highly enriched near the TSS of these vernalization-induced genes. In addition, the binding of WRKY63 on these genes was substantially increased after vernalization (Figure 7A). We also identified a subset of genes with decreased H3K27me3 levels after vernalization from the published ChIP-seq datasets (Xi et al., 2020). Similarly, the binding of WRKY63 to these genes with decreased H3K27me3 was substantially increased after vernalization (Figure 7B). Compared to the non-vernalization-specific targets of WRKY63, the vernalization-specific targets of WRKY63 tend to have high degrees of overlap with the vernalization-induced genes and the H3K27me3 decreased genes (Supplemental Figure S10).
Figure 7.
WRKY63 is associated with vernalization-induced gene activation and H3K27me3 demethylation. A and B, Metagene ChIP-seq binding profiles of WRKY63 among the vernalization-induced genes (A) and the H3K27me3-decreased genes after vernalization (B). The profile is from 2-kb upstream of the TSS to 1-kb downstream of the TTS, and the gene lengths were scaled to the same size. C, The mean density of H3K27me3 enrichment in the WRKY63 occupied regions under non-vernalization (WRKY63 nV) or vernalization (WRKY63 +V) treatments. The average H3K27me3 signal under vernalization (H3K27me3 +V) or non-vernalization (H3K27me3 nV) within 0.5-kb genomic regions flanking the center of WRKY63 peaks is shown. D, X–Y scatter plots showing the relative enrichment of H3K27me3 levels (+V: nV) and the binding level of WRKY63 under vernalization with the vernalization-specific WRKY63 targeted genes. E, Boxplot showing the relative H3K27me3 level (+V: nV) of the vernalization-specific WRKY63 targeted genes. Center line: median value; box limits: central 50% of the data, upper/lower quartiles: first/last 25% of the data; points: value of the outliers. **P < 0.001 (Student’s t test).
To further analyze the function of WRKY63 in association with H3K27me3, we compared the H3K27me3 level in the WRKY63 targeted genomic regions under vernalization (WRKY63 +V) or non-vernalization (WRKY63 nV) treatments. Interestingly, we found that the H3K27me3 level on the targeted regions of WRKY63+V was substantially decreased after vernalization (Figure 7C). By contrast, the H3K27me3 level on the binding regions of WRKY63-nV was not changed (Figure 7C). Furthermore, the relative H3K27me3 levels of the vernalization-specific target genes of WRKY63 were substantially decreased after vernalization (Figure 7, D and E). Collectively, these results indicated that WRKY63 is involved in both gene activation and H3K27me3 decreasing induced by vernalization. However, a subset of vernalization-specific WRKY63 targets did not show decreased H3K27me3 after vernalization (Supplemental Figure S10), suggesting that in addition to H3K27 demethylation, other histone modifications may also be involved in WRKY63-regulated genes.
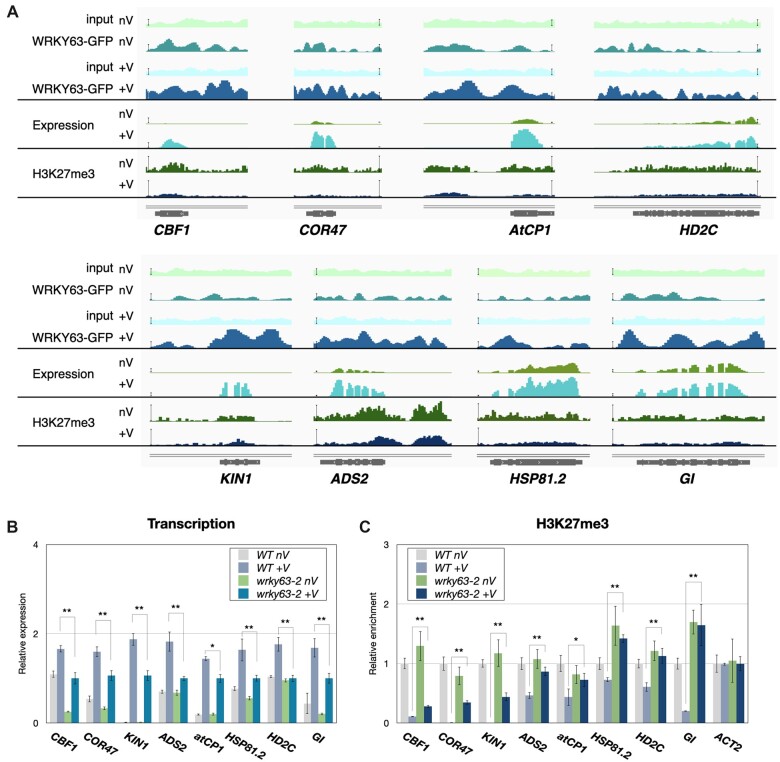
We selected several vernalization-induced genes that are targeted by WRKY63 with decreased H3K27me3 after vernalization for further analysis. Genome browser views of the RNA-seq data also indicated that the expression of these genes was increased after vernalization (Figure 8A). Furthermore, these vernalization-induced genes were also directly targeted by WRKY63, and the binding of WRKY63 was increased after vernalization (Figure 8A). We found that the expression of several cold-response genes was also decreased in wrky63 compared to WT after vernalization, such as C-REPEAT/DRE BINDING FACTOR 1 (CBF1) (Stockinger et al., 1997), COLD-REGULATED 47 (COR47) (Guo et al., 1992), KIN1 (Knight et al., 1996), ACYL-LIPID DESATURASE2 (ADS2) (Chen and Thelen, 2013), and CA2+-BINDING PROTEIN 1 (AtCP1) (Knight et al., 1996; Delk et al., 2005) (Figure 8B). Recent studies indicate that the HD2-type histone deacetylase HD2C and the flowering regulator GIGANTEA (GI) are involved in cold responses (Fornara et al., 2015; Park et al., 2018; Lim et al., 2020). We found that vernalization-induced expression of HD2C and GI was decreased in wrky63 (Figure 8B). Together, these results indicated that WRKY63 is required for the activation of vernalization-induced genes. In addition, the increased expression of these vernalization-induced genes was associated with decreased H3K27me3 after vernalization (Figure 8A). We further analyzed the H3K27me3 levels of these genes in wrky63 using ChIP-qPCR. Vernalization reduced H3K27me3 levels in WT (Figure 8C). However, the H3K27me3 levels of these genes in wrky63 were significantly higher compared to WT after vernalization (Figure 8C). Collectively, these results indicated that WRKY63 is associated with the decreased H3K27me3 of the vernalization-induced genes.
Figure 8.
The WRKY63 binding, expression and H3K27me3 level of WRKY63 regulated genes. A, IGV showing the WRKY63 binding, expression, and H3K27me3 level of selected genes. Bars: normalized reads = 25. B and C, Expression (B) and H3K27me3 patterns (C) in WT and wrky63 mutants. Expression of the indicated genes was analyzed by RT-qPCR. UBQ10 was used as an internal control. ChIP assays were performed with the H3K27me3 antibody. The amounts of DNA after ChIP were quantified by qPCR and normalized to TA3. Error bars correspond to standard derivations. *P < 0.05; **P < 0.005 (Student’s t test). At least three independent biological replicates were performed with similar results.
Discussion
Arabidopsis WRKY63 is a member of group-IIIa WRKY transcription factors (Eulgem et al., 2000). Previous studies have demonstrated that WRKY63 plays a role in ABA response and drought tolerance by directly targeting to the W-boxes of ABF2 and AOX1a (Ren et al., 2010; Van Aken et al., 2013). In this study, we identified that WRKY63 functions as a transcriptional activator of FLC and is involved in the vernalization-induced flowering. The transcript and protein of WRKY63 are increased after vernalization. Furthermore, a subset of vernalization-induced genes can be activated by WRKY63. In addition to flowering, WRKY63-regulated genes are also involved in the other development processes as well as hormone and stress responses. Recently, the genomic binding patterns of WRKY18, WRKY33, and WRKY40 were also investigated by using ChIP-seq (Birkenbihl et al., 2017). It was found that WRKY18 and WRKY40 are redundantly involved in flg22-induced defense responses (Birkenbihl et al., 2017). Our ChIP-seq analyses indicate that the enrichment of WRKY63 was higher in the promoter regions, but lower in the exon regions. In addition, the binding of WRKY63 is highly enriched in regions near the TSS. Similar binding patterns were also observed for WRKY18, WRKY33, and WRKY40 (Birkenbihl et al., 2017).
lncRNAs are involved in diverse gene regulation processes, including chromatin remodeling, histone modification, alternative splicing, and transcriptional activation (Wang et al., 2011; Bardou et al., 2014; Liu et al., 2015; Hung et al., 2020a). In Arabidopsis, the lncRNAs COOLAIR and COLDAIR are necessary for vernalization-induced FLC repression by increasing the occupancy of PRC2 at FLC chromatin (Swiezewski et al., 2009; Heo and Sung, 2011; Marquardt et al., 2014; Kim et al., 2017). The expression of COOLAIR and COLDAIR is highly increased under prolonged cold treatment (Swiezewski et al., 2009; Heo and Sung, 2011; Marquardt et al., 2014; Kim et al., 2017). However, the molecular mechanism of how COOLAIR and COLDAIR are transcriptionally activated remains elusive. In this study, we found that WRKY63 can directly bind to the FLC locus. Furthermore, WRKY63 binding on the promoters of COOLAIR and COLDAIR is significantly increased after vernalization. We also found that the vernalization-induced expression of COOLAIR and COLDAIR is decreased in the wrky63 mutant compared to WT. In addition, WRKY63 can also activate the LUC reporter gene driven by COOLAIR and COLDAIR promoters in the transactivation assays. These results indicated that WRKY63 can act as a transcriptional activator of COOLAIR and COLDAIR. Our recent study has revealed that the expression of lncRNAs is associated with H3Ac/H3K4me2 changes regulated by the HDA6-LDL1/2 histone modification complex in Arabidopsis (Hung et al., 2020a). In addition, COOLAIR interacts with the FRI super-complex and was predicted to be involved in forming a loop on the FLC transcript during vernalization (Sun et al., 2013). It remains to be determined whether COOLAIR and COLDAIR activation mediated by WRKY63 is related to these processes. Intriguingly, we found that WRKY63 can target different genomic regions of the FLC locus. Although WRKY63 can bind to the FLC promoter and first exon without vernalization, the binding of WRKY63 on COOLAIR and COLDAIR promoters is increased after vernalization. These results suggested that WRKY63 can activate FLC under non-vernalization conditions and also activate COOLAIR/COLDAIR transcription to suppress the expression of FLC during vernalization. However, the mechanism underlying the binding of WRKY63 to different regions of the FLC locus is still unclear. It is possible that WRKY63 may associate with different protein complexes under vernalization and non-vernalization conditions. It has been reported that the protein components of the PRC2 complex are different and can target different regions of FLC under vernalization and non-vernalization conditions (Wood et al., 2006; Swiezewski et al., 2009; Heo and Sung, 2011; Marquardt et al., 2014; Kim et al., 2017). Changed binding of WRKY transcription factors under specific stress conditions has also been reported (Rushton et al., 2010; Llorca et al., 2014; Li et al., 2016b). Genome-wide analyses of WRKY63 occupancy indicated that WRKY63-targeted genes are substantially changed after vernalization, suggesting that WRKY63 may regulate gene expression under different environmental conditions by targeting different genomic regions.
There are 21 JmjC domain-containing proteins in Arabidopsis, which can act as demethylases to remove methyl-group on lysine residues of histone proteins and are involved in developmental processes (Lu et al., 2008; Luo et al., 2014; Hung et al., 2020b, 2021). The KDM4 group of JmjC-demethylases is specific for H3K27me3 demethylation (Lu et al., 2008; Luo et al., 2014). EARLY FLOWERING 6 (ELF6/JMJ11) is an activator of FLC (Noh et al., 2004), but RELATIVE OF EARLY FLOWERING 6 (REF6/JMJ12) positively regulates FT by removing H3K27me3 (Lu et al., 2011). In addition, the H3K27 demethylase REF6 can directly bind to DNA by its own DNA-binding domain (Li et al., 2016a). In this study, we found that WRKY63-activated gene expression in vernalization is associated with decreased H3K27me3 level, suggesting that WRKY63 might be functionally associated with H3K27me3 demethylation to regulate gene expression in Arabidopsis. Further studies are needed to investigate the functional association between H3K27me3 demethylases and WRKY63. However, since the removing of H3K27me3 can be directly caused by the transcriptional activation (Holoch et al., 2021), the decreased H3K27me3 might be indirectly caused by the transcriptional activation of WRKY63.
In addition to COOLAIR and COLDAIR, we found that WRKY63 can also activate the expression of other vernalization-induced genes, such as CBF1, COR47, KIN1, ADS2, AtCP1, HD2C, and GI. Intriguingly, the expression of these genes is increased after vernalization. By contrast, FLC expression is decreased after vernalization. In addition, WRKY63 can activate FLC under non-vernalization conditions. Since there is no difference in the expression of KIN1, ADS2, AtCP1, and HD2C between wrky63 and WT before vernalization treatment, WRKY63 may not affect these genes under non-vernalization conditions. These results support our hypothesis that WRKY63 may associate with different protein complexes to regulate gene expression under vernalization and non-vernalization conditions. Interestingly, WRKY63 functions as a transcriptional activator of FLC under non-vernalization condition. However, vernalization increases the binding of WRKY63 to the promoters of the lncRNA COOLAIR/COLDAIR, resulting in the activation of COOLAIR/COLDAIR and repression of FLC during vernalization. Therefore, WRKY63 acts as a dual regulator that activates FLC directly under non-vernalization conditions but represses FLC indirectly during vernalization through COOLAIR/COLDAIR.
According to the GO-BP analysis, we found that the WRKY63 target genes are involved in hormone and stress responses as well as developmental processes, suggesting that in addition to vernalization and flowering regulation, WRKY63 may also be involved in other developmental processes. For example, we found that CBF1, COR47, and GI can be regulated by WRKY63 under both vernalization and non-vernalization conditions. Similar to WRKY63, CBF1, COR47, and GI have also been reported to be involved in drought stress response (Welin et al., 1994; Haake et al., 2002; Riboni et al., 2013). In addition, GI is one of the core regulators in the evening-loop of circadian rhythm and is involved in photoperiod-mediated flowering (Fowler et al., 1999). CBF1 is also regulated by the circadian rhythm and PHYTOCHROME INTERACTING FACTOR 7 (PIF7) (Kidokoro et al., 2009). Further research is required to analyze whether WRKY63 is involved in these processes.
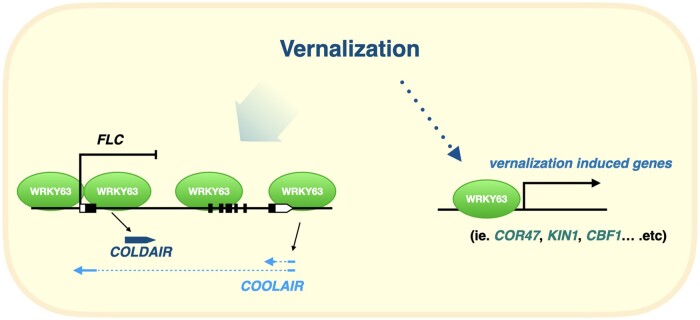
In conclusion, we demonstrated that the transcription factor WRKY63 is involved in the regulation of vernalization-induced flowering. WRKY63 binds to the FLC locus and activates the lncRNAs COOLAIR and COLDAIR under vernalization. Furthermore, WRKY63 can also activate the expression of a subset of vernalization-induced genes and the activation of gene expression mediated by WRKY63 is associated with decreased H3K27me3 (Figure 9).
Figure 9.
A model of how Arabidopsis WRKY63 is involved in vernalization-induced flowering. Without vernalization, WRKY63 can activate FLC expression by direct targeting. During vernalization, the binding of WRKY63 is increased in the promoters of the lncRNAs COOLAIR and COLDAIR, resulting in COOLAIR and COLDAIR activation. Therefore, the expression of FLC is repressed by increased COOLAIR and COLDAIR during vernalization. Furthermore, WRKY63 can also activate the expression of other vernalization-induced genes.
Materials and methods
Plant materials and growth conditions
In this study, the WT Arabidopsis (A.thaliana) Columbia (Col-0) ecotype was used. Plants were grown in growth chambers under 16/8 h light/dark LD or 8/16 h light/dark SD conditions at 22°C. For vernalization treatment, seeds were germinated and grown for 10 days under standard conditions (LD, 22°C), and then transferred to 4°C for 2, 4, or 6 weeks. The wrky63-2 (abo3) mutant (SALK_007496) has the T-DNA insertion in the third exon (Ren et al., 2010), and wrky63-1 (SALK_068280) has the T-DNA insertion in the promoter region. These wrky63 mutants were obtained from TAIR (www.arabidopsis.org). The wrky63/ft and wrky63/flc mutant plants were generated by crossing wrky63-2 (abo3) with ft-10 and flc-3, respectively. All the above mutants used in this study are in the Col-0 background. FRI/wrky63 mutant plant was generated by crossing wrky63-2 (abo3) with FRIcol (Col-0 with a functional FRI allele) (Gazzani et al., 2003).
Plasmid construction and plant transformation
The full-length CDS fragments of WRKY63 were PCR-amplified and cloned into the pCR8/GW/TOPO vector (Invitrogen). Point mutation constructs were constructed by specific primer sets then cloned into the pCR8/GW/TOPO vector (Invitrogen). WRKY63 was recombined into the PK7WGF2 (Invitrogen) to generate the GFP:WRKY63 plasmid. To construct WRKY63pro::WRKY63:GFP, approximately 2-kb promoter with the genome sequence of WRKY63 was cloned into the pCR8/GW/TOPO vector (Invitrogen), then recombined into the pMDC107 binary vector. The transgenic plants were generated by using the floral dip method.
LUCIFERASE transient assays
The analysis of LUCIFERASE transient expression was performed as described previously (Fujimoto et al., 2000; Ohta et al., 2000).The reporter construct [GAL4BS(4X)-GCC box(4X)-TATA box-Translational enhancer::Firefly LUC] (Fujimoto et al., 2000; Ohta et al., 2000) contains the binding site (GAL4BS) of the yeast transcription factor GAL4 (Ma and Ptashne, 1987). The effector construct contains the GAL4 DNA binding domain (GAL4-BD) fussed at the N-terminus of WRKY63 (GALBD-WRKY63). GAL4BD was used as a negative control, and GAL4BD fused with the VP16 activation domain (GAL4BD-VP16) (Triezenberg et al., 1988) was used as a positive control. The analyses of LUCIFERASE transient expression in protoplasts were described in previous studies (Ohta et al., 2000; Ding et al., 2018). Four micrograms of reporter constructs and 5 μg of effector constructs were used for each co-transfection assays. To analyze the promoter activity of COOLAIR and COLDAIR, 1.2 kb of COOLAIR promoter and 1.5 kb of COLDAIR promoter were PCR-amplified, then cloned into the pCR8/GW/TOPO vector (Invitrogen). The COOLAIR promoter and COLDAIR promoter were recombined into pKGWL7 with the firefly LUCIFERASE (LUC) as reporter constructs. Effector constructs 35Spro::WRKY63 and 35Spro::mcherry control were co-transformed as described previously (Ohta et al., 2000; Ding et al., 2018) for transcriptional activity assays. The reporter luciferase activities were standardized by activities of the co-expressed Renilla LUCIFERASE (rLUC), and relative reporter activities were calculated. Experiments were repeated at least three times for each reporter–effector combination. Signals of Firefly and Renilla luciferase were assayed with the dual luciferase assay reagents (Promega).
Fluorescence imaging and western blot
The vernalization or non-vernalization treated WRKY63pro::WRKY63:GFP transgenic plants were used for the fluorescence imaging. GFP fluorescence was visualized by using the Zeiss Axio Imager.Z2 microscope system (https://www.zeiss.com/microscopy). The GFP signal was quantified in the middle section of cotyledons for analyzing GFP intensity by the Zeiss ZEN software (https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html). GFP fluorescence was observed through the filter sets 46 HE (excitation at 500 nm, beamsplitter at 515 nm, and emission at 535 nm). Images were captured using a 5× objective with tile scan mode. The vernalization and non-vernalization samples were captured in the same field together and then processed and analyzed using the Zeiss ZEN software as described above.
Western blot assays were performed as previously described (Hung et al., 2018). Anti-GFP (Santa Cruz Biotechnologies, catalog no. SC-9996; 1:3,000 dilution), anti-FLAG (sigma, M2; 1:3,000 dilution), and anti-H3 (Abcam, ab1791; 1:3,000 dilution) antibodies were used as primary antibodies for Western blot; the resulting signals were detected by using an Advansta ECL Western blotting kit (Advansta, K-12045-D20).
Reverse transcription quantitative PCR analysis
Total RNA was isolated using TRIZOL reagent (Invitrogen, 15596026) according to the manufacturer’s instructions. Two micrograms of DNAse (Promega, RQ1 #M6101) treated total RNA were used to synthesize cDNA (Promega, #1012891). RT-qPCR was performed using iQ SYBR Green Supermix solution (Bio-Rad, #170-8880). The CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.) was used with the following cycling conditions: 95°C for 10 min, followed by 45 cycles of 95°C for 15 s, 60°C for 30s, and then fluorescent detection. This was immediately followed by a melting curve (65°C–95°C, incrementing 0.5°C for 5 s, and plate reading). The melting curve analysis confirmed the absence of non-specific products. Each sample was quantified at least in triplicate and normalized by calculating delta Cq (quantification cycle) to the expression of the internal control Ubiquitin10 (UBQ10). The Cq and relative expression level are calculated by the Biorad CFX Manager 3.1 based on the MIQE guidelines. Standard deviations represent at least three technical and two biological replicates. The variance in average data is represented by standard error of the mean (SEM). The standard deviation (sd), SEM determination, and P-value were calculated using Student's paired t test. The gene-specific primers used for RT-qPCR are listed in Supplemental Table S1.
Chromatin immunoprecipitation assays
ChIP assays were performed as previously described (Hung et al., 2018, 2021). Chromatin extracts were prepared from seedlings treated with 1% formaldehyde (v/v). The chromatin was sheared to the mean length of 500 bp by sonication, proteins and DNA fragments were then immunoprecipitated using antibodies against GFP (Abcam, catalog no. ab290), H3K27me3 (Diagenode, catalog no. C15410195), or H3Ac (Millipore, catalog no. 06-599). The DNA cross-linked to immunoprecipitated proteins were reversed and then analyzed by qPCR using specific primers (Supplemental Table S1). Percent input was calculated as follows: 2∧(Cq(IN)-Cq(IP))X100. Cq is the quantification cycle as calculated by the Biorad CFX Manager 3.1 based on the MIQE guidelines. Standard deviations represent at least three technical and two biological replicates. The variance in average data is represented by SEM . The sd, SEM determination, and P-value were calculated using Student’s paired t test.
ChIP-seq and data analyses
ChIP-seq assays were performed based on previous research (Hung et al., 2018, 2021). Two nanograms of DNA from ChIP was pooled to ensure that there are enough starting DNA for library construction. Two biological replicates were prepared and sequenced for each ChIP-seq experiment. The ChIP DNA was first tested by RT-qPCR and then used to prepare ChIP-seq libraries. End repair, adaptor ligation, and amplification were carried out using the NEBNext Ultra II DNA Library Prep kit (cat no. E7103) according to the manufacturer’s protocol. The Novoseq PE150 was used for high-throughput sequencing of the ChIP-seq libraries. The raw sequence data were processed using the GAPipeline Illumina sequence data analysis pipeline. Reads were mapped to the TAIR10 Arabidopsis genome (Lamesch et al., 2012) using Qiagen CLC Genomics Workbench (https://www.qiagenbioinformatics.com) with default settings. Reads that mapped to multiple regions were discarded. The data were then imported into the Integrated Genome Viewer (IGV) (Thorvaldsdottir et al., 2013) for visualization. The distribution of the ChIP binding peaks was analyzed with ChIPseeker (Yu et al., 2015), and a high-read random Arabidopsis genomic region subset (1,350,000 regions) was used to represent the ratio of the total Arabidopsis genomic regions. The regions with peak value less than 15 were excluded to locate the target genes (Supplemental Table S2). To identify DNA motifs enriched sites, 400-bp sequences encompassing each peak summit (200-bp upstream and 200-bp downstream) were extracted and searched for enriched DNA motifs using MEME-ChIP with the default parameters (Machanick and Bailey, 2011). To compare the association between RNA expression and histone modifications, the ChIP-seq results as well as previously published Arabidopsis RNA-seq (GSE123459), H3K27me3 (GSE130291), and Pol II (GSE95301) ChIP-seq data (Gibbs et al., 2018; Liu et al., 2018; Xi et al., 2020) were analyzed by using deepTools. The WRKY63 ChIP-seq short read data have been submitted to the NCBI Gene Expression Omnibus (GEO) database (GSE177884).
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers (Supplemental Table S1). Short read data of WRKY63 ChIP-seq have been submitted to the NCBI-Gene Expression Omnibus (GEO) database (GSE177884).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Identification of wrky63 T-DNA insertion mutants.
Supplemental Figure S2. Flowering phenotypes of WT, wrky63, ft and wrky 63 ft.
Supplemental Figure S3. Flowering phenotypes of WT, wrky63, flc and wrky63 flc.
Supplemental Figure S4. Flowering time of 35Spro::GFP:WRKY63 and pro::WRKY63:GFP transgenic plants.
Supplemental Figure S5. The flowering phenotype of WT, wrky63-1 and wrky63-2 under vernalization in LD.
Supplemental Figure S6. The protein accumulation of WRKY63 is induced by vernalization.
Supplemental Figure S7. The flowering phenotype and gene expression patterns of FRIcol and FRI/wrky63.
Supplemental Figure S8. H3Ac level on the FLC locus in vivo.
Supplemental Figure S9. Gene Ontology Biological Processes (GO-BP) analysis of WRKY63 targeted genes.
Supplemental Figure S10. WRKY63 is associated with vernalization-induced gene activation and demethylation of H3K27me3.
Supplemental Table S1. Primers used in this study and the gene accession numbers.
Supplemental Table S2. ChIP-seq data of WRKY63 targeted genes.
Supplementary Material
Acknowledgments
We thank Technology Commons, College of Life Science, National Taiwan University for the convenient use of the Zeiss Axio Imager.Z2 microscope system and Bio-Rad real-time PCR system. We also thank to Pei-Yin, Wu in Technology Commons for technical support.
Funding
This work was supported by the Ministry of Science and Technology of the Republic of China (108-2311-B-002-013-MY3 and 110-2311-B-002-027 to K.W.) and National Taiwan University (NTUCC-111L893001 to K.W.).
Conflict of interest statement. None declared.
Contributor Information
Fu-Yu Hung, Institute of Plant Biology, National Taiwan University, Taipei 10617, Taiwan.
Yuan-Hsin Shih, Institute of Plant Biology, National Taiwan University, Taipei 10617, Taiwan.
Pei-Yu Lin, Institute of Plant Biology, National Taiwan University, Taipei 10617, Taiwan.
Yun-Ru Feng, Institute of Plant Biology, National Taiwan University, Taipei 10617, Taiwan.
Chenlong Li, State Key Laboratory of Biocontrol and Guangdong Key Laboratory of Plant Resource, School of Life Sciences, Sun Yat-sen University, Guangzhou 510275, China.
Keqiang Wu, Institute of Plant Biology, National Taiwan University, Taipei 10617, Taiwan.
F.-Y.H., Y.-H.S., and K.W. designed research. Y.-H.S., P.-Y.L., F.-Y.H., Y.-R.F., and C.L. performed research. F.-Y.H., Y.-H.S., P.-Y.L., C.L., and K.W. analyzed data. F.-Y.H., Y.-H.S., P.-Y.L., and K.W. wrote the article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Keqiang Wu (kewu@ntu.edu.tw).
References
- Bardou F, Ariel F, Simpson CG, Romero-Barrios N, Laporte P, Balzergue S, Brown JWS, Crespi M (2014) Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev Cell 30: 166–176 [DOI] [PubMed] [Google Scholar]
- Birkenbihl RP, Kracher B, Roccaro M, Somssich IE (2017) Induced genome-wide binding of three Arabidopsis WRKY transcription factors during early MAMP-triggered immunity. Plant Cell 29: 20–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MJ, Thelen JJ (2013) ACYL-LIPID DESATURASE2 is required for chilling and freezing tolerance in Arabidopsis. Plant Cell 25: 1430–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia F, Crevillen P, Jones AM, Greb T, Dean C (2008) A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci USA 105: 16831–16836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delk NA, Johnson KA, Chowdhury NI, Braam J (2005) CML24, regulated in expression by diverse stimuli, encodes a potential Ca2+ sensor that functions in responses to abscisic acid, daylength, and ion stress. Plant Physiol 139: 240–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Sun T, Ao K, Peng Y, Zhang Y, Li X, Zhang Y (2018) Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 173: 1454–1467 e1415 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Fornara F, de Montaigu A, Sanchez-Villarreal A, Takahashi Y, van Themaat EVL, Huettel B, Davis SJ, Coupland G (2015) The GI-CDF module of Arabidopsis affects freezing tolerance and growth as well as flowering. Plant J 81: 695–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J (1999) GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18: 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzani S, Gendall AR, Lister C, Dean C (2003) Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol 132: 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendall AR, Levy YY, Wilson A, Dean C (2001) The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107: 525–535 [DOI] [PubMed] [Google Scholar]
- Gibbs DJ, Tedds HM, Labandera AM, Bailey M, White MD, Hartman S, Sprigg C, Mogg SL, Osborne R, Dambire C, et al. (2018) Oxygen-dependent proteolysis regulates the stability of angiosperm polycomb repressive complex 2 subunit VERNALIZATION 2. Nat Commun 9(1): 5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo WW, Ward RW, Thomashow MF (1992) Characterization of a cold-regulated wheat gene related to Arabidopsis Cor47. Plant Physiol 100: 915–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ (2002) Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol 130: 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JB, Sung S (2011) Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79 [DOI] [PubMed] [Google Scholar]
- Hepworth J, Dean C (2015) Flowering locus C's lessons: Conserved chromatin switches underpinning developmental timing and adaptation. Plant Physiol 168: 1237–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G (2002) Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J 21: 4327–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch D, Wassef M, Lovkvist C, Zielinski D, Aflaki S, Lombard B, Hery T, Loew D, Howard M, Margueron R (2021) A cis-acting mechanism mediates transcriptional memory at Polycomb target genes in mammals. Nat Genet 53: 1686–1697 [DOI] [PubMed] [Google Scholar]
- Hu XY, Kong XX, Wang CT, Ma L, Zhao JJ, Wei JJ, Zhang XM, Loake GJ, Zhang TC, Huang JL, et al. (2014) Proteasome-mediated degradation of FRIGIDA modulates flowering time in Arabidopsis during vernalization. Plant Cell 26: 4763–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung FY, Chen C, Yen MR, Hsieh JWA, Li CL, Shih YH, Chen FF, Chen PY, Cui YH, Wu KQ (2020a) The expression of long non-coding RNAs is associated with H3Ac and H3K4me2 changes regulated by the HDA6-LDL1/2 histone modification complex in Arabidopsis. NAR Genom Bioinform 2(3): lqaa066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung FY, Chen FF, Li CL, Chen C., Lai YC, Chen JH, Cui YH, Wu KQ (2018) The Arabidopsis LDL1/2-HDA6 histone modification complex is functionally associated with CCA1/LHY in regulation of circadian clock genes. Nucleic Acids Res 46: 10669–10681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung FY, Chen JH, Feng YR, Lai YC, Yang SG, Wu KQ (2020b) Arabidopsis JMJ29 is involved in trichome development by regulating the core trichome initiation gene GLABRA3. Plant J 103: 1735–1743 [DOI] [PubMed] [Google Scholar]
- Hung FY, Lai YC, Wang J, Feng YR, Shih YH, Chen JH, Sun HC, Yang S, Li C, Wu K (2021) The Arabidopsis histone demethylase JMJ28 regulates CONSTANS by interacting with FBH transcription factors. Plant Cell 33(4): 1196–1211 [DOI] [PubMed] [Google Scholar]
- Kalde M, Barth M, Somssich IE, Lippok B (2003) Members of the Arabidopsis WRKY group III transcription factors are part of different plant defense signaling pathways. Mol Plant Microbe Interact 16(4): 295–305 [DOI] [PubMed] [Google Scholar]
- Kidokoro S, Maruyama K, Nakashima K, Imura Y, Narusaka Y, Shinwari ZK, Osakabe Y, Fujita Y, Mizoi J, Shinozaki K, et al. (2009) The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol 151: 2046–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sung S (2013) Coordination of the vernalization response through a VIN3 and FLC gene family regulatory network in Arabidopsis. Plant Cell 25: 454–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Xi Y, Sung S (2017) Modular function of long noncoding RNA, COLDAIR, in the vernalization response. PLoS Genet 13: e1006939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H, Trewavas AJ, Knight MR (1996) Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 8: 489–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li DH, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. (2012) The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res 40: D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CL, Gu LF, Gao L, Chen C, Wei CQ, Qiu Q, Chien CW, Wang SK, Jiang LH, Ai LF, et al. (2016a) Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat Genet 48(6): 687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wang HP, Yu DQ (2016b) Arabidopsis WRKY transcription factors WRKY12 and WRKY13 oppositely regulate flowering under short-day conditions. Mol Plant 9: 1492–1503 [DOI] [PubMed] [Google Scholar]
- Lim CJ, Park J, Shen M, Park HJ, Cheong MS, Park KS, Baek D, Bae MJ, Ali A, Jan M, et al. (2020) The histone-modifying complex PWR/HOS15/HD2C epigenetically regulates cold tolerance. Plant Physiol 184: 1097–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Xin Y, Xu L, Cai ZK, Xue YC, Liu Y, Xie DX, Liu YL, Qi YJ (2018) Arabidopsis ARGONAUTE 1 binds chromatin to promote gene transcription in response to hormones and stresses. Dev Cell 44(3): 348–361 [DOI] [PubMed] [Google Scholar]
- Liu X, Hao LL, Li DY, Zhu LH, Hu SN (2015) Long non-coding RNAs and their biological roles in plants. Genom Proteom Bioinf 13: 137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca CM, Potschin M, Zentgraf U (2014) bZIPs and WRKYs: Two large transcription factor families executing two different functional strategies. Front Plant Sci 5: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu FL, Cui X, Zhang SB, Jenuwein T, Cao XF (2011) Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat Genet 43: 715–719 [DOI] [PubMed] [Google Scholar]
- Lu FL, Li GL, Cui X, Liu CY, Wang XJ, Cao XF (2008) Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J Integr Plant Biol 50: 886–896 [DOI] [PubMed] [Google Scholar]
- Luo M, Hung FY, Yang SG, Liu XC, Wu KQ (2014) Histone lysine demethylases and their functions in plants. Plant Mol Biol Rep 32: 558–565 [Google Scholar]
- Ma J, Ptashne M (1987) Deletion analysis of GAL4 defines two transcriptional activating segments. Cell 48: 847–853 [DOI] [PubMed] [Google Scholar]
- Machanick P, Bailey TL (2011) MEME-ChIP: Motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeo K, Hayashi S, Kojima-Suzuki H, Morikami A, Nakamura K (2001) Role of conserved residues of the WRKY domain in the DNA-binding of tobacco WRKY family proteins. Biosci Biotech Biochem 65: 2428–2436 [DOI] [PubMed] [Google Scholar]
- Marquardt S, Raitskin O, Wu Z, Liu F, Sun Q, Dean C (2014) Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol Cell 54: 156–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (2001) Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13: 935–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh B, Lee SH, Kim HJ, Yi G, Shin EA, Lee M, Jung KJ, Doyle MR, Amasino RM, Noh YS (2004) Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16: 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Ohme-Takagi M, Shinshi H (2000) Three ethylene-responsive transcription factors in tobacco with distinct transactivation functions. Plant J 22: 29–38 [DOI] [PubMed] [Google Scholar]
- Park J, Lim CJ, Shen MZ, Park HJ, Cha JY, Iniesto E, Rubio V, Mengiste T, Zhu JK, Bressan RA, et al. (2018) Epigenetic switch from repressive to permissive chromatin in response to cold stress. Proc Natl Acad Sci USA 115: E5400–E5409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XZ, Chen ZZ, Liu Y, Zhang HR, Zhang M, Liu QA, Hong XH, Zhu JK, Gong ZZ (2010) ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J 63: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riboni M, Galbiati M, Tonelli C, Conti L (2013) GIGANTEA enables drought escape response via abscisic acid-dependent activation of the florigens and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS. Plant Physiol 162: 1706–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Macdonald H, Huttly AK, Lazarus CM, Hooley R (1995) Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of alpha-Amy2 genes. Plant Mol Biol 29: 691–702 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Krober S, Amasino RA, Coupland G (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun QW, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C (2013) R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 340: 619–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462: 799–802 [DOI] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP (2013) Integrative genomics viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triezenberg SJ, Kingsbury RC, McKnight SL (1988) Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev 2: 718–729 [DOI] [PubMed] [Google Scholar]
- Van Aken O, Zhang BT, Law S, Narsai R, Whelan J (2013) AtWRKY40 and AtWRKY63 modulate the expression of stress-responsive nuclear genes encoding mitochondrial and chloroplast proteins. Plant Physiol 162: 254–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. (2011) A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472: 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welin BV, Olson A, Nylander M, Palva ET (1994) Characterization and differential expression of dhn/lea/rab-like genes during cold acclimation and drought stress in Arabidopsis thaliana. Plant Mol Biol 26: 131–144 [DOI] [PubMed] [Google Scholar]
- Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES, Helliwell CA (2006) The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci USA 103: 14631–14636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Park SR, Kim DH, Kim ED, Sung S (2020) Transcriptome and epigenome analyses of vernalization in Arabidopsis thaliana. Plant J 103: 1490–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu GC, Wang LG, He QY (2015) ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31: 2382–2383 [DOI] [PubMed] [Google Scholar]
- Yu YC, Liu ZH, Wang L, Kim SG, Seo PJ, Qiao M, Wang N, Li S, Cao XF, Park CM, et al. (2016) WRKY71 accelerates flowering via the direct activation of FLOWERING LOCUS T and LEAFY in Arabidopsis thaliana. Plant J 85: 96–106 [DOI] [PubMed] [Google Scholar]
- Zhang LP, Chen LG, Yu DQ (2018) Transcription factor WRKY75 interacts with DELLA proteins to affect flowering. Plant Physiol 176: 790–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.